Abstract
The de novo design of molecular structures using deep learning generative models introduces an encouraging solution to drug discovery in the face of the continuously increased cost of new drug development. From the generation of original texts, images, and videos, to the scratching of novel molecular structures the creativity of deep learning generative models exhibits the height machine intelligence can achieve. The purpose of this paper is to review the latest advances in generative chemistry which relies on generative modeling to expedite the drug discovery process. This review starts with a brief history of artificial intelligence in drug discovery to outline this emerging paradigm. Commonly used chemical databases, molecular representations, and tools in cheminformatics and machine learning are covered as the infrastructure for generative chemistry. The detailed discussions on utilizing cutting-edge generative architectures, including recurrent neural network, variational autoencoder, adversarial autoencoder, and generative adversarial network for compound generation are focused. Challenges and future perspectives follow.
Keywords: Drug discovery, Deep learning, Generative model, Recurrent neural network, Variational autoencoder, Adversarial autoencoder, Generative adversarial network
Introduction
Drug discovery is expensive. The cost for the development of a new drug now can hit 2.8 billion USD and the overall discovery process takes over 12 years to finish [1, 2]. Moreover, these numbers keep increasing. It is critical to think and explore efficient and effective strategies to confront the growing cost and to accelerate the discovery process. The progression in the high-throughput screening (HTS) dramatically speeded up the task of lead identification by screening candidate compounds in large volume [3, 4]. When it comes to the lead identification, the concept can be further classified into two divisions, the structure-based approach [5, 6] and the ligand-based approach [7]. Combined with the significant progress in computation, the development of these two approaches has resulted in constructive virtual screening (VS) methodologies. Traditionally, with the structure of the target protein available, structure-based approaches including molecular docking studies [8–10], molecular dynamic simulations [11–13], and fragment-based approach [10, 14] can be applied to explore the potential receptor-ligand interactions and to virtually screen a large compound set for finding the plausible lead. Then, with the identified active molecules for the given target, ligand-based approaches such as pharmacophore modeling [15, 16], scaffolding hopping [17, 18], and molecular fingerprint similarity search [19] can be conducted for modifying known leads and for finding future compounds. The rapid advancement in computational power and the blossom of machine learning (ML) algorithms brought the ML-based decision-making model [20, 21] as an alternative path to the VS campaigns in the past decades. There is increased availability of data in cheminformatics and drug discovery. The capability of dealing with large data to detect hidden patterns and to facilitate future data prediction in a time-efficient manner favored ML in building VS pipelines.
It is encouraging to note the successful applications of the abovementioned computational chemistry approaches and ML-based VS pipelines on drug discovery these days. The conventional methods are effective. However, the challenge remains on developing pioneering methods, techniques, and strategies in the confrontation of the costly procedure of drug discovery. The flourishing of deep learning generative models brings fresh solutions and opportunities to this field. From the generated human faces that are indistinguishable from real people [22], to the text generation tools that mimic the tone and vocabulary of certain authors [23], the creativity of deep learning generative models brings our understanding of machine intelligence to a new level. In recent years, the expeditions toward generative chemistry mushroomed, which explored the possibility of utilizing generative models to effectively and efficiently design molecular structures with desired properties. Promising and compelling outcomes including the identification of DDR1 kinase inhibitors within 21 days using deep learning generative models [24] may indicate that we are probably at the corner of an upcoming revolution of drug discovery in the artificial intelligence (AI) era. There are published reviews on applying AI technology to different drug discovery stages from preclinical research to clinical studies [25–30]. This review article gives a major focus on applying deep learning generative modeling for the de novo molecular design. This review article starts with a brief evolution of AI in drug discovery and the infrastructures in both cheminformatics and machine learning. The state-of-the-art generative models including recurrent neural networks (RNNs), variational autoencoders (VAEs), adversarial autoencoders (AAEs), and generative adversarial networks (GANs) are focused on to discuss their fundamental architectures as well as their applications in the de novo drug design.
Artificial intelligence in drug discovery
Artificial intelligence (AI) is the study of developing and implementing techniques that enable the machine to behave with intelligence [31]. The concept of AI can be traced back to the 1950s when researchers questioned whether computers can be made to handle automated intelligence tasks which are commonly fulfilled by humans [32]. Thus, AI is a broad area of research that includes both (1) methodologies employing learning processes and (2) approaches that no learning process is involved in. At the early stage, researchers believed that by defining a sufficient number of explicit rules to maneuver knowledge, the human-level AI can be expected (Fig. 1a). In the face of a specific problem, the human studying process on existing observations can contribute to the accumulation of knowledge. Explicit rules were expected to describe knowledge. By programming and applying these rules, the answers for future observations are anticipated. This strategy is also known as symbolic AI [33]. Symbolic AI is an efficient solution to logical problems, for instance, chess playing. However, when handling problems with blurry, unclear, and distorted knowledge, such as image recognition, language translation, and to our topic, the classification of active compounds from decoys for a therapeutic target, symbolic AI turned out to show limited capability. We may define explicit rules to guide the selection of general drug-like compounds, Lipinski’s rule of five [34], for example, but it is almost impossible to exhaust specified rules for guiding the selection of agonists to cannabinoid receptor 2 or other targets [35]. Machine learning (ML) took over symbolic AI’s position as a novel method with the ability to learn on its own.
Fig. 1.
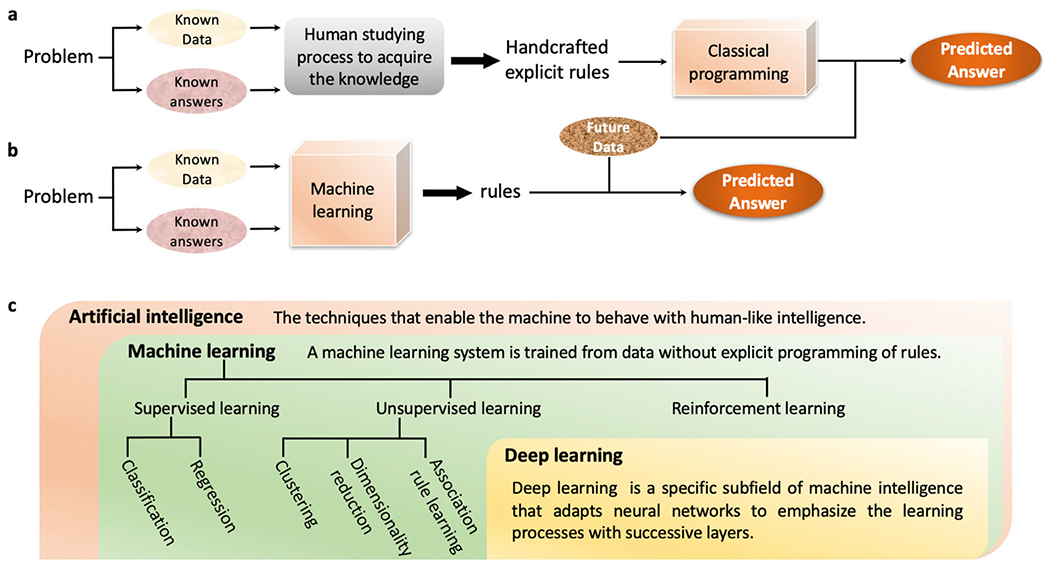
From artificial intelligence to deep learning. a The programming paradigm for symbolic AI. b The programming paradigm for ML. c The relationship among artificial intelligence, machine learning, and deep learning
ML allows computers to solve specific tasks by learning on their own [36, 37]. Through directly looking at the data, computers can summarize the rules instead of waiting for programmers to craft them (Fig. 1b). In the paradigm of ML-based problem solving, data and the answers to the data are functioned as input with rules as the outcome. The produced rules can then be applied to predict answers for future data. Statistical analysis is always associated with ML, while they can be distinguished at several aspects [38]. The application of ML is usually toward large and complex datasets, such as a dataset with millions of small molecules that cover a relatively big chemical space with diversified scaffolds, which statistical analysis can be incapable of dealing with [39]. The flourish of ML started in the 1990s [40]. The method rapidly became a dominant player in the field of AI. Commonly used ML systems in drug discovery can be categorized into supervised learning, unsupervised learning, and reinforcement learning (Fig. 1c). In supervised learning, the algorithms are fed with both the data and the answers to these data (label). Protein family subtypes selectivity prediction is an example for classification: the classifier is trained with numbers of sample molecules along with their labels (the specific protein family member they interact with) and the well-trained classifier should be able to classify the future molecules [20, 35, 41, 42]. Quantitative structure-activity relationship analysis is an example for regression: the regressor is trained with molecules sharing a similar scaffold along with their biological activity data (Ki, IC50, and EC50 values for example), and the well-trained regressor should be able to predict the numeric activity values for future molecules with the similar scaffold [10, 43]. In unsupervised learning, the algorithms are trained with unlabeled data. For instance, a high-throughput screening campaign may preselect a smaller representative compound set from a large compound database using the clustering method to group molecules with similar structures into clusters [44, 45]. A subset of molecules selected from different clusters can then offer improved structural diversity to cover a bigger chemical space than a random pickup. In reinforcement learning, the learning system can choose actions according to its observation of the environment and get a penalty (or reward) in return [46]. To achieve the lowest penalty (or highest reward), the system must learn and choose the best strategy by itself.
Deep learning (DL) is a specific subfield of ML that adapts neural networks to emphasize the learning processes with successive layers (Fig. 1c). DL methods can transfer the representation at one level to a higher and more abstract level [47]. The feature of representation learning enables DL methods to discover representations from the raw input data for tasks such as detection and classification. The word “deep” in DL reflects this character of successive layers of representations, and the number of layers determines the depth of a DL model [48]. In contrast, conventional ML methods that transform the input data into one or two successive representation spaces are sometimes referred to as shallow learning methods. The vast development in the past decades brought DL great flexibility on the selection of architectures, such as the fully connected artificial neural network (ANN) or multi-layer perceptron (MLP) [49], convolutional neural network (CNN) [50], and recurrent neural network (RNN) [51]. The rise of generative chemistry is largely benefited from the extensive advancement of generative modeling, which predominantly depends on the flourishing of DL architectures. The successful application of the long short-term memory (LSTM) model [52], a special type of RNN model, on text generation inspired the simplified molecular-input line-entry system (SMILES)-based compound design. And, the promising exercise of using the generative adversarial network (GAN) model [53] for image generation motivated the fingerprint and graph centered molecular structural scratch. The major reason for DL to bloom rapidly can be that the very method provides solutions to previously unsolvable problems and outperforms the competitors with a simplified representation learning process [32, 47]. It is foreseen that the process of molecule design can evolve into a more efficient and effective manner with the proper fusion with DL.
Data sources and machine learning infrastructures
Deep learning campaigns start with high-quality input data. The successful development of generative chemistry models relies on cheminformatics and bioinformatics data for the molecules and biological systems. Table 1 exhibits some routinely used databases in drug discovery for both small and large biological molecules. In a typical case of structure-based drug discovery, a 3D model of the protein (or DNA/RNA) target is critical for the following steps on evaluating potential receptor-ligand interactions. The PDB database [54] is a good source for accessing structural information for large biological systems, and the UniProt database [55] will be a convenient source for sequence data. Regarding chemicals, PubChem [56] can be a go-to place. PubChem is comprehensive. It currently contains ~ 103 million compounds (with unique chemical structures) and ~ 253 million substances (information about chemical entities). If the major focus is on bioactive molecules, ChEMBL [57] can be an efficient database to interact. ChEMBL currently documents ~ 2 million reported drug-like compounds with bioactivity data for 13,000 targets. Supposing that the interest is more on studying the existing drugs on the market instead of drug-like compounds, the DrugBank [58] can serve. To date, DrugBank records ~ 14,000 drugs, including approved small molecule drugs and biologics, nutraceuticals, discovery-phase drugs, and withdrawn drugs. With virtual screening campaigns, adding some commercially available compounds to in-house libraries are preferred as they may further increase the structural diversity and expand the coverage of the chemical space. Once potential hits were predicted to be among these compounds, the commercial availability gives them easy access for future experimental validations. The Zinc database [59] now archives ~ 230 million purchasable compounds in ready-to-dock format. It is worth mentioning that constructing topic-specific and target-specific databases is trending. ASD [60] is one example that files allosteric modulators and related macromolecules to facilitate the research on allosteric modulation. The rising of Chemogenomics databases [61, 62] for certain diseases and druggable targets is another example that these libraries focus on particular areas of research. Usually, these target and disease-focused Chemogenomics databases integrate both target and chemical information with computing tools for systems pharmacology research. Pain-CKB [61], for example, it is a pain-domain-specific knowledgebase describes the chemical molecules, genes, and proteins involved in pain regulation.
Table 1.
Well-established cheminformatics databases available for drug discovery
| Database | Description | Web linkage | Examples of usage |
|---|---|---|---|
| UniProt [55] | The Universal Protein Resource (UniProt) is a resource for protein sequence and annotation data | https://www.uniprot.org | Protein sequence homology search, alignment, and protein ID retrieving especially for structural-based drug discovery |
| RCSB PDB [54] | The Protein Data Bank (PDB) provides access to 3D structure data for large biological molecules, including protein, DNA, and RNA. | https://www.rcsb.org | Protein 3D structures are fundamental for hot spot identification, docking simulation, and molecular dynamics simulation in structural-based drug discovery. |
| PDBbind [72] | PDBbind provides a collection of the experimentally measured binding affinity data for all types of biomolecular complexes deposited in the PDB. | http://www.pdbbind.org.cn | The receptor-ligand binding data for resolved protein structures can function as the benchmark to evaluate future simulations |
| PubChem [73] | PubChem is a key chemical information resource for the biomedical research community. | https://pubchem.ncbi.nlm.nih.gov | To acquire comprehensive chemical information ranging from NMR spectra, physical-chemical properties, to biomolecular interactions. |
| ChEMBL [57] | ChEMBL is a manually curated database of bioactive molecules with drug-like properties. | https://www.ebi.ac.uk/chembl/ | To collect cheminformatics data of reported molecules for a given target. A high-quality compound collection is the key to the ligand-based drug discovery |
| SureChEMBL [74] | SureChEMBL is a resource containing compounds extracted from patent literature. | https://www.surechembl.org/search/ | Compound-patent associations |
| BindingDB [75] | BindingDB is a database of measured binding affinities for the interactions of protein considered to be drug targets with small, drug-like molecules. | https://www.bindingdb.org/bind/index.jsp | To retrieve compound sets for a specific target similar to ChEMBL but with the focus on experimental binding affinities. |
| DrugBank [58] | The DrugBank database combines detailed drug data with comprehensive drug target information | https://www.drugbank.ca | Drug repurposing study for existing drugs. On-target and off-target analysis for a compound. |
| ZINC [59] | Zinc is a database of commercially available compounds | https://zinc.docking.org | Zinc database is good for virtual screening on hit identification as the compounds are commercially available for quick biological validations afterwards. |
| Enamine | Enamine provides an enumerated database of synthetically feasible molecules for purchase | https://enamine.net | The establishment of a target-specific compound library. Fragment-based drug discovery. |
| ASD [60] | Allosteric Database (ASD) provides a resource for structure, function, disease, and related annotation for allosteric macromolecules and allosteric modulators | http://mdl.shsmu.edu.cn/ASD/ | To facilitate the research on allosteric modulation with enriched chemical data on allosteric modulators. |
| GDB [76] | GDB databases provide multiple subsets of combinatorially generated compounds following chemical stability and synthetic feasibility rules | http://gdb.unibe.ch/downloads/ | Using combinatorial chemistry is a good way to largely expand the chemical space. |
With the input data available, the next issue is tidying data as there are often inconsistencies and sometimes errors in databases. Collected molecules are needed to be transformed into machine-readable representations. Table 2 lists commonly used molecular representations. SMILES [63] describes molecular structures in a text-based format using short ASCII strings. Multiple SMILES strings can be generated for the same molecule with different starting atoms. This ambiguity led to the effects of canonicalization that determines which of all possible SMILES will be used as the reference SMILES for a molecule. Popular cheminformatics packages such as OpenEye [64] and RDKit [65] are possible solutions for standardizing the canonical SMILES [66]. The canonical SMILES is a type of well-liked molecular representation in generative chemistry models as it packs well with language processing and sequence generation techniques like RNNs. Usually, the SMILES strings are first converted with one-hot encoding. The categorical distribution for each element can then be produced by the generative models. Fingerprints are another vital group of molecular representations, using extended-connectivity fingerprint (ECFP) as one example. It is a circular fingerprint that represents the presence of particular substructures [67]. ECFP is not predefined and can be rapidly calculated to represent different molecular features. Fingerprints can be calculated through different approaches. By enumerating circular fragments, linear fragments, and tree fragments from the molecular graph, Circular [67], Path, and Tree fingerprints [68] can be created. Using fingerprints as representations may suffer from inconvertibility in that the complete structure of a molecule cannot be reconstructed directly from the fingerprints [69]. To have fingerprints calculated for large enough compounds, the library to function as a look-up index may be a compromised solution [29]. Despite this difficulty, fingerprints are popular among ML classification models for tasks like distinguishing active compounds from inactive ones for a given target. Graph-based [70] and tensorial [71] representations are popular alternative options. Most molecules can be easily represented in 2D graphs. Neural networks such as CNNs may operate directly on molecular graphs. Tensorial representation is another approach that stores molecular information into atom types, bond types, and connectivity information in tensors.
Table 2.
Examples of commonly used molecular representations
| Representation | Description | |
|---|---|---|
| String based | SMILES [63] | The simplified molecular-input line-entry system (SMILES) is a specification in the form of a line notation for describing the structure of chemical species using short ASCII strings. SMILES is readable by humans. |
| Canonical SMILES | Canonicalization is a way to determine which of all possible SMILES will be used as the reference SMILES for a molecular graph. | |
| InChI [77] | The International Chemical Identifier (InChI) is a textual identifier for chemical substances, designed to provide a standard way to encode molecular information. InChIs are much more convoluted but offer tautomer-invariant notation and are preferred for duplicate removal. | |
| InChI Key | The condensed, 27-character InChI Key is a hashed version of the full InChI. | |
| Fingerprints | MACCS Keys [78] | MACCS keys are 166 bits structural key descriptors in which each bit is associated with a SMARTS pattern. |
| Circular [67, 79] | Circular fingerprints are created by exhaustively enumerating all circular fragments grown radially from each heavy atom of the molecule up to the given radius. | |
| Path [68] | Path fingerprints are created by exhaustively enumerating all linear fragments of a molecular graph up to a given size. | |
| Tree [68] | Tree fingerprints are generated by exhaustively enumerating all tree fragments of a molecular graph up to a given size. | |
| Atom Pair [80] | Atom Pair fingerprints encode each atom as a type, enumerate all distances between pairs, and then hash the results. |
After collecting the high-quality data and transforming the data into the appropriate format, it is time to apply data science to the development of predictive models. Table 3 illustrates examples of frequently considered cheminformatics toolkits and machine learning packages. RDKit, Open Babel [81], and CDK [82] are cheminformatics toolkits that are comprised of a set of libraries with source codes for various functions, such as chemical files I/O formatting, substructure and pattern search, and molecular representations generation. The typical applications of deploying these toolkits can contribute to virtual screening, structural similarity search, structure-activity relationship analysis, etc [83]. The workflow environment is not unique to the cheminformatics research but can facilitate the automation of data processing with a user-friendly interface. The workflow systems like KNIME [84, 85] can execute tasks in succession and perform recurring tasks efficiently, such as iterative fingerprints calculation for a compound library. The strategy of integrating cheminformatics toolkits as nodes into a workflow and connecting them with edges is gaining popularity and is increasingly employed [86–88]. When it comes to ML and DL modeling, TensorFlow [89], CNTK [90], Theano [91], and PyTorch [92] are well-recognized packages for employment. These packages handle low-level operations including tensor manipulation and differentiation. In contrast, Keras [93] is a model-level library that deals with tasks in a modular way. As a high-level API, Keras is running on top of TensorFlow, CNTK, and Theano. Scikit-Learn [94] is an efficient and straightforward tool for predictive data analysis. It is known more for its role in conventional ML modeling as the library comprehensively integrates algorithms like support vector machine (SVM), random forest (RF), logistic regression, and naïve Bayes (NB).
Table 3.
Commonly used cheminformatics and machine learning packages
| Package | Description | Web linkage |
|---|---|---|
| RDKit [65] | RDKit is an open-source toolkit for cheminformatics. Features include 2D and 3D molecular operations, descriptor generation, molecular database cartridge, etc. | https://www.rdkit.org |
| Open Babel [81] | Open Babel is an open chemical toolbox to search, convert, analyze, or store data from molecular modeling, chemistry, solid-state materials, biochemistry, or related areas. | http://openbabel.org/wiki/Main_Page |
| CDK [82] | The Chemistry Development Kit (CDK) is a collection of modular Java libraries for processing cheminformatics. | https://cdk.github.io |
| KNIME [84] | KNIME is a workflow environment in data science that can be integrated to automate certain cheminformatics operations. | https://www.knime.com |
| TensorFlow [89] | TensorFlow is an open-source platform for machine learning. It has a set of tools, libraries, and community resources that enable researchers to build and deploy ML applications. | https://www.tensorflow.org |
| CNTK [90] | The Cognitive Toolkit (CNTK) is an open-source toolkit for commercial-grade distributed deep learning. It describes neural networks as a series of computational steps via a directed graph. | https://github.com/microsoft/CNTK |
| Theano [91] | Theano is a Python library for defining, optimizing, and evaluating mathematical expressions. | http://deeplearning.net/software/theano/ |
| PyTorch [92] | PyTorch is an open-source machine learning library based on the Torch library. | https://pytorch.org |
| Keras [93] | Keras is a high-level neural networks API, written in Python and capable of running on top of TensorFlow, CNTK, or Theano. It was developed with a focus on enabling fast experimentation. | https://keras.io |
| Scikit-Learn [94] | Scikit-learn is a free software machine learning library for the Python programming language. | https://scikit-learn.org/stable/ |
Generative chemistry with the recurrent neural network
RNN is a widely used neural network architecture in generative chemistry for proposing novel structures. As a type of powerful generative model especially in natural language processing, RNNs usually use sequences of words, strings, or letters as the input and output [51, 95–97]. In this case, the SMILES strings are usually employed as a molecular representation. Different from ANNs and CNNs which do not have memories, RNNs iteratively process sequences and store a state holding current information. On the contrary, ANNs and CNNs process each input independently without stored information between them. When describing an RNN, it can be considered as a network with an internal loop that loops over the sequence elements instead of processing in a single step (Fig. 2a). The state that stored information will be updated during each loop. For simplicity, the process of computing the output y can follow the equation: y = activation (Wox + Uoh + bo), where Wo and Uo are weight matrices for the input x and state h, and bo as a bias vector. Figure 2 a can represent the structure of a simple RNN model. However, this structure can suffer severely from the vanishing gradient problem which makes neural networks untrainable after adding more layers. Even though the state h is supposed to hold the information from the sequence elements previously seen, the long-term dependencies make the learning process impossible [98, 99]. The long short-term memory (LSTM) algorithm [52] was developed to overcome this shortcoming. The LSTM layer attaches a carry track to carry information across the learning process to counter the loss of signals from gradual vanishing (Fig. 2b). With this carry track, the information learned from each sequence element can be loaded, and the loaded information can be transported and accessed at a later stage. The process of computing the output y for LSTM is similar to the previous equation but adding the contribution of the carry track: y = activation (Wox + Uoh + Voc + bo), where Wo, Uo, and Vo are weight matrices for the input x, state h, and carry c, and bo as a bias vector. In certain cases, multiple recurrent layers in a model can be stacked to enhance representational power.
Fig. 2.
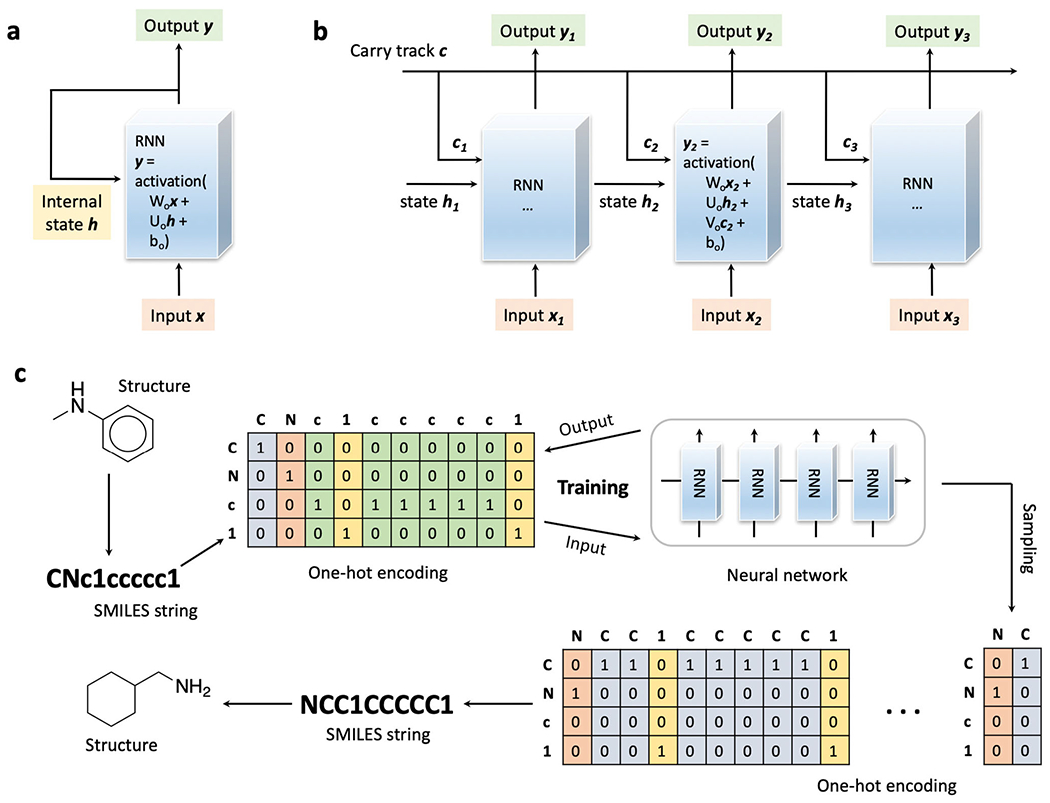
The RNN, the LSTM, and their application in generative chemistry. a The schematic illustration of the RNN, the neural network with an internal loop. b The schematic illustration of data processing with the LSTM. c The typical framework on building generative models applying RNN for molecules generation
A typical framework on generative modeling for molecule generation applying the LSTM algorithm (Fig. 2c) starts with the collection of training molecules. The RNN model can be fine-tuned through the transfer learning that first accumulates knowledge from the large compound datasets and then produces the novel structures by learning smaller focused datasets. When the collections of training molecules (for large sets or small focused sets) are ready, SMILES strings can be calculated for each molecule. One-hot encoding is a regular operation for processing molecular representations. In one-hot encoding, a unique integer index i is assigned to every character in the SMILES string. Then, a binary vector can be constructed of size C (the number of unique characters in the string) with all zeros but for the ith entry which is one. For instance, there are four (C = 4) unique characters, “C,” “N,” “c,” and “1” in SMILES strings, input “C” is transferred to (1, 0, 0, 0), “N” to (0, 1, 0, 0), “c” to (0, 0, 1, 0), and “1” to (0, 0, 0, 1) after one-hot encoding. In practice, usually, an additional starting character like “G” and an ending character like “E” will be added to the SMILES to denote a complete string. The neural network with LSTM layer(s) can be trained to predict the n+1th character given the input of string with n characters. The probability of distribution for the n+1th character is calculated as the loss to evaluate the model performance. With the trained model, the sampling process can start with the starting character or certain SMILES strings of molecular fragments to sample the next character until the ending character is hit. The SMILES strings are reversed from the generated binary matrices according to the previous one-hot encoding to construct the molecular graphs as the output for this generative model.
Representative case studies are discussed in this paragraph. All the case applications covered in this review are summarized in Table 4. Gupta et al. trained an LSTM-based generative model with transfer learning to generate libraries of molecules with structural similarity to known actives for PPARγ and trypsin [100]. The model was first trained with 550,000 SMILES strings of active compounds from ChEMBL and further fine-tuned with SMILES strings for 4367 PPARγ ligands and 1490 trypsin inhibitors. Among the valid generated molecules, around 90% are unique from the known ligands and are different from each other. The proposed model was assessed for fragment-based drug discovery as well. In fragment-based drug discovery, fragment growing is a strategy for novel compounds generation with the identified fragment lead. Substitutions can be added to the identified fragment with the consideration of pharmacophore features and proper physical-chemical properties to enhance the receptor-ligand interactions [101]. Instead of using the starting character to initiate the generative process, the SMILES string of the molecular fragment can be read and extended by calculating the probability of distribution for the next character. Segler et al. also reported their application of LSTM-based generative models for structure generation with transfer learning [102]. There was a good correlation between the generated structures and the molecules used for training. Notably, the complete de novo drug design cycle can be achieved with prediction models for scoring. As the scoring model can be a molecular docking algorithm or even robot synthesis and biotesting system, the drug design cycle does not require known active compounds to start. The chemical language model (CLM) proposed by Moret et al. is another example of applying LSTM-based generative models to work with chemical SMILES strings with transfer learning processes [103]. This approach enables the early-stage molecular design in a low data regime. When it comes to real-world validation, Merk et al. published their prospective study with experimental evaluations [104]. Using the SMILES strings as the input, the LSTM-based generative model was trained and fine-tuned with the transfer learning process for the peroxisome proliferator-activated receptor. Five top-ranked compounds designed by the model were synthesized and tested. Four of them have nanomolar to low micromolar activities in cell-based assays. Besides using the LSTM algorithm, some other RNN architectures such as implementing gated recurrent unit [105] (GRU) can also have promising applications. GRU layers work with the same principle as LSTM layers but may have less representational power. Zheng et al. developed a quasi-biogenic molecular generator with GRU layers [106]. As biogenic compounds and pharmaceutical agents are biologically relevant, over 50% of existing drugs result from drug discovery campaigns starting with biogenic molecules. Their generative model is an effort to explore greater biogenic diversity space. Similarly, focused compound libraries can be constructed with transfer learning processes.
Table 4.
Representative applications of generative chemistry covered in this review
| # | Generative architecture | Neural networks involved | Data source | Molecular representation | Note | Ref. |
|---|---|---|---|---|---|---|
| 1 | RNN | LSTM | ChEMBL | SMILES | The application was extended to fragment-based drug design. | [100] |
| 2 | RNN | LSTM | ChEMBL | SMILES | The design-synthesis-test cycle was simulated with target prediction models for scoring. | [102] |
| 3 | RNN | LSTM | ChEMBL | SMILES | A chemical language model (CLM) in low data regimes. | [103] |
| 4 | RNN | LSTM | ChEMBL | SMILES | A prospective application with experimental validations of top-ranking compounds. | [104] |
| 5 | RNN | GRU | ZINC ChEMBL |
SMILES | The generative model explored greater biogenic diversity space. | [106] |
| 6 | VAE | Encoder: CNN Decoder: GRU |
ZINC QM9 |
SMILES | An MLP model was jointed to predict property values. | [113] |
| 7 | VAE | Encoder: CNN Decoder: GRU |
ChEMBL | SMILES | An SVM classification model was added to evaluate the outcome. | [114] |
| 8 | VAE | Encoder: LSTM Decoder: LSTM |
ChEMBL | SMILES | A sequence-to-sequence VAE model was combined with generative topographic mapping (GTM) for molecular design. | [115] |
| 9 | VAE | Encoder: CNN Decoder: CNN |
ZINC QM9 |
Molecular graph | The nodes and edges in the graph of NeVAE represent atoms and bonds respectively. | [117] |
| 10 | VAE | Encoder: CNN Decoder: MLP |
ZINC QM9 |
Molecular graph | The central hypothesis of GraphVAE was to decode a probabilistic fully connected graph. | [118] |
| 11 | VAE | Encoder: GGNN# Decoder: GGNN |
ZINC CASF* |
Molecular graph | DeLinker was designed to incorporate two fragments into a new molecule. | [119] |
| 12 | VAE | Encoder: GRU Decoder: GRU |
ZINC | SMILES | A cross-domain latent space capturing both chemical and biological information | [116] |
| 13 | AAE | Encoder: MLP Decoder: MLP Discriminator: MLP |
MCF-7^ | MACCS fingerprints | Fingerprints cannot be directly converted to structures but can provide certain substructure information. | [121] |
| 14 | AAE | Encoder: LSTM Decoder: LSTM Discriminator: MLP |
ZINC | SMILES | The generated molecules targeting JAK3 were evaluated with in silico and in vitro methods. | [122] |
| 15 | AAE | Encoder: GRU Decoder: GRU Discriminator: MLP |
LINCS& ChEMBL |
SMILES | The combination of molecules and gene expression data was analyzed. | [123] |
| 16 | GAN | Discriminator: CNN Generator: LSTM |
ZINC | SMILES | Sequence generation with objective-reinforced generative adversarial networks (ORGAN). | [124] |
| 17 | GAN | Discriminator: MLP Generator: MLP |
ZINC | Molecular graph | The model operated in the latent space trained by the Junction Tree VAE. | [125] |
| 18 | GAN | Discriminator: MLP Generator: MLP |
LINCS& | SMILES | The compound design was connected to the systems biology. | [126] |
| 19 | GAN | Encoder: LSTM Decoder: LSTM Discriminator: MLP Generator: MLP |
ChEMBL | SMILES | The concept of the autoencoder and the generative adversarial network was combined to propose a latentGAN. | [127] |
GGNN represents the gated graph neural network
CASF is also known as PDBbind core set
MCF-7 represents a small data set of compounds profiled on the MCF-7 cell line
LINCS represents the LINCS L1000 dataset that collects gene expression profiles
Generative chemistry with the variational autoencoder
The principle aim of an autoencoder (AE) is to construct a low-dimensional latent space of compressed representations that each element can be reconstructed to the original input (Fig. 3a). The module that maps the original input data, which is in high dimension, to a low-dimensional representation is called the encoder, while the module that realizes the mapping and reconstructs the original input from the low-dimensional representation is called the decoder [48, 107]. The encoder and the decoder are usually neural networks with RNN and CNN architectures as SMILES strings and molecular graphs are commonly used molecular representations. With the molecular representations calculated, a typical data processing procedure with AE on molecule generation starts with encoding the input into a low-dimensional latent space. Within the latent space, the axis of variations from the input can be encoded. Using the variation of molecular weight (M.W.) as an example, while in practice, the features learned can be highly abstractive as the M.W. is used here for simplified illustration; the points along this axis are embedded representations of compounds with different M.W. These variations are termed concept vectors. With an identified vector, it makes molecular editing possible by exploring the representations in a relevant direction. The encoded latent space with compressed representations can then be sampled with the decoder to map them back to molecular representations. Novel structures alongside the original input can be expected.
Fig. 3.
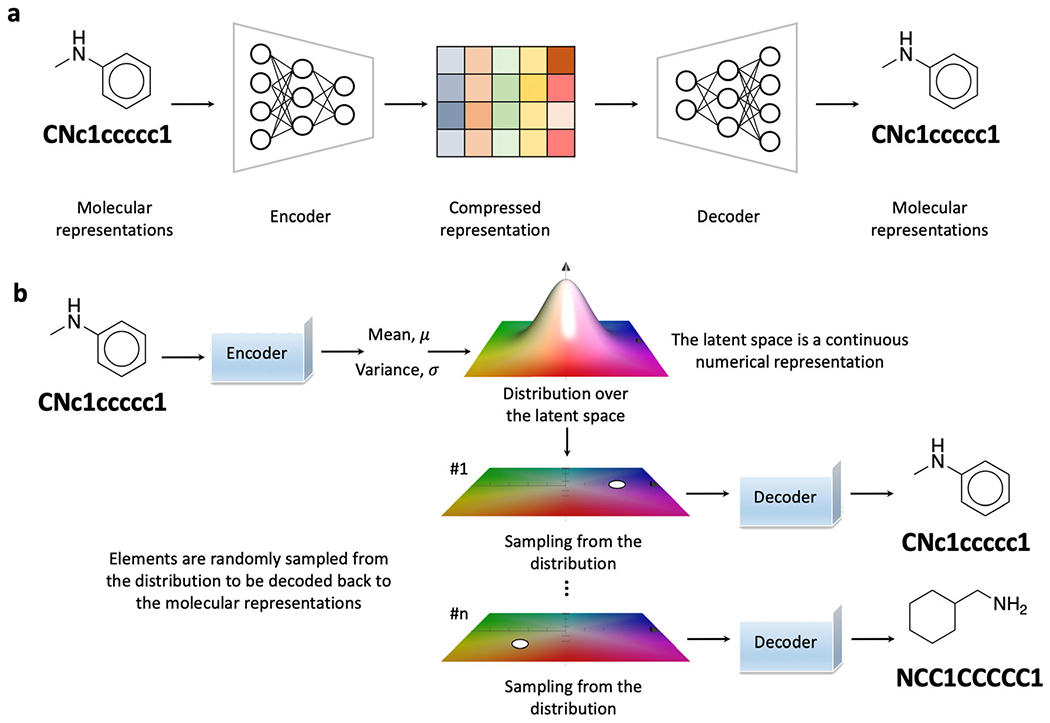
The autoencoder and the variational autoencoder. a An autoencoder encodes input molecules into compressed representations and decodes them back. b A variational autoencoder maps the molecules into the parameters of a statistical distribution as the latent space is a continuous numerical representation
The concept of VAE was first proposed by Kingma and Welling at the end of 2013 [108, 109]. This technique quickly gained popularity in building robust generative models for images, sounds, and texts [110–112]. The AE compresses a molecule x into a fixed code in the continuous latent space z and trends to summarize the explicit mapping rules as the number of adjustable parameters is often much more than the number of training molecules. These explicit rules make the decoding of random points in the continuous latent space challenging and sometimes impossible [32]. Instead, VAE maps the molecules into the parameters of a statistical distribution (Fig. 3b). With p(z) describing the distribution of prior continuous latent space, the probabilistic encoding distribution is qφ(z|x) and the probabilistic decoding distribution is pθ(x|z). The training iterations with back propagation will gradually optimize the parameters of both qφ(z\x) and pθ(x|z). VAE is fundamentally a latent variable model p(x,z) = pθ(x|z)p(z). The stochasticity of the training process enables the latent space to encode valid representations, which further results in a structured latent space [109]. Both the reconstruction loss and the regularization loss are often used for parameter optimization during the training process. The reconstruction loss evaluates whether the decoded samples match the input while the regularization loss investigates whether the latent space is overfitting to the training data.
Applications of VAE for generating chemical structures started in 2016 as Gomez-Bombarelli et al. developed a VAE-based automatic chemical design system [113]. In their practice, the ZINC database and QM9 dataset were referred to as the sources for collecting molecules. The QM9 dataset archives small molecules following three rules: (1) no more than 9 heavy atoms, (2) with 4 distinct atomic numbers, and (3) with 4 bond types. Canonical SMILES strings were calculated as the molecular representation. The encoder maps input SMILES strings into the continuous real-valued vectors, and the decoder reconstructs molecular representations from these vectors. The encoder was formed with three convolutional layers and one fully connected dense layer while the decoder contained three GRU layers. The architectures of CNNs and RNNs were compared for string encoding and convolutional layers achieved superior performance. The last layer of the decoder would report a probability distribution for characters of the SMILES string at each position. This stochastic operation allowed the same point in the latent space to have different decoded outcomes. Besides, they added one additional module for property prediction. An MLP was jointed to predict the property values from the continuous representation created by the encoder in order to optimize the desired properties for the new molecules. Thomas Blaschke et al. tested various generative AE models including VAE for compound design targeting dopamine receptor 2 (DRD2) [114]. Their study showed that the generated latent space preserved the chemical similarity principles. The generated molecules similar to known active compounds can be observed. In their VAE model, CNN layers were used for the encoder for pattern recognition and the RNN layers of GRU cells were adapted for the decoder. The ChEMBL database functioned as the data source for molecular structures. Canonical SMILES were prepared as the molecular representation. Similarly, an SVM classification model trained with extensive circular fingerprint (ECFP) of active and inactive DRD2 ligands was integrated to investigate the newly generated molecules. Sattarov et al. combined a sequence-to-sequence VAE model with generative topographic mapping (GTM) for molecular design [115]. Both the encoder and the decoder were RNN models containing two LSTM layers in their practice. SMILES strings with one-hot encoding for molecules from the ChEMBL database were prepared prior to the training. Their GTM module contributed to the selection of sampling points in the VAE latent space, which facilitated the generation of a focused library of compounds with desired properties. Mohammadi et al. introduced the penalized VAE using gated recurrent units with an effort to learn a cross-domain latent space apprehending chemical and biological information simultaneously [116]. A weight penalty term was added in the decoder to confront the imbalanced distribution of SMILES characters. The quality and the generalization ability to new chemistry of the latent space have been shown to be improved.
Besides the use of SMILES strings, molecular graphs have also been applied as a type of molecular representation to feed the VAE models. Samanta et al. proposed NeVAE, a VAE-based compound generative model employing molecular graphs [117]. The molecular structures are usually not grid like and come with an inconsistent number of nodes and edges, which impedes the use of molecular graphs as representations, In their work, the molecular graphs were prepared for drug-like compounds collected from the ZINC database and QM9 dataset. The nodes and edges in the graph represent atoms and bonds respectively. The node features are types of atoms with one-hot encoding and the edge weights are bond types (saturated bonds, unsaturated double/triple bonds, etc.). The purpose of training is to enable the VAE to create credible molecular graphs including node features and edge weights. Another example is GraphVAE. Simonovsky et al. proposed GraphVAE to facilitate the compound design using molecular graphs [118]. Their central hypothesis was to decode a probabilistic fully connected graph in which the existence of nodes, edges, and their attributes are independent random variables. The encoder was a feed-forward network with convolutional layers and the architecture for the decoder was an MLP. The model training and evaluation involved the molecules from the ZINC database and QM9 dataset. Some other generative applications can switch the topic to lead optimizations with methods such as scaffold hopping, substitutions design, and fragment-based approaches. One example is the DeLinker which was proposed by Imrie et al. to incorporate two fragments into a new molecule [119]. This method is VAE based, using molecular graphs as the input. The design process heavily relied on 3D structural information that considers relative distance and orientation between the starting fragments.
Generative chemistry with the adversarial autoencoder
The architecture of the AAE is comparatively similar to the VAE except for the appending of the additional discriminator network [120]. An AAE trains three modules, an encoder, a decoder, and a discriminator (Fig. 4). The encoder learns the input data and maps the molecule into the latent space following the distribution of qφ(z|x). The decoder reconstructs molecules through sampling from the latent space following the probabilistic decoding distribution of pθ(x|z). And, the discriminator distinguishes the distribution of the latent space z~qφ(z) from the prior distribution z′~p(z). During the training iterations, the encoder is modified consistently to have the output, qφ(z|x), follow a specific distribution, p(z), in an effort to minimize the adversarial cost of the discriminator. A simplistic prior, like Gaussian distribution, is assumed in VAE, while alternative priors can exist in real-world practices [121]. The AAE architecture with the additional discriminator module demonstrates improved adaptability.
Fig. 4.
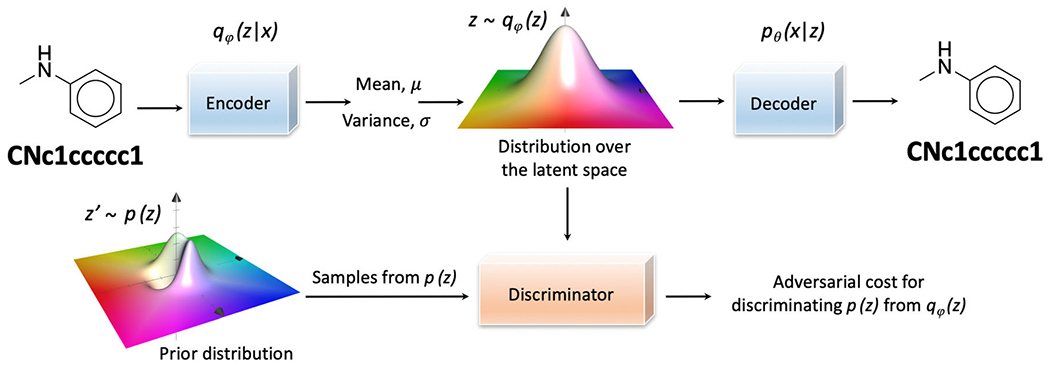
The illustrated architecture of an adversarial autoencoder. A discriminator network is appended to calculate the adversarial cost for discriminating p(z) from qφ(z). As a result, the outcome latent space from the encoder is driven to follow the prior distribution
Blaschke et al. summarized a three-step training process in their compound design practice with AAE: (1) the simultaneous training of both the encoder and the decoder to curtail the reconstruction loss of the decoder; (2) the training of the discriminator to distinguish the distribution of the latent space, qφ(z), from the prior distribution p(z) effectively; (3) the training of the encoder to minimize the adversarial cost for discriminating p(z) from qφ(z) [114]. The training iterations continue until the reconstruction loss converges. Kadurin et al. proposed the method of using a generative adversarial autoencoder model to identify fingerprints of new molecules with potential anticancer properties [121]. The input molecules come from a small data set of compounds profiled on the MCF-7 cell line. The MACCS fingerprints were used as the molecular representation and two fully connected dense layers with different dimensions were used as the network architecture for the encoder, decoder, and discriminator. One notable modification in this study was the removal of the batch normalization layers for the discriminator. Batch normalization is an optimization method that reduces the covariance shift among the hidden units and allows each layer to learn more independently. In the authors’ opinion, the noise from the generator can be masked into target random noise with the batch normalization layers, which prohibits the training of the discriminator. As each bit of the MACCS fingerprints represents certain substructure features, the learned structural information by machine can be beneficial to the design of chemical derivatives for identified leads. Polykovskiy et al. reported their work on building a conditional AAE for molecule design targeting Janus kinase 3 (JAK3) [122]. The contributions from a set of physical-chemical properties including bioactivity, solubility, and synthesizability were considered and the model was conditioned to produce molecules with specified properties. Clean lead molecules were collected from the ZINC database and encoded as SMILES strings. The LSTM layers are adapted for building the encoder and the decoder networks. Both in silico method (molecular docking) and in vitro assay (inhibition of JAK2 and JAK3) were conducted as the evaluation for the newly generated molecules. Shayakhmetov et al. reported a bidirectional AAE model that generates molecules with the capacity of inducing a desired change in gene expression [123]. The model was validated using LINCS L1000, a database that collects gene expression profiles. The molecular structures x and induced gene expression changes y contributed to a joint model p(x,y). In this specific conditional task, there is no direct association between x and y as certain changes in the gene expression are irrelevant to the drug-target interactions. The proposed bidirectional AAE model then learned the joint distribution and decomposed objects into shared features, exclusive features to x, and exclusive features to y. Therefore, the discriminator that divides the latent representations into shared and exclusive sections was constructed to secure the conditional generation to be consequential.
Generative chemistry with the generative adversarial network
The architecture of the convolutional neural network [50] (CNN) is briefly covered in this section as the convolutional layers are widely used in GAN modeling. The implementation of convolutional layers can also be found in case studies discussed above among autoencoder models. A convolutional layer does not learn an input globally but focuses on the local pattern within a receptive field, the kernel (Fig. 5a). The low-level patterns learned in a prior layer can then be concentrated on the high-level features at the subsequent layers [128, 129]. This characteristic allows the CNN to learn and summarize abstract patterns with complexity. Another characteristic that comes out from the local pattern learning is that the learned features can be recognized anywhere [128]. It enables the CNN to process input data with efficiency and powerfulness even with a smaller number of input sample representations. Meanwhile, multiple feature maps (filters) can be stacked to encode different aspects of the input data. Applying several filters capacitates a CNN model to detect distinct features anywhere among the input data. The pooling operation on the other hand subsamples the feature map to reduce the number of parameters and eventually, the computational load [130]. Using a max-pooling layer as one example, only the max input value in that pooling kernel will be kept. Alongside dropout layers and regularization penalties, the pooling layers also contribute to confronting the overfitting issues. Putting together, the convolutional layers, pooling layers, and dense layers are carefully selected and arranged to construct a sophisticated CNN architecture.
Fig. 5.
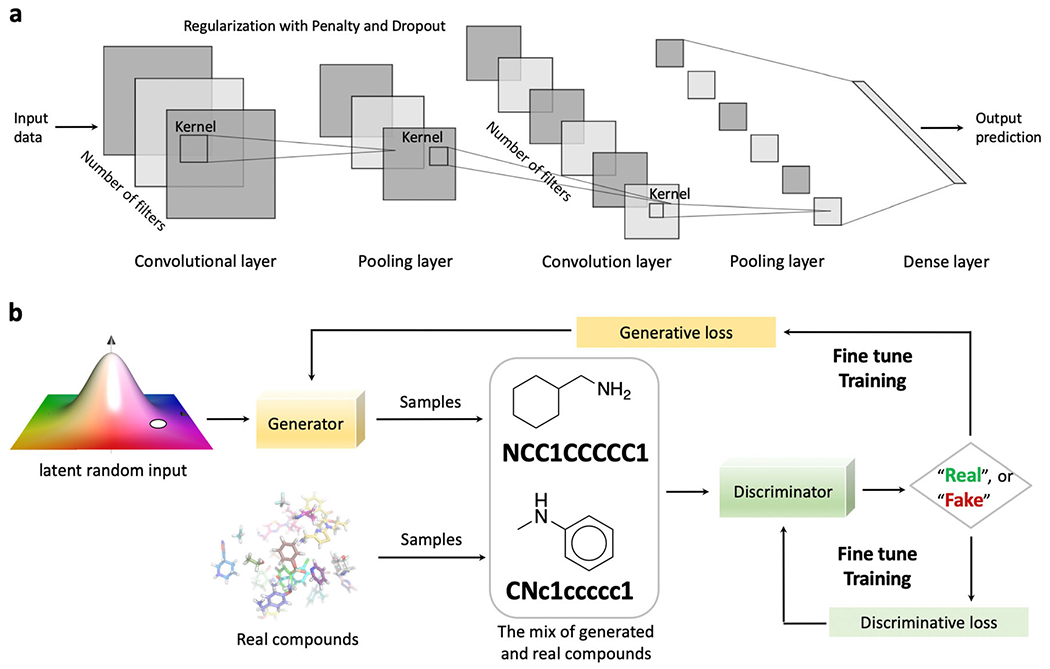
Sample architecture of the convolutional neural network and the framework of a generative adversarial network. a The careful selection and arrangement of convolutional layers, pooling layers, and dense layers, etc. constitute a convolutional neural network. b The generative adversarial network comprises two modules, the generator and the discriminator. Both the generative loss and discriminative loss are monitored during the training process
The concept of the GAN was first raised by Goodfellow in 2014 [53]. The method quickly gained popularity on generative tasks regarding image, video, and audio processing and related areas [131–133]. Two models, the discriminator and the generator, are trained iteratively and simultaneously during the adversarial training process [69]. The discriminator is supposed to discover the hidden patterns behind the input data and to make accurate discrimination of the authentic data from the ones generated by the generator. The generator is trained to keep proposing compelling data to fool the well-trained discriminator by consistently optimizing the data sampling process. The training process is a zero-sum noncooperative game with the purpose of achieving the Nash equilibrium [134] by the discriminator and the generator. In generative chemistry, the generator generates SMILES strings, molecular graphs, or fingerprints, depending on the selection of the molecular representation, using the latent random inputs (Fig. 5b). The generated molecules are mixed with the samples of real compounds to feed the discriminator after correct labeling. The discriminative loss is calculated to evaluate whether the discriminator can distinguish the real compounds from the generated ones, while the generative loss is computed to assess whether the generator can fool the discriminator by generating undistinguishable molecules. The constringency of both loss functions after the iterative training indicates that even a well-established discriminator can be misled to classify generated molecules as real, which further reflects that the generator has learned and accumulated authentic data patterns to create captivating compounds. However, it is worth mentioning that the simultaneous optimization of both loss functions is challenging as the instability can lead to the gradient of one part instead of both being favored (results in a stronger discriminator or generator, but not both). Another limitation may come from the restricted chemical space that is being covered by the generated molecules [29]. To confront the discriminator and minimize the generative loss, the generator can only explore a limited chemical space defined by the real compounds.
Guimaraes et al. presented a sequence-based GAN framework termed objective-reinforced generative adversarial network (ORGAN) [124] that includes domain-specific objectives to the training process besides the discriminator reward. The discriminator drove the generated samples to follow the distribution of the real data and the domain-specific objectives secured that the traits maximizing the specific heuristic would be selected. The drug-like and nondrug-like molecules were collected from ZINC databases. SMILES strings were calculated as the molecular representations. A CNN model was designed as the discriminator to classify texts, and an RNN model with LSTM units was used as the generator. Maziarka et al. introduced Mol-CycleGAN for derivatives design and compound optimization [125]. The model could generate structures with high similarity to the original input but improved values on considered properties. Molecular graphs of compounds extracted from the ZINC database were used as the molecular representation. The model operated in the latent space trained by the Junction Tree VAE. Dense layers and fully connected residual layers constituted the generator and the discriminator. Méndez-Lucio et al. reported a GAN model to connect the compound design with systems biology [126]. They have shown that active-like molecules can be generated given that the gene expression signature of the selected target is supplied. The architectures of both the discriminator and the generator were composed of dense layers. There were two stages of training: in stage I, the random noise was taken as the input, while in stage II, the output from stage I and the gene expression signature were taken. Prykhodko et al. combined the concept of AE with GAN and proposed a latent vector-based GAN model [127]. A heteroencoder mapped one-hot encoded SMILES strings into the latent space and the generator and discriminator would directly use the latent vector to focus on the optimization of the sampling process. A pre-trained heteroencoder was then used to transfer the generated vectors back to molecular structures. Both general drug-like compounds and target-biased molecules were generated as applications of the method.
Evaluation of generative models
Salimans et al. proposed an evaluation metric, the Inception Score (IS), for generative models in their study of improving architectural features and training procedures [135]. An Inception model is a neural network trained for classification. Every generated outcome is assessed to get the conditional label distribution. In the application of the molecular generation, the divergence between the distributions of generated molecules and the training compounds is computed to report as the IS. In short, the IS investigates whether the generated molecules can be correctly classified to cover the chemical space defined by the training set. Heusel et al. introduced the Fréchet inception distance (FID) to capture the similarity of generated data to real ones [136]. Different from the IS that compares label distributions, FID compares latent vectors from a specific layer of an Inception model. Molecules are embedded in latent vectors. A continuous multivariate Gaussian is then fit to the data and the distance is computed as the FID. Thus, the FID can be calculated on unlabeled data. One critical weakness of the IS and the FID has been pointed out that both methods report a one-dimensional score which is incapable of distinguishing different failure cases. Sajjadi et al. proposed the approach of using precision and recall for distributions (PRD) to evaluate generated outcomes from two separate dimensions [137]. Intuitively, the precision in the PRD measures the quality of generated data while the recall measures the coverage of reference training data. As a result, the divergence between distributions is disentangled into two components, precision and recall.
In the perspective of generative chemistry, the synthetic feasibility of generated molecules is a crucial consideration besides the evaluation of the model itself. Gao et al. pointed out that generative models can propose unrealistic molecules even with high performance scores on quantitative benchmarks [138]. Some existing methods of evaluating the synthesizability are based on synthetic routes and molecular structural data, which require heuristic definition to be complex and comprehensive [139], while the change of one single functional group to a scaffold can cause a distinctive synthetic pathway. The ignorance of synthesizability turns out to be an eminent hindrance to connecting generative models with medicinal chemistry synthesis. It is a common solution to add a filter for checking the synthetic feasibility after the generation process [140, 141]. Software such as ChemAxon Structure Checker can be applied to filter out undesired structures. Ertl et al. published a method to estimate synthetic accessibility score (SA score) based on fragment contributions and a complexity penalty [142]. In this method, the historical synthetic knowledge is acquired by studying already synthesized chemicals. Incorporating the SA score as a reward is an approach to enforce the generative model to sample synthetically feasible molecules [113, 143].
Conclusion and future perspectives
Besides the successful generative chemistry stories described above, challenges and opportunities can be found at the following four aspects: (1) the alternative molecular representations that can better portray a structure, (2) the generation of macromolecules, and (3) the close-loop automation in combination with experimental validations. The molecular representations such as SMILES strings and molecular fingerprints serve well in describing small molecules at the current stage. However, it will be appealing if the novel representations can be designed to also consider three-dimensional geometry data. Chiral compounds may exhibit divergent activities to the biological system [144], and even the conformational change of the same small molecule can alter the receptor-ligand interactions. The case studies that deployed molecular graphs as the representation illustrate the benefits of working with structures directly [117–119, 125]. The extended consideration of bond type, length, and angles improves the performance of feature extraction on spatial patterns. Peptides possess a superior advantage among protein subtype selectivity. The strategy of developing antibodies and peptides as therapeutic agents draws increasing attention from both the academia and industry. Deep learning is data-driven research. Current generative chemistry applications mainly focus on the design of small molecules as there is an increased availability of accessing chemical data [145]. As the construction of protein-related databases is rising, the attempts of de novo protein generation are expected [146]. Better representations are certainly required for describing protein, as the folding and its conformation are even more critical to determine the functionality. Lastly, it is noteworthy of how to integrate the generative chemistry into the drug design framework to close the loop of this automated process. Segler et al. mentioned a design-synthesis-test cycle in their application of using the RNN model to generate molecules [102]. Ideally, the HTS will first recognize some hit compounds for a given target. The identified hits will contribute to the iterative training of a deep learning generative model for novel compounds generation, and a machine learning-based target prediction model for virtual classification. The top molecules will be synthesized and tested with biological assays. The true new actives can then be appended to the identified hits, which closes the loop.
In a nutshell, this paper reviewed the latest advances of generative chemistry that utilizes deep learning generative models to expedite the drug discovery process. The review starts with a brief history of AI in drug discovery to outline this emerging paradigm. Commonly used chemical databases, molecular representations, and operating sources of cheminformatics and machine learning are covered as the infrastructure. The detailed discussions on RNN, VAE, AAE, and GAN are centered, which is followed by future perspectives. As a fast-growing area of research, we are optimistic to expect a boosting number of studies on generative chemistry. We are probably at the corner of an upcoming revolution of drug discovery in the AI era, and the good news is that we are witnessing the change.
Funding
The authors would like to acknowledge the funding support to the Xie laboratory from the NIH NIDA (P30 DA035778A1) and DOD (W81XWH-16-1-0490).
Footnotes
Materials availability N/A
Code availability N/A
Ethics approval and consent to participate N/A
Consent for publication N/A
Competing interest The authors declare no competing interest.
Data availability
N/A
References
- 1.Wouters OJ, McKee M, Luyten J (2020) Estimated research and development investment needed to bring a new medicine to market, 2009-2018. Jama 323:844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiMasi JA, Grabowski HG, Hansen RW (2016) Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ 47:20–33 [DOI] [PubMed] [Google Scholar]
- 3.Yasi EA, Kruyer NS, Peralta-Yahya P (2020) Advances in G protein-coupled receptor high-throughput screening. Curr Opin Biotechnol 64:210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blay V, Tolani B, Ho SP, Arkin MR (2020) High-Throughput Screening: today’s biochemical and cell-based approaches. Drug Discov Today 25:1807–1821 [DOI] [PubMed] [Google Scholar]
- 5.Kroemer RT (2007) Structure-based drug design: docking and scoring. Curr Protein Pept Sci 8:312–328 [DOI] [PubMed] [Google Scholar]
- 6.Blundell TL (1996) Structure-based drug design. Nature 384:23. [DOI] [PubMed] [Google Scholar]
- 7.Bacilieri M, Moro S (2006) Ligand-based drug design methodologies in drug discovery process: an overview. Curr Drug Discov Technol 3:155–165 [DOI] [PubMed] [Google Scholar]
- 8.Pagadala NS, Syed K, Tuszynski J (2017) Software for molecular docking: a review. Biophys Rev 9:91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bian Y-m, He X-b, Jing Y-k, Wang L-r, Wang J-m, Xie X-q (2019) Computational systems pharmacology analysis of cannabidiol: a combination of chemogenomics-knowledgebase network analysis and integrated in silico modeling and simulation. Acta Pharmacol Sin 40:374–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bian Y, Feng Z, Yang P, Xie X-Q (2017) Integrated in silico fragment-based drug design: case study with allosteric modulators on metabotropic glutamate receptor 5. AAPS J 19:1235–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174 [DOI] [PubMed] [Google Scholar]
- 12.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31:671–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge H, Bian Y, He X, Xie X-Q, Wang J (2019) Significantly different effects of tetrahydroberberrubine enantiomers on dopamine D1/D2 receptors revealed by experimental study and integrated in silico simulation. J Comput Aided Mol Des 33:447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hajduk PJ, Greer J (2007) A decade of fragment-based drug design: strategic advances and lessons learned. Nat Rev Drug Discov 6:211–219 [DOI] [PubMed] [Google Scholar]
- 15.Yang S-Y (2010) Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov Today 15:444–450 [DOI] [PubMed] [Google Scholar]
- 16.Wieder M, Garon A, Perricone U, Boresch S, Seidel T, Almerico AM, Langer T (2017) Common hits approach: combining pharmacophore modeling and molecular dynamics simulations. J Chem Inf Model 57:365–385 [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Chen H, Wang P, Li Y, Wold EA, Leonard PG, Joseph S, Brasier AR, Tian B, Zhou J (2020) Discovery of Orally Bioavailable Chromone Derivatives as Potent and Selective BRD4 Inhibitors: Scaffolding Hopping, Optimization and Pharmacological Evaluation. J Med Chem 63(10):5242–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y, Stumpfe D, Bajorath JR (2017) Recent advances in scaffold hopping: miniperspective. J Med Chem 60:1238–1246 [DOI] [PubMed] [Google Scholar]
- 19.Muegge I, Mukherjee P (2016) An overview of molecular fingerprint similarity search in virtual screening. Expert Opin Drug Discovery 11:137–148 [DOI] [PubMed] [Google Scholar]
- 20.Fan Y, Zhang Y, Hua Y, Wang Y, Zhu L, Zhao J, Yang Y, Chen X, Lu S, Lu T (2019) Investigation of machine intelligence in compound cell activity classification. Mol Pharm 16:4472–4484 [DOI] [PubMed] [Google Scholar]
- 21.Minerali E, Foil DH, Zorn KM, Lane TR, Ekins S (2020) Comparing Machine Learning Algorithms for Predicting Drug-Induced Liver Injury (DILI). Mol Pharm 17(7):2628–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karras T, Laine S, Aittala M, Hellsten J, Lehtinen J, Aila T (2019) Analyzing and improving the image quality of stylegan. arXiv preprint arXiv:1912.04958 [Google Scholar]
- 23.Wen T-H, Gasic M, Mrksic N, Su P-H, Vandyke D, Young S (2015) Semantically conditioned lstm-based natural language generation for spoken dialogue systems. arXiv preprint arXiv:1508.01745 [Google Scholar]
- 24.Zhavoronkov A, Ivanenkov YA, Aliper A, Veselov MS, Aladinskiy VA, Aladinskaya AV, Terentiev VA, Polykovskiy DA, Kuznetsov MD, Asadulaev A (2019) Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat Biotechnol 37:1038–1040 [DOI] [PubMed] [Google Scholar]
- 25.Rifaioglu AS, Atas H, Martin MJ, Cetin-Atalay R, Atalay V, Doğan T (2019) Recent applications of deep learning and machine intelligence on in silico drug discovery: methods, tools and databases. Brief Bioinform 20:1878–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen D, Liu S, Kingsbury P, Sohn S, Storlie CB, Habermann EB, Naessens JM, Larson DW, Liu H (2019) Deep learning and alternative learning strategies for retrospective real-world clinical data. NPJ Digit Med 2:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipinski C, Maltarollo V, Oliveira P, da Silva A, Honorio K (2019) Advances and perspectives in applying deep learning for drug design and discovery. Front Robot AI 6:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Lin K, Wang S, Wang L, Cai C, Song C, Lai L, Pei J (2019) Deep learning for molecular generation. Future Med Chem 11:567–597 [DOI] [PubMed] [Google Scholar]
- 29.Elton DC, Boukouvalas Z, Fuge MD, Chung PW (2019) Deep learning for molecular design—a review of the state of the art. Mol Syst Des Eng 4:828–849 [Google Scholar]
- 30.Hutchinson L, Steiert B, Soubret A, Wagg J, Phipps A, Peck R, Charoin JE, Ribba B (2019) Models and machines: how deep learning will take clinical pharmacology to the next level. CPT Pharmacometrics Syst Pharmacol 8:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turing AM (2009) Computing Machinery and Intelligence. In: Epstein R, Roberts G, Beber G (eds) Parsing the Turing Test: Philosophical and Methodological Issues in the Quest for the Thinking Computer. Springer, Netherlands: Dordrecht, pp 23–65 [Google Scholar]
- 32.Chollet F (2018) Deep learning with Python (Vol. 361). Manning, New York [Google Scholar]
- 33.Segler MH, Preuss M, Waller MP (2018) Planning chemical syntheses with deep neural networks and symbolic AI. Nature 555:604–610 [DOI] [PubMed] [Google Scholar]
- 34.Lipinski CA (2016) Rule of five in 2015 and beyond: target and ligand structural limitations, ligand chemistry structure and drug discovery project decisions. Adv Drug Deliv Rev 101:34–41 [DOI] [PubMed] [Google Scholar]
- 35.Bian Y, ring Y, Wang L, Ma S, Jun JJ Xie X-Q (2019) Prediction of orthosteric and allosteric regulations on cannabinoid receptors using supervised machine learning classifiers. Mol Pharm 16:2605–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lo Y-C, Rensi SE, Torng W, Altman RB (2018) Machine learning in chemoinformatics and drug discovery. Drug Discov Today 23:1538–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jing Y, Bian Y, Hu Z, Wang L, Xie X-QS (2018) Deep learning for drug design: an artificial intelligence paradigm for drug discovery in the big data era. AAPS J 20:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bzdok D, Altman N, Krzywinski M (2018) Points of significance: statistics versus machine learning. Nat Methods 15:233–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Wang Y, Byrne R, Schneider G, Yang S (2019) Concepts of artificial intelligence for computer-assisted drug discovery. Chem Rev 119:10520–10594 [DOI] [PubMed] [Google Scholar]
- 40.Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G, Li B, Madabhushi A, Shah P, Spitzer M (2019) Applications of machine learning in drug discovery and development. Nat Rev Drug Discov 18:463–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korotcov A, Tkachenko V, Russo DP, Ekins S (2017) Comparison of deep learning with multiple machine learning methods and metrics using diverse Drug discovery data sets. Mol Pharm 14:4462–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma XH, Jia J, Zhu F, Xue Y, Li ZR, Chen YZ (2009) Comparative analysis of machine learning methods in ligand-based virtual screening of large compound libraries. Comb Chem High Throughput Screen 12:344–357 [DOI] [PubMed] [Google Scholar]
- 43.Verma J, Khedkar VM, Coutinho EC (2010) 3D-QSAR in Drug design-a review. Curr Top Med Chem 10:95–115 [DOI] [PubMed] [Google Scholar]
- 44.Fan F, Warshaviak DT, Hamadeh HK, Dunn RT (2019) The integration of pharmacophore-based 3D QSAR modeling and virtual screening in safety profiling: A case study to identify antagonistic activities against adenosine receptor, A2A, using 1,897 known drugs. PLoS One 14(1):e0204378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gladysz R, Dos Santos FM, Langenaeker W, Thijs G, Augustyns K, De Winter H (2018) Spectrophores as one-dimensional descriptors calculated from three-dimensional atomic properties: applications ranging from scaffold hopping to multi-target virtual screening. J Cheminformatics 10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen TT, Nguyen ND, Nahavandi S (2020) Deep reinforcement learning for multiagent systems: A review of challenges, solutions, and applications. IEEE Trans Cybern 50(9):3826–3839 [DOI] [PubMed] [Google Scholar]
- 47.LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444 [DOI] [PubMed] [Google Scholar]
- 48.Goodfellow I, Bengio Y, Courville A (2016) Deep Learning. MIT Press, http://www.deeplearningbook.org [Google Scholar]
- 49.Hastie T, Tibshirani R, Friedman J (2009) The Elements of Statistical, Learning: Data Mining Inference and Prediction (second ed.). Springer [Google Scholar]
- 50.LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86:2278–2324 [Google Scholar]
- 51.Rumelhart DE, Hinton GE, Williams RJ (1986) Learning representations by back-propagating errors. Nature 323:533–536 [Google Scholar]
- 52.Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9:1735–1780 [DOI] [PubMed] [Google Scholar]
- 53.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2661) Generative adversarial nets. arXiv preprint arXiv:1406 [Google Scholar]
- 54.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28(1):235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.(2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA (2016) PubChem substance and compound databases. Nucleic Acids Res 44:D1202–D1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Beilis LJ, Cibrián-Uhalte E (2017) The ChEMBL database in 2017. Nucleic Acids Res 45:D945–D954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46:D1074–D1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sterling T, Irwin JJ (2015) ZINC 15–ligand discovery for everyone. J Chem Inf Model 55:2324–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang Z, Mou L, Shen Q, Lu S, Li C, Liu X, Wang G, Li S, Geng L, Liu Y (2014) ASD v2. 0: updated content and novel features focusing on allosteric regulation. Nucleic Acids Res 42:D510–D516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng Z, Chen M, Shen M, Liang T, Chen H, Xie X-Q (2020) Pain-CKB, A Pain-Domain-Specific Chemogenomics Knowledgebase for Target Identification and Systems Pharmacology Research. J Chem Inf Model 60(10):4429–4435 [DOI] [PubMed] [Google Scholar]
- 62.Feng Z, Chen M, Liang T, Shen M, Chen H, Xie X-Q (2020) Virus-CKB: an integrated bioinformatics platform and analysis resource for COVID-19 research. Brief Bioinform:bbaa155. 10.1093/bib/bbaa155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weininger D (1988) SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci 28:31–36 [Google Scholar]
- 64.OEChemTK (2010) version1.7.4.3;Open Eye Scientific Software Inc.: Santa Fe, NM [Google Scholar]
- 65.Landrum G, RDKit: Open-Source Cheminformatics Software, http://www.rdkit.org/
- 66.O’Boyle NM (2012) Towards a Universal SMILES representation-a standard method to generate canonical SMILES based on the InChI. J Cheminformatics 4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rogers D, Hahn M (2010) Extended-connectivity fingerprints. J Chem Inf Model 50:742–754 [DOI] [PubMed] [Google Scholar]
- 68.Hert J, Willett P, Wilton DJ, Acklin P, Azzaoui K, Jacoby E, Schuffenhauer A (2004) Comparison of fingerprint-based methods for virtual screening using multiple bioactive reference structures. J Chem Inf Comput Sci 44:1177–1185 [DOI] [PubMed] [Google Scholar]
- 69.Bian Y, Wang J, Jun JJ, Xie X-Q (2019) Deep convolutional generative adversarial network (dcGAN) models for screening and design of small molecules targeting cannabinoid receptors. Mol Pharm 16:4451–4460 [DOI] [PubMed] [Google Scholar]
- 70.Goh GB, Siegel C, Vishnu A, Hodas NO, Baker N (2017) Chemception: a deep neural network with minimal chemistry knowledge matches the performance of expert-developed QSAR/QSPR models. arXiv preprint arXiv:1706.06689 [Google Scholar]
- 71.De Cao N, Kipf T (2018) MolGAN: An implicit generative model for small molecular graphs. arXiv preprint arXiv:1805.11973 [Google Scholar]
- 72.Wang R, Fang X, Lu Y, Yang C-Y, Wang S (2005) The PDBbind database: methodologies and updates. J Med Chem 48:4111–4119 [DOI] [PubMed] [Google Scholar]
- 73.Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B (2019) PubChem 2019 update: improved access to chemical data. Nucleic Acids Res 47:D1102–D1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Papadatos G, Davies M, Dedman N, Chambers J, Gaulton A, Siddle J, Koks R, Irvine SA, Pettersson J, Goncharoff N (2016) SureChEMBL: a large-scale, chemically annotated patent document database. Nucleic Acids Res 44:D1220–D1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J (2016) BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 44:D1045–D1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruddigkeit L, Van Deursen R, Blum LC, Reynrond J-L (2012) Enumeration of 166 billion organic small molecules in the chemical universe database GDB-17. J Chem Inf Model 52:2864–2875 [DOI] [PubMed] [Google Scholar]
- 77.Heller SR, McNaught A, Pletnev I, Stein S, Tchekhovskoi D (2015) InChI, the IUPAC international chemical identifier. J Cheminformatics 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Durant JL, Leland BA, Henry DR, Nourse JG (2002) Reoptimization of MDL keys for use in drug discovery. J Chem Inf Comput Sci 42:1273–1280 [DOI] [PubMed] [Google Scholar]
- 79.Glen RC, Bender A, Arnby CH, Carlsson L, Boyer S, Smith J (2006) Circular fingerprints: flexible molecular descriptors with applications from physical chemistry to ADME. IDrugs 9:199. [PubMed] [Google Scholar]
- 80.Pérez-Nueno VI, Rabal O, Borrell JI, Teixidó J (2009) APIF: a new interaction fingerprint based on atom pairs and its application to virtual screening. J Chem Inf Model 49:1245–1260 [DOI] [PubMed] [Google Scholar]
- 81.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open babel: an open chemical toolbox. J Cheminformatics 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Willighagen EL, Mayfield JW, Alvarsson J, Berg A, Carlsson L, Jeliazkova N, Kuhn S, Pluskal T, Rojas-Chertó M, Spjuth O (2017) The Chemistry Development Kit (CDK) v2. 0: atom typing, depiction, molecular formulas, and substructure searching. J Cheminformatics 9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ambure P, Aher RB, Roy K (2014) Recent advances in the open access cheminformatics toolkits, software tools, workflow environments, and databases. Computer-Aided Drug Discovery:257–296 [Google Scholar]
- 84.Arabie P, Baier ND, Critchley CF, Keynes M (2006) Studies in classification, data analysis, and knowledge organization.
- 85.Warr WA (2012) Scientific workflow systems: pipeline pilot and KNIME. J Comput Aided Mol Des 26:801–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beisken S, Meinl T, Wiswedel B, de Figueiredo LF, Berthold M, Steinbeck C (2013) KNIME-CDK: workflow-driven cheminformatics. BMC Bioinf 14:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saubern S, Guha R, Baell J (2011) B., KNIME workflow to assess PAINS filters in SMARTS format. Comparison of RDKit and indigo cheminformatics libraries. Mol Inf 30:847–850 [DOI] [PubMed] [Google Scholar]
- 88.Roughley SD (2020) Five years of the KNIME vernalis cheminformatics community contribution. Curr Med Chem 27(38):6495–6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abadi M et al. (2016) TensorFlow: A system for large-scale machine learning. 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), 265–283 [Google Scholar]
- 90.Etaati L (2019) Deep Learning Tools with Cognitive Toolkit (CNTK). Machine Learning with Microsoft Technologies. Apress, Berkeley, pp 287–302 [Google Scholar]
- 91.Al-Rfou R, Alain G, Almahairi A, Angermueller C, Bahdanau D, Ballas N, Bastien F, Bayer J, Belikov A, Belopolsky A, Bengio Y, Bergeron A, Bergstra J, Bisson V, Bleecher Snyder J, Bouchard N, Boulanger-Lewandowski N, Bouthillier X, Zhang Y (2016) Theano: A Python framework for fast computation of mathematical expressions. arXiv e-prints, arXiv-1605 [Google Scholar]
- 92.Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L (2019) PyTorch: an imperative style, high-performance deep learning library. Advances in Neural Information Processing Systems, 2019, pp 8024–8035 [Google Scholar]
- 93.Chollet F (2015) “keras.” https://github.com/fchollet/keras
- 94.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830 [Google Scholar]
- 95.Mikolov T, Karafiat M, Burget L, Cernocky J, Khudanpur S (2010) Recurrent neural network based language model. INTERSPEECH-2010 1045–1048 [Google Scholar]
- 96.Mikolov T, Kombrink S, Burget L, Čemocky J, Khudanpur S Extensions of recurrent neural network language model, in: Acoustics, Speech and Signal Processing (ICASSP), 2011 IEEE International Conference on, 5528–5531 [Google Scholar]
- 97.Mikolov T, Zweig G (2012) Context dependent recurrent neural network language model. 2012 IEEE Spoken Language Technology Workshop (SLT), 234–239 [Google Scholar]
- 98.Hanson J, Yang Y, Paliwal K, Zhou Y (2017) Improving protein disorder prediction by deep bidirectional long short-term memory recurrent neural networks. Bioinformatics 33:685–692 [DOI] [PubMed] [Google Scholar]
- 99.Cheng J, Dong L, Lapata M (2016) Long short-term memory-networks for machine reading. arXiv preprint arXiv:1601.06733 [Google Scholar]
- 100.Gupta A, Müller AT, Huisman BJ, Fuchs JA, Schneider P, Schneider G (2018) Generative recurrent networks for de novo drug design. Mol Inf 37:1700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bian Y, Xie X-QS (2018) Computational fragment-based Drug design: current trends, strategies, and applications. AAPS J 20:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Segler MH, Kogej T, Tyrchan C, Waller MP (2018) Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent Sci 4:120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moret M, Friedrich L, Grisoni F, Merk D, Schneider G (2020) Generative molecular design in low data regimes. Nat Mach Intell 2:171–180 [Google Scholar]
- 104.Merk D, Friedrich L, Grisoni F, Schneider G (2018) De novo design of bioactive small molecules by artificial intelligence. Mol Inf 37:1700153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chung J, Gulcehre C, Cho K, Bengio Y (2014) Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv preprint arXiv:1412.3555 [Google Scholar]
- 106.Zheng S, Yan X, Gu Q, Yang Y, Du Y, Lu Y, Xu J (2019) QBMG: quasi-biogenic molecule generator with deep recurrent neural network. J Cheminformatics 11:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kramer MA (1991) Nonlinear principal component analysis using autoassociative neural networks. AICHE J 37:233–243 [Google Scholar]
- 108.Kingnra DP, Welling M (2013) Auto-encoding variational bayes. arXiv preprint arXiv:1312.6114 [Google Scholar]
- 109.Kingnra DP, Welling M (2019) An introduction to variational autoencoders. arXiv preprint arXiv:1906.02691 [Google Scholar]
- 110.Kingnra DP, Mohamed S, Rezende DJ, Welling M (2014) Semi-supervised learning with deep generative models. Advances in neural information processing systems, 2014, pp 3581–3589 [Google Scholar]
- 111.Khenrakhenr I, Kingnra DP, Hyvärinen A (2019) Variational autoencoders and nonlinear ica: a unifying framework. arXiv preprint arXiv:1907.04809 [Google Scholar]
- 112.Pu Y, Gan Z, Henao R, Yuan X, Li C, Stevens A, Carin L (2016) Variational autoencoder for deep learning of images, labels and captions. In Advances in neural information processing systems, arXiv preprint arXiv:1609.08976 [Google Scholar]
- 113.Gónrez-Bombarelli R, Wei JN, Duvenaud D, Hernández-Lobato JM, Sánchez-Lengeling B, Sheberla D, Aguilera-Iparraguirre J, Hirzel TD, Adams RP, Aspuru-Guzik A (2018) Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent Sci 4:268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Blaschke T, Olivecrona M, Engkvist O, Bajorath J, Chen H (2018) Application of generative autoencoder in de novo molecular design. Mol Inf 37:1700123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sattarov B, Baskin II, Horvath D, Marcou G, Bjerrum EJ, Varnek A (2019) De novo molecular design by combining deep autoencoder recurrent neural networks with generative topographic mapping. J Chem Inf Model 59:1182–1196 [DOI] [PubMed] [Google Scholar]
- 116.Mohammadi S, O’Dowd B, Paulitz-Erdmann C, Goerlitz L (2019) Penalized Variational Autoencoder for Molecular Design. ChemRxiv. 10.26434/chemrxiv.7977131.v2 [DOI] [Google Scholar]
- 117.Samanta B, De A, Jana G, Gómez V, Chattaraj P, Ganguly N, Gomez-Rodriguez M (2020) Nevae: A deep generative model for molecular graphs. J Mach Learn Res 21(114):1–3334305477 [Google Scholar]
- 118.Simonovsky M, Komodakis N (1802) GraphVAE: Towards Generation of Small Graphs Using Variational Autoencoders, 2018. arXiv:03480 [Google Scholar]
- 119.Imrie F, Bradley AR, van der Schaar M, Deane CM (2020) Deep generative models for 3D linker design. J Chem Inf Model 60:1983–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Makhzani A, Shlens J, Jaitly N, Goodfellow I, Frey B (2015) Adversarial autoencoders. arXiv preprint arXiv:1511.05644 [Google Scholar]
- 121.Kadurin A, Nikolenko S, Khrabrov K, Aliper A, Zhavoronkov A (2017) druGAN: an advanced generative adversarial autoencoder model for de novo generation of new molecules with desired molecular properties in silico. Mol Pharm 14:3098–3104 [DOI] [PubMed] [Google Scholar]
- 122.Polykovskiy D, Zhebrak A, Vetrov D, Ivanenkov Y, Aladinskiy V, Mamoshina P, Bozdaganyan M, Aliper A, Zhavoronkov A, Kadurin A (2018) Entangled conditional adversarial autoencoder for de novo drug discovery. Mol Pharm 15:4398–4405 [DOI] [PubMed] [Google Scholar]
- 123.Shayakhmetov R, Kuznetsov M, Zhebrak A, Kadurin A, Nikolenko S, Aliper A, Polykovskiy D (2020) Molecular generation for desired transcriptome changes with adversarial autoencoders. Front Pharmacol 11:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guimaraes GL, Sanchez-Lengeling B, Outeiral C, Farias PLC, Aspuru-Guzik A (2017) Objective-reinforced generative adversarial networks (organ) for sequence generation models. arXiv preprint arXiv:1705.10843 [Google Scholar]
- 125.Maziarka Ł, Pocha A, Kaczmarczyk J, Rataj K, Danel T, Warchoł M (2020) Mol-CycleGAN: a generative model for molecular optimization. J Cheminformatics 12:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Méndez-Lucio O, Baillif B, Clevert D-A, Rouquié D, Wichard J (2020) De novo generation of hit-like molecules from gene expression signatures using artificial intelligence. Nat Commun 11:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Prykhodko O, Johansson SV, Kotsias P-C, Arús-Pous J, Bjerrum EJ, Engkvist O, Chen H (2019) A de novo molecular generation method using latent vector based generative adversarial network. J Cheminformatics 11:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huang G, Liu Z, Weinberger KQ, van der Maaten L (2017) Densely connected convolutional networks. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit:2261–2269 [Google Scholar]
- 129.LeCun Y, Bengio Y (1995) Convolutional networks for images, speech, and time series. The handbook of brain theory and neural networks 3361:310 [Google Scholar]
- 130.Yu D, Wang H, Chen P, Wei Z (2014) Mixed pooling for convolutional neural networks. International conference on rough sets and knowledge technology, 2014. Springer, pp 364–375 [Google Scholar]
- 131.Radford A, Metz L, Chintala S (2015) Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv preprint arXiv:1511.06434 2015 [Google Scholar]
- 132.Zhang H, Goodfellow I, Metaxas D, Odena A (2018) Self-attention generative adversarial networks. arXiv preprint arXiv:1805.08318 [Google Scholar]
- 133.Li C, Wand M (2016) Precomputed real-time texture synthesis with markovian generative adversarial networks. European conference on computer vision, 2016. Springer, pp 702–716 [Google Scholar]
- 134.Holt CA, Roth AE (2004) The Nash equilibrium: a perspective. Proc Natl Acad Sci 101:3999–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, Chen X (2016) Improved techniques for training gans. arXiv preprint arXiv:1606.03498 [Google Scholar]
- 136.Heusel M, Ramsauer H, Unterthiner T, Nessler B, Hochreiter S (2017) Gans trained by a two time-scale update rule converge to a local nash equilibrium. Adv Neural Inf Proces Syst 2017:6626–6637 [Google Scholar]
- 137.Sajjadi MS, Bachem O, Lucic M, Bousquet O, Gelly S (2018) Assessing generative models via precision and recall. Adv Neural Inf Proces Syst 2018:5228–5237 [Google Scholar]
- 138.Gao W, Coley CW (2020) The synthesizability of molecules proposed by generative models. J Chem Inf Model 60(12):5714–5723 [DOI] [PubMed] [Google Scholar]
- 139.Coley CW, Rogers L, Green WH, Jensen KF (2018) SCScore: synthetic complexity learned from a reaction corpus. J Chem Inf Model 58:252–261 [DOI] [PubMed] [Google Scholar]
- 140.Popova M, Isayev O, Tropsha A (2018) Deep reinforcement learning for de novo drug design. Sci Adv 4:eaap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sumita M, Yang X, Ishihara S, Tamura R, Tsuda K (2018) Hunting for organic molecules with artificial intelligence: molecules optimized for desired excitation energies. ACS Cent Sci 4:1126–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ertl P, Schuffenhauer A (2009) Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J Cheminformatics 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sanchez-Lengeling B, Aspuru-Guzik A (2018) Inverse molecular design using machine learning: generative models for matter engineering. Science 361:360–365 [DOI] [PubMed] [Google Scholar]
- 144.Vargesson N (2015) Thalidomide-induced teratogenesis: history and mechanisms. Birth Defects Res C Embryo Today 105:140–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Polishchuk PG, Madzhidov TI, Varnek A (2013) Estimation of the size of drug-like chemical space based on GDB-17 data. J Comput Aided Mol Des 27:675–679 [DOI] [PubMed] [Google Scholar]
- 146.Alley EC, Khimulya G, Biswas S, AlQuraishi M, Church GM (2019) Unified rational protein engineering with sequence-based deep representation learning. Nat Methods 16:1315–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
N/A


