Abstract
MRL/lpr mice have been extensively used as a murine model of lupus. Disease progression in MRL/lpr mice can differ among animal facilities, suggesting a role for environmental factors. We noted a phenotypic drift of our in-house colony, which was the progeny of mice obtained from The Jackson Laboratory (JAX; stocking number 000485), that involved attenuated glomerulonephritis, increased splenomegaly, and reduced lymphadenopathy. To validate our in-house mice as a model of lupus, we compared these mice with those newly obtained from JAX, which were confirmed to be genetically identical to our in-house mice. Surprisingly, the new JAX mice exhibited a similar phenotypic drift, most notably the attenuation of glomerulonephritis. Interestingly, our in-house colony differed from JAX mice in body weight and kidney size (both sexes), as well as in splenic size, germinal center formation, and level of anti-dsDNA auto-IgG in the circulation (male only). In addition, we noted differential expression of microRNA (miR)-21 and miR-183 that might explain the splenic differences in males. Furthermore, the composition of gut microbiota was different between in-house and new JAX mice at early time points, which might explain some of the renal differences (e.g., kidney size). However, we could not identify the reason for attenuated glomerulonephritis, a shared phenotypic drift between the two colonies. It is likely that this was due to certain changes of environmental factors present in both JAX and our facilities. Taken together, these results suggest a significant phenotypic drift in MRL/lpr mice in both colonies that may require strain recovery from cryopreservation.
INTRODUCTION
Systemic lupus erythematosus (SLE) is an autoimmune disease that affects multiple organs of the body, including lymphoid organs as well as peripheral organs such as kidney and brain. The manifestations of SLE include skin rash, blood/serum abnormalities (e.g., IFN signature, anti-nuclear Abs), lupus nephritis, arthritis, splenomegaly, lung inflammation, and neurologic damage. A large proportion of patients also exhibit lymphadenopathy as a major side effect that may not be caused by SLE. Many mouse models of SLE have been developed over the years, including both spontaneous (e.g., NZB/W F1, MRL/lpr, BXSB) and inducible models (e.g., pristane-induced lupus). Among these, the MRL/lpr model (MRL/Mp-Faslpr/lpr) stands out as one that exhibits all of the manifestations and side effects of SLE described above. Compared to its parent strain MRL, MRL/lpr mice develop lupus-like disease early in life due to the Faslpr/lpr mutation. Importantly, however, note that their clinical signs do not solely depend on this mutation; rather, the exhibition of lupus-like disease mainly comes from the MRL background, where multiple SLE susceptibility loci are present. The Faslpr/lpr mutation simply accelerates the disease, thus making MRL/lpr a more efficient model than MRL.
We have studied the MRL/lpr model for the past 10 y. Female mice develop severe lupus-like disease (e.g., skin rash, glomerulonephritis, splenomegaly, lymphadenopathy, arthritis, brain inflammation) at around 16 wk of age. In recent years, however, we noticed the attenuation of kidney disease in our in-house colony, which had originated from The Jackson Laboratory (JAX; stock number 000485). This was not entirely surprising, as JAX had experienced a loss of disease phenotype in their colony ~15 y ago, when they had to recover the strain from cryopreservation to preserve the original phenotype. In that incident, the loss of phenotype was manifested by reduced splenomegaly and lymphadenopathy, as well as prolonged survival. Importantly, the single-nucleotide polymorphism (SNP) profile did not show any genetic drift, suggesting that the phenotypic drift may be due to environmental factors.
As we routinely use both the in-house colony and mice directly purchased from JAX for our studies, it became important to investigate whether our in-house colony had indeed lost the kidney phenotype compared with JAX mice. In this study, we report a phenotypic drift of MRL/lpr mice in both our and JAX colonies. Both exhibit attenuated glomerulonephritis; however, some clinical signs are different between the two types of mice in a sex-dependent manner, even though they are genetically identical. In addition, because MRL/lpr mice manifest signature changes in splenic microRNAs (miRNAs) and gut microbiota, these parameters are analyzed to investigate their potential contributions to the phenotypic drift.
MATERIALS AND METHODS
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of Virginia Tech College of Veterinary Medicine (Animal Welfare Assurance number A3208-01). All animal experiments were conducted under IACUC protocol 18-060. Euthanasia was performed with CO2 according to the IACUC protocol.
Mice
All MRL/lpr (MRL/Mp-Faslpr/lpr) mice originated from The Jackson Laboratory (JAX). Mice purchased in 2015 were bred and the colony was maintained in our animal facility at Virginia-Maryland College of Veterinary Medicine (VMCVM) for 3 y. Mice in the “in-house” group were descendants from this colony. As a comparison, 5-wk-old female and male were introduced from JAX in 2018 right before the initiation of this study and identified as the “JAX” group. All mice were monitored for proteinuria every week and euthanized at 15 wk of age. Urine samples were analyzed with a Pierce Coomassie protein assay kit (Thermo Scientific). Body and whole-tissue weights were recorded, and the level of anti-dsDNA IgG was determined as previously described (1-7). Tail clips were sent to JAX for SNP analysis to determine any genetic drift from the MRL background. An additional group of mice labeled “JAX-2015” were purchased in 2015 at 3–5 wk of age and euthanized at 15 wk of age. For this group, historical data were used.
Flow cytometry
Spleen was collected and mashed in 70-μm cell strainers with C10 medium (RPMI 1640, 10% FBS, 1 mM sodium pyruvate, 1% 100× MEM nonessential amino acids, 10 mM HEPES, 55 μM 2-ME, 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin, all from Life Technologies, Grand Island, NY). To isolate splenocytes, RBCs were lysed with RBC lysis buffer (eBioscience, San Diego, CA). For surface marker staining, cells were blocked by anti-mouse CD16/32 (eBioscience), stained with fluorochrome-conjugated Abs, and analyzed with a BD FACSAria Fusion flow cytometer (BD Biosciences, San Jose, CA). Anti-mouse Abs used in this study include: CD38-FITC, GL7-allophycocyanin, CD19-BV421, and CD138-PerCP-Cy5 (all from BioLegend, San Diego, CA). For analysis of germinal center B cells, the plots were pregated on CD19+ cells. Flow cytometry data were analyzed with FlowJo.
Immunohistochemistry
Splenic and kidney sections were embedded in Tissue-Tek OCT compound (Sakura Finetek) and rapidly frozen in a freezing bath of dry ice and 2-methylbutane. Frozen OCT samples were cryosectioned and unstained slides were stored at −80°C. Immunohistochemical staining procedures were performed as previously described (1, 2, 5). For detection of germinal centers in the spleen, the following monoclonal Abs were used: GL7-AF488, IgD-PE, and CD4-allophycocyanin (all from BioLegend). For detection of renal deposition, C3-FITC (Cedarlane, Burlington, NC) and IgG2a-PE (eBioscience) Abs were used. Images were captured with a Zeiss LSM 880 confocal microscope (Fralin Imaging Center, Virginia Tech). Corrected total cell fluorescence scores were calculated with ImageJ software (National Institutes of Health, Rockville, MD).
Histopathology
Kidneys were fixed in formalin immediately after isolation. Fixed tissues were paraffin-embedded, sectioned, and stained for periodic acid-Schiff at the Histopathology Laboratory at VMCVM. Glomerular lesions were graded for increased cellularity, increased mesangial matrix, necrosis, percentage of sclerotic glomeruli, and presence of crescents (8). Similarly, tubulointerstitial lesions were graded for interstitial mononuclear infiltration, tubular damage, interstitial fibrosis, and vasculitis. Slides were scored by a board-certified veterinary pathologist (V.O.) in a blinded fashion.
Analysis of splenic miRNAs
The spleen was removed immediately after the mouse was euthanized and mashed into a single-cell suspension. After flowing through a 70-μm nylon mesh (Fisher Scientific), the cells were treated with ACK (ammonium-chloride-potassium) lysis buffer to deplete erythrocytes. The freshly prepared splenic lymphocytes were pelleted, washed with cold PBS, and stored at −80°C. Total RNA containing small RNA was extracted from cells using the miRNeasy mini kit (Qiagen) following the manufacturer’s protocol. On-column DNA digestion was performed to remove any residual DNA in the RNA samples during RNA extraction. Concentration and purity of RNA were determined by a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), and samples with the OD260/280 ratio of ~2.0 were used. TaqMan miRNA assays (Applied Biosystems, Waltham, MA) were used to quantify miRNA expression levels. Briefly, the total RNAs were reverse transcribed into cDNA with the TaqMan miRNA reverse transcription kit, followed by quantitative real-time PCR with the TaqMan miRNA assay reagent using the 7500 Fast real-time PCR system (Applied Biosystems). The relative expression level of a specific miRNA was normalized to the endogenous small nucleolar RNA 202 (sno202) and calculated using the 2−ΔΔCt method.
16S rRNA sequencing analysis of gut microbiota
Fecal samples were collected weekly directly from the anus of the mouse. To avoid cross-contamination, each microbiota sample was collected by using a new pair of sterile tweezers. Samples were stored at −80°C until being processed at the same time. Sample homogenization, cell lysis, and DNA extraction were performed in-house. PCR analyses were performed and purified amplicons (V4 region) were sequenced bidirectionally on an Illumina MiSeq at Argonne National Laboratory. Data analysis was performed as previously described (3, 4, 6, 7, 9), and 16S rRNA sequences are available in the National Institutes of Health SRA database (PRJNA758126).
Statistical analysis
For the comparison of two groups, an unpaired Student t test was used. For the comparison of three groups, one-way ANOVA and a Tukey’s posttest were used. Two-way ANOVA was used to reveal time- and group-dependent effects. High-dimensional omics data were visualized by principal-coordinate analysis with permutational multivariate ANOVA. Results were considered statistically significant when p < 0.05. All analyses were performed with GraphPad Prism software.
RESULTS
Phenotypic drift in lupus-prone MRL/lpr mice
We routinely breed MRL/lpr mice in-house and only reintroduce the mouse strain from JAX every 2–3 y. Prior to this study, we had purchased female and male MRL/lpr mice in 2015 and started a breeding colony at VMCVM. Two years later, we noticed that the mice in our colony had delayed onset of proteinuria. By 2018, we could no longer detect a significant increase of proteinuria even at the endpoint of 15 wk of age. As lupus-prone mice were known to exhibit different levels of disease severity in different facilities (10), we hypothesized that the animal housing conditions at VMCVM led to the gradual loss of disease phenotype and particularly attenuated glomerulonephritis. To test the hypothesis, we purchased 5-wk-old female and male MRL/lpr mice from JAX that were age-matched to a group of mice in our in-house colony. Surprisingly, both groups of mice (in-house versus JAX) regardless of sex exhibited low levels of proteinuria at 15 wk of age (Fig. 1A). As a comparison we used historical data from mice purchased and analyzed in 2015, which had significantly higher levels of proteinuria at 15 wk of age, especially for females.
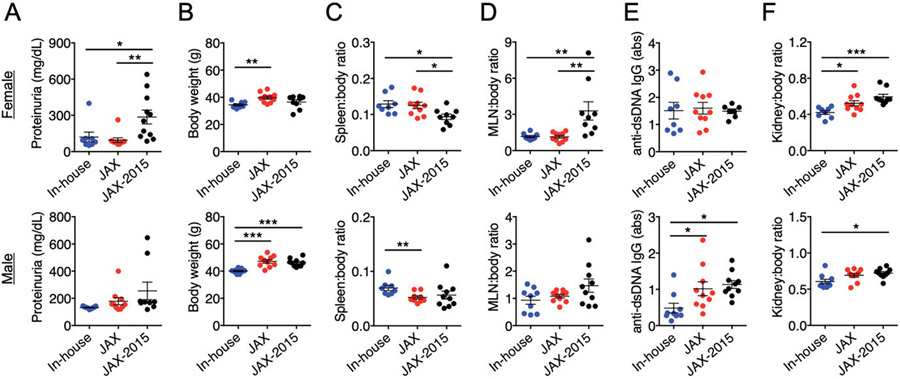
FIGURE 1. Phenotypic drift in MRL/lpr mice.
Female (top) and male (bottom) MRL/lpr from our in-house colony were compared with those newly purchased from JAX in 2018 (JAX) and those purchased 3 y prior (JAX-2015). In females, n = 8, 10, and 9 for in-house, JAX, and JAX-2015 groups, respectively. In males, n = 9, 10, and 10 for the three respective groups. Characterization of the disease phenotype was performed when mice were 15 wk of age. Historical data were used for JAX-2015. (A) Level of proteinuria. (B) Body weight. (C) Spleen-to-body weight ratio. (D) Mesenteric lymph node-to-body weight ratio. (E) Serum level of anti-dsDNA IgG. (F) Kidney-to-body weight ratio. Statistical significance (*p < 0.05, **p < 0.01, ***p < 0.001) is shown based on one-way ANOVA.
Interestingly, although the level of endpoint proteinuria was similarly low, the new JAX mice (purchased and analyzed in 2018) were considerably larger than the MRL/lpr mice in our in-house colony for both females and males (Fig. 1B). Upon euthanasia at 15 wk of age, we measured the weight of different organs including spleen, mesenteric lymph node (MLN), and kidney, and calculated the organ-to-body weight ratios. Our in-house male mice had significantly larger spleens than did the new JAX mice, whereas the size of the spleen was similar between female in-house and JAX mice (Fig. 1C). Both female groups sacrificed in 2018 had significantly larger spleens than female mice sacrificed back in 2015. This suggests that the housing conditions at VMCVM did not adversely affect splenomegaly, one of the key clinical signs of lupus-like disease in MRL/lpr mice. The weight of MLNs, in contrast, was significantly smaller in female in-house and new JAX mice than that recorded in 2015 (Fig. 1D). Both in-house and new JAX mice had similarly small MLNs, suggesting a phenotypic drift in lymphadenopathy at JAX as well. Another hallmark of lupus-like disease in MRL/lpr mice is the increase of autoantibodies, including anti-dsDNA IgG. Although there was no difference in female mice, the male mice in our colony had a significantly lower level of anti-dsDNA IgG in the blood than did both groups obtained from JAX (Fig. 1E), suggesting a loss of this disease parameter only in males. Moreover, we measured the weight of the kidney and observed that our in-house MRL/lpr mice had significantly smaller kidneys than those in the JAX mice (Fig. 1F).
These results indicate a significant phenotypic drift in our in-house MRL/lpr mice from mice purchased in 2015 that involves attenuated proteinuria and smaller MLNs and kidneys (both sexes), larger spleens (females), and smaller body weight and reduced circulatory autoantibodies (males). Surprisingly, female MRL/lpr mice at JAX also exhibited a phenotypic drift during 3 y that involved attenuated proteinuria, increased splenomegaly, and reduced lymphadenopathy. To exclude the contribution from genetic factors, we ordered SNP analysis that did not identify any genetic drift (100% identical between in-house and JAX mice). This suggests that the phenotypic drift in MRL/lpr mice, most notably the attenuated proteinuria, may be solely due to environmental factors present both in-house and at JAX.
Splenic differences between in-house and new JAX mice
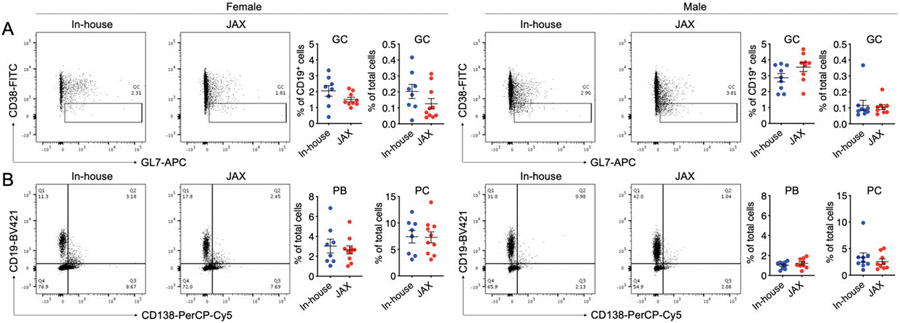
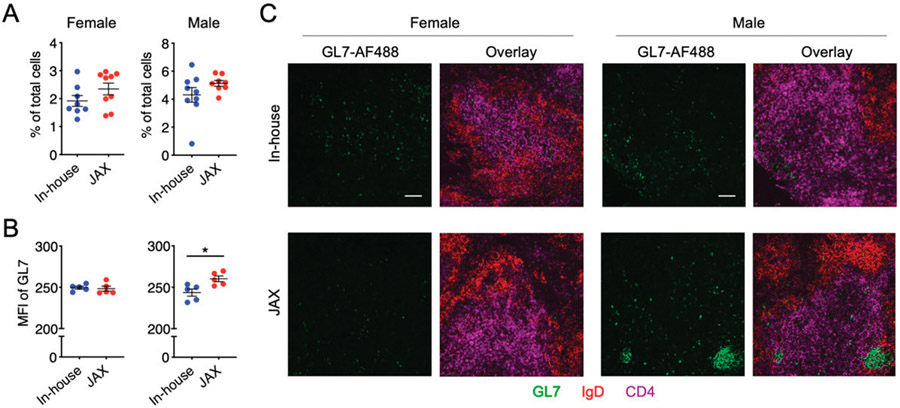
As splenomegaly was exacerbated in in-house male mice, we asked whether there were immunological changes in the spleen. Neither the frequencies of germinal center B cells (CD19+GL7+ CD38−/low) nor those of plasmablasts (CD19+CD138+) and plasma cells (CD19−CD138+) were different between in-house and new JAX mice, regardless of sex (Fig. 2A, 2B). However, we noted a small increase of total GL7-expressing cells (Fig. 3A), which prompted us to investigate the localization of germinal center B cells in relevance to splenic Th cells. Interestingly, GL7-expressing cells were scattered among Th cells in the splenic section, suggesting the lack of germinal center formation in both groups of females as well as in-house male mice. However, in the new JAX male mice, these cells were significantly brighter (Fig. 3B) and clustered (Fig. 3C). Although cells expressing a higher amount of GL7 have not been shown to exhibit enhanced functions, the clustering pattern suggests that the spleen of male mice newly obtained from JAX may contain more mature germinal centers even though the spleen size was smaller. This is consistent with the observation that the level of anti-dsDNA IgG was significantly higher in this group of male mice.
FIGURE 2. Splenic differences between in-house and new JAX mice as assessed with flow cytometry.
(A) Frequency of GL7+CD38− germinal center (GC) B cells in B cells or total live cells in the spleen of female (left) and male (right) MRL/lpr mice. The FACS plots were regated on CD19+ cells. (B) Frequencies of CD19+CD138+ plasmablasts (PBs) and CD19−CD138+ plasma cells (PCs) in total live cells in the spleen of female (left) and male (right) MRL/lpr mice.
FIGURE 3. Splenic differences between in-house and new JAX mice as assessed with immunohistochemistry.
(A) Frequency of GL7+ cells in total splenocytes based on flow cytometry. (B) Mean fluorescence intensity (MFI) of GL7 based on immunohistochemistry. Splenic sections from five randomly selected mice per group were stained and quantified. Statistical significance (*p < 0.05) is shown based on Student t test. (C) Representative images of splenic sections stained with fluorescent conjugated Abs (GL7, IgD, CD4). Scale bar, 100 μm.
Renal differences between in-house and new JAX mice
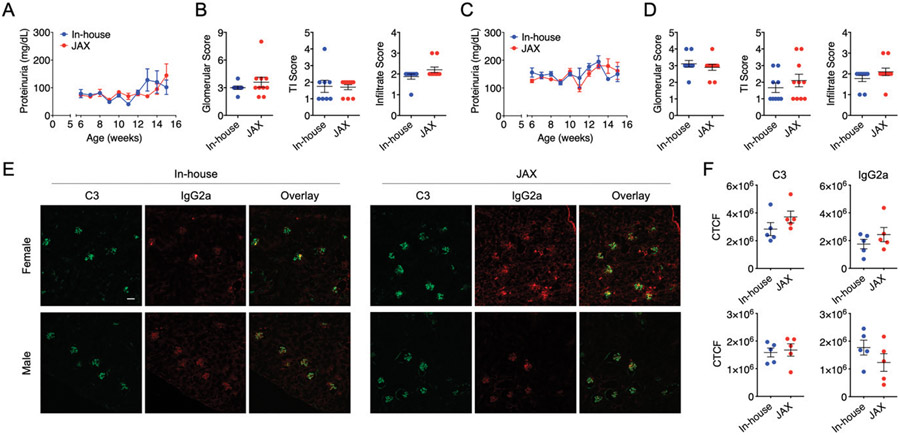
We were particularly interested in glomerulonephritis that made the MRL/lpr mice a classical model of lupus nephritis, a leading cause of mortality for SLE patients (11). Using weekly collected urine samples, we observed that the time course of proteinuria was not different between our in-house mice and new JAX mice for both females and males (Fig. 4A, 4C). There was a slight increase of proteinuria in both female groups close to the endpoint, but it was not statistically significant. Importantly, histopathological analysis (Fig. 4B, 4D) revealed no difference in glomerular and tubulointerstitial lesions as well as leukocyte infiltration between the in-house and new JAX mice. Immunohistochemical analysis also showed no difference in renal depositions of complement C3 and IgG2a, the pathogenic IgG in this mouse strain (Fig. 4E, 4F). These results suggest that the severity of renal damage is the same between in-house and new JAX mice, and that the environmental factors driving the attenuation of glomerulonephritis may be common between the two facilities.
FIGURE 4. Renal differences between in-house and new JAX mice.
(A–D) Time course of proteinuria [(A) for females, (C) for males)] and kidney histopathological scores [(B) for females, (D) for males)] are shown. TI, tubulointerstitial. (E) Representative images of kidney sections stained with fluorescent conjugated Abs (complement C3, IgG2a). Scale bar, 100 μm. (F) Quantification of kidney immunohistochemistry data for female (top) and male (bottom) MRL/lpr mice.
Analysis of lupus-associated miRNAs
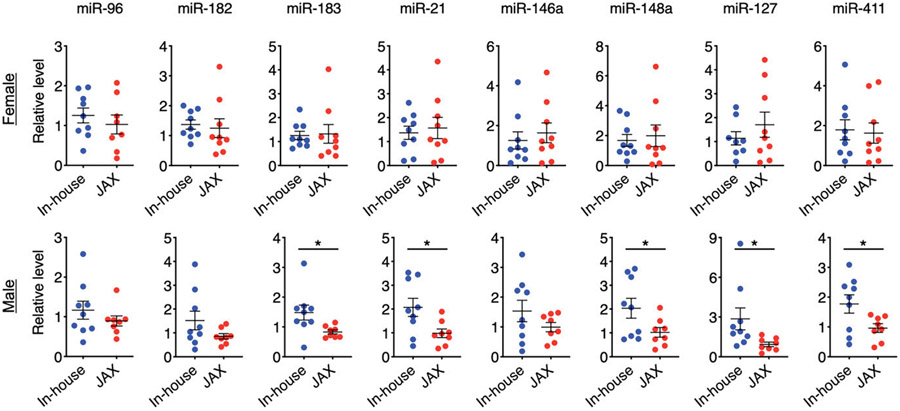
Although the severity of glomerulonephritis was the same between the two groups, the MRL/lpr mice in our colony did have significantly smaller kidneys and body weight (both sexes), and significantly larger spleens and lower blood levels of anti-dsDNA IgG (males) than did the mice newly purchased from JAX. We asked whether there were associations between the disease phenotype and differential expression of lupus-associated miRNAs in the spleen. We had previously identified several lupus-associated miRNAs that were increased in multiple murine models of SLE compared with healthy controls (12). While the levels of these miRNAs were not statistically different between JAX and in-house female mice due to large variations within groups, in-house males had significantly higher levels of miRNA (miR)-183, miR-21, miR-148a, miR-127, and miR-411 than did males purchased from JAX in 2018 (Fig. 5). A similar trend was observed for miR-182 and miR-146a in males, albeit no statistical significance was found. Notably, miR-146 has been shown to limit germinal center formation (13); thus, the lower level of this miRNA in new JAX mice may be associated with more mature germinal centers in these mice.
FIGURE 5. Analysis of lupus-associated miRNAs.
Relative levels of miRNA expression in the spleen of female (top) and male (bottom) MRL/lpr mice are shown. Statistical significance (*p < 0.05) is shown based on a Student t test.
Among the noted lupus-associated miRNAs, the expression of miR-21 has been shown to be positively correlated with splenic size in lupus-prone mice (14), consistent with our observation of larger spleens in male in-house MRL/lpr mice. Interestingly, although our previous studies point to a pathogenic role for miR-183 (12, 15), a recent report showed that i.p. injection of this miRNA attenuated several disease parameters in MRL/lpr mice, including the reduction of anti-dsDNA Abs (16). It is consistent with our observation that in-house males had less anti-dsDNA IgG in the circulation while expressing more miR-183 in the spleen. Taken together, these results suggest that the differential expression of miR-21 and miR-183 in the spleen may explain the phenotypic differences observed for male mice in our colony versus those newly purchased from JAX. In addition, the lack of miRNA expression difference in the female spleen is consistent with the lack of phenotypic difference in this tissue for females.
Analysis of gut microbiota
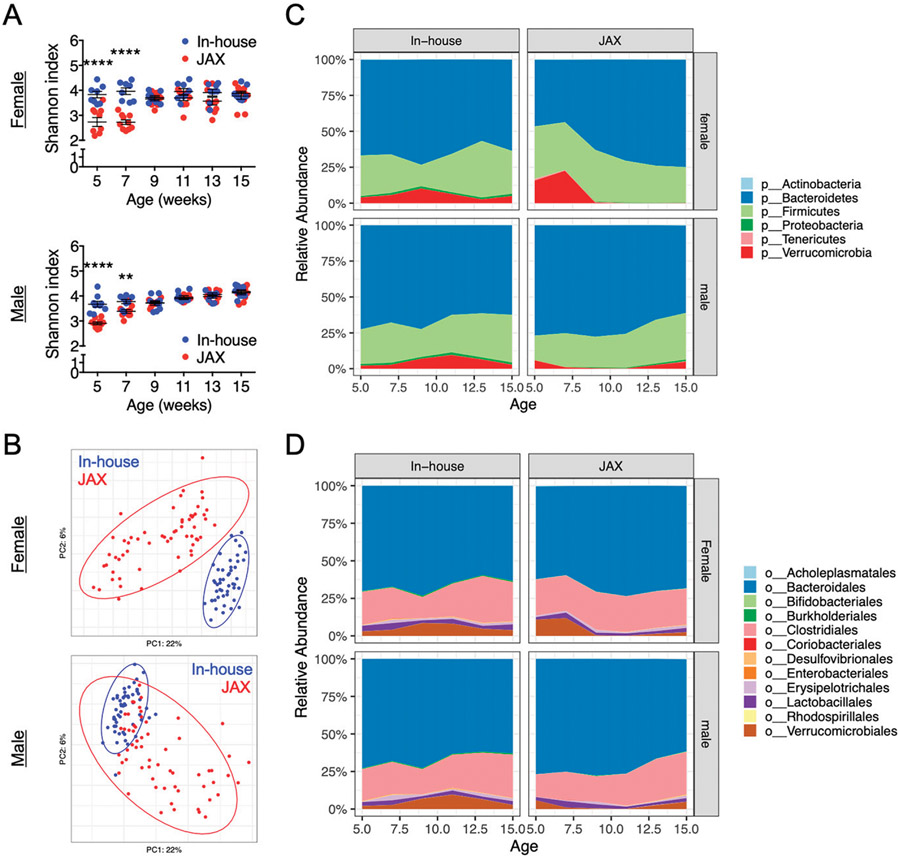
To establish associations between changes of disease phenotype with the composition of gut microbiota, we analyzed fecal samples collected from the two groups of mice harvested in 2018. It was evident that at the earlier time points of 5 and 7 wk of age, the gut microbiota of our in-house mice was significantly more diverse than that of the new JAX mice as indicated by the Shannon index (Fig. 6A). This was true for both female and male mice, suggesting that the housing conditions were indeed different between our facility and JAX. In addition, based on principal-coordinate analysis, the gut microbiotas were significantly distinct when comparing the two groups, especially for female mice (Fig. 6B). At the phylum level (Fig. 6C), it was consistent between sexes that Proteobacteria were significantly higher in the in-house mice than in the new JAX mice (Supplemental Fig. 1). Increased Proteobacteria has been found to be associated with human SLE (17). Other phyla that were different included significantly increased Verrucomicrobia in the in-house male mice, and significantly increased Tenericutes (females) and Bacteroidetes (males) in the new JAX mice (Supplemental Fig. 1). Verrucomicrobia, which is essentially Akkermansia muciniphila, is considered protective in the pathogenesis of some autoimmune diseases (18, 19). The opposite functions of proinflammatory Proteobacteria and anti-inflammatory Verrucomicrobia may neutralize each other, leading to similar levels of severity in glomerulonephritis between our in-house mice and the new JAX mice (Fig. 4). At the order level (Fig. 6D), Lactobacillales was significantly higher in female in-house mice (Supplemental Fig. 2). Erysipelotrichales, in contrast, was significantly higher in female JAX but significantly lower in male JAX mice than in our in-house mice. These two orders represented ~5–10% of the gut bacteria.
FIGURE 6. Analysis of gut microbiota composition.
(A) Shannon diversity index over time for the fecal microbiota of female (top) and male (bottom) MRL/lpr mice. Statistical significance between groups (**p < 0.01, ****p < 0.0001) is shown based on two-way ANOVA. (B) Principal coordinate analysis of fecal microbiota composition for female (top) and male (bottom) MRL/lpr mice. (C) Changes of relative bacterial abundance over time at the phylum level in the fecal microbiota of female (top) and male (bottom) MRL/lpr mice. (D) Changes of relative bacterial abundance over time at the order level in the fecal microbiota of female (top) and male (bottom) MRL/lpr mice.
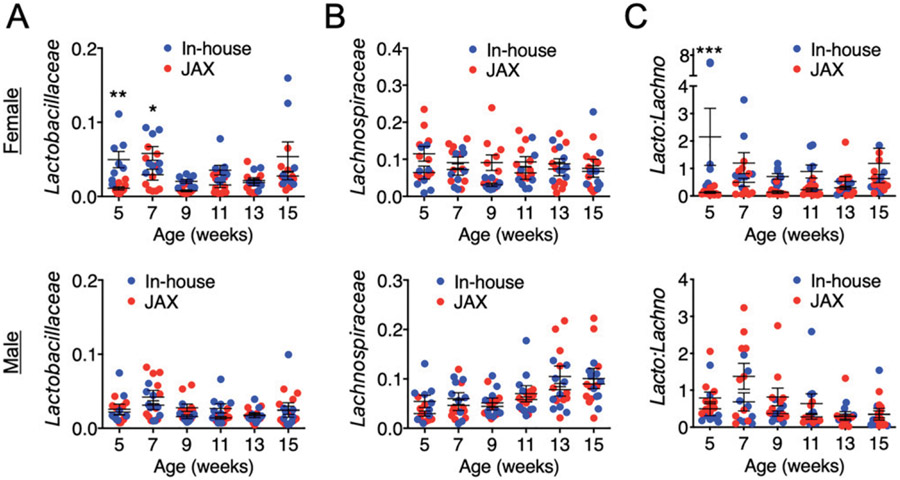
We paid special attention to the families Lactobacillaceae and Lachnospiraceae, which we had previously shown to be associated with disease outcomes in female MRL/lpr mice (20). A higher level of Lactobacillaceae and a lower level of Lachnospiraceae were associated with less severe lupus-like disease, including glomerulonephritis, splenomegaly, lymphadenopathy, and autoantibodies. In this study, we found time-dependent differences of Lactobacillaceae, with an increased abundance of these bacteria in our colony than in the new JAX mice only in females at the earlier time points of 5 and 7 wk of age (Fig. 7A). However, it is difficult to establish associations with disease because the phenotypic differences in females were not obvious between the two groups except the body weight and kidney size. The levels of Lachnospiraceae were not different for both sexes (Fig. 7B). Therefore, the difference of the ratio of Lactobacillaceae to Lachnospiraceae resembled the difference in Lactobacillaceae (Fig. 7C).
FIGURE 7. Analysis of gut microbiota with a focus on Lactobacillaceae and Lachnospiraceae.
(A) Relative abundance of Lactobacillaceae over time in the fecal microbiota of female (top) and male (bottom) MRL/lpr mice. (B) Relative abundance of Lachnospiraceae over time in the fecal microbiota of female (top) and male (bottom) MRL/lpr mice. (C) The ratio of Lactobacillaceae to Lachnospiraceae. Statistical significance between groups (*p < 0.05, **p < 0.01, ***p < 0.001) is shown based on two-way ANOVA.
Many groups of bacteria at the genus level were altered comparing in-house versus JAX mice (Supplemental Table I). Consistent for both females and males, Odoribacter and Parabacteroides from the family of Porphyromonadaceae and Flavonifractor in Ruminococcaceae were significantly upregulated in our in-house colony. In contrast, the following genera were significantly upregulated in the new JAX mice: Eubacterium of Lachnospiraceae, Romboutsia of Peptostreptococcaceae, and Acutalibacter of Ruminococcaceae. One genus of interest was Enterococcus under the order Lactobacillales, family Enterococcaceae. It has been recently discovered that Enterococcus gallinarum can translocate into the liver of lupus-prone mice to facilitate the development of autoimmunity (21). Enterococcus was significantly higher in female mice from JAX, which might be associated with the significant increase of kidney size comparing female JAX to female in-house mice.
Taken together, these results indicate that the gut microbiota is different between in-house and new JAX mice at early time points, suggesting significant housing/environmental differences in the two facilities. They also suggest that the difference in gut microbiota may explain some of the phenotypic differences (e.g., kidney size) but not the attenuated glomerulonephritis, which is a shared phenotype between in-house and new JAX mice. The latter may be due to certain changes of environmental factors common for both our and JAX facilities.
DISCUSSION
The MRL (Murphy Roths large) background, which came from various mouse strains including LG/J, AKR/J, C3H/Di and C57BL/6J, distinguishes MRL/lpr mice from other mouse strains with the Faslpr mutation. Genomic mapping identified four SLE susceptibility loci in mice with the MRL background that contribute to splenomegaly, lymphadenopathy, and glomerulonephritis (22). The loci responsible for splenomegaly and lymphadenopathy (chromosomes 4, 5, and 7) are different from those important for glomerulonephritis (chromosome 10). Notably, our in-house mice are different from the new JAX mice in the spleen (splenic size, germinal center formation, and level of anti-dsDNA autoantibodies; males only), but similar to JAX in terms of attenuated glomerulonephritis (both sexes). Although we have confirmed that the in-house mice were genetically identical to the new JAX mice, it remains unclear whether there had been a genetic drift in both colonies during a period of 3 y (2015–2018), particularly within chromosome 10. Notably, female JAX mice from 2015 exhibited a significantly higher level of proteinuria than female JAX mice purchased in 2018. In addition, although both were from JAX, female mice purchased in 2015 had significantly smaller spleens and significantly larger MLNs than did female mice purchased in 2018. These results suggest a significant phenotypic drift in MRL/lpr mice that may require JAX to perform strain recovery from cryopreservation.
We next explored the potential associations between the phenotypic changes and splenic expression of lupus-associated miRNAs. We have previously reported a positive correlation between miR-183-96-182 cluster miRNAs (miR-183C) and murine lupus (12, 15). Previous studies from another laboratory also showed the pathogenic role of miR-182 in lupus, as inhibition of miR-182 in vivo with antagomir-182 was able to attenuate lupus nephritis in MRL/lpr mice (23). In addition, our unpublished data showed that conditional deletion of the whole miR-183C cluster specifically in lymphocytes significantly reduced the serum level of anti-dsDNA as well as IgG deposition in the kidney, but not renal function in C57BL/6.MRL-Faslpr mice. Lymphocyte-specific deletion of miR-182 alone, in contrast, reduced renal deposition of IgG and slightly suppressed the serum level of anti-dsDNA. Consistent with these results, positive correlations between miR-182/miR-183c have been observed in murine models of rheumatoid arthritis (24) and multiple sclerosis (25, 26). Notably, although these studies showed potential pathogenic roles of miR-182 and miR-183C in lupus nephritis and other autoimmune diseases, contradictory data exist regarding the role of miR-183 in lupus nephritis. miR-183 was found to be reduced in the renal biopsy of MRL/lpr mice and human patients with lupus nephritis (16), suggesting a potential protective role of this miRNA. In addition, i.p. delivery of miR-183 mimics reduced anti-dsDNA Abs and attenuated lupus nephritis in MRL/lpr mice (16). If this miRNA indeed plays a protective role in lupus, our results of reduced miR-183 expression in male (but not female) JAX mice may explain the increase of anti-dsDNA autoantibodies specifically in male JAX mice. Female in-house and JAX mice, in contrast, did not differ in either the phenotype or miRNA expression in the spleen.
Recent studies have started to establish the effects of different miRNAs on gut microbiota composition. Without miR-21, for example, Proteobacteria (27) and Lactobacillus spp. (28) flourished in the mouse gut microbiota. This is opposite to our observations for splenic expression of miR-21 and the relative abundances of Proteobacteria and Lactobacillus spp. in male in-house versus JAX mice. However, deletion of miR-21 also led to a reduction of Verrucomicrobia (27), which is consistent with our observation that JAX mice with lower splenic expression of miR-21 had significantly lower Verrucomicrobia in the gut microbiota. This suggests a possible connection between splenic miRNA expression and the composition of gut microbiota.
Importantly, note that once both types of mice were in our facility, they were housed in the same room on the same rack. Therefore, starting at 5 wk of age, the housing conditions were essentially the same including bedding, water, feed, light/dark hours, noise, and room temperature and humidity (29). However, the JAX mice had been exposed to different housing conditions as detailed in Table I. MRL/lpr mice were housed in maximum barrier rooms at JAX with acidified water and more excluded pathogens or opportunistic agents. In addition, a quaternary ammonium compound (QAC) disinfectant was used at JAX. We have previously shown that QAC disinfectants decrease splenomegaly in female MRL/lpr mice (30). In this study, the splenic size was not different between JAX and in-house female mice, probably because our facility shared a cage washing machine with adjacent facilities that did use QAC disinfectants, leading to ambient exposure of in-house mice to QACs. However, although we did not investigate male mice in the previous QAC study (30), our observation that the male splenic size was significantly larger in in-house versus new JAX mice may be explained by a QAC-associated decrease in splenomegaly. Furthermore, the differences in previous housing conditions may explain why JAX mice after arriving at our facility harbored significantly different gut microbiotas from our in-house mice during the first month.
TABLE I.
Comparison of housing conditions between JAX and our facilities
| Housing Conditions | VMCVM (in-house) | JAX |
|---|---|---|
| Caging | Microisolator cages on individually ventilated caging racks; air-exchange rate of 60 per hour; covered with filter lids; sanitization in a mechanical cage washer | Microisolator cages on individually ventilated caging racks; air-exchange rate of 60 per hour; covered with filter lids; washed cages are bedded and steam-autoclaved |
| Bedding | Teklad diamond dry cellulose bedding; red shelter huts and diamond twists for enrichment | Equal parts of Aspen shavings and Aspen Sani-Chips; shelter-type enrichment or additional nesting material |
| Cage density | Five mice per cage | Five mice per cage |
| Feed | Teklad diet formulation 2918, a natural ingredient, irradiated feed (6% fat, 18% protein) ad libitum | LabDiet 5K52 formulation (6% fat, 19% protein) ad libitum |
| Water | Supplied from a municipal water source; water is not treated but is tested quarterly for microbial contamination; dispensed into bottles | Supplied from a municipal water source and then treated with ultraviolet light, filtered, and acidified using hydrochloric acid to pH 2.5–3.0 to control bacterial growth; dispensed into bottles and then autoclaved |
| Temperature | 72 ± 2°F | 65–75°F |
| Humidity | 30–70% | 30–70% |
| Light cycle | 14 h of light, 10 h of dark | 14 h of light, 10 h of dark |
| Access | Restricted to only staff and research personnel | Maximum barrier room with restricted access that requires an air shower prior to entry |
| Personal protective equipment | Gown, shoe covers, gloves, mask, hair bouffant | Clean room processed scrubs, smock and shoes, gloves, air hat or mask, cap, face shield |
| Cage change | Under an animal transfer station using proper microisolator techniques | Under a clean transfer station; mice are handled with smooth-tipped forceps that have been soaked in a sporicidal disinfectant |
| Disinfectant | Peroxigard, an accelerated hydrogen peroxide | Quaternary ammonium-based disinfectant |
| Excluded murine pathogens | Ectoparasites (fur mites and lice), pinworms (Aspiculuris and Syphacia spp.), giardia, cilia-associated respiratory bacillus, Citrobacter rodentium, Clostridium piliforme, Encephalitozoon cuniculi, Mycoplasma pulmonis, Pneumocystis carinii (muris), Salmonella species, Streptobacillus moniliformis, lymphocytic choriomeningitis virus, minute virus of mice, mouse adenovirus, mouse hepatitis virus, mouse parvovirus (MPV1, MPV2, MPV3), mouse rotavirus/epizootic diarrhea of infant mice, mousepox/ectromelia virus, pneumonia virus of mice, reovirus, Sendai virus, Theiler’s murine encephalomyelitis virus (GDVII) | Ectoparasites (fleas, fur mites, and lice), pinworms (Aspiculuris and Syphacia spp.), giardia, Spironucleus, Toxoplasma gondii, Encephalitozoon cuniculi, cilia-associated respiratory bacillus, Bordetella, Citrobacter rodentium, Clostridium piliforme, Corynebacterium bovis and Corynebacterium kutscheri, Mycoplasma spp., Pneumocystis carinii (muris), Salmonella spp., Streptobacillus moniliformis, lymphocytic choriomeningitis virus, minute virus of mice, mouse adenovirus, mouse norovirus, mouse CMV, mouse hepatitis virus, Hantaan virus, mouse parvovirus (MPV1, MPV2, MPV3), murine chapparvovirus, epizootic diarrhea of infant mice, mousepox/ectromelia virus, pneumonia virus of mice, reovirus, Sendai virus, Theiler’s murine encephalomyelitis virus (GDVII), lactate dehydrogenase elevating virus, polyoma virus, mouse thymic virus Excluded opportunistic agents include Helicobacter spp., Klebsiella spp., Pasteurella spp., Pneumocystis murina, Proteus mirabilis, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus pneumoniae, β-hemolytic Streptococcus spp., Yersinia spp., dermatophytes, trichomonads |
In summary, we have characterized a phenotypic drift in MRL/lpr mice housed in our facility as well as at JAX. Some phenotypic changes may be associated with differential expression of miRNAs in the spleen and/or changes of the gut microbiota composition. Future studies will involve modifications of specific miRNAs (e.g., miR-21 or miR-183) or commensal bacteria (e.g., Enterococcus) followed by phenotypic characterizations of MRL/lpr mice to provide more mechanistic insight.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sarah Owens for assistance on Illumina MiSeq sequencing, and Husen Zhang for bioinformatics analysis of 16S rRNA sequences. We also thank Melissa Makris for assistance on flow cytometry.
This work was supported by internal funding from Virginia-Maryland College of Veterinary Medicine and National Institutes of Health Grants AR067418 and AR073240 (X.M.L.).
Abbreviations used in this article:
- IACUC
Institutional Animal Care and Use Committee
- JAX
The Jackson Laboratory
- miR/miRNA
microRNA
- MLN
mesenteric lymph node
- QAC
quaternary ammonium compound
- SLE
systemic lupus erythematosus
- SNP
single-nucleotide polymorphism
- VMCVM
Virginia-Maryland College of Veterinary Medicine.
Footnotes
The 16S rRNA sequences presented in this article have been submitted to the National Institutes of Health SRA database (https://www.ncbi.nlm.nih.gov/bioprojectPRJNA758126/) under accession number PRJNA758126.
DISCLOSURES
The authors have no financial conflicts of interest.
Supplementary Material http://www.immunohorizons.org/content/suppl/2022/01/17/immunohorizons.2100082.DCSupplemental
REFERENCES
- 1.Liao X, Ren J, Wei CH, Ross AC, Cecere TE, Jortner BS, Ahmed SA, and Luo XM. 2015. Paradoxical effects of all-trans-retinoic acid on lupus-like disease in the MRL/lpr mouse model. PLoS One 10: e0118176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao X, Ren J, Reihl A, Pirapakaran T, Sreekumar B, Cecere TE, Reilly CM, and Luo XM. 2017. Renal-infiltrating CD11c+ cells are pathogenic in murine lupus nephritis through promoting CD41 T cell responses. Clin. Exp. Immunol 190: 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mu Q, Tavella VJ, Kirby JL, Cecere TE, Chung M, Lee J, Li S, Ahmed SA, Eden K, Allen IC, et al. 2017. Antibiotics ameliorate lupus-like symptoms in mice. Sci. Rep 7: 13675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mu Q, Zhang H, Liao X, Lin K, Liu H, Edwards MR, Ahmed SA, Yuan R, Li L, Cecere TE, et al. 2017. Control of lupus nephritis by changes of gut microbiota. Microbiome 5: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdelhamid L, Cabana-Puig X, Swartwout B, Lee J, Li S, Sun S, Li Y, Ross AC, Cecere TE, LeRoith T, et al. 2020. Retinoic acid exerts disease stage-dependent effects on pristane-induced lupus. Front. Immunol 11: 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mu Q, Edwards MR, Swartwout BK, Cabana Puig X, Mao J, Zhu J, Grieco J, Cecere TE, Prakash M, Reilly CM, et al. 2020. Gut microbiota and bacterial DNA suppress autoimmunity by stimulating regulatory B cells in a murine model of lupus. Front. Immunol 11: 593353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mu Q, Cabana-Puig X, Mao J, Swartwout B, Abdelhamid L, Cecere TE, Wang H, Reilly CM, and Luo XM. 2019. Pregnancy and lactation interfere with the response of autoimmunity to modulation of gut microbiota. Microbiome 7: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez de Lema G, Lucio-Cazaña FJ, Molina A, Luckow B, Schmid H, de Wit C, Moreno-Manzano V, Banas B, Mampaso F, and Schlöndorff D. 2004. Retinoic acid treatment protects MRL/lpr lupus mice from the development of glomerular disease. Kidney Int. 66: 1018–1028. [DOI] [PubMed] [Google Scholar]
- 9.Mu Q, Swartwout BK, Edwards M, Zhu J, Lee G, Eden K, Cabana-Puig X, McDaniel DK, Mao J, Abdelhamid L, et al. 2021. Regulation of neonatal IgA production by the maternal microbiota. Proc. Natl. Acad. Sci. USA 118: e2015691118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei Y, Sehnert B, Voll RE, Jacobs-Cachá C, Soler MJ, Sanchez-Niño MD, Ortiz A, Bülow RD, Boor P, and Anders HJ. 2021. A multicenter blinded preclinical randomized controlled trial on Jak1/2 inhibition in MRL/MpJ-Faslpr mice with proliferative lupus nephritis predicts low effect size. Kidney Int. 99: 1331–1341. [DOI] [PubMed] [Google Scholar]
- 11.Tsokos GC 2011. Systemic lupus erythematosus. N. Engl. J. Med 365: 2110–2121. [DOI] [PubMed] [Google Scholar]
- 12.Dai R, Zhang Y, Khan D, Heid B, Caudell D, Crasta O, and Ahmed SA. 2010. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PLoS One 5: e14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pratama A, Srivastava M, Williams NJ, Papa I, Lee SK, Dinh XT, Hutloff A, Jordan MA, Zhao JL, Casellas R, et al. 2015. MicroRNA-146a regulates ICOS-ICOSL signalling to limit accumulation of T follicular helper cells and germinal centres. Nat. Commun 6: 6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garchow BG, Bartulos Encinas O, Leung YT, Tsao PY, Eisenberg RA, Caricchio R, Obad S, Petri A, Kauppinen S, and Kiriakidou M. 2011. Silencing of microRNA-21 in vivo ameliorates autoimmune splenomegaly in lupus mice. EMBO Mol. Med 3: 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi EW, Lee M, Song JW, Shin IS, and Kim SJ. 2016. Mesenchymal stem cell transplantation can restore lupus disease-associated miRNA expression and Th1/Th2 ratios in a murine model of SLE. Sci. Rep 6: 38237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Luo F, Li J, and Luo C. 2019. miR-183 delivery attenuates murine lupus nephritis-related injuries via targeting mTOR. Scand. J. Immunol 90: e12810. [DOI] [PubMed] [Google Scholar]
- 17.Luo XM, Edwards MR, Mu Q, Yu Y, Vieson MD, Reilly CM, Ahmed SA, and Bankole AA. 2018. Gut microbiota in human systematic lupus erythematosus and a mouse model of lupus. Appl. Environ. Microbiol 84: e02288–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hänninen A, Toivonen R, Pöysti S, Belzer C, Plovier H, Ouwerkerk JP, Emani R, Cani PD, and De Vos WM. 2018. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut 67: 1445–1453. [DOI] [PubMed] [Google Scholar]
- 19.Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sørensen SJ, Buschard K, and Hansen AK. 2012. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 55:2285–2294. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Liao X, Sparks JB, and Luo XM. 2014. Dynamics of gut microbiota in autoimmune lupus. Appl. Environ. Microbiol 80: 7551–7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, Costa FRC, Tiniakou E, Greiling T, Ruff W, et al. 2018. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 359:1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vidal S, Kono DH, and Theofilopoulos AN. 1998. Loci predisposing to autoimmunity in MRL-Fas lpr and C57BL/6-Faslpr mice. J. Clin. Invest 101: 696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Wang G, Zhang X, Dou Y, Dong Y, Liu D, Xiao J, and Zhao Z. 2018. Inhibition of microRNA-182-5p contributes to attenuation of lupus nephritis via Foxo1 signaling. Exp. Cell Res 373: 91–98. [DOI] [PubMed] [Google Scholar]
- 24.Stittrich AB, Haftmann C, Sgouroudis E, Kühl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann F, et al. 2010. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat. Immunol 11: 1057–1062. [DOI] [PubMed] [Google Scholar]
- 25.Wan C, Ping CY, Shang XY, Tian JT, Zhao SH, Li L, Fang SH, Sun W, Zhao YF, Li ZY, et al. 2016. MicroRNA 182 inhibits CD4+CD25+Foxp3+ Treg differentiation in experimental autoimmune encephalomyelitis. Clin. Immunol 173: 109–116. [DOI] [PubMed] [Google Scholar]
- 26.Ichiyama K, Gonzalez-Martin A, Kim BS, Jin HY, Jin W, Xu W, Sabouri-Ghomi M, Xu S, Zheng P, Xiao C, and Dong C. 2016. The microRNA-183-96-182 cluster promotes T helper 17 cell pathogenicity by negatively regulating transcription factor Foxo1 expression. Immunity 44: 1284–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston DGW, Williams MA, Thaiss CA, Cabrera-Rubio R, Raverdeau M, McEntee C, Cotter PD, Elinav E, O’Neill LAJ, and Corr SC. 2018. Loss of microRNA-21 influences the gut microbiota, causing reduced susceptibility in a murine model of colitis. J. Crohn’s Colitis 12: 835–848. [DOI] [PubMed] [Google Scholar]
- 28.Santos AA, Afonso MB, Ramiro RS, Pires D, Pimentel M, Castro RE, and Rodrigues CMP. 2020. Host miRNA-21 promotes liver dysfunction by targeting small intestinal Lactobacillus in mice. Gut Microbes 12: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards MR, Dai R, Heid B, Cecere TE, Khan D, Mu Q, Cowan C, Luo XM, and Ahmed SA. 2017. Commercial rodent diets differentially regulate autoimmune glomerulonephritis, epigenetics and microbiota in MRL/lpr mice. Int. Immunol 29: 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelhamid L, Cabana-Puig X, Mu Q, Moarefian M, Swartwout B, Eden K, Das P, Seguin RP, Xu L, Lowen S, et al. 2020. Quaternary ammonium compound disinfectants reduce lupus-associated splenomegaly by targeting neutrophil migration and T-cell fate. Front. Immunol 11: 575179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.