Abstract
ERV-3 is an evolutionarily conserved single-copy human endogenous retrovirus with a coding envelope gene potentially involved in important placental functions. We have investigated the sequence variability of this gene among 150 unrelated Caucasian individuals and found eight polymorphic sites. One of them corresponds to the introduction of a stop codon resulting in the production of a severely truncated ERV-3 envelope protein lacking both the fusion peptide and the immunosuppressive domain of the protein. The stop codon is observed in a homozygous state in approximately 1% of Caucasian individuals without evidence for counterselection, thus precluding the involvement of any essential function of the gene in placental implantation and development. This natural knockout provides a mean to investigate other potential roles for this otherwise highly conserved gene.
ERV-3 (HERV-R) is a single-copy human endogenous retrovirus (reviewed in references 10, 16, and 18) that is present in the genomes of all great apes and Old World monkeys and has also been identified in the genome of one New World monkey (3, 11, 15). This provirus, located close to the chromosome 7 centromere, comprises gag, pol, and env genes bordered by 5′ and 3′ long terminal repeats. The gag and pol genes both contain in-frame termination codons, but the env gene has an intact open reading frame capable of encoding a retroviral envelope glycoprotein. This protein would comprise the surface unit (SU) domain and a transmembrane (TM) moiety lacking a hydrophobic domain (4). Envelope-associated mRNAs are expressed at a low level in most human tissues and at a high level in placental syncytiotrophoblastic cells (i.e., trophoblastic cells which have fused to form a syncytial layer), in embryonic tissues such as the adrenal glands and nervous tissues, in the fetal heart, in sebaceous glands, and in some tumor cell lines and tissues (1, 2, 6, 8, 9). Antibodies raised against part of the TM domain demonstrate expression in the syncytiotrophoblast of a 65-kDa protein, as revealed by immunofluorescence and by Western blot analyses (17).
The persistence of an open reading frame within a sequence present in primates for more than 30 million years (3, 11, 15) strongly suggests a biological function for this protein. Taking into account established properties of retroviral envelopes, several hypotheses (reviewed in reference 17) have been proposed. (i) The ERV-3 envelope contains a putative immunosuppressive domain, whose expression at the placental barrier could participate in protection of the fetus from the maternal immune system. (ii) The fusion peptide within the ERV-3 TM domain could be involved in cell fusion and therefore participate in the formation of the syncytiotrophoblast, where ERV-3 is specifically expressed. (iii) Expression of the ERV-3 envelope could prevent retroviral infections, through receptor interference.
A way to evaluate the relevance of such hypotheses is to investigate the possible existence of polymorphisms among individuals within the ERV-3 envelope coding region. Polymorphisms have already been described for the ERV-3 provirus, but in noncoding regions (13, 14). One is a restriction polymorphism located at the 3′ end of pol, revealed upon MspI digestion (14), and eight other polymorphisms were identified in the 5′ and 3′ long terminal repeats, the linkage of which defined three allelic forms of ERV-3 (13). In the present study, we found that polymorphisms within the coding region of the ERV-3 envelope gene do exist. We further found that one of them results in the introduction of a stop codon, leading to the production of a severely truncated ERV-3 glycoprotein. Screening of a large panel of individuals shows that this knockout is present in a homozygous state in approximately 1% of the Caucasian population.
Polymorphism within the env coding region.
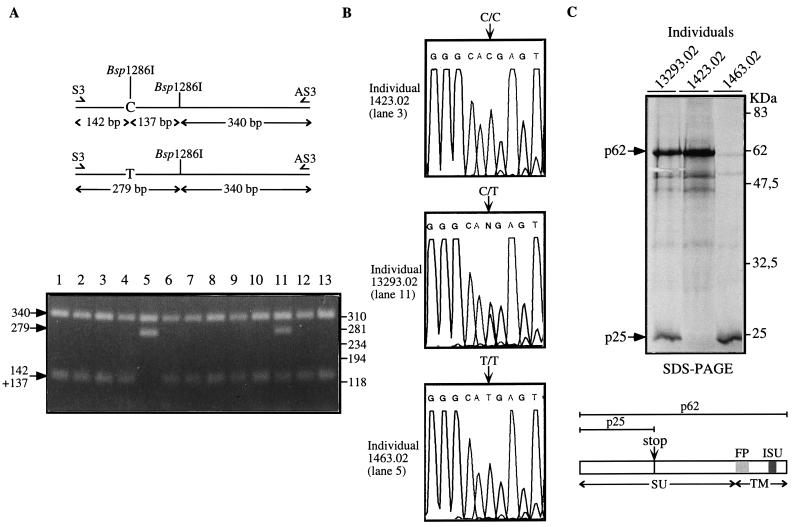
The entire ERV-3 envelope gene of five unrelated healthy Caucasian donors was PCR amplified from genomic DNA by using the 5′ S2 and the 3′ AS primers indicated in Fig. 1. A PCR was carried out for 35 cycles (1 min at 94°C, 1 min at 60°C, 4 min at 72°C) with 50 ng of genomic DNA, 50 pmol of each PCR primer, 1.5 mM magnesium acetate, and 0.2 mM each deoxynucleoside triphosphate in 50 μl of PCR buffer (Perkin-Elmer Tth polymerase and buffer). Direct sequencing of the complete gene was performed with 200 ng of the PCR product by using the set of primers shown in Fig. 1 and the Applied Biosystems Taq DyeDeoxy Terminator Cycle Sequencing kit with an ABI 373A sequencer (Perkin-Elmer). As illustrated in Fig. 1, single nucleotide differences among the sequences from the five individuals (i.e., polymorphic sites) were detected, clustered in two regions of the gene. Comparison with the published ERV-3 sequence (4) further revealed five differences common to the DNAs of all five individuals, suggesting that they correspond to errors in the original report rather than to polymorphisms (as confirmed below by partial sequencing of 24 more individuals). Altogether, eight polymorphic sites can therefore be defined, seven of which correspond to amino acid substitutions (Table 1). Strikingly, one of these substitutions (a C-to-T transition at position 1354) results in the introduction of a stop codon, which very prematurely closes the envelope reading frame. This was observed for one of the five individuals (no. 618) in a heterozygous state. To analyze the actual distribution of this unexpected polymorphism, we screened a much larger panel of unrelated Caucasian individuals, including 23 additional healthy donors and 122 parents from the panel of the Centre d’Etude du Polymorphisme Humain (CEPH). DNAs were amplified by using the S3 and AS3 primers (Fig. 1) and slightly modified PCR conditions (30 s at 94°C, 30 s at 60°C, 1 min at 72°C). As the C→T transition results in the loss of a Bsp1286I site (see Fig. 2A), 200 ng of the S3-AS3 amplimers was digested with Bsp1286I and analyzed by agarose gel electrophoresis and ethidium bromide staining. Figure 2A shows a series of 13 restriction patterns, disclosing one individual with a T/T profile (lane 5), one with a C/T profile (lane 11), and the others with a C/C profile. As the C→T transition also predicts the gain of an NlaIII site, the C/T and T/T amplified fragments were digested with NlaIII to confirm the allelic profile (data not shown). Direct sequencing of the S3-AS3 amplimers was also performed by using S3 as a sequencing primer for 29 individuals to further verify the result of the restriction analysis. Chromatograms of the three different allelic profiles at nucleotide 1354 are shown in Fig. 2B for individual 1423.02 with a C/C profile, individual 13293.02 with a C/T profile, and individual 1463.02 with a T/T profile. The consequence of this polymorphism for the encoded proteins was finally assessed by a direct coupled transcription-translation assay performed on the DNA from individuals with the stop codon (together with control individuals) PCR amplified by using a T7 promoter and a Kozak ATG-containing primer (T7atg 5′ primer and AS 3′ primer pair). PCR amplification of the complete envelope gene was performed under the same conditions as for the S2-AS primer pair. Two hundred nanograms of the amplification products was used, after ethanol precipitation, in the TNT Coupled Reticulocyte Lysate System (Promega) in accordance with the manufacturer’s instructions, with [3H]methionine (ICN) for protein labelling. After electrophoresis of the translation products, the sodium dodecyl sulfate-polyacrylamide gels were impregnated with En3Hance (Dupont de Nemours), rinsed with water, dried, and autoradiographed. As illustrated in Fig. 2C, sodium dodecyl sulfate-polyacrylamide gel electrophoretic analysis of the translational products for the three allelic profiles shows different patterns, 62- and 25-kDa proteins for individual 13293.02, a single 62-kDa protein for individual 1423.02, and a truncated 25-kDa product for individual 1463.02, as expected from the genotypes of these individuals.
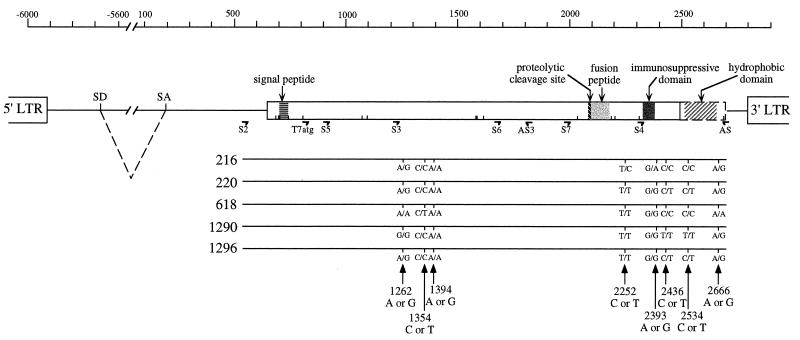
FIG. 1.
Identification of polymorphic sites within the sequences of ERV-3 envelope genes from five unrelated individuals. The ERV-3 provirus with the envelope open reading frame (open rectangle) and its expected functional domains, including the proteolytic cleavage site, the fusion peptide, and the immunosuppressive domain, are schematically represented at the top. The putative hydrophobic domain is shown in a dotted rectangle as an extension of the open reading frame if the stop codon at nucleotide 2500 is read through. The G insertion at position 796 (see Table 1) leads to an upstream shift in the frame, unlike that of the previously published ERV-3 sequence, and introduces a potential signal peptide and four new Met codons (represented by short vertical lines). LTR, long terminal repeat; SD, splice donor site; SA, splice acceptor site. The primers used in this study are indicated, and 5′-3′ sequences are given for S2 (CGGCCGAAGCTTGAGTCATCATCAGGG), T7atg (GCTAATACGACTCACTATAGGAACAGACCACCatgACTAAAACCCTGTTGTATCA), S3 (AACCAACAATCACTAGGGCC), AS3 (TGCCCCTCCATAAAGTCTTTCCTAG), and AS (GTTAATACTTAGTTAGGGCC). At the bottom, the envelope gene sequences of five individuals and the corresponding polymorphic sites within the ERV-3 consensus sequence are represented; both alleles for each site are indicated. Numbers refer to the ERV-3 pol-env published sequence (4; accession no. M12140).
TABLE 1.
Identification and distribution of polymorphic sites within the ERV3 envelope
| Position | Nucleotide | Amino acid change | No. of chromo- somes testeda | Distribution |
|---|---|---|---|---|
| 796b | G insertion | 10 | 10/0 | |
| 1262 | A/G | Cys/Tyr | 58 | 36/22 |
| 1338b | A/G | None | 58 | 58/0 |
| 1339b | G/A | Ala/Thr | 58 | 58/0 |
| 1354 | C/T | Arg/Opl | 300 | 271/29 |
| 1364b | G/A | Gly/Asp | 58 | 58/0 |
| 1394 | A/G | Tyr/Cys | 58 | 56/2 |
| 1440b | C/G | None | 58 | 58/0 |
| 2252 | T/C | Leu/Pro | 10 | 9/1 |
| 2393 | G/A | Ser/Asn | 10 | 9/1 |
| 2436 | C/T | None | 10 | 6/4 |
| 2534 | C/T | Ser/Leu | 10 | 6/4 |
| 2666 | A/G | Gln/Arg | 10 | 6/4 |
Compilation of data obtained by sequencing and restriction site analysis (see text).
Differences from the published sequence observed in all individuals tested, most probably corresponding to errors in the original report rather than to polymorphisms.
FIG. 2.
(A) Expected Bsp1286I restriction map of the ERV-3 S3-AS3 amplimer and restriction analysis of a series of 13 individuals. The arrows indicate the restriction fragments obtained after electrophoresis in a 2% agarose gel. Molecular size markers are shown on the right. (B) Nucleotide sequence analysis of PCR-amplified genomic fragments from three individuals for the polymorphism at nucleotide 1354. Lane numbers refer to those in A. (C) Direct in vitro transcription-translation assay for ERV-3 envelope gene products from the three individuals in B. The arrows indicate the translational products after migration in a 14% polyacrylamide gel. The two ERV-3 envelope proteins obtained are represented, and the SU, the TM, the FP (fusion peptide), and the ISU (immunosuppressive domain) are indicated. SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Distribution of the stop codon among individuals.
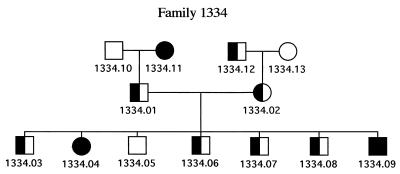
As summarized in Table 2, among the 150 individuals tested for the stop codon polymorphism, we detected C/C homozygotes in 123 cases (82%) and C/T heterozygotes in 25 cases (16.7%), and 2 individuals (1.3%) were homozygous for the stop codon (T/T profile). The relative proportion of homozygotes and heterozygotes for the stop codon is compatible with the Hardy-Weinberger equilibrium, suggesting that no counterselection is associated with the T/T genotype. This was further strengthened by the proportion of the T/T genotype among the children from two parents of the CEPH panel that we had identified as being both heterozygous for the stop polymorphism. As illustrated in Fig. 3, determination of the genotypes of the individuals of the corresponding family (including four grandparents and seven children not included in the previous analysis) showed that two children harbor the T/T genotype, one child has the C/C genotype, and four children have the C/T genotype. Altogether, these data strongly suggest that no essential function is associated with the expression of the envelope protein. More specifically, among the plausible roles that had been hypothesized for the ERV-3 gene (see the introductory paragraph), those involving immunosuppression at the placental barrier or differentiation of the placental cells seem to be precluded by the existence of T/T homozygotes at an expected frequency. Indeed, for those individuals, fetal development must have been associated with a placenta expressing a severely truncated ERV-3 envelope protein (the placenta is of fetal origin and thus has the same T/T genotype), i.e., lacking both the fusion peptide and the immunosuppressive domain of ERV-3. Thus, the reasons for the conservation of the ERV-3 2-kb open reading frame are not to be found in a selective pressure imposed by essential placental functions. It can be noted, in addition, that the two individuals in the CEPH panel with a T/T genotype (a man and a woman) were parents of 10 children each, also suggesting that the ERV-3 knockout of one of the parents has no effect on procreative function.
TABLE 2.
Frequency of the Opl stop mutation within the ERV3 envelope among unrelated Caucasian individuals
| Genotype (amino acid) | No. (%) of individuals |
|---|---|
| C/C (Arg/Arg) | 123 (82) |
| C/T (Arg/Opl) | 25 (16.7) |
| T/T (Opl/Opl) | 2 (1.3) |
FIG. 3.
Pedigree of a CEPH family whose parents are heterozygous for the stop mutation. Open symbols represent C/C homozygous individuals; filled symbols represent T/T homozygous individuals, and mixed symbols represent C/T heterozygous individuals. Squares, males; circles, females.
Conclusion.
In this study, we have shown that the ERV-3 envelope gene is subject to important genetic variations among members of the Caucasian population. Eight polymorphic sites have been found in this gene, seven of which correspond to an amino acid change. One of them introduces a stop codon, resulting in a physiological knockout of the ERV-3 envelope in 1% of the Caucasian population. This already permits rejection of previously proposed roles for this protein in relation to essential functions in placental development. Yet, the polymorphisms we describe here—which are superimposed on strong sequence conservation of the gene in evolution (confirmed by partial sequencing of the chimpanzee gene which discloses >99% homology at the amino acid level in the regions of the two polymorphic clusters [5a])—now provide a means to investigate other possible roles for this conserved open reading frame. Among them, involvement of the ERV-3 envelope as a protection against infectious retroviruses remains plausible, all the more so as it would actually impose only limited selective pressure among members of the human population. Other plausible roles concern the involvement of this protein in autoimmune diseases or cancer. Several lines of evidence establish a correlation between expression of TM retroviral proteins by tumor cells and enhanced tumorigenicity (reviewed in reference 12), and it is tempting to speculate that individuals homozygous for the truncated ERV-3 envelope protein might be less prone to certain forms of cancer. A possible role for endogenous retroviruses in autoimmune diseases has also been suggested (7, 16) and was actually recently demonstrated in type I diabetes (5). As far as ERV-3 is concerned, antibodies against the TM protein of the envelope were found in patients with Sjögren’s syndrome and systemic lupus erythematosus at a significantly higher level than in healthy donors (9), and anti-ERV-3 TM antibodies are also elevated in pregnant women, the highest level being detected in women whose babies are subject to congenital heart block, suggesting a role of this protein in the pathogenesis of this pregnancy-associated autoimmune disease (9). These hypotheses can now be tested by examination of the ERV-3 protein polymorphic sites identified here.
Acknowledgments
We are grateful to the CEPH for access to their panel of individuals, to J. Weissenbach and J. Feunteun for the gift of DNAs from healthy donors and helpful discussions, and to C. Lavialle for critical reading of the manuscript.
This work was supported by the CNRS and by grants from the ARC (contract 6552 awarded to T.H. and a fellowship awarded to N.P.) and from the Ligue Nationale contre le Cancer.
REFERENCES
- 1.Andersson A C, Merza M, Venables P, Ponten F, Sundström J, Cohen M, Larsson E. Elevated levels of the endogenous retrovirus erv3 in human sebaceous glands. J Invest Dermatol. 1996;106:125–128. doi: 10.1111/1523-1747.ep12329612. [DOI] [PubMed] [Google Scholar]
- 2.Boyd M T, Bax C M, Bax B E, Bloxam D L, Weiss R A. The human endogenous retrovirus ERV-3 is upregulated in differentiating placental trophoblast cells. Virology. 1993;196:905–909. doi: 10.1006/viro.1993.1556. [DOI] [PubMed] [Google Scholar]
- 3.Cohen M, Larsson E. Human endogenous retroviruses. Bioessays. 1988;9:191–196. doi: 10.1002/bies.950090603. [DOI] [PubMed] [Google Scholar]
- 4.Cohen M, Powers M, O’Connell C, Kato N. The nucleotide sequence of the env gene from the human provirus erv3 and isolation and characterization of an erv3-specific cDNA. Virology. 1985;147:449–458. doi: 10.1016/0042-6822(85)90147-3. [DOI] [PubMed] [Google Scholar]
- 5.Conrad B, Weissmahr R N, Böni J, Arcari R, Schüpbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–313. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 5a.de Parseval, N. Unpublished data.
- 6.Kato N, Larsson E, Cohen M. Absence of expression of a human endogenous retrovirus is correlated with choriocarcinoma. Int J Cancer. 1988;41:380–385. doi: 10.1002/ijc.2910410310. [DOI] [PubMed] [Google Scholar]
- 7.Krieg A M, Gourley M F, Perl A. Endogenous retroviruses: potential etiologic agents in autoimmunity. FASEB J. 1992;6:2537–2544. doi: 10.1096/fasebj.6.8.1592206. [DOI] [PubMed] [Google Scholar]
- 8.Larsson E, Andersson A C, Nilsson B O. Expression of an endogenous retrovirus (erv3 HERV-R) in human reproductive and embryonic tissues—evidence for a function for envelope gene products. Ups J Med Sci. 1994;99:113–120. doi: 10.3109/03009739409179354. [DOI] [PubMed] [Google Scholar]
- 9.Li J-M, Fan W S, Horsfall A C, Anderson A C, Rigby S, Larsson E, Venables P J W. The expression of human endogenous retrovirus-3 in fetal cardiac tissue and antibodies in congenital heart block. Clin Exp Immunol. 1996;104:388–393. [PubMed] [Google Scholar]
- 10.Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connell C, O’Brien S, Nash W G, Cohen M. Erv3, a full length human endogenous provirus: chromosomal localization and evolutionary relationship. Virology. 1984;138:225–235. doi: 10.1016/0042-6822(84)90347-7. [DOI] [PubMed] [Google Scholar]
- 12.Oostendorp R A J, Meijer C J L M, Scheper R J. Immunosuppression by retroviral-envelope-related proteins, and their role in non-retroviral human disease. Crit Rev Oncol Hematol. 1993;14:189–206. doi: 10.1016/1040-8428(93)90009-s. [DOI] [PubMed] [Google Scholar]
- 13.Rassmussen H B, Heltberg A, Lisby G, Clausen J. Three allelic forms of the human endogenous retrovirus erv3, and their frequencies in multiple sclerosis patients and healthy individuals. Autoimmunity. 1996;23:111–117. doi: 10.3109/08916939608995334. [DOI] [PubMed] [Google Scholar]
- 14.Rubin, L. A., K. A. Siminovitch, M. H. Shi, and M. Cohen. 1991. A novel retroviral gene association with rheumatoid arthritis. Arthritis Rheum. 34(Suppl.):S60.
- 15.Shih A, Coutavas E, Rush M G. Evolutionary implications of primate endogenous retroviruses. Virology. 1991;182:495–502. doi: 10.1016/0042-6822(91)90590-8. [DOI] [PubMed] [Google Scholar]
- 16.Urnovitz H B, Murphy W H. Human endogenous retroviruses: nature, occurrence, and clinical implications in human disease. Clin Microbiol Rev. 1996;9:72–99. doi: 10.1128/cmr.9.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venables S, Brookes M, Fan W, Larsson E, Maini R N, Boyd M T. Abundance of an endogenous retroviral envelope protein in placental trophoblast suggests a biological function. Virology. 1995;211:589–592. doi: 10.1006/viro.1995.1442. [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson D A, Mager D L, Leong J-A C. Endogenous human retroviruses, 465–535. In: Levy J A, editor. The Retroviridae. New York, N.Y: Plenum Press; 1994. [Google Scholar]