Abstract
The hepatitis A virus cellular receptor 1 (HAVcr-1) cDNA was isolated from a cDNA expression library of African green monkey kidney (AGMK) cells by using protective monoclonal antibody (MAb) 190/4, which blocks the binding of hepatitis A virus (HAV) to AGMK cells. The HAVcr-1 cDNA codes for havcr-1, a 451-amino-acid class I integral-membrane mucin-like glycoprotein of unknown natural function. To determine the existence of a human homolog(s) of HAVcr-1 (huHAVcr-1), we used HAVcr-1-specific primers to amplify cDNAs from human liver and kidney mRNA by reverse transcription-PCR. Nucleotide sequence analysis revealed that the amplified liver and kidney huHAVcr-1 cDNAs were identical and that they coded for a 359-amino-acid glycoprotein, termed huhavcr-1, which was approximately 79% identical to havcr-1. The six Cys residues of the extracellular domain of havcr-1 and its first N-glycosylation site were conserved in huhavcr-1. However, the number of hexameric repeats of the mucin-like region was reduced from 27 in havcr-1 to 13 in huhavcr-1. In addition, 12 C-terminal amino acids in the cytoplasmic domain of huhavcr-1 were deleted. Northern blot analysis of poly(A) RNA showed that huhavcr-1 is expressed in every organ analyzed, including the liver, small intestine, colon, and spleen, and that it is expressed at higher levels in the kidney and testis. Although dog cells transfected with the huHAVcr-1 cDNA did not express the protective 190/4 epitope, they bound hepatitis A virus (HAV) and gained limited susceptibility to HAV infection. Treatment with MAb 190/4 did not protect AGMK cell transfectants expressing huhavcr-1 against HAV, suggesting that HAV infected these cells via the huhavcr-1 receptor and not the endogenously expressed havcr-1, which was blocked by MAb 190/4. Our data demonstrate that huhavcr-1 is a binding receptor for HAV and suggest that it is also a functional receptor for HAV.
Hepatitis A virus (HAV), a hepatotropic picornavirus, causes a medically important acute hepatitis. The first step in the life cycle of HAV is binding to a cell surface receptor that in African green monkey kidney (AGMK) cells is coded by HAVcr-1 (7). Monoclonal antibody (MAb) 190/4, which was used as a probe to molecularly clone the HAVcr-1 cDNA from an expression cDNA library of AGMK cells, blocks the binding of HAV and protects AGMK cells against HAV infection (7). Nucleotide sequence analysis showed that HAVcr-1 cDNA codes for a class I integral membrane glycoprotein, termed havcr-1, of unknown natural function. The extracellular domain of havcr-1 consists of an N-terminal cysteine-rich (Cys-rich) region followed by a threonine-, serine-, and proline-rich (TSP-rich) region. The havcr-1 Cys-rich region displays homology to members of the immunoglobulin (Ig) superfamily (7), and the TSP-rich region has the characteristics of mucin-like glycoproteins (15). The putative “lollipop-on-a-stick” structure (6) of havcr-1 suggested that the extended O-glycosylated TSP-rich region presents the Cys-rich globular domain above the cell surface and makes it accessible for interactions with extracellular molecules. We have recently shown that the Cys-rich region of havcr-1 and its first N-glycosylation site are required for the binding of HAV and protective MAb 190/4 (19), and we are currently analyzing whether the Cys-rich region of havcr-1 is sufficient for HAV receptor function.
Receptor-negative cell lines that are otherwise fully susceptible to HAV replication have not yet been identified. Although we devoted considerable effort to isolating one, we have not been able to do so (4). We have previously shown that mouse and dog cells, which contain internal blocks to HAV replication, gain limited susceptibility to HAV infection upon transfection with the HAVcr-1 cDNA (7, 19). Infection of these mouse and dog cell transfectants with HAV resulted in low levels of the characteristic granular cytoplasmic fluorescence of HAV-infected cells, which lasted for several days but became undetectable after 1 month postinfection. This limited level of susceptibility of the mouse and dog cell transfectants to HAV infection resulted in only a <10-fold increase in HAV titers and a <2-fold increase in HAV-specific RNA (7, 19); therefore, its is likely that the input virus internalized through havcr-1 contributed to the HAV-specific fluorescence observed in the mouse and dog cell transfectants. Although further analysis of the HAV receptor function of havcr-1 awaits the isolation of nonsusceptible cells that could fully support HAV replication upon transfection of the HAVcr-1 cDNA, the mouse and dog cell transfectants allowed us to characterize the havcr-1-mediated binding of HAV, its internalization, and the limited level of susceptibility to HAV infection (5, 7, 19).
Initial studies revealed that protective MAb 190/4 reacted with the cell surfaces of clone GL37 AGMK cells but not with the cell surfaces of HeLa cells (7). Therefore, it was of great interest to ascertain the existence of the human homolog of HAVcr-1 and determine its function as an HAV receptor and its role in the pathogenesis of HAV in humans. In this report, we describe the molecular cloning of the cDNA coding for the human homolog of HAVcr-1 (huHAVcr-1). Nucleotide sequence analysis revealed that the huHAVcr-1 cDNA codes for a glycoprotein, termed huhavcr-1, that is 79% identical to havcr-1. Northern blot analysis showed that huHAVcr-1 is expressed in every human organ analyzed, including the liver, small intestine, colon, and spleen, and that it is expressed at higher levels in the kidney and testis. Dog cells transfected with the huHAVcr-1 cDNA gained limited susceptibility to HAV infection, whereas dog cells transfected with vector alone or HAVcr-1 cDNA with the Cys-rich region deleted were resistant to HAV infection. These results suggest that huhavcr-1 is a receptor for HAV which may play a role in the pathogenesis of HAV in humans.
MATERIALS AND METHODS
Cells and viruses.
Continuous clone GL37 AGMK cells, termed GL37 cells, were selected for supporting optimal growth of HAV (20). Canine osteogenic sarcoma D-17 cells, obtained from the American Type Culture Collection, were cotransfected with pCMVEBNA and pSV2neo (Clontech Laboratories). A G418-resistant cell clone, termed Perro6D, that had 10- to 100-times-higher transfection efficiency with pDR2 (14, 18) than parental D-17 cells was isolated (19) and used for transfection with HAVcr-1 cDNA constructs cloned into the pDR2 vector. Cell lines were grown in Eagle’s minimal essential medium (EMEM) containing 10% fetal bovine serum (FBS) at 37°C in a CO2 incubator.
Human tissue culture-adapted HAV strain HM175 of genotype 1B was derived from infectious cDNA (3) and grown in GL37 cells.
Antisera.
Anti-havcr-1 antiserum was obtained from rabbits immunized with recombinant protein GST-2 consisting of the mucin-like region of havcr-1 fused to the C terminus of glutathione S-transferase expressed in Escherichia coli (19).
Murine IgG1 subtype MAbs P1B5 raised against the human α3 integrin (Gibco BRL, Inc.), 190/4 directed against havcr-1 (7), and M2 directed against the FLAG peptide DTKDDDDK (IBI, Inc.) were purified through protein A-agarose columns. Unlabeled and 125I-labeled human anti-HAV polyclonal antisera were obtained from the HAVAB kit (Abbott Laboratories). Fluorescein isothiocyanate (FITC)-labeled goat anti-human IgG and IgM Abs (Accurate Inc.) were used to detect HAV by indirect immunofluorescence (IF). Alkaline phosphatase-labeled goat anti-rabbit Abs and peroxidase-labeled goat anti-mouse Abs were used as suggested by the manufacturer (Kirkegaard and Perry Laboratories, Inc.).
Indirect IF analysis.
Growth of HAV was assessed by indirect IF analysis. Monolayers of GL37 cells and dog cell transfectants grown in eight-well Permanox culture slides (Nunc, Inc.) were fixed with cold acetone for 20 min, treated with a 1:1,000 dilution of human anti-HAV Ab for 1 h at room temperature, and stained with a 1:400 dilution of FITC-labeled goat anti-human IgG and IgM. IF micrographs were taken with a Zeiss Axioscope microscope at ×1,000 with an oil immersion objective.
Southern blot analysis.
Genomic DNA extracted from cell lines and human leukocytes was digested with restriction enzyme PstI, fractionated in a 1% TAE-agarose gel, transferred to a nylon membrane (Zeta-Probe; BioRad, Inc.), dried, and irradiated with 120 mJ of UV light (254 nm) in an auto-cross-linker (Stratagene, Inc.). Blots were hybridized in 50% formamide–5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 45°C with a full-length HAVcr-1 cDNA probe which had been cut from pDR2GL37/5 (7) with restriction endonucleases BamHI and XbaI, purified by 1% TAE-agarose gel electrophoresis, and labeled with 32P by random priming with the Prime-a-Gene kit as recommended by the manufacturer (Promega Corp.). After 20 h of hybridization, all blots were washed with 2× SSC–0.1% SDS at 65°C and exposed for 48 h to X-ray film by using intensifying screens.
Northern blot analysis.
Human multiple-tissue Northern blots (MTN blots; Clontech Laboratories), which contain poly(A) RNA (2 μg per well) purified from different normal human tissues and blotted onto a nylon membrane, were hybridized in 50% formamide–5× SSC at 42°C with the above-mentioned 32P-labeled full-length huHAVcr-1 cDNA probe. MTN blots were washed with 2× SSC–0.1% sodium dodecyl sulfate (SDS) at 65°C and autoradiographed for 1 day or 3 weeks in the presence of intensifying screens. MTN blots were stripped and rehybridized under the same conditions to a β-actin probe (Clontech Laboratories) labeled with 32P as recommended by the manufacturer.
Slot blot analysis.
Confluent monolayers of GL37 cells in six-well plates were treated with 0.5 ml (10 μg/ml) of MAb 190/4 or MAb P1B5 in EMEM–5% FBS for 1 h at 37°C and inoculated at a multiplicity of infection (MOI) of 0.1 50% tissue culture infectious doses (TCID50) of HAV per cell for 1 h at room temperature. It should be pointed out that both MAbs react against the cell surfaces of GL37 cells. After being washed three times, monolayers were placed in a 35°C incubator under 5% CO2, and cytoplasmic extracts were prepared at 72 h postinfection. Total cell RNA was extracted with phenol–chloroform–1% SDS, applied to nitrocellulose with a slot blotter (Schleicher & Shuell, Inc.), baked at 80°C for 2 h, and hybridized with a 32P-labeled HAV cDNA probe (4). To control for loading, the same blots were stripped and rehybridized under the same conditions with the 32P-labeled β-actin probe described above.
PCR.
The huHAVcr-1 cDNA was amplified from 1 ng of human kidney or liver cDNA (Quick-Clone; Clontech, Inc.) by using 1 μg of synthetic oligonucleotides cr196-218(+) and cr1548-1525(−) (Table 1) and a mixture of Taq and Pwo DNA polymerases in 30 cycles as recommended by the manufacturer (Expand High Fidelity PCR System; Boehringer Mannheim). PCR was initiated by the hot-start technique in a 50-μl reaction mixture without MgCl2 but containing wax beads which, upon melting, provided a final concentration of 1.5 mM MgCl2 (HotWax Mg+ beads; Invitrogen). A 1.1-kb cDNA fragment was amplified from both kidney and liver cDNA and purified by TAE–1% low-melting-point agarose gel electrophoresis. To determine the sequence of the 5′ untranslated region (5′UTR), huHAVcr-1 cDNA was amplified with synthetic oligonucleotides 136(+) and cr425-403(−) (Table 1) under the above-mentioned PCR conditions. Similarly, the huhavcr-1 mRNA 3′UTR was amplified with synthetic oligonucleotides cr487-509(+) and 1794(−) (Table 1) under the above-mentioned PCR conditions. As expected, we did not detect a poly(A) tract in any of the huHAVcr-1 PCR fragments, which confirmed that the very 3′ end of the huHAVcr-1 mRNA was not amplified.
TABLE 1.
Oligonucleotides used in this study
| Namea | Sequence (5′ to 3′) |
|---|---|
| cr196-218(+) | ATGCATCTTCAAGTGGTCATCTT |
| cr487-509(+) | TCTGACAGTGGCATATATTGTTG |
| cr1548-1525(−) | TGGGCGTAAACTCTCAAAGAGCAC |
| cr425-403(−) | TAGCGTGTCTCCTTCCGATAGG |
| 136(+) | AGAGAACTCAGGAAGG |
| 1794(−) | CAGTGACTATTCTCTCTGATGG |
| FLAG4(+) | CGAGACTACAAGGACGACGATGACAAGATCTCG |
| FLAG4(−) | CGCGAGATCTTGTCATCGTCGTCCTTGTAGTCT |
The + or − in the name indicates the polarity.
Cycle sequencing analysis.
The nucleotide sequences of the PCR fragments, cloned cDNAs, and plasmid constructs were obtained by automatic sequencing using an ABI Prism model 377 automatic sequencer and the ABI PRISM Dye terminator cycle sequencing ready reaction kit (Perkin-Elmer Cetus, Inc.). Both strands of the PCR products were sequenced by using positive- and negative-sense synthetic oligonucleotides spaced 300 to 400 bases apart.
Plasmid constructs.
Recombinant DNA manipulations were done by standard methods (17). Constructions were verified by automatic nucleotide sequencing. All plasmids were grown in E. coli DH5α and purified by chromatography with plasmid preparation kits as recommended by the manufacturer (Qiagen, Inc.). The nucleotide positions of the HAVcr-1 cDNA and the corresponding amino acid positions are in accordance with the previously published sequence (7), and those of the huHAVcr-1 cDNA and corresponding amino acids are in accordance with the sequence obtained from the PCR products (Fig. 2).
FIG. 2.
cDNA sequence and predicted amino acid composition of huhavcr-1. The nucleotide sequence of the huHAVcr-1 cDNA was determined on both strands by automatic sequencing using specific oligonucleotides. The white box indicates the putative signal sequence. The six cysteines of the Cys-rich region are marked with solid lines. Potential N-linked glycosylation sites are marked with dashed lines. The arrowhead indicates the putative beginning of the TSP-rich region. The putative transmembrane domain is boxed in grey. The termination codon is indicated by asterisks.
(i) pDR2huHAVcr.
The 1.1-kb PCR cDNA fragment amplified from human kidney cDNA with synthetic oligonucleotides cr196-218(+) and cr1548-1525(−) was purified by TAE–1% low-melting-point agarose gel electrophoresis, phosphorylated with ATP and T4 polynucleotide kinase (NEB, Inc.), and cloned into the SmaI site of pUC19. It should be pointed out that due to the use of synthetic oligonucleotide cr196-218(+) in the PCR, the amplified huHAVcr-1 cDNA codes for a P-to-L change at amino acid position 3 of the signal sequence which corresponds to the sequence found in havcr-1. The nucleotide sequences of three independent clones were determined, and one of the clones, whose corresponding amino acid sequence was identical to that of the uncloned huhavcr-1 PCR cDNA product except for the P-to-L change at amino acid residue 3, was named pUChuHAVcr. This plasmid was linearized with EcoRI, filled in with DNA polymerase I Klenow fragment (Klenow enzyme), and cut with SalI. The resulting 1.1-kb huHAVcr-1 cDNA fragment was subcloned into the XbaI and filled-in BamHI sites of pDR2. This construct, termed pDR2huHAVcr, contains the full-length huHAVcr-1 cDNA under the control of a Rous Sarcoma Virus promoter.
(ii) pDR2huHAVcrFlagBstBI.
To introduce a tag epitope into the extracellular domain of huhavcr-1, synthetic oligonucleotides FLAG4(+) and FLAG(4−) (Table 1) were treated with polynucleotide kinase and ATP, annealed, and inserted into the unique BstBI site of pUChuHAVcr at nucleotide 482 of the huHAVcr-1 cDNA. The nucleotide sequences of several clones were analyzed, and a clone containing the inserted oligonucleotides in the correct orientation was selected and termed pUChHAVcrFlagBstBI. This plasmid was cut with PvuII, and the resulting 943-bp fragment was subcloned into the PvuII sites of pDR2huHAVcr. The resulting construct, pDR2huHAVcrFlagBstBI, codes for a FLAG-tagged receptor termed huflagBst under the control of a Rous sarcoma virus promoter.
(iii) pHook3hHAVcrFlagBstBI.
The 1.1-kb KpnI-XbaI fragment of pDR2huHAVcrFlagBstBI coding for huflagBst was subcloned into KpnI-XbaI-cut pHook3 (Invitrogen, Inc.). The resulting plasmid, pHook3hHAVcrFlagBstBI, codes for huflagBst under the control of a cytomegalovirus promoter.
Transfections and selection of antibiotic-resistant cells.
Cell monolayers grown in 25-cm2 flasks were transfected with 1 μg of plasmid and 10 μl of DOSPER as recommended by the manufacturer (Boehringer Mannheim, Inc.) in a final volume of 3 ml of OptiMEM (Gibco BRL, Inc.). After overnight incubation at 37°C under 5% CO2, 3 ml of EMEM–10% FBS was added to each 25-cm2 flask.
Perro6D cells were transfected with constructs based in pDR2, a shuttle vector which contains the Epstein-Barr virus P1 origin of replication that allows the episomal maintenance of these plasmids in dog cells and that codes for a hygromycin resistance selectable marker. At 48 h posttransfection, the medium was changed to EMEM–10% FBS containing 250 μg of hygromycin (Boehringer Manheim, Inc.) per ml. After 7 days of treatment with hygromycin, approximately 20 to 30% of the transfected cells survived, whereas none of the mock-transfected cells resisted the antibiotic selection. Hygromycin-resistant Perro6D cells transfected with pDR2HAVcrFlag, which codes for a FLAG-tagged havcr-1, and pDR2HAVcrD1−, which codes for a FLAG-tagged havcr-1 with the Cys-rich region deleted, were described previously (19) and were named flag and d1− cells, respectively. Hygromycin-resistant Perro6D cells transfected with vector pDR2 alone were named DR2 cells. Hygromycin-resistant Perro6D cells transfected with pDR2huHAVcr and pDR2huHAVcrFlagBstBI, constructs which are described in this paper, were named huhavcr-1 and huflagBst cells, respectively. The expression of huhavcr-1 and huflagBst in the dog cell transfectants grown in EMEM–10%FBS–250 μg of hygromycin per ml was stable for 2 weeks but decreased thereafter to undetectable levels as judged by Western blot analysis using an anti-GST2 Ab.
GL37 cells were transfected with constructs based in pHook3, a shuttle vector which contains the simian virus 40 origin of replication and which codes for a zeomycin resistance selectable marker. At 48 h posttransfection, the transfection medium was changed to EMEM–10% FBS containing 500 μg of zeomycin (Invitrogen, Inc.) per ml. Two zeomycin-resistant cell clones, GL37huflagBst 2 and GL37huflagBst 3, which expressed the FLAG epitope as determined by a cell surface enzyme-linked immunosorbent assay (ELISA) and Western blot analysis using MAb M2 (data not shown), were isolated.
Cell surface ELISA.
Expression of HAVcr-1 in GL37 cells and dog cell transfectants was analyzed by a cell surface ELISA as described previously (7). Briefly, duplicate wells of unfixed cells grown in 96-well plates were treated with twofold dilutions of 190/4 or M2 MAbs for 1 h at room temperature, washed extensively, and treated with a 1:1,000 dilution of affinity-purified peroxidase-labeled anti-mouse Ab for 1 h at room temperature. After the cells were washed extensively, 100 μl of the one-component tetramethyl-benzidine (TMB) substrate (Kirkegaard and Perry Laboratories, Inc.) was added per well. The reaction was stopped with 1% H2SO4, and absorbance was read at 450 nm. The difference in the optical densities at 450 nm (OD450) of the duplicate wells were within the experimental error of 5 to 10%, the average values were highly reproducible, and backgrounds were below 0.1 OD450 units. The mean OD450 of duplicate wells was plotted versus the log10 of the antibody dilution.
Western blot analysis.
Confluent monolayers of dog cell transfectants grown in 25-cm2 flasks were scraped into 1 ml of phosphate-buffered saline (PBS), pelleted, resuspended in 0.2 ml of reticulocyte standard buffer (RSB; 10 mM NaCl, 10 mM Tris-HCl [pH 7.2]) containing 1% Nonidet P-40 (NP-40), and incubated for 2 min at room temperature. After the nuclei were removed by centrifugation at 12,000 × g, the total amount of protein in the supernatant was determined by the Bradford method using the Bio-Rad protein assay kit. The cytoplasmic extracts were used immediately or stored at −70°C. Approximately one-third of the cytoplasmic extract obtained from a 25-cm2 cell monolayer (20 to 25 μg of total protein) was loaded per well and fractionated in SDS–10% polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Inc.), probed with a 1:1,000 dilution of rabbit anti-GST2 Ab, and stained with a 1:5,000 dilution of alkaline phosphatase-labeled goat anti-rabbit Ab. The substrate 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (BCIP-NBT substrate) was used as recommended by the manufacturer (Kirkegaard & Perry Laboratories).
HAV binding assay.
Binding of HAV to the dog cell transfectants was quantitated by radioimmunoassay as reported previously (7) but with minor modifications. HAV was purified from 20 15-cm2 dishes containing confluent monolayers of AGMK cells infected at a MOI of 1 TCID50 of HAV per cell for 1 week at 35°C in a CO2 incubator. Cytoplasmic extracts were prepared as described above in 5 ml of RSB–1% NP-40 and treated with 1% SDS and 1% Sarkosyl overnight at room temperature. HAV present in the cytoplasmic extracts was pelleted through a 4-ml-thick cushion of 40% sucrose–NTE (150 mM NaCl, 10 mM Tris-HCl [pH 7.4], 1 mM EDTA [pH 8.0]) by centrifugation at 40,000 rpm for 4 h at 16°C in a Beckman SW40 rotor. The pelleted virus was resuspended in 2 ml of NTE, aliquoted, and stored at −70°C. Purified HAV had a titer of approximately 1010 TCID50/ml, as assessed in 96-well plates containing confluent monolayers of FRhK-4 cells (4). The purified HAV was bound to 80%-confluent monolayers of dog cell transfectants grown in 96-well plates. Duplicate wells were treated with 50 μl of 1:10, 1:20, and 1:40 dilutions of purified HAV in EMEM–10% FBS for 1 h at 35°C in a CO2 incubator. Monolayers were washed four times with EMEM–10% FBS, fixed with 80% methanol, blocked with 5% bovine serum albumin in PBS, incubated with 50 μl of 125I-labeled human anti-HAV Ab, washed four times with PBS, and exposed to X-ray film (XAR-2; Kodak) with an intensifying screen for 24 to 96 h. After exposure, the 96-well plates were stained with 1% crystal violet and absorbance at 595 nm was determined with an ELISA plate reader (Bio-Rad Laboratories); this assay indicated that similar numbers of cells, within a 5 to 10% range, were present in each well. Densitometric analysis of the autoradiography was performed on a Macintosh Quadra950 computer by using the public-domain NIH Image program (written by Wayne Rasband at the National Institutes of Health).
HAV infectivity assay.
Dog cell transfectants and GL37 cells grown in eight-well Permanox culture slides (Nunc, Inc.) were infected with 107 to 108 TCID50 of HAV purified as described above or were mock infected for 6 h at 35°C under 5% CO2. After being washed three times with EMEM–10% FBS, cells were incubated for 3 days at 35°C under 5% CO2. Cell monolayers were fixed with cold acetone and analyzed by indirect IF as described above.
Nucleotide sequence accession number.
GenBank accession no. AF043724 was obtained for the huHAVcr-1 cDNA.
RESULTS
Molecular cloning of the human homolog of HAVcr-1.
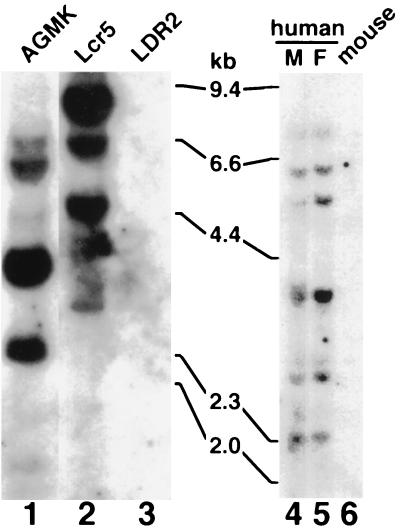
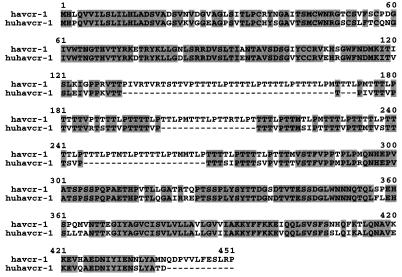
Preliminary results indicated that the 190/4 epitope was not expressed in HeLa cells (7); therefore, it was of interest to determine whether human cells coded for an HAVcr-1 homolog(s). To do so, genomic DNA was extracted from clone GL37 AGMK cells, from Ltk− cells transfected with HAVcr-1 cDNA (Lcr5 cells) or pDR2 vector (LDR2 cells), from human male and female peripheral blood leukocytes, and from mouse cells. Southern blot analysis of PstI-digested genomic DNA hybridized with a 32P-labeled full-length HAVcr-1 cDNA probe (Fig. 1) revealed the presence of several HAVcr-1-specific bands in AGMK (lane 1), Lcr5 (lane 2), and human (lanes 4 and 5) cells but not in vector-transfected LDR2 cells (lane 3) and untransfected mouse cells (lane 6). To assess whether these HAVcr-1 homolog sequences were expressed in human cells, we performed an reverse transcription-PCR analysis of mRNA extracted from human kidney and liver. Using synthetic oligonucleotides cr196-218(+) and cr1548-1525(−) corresponding to the N- and C-terminal sequences of havcr-1, respectively, we amplified cDNA fragments of 1.1 kb from human kidney and liver cDNA. Nucleotide sequence analysis revealed that the 1.1-kb PCR cDNA fragments amplified from human kidney and liver cDNA were identical and contained the complete coding sequence of a human homolog of HAVcr-1 that we termed huHAVcr-1 (Fig. 2). To verify that the initiation and termination codons of the amplified 1.1-kb huHAVcr-1 cDNA were not artificially introduced by the synthetic oligonucleotides used in the PCR, we amplified cDNA fragments corresponding to the 5′ and 3′ ends of the huHAVcr-1 mRNA with synthetic oligonucleotides 136(+) and cr425-403(−) or cr487-509(+) and 1794(−), respectively. Nucleotide sequence analysis of the 5′-end PCR product showed that the ATG initiation codon was present in the huHAVcr-1 mRNA but that synthetic oligonucleotide cr196-218(+) introduced a C-to-T change in the huHAVcr-1 cDNA which resulted in an P-to-L amino acid change at position 3 in the signal sequence. Nucleotide sequence analysis of the 3′-end PCR product showed that PCR primer cr1548-1525(−) did not introduce any change into the huHAVcr-1 cDNA. Nucleotide sequence analysis (Fig. 2) revealed that the huHAVcr-1 cDNA encodes a polypeptide of 359 amino acids, termed huhavcr-1, which is 79.11% identical to havcr-1 (Fig. 3). Like havcr-1, huhavcr-1 has the typical features of a type I integral-membrane glycoprotein, with two distinctive hydrophobic regions: a putative 17-amino-acid signal sequence with a hydrophobic core following the initiating methionine and a putative transmembrane domain of 22 residues between amino acids 290 and 311, which is the major hydrophobic region of the protein. A conserved cysteine residue (Cys296) found within the transmembrane domain of huhavcr-1 is possibly used for the addition of fatty acids that may stabilize the receptor attachment to the membrane (10). Between the signal sequence and the transmembrane domain, there is a predicted extracellular domain of 272 residues comprising two distinctive regions: a Cys-rich N-terminal region of 109 residues and a C-terminal segment of 163 residues containing a TSP-rich region. The six Cys residues and the length of the Cys-rich region of havcr-1 are conserved in huhavcr-1. The first but not the second N-glycosylation site of the havcr-1 Cys-rich region is conserved in huhavcr-1. Only 13 of the 27 consecutive repeats of the consensus sequence PTTTTL of the TSP-rich region of havcr-1 remained in huhavcr-1 (Fig. 3). The two N-glycosylation sites of the TSP-rich region of havcr-1 are conserved in huhavcr-1, which also contains a third putative N-glycosylation site at amino acid residues 258 to 260. The huhavcr-1 intracellular domain contains 48 amino acids and is 12 residues shorter than the intracellular domain of havcr-1. Consequently, the havcr-1 and huhavcr-1 putative structures are very similar, resembling a lollipop-on-a-stick model (6) in which the Cys-rich region of havcr-1 will most likely be extended further above the cell surface than that of huhavcr-1.
FIG. 1.
Southern blot analysis of the huhavcr-1 gene. Genomic DNA extracted from simian AGMK cells, mouse Ltk− cells transfected with pDR2GL37/5 (Lcr5 cells) or pDR2 (LDR2 cells), human male (M) and female (F) peripheral blood leukocytes, and mouse cells was digested with PstI and examined by Southern blot analysis using a full-length 32P-labeled HAVcr-1 cDNA probe. The positions of size markers in kilobase pairs (kb) are illustrated.
FIG. 3.
Alignment of havcr-1 and huhavcr-1. Alignment of amino acid sequences predicted from the HAVcr-1 and huHAVcr-1 cDNAs was done with the Clustal W program. Gaps introduced in the sequences for the alignment are indicated by dashes; numbers of residues starting with the respective initiating methionine codons are indicated. Amino acids identical to those of the havcr-1 sequence are in grey background.
Expression of huhavcr-1 in different human tissues.
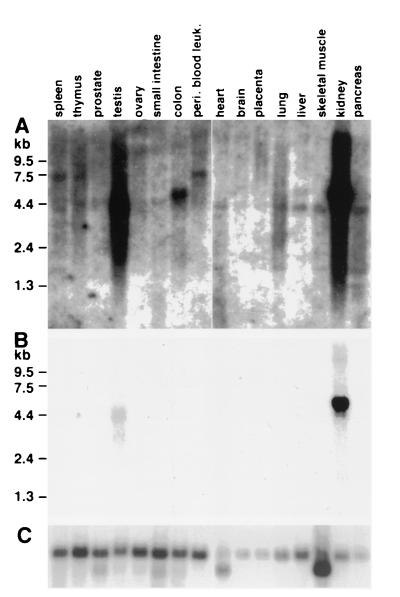
To study the pattern of expression of huHAVcr-1 in different human tissues, MTN blots were hybridized to a 32P-labeled full-length huHAVcr-1 cDNA probe (Fig. 4). After 3 weeks of exposure, the autoradiogram of the MTN blots (Fig. 4A) showed that huHAVcr-1 was expressed in every organ analyzed and expressed at higher levels in the kidney and testis than in other organs. An huHAVcr-1-specific 4.4-kb band was present for almost every organ, a 5.5-kb band was observed for the colon and liver, and a 7.5-kb band was observed for the spleen and thymus and for peripheral blood leukocytes. Several bands of lower intensity and smaller than 4.4 kb were also observed for the spleen, lung, skeletal muscle, and pancreas. A shorter exposure of the MTN blots for 24 h (Fig. 4B) revealed that the kidney expressed 5.5-kb huHAVcr-1 mRNA at higher levels than those of the 3- and 4.4-kb huHAVcr-1-specific mRNAs expressed in the testis. Since the huHAVcr-1 1.1-kb cDNA fragments amplified from the kidney and liver contain the whole coding sequence of huhavcr-1, it is possible that the huHAVcr-1 mRNA species detected in the MTN blots code for long 3′ and/or 5′ nontranslated regions. However, we cannot rule out the possibility that additional coding sequences are contained in these huHAVcr-1 mRNAs. To control for loading and integrity of the mRNA, MTN blots were rehybridized to a 32P-labeled β-actin probe (Fig. 4C), which showed that similar levels of undegraded mRNA were loaded into each lane except for lanes for the pancreas, placenta, and brain, which contained three- to fourfold less mRNA.
FIG. 4.
Expression of huhavcr-1 in human tissues. MTN blots containing poly(A) RNA from different human tissues were probed with 32P-labeled full-length huHAVcr-1 cDNA at high stringency and autoradiographed for 3 weeks (A) or 1 day (B). (C) The same blots were stripped and rehybridized with a 32P-labeled β-actin probe. The positions of size markers in kilobases are illustrated. peri. blood leuk., peripheral blood leukocytes.
Characterization of the receptor encoded by the huHAVcr-1 cDNA.
To characterize huhavcr-1, we transfected Perro6D cells with pDR2huHAVcr and selected hygromycin-resistant cells that expressed huhavcr-1. Because there are no available Abs capable of detecting the expression of huhavcr-1 at the cell surface, we introduced a tag epitope into the mucin-like region of huhavcr-1 to monitor its expression at the cell surfaces of the dog cell transfectants. The resulting construct, termed huflagBst, contains the FLAG octapeptide DTKDDDDK inserted between amino acid residues 145 and 146 of the TSP-rich region of huhavcr-1. Dog cells transfected with pDR2HAVcrFlag (flag cells), which express a FLAG-tagged havcr-1, or vector pDR2 (DR2 cells) were previously characterized (19) and were used as controls for our studies.
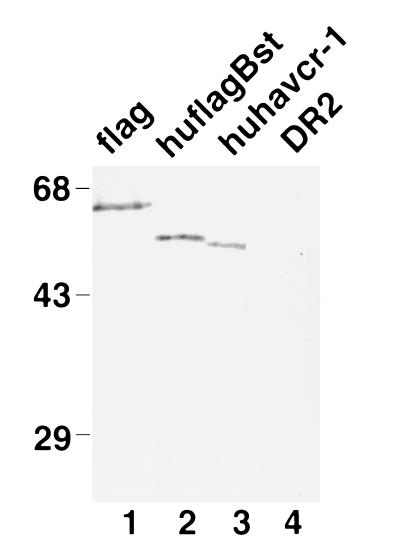
Western blot analysis showed that the anti-GST2 Ab (Fig. 5) reacted with a 64-kDa protein in flag cells (lane 1), corresponding to the major form of the FLAG-tagged havcr-1 (19), and cross-reacted with a 54-kDa protein in huflagBst cells (lane 2) and a 53-kDa protein in huhavcr-1 cells (lane 3). The anti-GST2 Ab did not recognize havcr-1-specific bands in DR2 cells (lane 4), which indicated that the 64-, 54-, and 53-kDa proteins corresponded to HAV cellular receptors. It should be pointed out that the 54-kDa huflagBst band migrated with the expected molecular weight of a FLAG-tagged huhavcr-1.
FIG. 5.
Western blot analysis of the expression of huhavcr-1 in dog cell transfectants. Cytoplasmic extracts of dog cell transfectants expressing FLAG-tagged havcr-1 (flag), FLAG-tagged huhavcr-1 (huflagBst), and untagged huhavcr-1 (huhavcr-1) and dog cells transfected with vector pDR2 alone (DR2) were prepared in RSB–1% NP-40, resolved by SDS–10% polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and probed with rabbit anti-GST2 Ab directed against the TSP-rich region of huhavcr-1. The positions of prestained molecular weight markers and their sizes in kilodaltons are shown on the left.
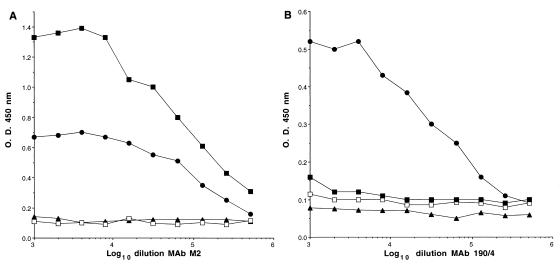
To determine whether huhavcr-1 was presented on the cell surfaces of the dog cell transfectants, we performed a cell surface ELISA of huhavcr-1, huflagBst, flag, and DR2 cells with the anti-FLAG M2 MAb and anti-havcr-1 190/4 MAb (Fig. 6). As expected, flag cells reacted with both MAbs and DR2 cells reacted with neither MAb, which confirmed that the cell surface ELISA specifically recognized the M2 and 190/4 epitopes. MAb M2 did not react with huhavcr-1 cells but reacted with huflagBst cells (Fig. 6A), which indicated that the FLAG-tagged huhavcr-1 was expressed at the cell surfaces of the dog cell transfectants and suggested the same for untagged huhavcr-1. Unfortunately, we could not confirm this with the anti-GST2 Ab because this Ab recognized neither havcr-1 nor huhavcr-1 when they were presented at the cell surface. MAb 190/4 did not react with the huhavcr-1 and huflagBst cells (Fig. 6B), which confirmed that huhavcr-1 does not code for the 190/4 epitope.
FIG. 6.
Expression of huhavcr-1 at the cell surfaces of dog cell transfectants. Expression of the M2 (A) and 190/4 (B) epitopes at the cell surfaces of dog cell transfectants expressing FLAG-tagged huhavcr-1 (flag cells; solid circles), FLAG-tagged huhavcr-1 (huflagBst cells; solid squares); and untagged huhavcr-1 (huhavcr-1 cells; open squares) and dog cells transfected with vector pDR2 (DR2 cells; solid triangles) was determined by ELISA using twofold dilutions of the MAbs. Absorbance at 450 nm was plotted versus the log10 of the MAb dilution. Data are means of duplicate wells; duplicate values varied by less than 10%. The results correspond to one experiment which was repeated two times with approximately 5 to 10% experimental error.
Although similar levels of flag and huflagBst were detected by Western blot analysis of the dog cell transfectants (Fig. 5, compare lanes 1 and 2), the cell surface ELISA showed that huflagBst cells expressed higher levels of the M2 epitope than flag cells (Fig. 6A). Therefore, it is possible that the human homolog of havcr-1 is expressed at a higher level at the cell surface than the simian havcr-1 receptor. However, since the FLAG tag was inserted at different positions in the flag and huflagBst receptors, we cannot rule out the possibility that the difference in the level of reaction of the dog cell transfectants with MAb M2 was due to steric hindrance.
Binding of HAV to dog cell transfectants.
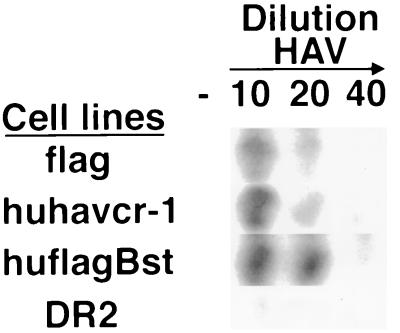
To determine whether HAV binds specifically to huhavcr-1, 96-well plates containing subconfluent monolayers of flag, huhavcr-1, huflagBst, and DR2 cells were infected with 1:10, 1:20, and 1:40 dilutions of purified HAV or were mock infected for 1 h at 35°C. After being washed extensively, monolayers were fixed, and bound HAV was detected with 125I-labeled human anti-HAV antibody. This assay showed a concentration-dependent binding of HAV to flag, huhavcr-1, and huflagBst cells and a low background level of binding of HAV to DR2 cells (Fig. 7). No signal was observed for mock-infected cells, which indicated that the 125I-labeled Ab reacted specifically against HAV bound to the cells. These results indicated that huhavcr-1 is a binding receptor for HAV and that the insertion of the FLAG tag into its TSP-rich region did not affect the binding of HAV.
FIG. 7.
Binding of HAV to dog cells transfected with huHAVcr-1 cDNA. Dog cell transfectants expressing FLAG-tagged huhavcr-1 (flag), FLAG-tagged huhavcr-1 (huflagBst), and untagged huhavcr-1 (huhavcr-1) and dog cells transfected with vector pDR2 (DR2) were grown in 96-well plates and infected with 1:10, 1:20, and 1:40 dilutions of purified HAV or were mock infected (−) for 1 h at 35°C. After being extensive washed, monolayers were fixed, and HAV bound to the cells was detected with 125I-labeled human anti-HAV Ab. Autoradiography of the 96-well plate showing a single well for each treatment is presented.
Susceptibility of dog cells expressing huHAVcr-1 to HAV infection.
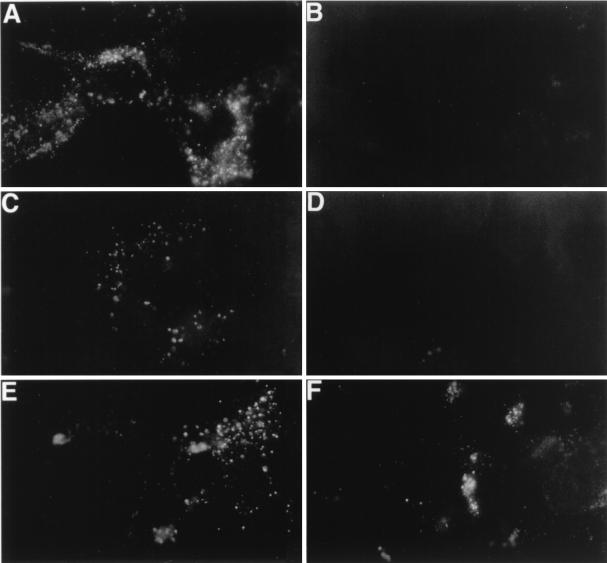
Receptor-negative but otherwise fully HAV-susceptible cell lines have not been identified yet. We previously showed that dog cells do not support HAV growth upon inoculation or transfection of infectious HAV RNA, which indicated that dog cells contain an internal block(s) to HAV replication (4). However, we have recently shown that HAV inoculation of dog cell transfectants expressing havcr-1 resulted in the characteristic granular cytoplasmic fluorescence of HAV-infected cells, which lasted approximately 1 week (19), indicating that these dog cell transfectants gained a limited level of susceptibility to HAV infection. Therefore, we used this dog cell system to analyze the HAV receptor function of huhavcr-1. To do so, we infected monolayers of dog cell transfectants with a MOI of 100 to 1,000 TCID50 of HAV per cell and analyzed the presence of HAV antigen by indirect IF staining with human anti-HAV Ab (Fig. 8). At 3 days postinfection, HAV-specific IF was detected in control GL37 (Fig. 8A) and flag (Fig. 8C) cells but not in DR2 cells (Fig. 8B) and d1− cells (which contain FLAG-tagged havcr-1 constructs with the Cys-rich regions deleted; Fig. 8D) (19). HAV-specific IF was also detected in huhavcr-1 and huflagBst cells (Fig. 8E and F), which indicated that huhavcr-1 functions as an HAV receptor that mediates a limited level of susceptibility to HAV infection, similar to what was previously reported for havcr-1 (7, 19). It should be pointed out that in huhavcr-1 and huflagBst cells we detected a low level of HAV-specific IF at 0 days postinfection; this level increased approximately twofold at 3 days postinfection, as judged by microscopic examination (data not shown). Mock-infected GL37 cells and dog cell transfectants did not fluoresce (data not shown), which showed that the human anti-HAV Ab specifically detected the HAV antigen present in the infected cells.
FIG. 8.
Detection of HAV antigen in the cytoplasm of dog cell transfectants by indirect IF analysis. Monolayers of GL37 (A), DR2 (B), flag (C), d1− (D), huhavcr-1 (E), and huflagBst (F) cells grown in eight-well slides were infected with a MOI of 100 to 1,000 TCID50 of purified HAV per cell for 6 h, washed extensively, and incubated at 35°C for 3 days. Cells were fixed with cold acetone and stained with human anti-HAV Ab and FITC-labeled goat anti-human IgG and IgM Abs. Mock-infected cells did not immunofluoresce (data not shown).
MAb 190/4 protects GL37 cells but not GL37 transfectants expressing huhavcr-1 against HAV infection.
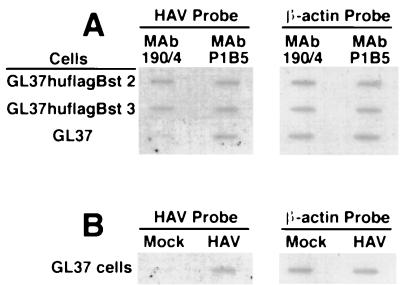
To further analyze the function of huhavcr-1 as a receptor for HAV, we used protective MAb 190/4 to block the endogenous havcr-1 expressed at the cell surfaces of GL37 cell transfectants. Since huhavcr-1 does not contain the 190/4 epitope (Fig. 6), we reasoned that MAb 190/4 will not protect the GL37 cell transfectants expressing huhavcr-1 against HAV infection. To test this hypothesis, we treated GL37hflagBst 2 and GL37huflagBst 3 cells, two zeomycin-resistant GL37 cell clones that expressed a FLAG-tagged huhavcr-1 construct at the cell surface, and GL37 cells with control MAb P1B5 or MAb 190/4 for 1 h and then inoculated these cells at a MOI of 0.1 TCID50 of HAV per cell. Three days after infection, the protective effect of the MAb treatment was determined by slot blot analysis of total cellular RNA probed with 32P-labeled HAV cDNA (Fig. 9A). Treatment of GL37 cells with MAb 190/4 resulted in a lower level of HAV-specific signal than treatment with MAb P1B5, which indicated that MAb 190/4 protected GL37 cells against HAV infection (7). Similar levels of HAV-specific signal were observed in GL37huflagBst 2 and GL37huflagBst 3 cells treated with MAbs 190/4 or P1B5, which indicated that MAb 190/4 did not protect these cell lines against HAV infection. Untreated GL37 cells were infected with HAV or were mock infected in parallel under the same conditions mentioned above. Samples of total cellular RNA extracted from these control cells were included in the slot blot analysis (Fig. 9B), which showed that only HAV-inoculated cells reacted with the HAV probe. To control for RNA loading, the same blot was stripped and rehybridized with a 32P-labeled β-actin cDNA probe, which showed that similar levels of total cellular RNA were loaded in each slot (Fig. 9A and B). Considered together, these results further suggested that huhavcr-1 is a functional receptor for HAV.
FIG. 9.
Slot blot analysis of MAb 190/4-mediated protection of GL37 cell transfectants against HAV infection. (A) Slot blot analysis of protection of GL37 cells and GL37 cell transfectants expressing FLAG-tagged huhavcr-1 (GL37huflagBst 2 and GL37hflaBst 3 cells) against HAV infection. Confluent monolayers of GL37 cells in six-well plates were treated with MAb 190/4 or anti-human α3 integrin MAb P1B5 for 1 h at 35°C, washed, and inoculated at a MOI of 0.1 TCID50 of HAV per cell for 1 h at 35°C. Monolayers were washed and incubated at 35°C, cytoplasmic extracts were prepared at 3 days postinfection, and total RNA was extracted. HAV-specific RNA was detected with a 32P-labeled HAV cDNA probe. To control for RNA loading, the slot blot was stripped and rehybridized with a 32P-labeled β-actin cDNA probe. (B) Control GL37 cells were infected with HAV or were mock infected (Mock) without prior MAb treatment. RNA samples were prepared in parallel with the samples shown in panel A and hybridized under the same conditions. The autoradiogram was exposed for 2.5 h with intensifying screens.
DISCUSSION
Our initial studies on the identification of cellular receptors for HAV showed that the protective epitope 190/4 was not expressed in HeLa cells (7) and raised the possibility that the HAVcr-1 gene itself was not conserved in humans. In this work, we identified a human homolog of havcr-1 that has approximately 79% homology with its simian counterpart and showed that it did not express the 190/4 epitope. This antigenic variability among primates of protective epitope 190/4 contrasts to the high degree of conservation of protective epitope D171 of the poliovirus receptor, which has been found in all the cell lines of primate origin tested (13, 16). Moreover, we recently showed that antigenic variants of havcr-1 expressed in BS-C-1 and CV-1 cells, two widely used AGMK cell lines, contain a K108Q change in the Cys-rich region that is responsible for their lack of reaction with MAb 190/4 (5). Taken all together, these data suggested that the HAV–havcr-1 interaction tolerates some degree of variability in protective epitope 190/4. Mutagenesis of havcr-1 and further mapping of the 190/4 epitope will allow us to understand how HAV interacts with its cellular receptor.
The receptor function of huhavcr-1 was analyzed by a binding assay which showed that dog cell transfectants expressing huhavcr-1 bound HAV (Fig. 7). Further analysis of the receptor function of huhavcr-1 is complicated by the lack of receptor-negative fully HAV-susceptible cell lines (4, 7, 19). To overcome this limitation, we performed two kinds of experiments, which provided further evidence that huhavcr-1 is a functional receptor for HAV. First, we showed that dog cell transfectants expressing huhavcr-1 developed the characteristic cytoplasmic granular fluorescence of HAV-infected cells, whereas dog cell transfectants expressing an havcr-1 construct with the Cys-rich region deleted and vector-transfected dog cells did not fluoresce. This experiment suggested that dog cell transfectants expressing huhavcr-1 gained limited susceptibility to HAV infection (Fig. 8). Second, MAb 190/4 did not protect GL37 cell transfectants expressing huhavcr-1 against HAV infection (Fig. 9), which suggested that HAV entered the cells via huhavcr-1 and not the endogenous havcr-1 blocked by MAb 190/4. Although these two lines of evidence strongly suggested that huhavcr-1 is a functional receptor for HAV, further confirmation of its functionality will have to wait for the isolation of a receptor-negative fully HAV-susceptible cell lines.
The pathogenesis of HAV is poorly understood, and extrahepatic sites of HAV replication are not well defined. HAV is transmitted through the fecal/oral route and, after ingestion, it probably travels from the gut to the liver where it replicates before being excreted with bile to the intestine. Detection of HAV in saliva and in the throat of an experimentally infected chimpanzee suggested that the initial viral replication might occur in the oropharynx and salivary glands (2). Replication of HAV in the gastrointestinal tract has been difficult to determine (9, 11, 12), but HAV antigen was found in the intestinal mucosa of two marmosets inoculated intravenously with HAV (8). HAV antigen was also detected in the kidneys, spleens, and lymph nodes of experimentally infected primates (2, 8, 11). Recently, HAV has been detected by IF analysis in the epithelial cells of the intestinal crypts and in cells of the lamina propria of the small intestines of orally inoculated owl monkeys (1). The same study showed that HAV antigen, besides being detected in the liver, was also detected in the kidneys and spleen but not in pharyngeal tissues of the owl monkey. These data on the detection of HAV antigen in different organs of nonhuman primates correlated well with our data on the ubiquitous expression of huHAVcr-1-specific messages in humans.
ACKNOWLEDGMENTS
We thank Stephen Feinstone for encouragement and helpful advice and Barry Falgout and Robin Levis for comments on the manuscript. We also thank Michael Klutch for automatic sequencing of DNA samples.
This research was supported in part by the appointment of D.F. to the Postgraduate Research Participation Program at the Center for Biologics Evaluation and Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
REFERENCES
- 1.Asher L V, Binn L N, Mensing T L, Marchwicki R H, Vassell R A, Young G D. Pathogenesis of hepatitis A in orally inoculated owl monkeys (Aotus trivirgatus) J Med Virol. 1995;47:260–268. doi: 10.1002/jmv.1890470312. [DOI] [PubMed] [Google Scholar]
- 2.Cohen J I, Feinstone S, Purcell R H. Hepatitis A virus infection in a chimpanzee: duration of viremia and detection of virus in saliva and throat swabs. J Infect Dis. 1989;160:887–890. doi: 10.1093/infdis/160.5.887. [DOI] [PubMed] [Google Scholar]
- 3.Cohen J I, Ticehurst J R, Feinstone S M, Rosenblum B, Purcell R H. Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. J Virol. 1987;61:3035–3039. doi: 10.1128/jvi.61.10.3035-3039.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dotzauer A, Feinstone S M, Kaplan G. Susceptibility of nonprimate cell lines to hepatitis A virus infection. J Virol. 1994;68:6064–6068. doi: 10.1128/jvi.68.9.6064-6068.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feigelstock D, Thompson P, Mattoo P, Kaplan G G. Polymorphisms of the hepatitis A virus cellular receptor 1 in African green monkey kidney cells result in antigenic variants that do not react with protective monoclonal antibody 190/4. J Virol. 1998;72:6218–6222. doi: 10.1128/jvi.72.7.6218-6222.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jentoft N. Why are proteins O-glycosylated? Trends Biochem Sci. 1990;15:291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan G, Totsuka A, Thompson P, Akatsuka T, Moritsugu Y, Feinstone S M. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J. 1996;15:4282–4296. [PMC free article] [PubMed] [Google Scholar]
- 8.Karayiannis P, Jowett T, Enticott M, Moore D, Pignatelli M, Brenes F, Scheuer P J, Thomas H C. Hepatitis A virus replication in tamarins and host immune response in relation to pathogenesis of liver cell damage. J Med Virol. 1986;18:261–276. doi: 10.1002/jmv.1890180308. [DOI] [PubMed] [Google Scholar]
- 9.Krawczynski K K, Bradley D W, Murphy B L, Ebert J W, Anderson T E, Doto I L, Nowoslawski A, Duermeyer W, Maynard J E. Pathogenetic aspects of hepatitis A virus infection in enterally inoculated marmosets. Am J Clin Pathol. 1981;76:698–706. doi: 10.1093/ajcp/76.5.698. [DOI] [PubMed] [Google Scholar]
- 10.Magee A I, Gutierrez L, Marshall C J, Hancock J F. Targeting of oncoproteins to membranes by fatty acylation. J Cell Sci Suppl. 1989;11:149–160. doi: 10.1242/jcs.1989.supplement_11.12. [DOI] [PubMed] [Google Scholar]
- 11.Mathiesen L R, Drucker J, Lorenz D, Wagner J A, Gerety R J, Purcell R H. Localization of hepatitis A antigen in marmoset organs during acute infection with hepatitis A virus. J Infect Dis. 1978;138:369–377. doi: 10.1093/infdis/138.3.369. [DOI] [PubMed] [Google Scholar]
- 12.Mathiesen L R, Feinstone S M, Purcell R H, Wagner J A. Detection of hepatitis A antigen by immunofluorescence. Infect Immun. 1977;18:524–530. doi: 10.1128/iai.18.2.524-530.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minor P D, Pipkin P A, Hockley D, Schild G C, Almond J W. Monoclonal antibodies which block cellular receptors of poliovirus. Virus Res. 1984;1:203–212. doi: 10.1016/0168-1702(84)90039-x. [DOI] [PubMed] [Google Scholar]
- 14.Murphy A J M, Kung A L, Swirski R A, Schimke R T. cDNA expression cloning in human cells using the pλDR2 episomal vector system. Methods (Orlando) 1992;4:111–131. [Google Scholar]
- 15.Neutra M R, Forstner J F. Gastrointestinal mucus: synthesis, secretion, and function. In: Johnson L R, editor. Physiology of the gastrointestinal tract. New York, N.Y: Raven Press; 1987. pp. 975–1009. [Google Scholar]
- 16.Nobis P, Zibirre R, Meyer G, Kühne J, Warnecke G, Koch G. Production of a monoclonal antibody against an epitope of HeLa cells that is the functional poliovirus biding site. J Gen Virol. 1985;66:2563–2569. doi: 10.1099/0022-1317-66-12-2563. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 18.Swirski R A, Van Den Berg D, Murphy A J M, Lambert C M, Friedberg E C, Schimke R T. Improvements in the Epstein-Barr-based shuttle vector system for direct cloning in human tissue culture cells. Methods (Orlando) 1992;4:133–142. [Google Scholar]
- 19.Thompson P, Lu J, Kaplan G G. The Cys-rich region of the hepatitis A virus cellular receptor 1 (HAVcr-1) is required for binding of hepatitis A virus and protective monoclonal antibody 190/4. J Virol. 1998;72:3751–3761. doi: 10.1128/jvi.72.5.3751-3761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Totsuka A, Moritsugu Y. Hepatitis A vaccine development in Japan. In: Nishioka K, Suzuki H, Mishiro S, Oda T, editors. Viral hepatitis and liver disease. Tokyo, Japan: Springer-Verlag; 1994. pp. 509–513. [Google Scholar]