Abstract
Skeletal muscle is not only the largest organ in the body that is responsible for locomotion and exercise but also crucial for maintaining the body’s energy metabolism and endocrine secretion. The trimethylation of histone H3 lysine 27 (H3K27me3) is one of the most important histone modifications that participates in muscle development regulation by repressing the transcription of genes. Previous studies indicate that the RASGRP1 gene is regulated by H3K27me3 in embryonic muscle development in pigs, but its function and regulatory role in myogenesis are still unclear. In this study, we verify the crucial role of H3K27me3 in RASGRP1 regulation. The gain/loss function of RASGRP1 in myogenesis regulation is performed using mouse myoblast C2C12 cells and primarily isolated porcine skeletal muscle satellite cells (PSCs). The results of qPCR, western blot analysis, EdU staining, CCK-8 assay and immunofluorescence staining show that overexpression of RASGRP1 promotes cell proliferation and differentiation in both skeletal muscle cell models, while knockdown of RASGRP1 leads to the opposite results. These findings indicate that RASGRP1 plays an important regulatory role in myogenesis in both mice and pigs.
Keywords: RASGRP1, H3K27me3, cell proliferation, myoblasts, skeletal muscle
Introduction
Skeletal muscle is not only the largest organ responsible for animal locomotion and exercise capacity but also crucial for maintaining the body’s energy metabolism and endocrine secretion [ 1– 3]. In livestock-producing fields, skeletal muscle is of particular importance, as meat is the main animal product, in addition to animal fur, milk and others. Increasing evidence indicates that epigenetic regulators participate in the development of skeletal muscle.
Epigenetic regulators such as DNA methylation, histone methylation, microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) play vital roles in regulating myogenesis by affecting the transcription process or posttranscriptional modifications of RNAs [ 4– 7]. Among them, the trimethylation of histone H3 lysine 27 (H3K27me3) is a well-known repressive marker involved in various biological processes [ 8– 10]. H3K27me3 participates in the regulation of myogenic differentiation by silencing muscle-specific genes and cell cycle genes [ 11– 14]. The loss of H3K27me3 in the gene body, especially in the promoter of genes, repressed the differentiation of skeletal muscle cells [ 15– 17]. The methyltransferase of H3K27me3, named enhancer of zeste homolog 2 (EZH2), which is a subunit of polycomb repressive complex 2 (PRC2), can also directly or indirectly regulate the expressions of myogenic genes [ 18, 19]. During myogenic differentiation, the phosphorylated EZH2 enhancer induces a shift from H3K4me3 to H3K27me3 on the Pax7 promoter to downregulate gene expression [20]. In contrast, the decrease in H3K27me3 is related to genes that are more actively transcribed [21] .
The RASGRP1 gene is a member of the Ras gene family, which is highly expressed in T cells [22]. It has been reported that dysregulation of RASGRP1 leads to the occurrence of cancers [ 23 – 28] and other diseases [ 29– 33] related to the body’s immunity [ 34 , 35]. Our previous study found that RASGRP1 may regulate pig embryonic myogenesis, which was regulated by H3K27me3 through chromatin immunoprecipitation-sequencing (ChIP-seq) and RNA-sequencing of the longissimus dorsi muscle in Duroc pig embryos at gestation days 33 (E33), 65 (E65), and 90 (E90) [36]. However, the detailed function of this gene in regulating myogenesis is still unclear.
In this study, we first confirmed that the expression of the RASGRP1 gene was regulated by H3K27me3 enrichment in its promoter during skeletal muscle development. Then, we studied the function of RASGRP1 during cell proliferation and differentiation in both mouse C2C12 myoblasts and PSCs. Our results demonstrated that RASGRP1 is regulated by H3K27me3 and promotes myogenesis in pigs and mice.
Materials and Methods
Animals
Pigs, including Duroc sows for embryonic skeletal muscle collection and newborn piglets for isolating porcine skeletal muscle satellite cells, were purchased from Guangdong Wen’s Foodstuffs Group (Yunfu, China). All animal experiments were conducted following the requirements for the Care and Use of Laboratory Animals by the Ethics Committee of the Laboratory Animal Center of South China Agricultural University, Guangzhou, China (Permit Number 2021F036, Permit Date 2 March 2021) [37].
Cell culture
C2C12 cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Dulbecco’s modified Eagle’s medium (DMEM) high glucose (Gibco, Grand Island, USA) and 10% fetal bovine serum (FBS; Gibco) was used for cell culture. For cell differentiation, DMEM high glucose with 2% horse serum (Gibco) was used. The cells were cultured with 5% CO 2 at 37°C.
Isolation and culture of porcine skeletal muscle satellite cells
Porcine skeletal muscle satellite cells (PSCs) were isolated from one-week-old piglets and cultivated according to protocols described in previous studies [ 38, 39]. The tissues were digested with 2 mg/mL type II collagenase (Sigma-Aldrich, Darmstadt, Germany) and then placed in a 37°C incubator for 2.5 h. Then, the digestion was stopped with an equal volume of RPMI 1640 medium (Gibco) containing 1% penicillin-streptomycin (P-S; Gibco). Then, 100, 200, and 400-mesh sieves were used to filter the cell suspension. Cell proliferation medium was prepared according to a previous study and used to resuspend cells [39]. Differential adhesion was used to obtain purified cells after 2 h of culture. When cells reached 70%‒80% confluence, differentiation medium consisting of DMEM high glucose with 2% horse serum and 1% P-S was used to induce cell differentiation.
Plasmid construction, small interfering RNA synthesis, and transfection
The coding sequence of the porcine RASGRP1 gene was cloned in vitro by PCR amplification and connected to the linear pcDNA3.1(+) vector (Sangon, Shanghai, China) to produce the pcDNA3.1- RASGRP1 plasmid. The primers for the RASGRP1 CD sequence were as follows: forward primer 5′-TACCGAGCTCGGATCCATGGGCACCCTGGGCAAG-3′ and reverse primer 5′-GATATCTGCAGAATTCCTAAGAACAGTCACCGTGCTCCATC-3′. Small interfering RNAs (siRNAs) against pig RASGRP1 and mouse RASGRP1 genes were designed and synthesized by GenePharma (Shanghai, China). The siRNA sequences were as follows: Sus-siRNA- RASGRP1 (sense 5′-CCCAGUGGGUUCAACUCAUTT, antisense 5′-AUGAGUUGAACCCACUGGGTT-3′), Mus-siRNA- RASGRP1 (sense 5′-GGACCUCAUAUCCCUGUAUTT-3′, antisense 5′-AUACAGGGAUAUGAGGUCCTT-3′), and siRNA negative control (sense: 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense: 5′-ACGUGACACGUUCGGAGAATT-3′). Lipofectamine 3000 (Invitrogen, Carlsbad, USA) was used to conduct cell transfection following the manufacturer’s protocol.
Quantitative real-time polymerase chain reaction (qPCR)
Total RNA kit ll (Omega Biotek, Norcross, USA) was used to harvest the total RNA following the manufacturer’s protocol. cDNA was prepared using a PrimeScript RT reagent kit (TaKaRa, Tokyo, Japan) and used to perform qPCR in a Quant Studio 7 Flex system (Thermo Fisher, Scientific, Waltham, USA). All primers used are presented in Supplementary Table S1.
Chromatin immunoprecipitation (ChIP)
ChIP was conducted according to the protocol in a previous study [40]. Briefly, micrococcal nuclease was used to fragment chromatin. After that, the chromatin fragments were incubated with magnetic beads (Biogle, Wuxi, China). The DNA-bead complex was immunoprecipitated with anti-H3K27me3 antibody (Millipore, Billerica, USA) or negative control IgG. The primers for the promoter of the RASGRP1 gene used for ChIP-qPCR were as follows: forward primer 5′-CTCTCCGAATTCCCCATTGTG-3′ and reverse primer 5′-AAATCAGAGCTGCATCCAC-3′.
Western blot analysis
The concentration of protein harvested from cells by RIPA buffer with 1% PMSF (Beyotime, Shanghai, China) was determined using a BCA Assay Kit (Thermo Fisher Scientific). Western blot analysis was performed following a previous study [39]. The antibodies used were as follows: Ki67 (ab16667; 1:1000; Abcam, Cambridge, UK), CDK2 (PA1547; 1:1000; Boster, Pleasanton, USA), MyoG (sc-12732; 1:500; Santa Cruz Biotechnology, Santa Cruz, USA), MyoD (sc-377460; 1:500; Santa Cruz Biotechnology), MyHC (sc-376157; 1:1000; Santa Cruz Biotechnology), β-Tubulin (GB11017; 1:1000; Servicebio, Wuhan, China), goat anti-mouse IgG (A0216; 1:3000; Beyotime) and goat anti-rabbit IgG (A0208; 1:3000; Beyotime).
Cell proliferation and cell cycle assays
For flow cytometry analysis, C2C12 cells stored at ‒20°C were fixed in 70% (v/v) ethanol overnight and treated at 4°C in 50 mg/mL propidium iodide for 30 min. Cell cycle detection was conducted in accordance with a previous study [39] by using a flow cytometer (Becton Dickinson, Franklin Lakes, USA). The cells were transfected with RASGRP1 siRNA or overexpression vectors when they reached approximately 50% confluence. CCK-8 assays (Yeasen, Shanghai, China) were conducted following the manufacturer’s protocol after transfection. EdU staining was performed following the manufacturer’s protocol (RiboBio, Guangzhou, China). Cells were to fixed and permeabilized separately with 4% paraformaldehyde and 0.5% Triton X-100, respectively. DAPI was used to stain cell nuclei. All images were captured with the TE2000U fluorescence microscope imaging system (Nikon, Tokyo, Japan).
Immunofluorescence staining
Cells seeded in a 24-well plate were transfected with RASGRP1 siRNA or overexpression vectors when they reached approximately 50%–70% confluence. Then, 4% paraformaldehyde and 0.5% Triton X-100 were separately used to fix and permeabilize the cells for 30 min three days after transfection. Then, the cells were blocked in QuickBlock Buffer (Beyotime) at 37°C for 2 h and treated with the anti-MyHC antibody (sc-376157; 1:200; Santa Cruz Biotechnology) at 4°C overnight. Then, the cells were incubated with Alexa Fluor 488 fluorescent antibody (1:500; Thermo Fisher Scientific) after wash with PBS (Servicebio). DAPI was used to stain cell nuclei. All images were captured with the TE2000U fluorescence microscope imaging system.
Statistical analysis
Data are presented as the mean±standard error of the mean (SEM). The comparative threshold cycle (2 –ΔΔct) method was used to quantify the relative mRNA levels [ 38, 41]. Statistical analysis between different groups was performed by using two-tailed Student’s t-test or one-way analysis of variance (ANOVA) in SPSS software (version 22.0). ImageJ software was used for the quantification of positively-stained cells and the visualization of western blots. All experiments were performed in triplicate. P<0.05 indicated significant difference.
Results
RASGRP1 is targeted by H3k27me3
We collected longissimus dorsi muscles on gestation days 33, 65, and 90, which represent three critical developmental time points for embryonic myogenesis [ 42– 45]. ChIP-qPCR assay in skeletal muscles at E33, E65, and E90 showed that the enrichment of H3K27me3 in the RASGRP1 promoter was highest at E33 and lowest at E90 ( Figure 1A). The mRNA expression of the RASGRP1 gene significantly increased during embryonic development ( Figure 1B and Supplementary Figure S1), while its expression was pretty low in adult skeletal muscle. These results indicated that during embryonic porcine myogenesis, increased RASGRP1 expression might be specifically regulated by H3K27me3.
Figure 1 .

RASGRP1 is targeted by H3K27me3 and upregulated in the longissimus dorsi muscle of Duroc pig embryos at gestation days 33 (E33), 65 (E65), and 90 (E90)
(A) The enrichment of H3K27me3 in the RASGRP1 promoter at E33, E65, and E90. (B) The expression level of RASGRP1 in pig skeletal muscle at E33, E65, E90 and the adult period. β-Actin was used as a control. *P<0.05, **P<0.01.
RASGRP1 promotes the proliferation of C2C12 cells
To investigate the role of the RASGRP1 gene in C2C12 cell proliferation, we performed gain- and loss-of-function analyses of the RASGRP1 gene. We overexpressed the RASGRP1 gene in C2C12 cells, and the qPCR results demonstrated that the relative expression of the RASGRP1 gene and the proliferation marker genes CDK2 and CDK14 were significantly increased ( Figure 2A). CCK-8 assay demonstrated that RASGRP1 overexpression also significantly promoted the proliferation of C2C12 cells ( Figure 2B). The relative protein expressions of the proliferation marker genes Ki67 and CDK2 were also increased, according to western blot analysis ( Figure 2C). EdU staining revealed that RASGRP1 overexpression dramatically increased the percentage of EdU-positive (EdU +) cells ( Figure 2D). In addition, flow cytometry analysis showed that there were 5.33% more S-phase cells among the cells overexpressing RASGRP1 than in the pcDNA3.1(+) control group ( Figure 2E). All the above results indicated that RASGRP1 promoted C2C12 cell proliferation.
Figure 2 .
RASGRP1 overexpression promotes the proliferation of C2C12 cells
(A) The relative expression levels of RASGRP1 and proliferation marker genes after RASGRP1 overexpression. (B) Detection of C2C12 cell viability by CCK-8 assay at 18 h, 24 h and 30 h after RASGRP1 overexpression. (C) The relative protein levels of the Ki67 and CDK2 genes after RASGRP1 overexpression. (D) EdU staining after RASGRP1 overexpression in C2C12 cells. Scale bar: 100 μm. (E) Detection of the number of S-phase cells by flow cytometry after RASGRP1 overexpression. *P<0.05, **P<0.01.
To verify the effect of RASGRP1 overexpression on C2C12 cell proliferation, we then knocked down RASGRP1 by siRNA in C2C12 cells and detected its effect on C2C12 cell proliferation. The qPCR results showed that the relative expression of RASGRP1 as well as CDK2 and CDK14 was significantly reduced after RASGRP1 knockdown ( Figure 3A). CCK-8 assay demonstrated that RASGRP1 knockdown also dramatically reduced the proliferation activity of C2C12 cells ( Figure 3B). The relative protein expression of the Ki67 gene was markedly decreased ( Figure 3C). EdU staining revealed that RASGRP1 knockdown dramatically reduced the percentage of EdU + cells ( Figure 3D). These results indicated that RASGRP1 knockdown inhibited the proliferation of C2C12 cells, which was opposite to the positive effect of RASGRP1 overexpression on the proliferation of C2C12 cells.
Figure 3 .
RASGRP1 knockdown inhibits the proliferation of C2C12 cells
(A) The relative expression levels of the RASGRP1 gene and proliferation marker genes after RASGRP1 knockdown. (B) Detection of C2C12 cell viability by CCK-8 assay at 18 h and 24 h after RASGRP1 knockdown. (C) The relative protein expression levels of the Ki67 and CDK2 genes after RASGRP1 knockdown. (D) EdU staining after RASGRP1 knockdown in C2C12 cells. Scale bar: 100 μm. *P<0.05, **P<0.01.
RASGRP1 promotes the differentiation of C2C12 cells
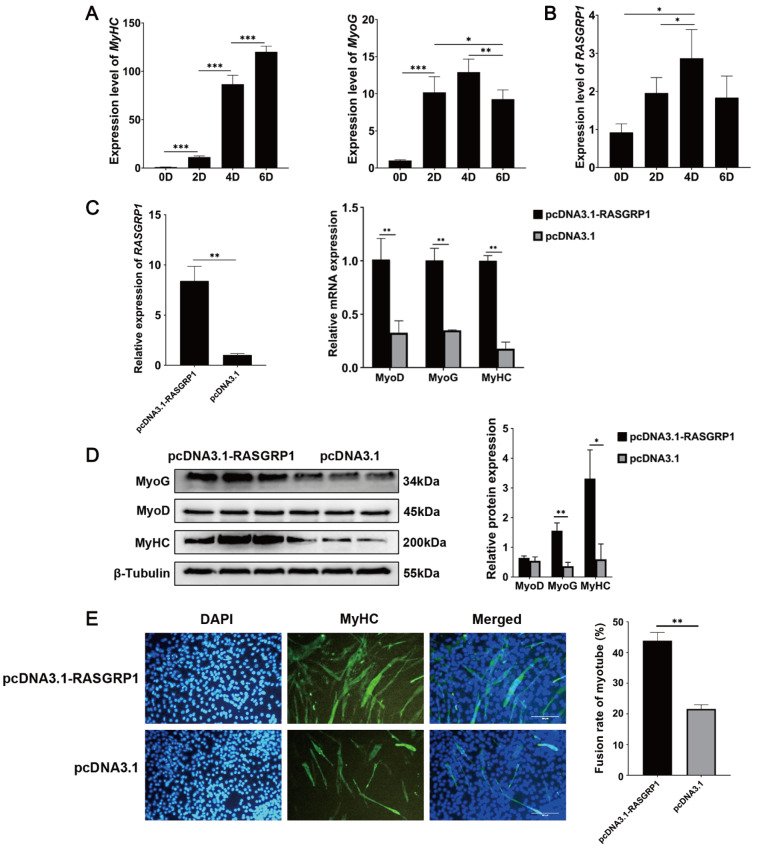
To explore the function of RASGRP1 in C2C12 cell differentiation, we examined the expression profiles of RASGRP1 as well as the MyHC and MyoG genes during myogenic differentiation. The qPCR results demonstrated that the expression of the RASGRP1 gene was significantly upregulated during the differentiation of C2C12 cells, while the expressions of the MyHC gene and MyoG gene were also upregulated significantly ( Figure 4A,B), indicating that RASGRP1 regulated the differentiation of C2C12 cells. Then, we overexpressed RASGRP1 to explore its regulatory effect on C2C12 cell differentiation. qPCR results demonstrated that the expression of RASGRP1 and differentiation marker genes ( MyoG, MyoD and MyHC) was significantly increased after RASGRP1 overexpression ( Figure 4C). The relative protein expressions of MyoG and MyHC were markedly increased after RASGRP1 overexpression ( Figure 4D). The MyHC immunofluorescence assay results showed that 22.23% more fusion myotubes were present in the cells overexpressing RASGRP1 than in the control group ( Figure 4E). These results suggested that RASGRP1 enhanced the differentiation of C2C12 cells.
Figure 4 .
RASGRP1 overexpression promotes the differentiation of C2C12 cells
(A) Expression levels of differentiation marker genes during differentiation. (B) The relative expression level of RASGRP1 was increased significantly and then decreased during differentiation. (C) The relative expression levels of RASGRP1 and differentiation marker genes after RASGRP1 overexpression. (D) The relative protein expression of MyHC, MyoD and MyoG after RASGRP1 overexpression. (E) MyHC immunofluorescence assay after RASGRP1 overexpression in C2C12 cells. Scale bar: 50 μm. *P<0.05, **P <0.01.
To further verify the effect of RASGRP1 in regulating skeletal cell differentiation, we performed RASGRP1 knockdown experiments by siRNA in C2C12 cells. After transfection with siRNA for three days, total RNA and protein were extracted. The qPCR results demonstrated that the relative expressions of the RASGRP1 and MyHC genes were significantly decreased after siRNA transfection ( Figure 5A). The relative protein expressions of the MyoD and MyoG genes were also decreased after RASGRP1 knockdown ( Figure 5B). The MyHC immunofluorescence assay results showed that there were 8.14% fewer fusion myotubes in the cells with RASGRP1 gene knockdown than in the control group ( Figure 5C). All these results indicated that RASGRP1 knockdown inhibited C2C12 cell differentiation.
Figure 5 .
RASGRP1 knockdown inhibits the differentiation of C2C12 cells
(A) The relative expressions of RASGRP1 and differentiation marker genes after RASGRP1 knockdown. (B) The relative protein expression levels of MyHC, MyoD and MyoG after RASGRP1 knockdown. (C) MyHC immunofluorescence assay after RASGRP1 knockdown in C2C12 cells. Scale bar: 50 μm. *P<0.05, **P<0.01.
RASGRP1 promoted the proliferation and differentiation of PSCs
To investigate the role of RASGRP1 in PSC proliferation and differentiation, we also studied the function of RASGRP1 in pigs. After RASGRP1 overexpression, the qPCR results demonstrated that the relative expressions of the RASGRP1 gene and proliferation marker genes were significantly increased in PSCs ( Figure 6A). CCK-8 assay demonstrated that RASGRP1 overexpression also significantly promoted the proliferation of PSCs ( Figure 6B). EdU staining showed that RASGRP1 overexpression dramatically increased the percentage of EdU + cells ( Figure 6C). Knockdown of RASGRP1 led to a significant decrease in the expressions of RASGRP1 gene and proliferation marker genes ( Figure 6D). CCK-8 assay demonstrated that RASGRP1 knockdown significantly reduced the proliferative activity of PSCs ( Figure 6E). EdU staining showed that RASGRP1 knockdown significantly reduced the percentage of EDU + cells ( Figure 6F). We also detected the role of RASGRP1 in PSC differentiation. In PSCs, the expression level of the RASGRP1 gene was significantly increased during differentiation ( Figure 6G ). MyHC immunofluorescence assay results revealed that the fusion rate of myotubes was markedly increased/decreased after RASGRP1 overexpression/knockdown ( Figure 6H,I). These findings suggested that RASGRP1 stimulated the proliferation and differentiation of PSCs.
Figure 6 .
RASGRP1 promotes the proliferation and differentiation of PSCs
(A) The relative expression levels of RASGRP1 and proliferation marker genes after RASGRP1 overexpression. (B) Detection of PSC viability by CCK-8 assay at 6 h, 48 h and 72 h after RASGRP1 overexpression. (C) EdU staining after RASGRP1 overexpression in PSCs. Scale bar: 100 μm. (D) The relative expression levels of RASGRP1 and proliferation marker genes after RASGRP1 knockdown. (E) Detection of PSC viability at 6 h, 48 h and 72 h after RASGRP1 knockdown. (F) EdU staining after RASGRP1 knockdown in PSCs. Scale bar: 100 μm. (G) The expression of RASGRP1 at different differentiation stages of PSCs. (H) MyHC immunofluorescence assay after RASGRP1 overexpression in PSCs. Scale bar: 100 μm. (I) MyHC immunofluorescence assay after RASGRP1 knockdown in PSCs. Scale bar: 100 μm. *P<0.05, **P<0.01.
Discussion
The RASGRP1 gene is one of the Ras gene families whose proteins can coordinate responses within each cell by detecting signals from other parts of the body [ 46– 48] and self-regulate once their activity is altered, which may lead to the occurrence of cancer and developmental diseases [49]. Increasing numbers of studies have revealed a connection between the RASGRP1 gene and muscle growth. RASGRP1 can affect the body size trait of pigs and exhibits a strong positive correlation with the body height and tube circumference traits of Suhuai pigs [50]. In addition, deletion of the RASGRP1 gene inhibits the activation of ERK, which is a part of the classic MAPK/ERK pathway for regulating skeletal muscle proliferation [51]. Furthermore, RASGRP1 was reported to be related to the regulation of p-ERK in the PPAR β/δ signaling pathway [52], which is widely involved in regulating myogenic and adipogenic differentiation [53]. These results indicated that RASGRP1 may be involved in muscle development. However, the detailed function of RASGRP1 in myogenesis is largely unknown.
H3K27me3 modification is widely distributed throughout the genome of myoblasts or myotubes and regulates muscle development as a silent marker mainly located at the promoter of genes [ 54, 55]. Previous studies also demonstrated that myogenic transcription factors were repressed by H3K27me3 during cell proliferation in PSCs, while H3K27me3 depletion promoted myogenic differentiation [55]. Our previous sequencing results [36] revealed that RASGRP1 has a significant H3K27me3 enrichment peak in its promoter. Thus, we aimed to investigate the function of RASGRP1 in myogenesis and explore the regulatory function of H3K27me3 in RASGRP1. In this study, we confirmed that the enrichment level of H3K27me3 on the RASGRP1 gene decreased in pig embryonic skeletal muscle at E33, E65 and E90, which showed a significant negative correlation with RASGRP1 gene expression during pig embryonic muscle development.
We then examined the functions of the RASGRP1 gene in myogenesis. Since cell maturation is based on myoblast proliferation and myoblast differentiation is essential for muscle development and maturity, we examined the expression changes of some proliferation markers after loss/gain of RASGRP1 both in C2C12 cells and PSCs, such as Ki67, cyclin-dependent kinases (CDKs) and proliferating cell nuclear antigen (PCNA). Ki67 is often used as a key indicator and marker gene for cell proliferation in clinical studies [56], and CDKs participate in the regulation of the cell proliferation cycle [57]. PCNA plays an important role in cell replication and promotes cell proliferation. Our results showed that RASGRP1 significantly promoted the relative expression of the above proliferation marker genes, which indicated its positive effect on cell proliferation. As expected, consistent results were obtained in cell proliferation assays. We found that the expression of the RASGRP1 gene increased continuously in the process of myoblast differentiation, as did that of myocyte differentiation marker genes. After overexpression of RASGRP1, the relative expressions of differentiation marker factors were also upregulated significantly. The results of the MyHC immunofluorescence experiment showed that the number of differentiated myotube fusions was increased, indicating the promotion of cell differentiation. Consistently, the opposite trend was observed after RASGRP1 knockdown. These results suggested that RASGRP1 regulates skeletal muscle development by affecting the expressions of proliferation and differentiation factors, and this function may be conserved in mice and pigs.
In conclusion, we found that the promoter of the RASGRP1 gene is enriched by H3K27me3. The loss of H3K27me3 enrichment in RASGRP1 can promote its transcriptional activity during the development of skeletal muscle. Meanwhile, up/downregulation of RASGRP1 expression can promote/repress the proliferation and differentiation of PSCs and C2C12 cells. These results suggest that RASGRP1 has a positive regulatory role in myogenesis.
Supporting information
Supplementary Data
Supplementary data is available at Acta Biochimica et Biphysica Sinica online.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the 2023 Key Areas Research and Development Programs - Modern Seed Industry of Guangdong Province, China (No. 2022B0202090002) and the Provincial Rural Revitalization Strategy Special Project on the Protection and Development and Utilization of Local Livestock and Poultry of Guangdong Province, China.
References
- 1.Kissane RWP, Charles JP, Banks RW, Bates KT. Skeletal muscle function underpins muscle spindle abundance. Proc R Soc B. . 2022;289:20220622. doi: 10.1098/rspb.2022.0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argilés JM, Campos N, Lopez-Pedrosa JM, Rueda R, Rodriguez-Mañas L. Skeletal muscle regulates metabolism via interorgan crosstalk: roles in health and disease. J Am Med Directors Assoc. . 2016;17:789–796. doi: 10.1016/j.jamda.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 3.Pedersen BK, Febbraio MA. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat Rev Endocrinol. . 2012;8:457–465. doi: 10.1038/nrendo.2012.49. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Jiang J, Liu W, Wang H, Zhao L, Liu S, Li P, et al. microRNA-378 promotes autophagy and inhibits apoptosis in skeletal muscle. Proc Natl Acad Sci USA. . 2018;115:E10849–E10858. doi: 10.1073/pnas.1803377115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Zhang Y, Hu Q, Egranov SD, Xing Z, Zhang Z, Liang K, et al. Functional significance of gain-of-function H19 lncRNA in skeletal muscle differentiation and anti-obesity effects. Genome Med. . 2021;13:137. doi: 10.1186/s13073-021-00937-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin W, Peng J, Jiang S. The epigenetic regulation of embryonic myogenesis and adult muscle regeneration by histone methylation modification. Biochem Biophys Rep. . 2016;6:209–219. doi: 10.1016/j.bbrep.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Translating the histone code. Science. . 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. . 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 9.Duan R, Du W, Guo W. EZH2: a novel target for cancer treatment. J Hematol Oncol. . 2020;13:104. doi: 10.1186/s13045-020-00937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. . 2013;58:439–445. doi: 10.1038/jhg.2013.66. [DOI] [PubMed] [Google Scholar]
- 11.Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, Bowman C, et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci USA. . 2011;108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blais A, van Oevelen CJC, Margueron R, Acosta-Alvear D, Dynlacht BD. Retinoblastoma tumor suppressor protein–dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol. . 2007;179:1399–1412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vella S, Pomella S, Leoncini PP, Colletti M, Conti B, Marquez VE, Strillacci A, et al. MicroRNA-101 is repressed by EZH2 and its restoration inhibits tumorigenic features in embryonal rhabdomyosarcoma. Clin Epigenet. . 2015;7:82. doi: 10.1186/s13148-015-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andresini O, Rossi MN, Matteini F, Petrai S, Santini T, Maione R. The long non-coding RNA Kcnq1ot1 controls maternal p57 expression in muscle cells by promoting H3K27me3 accumulation to an intragenic MyoD-binding region. Epigenet Chromatin. . 2019;12:8. doi: 10.1186/s13072-019-0253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faralli H, Wang C, Nakka K, Benyoucef A, Sebastian S, Zhuang L, Chu A, et al. UTX demethylase activity is required for satellite cell–mediated muscle regeneration. J Clin Invest. . 2016;126:1555–1565. doi: 10.1172/JCI83239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong SH, Blais A, et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. . 2010;29:1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Zhang YS, Li N, Hu XX, Shi GQ, Liu SR, Liu N. Expression of Myogenin and MCK genes regulated by PI3K/AKT pathway. Hereditas (Beijing) . 2013;35:637–642. doi: 10.3724/SP.J.1005.2013.00637. [DOI] [PubMed] [Google Scholar]
- 18.Zoroddu S, Marchesi I, Bagella L. PRC2: an epigenetic multiprotein complex with a key role in the development of rhabdomyosarcoma carcinogenesis. Clin Epigenet. . 2021;13:156. doi: 10.1186/s13148-021-01147-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchesi I, Giordano A, Bagella L. Roles of enhancer of zeste homolog 2: from skeletal muscle differentiation to rhabdomyosarcoma carcinogenesis. Cell Cycle. . 2014;13:516–527. doi: 10.4161/cc.27921. [DOI] [PubMed] [Google Scholar]
- 20.Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, et al. TNF/p38α/polycomb signaling to pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. . 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papait R, Cattaneo P, Kunderfranco P, Greco C, Carullo P, Guffanti A, Viganò V, et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc Natl Acad Sci USA. . 2013;110:20164–20169. doi: 10.1073/pnas.1315155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebinu JO, Stang SL, Teixeira C, Bottorff DA, Hooton J, Blumberg PM, Barry M, et al. RasGRP links T-cell receptor signaling to Ras. Blood. . 2000;95:3199–3203. doi: 10.1182/blood.V95.10.3199. [DOI] [PubMed] [Google Scholar]
- 23.Diez FR, Garrido AA, Sharma A, Luke CT, Stone JC, Dower NA, Cline JM, et al. RasGRP1 transgenic mice develop cutaneous squamous cell carcinomas in response to skin wounding. Am J Pathol. . 2009;175:392–399. doi: 10.2353/ajpath.2009.090036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luke CT, Oki-Idouchi CE, Cline JM, Lorenzo PS. RasGRP1 overexpression in the epidermis of transgenic mice contributes to tumor progression during multistage skin carcinogenesis. Cancer Res. . 2007;67:10190–10197. doi: 10.1158/0008-5472.CAN-07-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gbenedio OM, Bonnans C, Grun D, Wang CY, Hatch AJ, Mahoney MR, Barras D, et al. RasGRP1 is a potential biomarker for stratifying anti-EGFR therapy response in colorectal cancer. JCI Insight. . 2019;4:e127552. doi: 10.1172/jci.insight.127552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Zhuang H, Han F, Shao X, Liu Y, Ma X, Wang Z, et al. Sp1-regulated transcription of RasGRP 1 promotes hepatocellular carcinoma (HCC) proliferation. Liver Int. . 2018;38:2006–2017. doi: 10.1111/liv.13757. [DOI] [PubMed] [Google Scholar]
- 27.Hartzell C, Ksionda O, Lemmens E, Coakley K, Yang M, Dail M, Harvey RC, et al. Dysregulated rasgrp1 responds to cytokine receptor input in T cell leukemogenesis. Sci Signal. . 2013;6:ra21. doi: 10.1126/scisignal.2003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karra L, Romero-Moya D, Ksionda O, Krush M, Gu Z, Mues M, Depeille P, et al. Increased baseline RASGRP1 signals enhance stem cell fitness during native hematopoiesis. Oncogene. . 2020;39:6920–6934. doi: 10.1038/s41388-020-01469-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Gan W, Lu L, Dong X, Han X, Hu C, Yang Z, et al. A Genome-wide association study identifies GRK5 and RASGRP1 as type 2 diabetes loci in Chinese Hans . Diabetes. . 2013;62:291–298. doi: 10.2337/db12-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu HQ, Grant SFA, Bradfield JP, Kim C, Frackelton E, Hakonarson H, Polychronakos C. Association of RASGRP1 with type 1 diabetes is revealed by combined follow-up of two genome-wide studies. J Med Genet. . 2009;46:553–554. doi: 10.1136/jmg.2009.067140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plagnol V, Howson JM, Smyth DJ, Walker N, Hafler JP, Wallace C, et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011, 7: e1002216 . [DOI] [PMC free article] [PubMed]
- 32.Yasuda S, Stevens RL, Terada T, Takeda M, Hashimoto T, Fukae J, Horita T, et al. Defective expression of ras guanyl nucleotide-releasing protein 1 in a subset of patients with systemic lupus erythematosus. J Immunol. . 2007;179:4890–4900. doi: 10.4049/jimmunol.179.7.4890. [DOI] [PubMed] [Google Scholar]
- 33.Golinski ML, Vandhuick T, Derambure C, Fréret M, Lecuyer M, Guillou C, Hiron M, et al. Dysregulation of RasGRP1 in rheumatoid arthritis and modulation of RasGRP3 as a biomarker of TNFα inhibitors. Arthritis Res Ther. . 2015;17:382. doi: 10.1186/s13075-015-0894-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ksionda O, Melton AA, Bache J, Tenhagen M, Bakker J, Harvey R, Winter SS, et al. RasGRP1 overexpression in T-ALL increases basal nucleotide exchange on Ras rendering the Ras/PI3K/Akt pathway responsive to protumorigenic cytokines. Oncogene. . 2016;35:3658–3668. doi: 10.1038/onc.2015.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzer E, Cagdas D, Hons M, Mace EM, Garncarz W, Petronczki ÖY, Platzer R, et al. RASGRP1 deficiency causes immunodeficiency with impaired cytoskeletal dynamics. Nat Immunol. . 2016;17:1352–1360. doi: 10.1038/ni.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan B, Wang S, Wang S, Zeng J, Hong L, Li Z, Yang J, et al. Genome-wide analysis of H3K27me3 in porcine embryonic muscle development. Front Cell Dev Biol. . 2021;9:739321. doi: 10.3389/fcell.2021.739321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Tan B, Xiao L, Zhao X, Zeng J, Hong L, Yang J, et al. Comprehensive analysis of long noncoding RNA modified by m6A methylation in oxidative and glycolytic skeletal muscles. Int J Mol Sci. . 2022;23:4600. doi: 10.3390/ijms23094600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, Xu X, Liu Y, Jin J, Zhu F, Bai W, Guo Y, et al. RIP-Seq of EZH2 identifies TCONS-00036665 as a regulator of myogenesis in pigs. Front Cell Dev Biol. . 2020;8:618617. doi: 10.3389/fcell.2020.618617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiao J, Wang S, Zhou J, Tan B, Li Z, Zheng E, Cai G, et al. ITGB6 inhibits the proliferation of porcine skeletal muscle satellite cells . Cell Biol Int. . 2022;46:96–105. doi: 10.1002/cbin.11702. [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Brind′Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, Sasaki H, et al. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells . Genes Dev. . 2014;28:2041–2055. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. . 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Tang Z, Li Y, Wan P, Li X, Zhao S, Liu B, Fan B, et al. LongSAGE analysis of skeletal muscle at three prenatal stages in Tongcheng and Landrace pigs. Genome Biol. . 2007;8:R115. doi: 10.1186/gb-2007-8-6-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao X, Mo D, Li A, Gong W, Xiao S, Zhang Y, et al. Comparative analyses by sequencing of transcriptomes during skeletal muscle development between pig breeds differing in muscle growth rate and fatness. PLoS One. 2011, 6: e19774 . [DOI] [PMC free article] [PubMed]
- 44.Yue J, Hou X, Liu X, Wang L, Gao H, Zhao F, Shi L, et al. The landscape of chromatin accessibility in skeletal muscle during embryonic development in pigs. J Anim Sci Biotechnol. . 2021;12:56. doi: 10.1186/s40104-021-00577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali A, Murani E, Hadlich F, Liu X, Wimmers K, Ponsuksili S. Prenatal skeletal muscle transcriptome analysis reveals novel microRNA-mRNA networks associated with intrauterine growth restriction in pigs. Cells. . 2021;10:1007. doi: 10.3390/cells10051007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Depeille P, Henricks LM, van de Ven RAH, Lemmens E, Wang CY, Matli M, Werb Z, et al. RasGRP1 opposes proliferative EGFR–SOS1–Ras signals and restricts intestinal epithelial cell growth. Nat Cell Biol. . 2015;17:804–815. doi: 10.1038/ncb3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myers DR, Norlin E, Vercoulen Y, Roose JP. Active tonic mTORC1 signals shape baseline translation in naive T cells. Cell Rep. . 2019;27:1858–1874.e6. doi: 10.1016/j.celrep.2019.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poltorak M, Meinert I, Stone JC, Schraven B, Simeoni L. S os1 regulates sustainedTCR ‐mediatedE rk activation. Eur J Immunol. . 2014;44:1535–1540. doi: 10.1002/eji.201344046. [DOI] [PubMed] [Google Scholar]
- 49.Iwig JS, Vercoulen Y, Das R, Barros T, Limnander A, Che Y, Pelton JG, et al. Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1. ELife. . 2013;2:e00813. doi: 10.7554/eLife.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu H, Huang R, Li P, Zhou L, Fu D, Li Q, et al. Association analysis between RasGRP1 gene polymorphism and body size traits of Suhuai pigs. Swine Industry Science. 2017, 34: 112–114
- 51.Roose JP, Mollenauer M, Ho M, Kurosaki T, Weiss A. Unusual interplay of two types of ras activators, RasGRP and SOS, establishes sensitive and robust ras activation in lymphocytes. Mol Cell Biol. . 2007;27:2732–2745. doi: 10.1128/MCB.01882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu M, Fuller DM, Zhang W. The role of ras guanine nucleotide releasing protein 4 in FcϵRI-mediated signaling, mast cell function, and T cell development. J Biol Chem. . 2012;287:8135–8143. doi: 10.1074/jbc.M111.320580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez-Schindler J, Philp A. Regulation of skeletal muscle mitochondrial function by nuclear receptors: implications for health and disease. Clin Sci. . 2015;129:589–599. doi: 10.1042/CS20150246. [DOI] [PubMed] [Google Scholar]
- 54.Zhang G, Pradhan S. Mammalian epigenetic mechanisms. IUBMB Life. . 2014;66:240–256. doi: 10.1002/iub.1264. [DOI] [PubMed] [Google Scholar]
- 55.Wang S, Sun Y, Ren R, Xie J, Tian X, Zhao S, Li X, et al. H3K27me3 depletion during differentiation promotes myogenic transcription in porcine satellite cells. Genes. . 2019;10:231. doi: 10.3390/genes10030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller I, Min M, Yang C, Tian C, Gookin S, Carter D, Spencer SL. Ki67 is a graded rather than a binary marker of proliferation versus quiescence. Cell Rep. . 2018;24:1105–1112.e5. doi: 10.1016/j.celrep.2018.06.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. . 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.