Abstract
Background
Imaging is vital for assessing rectal cancer, with endoanal ultrasound (EAUS) being highly accurate in large tertiary medical centers. However, EAUS accuracy drops outside such settings, possibly due to varied examiner experience and fewer examinations. This underscores the need for an AI-based system to enhance accuracy in non-specialized centers. This study aimed to develop and validate deep learning (DL) models to differentiate rectal cancer in standard EAUS images.
Methods
A transfer learning approach with fine-tuned DL architectures was employed, utilizing a dataset of 294 images. The performance of DL models was assessed through a tenfold cross-validation.
Results
The DL diagnostics model exhibited a sensitivity and accuracy of 0.78 each. In the identification phase, the automatic diagnostic platform achieved an area under the curve performance of 0.85 for diagnosing rectal cancer.
Conclusions
This research demonstrates the potential of DL models in enhancing rectal cancer detection during EAUS, especially in settings with lower examiner experience. The achieved sensitivity and accuracy suggest the viability of incorporating AI support for improved diagnostic outcomes in non-specialized medical centers.
Keywords: Rectal cancer; Endoanal ultrasound,deep learning; Convolutional neural network
Introduction
Imaging is crucial in evaluating rectal cancer for staging, therapeutic strategy planning, treatment response assessment, and follow-up. The local and distal extent of the disease is typically evaluated with endoanal ultrasound (EAUS) to facilitate the staging of rectal cancer [1]. In an EAUS, the rectal tumor displays as a hypoechoic mass that disrupts the normal echo-layer pattern of the rectal wall [2]. Meta-analysis of EAUS staging data has demonstrated pooled sensitivity estimates of 80.5–96.4% and pooled specificity estimates of 90.6% to 98.3% for cancer detection [3]. Moderate but acceptable accuracy has been demonstrated for lymph node detection by EAUS [3]. However, data on the accuracy of EAUSs outside large tertiary medical centers have demonstrated lower correlation rates between EAUSs and surgical staging [4]. This finding is probably related to the higher levels of experience among examiners and the higher number of examinations performed within large medical centers [5, 6]. Therefore, an operator-supporting system using artificial intelligence to detect rectal cancer may increase the accuracy of EAUSs performed by less experienced professionals.
Advances in computerized image processing support the development of deep learning (DL) models for image classification. DL techniques have shown great promise in medical image analysis. In the last few years, the use of DL in image segmentation, recognition, and registration has accelerated, and DL algorithms have been demonstrated to learn which feature space is most appropriate for the task at hand [7]. Moreover, DL models are particularly useful in diagnosing and treating colorectal cancer [8]. A convolutional neural network (CNN) architecture is a DL model with an artificial neural network that uses images as input. The use of CNN architecture is rapidly expanding in the field of medicine and has been used in the field of rectal cancers for automatic T-staging of rectal cancers in MRI images [9, 10].
A particularly promising direction in using DL models for medical imaging is self-supervised learning (SSL) [11]. SSL is a type of machine learning that trains a model on unlabeled data. One SSL method that is especially relevant to medical imaging is contrastive learning. The goal of contrastive learning is to learn a feature representation space where similar patches of an image are brought closer together while dissimilar samples are pushed farther apart. Recent results show the particular applicability of this method to ultrasound classification tasks [12].
While previous studies on the DL-assisted diagnosis of colorectal cancer have concentrated on MRI and CT imaging [9], the purpose of this study was to develop and validate DL models that can distinguish rectal cancer from standard rectal EAUS images since ultrasound imaging is a significantly more affordable technique than MRI and CT. The acceptable accuracy of EAUSs may significantly improve the outreach of medical services.
Methods
A prospective EAUS image database was reviewed to identify examinations performed for primary stage rectal cancer. We included all examinations performed between February 1, 2021, and December 31, 2021. A total of 40 patients were included in this study, and 294 two-dimensional (2D) images were extracted for analysis (161 abnormal, 133 normal) (Table 1). The study was approved by the Sheba Medical Center ethics committee.
Table 1.
Cohort and disease characteristics
| Male | 24 (60%) |
| Female | 16 (40%) |
| Age (Ys) | |
| Tumor invasiveness (T stage) | |
| T0 | 11 (27.5%) |
| T1 | 13 (32.5%) |
| T2 | 4 (10%) |
| T3 | 10 (25%) |
| T4 | 2 (5%) |
Endoanal ultrasound
All examinations were performed using a single ultrasound machine (BK 300, Peabody, USA) by two experienced examiners ( > 1000 examinations each) using the same technique. Examinees were instructed to perform two cleansing enemas, one at 2 h before the examination and one at 1 h prior. The rectum was inflated by inserting 250–300 cc of water before inserting the rectal us probe. Images were taken using an automatized 360° radial transrectal transducer with a frequency range of 3–20 MHz (3D 20R3) and were stored on the ultrasound machine (Fig. 1). The ultrasound imaging parameters, such as gain, frequency, and gain compensation, were left to the examiner’s discretion.
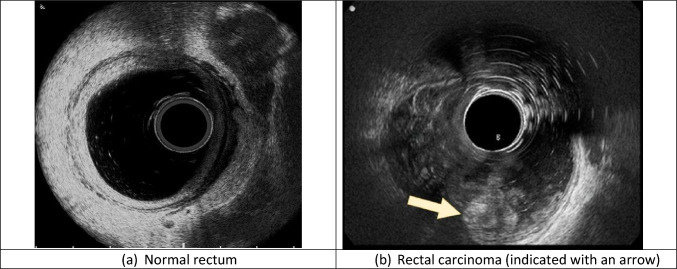
Fig. 1.
Endoanal ultrasound images of a water-filled rectum
General approach to artificial intelligence
This paragraph explains the basics of the scientific computing performed in this study. A detailed explanation will follow in the next paragraphs.
After the extraction of ultrasound images, all images were categorized as normal (no cancer) or abnormal (cancer) by an experienced sonographer. This step was followed by anonymizing the images by removing any data not related to the images. Image quality was then improved automatically by reprocessing the images to delineate the area of interest within the image. Because we had a limited number of images, we augmented the training dataset by randomly resizing, cropping, and rotating the images. To evaluate the performance of a Convolutional Neural Network (CNN) classifier, a tenfold cross-validation technique was utilized. This technique divided all the images into 10 groups, where each group contained an approximately equal number of images. For every iteration, a single group was used as the test set, while the remaining 9 groups were used for training and validation. This process was repeated 10 times. We used different types of CNN architectures and looked for the one with the best sensitivity for the detection of abnormal images.
Image extraction and cropping
For each patient, all of the images of the rectum were extracted, including normal and abnormal images. Images were reviewed by a single experienced ultrasonographer (DC) and categorized into two groups based on the positive or negative visualization of the tumor in the image. All patient data was anonymized by cropping the metadata areas to avoid ethical issues. The image resolution of the final output was 797×657 pixels.
Image preprocessing
Various preprocessing operations were applied to enhance each image for further analysis. First, a mask was created to remove colored marks from the image. The grayscale image was then masked to remove specific pixel values and colored marks, and inpainting was performed on the masked image to fill in missing values. The resulting image was then thresholded (using the value 8 as the threshold) to create a binary image. Next, separated objects were labeled, and the largest bounding box was determined to crop the image.
Further processing involved removing small spots and finding a solid circle, which was masked out from the cropped image. This cropped and processed grayscale image served as the first stage of the preprocessed image. The next step aims to enhance the visual contrast of the images by amplifying the difference between bright and dark areas. To do so, a combination of top hat and bottom hat morphological operations was used. The goal of the top hat operation is to detect bright regions of an image that appear on a darker background, whereas the bottom hat has the opposite goal. Adding the result of the top hat to the original image enhances the bright regions, and subtracting the result of the bottom hat enhances the dark regions. Combining these two operations (therefore making the bright regions brighter and the dark regions darker) increases the contrast in the image. The size of the structuring element used for top hat and bottom hat operations is selected iteratively using the contrast improvement ratio (CIR) measure, which compares the contrast of the original image to the contrast of the final image [13, 14]. The iterative process continues until convergence, meaning that an optimal contrast improvement is achieved. The resulting enhanced grayscale image was then used for classification purposes.
DL models
One main drawback of CNN architecture is the significant number of labeled images required during the learning process. The general approach to overcome this limitation during classification tasks in medical imaging is to combine transfer learning with fine-tuning [15]. Therefore, our proposed method used state-of-the-art (SOTA) models that were pre-trained to provide feature extraction and further fine-tuned with a relatively small ultrasound image dataset.
CNN Models
The CNN network cannot be directly trained due to overfitting caused by the limited size of the available database. To enhance performance, transfer learning from a pre-trained CNN was used, followed by fine-tuning. The DL models evaluated in this study are based on the recent SOTA (State of the art) CNNs architectures: Xception [16], InceptionV3 [17], EfficientNet [18], NasNetLarge [19], InceptionResNetV2 [20] and ConvNeXt [21]. The weights of publicly available models of these CNNs were pre-trained on ImageNet. The images in the database were resized based on the input dimensions of the pre-trained CNN.
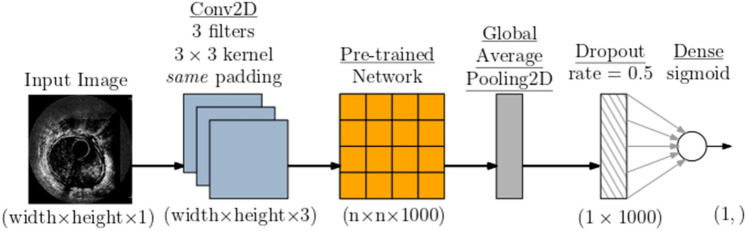
One of the main differences between the common transfer learning procedure and our particular methodology is that CNNs are pre-trained on RGB images, while our images are grayscale. In order to facilitate matching between input grayscale images and pre-trained Convolutional Neural Networks (CNNs), we employed a specialized input layer positioned at the bottom of the pre-trained CNN. This input layer was realized through a Convolutional 2D (Conv2D) layer, consisting of three filters with dimensions of 3 × 3, accompanied by same-size padding [22]. This approach enabled seamless integration of grayscale images with CNN models designed for RGB inputs without the need for any additional modifications to the pre-trained model. The top layer of the pre-trained network was replaced with the global average pooling layer. To reduce overfitting, an aggressive dropout rate of 0.5 was applied. We also tested a lower dropout rate, which resulted in significant overfitting of a training set in the preliminary experiments. Finally, the dropout output was connected to a fully connected layer with a sigmoid activation function. Figure 2 presents the resulting network architecture.
Fig. 2.
CNN ( convolutional neural network) model architecture
In order to facilitate matching between input grayscale images and pre-trained Convolutional Neural Networks (CNNs), we employed a specialized input layer positioned at the bottom of the pre-trained CNN. This input layer was realized through a Convolutional 2D (Conv2D) layer, consisting of three filters with dimensions of 3 × 3, accompanied by same-size padding. Additionally, the Conv2D layer was equipped with a learnable grayscale-to-RGB conversion feature. This approach enabled seamless integration of grayscale images with CNN models designed for RGB inputs, without the need for any additional modifications to the pre-trained model.
The first training stage was transfer learning, in which layers of pre-trained CNN were frozen, and only the added layers were trained. The second stage was fine-tuning with end-to-end training and a significantly decreased learning rate. An early stopping strategy was applied for both stages based on the classification performance on the validation dataset. Adam optimizer was used for training, in which the original images were resampled to the height and width of the pre-trained network. To enhance the learning progress, the training dataset (but not the test dataset) was randomly augmented with small brightness changes, zoom changes, and an arbitrary rotation angle.
A tenfold cross-validation was used to train the CNN and test classification performance. Specifically, in each round of cross-validation, 10% was used for testing, 15% for validation, and 75% for training. To ensure optimal performance, the early stopping strategy was implemented by monitoring the classification accuracy of the validation dataset. If there was no improvement in accuracy for four consecutive epochs, the training process was stopped, and model weights with the best performance were restored.
Beyond the standard CNN models, this study also used an ultrasound semi-supervised contrastive learning model [13]. The evaluation results are based on the publicly available pre-trained ResNet18 model and the supplementary code. The layers of pre-trained CNN were frozen, except the last three layers. Only these layers and the added fully-connected layer were trained. The training dataset was randomly augmented with resize, crop, and rotate operations, and the test dataset was not augmented. The model was evaluated with tenfold cross-validation, with 10% for testing, 15% for validation, and 75% for training. Significantly, the original code [13] used the same images for validation and testing, but this approach is inappropriate for small databases, such as the one discussed in this paper.
Development environment
The development environment used to train and test the models was a PC running the Windows operating system on an Intel Core i7-10,700, 2.90-GHz processor CPU, with 64 GB of RAM and an Nvidia RTX A4000 16 GB GPU. The Python 3.9 programming language is used in combination with extension packages, including Tensorflow 2.9, numpy 1.25, scipy 1.3, and confidence interval.
Results
After the model generated an output for each round of cross-validation, all outputs were accumulated into a probabilistic prediction vector for all the images. Since the outputs appear as a probability, this was converted into a binary decision by comparing it with a classification threshold of 0.5.
Among the evaluated models, the best results were for the EfficientNet model [18] of two different sizes, namely EfficientNetV2M and EfficientNetV2L. The achieved sensitivity for detecting a rectal tumor using the machine learning model was about 0.78, with a specificity of about 0.78 and an accuracy of 0.78. Table 2 displays the corresponding confusion matrices of these models.
Table 2.
Confusion matrices of the EfficientNetV2L and EfficientNetV2M models that compare actual and predicted values
| (a) EfficientNetV2L | |||
| Predicted | |||
| Yes | No | ||
| Actual | Yes | 127 | 34 |
| No | 28 | 105 | |
| (b) EfficientNetV2M | |||
| Predicted | |||
| Yes | No | ||
| Actual | Yes | 125 | 36 |
| No | 29 | 104 | |
The classification performance of all the evaluated models is summarized in Table 3. The confidence intervals were evaluated with the confidence interval package [23]. The contrastive learning model produced slightly lower results than the best CNN model.
Table 3.
Classification performance for different models
| Model | AUC | Accuracy | Sensitivity | Specificity | Precision |
|---|---|---|---|---|---|
| EfficientNetV2M | 0.853 (0.809, 0.897) | 0.779 (0.728, 0.823) | 0.782 (0.704, 0.844) | 0.776 (0.706, 0.834) | 0.743 (0.665, 0.808) |
| EfficientNetV2L3 | 0.848 (0.802, 0.893) | 0.789 (0.739, 0.832) | 0.789 (0.713, 0.85) | 0.789 (0.719, 0.845) | 0.755 (0.678, 0.819) |
| EfficientNetV2S | 0.74 (0.683, 0.797) | 0.69 (0.635, 0.741) | 0.729 (0.648, 0.798) | 0.658 (0.582, 0.727) | 0.638 (0.559, 0.71) |
| Contrastive learning | 0.827 (0.781, 0.874) | 0.735 (0.681, 0.782) | 0.662 (0.578, 0.737) | 0.795 (0.726, 0.85) | 0.727 (0.642, 0.799) |
| InceptionResNetV2 | 0.798 (0.747, 0.849) | 0.724 (0.671, 0.772) | 0.699 (0.617, 0.771) | 0.745 (0.673, 0.806) | 0.694 (0.612, 0.766) |
| Xception | 0.796 (0.744, 0.847) | 0.718 (0.664, 0.766) | 0.662 (0.578, 0.737) | 0.764 (0.693, 0.823) | 0.698 (0.613, 0.772) |
| ConvNeXtLarge | 0.791 (0.739, 0.843) | 0.707 (0.653, 0.757) | 0.797 (0.721, 0.857) | 0.634 (0.557, 0.704) | 0.642 (0.567, 0.712) |
| NASNetLarge | 0.716 (0.656, 0.776) | 0.694 (0.639, 0.744) | 0.639 (0.555, 0.716) | 0.739 (0.666, 0.801) | 0.669 (0.584, 0.745) |
Each value represents the mean and the resulting 95% confidence bounds. The results approximately reflect the Top-1 and/or Top-5 ranking of pre-trained CNNs’ performance on the ImageNet dataset
The receiver operating characteristic (ROC) plot was used for the performance analysis of a model at all classification thresholds. This curve plots the True Positive Rate (TPR) and False Positive Rate (FPR) at different classification thresholds. Figure 3 shows the ROC plot for EfficientNetV2M and EfficientNetV2L models. EfficientNetV2L may have achieved slightly higher accuracy, sensitivity, and specificity, but EfficientNetV2M performs slightly better regarding the area under the curve (AUC).
Fig. 3.
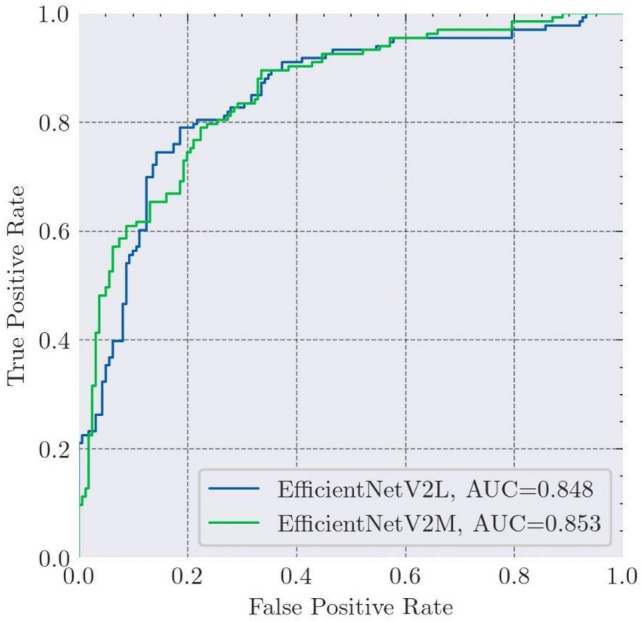
ROC plots of the EfficientNetV2M and EfficientNetV2L models
Discussion
While CNN has been previously used for various applications in the field of gastroenterology [24–28], our data demonstrates a novel use for a CNN deep learning model: the detection of rectal cancer in EAUS images.
DL models are typically used in gastroenterology to detect and classify pathological processes in the gastrointestinal tract. While applying different DL models to our EAUS image dataset, we achieved sustainable accuracy in detecting rectal cancer and differentiating it from normal rectal images. This has potential clinical implications. First, using an automatized transducer to perform an EAUS is relatively easy from a technical standpoint, but interpreting the images is demanding and requires expertise. Therefore, applying an automated system that can diagnose rectal cancer as a first stage of assessment of rectal cancer can potentially enable the use of EAUSs by less experienced medical personnel. A positive result, however, would require further assessment by an experienced ultra-sonographer. Another potential use case for this system is as an assistant to decision-making for examiners with limited or no experience with EAUSs. The fact that sustainable accuracy was achieved despite a relatively small image dataset may indicate that developing an AI system for detecting and staging rectal cancer is an achievable goal.
Our study has some limitations, however. Staging of rectal cancer is the main application of EAUSs, but this study only represents the first step in achieving this goal. The use of a small size population and a relatively restricted image dataset limited the ability to teach the model to differentiate between different stages of tumor invasion. Therefore, our group intends to develop a similar AI system for the staging of rectal cancer on a larger image dataset.
Nevertheless, this study demonstrated the feasibility of detecting rectal cancer in EAUS images using DL. Further studies are needed to validate our results during real-life EAUS examinations and to develop an AI module for rectal cancer staging.
Abbreviations
- EAUS
Endoanal ultrasound
- DL
Deep learning
- CNN
Convolutional neural network
- SSL
Self-supervised learning
- MRI
Magnetic resonance imaging
- CT
Computed tomography
- CIR
Contrast improvement ratio
- SOTA
State-of-the-art
- ROC
Receiver operating characteristic
- TPR
True Positive Rate
- FPR
False Positive Rate
- AUC
Area under the curve
Author contributions
D.C. planned the study, performed the US, collected and interpreted data (including categorizing), and wrote the manuscript; D.B. interpreted data and wrote the manuscript; A.H. and I.M. collected and/or interpreted data; E.R. and Y.Z. drafted the manuscript, O.H. planned and conducted the study, interpreted data and wrote the manuscript. All authors reviewed the manuscript.
Funding
Open access funding provided by Tel Aviv University.
Data availability
The anonymized data used in this study will be made available by contacting the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Conflict of interests
The authors declare no competing interests.
Ethical approval
All methods were performed in accordance with the relevant guidelines and regulations. Due to the retrospective design of the study, informed consent was waived by the ethical committee.
Informed consent
Informed consent was waived by the ethical committee due to the retrospective design of the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ahuja NK, Sauer BG, Wang AY, White GE, Zabolotsky A, Koons A, Leung W, Sarkaria S, Kahaleh M, Waxman I et al (2015) Performance of endoscopic ultrasound in staging rectal adenocarcinoma appropriate for primary surgical resection. Clin Gastroenterol Hepatol 13(2):339–344 [DOI] [PubMed] [Google Scholar]
- 2.Nuernberg D, Saftoiu A, Barreiros AP, Burmester E, Ivan ET, Clevert D-A, Dietrich CF, Gilja OH, Lorentzen T, Maconi G et al (2019) EFSUMB recommendations for gastrointestinal ultrasound part 3: endorectal, endoanal and perineal ultrasound. Ultrasound Int Open 5(01):34–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puli SR, Bechtold ML, Reddy JBK, Choudhary A, Antillon MR, Brugge WR (2009) How good is endoscopic ultrasound in differentiating various t stages of rectal cancer? Meta-analysis and systematic review. Ann Surg Oncol 16:254–265 [DOI] [PubMed] [Google Scholar]
- 4.Marusch F, Ptok H, Sahm M, Schmidt U, Ridwelski K, Gastinger I, Lippert H (2011) Endorectal ultrasound in rectal carcinoma–do the literature results really correspond to the realities of routine clinical care? Endoscopy 43(05):425–431 [DOI] [PubMed] [Google Scholar]
- 5.Morris OJ, Draganic B, Smith S (2011) Does a learning curve exist in endorectal two-dimensional ultrasound accuracy? Tech Coloproctol 15:301–311 [DOI] [PubMed] [Google Scholar]
- 6.Carmody BJ, Otchy DP (2000) Learning curve of transrectal ultrasound. Dis Colon Rectum 43:193–197 [DOI] [PubMed] [Google Scholar]
- 7.Carter D, Albshesh A, Shimon C, Segal B, Yershov A, Kopylov U, Meyers A, Brzezinski RY, Horin SB, Hoffer O (2023) Automatized detection of crohn s disease in intestinal ultrasound using convolutional neural network. Inflamm Bowel Dis 29:014 [DOI] [PubMed] [Google Scholar]
- 8.Yin Z, Yao C, Zhang L, Qi S (2023) Application of artificial intelligence in diagnosis and treatment of colorectal cancer: a novel prospect. Front Med 10:1128084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, Ji Eun Oh, Lee J, Kim MJ, Hur BY, Sohn DK, Lee B (2019) Rectal cancer: toward fully automatic discrimination of t2 and t3 rectal cancers using deep convolutional neural network. Int J Imaging Syst Technol 29(3):247–259 [Google Scholar]
- 10.Qing-Yao Wu, Liu S-L, Sun P, Li Y, Liu G-W, Liu S-S, Ji-Lin Hu, Niu T-Y, Yun Lu (2021) Establishment and clinical application value of an automatic diagnosis platform for rectal cancer t-staging based on a deep neural network. Chin Med J 134(07):821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S-C, Pareek A, Jensen M, Lungren MP, Yeung S, Chaudhari AS (2023) Self-supervised learning for medical image classification: a systematic review and implementation guidelines. NPJ Digit Med 6(1):74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Zhang C, Liu Li, Feng C, Dong C, Luo Y, Wan X (2021) USCL: pretraining deep ultrasound image diagnosis model through video contrastive representation learning. Medical image computing and computer assisted intervention–MICCAI 2021: 24th International conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part VIII 24. Springer, pp 627–637 [Google Scholar]
- 13.Wang YP, Wu Q, Castleman KR, Xiong Z (2003) Chromosome image enhancement using multiscale differential operator. IEEE Trans Med Imaging 22(5):685–693. 10.1109/TMI.2003.812255 [DOI] [PubMed] [Google Scholar]
- 14.Kimori Y (2013) Morphological image processing for quantitative shape analysis of biomedical structures: effective contrast enhancement. J Synchrotron Radiat 20(Pt 6):848–853. 10.1107/S0909049513020761. (Epub 2013 Sep 25. PMID: 24121326; PMCID: PMC3795542) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HE, Cosa-Linan A, Santhanam N, Jannesari M, Maros ME, Ganslandt T (2022) Transfer learning for medical image classification: a literature review. BMC Med Imaging 22(1):1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chollet F (2017) Xception: deep learning with depthwise separable convolutions. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Honolulu, HI, USA, pp 1800–1807 [Google Scholar]
- 17.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition. IEEE Computer Society, Las Vegas, June 27–30, pp 2818–2826
- 18.Tan M, Le Q (2021) Efficientnetv2: smaller models and faster training. International conference on machine learning. PMLR, pp 10096–10106 [Google Scholar]
- 19.Zoph B, Vasudevan V, Shlens J, Le QV (2018) Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. IEEE Computer Society, Salt Lake City, June 18–22, pp 8697–8710
- 20.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-ResNet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence association for the advancement of artificial intelligance. San Francisco, California, February 4–9, 2017
- 21.Liu Z, Mao H, Wu C-Y, Feichtenhofer C, Darrell T, Xie S (2022) A convnet for the 2020s. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. IEEE Computer Society, New Orleans, LA, June 18–22, 2022, pp 11976–11986
- 22.Chris Hughes. Transfer learning on greyscale images: How to fine-tune pretrained models on black-and-white datasets. https://towardsdatascience.com/transfer-learning-on-greyscale-images-how-to-fine-tune-pretrained-models-on-black-and-white-9a5150755c7a, Feb. 2022. Accessed 20 Sep 2023
- 23.Jacob Gildenblat. A Python library for confidence intervals. https://github.com/jacobgil/confidenceinterval, 2023. Accessed 20 Sep 2023
- 24.Soffer S, Klang E, Shimon O, Nachmias N, Eliakim R, Ben-Horin S, Kopylov U, Barash Y (2020) Deep learning for wireless capsule endoscopy: a systematic review and meta-analysis. Gastrointest Endosc 92(4):831–839 [DOI] [PubMed] [Google Scholar]
- 25.Hong Xu, Tang RSY, Lam TYT, Zhao G, Lau JYW, Liu Y, Qi Wu, Rong L, Weiran Xu, Li X et al (2023) Artificial intelligence–assisted colonoscopy for colorectal cancer screening: a multicenter randomized controlled trial. Clin Gastroenterol Hepatol 21(2):337–346 [DOI] [PubMed] [Google Scholar]
- 26.Saqib Mahmood, Mian Muhammad Sadiq Fareed, Gulnaz Ahmed, Farhan Dawood, Shahid Zikria, Ahmad Mostafa, Syeda Fizzah Jilani, Muhammad Asad, and Muhammad Aslam. A robust deep model for classification of peptic ulcer and other digestive tract disorders using endoscopic images. Biomedicines, 10(9):2195, 2022. [DOI] [PMC free article] [PubMed]
- 27.Dumoulin FL, Rodriguez-Monaco FD, Ebigbo A, Steinbrück I (2022) Artificial intelligence in the management of barrett s esophagus and early esophageal adenocarcinoma. Cancers 14(8):1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visaggi P, Barberio B, Gregori D, Azzolina D, Martinato M, Hassan C, Sharma P, Savarino E, de Bortoli N (2022) Systematic review with meta-analysis: artificial intelligence in the diagnosis of oesophageal diseases. Aliment Pharmacol Ther 55(5):528–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The anonymized data used in this study will be made available by contacting the corresponding author upon reasonable request.