Abstract
Conventional tumor models have critical shortcomings in that they lack the complexity of the human stroma. The heterogeneous stroma is a central compartment of the tumor microenvironment (TME) that must be addressed in cancer research and precision medicine. To fully model the human tumor stroma, the deconstruction and reconstruction of tumor tissues have been suggested as new approaches for in vitro tumor modeling. In this review, we summarize the heterogeneity of tumor-associated stromal cells and general deconstruction approaches used to isolate patient-specific stromal cells from tumor tissue; we also address the effect of the deconstruction procedure on the characteristics of primary cells. Finally, perspectives on the future of reconstructed tumor models are discussed, with an emphasis on the essential prerequisites for developing authentic humanized tumor models.
Subject terms: Cancer microenvironment, Cancer models
Revolutionizing cancer research: the power of reconstructed tumor models
Cancer research usually focuses on cancer cells, but the importance of the tumor microenvironment (TME-the environment around a tumor) is becoming more recognized. The TME, especially the tumor stroma, significantly influences cancer cell behavior. The tumor stroma consists of different cell types like fibroblasts, macrophages, and endothelial cells. The interactions between these cells aid tumor growth and development. However, current research models often don’t accurately represent the TME’s complexity and diversity. This study suggests a new method to recreate the TME in a lab, by breaking down patient tumors and rebuilding them using components from those tumors. This could offer a more precise model for cancer research and personalized treatment development. The study also emphasizes the need for standard methods to extract primary cells from tumor tissues, to maintain their original features.
This summary was initially drafted using artificial intelligence, then revised and fact-checked by the author.
Introduction
In cancer research, the primary focus has traditionally centered on neoplastic cells. However, the importance of the tumor microenvironment (TME) is receiving increasing amounts of attention. The concept of the TME is far from new; Stephen Paget’s seminal “seed and soil” hypothesis in the 1880s first posited that specific organs or ‘soils’ offer a more conducive environment for certain cancer cells or ‘seeds’ to flourish1. Cancer research has since validated and expanded upon this theory, illuminating the multifaceted role of the TME—especially the tumor stroma—in modulating the behavior of cancer cells.
The tumor stroma is composed of noncellular compartments, such as the extracellular matrix (ECM), and cellular compartments, including a wide range of invading and resident cells, such as fibroblasts, macrophages, endothelial cells, pericytes, adipocytes, and immune cells (T cells, B cells, natural killer cells (NK cells), and dendritic cells (DCs))2. The dynamic interactions between tumor cells and stromal cells contribute to tumor hallmarks3, for instance, sustained proliferative signaling4–6, evasion of growth suppressors4,7,8, immune evasion4, replicative immortality9, tumor-promoting inflammation10–13, activating invasion and metastasis14,15, enhanced vasculature16,17, genome instability18,19, resistance to cell death20,21, and deregulated cellular metabolism22,23. Given the importance of such tumor-stromal interactions, cancer research has shifted its emphasis to the TME, leading to the exploration of TME-targeted treatment approaches.
With the emergence of TME-focused cancer research and the development of TME-targeting therapies, numerous attempts have been made to develop tumor models that incorporate the stroma (Fig. 1)24–26. Cancer cell lines and patient-derived tumor organoids (PDOs) are widely used in vitro tumor models to understand cancer biology and develop therapeutic targets. Although they are easy to maintain and amenable to high-throughput assays, they are limited by the exclusive cultivation of cancer cells27. To overcome this limitation, the incorporation of stromal cells is being pursued to recapitulate cancer cell-stromal cell interactions in vitro28–30. Although partial recapitulations of the tumor stroma affect the tumor cell phenotype and behavior, these processes involve only a few stromal cell types, and the translation of observations from in vitro to in vivo tumors requires further validation.
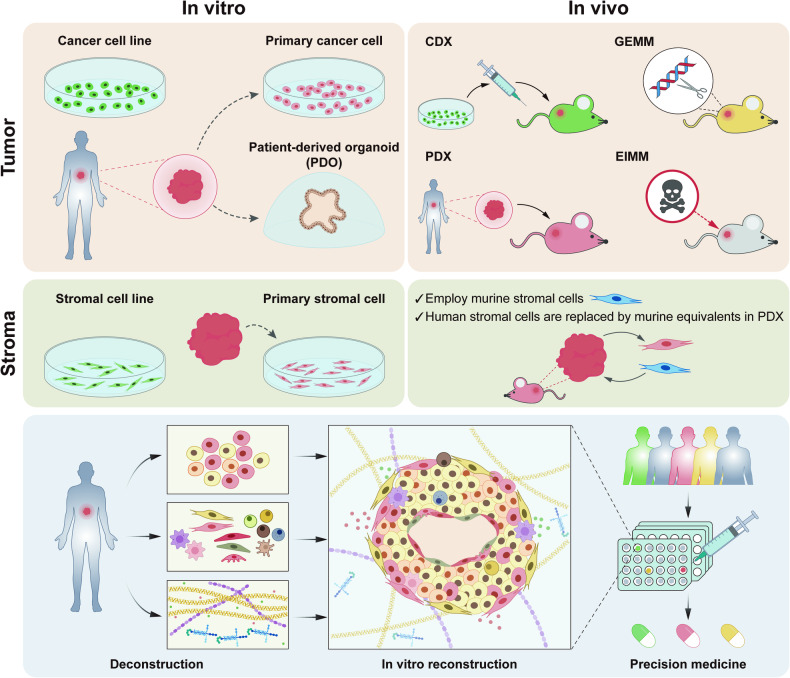
Fig. 1. Sources employed for in vitro and in vivo tumor modeling.
This figure outlines the major sources employed in tumor modeling. For in vitro representations of tumor cells, both established cancer cell lines and patient-derived tumor cells were used. Emerging models, such as patient-derived tumor organoids (PDOs), which are developed from patient-derived stem cells, serve to recapitulate the heterogeneity intrinsic to tumor cells more faithfully. In the context of in vitro stroma reconstruction, only stromal cell lines and primary stromal cells derived from tumor tissue can accurately represent the tumor stroma. For in vivo studies, mouse models are the most commonly utilized systems. These include cell line-derived xenograft (CDX) models that are generated by inoculating tumor cell lines into immunocompromised mice; patient-derived xenograft (PDX) models that are established through the implantation of patient tumor cells or tissue into immunocompromised mice; genetically engineered mouse models (GEMMs) that feature genomes altered to mimic in vivo genetic events; and environmentally induced mouse models (EIMMs) that are developed through the administration of carcinogens to immunocompetent mice. Notably, these models have limitations in their ability to incorporate a human stromal cell compartment, often relying on mouse stromal cells or substituting human stromal cells. Strategies focused on the deconstruction and subsequent reconstruction of the tumor microenvironment may offer more accurate representations of the heterogeneous tumor stroma.
Animal models are alternatives for understanding human tumor biology and evaluating therapeutic responses as they provide an in vivo microenvironment. Mouse models, including genetically engineered mouse models (GEMMs), cell line-derived xenograft (CDX) models, environmentally induced mouse models (EIMMS), and patient-derived xenograft (PDX) models, are commonly used31,32. Among them, the PDX model, which is established by the implantation of patient-derived tumors, is preferred for studying human tumors because it represents the heterogeneity of original patient tumors and allows for the capture of complex tumor-stoma interactions33. However, the major concern with the PDX model is that the model adopts stromal components of mice. Even though human stroma is present after engraftment, it is rapidly lost and eventually replaced by mouse stroma, which alters tumor-stroma interactions34,35. The stroma replacement limits the applicability of mouse models in identifying human-specific mechanisms underlying stoma-driven tumor malignancy and assessing human-specific TME-targeting therapies. For these reasons, there is an urgent need to develop humanized in vitro models or in vivo model systems for studying human cancer.
One possible breakthrough toward developing a tumor model resembling the patient-specific TME involves deconstructing patient tumors and reconstructing them in vitro using components isolated from those tumors36. The stromal cell subpopulations and their proportions are unique to each patient, even within patients with the same type of cancer37,38. Therefore, dissecting patient tumors to identify the characteristics of stromal cell subpopulations and then utilizing them to construct tumor models could be beneficial for understanding intricate mutual interactions and advancing precision medicine.
Primary tumor cells and patient-derived organoids (PDOs) are predominantly used as in vitro models. Patient-derived primary tumor cells (PTCs) are preferred model systems for acquiring more reliable information for translating basic research to clinical application. The general primary cell isolation process can easily be utilized to obtain primary tumor epithelial cells39. In contrast, PDOs require specific cell sources, such as primary adult stem cells (ASCs) or pluripotent stem cells (PSCs), and a culture matrix, especially basement membrane extract40.
In addition to PTCs, stromal cells are isolated from tumor tissues to reconstruct the tumor niche. Although patient-derived primary stromal cells are already being extensively utilized in cancer research, most previous studies neglected to evaluate the subtypes of isolated stromal cells. For example, myofibroblastic cancer-associated fibroblasts (myCAFs), inflammatory CAFs (iCAFs), and antigen-presenting CAFs (apCAFs) are well-recognized CAF subtypes in pancreatic ductal cancer41. Each subtype of cancer has distinct effects42; however, many studies have used primary fibroblasts without considering CAF subtypes43,44. This subtype-neglecting approach could limit the understanding and recapitulation of stromal heterogeneity.
In this review, we provide an overview of the different types of stromal cells that have been identified and the current methods for isolating primary stromal cells from tumor tissues. We also summarize how the current isolation methods affect the primary stromal cell phenotype and key features. Finally, we discuss potential breakthroughs and considerations for overcoming the obstacles of conventional tumor models.
Stromal cell heterogeneity in the tumor microenvironment
Many stromal cells make up the TME and actively engage with tumor cells, thus reshaping the tumor niche into a favorable niche for tumor progression. The stromal cells present in the TME include CAFs, tumor-associated macrophages (TAMs), tumor-associated endothelial cells (TECs), pericytes, adipocytes, and immune cells such as T cells, B cells, NK cells, DCs, and myeloid-derived suppressor cells (MDSCs) (Fig. 2)2. In Table 1, we summarize the three major types of stromal cells and outline the currently recognized subpopulations within each type.
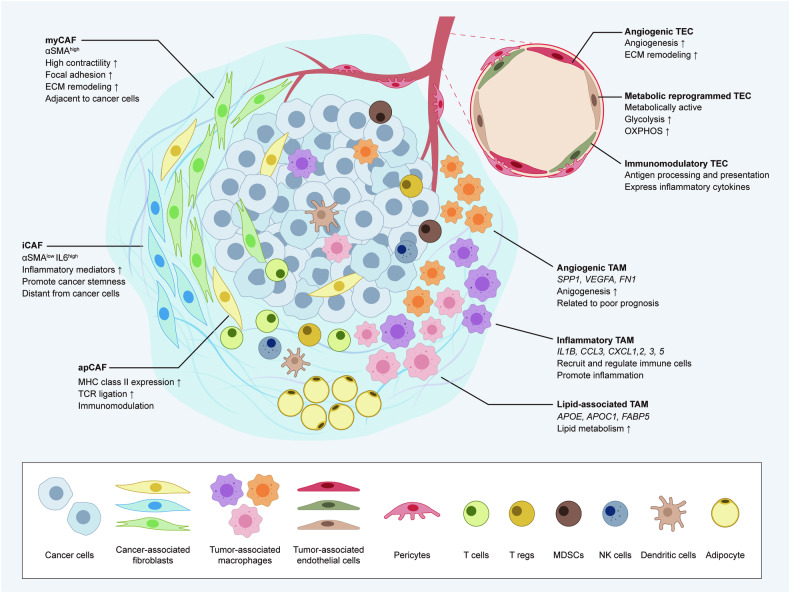
Fig. 2. Heterogenous cell subpopulations in the tumor microenvironment.
Schematic representation of TME heterogeneity. Several subpopulations of CAFs, TAMs, and TECs have been described, and the distinct markers and features of each cell type have been highlighted. The TME comprises various stromal cells in addition to tumor cells. Stromal cell heterogeneity is identified using cutting-edge technologies for single-cell analysis.
Table 1.
Previously identified stromal cell subpopulations in individual studies.
| Stromal cell | Cancer type | Subpopulation | Markers | Feature | Ref |
|---|---|---|---|---|---|
| CAF | Breast cancer | CAF-S1 | CD29Med FAPHi FSP1Low-Hi αSMAHi PDGFRbMed-Hi CAV1Low |
- High expression of CCL11, CXCL12, 13, 14 - Myofibroblastic subset - Enriched in TNBC - Immunosuppressive - Increase T lymphocyte survival and differentiation - Associated with accumulation of FOXP3 + T lymphocytes - Correlated with CD45+ hematopoietic cells and macrophages - Anti-correlated with CD8 + T lymphocytes - Enhance Treg capacity to inhibit effector T cells |
136 |
| CAF-S2 | CD29Low FAPNeg FSP1Neg-Low αSMANeg PDGFRbNeg CAV1Neg | - Enriched in LumA BC | |||
| CAF-S3 | CD29Med FAPNeg FSP1Med-Hi αSMANeg-Low PDGFRbMed CAV1Neg-Low | - Associated with Juxta-tumor | |||
| CAF-S4 | CD29Hi FAPNeg FSP1Low-Med αSMAHi PDGFRbLow-Med CAV1Neg-Low |
- Enriched in TNBC and HER2 BC - Myofibroblastic subset - Associated with CD8 + T lymphocytes - Anti-correlated with FOXP3 + T lymphocyte |
|||
| Breast cancer |
CAF-S1 ECM-myCAF |
GJB2, LRRC15 |
- ECM-myofibroblastic CAF - Enriched in LumA BC - Involved in collagen synthesis and ECM organization |
38 | |
|
CAF-S1 Detox-iCAF |
ADH1B, GPX3 |
- Detoxification-inflammatory CAF - Enriched in TNBC - Involved in detoxification and inflammatory response |
|||
|
CAF-S1 IL-iCAF |
RGMA, SCARA5 |
- Response to stimuli - Enriched in TNBC - Involved in the response to growth factor, TNF signaling, and IL pathway |
|||
|
CAF-S1 TGFβ-myCAF |
CST1, TGFB1 |
- TGFβ-myofibroblastic CAF - Enriched in LumA BC - Involved in response to TGFβ stimulus and matrisome |
|||
|
CAF-S1 Wound-myCAF |
SEMA3C, SFRP4 |
- Wound healing-myofibroblastic CAF - Enriched in LumA BC - Involved in the assembly of collagen fibrils and wound healing - Correspond to apCAF |
|||
|
CAF-S1 IFNγ-iCAF |
CCL19, CCL5 | - IFNγ and cytokines | |||
|
CAF-S1 IFNαβ-iCAF |
IFIT3, IRF7 | - IFNαβ-inflammatory CAF | |||
|
CAF-S1 Acto-myCAF |
GGH, PLP2 | - Actomyosin-myofibroblastic CAF | |||
| CAF-S2 | FAPNeg CD29Low SMANeg | - Abundant in healthy tissue | |||
| CAF-S3 | FAPNeg CD29Med SMANeg | - Abundant in healthy tissue | |||
| CAF-S4 | FAPNeg SMAHi CD29Hi MCAMHi |
- Restricted to cancer and metastatic lymph nodes - Characterized by a perivascular signature - Pro-metastatic function - Contractile |
|||
| Breast cancer | vCAF | Rgs5 |
- Vascular CAF - Upregulated vascular development and angiogenesis genes - Enriched in tumor core - Localized in proximity to vasculature |
51 | |
| mCAF | Pdgfra, Mfap5, Dcn |
- Matrix CAF - Strong ECM signature - Upregulated ECM, matrisome, and EMT associated genes - Low abundance in the tumor core |
|||
| cCAF | Nuf2, Mki67 |
- Cycling CAF - Upregulated cell-cycle-related genes - Proliferative segments of vCAF |
|||
| dCAF | Scrg1, Sox9, Sox10 |
- Developmental CAF - Upregulated development and morphogenesis of tissue-associated genes - Originate from tumor cells that have undergone EMT |
|||
| PDAC | myCAF | ACTA2, TAGLN, MMP11, MYL9, HOPX, POSTN, TPM1, TPM2, ACTA2high |
- Myofibroblastic CAF - Adjacent to cancer cells - Associated with smooth muscle contraction, focal adhesion, ECM organization, and collagen formation |
137 | |
| iCAF | IL6, PDGFRA, CXCL12, CFD, DPT, LMNA, AGTR1, HAS1, CXCL1, CXCL2, CCL2, IL8, ACTA2low Ly6Chigh |
- Inflammatory CAF - High inflammatory mediators: IL6, IL11, LIF - Located in the desmoplastic areas of the tumor - Distant from cancer cells - Associated with the synthesis of hyaluronan and the complement pathway |
|||
| apCAF | H2-Ab1, Cd74, Saa3, Slpi |
- Antigen-presenting CAF - Express MHC class II-related genes - Induce TCR ligation in CD4 + T cells in an antigen-dependent manner |
|||
| Lung cancer | Cluster 1 | COL10A1 |
- Highly enriched in tumor - EMT-related signal - ECM phenotype - HOXB2 and FOXO1 are highly upregulated |
53 | |
| Cluster 2 | COL4A1 |
- The highest expression of ACTA2, a myofibroblast marker - Involved in myogenesis, NOTCH pathway, and angiogenesis - Myogenesis phenotype |
|||
| Cluster 4 | PLA2G2A |
- Similar to Cluster 1 - COL14A1high |
|||
| Cluster 5 | MMP3 |
- Low expression of myogenesis signature - High expression of mTOR signature and glycolysis genes |
|||
| Cluster 6 | FIGF |
- Nonmalignant fibroblast - High expression of elastin - Low expression of some collagens: collagen type I, III, V, and VIII |
|||
| Cluster 7 | CCL2 |
- Present in NSCLC patients - Similar to Cluster 5 but with low expression of glycolysis genes |
|||
| Gastric cancer | myCAF | TPM1, TPM2, MYL9, TAGLN, POSTN |
- Myofibroblastic CAF - Prevalent in intestinal-type GC - Negatively correlated with tumor stemness |
138 | |
| iCAF | IL6, IL11, IL24, CXCL1, CXCL2, CXCL5, CXCL6, MMP1, MMP3, MMP10 |
- Inflammatory CAF - Prevalent in diffuse-type GC - Associated with GC invasion - Promote stemness of tumor cells, high stemness score |
|||
| inCAF | PDGFRA, POSTN, ID1, ID3 |
- Intermediate CAF - Negatively correlated with tumor stemness - inCAF signal is increased with tumor progression from the premalignant state |
|||
| Colorectal cancer | CAF-A | FAP, MMP2, LUM, COL1A2 |
- Involved in ECM remodeling - Intermediate state between NMFs and CAF-B |
139 | |
| CAF-B | ACTA2, TAGLN, PDGFA | - Express cytoskeletal genes known for activated myofibroblast markers | |||
| NMFs | SFRP1/2, MFAP5, DPT, S100A4 | - Normal mucosa fibroblasts | |||
| Pancancer | myoCAF | ACTA2, MYH11 |
- Myofibroblastic CAF - Enriched tumorigenesis and myogenic regulons (TBX2, MEF2C each) |
48 | |
| inflaCAF | MMP11, CTHRC1, FAP, TGFB1 |
- Inflammatory CAF - Associated with dedifferentiation regulon |
|||
| adiCAF | CFD |
- Adipogenic CAF - Associated with EMT regulon |
|||
| EndMT-CAF | RGS5, ACTA2, PLVAP, VWF |
- Endothelial to Mesenchymal Transition CAF - Associated with angiogenesis and endothelial differentiation |
|||
| PN-CAF | MPZ, S100B, LGI4, PLP1 | - Peripheral nerve-like CAF | |||
| apCAF | ACTA2, HLA-DRA, CD74, HLA-DRB1 |
- Antigen-presenting CAF - Enriched in PDAC - Interaction with tumor-infiltrating T-cell clusters |
|||
| TAM | Breast cancer | Stromal Macrophage | CD11b + MRC1+ | - Harbor potent T-cell activation capacity | 140 |
| Hyperplastic Ductal Macrophage | CD11b- |
- Associated with an advanced tumor stage - Potent phagocytes - Not efficient for activating CD8 T cells - Locally accumulated through the active proliferation |
|||
| Malignant ductal TAM | CD11b + SPP1+ |
- Associated with poor prognosis - Regulate immunosuppressive functions of TAMs of monocytic origin |
|||
| Breast cancer | LAM1:FABP5 TAM | SPP1, FABP5 |
- Similar to lipid-associated macrophages (LAM) - High expression of TREM2 and lipid/fatty acid metabolic genes - Reduced proportion in HER2 + BC - Correlation with worse survival |
141 | |
| LAM2:APOE TAM | APOE |
- Similar to lipid-associated macrophages - High expression of TREM2 and lipid/fatty acid metabolic genes |
|||
| CXCL10 TAM | CXCL10, CXCL11 | - M1-like phenotype | |||
| EGR1, SIGLEC1 TAM | IL2RA, CD209 | - M2-like phenotype | |||
| Gastric cancer | HSP + TAM | HSPA6, HSPA1B, HSPB1 | - Increased HSP-associated genes | 142 | |
| THBS1 + TAM | THBS1 | - N/A | |||
| Chemokine-TAM | CCL3, CCL18, CCL20 | - Increased expression of chemokines | |||
| MMP-TAM | MMP9, MMP12 | - MMP genes | |||
| Complement-TAM | C1QA, C1QB, C1QC | - Complement family | |||
| Cell cycle-TAM | TOP2A, STMN1 | - Cell-cycle regulation genes | |||
| Colon cancer | C1QC + TAM | C1Q genes, TREM2, MERTK, CD80 |
- Derived from IL1B+ Tissue-resident macrophage (TRM) - Expression of MAF/MAFB and JUN/FOS - Increased inflammatory signatures, including complement activation, antigen processing, and presentation pathways |
59 | |
| SPP1 + TAM | SPP1, MARCO, VEGFA |
- Derived from NLRP3 + TRM - Expression of the level of HLA-DRs, CEBPB, and ZEB2 - Angiogenic signatures: enriched tumor angiogenesis, ECM receptor interaction, and tumor vasculature pathways |
|||
| Colorectal cancer | C1QC + MRC1- TAM | C1QC | - Closely related to CD14/CD16 monocytes in blood | 60 | |
| SPP1 + TAM | C1QC, MRC1, STAT1, PPARG |
- Tumor-specific filtration - Originated from THBS1 + TAM - Exhibit shorter progression-free survival |
|||
| THBS1 + TAM | THBS1 |
- Promote malignant migration of cancer - Capable of performing antigen processing and presentation and regulating intestinal immune network for IgA production - Can differentiate into SPP1+ macrophages |
|||
| VCAN + TAM | VCAN | - N/A | |||
| Colorectal cancer | Proinflammatory macrophage | IL1B, IL6, S100A8, S100A9 | - Upregulation of genes associated with cytokines | 143 | |
| SPP1+ macrophage A | SPP1, IL6 |
- Enriched in tumor core and border - Proinflammatory phenotype - Association with CMS type 4 |
|||
| SPP1+ macrophage B | SPP1, CD163, SEPP1, APOE, MAF |
- Enriched in tumor core and border - Anti-inflammatory phenotype |
|||
| Proliferating macrophage | MKI67, KIAA0101 | - Upregulation of genes associated with cell cycle | |||
| Hepatocellular carcinoma | TAM1 | FOLR2, CD163, C1QB, SEPP1, CD163high CD206high |
- Fetal-like TAM - FOLR2 expressing TAM - Exhibit immunosuppressive interactions - Higher expression of immunomodulatory chemokines - Enrichment with TIGIT+ cells |
144 | |
| TAM2 | SPP1, TREM2, FABP5, NUPR1, CD163low CD206low | - SPP1 + TAM | |||
| TAM3 | MT1G, MT2A, MT1X, CD163low CD206low | - MT1G-enriched TAM | |||
| Pancancer | C1QC + TAM | C1QC |
- Tumor enriched macrophage - Higher M2 signature and phagocytosis scores |
145 | |
| SPP1 + TAM | SPP1 |
- Tumor enriched macrophage - Higher M2 signature and angiogenesis signature |
|||
| ISG15 + TAM | ISG15 |
- Tumor-enriched macrophage - Upregulated IFN inducible genes - Higher expression of canonical M1 signature |
|||
| FN1 + TAM | FN1 |
- Tumor-enriched macrophage - Mainly present in kidney cancer - Proangiogenic TAM |
|||
| INHBA + TAM | INHBA | - Compensate SPP1 + TAM in stomach cancer with a proangiogenic signature | |||
| VCAN + TAM | VCAN | - Compensate SPP1 + TAM in BC with a proangiogenic signature | |||
| LYVE1 Macrophage | LYVE1 |
- Identified within multiple cancer types - Enriched in noncancer tissue - Tissue-resident interstitial macrophage |
|||
| NLRP3 Macrophage | NLRP3 |
- Enriched in noncancer tissue - Represent proinflammatory TRM (Tissue-resident macrophage) |
|||
| Pancancer | HES1 + TAM | C1QA, C1QB, C1QC, IGF1, CCL3, CCL4 | - Harbor an embryonic signature | 146 | |
| TREM2 + TAM | APOC1, APOE, SPP1, FABP5 |
- Accumulated only in tumor tissue - Involved in metabolic disorders - Potentially immunosuppressive role |
|||
| IL4I1 + TAM |
IL4I1 + PD-L1 + IDO1+ CD38, IDO1, CXCL9, CXCL10, CXCL11 |
- Antigen presentation, interaction with both Th2 and Th1 T cells, T-cell exhaustion, and tryptophan degradation - Suppress T cells and attract Tregs into the tumor by producing chemokine, expressing PD-L1 and PD-L2, and degrading IL4I1/AHR tryptophan - Exhibit immunosuppressive - Promote the entry of Treg into the tumor |
|||
| Proliferating TAM | TOP2A, MKI67, IDO1 | - Accumulated in all cancer types | |||
| TEC | PDAC | Endothelial 1 | IGFBP3, SPP1, CFH, IGLL5, TIMP1 |
- Higher expression of HIF1A - Enriched for ECM organization, regulation of vasculature development, regulation of angiogenesis, cell junction assembly and epithelial cell migration |
147 |
| Endothelial 2 | CLPS, PRSS1, CTRB1, CA4, CELA3A | - Represent normal pancreatic tissue | |||
| Lung cancer | Cluster1 | MT2A | - Normal EC | 53 | |
| Cluster 3 | IGFBP3 |
- Tumor EC - Enrichment of Myc target, nucleotide metabolism, OXPHOS-associated genes - Immune activation-associated genes are downregulated |
|||
| Cluster 4 | SPRY1 |
- Tumor EC - Enrichment of Myc target, nucleotide metabolism, OXPHOS-associated genes - Immune activation-associated genes are downregulated |
|||
| Cluster 5 | EDNRB | - Normal EC | |||
| Cluster 6 | PDPN, PROX1 | - Lymphatic EC | |||
| Gastric cancer | E0 | IGFBP5, STC1, IGFBP3 |
- Influence angiogenic sprouting - Upregulation of mTOR and IGF-1 signaling - Increase the invasion and migration of tumor cells |
148 | |
| E1 | FOXO1, FOXP1, JUN |
- Associated with the regulation of T-cell exhaustion signaling pathway - Suppress immune response |
|||
| E2 | N/A |
- Low activity - Normal endothelial cells |
|||
| E3 | NRP1, FGFR1 |
- VEGF receptor encoding genes are significantly upregulated - White adipose tissue browning pathway and STAT3 pathways are activated - Promote cancer cell invasion and angiogenesis |
|||
| Gastric cancer | Endo1 | COL4A1, COL4A2, PROS1 |
- Predominantly enriched in tumor - Downregulated MHC class II genes - Limited antigen presentation function - Strong activation of TNF, VEGF, PDGF, PGF, and Notch signaling - Involved in angiogenesis |
149 | |
| Endo2, Endo3, Endo4 | N/A | - N/A | |||
| Colorectal cancer | Tip-like EC | RGCC, RAMP3 |
- Overrepresentation of regulators of angiogenesis in tumor - Overrepresentation of antigen processing and presentation in normal |
143 | |
| Stalk-like EC | ACKR1, SELP | - Associated with apoptosis inhibition and proliferation | |||
| Proliferative EC | BIRC5, CKS1B | - Overexpression of BIRC5 and CKS1B | |||
| Lymphatic EC | LYVE, PROX1 | - Found both in normal and tumor | |||
| Hepatocellular carcinoma | PLPP3 + TEC | PLPP3 | - Enriched in tumor tissue | 144 | |
| PLVAP + TEC | PLVAP, HLA-DRA, |
- Enriched in tumor tissue - Facilitate the emergence of fetal-like macrophages - Mainly enriched in fetal and tumor tissues - Major subset expressing the receptor for VEGF |
|||
| IGFBP3 + TEC | IGFBP3 | - Enriched in tumor tissue | |||
| Clear cell renal cell carcinoma | AVR1 TEC | PLVAP, FLT1, KDR, FLT4, EDNRB, VWF, HSPG2 |
- Higher expression of VEGF receptor - Upregulation of genes involved in hemostasis, angiogenesis, and stimulation of endothelial growth and regeneration |
63 | |
| AVR2 TEC | ACKR1, SELP | - Evade angiogenesis inhibitors | |||
| Glioblastoma | Pe1 EC | KLF2, TIMP3, SLC2A1, SLCO1A2, TSC22D1, DEGS2, CAVIN2 |
- Quiescent endothelial cells derived from nonmalignant tissue - Associated with vascular integrity and BBB function |
150 | |
| Co1 EC | COL4A1, COL4A2, HSPG2, INSR, KDR |
- Derived from tumor core - Angiogenic phenotype - Associated with developmental and tumor angiogenesis, vascular basement membrane remodeling, cytoskeletal rearrangements, angiogenic sprouting, and endothelial tip cell formation |
|||
| Co2 EC | TMSB4X, RPLP2, RPL39, GAPDH, VIM, ACTB |
- Derived from tumor core - Intermediate phenotype - Associated with cytoskeletal and ribosomal protein expression |
|||
| Pe2 EC | CCL3, CCL4, CCL4L2, HLA-DRB1, HLA-DRA, HLA-DPA1, HLA-DPB1, HLA-DQB1 |
- Immune-activated phenotype derived from nonmalignant tissue - Expression of inflammatory cytokines and MHC II-mediated antigen presentation genes |
|||
| Co3 EC | NR4A3, IL1B, IL1R1, SELE, SELP, VACM1 |
- Derived from tumor core - Upregulation of immune-activated genes - Associated with inflammation and immune cell recruitment |
|||
| Pancancer | ESM1 tip EC | ESM1, NID2 |
- Only resided in malignant tissue - Upregulation of glycolysis and OXPHOS |
37 | |
| ACKR1high HEV and venous EC | ACKR1, SELP |
- ACKR1high endothelial venules and venous EC - Enriched in tumor |
|||
| CA4 capillary EC | CA4, CD36 | - Characterized by PLVAP and IGFBP7 | |||
| FBLN5 arterial EC | FBLN5, GJA5 | - Upregulated fatty acid biosynthesis | |||
| PROX1 lymphatic EC | PROX1, PDPN | - Increased fatty acid oxidation | |||
| TECs | PLVAP, IGFBP7 |
- Activation of HOXB pathways - Reduced carbonic acid metabolism |
AVR aortic valve replacement, BBB brain‒blood barrier, CMS consensus molecular subtype, EMT epithelial–mesenchymal transition, GC gastric cancer, HER2, BC Her2-positive breast cancer, LumA, BC luminal A breast cancer, NMF normal mucosa fibroblasts, NSCLC non-small cell lung cancer, OXPHOS oxidative phosphorylation, PDAC pancreatic ductal adenocarcinoma, TNBC triple-negative breast cancer, TRM tissue-resident macrophage. ACKR1 atypical chemokine receptor 1 (Duffy blood group), ACTA2 actin alpha 2, smooth muscle, ACTB actin beta, ADH1B alcohol dehydrogenase 1B (class I), beta polypeptide, AGTR1 angiotensin II receptor type 1, AHR aryl hydrocarbon receptor, APOC1 apolipoprotein C1, APOE apolipoprotein E, ASMA actin alpha 1, skeletal muscle, BIRC5 baculoviral IAP repeat containing 5, C1QC complement C1q C chain, CA4 carbonic anhydrase 4, CAV1 caveolin 1, CAVIN2 caveolae associated protein 2, CCL2 C-C motif chemokine ligand 2, CCL4L2 C-C motif chemokine ligand 4 like 2, CD4 CD4 molecule, CEBPB CCAAT enhancer binding protein beta, CELA3A chymotrypsin like elastase 3A, CFD complement Factor D, CFH complement Factor H, CKS1B CDC28 protein kinase regulatory subunit 1B, CLPS colipase, COL10A1 collagen type X alpha 1 chain, COL14A1 collagen type XIV alpha 1 chain, COL4A1 collagen type IV alpha 1 chain, COL4A2 collagen type IV alpha 2 chain, CST1 cystatin SN, CTHRC1 collagen triple helix repeat containing 1, CTRB1 chymotrypsinogen B1, CXCL1 C-X-C motif chemokine ligand 1, Dcn Decorin, DEGS2 delta 4-desaturase, sphingolipid 2, DPT dermatopontin, EDNRB endothelin receptor type B, EGR1 early growth response 1, ESM1 endothelial cell specific molecule 1, FABP5 fatty acid binding protein 5, FAP fibroblast activation protein alpha, FBLN5 fibulin 5, FGFR1 fibroblast growth factor receptor 1, FIGF vascular endothelial growth factor D, FLT1 Fms related receptor tyrosine kinase 1, FN1 fibronectin 1, FOLR2 folate receptor beta, FOS Fos proto-oncogene, AP-1 transcription factor subunit, FOXO1 forkhead Box O1, FOXP1 Forkhead Box the P1, FSP1 S100 calcium binding protein A4, GAPDH glyceraldehyde-3-phosphate dehydrogenase, GGH gamma-glutamyl hydrolase, GJA5 gap junction protein alpha 5, GJB2 gap junction protein beta 2, GPX3 glutathione peroxidase 3, H2AB1 H2A. B variant histone 1, HAS1 hyaluronan synthase 1, HER2 Erb-b2 receptor tyrosine kinase 2, HES1 Hes family bHLH transcription Factor 1, HIF1A hypoxia inducible factor 1 subunit alpha, HLA-DPB1 major histocompatibility complex, class II, DP beta 1, HLA-DQB1 major histocompatibility complex, class II, DQ beta 1, HLA-DR human leukocyte antigen - DR isotype, HLA-DRA major histocompatibility complex, class II, DR alpha, HLA-DRB1 major histocompatibility complex, class II, DR beta 1, HOPX HOP homeobox, HOXB homeobox B, HOXB2 Homeobox B2, HSPG2 Heparan sulfate proteoglycan 2, ID1 Inhibitor of DNA binding 1, IDO1 indoleamine 2,3-dioxygenase 1, IFIT3 interferon induced protein with tetratricopeptide repeats 3, IGF1 insulin like growth factor 1, IGFBP3 insulin like growth factor binding protein 3, IGLL5 immunoglobulin lambda like polypeptide 5, IL1B interleukin 1 beta, IL1R1 interleukin 1 receptor type 1, IL2RA interleukin 2 receptor subunit alpha, IL4I1 interleukin 4 induced 1, INHBA inhibin subunit beta A, INSR insulin receptor, IRF7 interferon regulatory factor 7, ISG15 ISG15 ubiquitin like modifier, JUN Jun proto-oncogene, AP-1 transcription factor subunit, KDR kinase insert domain receptor, KIAA0101 PCNA clamp associated factor, KLF2 KLF transcription factor 2, LGI4 leucine rich repeat LGI family member 4, LIF LIF interleukin 6 family cytokine, LMNA lamin A/C, LRRC15 leucine rich repeat containing 15, Ly6C lymphocyte antigen 6 family member C 1, LYVE1 lymphatic vessel endothelial hyaluronan receptor 1, MAF MAF bZIP transcription factor, MAFB MAF bZIP transcription factor B, MARCO macrophage receptor with collagenous structure, MEF2C myocyte enhancer Factor 2C, MERTK MER proto-oncogene, tyrosine kinase, Mfap5 microfibril associated protein 5, Mki67 antigen identified by monoclonal antibody Ki 67, MMP1 matrix metallopeptidase 1, MPZ myelin protein zero, MRC1 mannose receptor C-type 1, MT1B metallothionein 1B, MT1X metallothionein 1X, MT2A metallothionein 2A, MYH11 myosin heavy chain 11, MYL9 myosin light chain 9, NID2 nidogen 2, NLRP3 NLR family pyrin domain containing 3, NR4A3 nuclear receptor subfamily 4 group A member 3, NRP1 neuropilin 1, Nuf2 NUF2, NDC80 kinetochore complex component, NUPR1 Nuclear protein 1, transcriptional regulator, PDGF platelet-derived growth factor, Pdgfra platelet derived growth factor receptor alpha, PDGFRB platelet derived growth factor receptor beta, PD-L1 CD274 molecule, PDPN podoplanin, PGF placental growth factor, PLA2G2A phospholipase A2 group IIA, PLP1 proteolipid protein 1, PLPP3 phospholipid phosphatase 3, PLVAP plasmalemma vesicle-associated protein, POSTN periostin, PPARG peroxisome proliferator activated receptor gamma, PROS1 protein S, PROX1 prospero homeobox 1, PRSS1 serine protease 1, RAMP3 receptor activity modifying protein 3, RGCC regulator of cell cycle, RGMA repulsive guidance molecule BMP coreceptor a, Rgs5 regulator of G-protein signaling 5, RPLP2 Ribosomal protein lateral stalk subunit the P2, S100A8 S100 calcium binding protein A8, S100B S100 calcium binding protein B, Saa3 serum amyloid A 3, SCARA5 scavenger receptor class A member 5, Scrg1 scrapie responsive gene 1, SELE selectin E, SELP selectin P, SEMA3C semaphorin 3C, SEPP1 selenoprotein P, SFRP4 secreted frizzled related protein 4, SIGLEC1 sialic acid binding Ig like lectin 1, SLC2A1 solute carrier family 2 member 1, SLCO1A2 solute carrier organic anion transporter family member 1A2, Slpi secretory leukocyte peptidase inhibitor, Sox9 SRY (sex determining region Y)-Box 9, SPP1 CXXC finger protein 1, SPRY1 Sprouty RTK signaling antagonist 1, STAT1 signal transducer and activator of transcription 1, STC1 stanniocalcin 1, TAGLN transgelin, TBX2 T-box transcription factor 2, TFF3 Trefoil factor 3, TGFB1 transforming growth factor beta 1, THBS1 thrombospondin 1, TIGIT T-cell immunoreceptor with Ig and ITIM domains, TIMP1 TIMP metallopeptidase inhibitor 1, TMSB4X thymosin beta 4 X-linked, TNF tumor necrosis factor, TOP2A DNA topoisomerase II alpha, TPM1 tropomyosin 1, TREM2 triggering receptor expressed on myeloid cells 2, TSC22D1 TSC22 domain family member 1, VACM1 Cullin 5, VCAN versican, VEGFA vascular endothelial growth Factor A, VIM Vimentin, VWF Von Willebrand factor, ZEB2 Zinc finger E-box binding homeobox 2.
Cancer-associated fibroblasts (CAFs)
CAFs are the most abundant stromal cell type within the TME. For decades, CAFs were considered as a single homogenous population. In addition, definitive markers of CAFs were absent45. This lack of awareness about CAFs led to conflicting outcomes in previous CAF research. For example, S100 calcium binding protein A4 (S100A4) is a known marker of CAFs. Vascular endothelial growth factor A (VEGF-A) and tenascin-C (TNC), which are produced by S100A4-positive CAFs (S100A4+ CAFs), promote tumor metastasis46. However, in another study, S100A4+ CAFs contributed to tumor suppression by depositing collagen around carcinogens and preventing DNA damage to epithelial cells47. These findings further obscure the comprehensive understanding of the distinctive attributes of CAFs. Recent studies have revealed that CAFs exist in different compositions depending on the tumor and patient, and these CAFs have different molecular characteristics38,48. At the pancancer level, various CAF subsets were observed. For instance, myoCAFs, inflaCAFs, and apCAFs were discovered via pancancer analyses, and each share similar transcriptomic patterns of myCAFs, iCAFs, and apCAFs identified in pancreatic ductal adenocarcinoma (PDAC)48. In PDAC, three major types of CAFs were identified. myCAFs constitute the majority of CAFs in tumor samples and express a high level of αSMA. This subtype is characterized by the significant expression of genes related to smooth muscle contraction, focal adhesion, and ECM remodeling, these genes are conventionally regarded as phenotypes of activated fibroblasts. In contrast, iCAFs are characterized by the increased expression of inflammatory factors such as IL6, IL8, and chemokines. apCAFs are a distinct CAF subtype from myCAFs and iCAFs, and they express high levels of MHC class II family-associated genes49.
Furthermore, the spatial distribution of CAF subsets within the TME adds to their heterogeneity50. For example, myCAFs and iCAFs are differentially distributed in the tumor niche of PDAC. myCAFs are located adjacent to tumors, while iCAFs are located at a much farther distance from tumors25. Bartoschek et al. also confirmed the distinct location of CAF subsets in breast cancer. Vascular CAFs (vCAFs) exhibit angiogenic signatures and are localized near the vasculature and tumor core. Unlike vCAFs, matrix CAFs (mCAFs) with strong ECM signatures are found at low levels in the tumor core51. Therefore, to develop in vitro tumor models with high-fidelity, the spatial distribution of stromal cells should be considered because spatial intercellular communication between tumor cells and stromal cells occurs and impacts the prognosis and response to antitumor therapies.
Tumor-associated macrophages (TAMs)
TAMs are generally regarded as contributors to tumor progression due to their involvement in ECM remodeling, angiogenesis, and immunosuppression52. These functions foster the development of a microenvironment conducive to tumor growth. However, recent research highlights the fact that TAMs also have antitumor activities that restrain tumor progression and improve patient prognosis53,54. The inherent plasticity of macrophages allows them to sense and adapt to cues from the microenvironment, which leads to altered macrophage phenotypes related to tumor-promoting or tumor-restraining functions55.
Macrophages have been classified into two polarized states: M1 and M2. M1 macrophages are characterized by a classically activated phenotype triggered by interferon-gamma (IFN-γ) or lipopolysaccharide (LPS). M1 macrophages exhibit proinflammatory properties. In contrast, M2 macrophages are in an alternatively activated state. M2 macrophages are induced by certain cytokines, such as IL-4, IL-10, and IL-13, and exhibit anti-inflammatory phenotypes56.
However, this conventional classification system oversimplifies the diverse nature of macrophages57,58. The diverse states of TAMs are being continuously identified (Table 1). For example, Zhang et al. classified TAMs in colon cancer into two main subsets: complement C1q C chain-positive TAMs (C1QC + TAMs) and secreted phosphoprotein 1-positive TAMs (SPP1 + TAMs). SPP1 + TAMs were characterized by increased expression of SPP1, macrophage receptor with collagenous structure (MARCO), and vascular endothelial growth factor A (VEGFA) and were associated with tumor angiogenesis. Signatures associated with colorectal adenoma and metastatic liver cancer pathways were also increased in this subset, which implies that the SPP1+ subset may possess protumorigenic and prometastatic functions in colon cancer59. In a separate study on colorectal cancer (CRC), SPP1 + TAMs were also recognized as a tumor-promoting subset. Among the four subpopulations of TAMs with unique markers, SPP1 + TAMs are a specific subset that infiltrates tumors, and CRC patients with a greater infiltration of this subset have a poor prognosis60. Tu et al. discovered a unique macrophage subset in osteosarcoma. C1Q + TAMs were identified in patients with high levels of immune infiltration. Patients with high levels of immune infiltration had an improved prognosis compared to those with low levels of immune infiltration. Although C1Q + TAMs are a tumor-restraining subset, they do not specifically express M1 or M2 signature genes54. Likewise, recent research has highlighted the existence of protumor and antitumor TAM subsets beyond the conventional M1 and M2 classifications. The distribution and characteristics of TAM subsets vary according to their origins and specific tumor microenvironments58. For a more precise representation of heterogeneous TAMs in vitro, a comprehensive understanding of TAM subsets and their biology is crucial. Such insights enable the construction of a more authentic TME, thereby facilitating studies on the subset-specific polarization of TAMs.
Tumor endothelial cells (TECs)
Multiple studies have recently revealed the presence of TEC heterogeneity at the single-cell level61,62. Heterogeneous subsets of TECs have been identified across various organs, and each subset may have a distinct molecular phenotype and function. For example, renal cell TECs are classified into two distinct subsets: the AVR-1 and AVR-2 groups63. The AVR-1 subset is positive for plasmalemma vesicle-associated protein (PLVAP), von Willebrand factor (VWF), heparan sulfate proteoglycan 2 (HSPG2), and endothelin receptor type B (EDNRB), while the AVR-2 subset is characterized by atypical chemokine receptor 1 (ACKR1) and selectin P (SELP). The two subsets differ not only in molecular expression but also in clinical benefit. PLVAP + ACR-1 was predominantly abundant in tumor tissues and negatively correlated with patient survival. However, ACKR + ACR-2 cells exhibited decreased expression of VEGF receptors, such as kinase insert domain receptor (KDR) and fms-related receptor tyrosine kinase 1 (FLT1), indicating that this subset may contribute to antiangiogenic therapy (AAT) evasion63. As demonstrated in previous studies, TECs are key stromal cells in the TME, and they can predict therapeutic response. Although many researchers have struggled to develop vascularized tumor models and mimic the near-native features of solid tumors in vitro, most studies have only focused on structural reconstruction of the microvasculature64,65. By incorporating affiliated TEC subsets and reconstructing microvessels in tumor models, it would be possible to generate high-fidelity tumor models.
General process of primary cell isolation and expansion
Unlike peripheral blood cells, which can be readily isolated by centrifugation or magnetic beads66,67, tissue-embedded cells require more complex procedures to isolate the desired cell types. The general procedure for isolating and cultivating primary cells from solid tissue involves five steps: tissue acquisition, dissection, tissue disaggregation, incubation/cell expansion, and cell separation or purification (Fig. 3). Here, we summarize the purpose of each step and the methods currently employed in the process (Table 2).
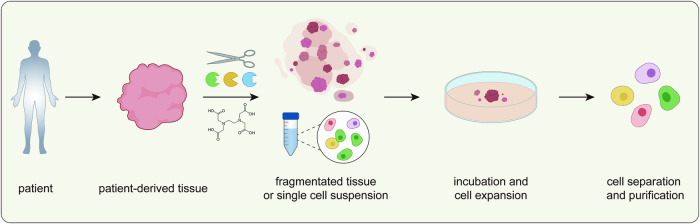
Fig. 3. General isolation process for primary cells.
A schematic illustration of the primary cell isolation process. Mechanical, enzymatic, and chemical methods can be utilized to dissociate tumor tissue acquired from patients. The tissue is divided into small pieces or single-cell suspensions, followed by incubation and expansion of the primary cells. Finally, the target cells are purified from the cell mixtures and utilized for downstream processes.
Table 2.
Isolation methods for various stromal cells in individual studies.
| Cell/ Cancer type | Dissociation | Incubation | Purification/Identification | Ref | ||
|---|---|---|---|---|---|---|
| Mechanical | Chemical/Enzymatic | Media | Substrate | |||
| Cancer-associated fibroblast (CAF) | ||||||
| Head and neck cancer | Not used | 10 mg/ml Collagenase P (2 hr, 37 °C) |
Fibroblast growth medium, 20% FCS, 1 ng/ml BFGF, 5 µg/ml Insulin, 1% P/S, 1% Gentamycin |
Tissue culture flask |
- VIM + αSMA + CD45- CD68- MelanA- HMB45- KRT- CD34- - Identified by flow cytometry and immunocytochemistry |
93 |
| Melanoma |
- Mincing - Scalpel - 1–4 mm2 |
0.25% Trypsin + 0.02% EDTA (20 min, 37 °C) |
High glucose DMEM, 10% FBS, 100U/ml penicillin, 100 µg/ml streptomycin, 100 µg/ml gentamycin |
6 well plate |
- Fibroblast: VIM - Myofibroblast: αSMA - Immunofluorescence - Minimize keratinocyte contamination by earlier detachment of fibroblasts after trypsin treatment |
78 |
| Breast cancer | - Dissection | Not used |
DMEM, 20% FBS, 1% Nonessential amino acid |
Cell culture plate |
- Spidle shaped morphology - Fibroblasts outnumbered tumor cells, and epithelial cells disappeared after passage 3 |
151 |
| Gastric cancer |
- Mincing - Scissors, scalpel - 2–4 mm |
Enzyme H, R, A of tumor dissociation kit (from Miltenyi Biotec) | RPMI1640, 10% FBS | Cell culture dish |
- CD90 - Morphology and FACS |
152 |
| Colorectal cancer |
- Mincing - 1 mm |
1 mg/ml Collagenase type I (1 h, 37 °C) | EGM2-MV | Tissue culture flask | - Proteomic and secretome analysis | 153 |
| Colorectal cancer |
- Mincing - 1 mm3 |
1.5 mg/ml Collagenase IV + 20 µg/ml Hyaluronidase (1 h, 37 °C) | DMEM, 10% FBS | Cell culture dish |
- 40 µm cell strainer - Red blood cell lysis buffer treatment - Wash away nonadherent cells after 3 h incubation - αSMA, VIM, and FAP - Immunofluorescence |
154 |
| Pancreatic cancer |
- Mincing - Scissors, scalpel |
Not used | DMEM, 10% FBS, 1X Glutamax, 1X P/S | 12 well plate |
- αSMA, VIM - myCAF: αSMA - iCAF: IL6 - apCAF: MHCII - Immunofluorescence |
155 |
| Liver cancer | - Cross sectioning | Enzyme H, R, A of tumor dissociation kit (from Miltenyi Biotec) | DMEM/F12 (1:1), 10% FBS | Cell culture dish |
- FAP + αSMA + EpCAM- ECad- PECAM1- - Spindle-shaped morphology and FACS |
156 |
| Ovarian cancer |
- Mincing - Scissors, scalpel - 3–5 mm2 |
- 0.25% Trypsin + 0.1% EDTA (30 min, 37 °C) - 0.5 mg/ml Hyaluronidase + 3 mg/ml Collagenase type 3 (6 h, 37 °C) |
RPMI1640, 20% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin | Cell culture dish |
- αSMA, VIM, FAP, PDGFRα, Desmin, DDR2, S100A4 - Mitotic figure - Immunofluorescence |
157 |
| Tumor-associated macrophage (TAM) | ||||||
| Breast cancer |
- Chopping - razor blade |
14 U/ml LiberaseTL + 28U/ml LiberaseDL + 15 mg/ml DNase I (1–18 h, 37 °C) | No cultivation | No cultivation |
- 100 µm cell strainer - CD45 + CD11b + CD14 + CD163 + CD3- CD19- CD56- - FACS |
158 |
| Breast cancer |
- Mincing - Scalpel - 2–4 mm |
0.1% Collagenase type I + 0.2% Dispase type I + 1% DNase I (30 min, 37 °C) | DMEM, 10% FBS, 100 units/ml penicillin, 10 µg/ml streptomycin | 96 well plate |
- 70 µm cell strainer - Red blood cell lysis buffer treatment - CD11b + F4/80+ |
159 |
| Breast cancer | - Mincing | LiberaseDH + DNase I (2 h, 37 °C) | DMEM, 5% FBS, 100 units/ml penicillin, 100 µg/ml streptomycin | 24 well plate | - CD11b + F4/80+ | 160 |
| Breast cancer | Not used | Collagenase IV + Hyaluronidase + Dispase II + DNase IV (15 min, 37 °C) | DMEM | Cell culture plate | - Isolate CD11b + Ly6G- cells | 161 |
| Breast cancer |
- Mincing - razor blade |
0.29 U/ml LiberaseDL + 0.56 U/ml LiberaseTL + 150 µg/ml DNase I (45 min, 37 °C) | No cultivation | No cultivation |
- 100 µm cell strainer - Red blood cell lysis buffer treatment - DAPI- CD45 + CD11b + CD14 + CD163 + CX3CR1 + HLA-DR + CD3- CD19- CD56- - FACS |
79 |
| Mammary carcinoma |
- Cutting, mincing - razor blade |
1X Collagenase + 1X Hyaluronidase + 10U/ul DNase I (45 min, 37 °C) |
RPMI, 20% FBS, 2 mM L-glutamine, 2 mM Sodium pyruvate, 1X Penicillin‒streptomycin, 55 µM 2-mercaptoethanol, 10 ng/ml mouse M-CSF |
10 µg/ml Fibronectin-coated glass bottom plate |
- Isolate CD11b+ cells by MACS - Determine purity of F4/80 + CD45+ cells by FACS |
91 |
| Colorectal cancer | - Slicing | Accumax (2 h, 37°C) | DMEM, 10% FBS | 6 well plate | - CD68 | 162 |
| Ovarian cancer | Not mentioned | Not mentioned | RPMI 1640, 10% FBS, 100 U/ml penicillin, 100 U/ml streptomycin | Cell culture plate |
- Isolate CD14+ cells by MACS - Identified with CD206high HLA-DRlow by FACS |
163 |
| Mouse solid tumor |
- Mincing - Scissors, scalpel - 1–1.5 mm |
10 U/ml Collagenase I + 400 U/ml Collagenase IV + 30 U/ml DNase I (25 min, 37 °C) |
RPMI 1640, 10% FCS, 300 µg/ml L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 0.02 mM beta-mercaptoethanol, 1 mM sodium pyruvate, 1 mM nonessential amino acids |
No cultivation |
- 70 µm filter - Erythrocyte lysis buffer treatment - CD11b + Ly6G- SiglecF- Ly6Clow - MACS, FACS |
95 |
| Tumor endothelial cell (TEC) | ||||||
| Mammary cancer |
- Mincing - Scissors - 5 mm |
2 mg/ml Collagenase + 1 mg/ml Dispase + 1 mg/ml DNase (75 min, 37 °C) | Low glucose DMEM, 10% FBS, 10% Nu-Serum IV, 1% antibiotic-antimycotic, hFGF, VEGF, hEGF, R3-IGF-1, heparin | 0.5% Gelatin-coated cell culture dish |
- Scrape off nonspecific cells surrounding the identified EC colonies - Isolate CD31+ cells by MACS - Identified with CD31+ by immunofluorescence and FACS |
94 |
| Melanoma |
- Cutting - Scalpel - 1 mm3 |
10 mg/ml Collagenase type II + 25 µg/ml DNase I (30 min, 37 °C) | Endothelial cell growth medium MV2 | Collagenase type I- coated cell culture dish |
- Density gradient centrifugation - Isolate CD31+ cells by MACS - Determine the purity and sort cells with CD31 + CD146+ by FACS |
164 |
| Melanoma |
- Cutting - Scissors - 5 mm3 |
Collagenase type II + Collagenase type IV + DNase I (30 min, 37 °C) | No cultivation | No cultivation |
- 70 µm cell strainer - Isolate CD45- EpCAM- CD31+ cells by MACS |
165 |
| Melanoma | - Mincing | Collagenase II | EGM-2MV, 15% FBS | 1.5% gelatin-coated cell culture dish |
- Remove blood cells by single sucrose step-gradient centrifugation - Isolate CD31+ cells by MACS - Treatment of 500 ng/ml Diphtheria toxin to eliminate remaining tumor cells - Further purification with FITC-BS1-B4-lectin by FACS |
166 |
| Lung carcinoma |
- Mincing - Scalpel - 0.1 cm3 |
0.5 mg/ml Collagenase type I, 60 min, RT | Endothelial cell medium | Cell culture dish and Matrigel bed | - Isolate CD105+ cells by MACS | 167 |
| Renal carcinoma |
- Mincing - Scissors |
Collagenase type II (1 h, 37 °C) |
MCDB131 medium, 10 ng/ml EGF, 1 µg/ml Hydrocortisone, bovine brain extract, 20% FCS |
1% Gelatin or Endothelial cell attachment factor |
- CD105+ cells by MACS - Identified with CD105, CD31, and VWF by immunofluorescence and FACS |
168 |
| Cancer-associated adipocyte (CAA) | ||||||
| Breast cancer | Not used | 250 U/ml Collagenase type I (30 min, 37 °C) |
2D: DMEM, 10% FCS, 1% P/S 3D: DMEM, 25 mM glucose, 10% FBS, 1% P/S |
2D: 6 well plate 3D: 6 or 4.2 mg/ml fibrin matrix |
- 200 µm cell strainer - Wash with Krebs-Ringer Bicarbonate buffer for purification - Identified with Bodipy 493/503 by immunofluorescence |
169 |
| Breast cancer | - Mincing | 1 mg/ml Collagenase I (1 h, 37 °C) | DMEM/F12, 10% FBS | Cell culture plate |
- 100 µm cell sieve - Centrifugation |
170 |
| Omentum cancer |
- Mincing - Scissors, scalpel - mm size |
0.2% Collagenase I (1 h, 37 °C) |
DMEM/F12 (1:1), 1% Penicillin, 1% Streptomycin, 0.1% BSA |
Cell culture flask |
- Filter using nylon mesh - Identified with Bodipy 493/503 by immunofluorescence |
171 |
| Tumor pericyte | ||||||
| Hemangioma |
- Mincing, homogenize - Scalpel, pestle - 2 mm3 |
50 µg/ml LiberaseTM + 5 U/ml Dispase (40–50 min, 37 °C) | EGM2-medium, 10% hiFBS, 1% GPS, EGM-2 Single Quot supplements except hydrocortisone | 1 µg/cm2 FN-coated cell culture dish |
- 100 µm cell strainer - Isolate GLUT1- CD31- PDGFRβ+ cells by MACS |
172 |
| Colon carcinoma | - Mincing | Collagenase IA + Collagenase II + Collagenase IV + DNAse I (15–20 min, 37 °C) | No cultivation | No cultivation |
- 70 µm cell strainer - Isolate CD45- CD31- NG2 + CD140b+ cells by FACS |
173 |
| CRC/Lung cancer/Glioblastoma |
- Cutting - Scissors |
1.5 mg/ml Collagenase I + 1.5 mg/ml Collagenase II (40-60 min, 37 °C) | No cultivation | No cultivation | - Isolate NG2+ cells by MACS | 174 |
bFGF basic fibroblast growth factor, CD3 CD3 molecule, CX3CR1 C-X3-C motif chemokine receptor 1, DAPI 4′ 6-diamidino-2-phenylindole, DDR2 discoidin domain receptor tyrosine kinase 2, DMEM Dulbecco’s modified eagle medium, ECad E cadherin (CDH1), EDTA ethylenediaminetetraacetic acid, EGF epidermal growth factor, EGM2-MV endothelial cell growth medium MV2, EpCAM epithelial cell adhesion molecule, FACS fluorescence-activated cell sorting, FAP fibroblast activation protein alpha, FBS fetal bovine serum, FCS fetal calf serum, GPS guinea pig serum, hEGF human epidermal growth factor, hFGF human fibroblast growth factor, hiFBS heat inactivated fetal bovine serum, HMB45 premelanosome protein (PMEL), IL6 interleukin 6, KRT keratin, Ly6G lymphocyte antigen 6 family member G, M-CSF macrophage colony stimulating factor, MHCII major histocompatibility complex, class II (HLA-DR), NG2 neural/glial antigen 2, P/S penicilin/streptomycin, PDGFRα platelet derived growth factor receptor alpha, PECAM1 platelet and endothelial cell adhesion molecule 1 (CD31), R3-IGF-1 long Arg3 insulin like growth factor 1, RPMI1640 Roswell park memorial institute, S100A4 S100 calcium binding protein A4, VEGF vascular endothelial growth factor, VIM vimentin, VWF von willebrand factor, αSMA alpha smooth muscle actin.
Tissue acquisition
Tumor tissue samples from donors, including animal models and tumor patients, are collected through biopsy or surgical resection. Once the tissue specimen has been obtained from the donor, the sample is immediately immersed in a tissue preservation solution (transport medium), such as Hank’s balanced salt solution (HBSS), to avoid tissue necrosis or the initiation of apoptosis. The specimen is transported and stored at low temperature to preserve tissue integrity for further procedures.
Tissue dissection
After tissue acquisition, the specimen is dissected to remove unwanted components, such as necrotic or nontumor regions, and minimize contaminants. Tissue dissection ensures the exclusion of extraneous tissues by isolating specific regions of interest and reducing contamination from nontarget cell types. This step is particularly important when working with tumor samples to ensure that the isolated primary cells are derived from tumor tissue68.
Tissue disaggregation and dissociation
Dissociation is required to isolate individual cells. This procedure aims to isolate cells from the surrounding ECM or neighboring cells in preparation for subsequent cell expansion or analysis. Three main approaches are used for tissue dissociation: mechanical dissociation, enzymatic digestion, and chemical dissociation69. The three methods can be used independently or in combination during the dissociation process.
Mechanical dissociation
Mechanical dissociation is a simple approach for physically disrupting tumor tissues. This procedure entails tissue fragmentation with scissors, scalpels, and homogenizers routinely employed to disintegrate tissue samples. There are two major approaches: (1) tissue explant and culture of cells released from the fragmented tissue and (2) cell expansion using mechanical dissociation alone70. The tissue explant method involves simply culturing small tissue pieces in culture dishes and collecting cells that have migrated out of the pieces. This method decreases tissue size and reduces the risk of cell loss in later stages, such as the filtration of floating cells or enzymatic digestion71. The tissue explant method could provide additional benefits by preserving communication between outgrowing cells and tissue fragments and supplying cytokines or growth factors derived from tissue fragments72. However, cell outgrowth from explant tissues requires a relatively longer time to harvest cells, and usually, this method selectively isolates cell types with enhanced migratory functions72.
Another method using mechanical dissociation alone involves collecting the floating cells released from the loosened tissue fragments after the mechanical disruption of tissues73,74. Mechanical dissociation is generally followed by filtration or additional purification to remove remaining tissue fragments and separate the cell suspension for subsequent cell expansion or analysis. Although this approach can eliminate cells during filtering and purification steps, it is advantageous because this quick and simple method can be used for obtaining single viable cells75,76. Therefore, mechanical dissociation is advantageous because it is a straightforward method that does not introduce additional biological or chemical factors that could impact cell viability or phenotype. However, the outcomes of mechanical dissociation might be inconsistent and can be influenced by the skills of the individuals conducting the procedure77.
Enzymatic digestion
Enzymatic digestion is a widely employed method in which enzymes disintegrate tissues into individual cells. Cells embedded within a tissue adhere not only to ECM proteins but also to neighboring cells. Enzymatic approaches can be effective at disrupting cell adhesion and liberating cells from tissue70. The selection of enzymes for enzymatic digestion should be based on careful consideration of the isolated cell tissue type, target cell type, and downstream applications. Enzymatic digestion is usually combined with mechanical disaggregation to reduce the tissue size and increase the surface area accessible for enzyme action78,79. The use of this tandem strategy reduces the reaction time and increases cell yields80.
ECM-degrading enzymes have been frequently used to remove ECM macromolecules and encourage cell detachment. Collagenase is the most widely employed enzyme that breaks down native collagen, a prevalent ECM protein in connective tissues81. Dispase is also a suitable enzyme for degrading ECM proteins, especially collagen type IV and fibronectin. As a neutral protease, dispase is relatively less cytotoxic than other antimicrobial agents and prevents cell clumping without causing damage to cell membranes82,83. In addition, hyaluronidase is often used to degrade hyaluronic acid (HA), which is a ECM protein abundant in cancer tissues84. A variety of ECM-degrading enzymes, including elastase and Liberase, are available. While ECM-degrading enzymes primarily degrade ECM components, trypsin generally disrupts cell junctions and detaches cells completely from the tissue. Trypsin can effectively separate cell clumps into individual single cells, but it can also induce damage to proteins on cell membranes85. Deoxyribonuclease I (DNase I) often combines with other enzymes during enzymatic digestion. When cells dissociate from tissue, the free DNA released from dead cells can become entangled, impair proteolysis, and induce cell reaggregation. By combining DNase I with other enzymes, avoiding unwanted cell clumping caused by free DNA is possible83.
In addition to well-known enzymes, a commercial tumor dissociation kit from Miltenyi Biotec utilizes enzymatic digestion methods86. Although the dissociation kit guarantees that essential cell surface epitopes can be preserved, it does not explicitly disclose the specific enzyme components included. Thus, anticipating the potential biological effects of these enzymes on the primary cells of interest and their consequences is challenging.
Due to the efficacy of enzymatic digestion in yielding greater quantities of cells and diverse cell populations from tissues, it has been employed for decades as a method for isolating primary cells. Although enzymes with distinct specificities are advantageous for obtaining a single-cell suspension, they have detrimental effects on critical cell surface proteins and cell viability, depending on the circumstances. Therefore, identifying the optimal conditions for enzymatic digestion for the sample is crucial.
Chemical dissociation
In addition to proteolytic enzymes or glycosidases, chemical agents are also used to weaken cell-cell and cell-ECM interactions while isolating primary cells. Various cations, including calcium, potassium, and sodium, play essential roles in maintaining cellular integrity and cell adhesion. Chemical agents such as ethylene diamine tetraacetic acid (EDTA) and ethylene glycol tetraacetic acid (EGTA) are frequently utilized to perturb ion-dependent adhesion between cells and the ECM by chelating cations39,87. These agents are typically combined with trypsin to break intercellular bonds. Loosening these connections allows primary cells to be released and isolated from tissues88.
Incubation and cell expansion
Culture platform
After the tissue specimen is disaggregated, fragmented pieces or single cells isolated from the tissue are incubated for primary cell outgrowth and expansion. In many previous studies, primary cells have been isolated and cultured for expansion using tissue culture plates that provide a hydrophilic, attachable surface for anchorage-dependent cells89,90. In some studies, plastic dishes coated with ECM proteins such as collagen, laminin, and basement membrane proteins were often used to promote cell adhesion, growth, and differentiation91. However, conventional monolayer cell culture conditions cannot provide an in vivo mimetic microenvironment because of the absence of the natural structure of tumors and the presence of ECM proteins that constitute the TME92.
Culture media
Proper cell culture media supplemented with essential nutrients and growth factors is required to support the in vitro proliferation and maintenance of primary cells. Serum is one of the key components that provides growth factors, hormones, lipids, and minerals for cell culture. The most common serum used in cell culture includes fetal bovine serum (FBS), also known as fetal calf serum (FCS), and bovine calf serum (BCS). While various cytokines and growth factors are supplied by neighboring cells in the TME, these factors should be supplemented in culture media for proper cell growth and maintenance. For example, basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF) are used to culture fibroblasts and endothelial cells, respectively93,94.
Chemical compounds are introduced into the culture media. In the case of macrophage culture, the antioxidant 2-mercaptoethanol, also known as β-mercaptoethanol, is supplemented to reduce oxidative stress by removing free radicals91,95. In addition, antibiotics are frequently used during tissue acquisition, processing, and culture to prevent contamination as primary cells are directly isolated from human or animal tissues, and there are frequent sources of microbial contamination, such as commensal flora and subclinical infections85,96. The antibiotics frequently used in primary cell isolation and culture include penicillin, streptomycin, amphotericin B, gentamicin, and kanamycin. These supplementary components and the selection of appropriate basal media are crucial. Because the composition of each basal medium varies in terms of inorganic salts, amino acids, vitamins, glucose/carbohydrates, buffering agents, and other components, selecting suitable media formulations based on the specific cell type is important.
Purification and separation of specific cell types
To isolate a specific cell type, adequate purification and separation steps are essential. Cell type purification can be performed before and after cell incubation and expansion. Cell type-specific characteristics such as cell shape, size, and surface proteins should be defined to separate target cell types from heterogeneous cell mixtures. The most common cell purification and separation techniques for cancer include magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS)97,98.
MACS is an immunomagnetic cell separation method that relies on the surface proteins of cells99,100. Magnetic beads are conjugated with antibodies, lectins, or enzymes. They are used to tag target cells that express surface protein markers. The magnetic bead–cell complexes are transferred to a column, and target cells labeled with magnetic beads can be captured and separated by applying a magnetic field to the column. Target cells are further recovered, and the pure population of interest can be harvested. FACS is another approach in which cell type-specific markers are used101,102. Unlike in MACS, target cells are labeled with fluorophore-conjugated antibodies. When the labeled cells are passed through a laser beam, they are sorted according to fluorescence intensity and scattered light. MACS and FACS separate cells based on cell type-specific protein markers and can be utilized for both the positive and negative selection of target cells.
Additional approaches involve using the physical features of cells. Density gradient centrifugation is a technique for separating cells based on the physical properties of cells. The most common method is Percoll gradient centrifugation87,103. Percoll is a density gradient medium that contains particles with specific physical characteristics. By using this medium, a density gradient can be produced. While the Percoll gradient remains stable, cells of differing densities settle into the specific bands corresponding to their density upon centrifugation. While Percoll centrifugation improves cell viability, this purification method can induce massive cell loss87.
Cell culture media is another method for purifying and obtaining specific cell types. Selection media are specialized media that support the selective growth of the desired cell population while minimizing the growth of unwanted cells or contaminants104,105. The composition of the selection media is optimized by adjusting the concentration of various components to achieve the desired selectivity, selective media for various cell types are also commercially available. As several strategies exist to separate specific cell populations, determining the optimal conditions for obtaining highly viable and pure target cells is essential. After collecting the cells of interest, characterizing and authenticating the isolated primary cells are necessary to determine their origin, purity, viability, and other characteristics.
Challenges and potential breakthroughs in primary cell isolation
Phenotypic alterations induced by the isolation and culture of primary cells
Primary cells have long been utilized for in vitro research, especially in oncology, because they maintain patient-specific characteristics and can be further utilized for clinical research and precision medicine106. As we discussed in the previous section, there are numerous combinations of methods available for isolating and expanding target cells in vitro. Consequently, the isolation process for the same cell type differs among individual studies, and there are no consensus protocols for cell type-specific isolation. However, some previous studies have reported that the phenotype of primary cells is affected by the isolation process and in vitro culture conditions. Nichols et al. examined two different approaches to compare the impact of the isolation method on primary tendon cells: tissue explant culture and enzymatic digestion107. The two isolation processes substantially affected cell morphology, proliferation, and marker expression. Tendon cells isolated by the explant method exhibited a more activated and myofibroblast-like phenotype, while those isolated by enzymatic digestion exhibited an altered phenotype not observed in vivo. These results indicate that different isolation methods critically affect cell behavior and phenotype.
In addition to the isolation process, in vitro culture itself changes the cell phenotype. For example, human primary fibroblasts isolated by the general method, which includes enzymatic digestion, cell expansion on a substrate-coated plastic plate, and further purification by MACS, were utilized to assess the effect of established culture conditions108. This research indicated that merely culturing primary fibroblasts under common conditions could induce the differentiation of fibroblasts into myofibroblast-like phenotypes, regardless of the media composition or whether serum was included. Given that the sequential steps, from primary cell isolation to cultivation, impact the population and characteristics of isolated cells, carefully designing all the procedures to achieve the desired isolation of specific cell types while preserving their inherent characteristics is crucial. In the following section, we investigate the influence of each factor on the isolation and culture of primary cells, with a primary focus on enzymes, culture media, and culture platforms (Fig. 4, Table 3).
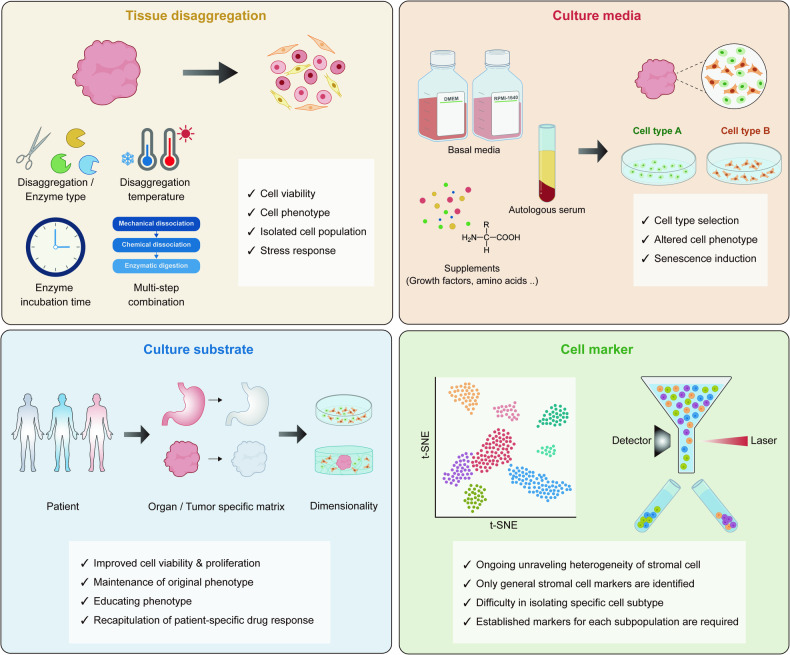
Fig. 4. Possible impacts of each isolation step on primary cells.
Summarized illustration showing how primary isolation procedures affect primary cells. Tissue disaggregation methods, cell culture media, cell culture substrates, and cell type selection markers should be carefully selected to minimize the deleterious effects of deconstruction.
Table 3.
Impact of each isolation step on primary cells.
| Process | Target | Condition | Findings | Ref |
|---|---|---|---|---|
| Disaggregation | Fibroblasts (lung tissue) |
Enzyme type and incubation time 1) Enzyme type: Liberase DL/TL/TM/Collagenase P 2) Incubation time: 15 min/60 min |
• Shorter incubation time lower protease-strength enzyme (Liberase) → Insufficient to isolate stromal cells → High yield of CD45+ immune cells • Longer incubation time with Collagenase P → A great diversity of cell types isolated → Low yield of CD45+ immune cells → Higher yield of CD45- EpCAM- CD31- CD90+ cells (fibroblasts) |
111 |
| Disaggregation | Single-cell suspension (breast and ovarian cancer) |
Enzyme type and incubation temperature 1) B. licheniformis serine protease, 6 °C 2) Collagenase + Hyaluronidase, 37 °C |
• Stress response induced by dissociation with Collagenase/Hyaluronidase at 37°C • Stress response-associated genes include FOS, FOSB, ATF3, and HSP • Upregulation of MHC-class I related genes by Collagenase/Hyaluronidase-based enzymatic digestion |
112 |
| Disaggregation | Microglial cell and astrocyte (brain tissue, glioblastoma) |
Disaggregation type 1) Mechanical disaggregation (MD): Dounce homogenizer, 4 °C 2) Enzymatic disaggregation (ED): Collagenase + DNase, 37 °C |
• Alterations in gene and protein expression induced by ED • Upregulation of immediate early genes by ED • ED lowers the yield of microglia detected by classical markers (CD45low/CD11b + ) • Cells isolated by ED were smaller in size |
110 |
| Disaggregation | Tumor lymphocytes (cutaneous T-cell lymphoma) |
Combination of MD and ED 1) Medimachine (automated mechanical disaggregation) 2) Collagenase 1 A + Medimachine |
• Decreased cell viability in the sample processed with Medimachine alone • Higher cell yields with collagenase pretreatment • Infiltrating T lymphocytes isolated with the collagenase pretreatment |
109 |
| Disaggregation | Single-cell suspension (gut mucosa) |
Multistep dissociation 1) One-step: Collagenase (37 °C) 2) Two-step: EDTA (37 °C) - Collagenase (37°C) 3) Three-step: EDTA (4 °C) - Protease (4 °C) - Collagenase(37 °C) |
• Balanced recovery of multiple cell types was achieved by one-step dissociation • Multistep protocols support the isolation of specific types (e.g., Three step protocols for epithelial cells) |
175 |
| Culture media | Tumor epithelial cells (PDAC) |
Cell line media and Organoid media 1) Minimal media 2) Cell line media (RP10) 3) Organoid media (OWRNA) |
• Cell state shift induced by culture media • PDAC cell line and organoid in the reciprocal media condition → Cell line lost scBasal features while Organoid gained scBasal expression |
176 |
| Culture media | Mammary epithelial cells |
Media type 1) MCDB 170: serum-free media 2) WIT-P: serum-free media 3) M87A: low stress media |
• Media type is important for the maintenance of lineage heterogeneity and cell growth • Rapid senescence and loss of heterogeneity induced by MCDB 170 • Impressive expansion of luminal cells in passage 2 and 3 caused by WIT-P • Maintenance of multiple lineages and robust growth supported by M87A |
115 |
| Culture media | Endothelial cells |
Media type 1) Vascular cell basal Media (ATCC) 2) VascuLife Basal Media (LifeLine Celltech) 3) EC Basal Media (Lonza) 4) EC Media MV (PromoCell) 5) Human microvasc. EC basal Media (Cell applications) 6) Endopan MV microvasc. EC Basal Media (PAN-Biotech) 7) MCDB 131 Media (Gibco) |
• Find optimal commercial media for successful primary cell isolation • Using MCDB131 medium failed to isolate pure and propagating endothelial cells • Isolation success and behavior of primary endothelial cells depend on the culture medium and the composition or nature of supplements |
114 |
| Culture media | Macrophages |
Essential supplements 1) RPMI 1640 2) DMEM 3) MEM 4) McCoys 5a 5) IMDM |
• Consider essential components for primary cell maintenance and function in vitro • Macrophages cultured in DMEM that lack nonessential amino acid (NEAA) → Smaller in size, less densely packed → Upregulated expression of TNFα, mLILRB1, and sLILRB1 → Impaired function of macrophage • NEAA supplementation in DMEM restores DMEM-induced changes in macrophages |
116 |
| Culture media | HNSCC and CRC cancer tissue slices | Use patient-derived autologous serum (AS) |
• Using AS induces a balanced induction of signal transduction pathways • Enhanced preservation of phenotypic and molecular features of patient tumors • Increased probability of predicting patient-specific chemotherapy outcomes |
118 |
| Culture platform | HNSCC and CRC cancer tissue slices |
Patient-specific matrix 1) Patient tumor-derived matrix protein (TMP) cocktail coating 2) Gelatin coating 3) Collagen coating 4) Matrigel coating |
• Tumor explants cultured on patient-specific TMP coating show improved maintenance of tissue morphology, proliferation, and cell viability • Noncoating: lost tumor architecture, decreased viability and proliferation, decreased activation of oncogenic pathways • Gelatin coating: similar to noncoated well • Collagen coating: only supports tumor proliferation • Matrigel coating: only improved cell viability |
118 |
| Culture platform | Mesenchymal stromal cells |
The dimensionality of culture substrate 1) 2D explant culture 2) 3D explant culture using PLMatrix |
• The 3D explant method increased cell yields • 3D PLMatrix provides matrix stiffness close to soft tissue (~0.1 kPa) • Cells isolated by 3D explant have the potential to differentiate into three lineages (adipogenic, chondrogenic, and osteogenic lineage) and maintain cell type-specific markers |
122 |
| Culture platform | Skeletal muscle, skin, and liver cells |
Tissue-specific ECM 1) Muscle ECM coating 2) Skin ECM coating 3) Liver ECM coating 4) Collagen coating 5) Noncoating |
• Tissue-specific ECM supports improved cell proliferation and the maintenance of cell type-specific phenotype | 177 |
| Culture platform | Endothelial colony-forming cells (Colon tumor) |
Tumor-specific ECM 1) Fibrin gel 2) Normal ECM gel 3) Tumor ECM gel |
• Tumor-specific ECM supports in vitro cells to mimic tumor-associated phenotypes • Tumor ECM promotes the formation of tumor-like vasculature • Tumor ECM includes additional components that are not present in normal ECM (e.g Fibronectin, Periostin, Versican, Thrombospondin-2, and Tenascin) |
178 |
| Culture platform | Monocyte/Macrophage (Ovarian cancer) |
Cell phenotype educated by ECM 1) Decellularized tissue slide of low disease tissue (LD) 2) Decellularized tissue slide of high disease tissue (HD) 3) Tissue culture plate |
• Tumor ECM alters the macrophage transcriptome • In vitro, ECM educated-Tumor associated macrophages (TAMs) show gene expression profiles of TAMs found in human tissue |
179 |
ATF3 activating transcription factor 3, ECM extracellular matrix, EpCAM epithelial cell adhesion molecule, FOS Fos proto-oncogene, AP-1 transcription factor subunit, FOSB FosB proto-oncogene, AP-1 transcription factor subunit, HSP heat shock protein, mLILRB1 leukocyte immunoglobulin-like receptor B1, PDAC pancreatic ductal adenocarcinoma, scBasal single-cell Basal, sLILRB1 leukocyte immunoglobulin-like receptor B1, TNFα tumor necrosis factor-alpha.
Enzyme effects
Enzymatic digestion is an inevitable step in detaching cells from tissue and making cell suspension. The types of enzymes used, along with conditions such as temperature and duration of treatment, significantly impact primary cells. The following changes in primary cells induced by enzymatic disaggregation have been identified: changes in cell yield, cell viability, isolated cell population, phenotype, and gene expression patterns.
As previously noted, enzymatic degradation of ECM proteins and cellular adhesions is critical for optimizing cell yield. For tumor lymphocyte isolation, the efficacy of mechanical disaggregation via Medimachine, an automated mechanical disaggregation instrument, was evaluated in conjunction with collagenase application109. Collagenase pretreatment followed by mechanical disaggregation improved cell viability and cell recovery. In addition, atypical lymphocyte populations that were highly infiltrative were also recovered by the combination method. Although enzyme treatment improved cell viability in these experiment, some conflicting studies have shown that enzymatic digestion induces cell death in specific cell populations, such as neurons and astrocytes, in brain tissue110. These inconsistent results imply that enzymatic digestion may enhance the isolation of viable cells with high yields of specific cell types. However, enzymes can also inflict cellular damage and have variable separation efficiency across different cells.
Depending on the cell type, isolating particular cells from tissues may be difficult; therefore, the isolation process must vary based on the target cells. For example, Waise et al. examined disaggregation enzymes and incubation times to explore the optimal conditions for isolating fibroblasts from primary human tissues111. Treatment with enzymes with lower proteolytic activity, such as Liberase, for shorter incubation time was insufficient for isolating stromal cells; however, this treatment also yielded a greater proportion of CD45+ immune cells. A longer incubation time with the collagenase P enzyme cocktail improved the diversity of the cell types while decreasing the proportion of CD45+ immune cells. In addition, the latter approach yielded a significantly greater proportion of fibroblasts. This study demonstrated that the types of enzymes employed and the duration of enzyme treatment should vary depending on the desired target cell type.
The optimal temperature for determining enzyme activity varies depending on the type of enzyme. The enzyme type and treatment temperature are coupled during enzymatic digestion. For instance, while collagenase and hyaluronidase are active at significantly higher temperatures, a serine protease from B. licheniformis exhibited activity at 6 °C. A notable difference in transcription was observed between the two enzyme-based dissociation methods112. The expression of stress response-associated genes, such as Fos proto-oncogene (FOS), FosB proto-oncogene (FOSB), activating transcription Factor 3 (ATF3), and heat shock protein (HSP), as well as major histocompatibility complex (MHC) class I-associated genes, was upregulated by the combination of collagenase and hyaluronidase at 37 °C. Like in previous findings, enzymatic digestion at higher temperatures led to the upregulated expression of specific genes, especially immediate early genes (IEGs)110. As FOS and FOSB are also categorized as IEGs, we can deduce that enzymatic digestion at elevated temperatures induces a shared transcriptional bias for certain genes. These altered gene expression patterns may indicate phenotypic changes in primary cells that could influence subsequent downstream experiments and analyses. Therefore, being aware that enzyme treatment can alter the expression of particular genes is crucial. Subsequently, during further analysis or experiments, assessing whether any biases were introduced by the enzymes is imperative.
Media effects
Owing to the intricate composition of cell culture media, determining the individual influence of each component on primary cells is a formidable challenge. Despite this complexity, pursuing optimal media formulations is crucial for the effective isolation and expansion of primary cells to promote cellular proliferation while maintaining intrinsic phenotypic attributes. Numerous studies have shown the impact of media formulations on primary cell growth, viability, morphology, gene expression patterns, and overall functionality113–117.
There are many typical culture media, such as Rosewell Park Memorial Institute (RPMI)-1640 medium and Dulbecco’s modified essential medium (DMEM), and they vary in terms of the composition and concentration of amino acids, vitamins, inorganic salts, and glucose. Leopold et al. studied seven types of commercially available primary endothelial cell isolation media114. Complex media containing endothelial cell growth supplements (ECGS) facilitated cell proliferation and viability, while defined media, which included specific growth factors such as EGF, FGF2, and VEGF, enhanced cell outgrowth and in vitro angiogenesis. In addition to the defined medium MCDB-131, other types of media enabled the isolation of endothelial cells without affecting the phenotype. Recognizing that using suboptimal culture media may result in the failure of primary cell isolation is important. Therefore, identifying an appropriate media formulation specific to the type of target cell is paramount.
Another crucial aspect is identifying the essential elements required for the expansion of target cells. For example, the cultivation of macrophages in DMEM, which lacks nonessential amino acids (NEAAs), altered the phenotype of macrophages116. The cell size was much smaller, and the expression of specific genes encoding tumor necrosis factor alpha (TNFα) and leukocyte immunoglobulin-like receptor subfamily B member 1 (mLILRB1 and sLILRB1) was upregulated without NEAAs. In addition to the alterations in morphological and transcriptional phenotypes, functional changes in macrophages were also observed. The release of soluble proteins, which is one of the major functions of immune cells, was impaired by using DMEM, and this effect was restored when NEAA was supplemented with DMEM.
Accurate in vitro emulation of patient-specific tumor-stroma interactions is contingent upon preserving the phenotypic and functional attributes of primary cells. If such characteristics are altered during the isolation and expansion phases, the resulting tumor model does not represent the biological complexities inherent in the patient’s original tumor. In this context, several methodologies have been proposed to enhance the preservation of tumor-specific features during in vitro cultivation. For example, supplementation of patient-derived autologous sera (AS) in culture media could be an alternative to the generally used animal-derived serum and supplementary factors118. Since AS reflects the enrichment level of growth factors that varies between individuals, it induced a balanced induction of signaling pathways, and the phenotypic and molecular features of the patient’s tumor were better preserved. The AS supplement also improved the prediction of patient-specific outcomes for chemotherapy. Patient-derived samples such as serum samples are generally difficult to obtain and limited in quantity; however, they are appealing and indispensable materials for reconstructing real tumors de novo.
Culture platform
Many researchers have attempted to improve in vitro cell culture techniques, particularly those mimicking the intricate in vivo microenvironment. The ECM is a known pivotal determinant of cellular behavior and phenotype119–121. Nevertheless, most studies focusing on the isolation and utilization of primary cells have predominantly employed two-dimensional (2D) monolayer cultures.
The environmental cues used in monolayer culture using plastic dishes differ substantially from those cues in in vivo tumor niches. Unlike in in vivo conditions where stromal cells are embedded within the ECM and actively engage with its proteins, monolayer cultures offer only a planar surface devoid of supportive ECM components. This discordance may lead to alterations in many aspects of primary cells. Two main approaches have been recently demonstrated to improve primary cell isolation and expansion: tissue-specific ECM and 3D explant-based cell isolation.
Utilizing tumor-specific ECM for primary cell expansion would facilitate the maintenance of the tumor-associated phenotypes of stromal cells. For example, Majumder et al. utilized four coating materials: gelatin, collagen, Matrigel, and patient tumor-derived matrix protein (TMP) cocktail118. While other coating materials supported only tumor proliferation or cell viability, TMP supported tumor proliferation, viability, and the maintenance of tumor morphology. By engineering a personalized ecosystem with TMP, the conservation of tumor characteristics was enabled during ex vivo tissue culture. Although TMP was used for tissue cultivation in this study, it could also be a promising material for isolating and expanding patient-specific stromal cells while preserving their original phenotype.
In addition to the specific ECM proteins, the dimensionality of the culture platform is also important for mimicking an in vivo-like ecosystem. Recent advances have included the implementation of 3D explant-based approaches for primary cell isolation. For instance, Egger et al. utilized a 3D human platelet lysate matrix to isolate primary mesenchymal stem/stromal cells (MSCs) by embedding adipose tissue122. MSCs isolated in this manner exhibit specific surface markers and can differentiate into adipogenic, chondrogenic, and osteogenic lineages. The 3D explant technique offers a more physiologically relevant environment for primary cell isolation. In contrast to those in conventional 2D methodologies, cells that migrate from tissue explants embedded in a 3D explant model integrate into a matrix that not only is enriched in ECM proteins but also mimics the mechanical properties of soft tissue, resulting in more effective retention of their original phenotypes.
Purification markers
Numerous studies have isolated primary cells from mixed cell populations by leveraging cell type-specific markers. Cell separation techniques such as FACS and MACS rely on the expression of marker proteins to segregate target cells. However, the recent recognition of stromal cell heterogeneity — facilitated by advancements in single-cell technologies — reveals that previous studies may have inadvertently selected markers without accounting for cellular subtypes, thereby leading to biased isolation of subpopulations. For example, Huang et al. purified CAFs from tumor tissues using FAP alone as a CAF marker123. However, the pancancer single-cell analysis revealed that CAFs can be classified into six main subpopulations: myoCAFs, inflaCAFs, adiCAFs, EndMT-CAFs, PN-CAFs, and apCAFs48. Among those subsets, FAP can only identify inflaCAFs. Thus, the findings from previous studies utilizing FAP+ CAFs might only be applicable to inflaCAFs.
In addition, different studies have often defined identical cell types using divergent molecular markers. This inconsistency complicates our understanding of cell behaviors and functions, as it conflates the characteristics of stromal cells identified through different markers as traits of a single, homogenized cell type. Consequently, further research is imperative for elucidating stromal cell heterogeneity. Future investigations should prioritize the utilization of a broad array of subtype-specific markers for the isolation and functional characterization of stromal cells.
Perspectives
We have reviewed stromal cell heterogeneity and strategies for isolating primary cells from patient-derived tissues, including their potential effects on primary cells. Although the tumor stroma plays a key role in the TME by dictating the behavior of tumor cells, most previous studies focused only on developing more advanced tumor cell-based models and neglected the importance of the tumor stroma. Current tumor models only partially incorporate human stromal cells or employ heterologous stromal counterparts for tumor modeling. Furthermore, even though stromal cells are known to be composed of heterogeneous populations, many studies use stromal cells without defining which subtypes they are. Although integrating all stromal cell types and subtypes into in vitro tumor models is challenging, certain cells are critical because they are found in all tissues and play a key role in shaping the microenvironment. These cells include fibroblasts and macrophages124. For instance, ASPN+ fibroblasts and SPP1+ macrophages are notably prevalent near tumors in various cancer types125. Their intense interactions with adjacent tumor cells contribute to immune exclusion within tumors by creating a desmoplastic niche, a factor that is closely associated with poor prognosis. These examples highlight the fact that certain subtypes are strongly linked to patient prognosis and treatment response.
Patient-derived explants (PDEs) are a viable alternative that mirror, to a certain extent, the stromal constitution of the original tumor microenvironment. This ex vivo culture of patient tumor specimens helps preserve the inherent stromal components and microarchitectural features, thereby maintaining critical tumor-stroma interactions. This highlights the utility in evaluating therapeutic responses and determining pharmacodynamic biomarkers. However, the temporal preservation of tissue integrity within PDEs is ephemeral, and the absence of their expandability limits their use in low-throughput applications and diminishes reproducibility126. Thus, humanized tumor models that accurately display the heterogeneity of tumor cells and stromal cells are urgently needed to better represent patient tumors for the development of precision medicine.
Advancements in tumor modeling now allow for detailed replication of both cancer cells and their complex surroundings, thanks to improved methods for breaking down and reassembling tumors. These methods include advanced sequencing and metabolomics for analyzing tumor cells and sophisticated techniques for studying the noncellular environment, such as ECM.
In vitro tumor reconstruction harnesses engineering methodologies to emulate the in vivo TME. Scaffolds, particularly those derived from decellularized ECM, offer a physiologically congruent 3D architecture for cell culture that supports cell adherence and facilitates cell-ECM interactions127. These scaffolds are particularly conducive to organoid culture and promote in vivo-like cellular dynamics. Additionally, to recapitulate complex cell-cell interactions within tumors, coculture systems incorporating stromal and cancer cells are utilized, underscoring the necessity of maintaining stromal cell integrity throughout in vitro processes128,129. The spatial arrangement of cellular constituents is addressed through innovative technologies such as 3D bioprinting and tumor-on-a-chip technology, with the goal of accurately reconstructing the intricate in vivo tumor architecture130,131.
After reconstruction, the fidelity of in vitro tumor models compared to their in vivo counterparts was ascertained through comprehensive multiomics analyses and immunohistochemical imaging, through which the in vitro constructs were compared with the original tumor samples. Such rigorous validation and ongoing refinement of deconstruction and reconstruction techniques are pivotal in generating robust, humanized in vitro tumor models, and will promote in vitro studies to clinical translation for precision medicine and drug discovery.
To develop a reliable humanized tumor model, we could consider a reconstructed tumor model as a potential platform that can accurately replicate heterogeneous human stroma. However, achieving such an ideal in vitro reconstruction poses challenges because the prerequisites need to be addressed. First, the specific functions and molecular markers of stromal cell subpopulations should be clearly defined across various cancer types. Many studies have obtained information about the diverse subpopulations of stromal cells through single-cell analysis. However, within a given cancer type, there is no consensus regarding the types and characteristics of these subpopulations. This information is imperative during tumor destruction, as it enables the isolation of desired stromal cell subtypes, comprehension of stromal cell attributes, and ultimately, the utilization of these cells.
Second, optimal protocols for primary cell isolation should be established according to stromal cell subpopulations. The isolation process, from disaggregation to cell expansion, may result in biased acquisition of specific cell populations or phenotypic alterations of isolated cells. Understanding stromal cell subtypes and standardizing isolation protocols for each cell subset should be performed to acquire cell populations of interest.
Third, the spatial distribution of stromal cells should be considered when evaluating tumor reconstruction. Tissue deconstruction inevitably accompanies the loss of information about cellular locations within the TME. As the spatial location of stromal cells determines the cell-cell and cell-ECM signals to which cells are exposed, the position of stromal cells in their original locations is crucial. In recent years, single-cell spatial technologies such as spatial transcriptomics have emerged; these methods help researchers understand the correlations between phenotypes and the locations of cells and characterize cell-cell interactions that affect tumor behavior and therapeutic responses. Locational information provided by spatial technologies can be further used to ensure the optimal distribution of stromal cells in the reconstructed tumor model.
Upon successfully developing an authentic humanized tumor model, this platform could serve as a potent preclinical model for investigating therapies targeting the stroma. The limitations of traditional preclinical tumor models, which often lack a complex human stromal component, have impeded the translation of promising therapeutic strategies into effective human cancer treatments. For instance, a preclinical study demonstrated the efficacy of saridegib, a Smoothened (SMO) inhibitor pivotal for the hedgehog pathway, in diminishing stromal compartments within tumors and increasing the density of blood vessels within the tumor core by targeting CAFs. These outcomes collectively contributed to a reduction in overall tumor size and an extension of survival in mice132,133. However, despite these promising results, saridegib failed to demonstrate therapeutic advantages in phase I and phase II clinical trials in humans134,135. This failure might be attributed to disparities between murine and human stromal responses. Reproducing a fully humanized ecosystem through the use of a reconstructed tumor model will minimize the disparities between in vitro and in vivo settings and effectively mimic biological events that occur in vivo. This deconstruction and reconstruction approach may provide a more precise representation of therapeutic responses, thereby facilitating the identification of the most appropriate therapeutic approach for each individual.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF), the National Cancer Center (NCC), and the Korean Fund for Regenerative Medicine (KFRM) funded by the Korean government (MSIP) (NRF-2022R1A4A502813112, NRF-RS-2023-00302584, NCC- HA22C005000, KFRM- RS-2023-00216047).
Author contributions
P.K. conceptualized the review; S.M. wrote the manuscript and prepared the figures; H.J.L. prepared the tables; P.K. edited, revised, and reviewed the manuscript. All the authors have read and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Langley RR, Fidler IJ. The seed and soil hypothesis revisited—the role of tumor-stroma interactions in metastasis to different organs. Int. J. Cancer. 2011;128:2527–2535. doi: 10.1002/ijc.26031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Visser KE, Joyce JA. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41:374–403. doi: 10.1016/j.ccell.2023.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan, D. In Cancer Discovery Vol. 12 31–46 (American Association for Cancer Research Inc., 2022).
- 4.Cohen N, et al. Fibroblasts drive an immunosuppressive and growth-promoting microenvironment in breast cancer via secretion of Chitinase 3-like 1. Oncogene. 2017;36:4457–4468. doi: 10.1038/onc.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler JM, Kobayashi H, Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat. Rev. Cancer. 2010;10:138–146. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. 2005;7:211–217. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Pontiggia O, et al. The tumor microenvironment modulates tamoxifen resistance in breast cancer: a role for soluble stromal factors and fibronectin through β1 integrin. Breast Cancer Res. Treat. 2012;133:459–471. doi: 10.1007/s10549-011-1766-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou W, et al. Microenvironment-derived FGF-2 stimulates renal cell carcinoma cell proliferation through modulation of p27Kip1: implications for spatial niche formation and functional intratumoral heterogeneity. Pathobiology. 2020;87:114–124. doi: 10.1159/000506709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, et al. Telomere length is positively associated with the expression of IL‑6 and MIP‑1α in bone marrow mesenchymal stem cells of multiple myeloma. Mol. Med. Rep. 2017;16:2497–2504. doi: 10.3892/mmr.2017.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeNardo DG, et al. CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoppmann SF, et al. Tumor-associated macrophages express lymphatic endothelial growth factors and are related to peritumoral lymphangiogenesis. Am. J. Pathol. 2002;161:947–956. doi: 10.1016/S0002-9440(10)64255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ershaid N, et al. NLRP3 inflammasome in fibroblasts links tissue damage with inflammation in breast cancer progression and metastasis. Nat. Commun. 2019;10:4375. doi: 10.1038/s41467-019-12370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, et al. IL-6 secreted by cancer-associated fibroblasts promotes epithelial-mesenchymal transition and metastasis of gastric cancer via JAK2/STAT3 signaling pathway. Oncotarget. 2017;8:20741. doi: 10.18632/oncotarget.15119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro R, et al. TGF‐β‐induced IGFBP‐3 is a key paracrine factor from activated pericytes that promotes colorectal cancer cell migration and invasion. Mol. Oncol. 2020;14:2609–2628. doi: 10.1002/1878-0261.12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li F, Zhao S, Guo T, Li J, Gu C. The nutritional cytokine leptin promotes NSCLC by activating the PI3K/AKT and MAPK/ERK pathways in NSCLC cells in a paracrine manner. BioMed Res. Int. 2019;2019:2585743. doi: 10.1155/2019/2585743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang B, Huang M, Li Q. Cancer-associated fibroblasts promote angiogenesis of hepatocellular carcinoma by VEGF-mediated EZH2/VASH1 pathway. Technol. Cancer Res. Treat. 2019;18:1533033819879905. doi: 10.1177/1533033819879905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huizer K, et al. Periostin is expressed by pericytes and is crucial for angiogenesis in glioma. J. Neuropathol. Exp. Neurol. 2020;79:863–872. doi: 10.1093/jnen/nlaa067. [DOI] [PubMed] [Google Scholar]
- 18.Koduru S, et al. Dendritic cell–mediated activation-induced cytidine deaminase (AID)–dependent induction of genomic instability in human myeloma. Blood J. Am. Soc. Hematol. 2012;119:2302–2309. doi: 10.1182/blood-2011-08-376236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perini T, et al. Bone marrow microenvironment induces genomic instability and enables clonal evolution in multiple myeloma. Blood. 2017;130:4408. [Google Scholar]
- 20.Zou F, et al. Cancer-associated-fibroblasts regulate the chemoresistance of lung cancer cell line A549 via SDF-1 secretion. Zhonghua Zhong Liu Za Zhi. 2017;39:339–343. doi: 10.3760/cma.j.issn.0253-3766.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Bian L, Sun X, Jin K, He Y. Oral cancer-associated fibroblasts inhibit heat-induced apoptosis in Tca8113 cells through upregulated expression of Bcl-2 through the Mig/CXCR3 axis. Oncol. Rep. 2012;28:2063–2068. doi: 10.3892/or.2012.2019. [DOI] [PubMed] [Google Scholar]
- 22.Ocaña MC, Martínez‐Poveda B, Quesada AR, Medina MÁ. Metabolism within the tumor microenvironment and its implication on cancer progression: An ongoing therapeutic target. Med. Res. Rev. 2019;39:70–113. doi: 10.1002/med.21511. [DOI] [PubMed] [Google Scholar]
- 23.Pavlides S, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 24.Rodrigues J, Heinrich MA, Teixeira LM, Prakash J. 3D in vitro model (R) evolution: unveiling tumor–stroma interactions. Trends Cancer. 2021;7:249–264. doi: 10.1016/j.trecan.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Öhlund D, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai S, et al. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer. 2018;18:1–13. doi: 10.1186/s12885-018-4238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dart A. Organoid 2.0. Nat. Rev. Cancer. 2019;19:126–127. doi: 10.1038/s41568-019-0108-x. [DOI] [PubMed] [Google Scholar]
- 28.Yuan J, Li X, Yu S. Cancer organoid co-culture model system: Novel approach to guide precision medicine. Front. Immunol. 2023;13:1061388. doi: 10.3389/fimmu.2022.1061388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magré L, et al. Emerging organoid-immune co-culture models for cancer research: from oncoimmunology to personalized immunotherapies. J. ImmunoTherapy of Cancer. 2023;11:e006290. doi: 10.1136/jitc-2022-006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subtil B, et al. Dendritic cell phenotype and function in a 3D co-culture model of patient-derived metastatic colorectal cancer organoids. Front. Immunol. 2023;14:1105244. doi: 10.3389/fimmu.2023.1105244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gengenbacher, N., Singhal, M. & Augustin, H. G. In Nature Reviews Cancer Vol. 17, 751–765 (Nature Publishing Group, 2017). [DOI] [PubMed]
- 32.Sajjad, H. et al. In Animal Models and Experimental Medicine Vol. 4, 87–103 (John Wiley and Sons Inc, 2021).
- 33.Invrea F, et al. Patient-derived xenografts (PDXs) as model systems for human cancer. Curr. Opin. Biotechnol. 2020;63:151–156. doi: 10.1016/j.copbio.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Shi, J., Li, Y., Jia, R. & Fan, X. In International Journal of Cancer Vol. 146, 2078–2088 (Wiley-Liss Inc., 2020).
- 35.Cassidy, J. W., Caldas, C. & Bruna, A. In Cancer Research Vol. 75, 2963–2968 (American Association for Cancer Research Inc., 2015).
- 36.Tang LJW, et al. Engineering stromal heterogeneity in cancer. Adv. Drug Deliv. Rev. 2021;175:113817. doi: 10.1016/j.addr.2021.05.027. [DOI] [PubMed] [Google Scholar]
- 37.Qian J, et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020;30:745–762. doi: 10.1038/s41422-020-0355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kieffer Y, et al. Single-cell analysis reveals fibroblast clusters linked to immunotherapy resistance in cancer. Cancer Discov. 2020;10:1330–1351. doi: 10.1158/2159-8290.CD-19-1384. [DOI] [PubMed] [Google Scholar]
- 39.Mitra A, Mishra L, Li S. Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol. 2013;31:347–354. doi: 10.1016/j.tibtech.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porter RJ, Murray GI, McLean MH. Current concepts in tumour-derived organoids. Br. J. Cancer. 2020;123:1209–1218. doi: 10.1038/s41416-020-0993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shinkawa T, Ohuchida K, Nakamura M. Heterogeneity of cancer-associated fibroblasts and the tumor immune microenvironment in pancreatic cancer. Cancers. 2022;14:3994. doi: 10.3390/cancers14163994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon T, Salhia B. Cancer-associated fibroblast subpopulations with diverse and dynamic roles in the tumor microenvironment. Mol. Cancer Res. 2022;20:183–192. doi: 10.1158/1541-7786.MCR-21-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sha M, et al. Isolation of cancer‐associated fibroblasts and its promotion to the progression of intrahepatic cholangiocarcinoma. Cancer Med. 2018;7:4665–4677. doi: 10.1002/cam4.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramaniam KS, et al. Cancer-associated fibroblasts promote proliferation of endometrial cancer cells. PloS One. 2013;8:e68923. doi: 10.1371/journal.pone.0068923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nurmik M, Ullmann P, Rodriguez F, Haan S, Letellier E. In search of definitions: cancer‐associated fibroblasts and their markers. Int. J. Cancer. 2020;146:895–905. doi: 10.1002/ijc.32193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Connell JT, et al. VEGF-A and tenascin-C produced by S100A4 + stromal cells are important for metastatic colonization. Proc. Natl Acad. Sci. USA. 2011;108:16002–16007. doi: 10.1073/pnas.1109493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, et al. Fibroblast-specific protein 1/S100A4-positive cells prevent carcinoma through collagen production and encapsulation of carcinogens. Cancer Res. 2013;73:2770–2781. doi: 10.1158/0008-5472.CAN-12-3022. [DOI] [PubMed] [Google Scholar]
- 48.Luo H, et al. Pan-cancer single-cell analysis reveals the heterogeneity and plasticity of cancer-associated fibroblasts in the tumor microenvironment. Nat. Commun. 2022;13:6619. doi: 10.1038/s41467-022-34395-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang T, Ren Y, Yang P, Wang J, Zhou H. Cancer-associated fibroblasts in pancreatic ductal adenocarcinoma. Cell Death Dis. 2022;13:897. doi: 10.1038/s41419-022-05351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cords L, et al. Cancer-associated fibroblast classification in single-cell and spatial proteomics data. Nat. Commun. 2023;14:4294. doi: 10.1038/s41467-023-39762-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bartoschek M, et al. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat. Commun. 2018;9:5150. doi: 10.1038/s41467-018-07582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, et al. Tumor-associated macrophages: an accomplice in solid tumor progression. J. Biomed. Sci. 2019;26:1–13. doi: 10.1186/s12929-019-0568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lambrechts D, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 54.Tu, J., Wang, D., Zheng, X. T. & Liu, B. Single-cell RNA datasets and bulk RNA datasets analysis demonstrated C1Q+ tumor-associated macrophage as a major and antitumor immune cell population in osteosarcoma. Front. Immunol.14, 10.3389/fimmu.2023.911368 (2023). [DOI] [PMC free article] [PubMed]
- 55.Ricketts, T. D., Prieto-Dominguez, N., Gowda, P. S. & Ubil, E. In Frontiers in Immunology Vol. 12 (Frontiers Media S.A., 2021). [DOI] [PMC free article] [PubMed]
- 56.Yunna C, Mengru H, Lei W, Weidong C. Macrophage M1/M2 polarization. Eur. J. Pharmacol. 2020;877:173090. doi: 10.1016/j.ejphar.2020.173090. [DOI] [PubMed] [Google Scholar]
- 57.Martinez, F. O. & Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Reports6, 10.12703/P6-13 (2014). [DOI] [PMC free article] [PubMed]
- 58.Ma, R. Y., Black, A. & Qian, B. Z. In Trends in Immunology Vol. 43, 546–563 (Elsevier Ltd, 2022). [DOI] [PubMed]
- 59.Zhang L, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020;181:442–459.e429. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 60.Qi, J. et al. Single-cell and spatial analysis reveal interaction of FAP + fibroblasts and SPP1 + macrophages in colorectal cancer. Nat. Commun.13, 10.1038/s41467-022-29366-6 (2022). [DOI] [PMC free article] [PubMed]
- 61.Goveia J, et al. An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell. 2020;37:21–36.e13. doi: 10.1016/j.ccell.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 62.Zeng Q, et al. Understanding tumour endothelial cell heterogeneity and function from single-cell omics. Nat. Rev. Cancer. 2023;23:544–564. doi: 10.1038/s41568-023-00591-5. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, et al. Single-cell analyses of renal cell cancers reveal insights into tumor microenvironment, cell of origin, and therapy response. Proc. Natl Acad. Sci. 2021;118:e2103240118. doi: 10.1073/pnas.2103240118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim, D. et al. Vascularized Lung cancer model for evaluating the promoted transport of anticancer drugs and immune cells in an engineered tumor microenvironment. Adv. Healthc. Mater.11, 10.1002/adhm.202102581 (2022). [DOI] [PMC free article] [PubMed]
- 65.Tatla, A. S., Justin, A. W., Watts, C. & Markaki, A. E. A vascularized tumoroid model for human glioblastoma angiogenesis. Sci. Rep.11, 10.1038/s41598-021-98911-y (2021). [DOI] [PMC free article] [PubMed]
- 66.Cui C, Schoenfelt KQ, Becker KM, Becker L. Isolation of polymorphonuclear neutrophils and monocytes from a single sample of human peripheral blood. STAR Protoc. 2021;2:100845. doi: 10.1016/j.xpro.2021.100845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rahmoune H, Guest PC. Studies of isolated peripheral blood cells as a model of immune dysfunction. Methods Appl. 2018;1735:221–229. doi: 10.1007/978-1-4939-7614-0_12. [DOI] [PubMed] [Google Scholar]
- 68.Walsh EM, Halushka MK. A comparison of tissue dissection techniques for diagnostic, prognostic, and theragnostic analysis of human disease. Pathobiology. 2023;90:199–208. doi: 10.1159/000525979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richter, M. et al. In Frontiers in Cell and Developmental Biology Vol. 9 (Frontiers Media S.A., 2021).
- 70.Freshney, R. I. Basic principles of cell culture. (Wiley Online Library, 2006).
- 71.Freshney, R. I. Culture of animal cells: a manual of basic technique and specialized applications. (John Wiley & Sons, 2015).
- 72.Hendijani, F. In Cell Proliferation Vol. 50 (Blackwell Publishing Ltd, 2017).
- 73.Herron, S., Delpech, J. C., Madore, C. & Ikezu, T. Using mechanical homogenization to isolate microglia from mouse brain tissue to preserve transcriptomic integrity. STAR Protoc.3, 10.1016/j.xpro.2022.101670 (2022). [DOI] [PMC free article] [PubMed]
- 74.Soteriou D, et al. Rapid single-cell physical phenotyping of mechanically dissociated tissue biopsies. Nat. Biomed. Eng. 2023 doi: 10.1038/s41551-023-01015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scheuermann S, et al. TissueGrinder, a novel technology for rapid generation of patient-derived single cell suspensions from solid tumors by mechanical tissue dissociation. Front. Med. 2022;9:721639. doi: 10.3389/fmed.2022.721639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scheuermann S, Schäfer A, Langejürgen J, Reis C. A step towards enzyme-free tissue dissociation. Curr. Dir. Biomed. Eng. 2019;5:545–548. doi: 10.1515/cdbme-2019-0137. [DOI] [Google Scholar]
- 77.Montanari M, et al. Automated—mechanical procedure compared to gentle enzymatic tissue dissociation in cell function studies. Biomolecules. 2022;12:701. doi: 10.3390/biom12050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dvořánková B, Lacina L, Smetana K. Isolation of normal fibroblasts and their cancer-associated counterparts (CAFs) for biomedical research. Skin Stem Cells Methods Protoc. 2019;1879:393–406. doi: 10.1007/7651_2018_137. [DOI] [PubMed] [Google Scholar]
- 79.Cassetta, L. et al. In Tumor Microenvironment: Study Protocols. 211–229 (Springer, 2016).
- 80.Alstrup T, Eijken M, Bohn AB, Møller B, Damsgaard TE. Isolation of adipose tissue–derived stem cells: enzymatic digestion in combination with mechanical distortion to increase adipose tissue–derived stem cell yield from human aspirated fat. Curr. Protoc. Stem Cell Biol. 2019;48:e68. doi: 10.1002/cpsc.68. [DOI] [PubMed] [Google Scholar]
- 81.Wu S, Zhou X, Jin Z, Cheng H. Collagenases and their inhibitors: a review. Collagen Leather. 2023;5:1–20. doi: 10.1186/s42825-023-00126-6. [DOI] [Google Scholar]
- 82.Stenn KS, Link R, Moellmann G, Madri J, Kuklinska E. Dispase, a neutral protease from Bacillus polymyxa, is a powerful fibronectinase and type IV collagenase. J. Investig. Dermatol. 1989;93:287–290. doi: 10.1111/1523-1747.ep12277593. [DOI] [PubMed] [Google Scholar]
- 83.Reichard A, Asosingh K. Best practices for preparing a single cell suspension from solid tissues for flow cytometry. Cytom. Part A. 2019;95:219–226. doi: 10.1002/cyto.a.23690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tan J-X, et al. Upregulation of HYAL1 expression in breast cancer promoted tumor cell proliferation, migration, invasion and angiogenesis. PLoS One. 2011;6:e22836. doi: 10.1371/journal.pone.0022836. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Wang F. Culture of animal cells: a manual of basic technique. In Vitro Cell. Dev. Biol. Anim. 2006;42:169–169. doi: 10.1290/BR090501.1. [DOI] [Google Scholar]
- 86.Okolo O, et al. Protocol for tumor dissociation and fluorescence-activated cell sorting of human head and neck cancers. STAR Protoc. 2023;4:102294. doi: 10.1016/j.xpro.2023.102294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Horner R, et al. Impact of Percoll purification on isolation of primary human hepatocytes. Sci. Rep. 2019;9:6542. doi: 10.1038/s41598-019-43042-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kurashina Y, et al. Enzyme-free release of adhered cells from standard culture dishes using intermittent ultrasonic traveling waves. Commun. Biol. 2019;2:393. doi: 10.1038/s42003-019-0638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kabacik, S. et al. Isolation of five different primary cell types from a single sample of human skin. STAR Protoc.3, 10.1016/j.xpro.2022.101378 (2022). [DOI] [PMC free article] [PubMed]
- 90.Zhang Y, et al. Efficient isolation, culture, purification, and stem cell expression profiles of primary tumor cells derived from uterine cervical squamous cell carcinoma. Am. J. Reprod. Immunol. 2020;84:e13251. doi: 10.1111/aji.13251. [DOI] [PubMed] [Google Scholar]
- 91.Sun, L., Han, X. & Egeblad, M. Isolation of mouse mammary carcinoma-derived macrophages and cancer cells for co-culture assays. STAR Protoc.3, 10.1016/j.xpro.2022.101833 (2022). [DOI] [PMC free article] [PubMed]
- 92.Kapałczyńska M, et al. 2D and 3D cell cultures–a comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018;14:910–919. doi: 10.5114/aoms.2016.63743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou, J. et al. Isolation and characterization of head and neck cancer-derived peritumoral and cancer-associated fibroblasts. Front. Oncol.12, 10.3389/fonc.2022.984138 (2022). [DOI] [PMC free article] [PubMed]
- 94.Xiao, L., McCann, J. V. & Dudley, A. C. Isolation and culture expansion of tumor-specific endothelial cells. J. Vis. Exp.2015, 10.3791/53072 (2015). [DOI] [PMC free article] [PubMed]
- 95.Laoui D. Purification of Tumor-Associated Macrophages (TAM) and Tumor-Associated Dendritic Cells (TADC) Bio-Protoc. 2014;4:e1294. doi: 10.21769/BioProtoc.1294. [DOI] [Google Scholar]
- 96.Koumenis, C., Coussens, L. M., Giaccia, A. & Hammond, E. Tumor Microenvironment: Study Protocols. Vol. 899 (Springer, 2016).
- 97.Pan J, Wan J. Methodological comparison of FACS and MACS isolation of enriched microglia and astrocytes from mouse brain. J. Immunol. Methods. 2020;486:112834. doi: 10.1016/j.jim.2020.112834. [DOI] [PubMed] [Google Scholar]
- 98.Amos PJ, Cagavi Bozkulak E, Qyang Y. Methods of cell purification: a critical juncture for laboratory research and translational science. Cells Tissues Organs. 2011;195:26–40. doi: 10.1159/000331390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Desikan SA, Chavan S, Ou P, Roman CA, Huan C. A MACS protocol for purification of untouched germinal center B cells from unimmunized or germinal center-induced mice. STAR Protoc. 2022;3:101388. doi: 10.1016/j.xpro.2022.101388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schreier S, Sawaisorn P, Udomsangpetch R, Triampo W. Advances in rare cell isolation: an optimization and evaluation study. J. Transl. Med. 2017;15:1–16. doi: 10.1186/s12967-016-1108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Basu S, Campbell HM, Dittel BN, Ray A. Purification of specific cell population by fluorescence activated cell sorting (FACS) J. Vis. Ex.p. 2010;10:1546. doi: 10.3791/1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu P, Zhang W, Xin H, Deng G. Single cell isolation and analysis. Front. Cell Dev. Biol. 2016;4:116. doi: 10.3389/fcell.2016.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jung Y, Zhao M, Svensson KJ. Isolation, culture, and functional analysis of hepatocytes from mice with fatty liver disease. STAR Protoc. 2020;1:100222. doi: 10.1016/j.xpro.2020.100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pretlow, T. G. & Pretlow, T. P. Cell separation: methods and selected applications. (Academic Press, 2014).
- 105.Ladke VS, et al. Isolation, culture and morphological assessment of primary cell lines from human primary oral squamous cell carcinoma using explant technique. Asian Pac. J. Cancer Prev. 2023;24:257. doi: 10.31557/APJCP.2023.24.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kodack DP, et al. Primary patient-derived cancer cells and their potential for personalized cancer patient care. Cell Rep. 2017;21:3298–3309. doi: 10.1016/j.celrep.2017.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nichols, A. E. C. et al. Impact of isolation method on cellular activation and presence of specific tendon cell subpopulations during in vitro culture. FASEB J.35, 10.1096/fj.202100405R (2021). [DOI] [PMC free article] [PubMed]
- 108.Baranyi, U. et al. Primary human fibroblasts in culture switch to a myofibroblast-like phenotype independently of TGF beta. Cells8, 10.3390/cells8070721 (2019). [DOI] [PMC free article] [PubMed]
- 109.Novelli M, et al. Collagenase digestion and mechanical disaggregation as a method to extract and immunophenotype tumour lymphocytes in cutaneous T-cell lymphomas. Clin. Exp. Dermatol. 2000;25:423–431. doi: 10.1046/j.1365-2230.2000.00680.x. [DOI] [PubMed] [Google Scholar]
- 110.Mattei D, et al. Enzymatic dissociation induces transcriptional and proteotype bias in brain cell populations. Int. J. Mol. Sci. 2020;21:1–20. doi: 10.3390/ijms21217944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Waise S, et al. An optimised tissue disaggregation and data processing pipeline for characterising fibroblast phenotypes using single-cell RNA sequencing. Sci. Rep. 2019;9:9580. doi: 10.1038/s41598-019-45842-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.O’Flanagan, C. H. et al. Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biol.20, 10.1186/s13059-019-1830-0 (2019). [DOI] [PMC free article] [PubMed]
- 113.MacPherson S, et al. Clinically relevant T cell expansion media activate distinct metabolic programs uncoupled from cellular function. Mol. Ther. Methods Clin. Dev. 2022;24:380–393. doi: 10.1016/j.omtm.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Leopold B, et al. Outgrowth, proliferation, viability, angiogenesis and phenotype of primary human endothelial cells in different purchasable endothelial culture media: feed wisely. Histochem. Cell Biol. 2019;152:377–390. doi: 10.1007/s00418-019-01815-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee, J. K. et al. Different culture media modulate growth, heterogeneity, and senescence in human mammary epithelial cell cultures. PLoS One13, 10.1371/journal.pone.0204645 (2018). [DOI] [PMC free article] [PubMed]
- 116.Antonsen, K. W. et al. Comparison of culture media reveals that non-essential amino acids strongly affect the phenotype of human monocyte-derived macrophages. Immunology, 10.1111/imm.13670 (2023). [DOI] [PubMed]
- 117.Hogenson TL, et al. Culture media composition influences patient-derived organoid ability to predict therapeutic responses in gastrointestinal cancers. JCI Insight. 2022;7:e158060. doi: 10.1172/jci.insight.158060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Majumder, B. et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat. Commun.6, 10.1038/ncomms7169 (2015). [DOI] [PMC free article] [PubMed]
- 119.Jang M, et al. Matrix stiffness epigenetically regulates the oncogenic activation of the Yes-associated protein in gastric cancer. Nat. Biomed. Eng. 2021;5:114–123. doi: 10.1038/s41551-020-00657-x. [DOI] [PubMed] [Google Scholar]
- 120.Yamada KM, Doyle AD, Lu J. Cell–3D matrix interactions: recent advances and opportunities. Trends Cell Biol. 2022;32:883–895. doi: 10.1016/j.tcb.2022.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stegemann JP, Hong H, Nerem RM. Mechanical, biochemical, and extracellular matrix effects on vascular smooth muscle cell phenotype. J. Appl. Physiol. 2005;98:2321–2327. doi: 10.1152/japplphysiol.01114.2004. [DOI] [PubMed] [Google Scholar]
- 122.Egger, D. et al. From 3D to 3D: Isolation of mesenchymal stem/stromal cells into a three-dimensional human platelet lysate matrix. Stem Cell Res. Ther.10, 10.1186/s13287-019-1346-2 (2019). [DOI] [PMC free article] [PubMed]
- 123.Huang, Y. et al. Isolation of fibroblast-activation protein-specific cancer-associated fibroblasts. BioMed Res. Int.2017, 10.1155/2017/4825108 (2017). [DOI] [PMC free article] [PubMed]
- 124.Buechler MB, Fu W, Turley SJ. Fibroblast-macrophage reciprocal interactions in health, fibrosis, and cancer. Immunity. 2021;54:903–915. doi: 10.1016/j.immuni.2021.04.021. [DOI] [PubMed] [Google Scholar]
- 125.Xun Z, et al. Reconstruction of the tumor spatial microenvironment along the malignant-boundary-nonmalignant axis. Nat. Commun. 2023;14:933. doi: 10.1038/s41467-023-36560-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Powley IR, et al. Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery. Br. J. Cancer. 2020;122:735–744. doi: 10.1038/s41416-019-0672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ferreira LP, Gaspar VM, Mano JF. Decellularized extracellular matrix for bioengineering physiomimetic 3D in vitro tumor models. Trends Biotechnol. 2020;38:1397–1414. doi: 10.1016/j.tibtech.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 128.Liu C, Mak M. Fibroblast-mediated uncaging of cancer cells and dynamic evolution of the physical microenvironment. Sci Rep. 2022;12:791. doi: 10.1038/s41598-021-03134-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guo YC, et al. Protocol for indirect and direct co-culture between human cancer cells and endothelial cells. Star Protocols. 2023;4:102177. doi: 10.1016/j.xpro.2023.102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Langer EM, et al. Modeling tumor phenotypes in vitro with three-dimensional bioprinting. Cell Rep. 2019;26:608–623.e606. doi: 10.1016/j.celrep.2018.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu X, et al. Tumor-on-a-chip: from bioinspired design to biomedical application. Microsyst. Nanoeng. 2021;7:50. doi: 10.1038/s41378-021-00277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Olive KP, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cortes JE, Gutzmer R, Kieran MW, Solomon JA. Hedgehog signaling inhibitors in solid and hematological cancers. Cancer Treat. Rev. 2019;76:41–50. doi: 10.1016/j.ctrv.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 134.Richards DA, et al. A phase Ib trial of IPI-926, a hedgehog pathway inhibitor, plus gemcitabine in patients with metastatic pancreatic cancer. J. Clin. Oncol. 2012;30:213–213. doi: 10.1200/jco.2012.30.4_suppl.213. [DOI] [Google Scholar]
- 135.Ko AH, et al. A phase Ib trial of FOLFIRINOX plus saridegib, an oral hedgehog (Hh) inhibitor, in pts with advanced pancreatic cancer (PDAC) J. Clin. Oncol. 2012;30:3105. doi: 10.1200/jco.2012.30.15_suppl.3105. [DOI] [Google Scholar]
- 136.Costa A, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463–479.e410. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 137.Elyada E, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9:1102–1123. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim, J. et al. Single-cell analysis of gastric pre-cancerous and cancer lesions reveals cell lineage diversity and intratumoral heterogeneity. npj Precis. Oncol.6, 10.1038/s41698-022-00251-1 (2022). [DOI] [PMC free article] [PubMed]
- 139.Li H, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 2017;49:708–718. doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- 140.Laviron, M. et al. Tumor-associated macrophage heterogeneity is driven by tissue territories in breast cancer. Cell Rep.39, 10.1016/j.celrep.2022.110865 (2022). [DOI] [PubMed]
- 141.Wu SZ, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021;53:1334–1347. doi: 10.1038/s41588-021-00911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sathe A, et al. Single-cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin. Cancer Res. 2020;26:2640–2653. doi: 10.1158/1078-0432.CCR-19-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lee HO, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 2020;52:594–603. doi: 10.1038/s41588-020-0636-z. [DOI] [PubMed] [Google Scholar]
- 144.Sharma A, et al. Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell. 2020;183:377–394.e321. doi: 10.1016/j.cell.2020.08.040. [DOI] [PubMed] [Google Scholar]
- 145.Cheng S, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184:792–809.e723. doi: 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 146.Mulder K, et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity. 2021;54:1883–1900.e1885. doi: 10.1016/j.immuni.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 147.Chen K, et al. Hypoxic pancreatic cancer derived exosomal miR-30b-5p promotes tumor angiogenesis by inhibiting GJA1 expression. Int. J. Biol. Sci. 2022;18:1220–1237. doi: 10.7150/ijbs.67675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jiang, H. et al. Revealing the transcriptional heterogeneity of organ‐specific metastasis in human gastric cancer using single‐cell RNA Sequencing. Clin. Transl. Med.12, 10.1002/ctm2.730 (2022). [DOI] [PMC free article] [PubMed]
- 149.Sun, K. et al. scRNA-seq of gastric tumor shows complex intercellular interaction with an alternative T cell exhaustion trajectory. Nat. Commun.13, 10.1038/s41467-022-32627-z (2022). [DOI] [PMC free article] [PubMed]
- 150.Xie, Y. et al. Key molecular alterations in endothelial cells in human glioblastoma uncovered through single-cell RNA sequencing. JCI Insight6, 10.1172/jci.insight.150861 (2021). [DOI] [PMC free article] [PubMed]
- 151.Busch, S. et al. Cellular organization and molecular differentiation model of breast cancer-associated fibroblasts. Mol. Cancer16, 10.1186/s12943-017-0642-7 (2017). [DOI] [PMC free article] [PubMed]
- 152.Yasuda, T. et al. Protocol to establish cancer-associated fibroblasts from surgically resected tissues and generate senescent fibroblasts. STAR Protoc.2, 10.1016/j.xpro.2021.100553 (2021). [DOI] [PMC free article] [PubMed]
- 153.Atanasova VS, et al. Mimicking tumor cell heterogeneity of colorectal cancer in a patient-derived organoid-fibroblast model. Cell Mol. Gastroenterol. Hepatol. 2023;15:1391–1419. doi: 10.1016/j.jcmgh.2023.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hu, Y. et al. Fibroblast-derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS One10, 10.1371/journal.pone.0125625 (2015). [DOI] [PMC free article] [PubMed]
- 155.Go, Y. H. et al. Modeling pancreatic cancer with patient-derived organoids integrating cancer-associated fibroblasts. Cancers14, 10.3390/cancers14092077 (2022). [DOI] [PMC free article] [PubMed]
- 156.Mun, K. et al. Isolation and characterization of cancer-associated fibroblasts in the tumor microenvironment of hepatocellular carcinoma. J. Liver Cancer, 10.17998/jlc.2023.04.30 (2023). [DOI] [PMC free article] [PubMed]
- 157.Bartlett, J. M. Ovarian cancer: methods and protocols. Vol. 39 (Springer Science & Business Media, 2008).
- 158.Cassetta L, et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell. 2019;35:588–602.e510. doi: 10.1016/j.ccell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bieniasz-Krzywiec, P., Martín-Pérez, R., Riera-Domingo, C. & Mazzone, M. Isolation and separation of murine tumor-associated macrophages (TAMs) subpopulations from orthotopic 4T1 breast tumors. STAR Protoc.2, 10.1016/j.xpro.2021.100481 (2021). [DOI] [PMC free article] [PubMed]
- 160.Hirano, R. et al. Tissue-resident macrophages are major tumor-associated macrophage resources, contributing to early TNBC development, recurrence, and metastases. Commun. Biol.6, 10.1038/s42003-023-04525-7 (2023). [DOI] [PMC free article] [PubMed]
- 161.Wettersten HI, et al. Arming tumor-associated macrophages to reverse epithelial cancer progression. Cancer Res. 2019;79:5048–5059. doi: 10.1158/0008-5472.CAN-19-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yu, J. et al. Tumor-associated macrophages (TAMs) depend on MMP1 for their cancer-promoting role. Cell Death Discov.7, 10.1038/s41420-021-00730-7 (2021). [DOI] [PMC free article] [PubMed]
- 163.Wu, Q. et al. Suppression of endothelial cell migration by tumor associated macrophage-derived exosomes is reversed by epithelial ovarian cancer exosomal lncRNA. Cancer Cell Int.17, 10.1186/s12935-017-0430-x (2017). [DOI] [PMC free article] [PubMed]
- 164.Taguchi K, et al. Isolation of tumor endothelial cells from murine cancer. J. Immunol. Methods. 2019;464:105–113. doi: 10.1016/j.jim.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 165.Verhoeven, J., Agostinis, P. & Agrawal, M. In Tumor Angiogenesis Assays: Methods and Protocols 45–54 (Springer, 2022).
- 166.Ohga N, et al. Heterogeneity of tumor endothelial cells: comparison between tumor endothelial cells isolated from high- and low-metastatic tumors. Am. J. Pathol. 2012;180:1294–1307. doi: 10.1016/j.ajpath.2011.11.035. [DOI] [PubMed] [Google Scholar]
- 167.Mao Q, et al. A novel method for endothelial cell isolation. Oncol. Rep. 2016;35:1652–1656. doi: 10.3892/or.2015.4490. [DOI] [PubMed] [Google Scholar]
- 168.Bussolati B, Deambrosis I, Russo S, Deregibus MC, Camussi G. Altered angiogenesis and survival in human tumor-derived endothelial cells. FASEB J. 2003;17:1159–1161. doi: 10.1096/fj.02-0557fje. [DOI] [PubMed] [Google Scholar]
- 169.Rebeaud, M. et al. A novel 3D culture model for human primary mammary adipocytes to study their metabolic crosstalk with breast cancer in lean and obese conditions. Sci. Rep.13, 10.1038/s41598-023-31673-x (2023). [DOI] [PMC free article] [PubMed]
- 170.Zhou C, et al. Cancer-associated adipocytes promote the invasion and metastasis in breast cancer through LIF/CXCLs positive feedback loop. Int. J. Biol. Sci. 2022;18:1363–1380. doi: 10.7150/ijbs.65227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mukherjee A. Isolation of primary normal and cancer-associated adipocytes from the omentum. Ovarian Cancer Methods Protoc. 2022;2424:167–175. doi: 10.1007/978-1-0716-1956-8_11. [DOI] [PubMed] [Google Scholar]
- 172.Huang, L. & Bischoff, J. Isolation of stem cells, endothelial cells and pericytes from human infantile hemangioma. Bio-Protoc.10, 10.21769/BioProtoc.3487 (2020). [DOI] [PMC free article] [PubMed]
- 173.Bose A, et al. Tumor-derived vascular pericytes anergize Th cells. J. Immunol. 2013;191:971–981. doi: 10.4049/jimmunol.1300280. [DOI] [PubMed] [Google Scholar]
- 174.Hosaka K, et al. Pericyte-fibroblast transition promotes tumor growth and metastasis. Proc. Natl Acad. Sci. USA. 2016;113:E5618–E5627. doi: 10.1073/pnas.1608384113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Uniken Venema, W. T. C. et al. Gut mucosa dissociation protocols influence cell type proportions and single-cell gene expression levels. Sci. Rep.12, 10.1038/s41598-022-13812-y (2022). [DOI] [PMC free article] [PubMed]
- 176.Raghavan S, et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell. 2021;184:6119–6137.e6126. doi: 10.1016/j.cell.2021.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang Y, et al. Tissue-specific extracellular matrix coatings for the promotion of cell proliferation and maintenance of cell phenotype. Biomaterials. 2009;30:4021–4028. doi: 10.1016/j.biomaterials.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Romero-López M, et al. Recapitulating the human tumor microenvironment: colon tumor-derived extracellular matrix promotes angiogenesis and tumor cell growth. Biomaterials. 2017;116:118–129. doi: 10.1016/j.biomaterials.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Puttock, E. H. et al. Extracellular matrix educates an immunoregulatory tumor macrophage phenotype found in ovarian cancer metastasis. Nat. Commun.14, 10.1038/s41467-023-38093-5 (2023). [DOI] [PMC free article] [PubMed]