Abstract
The dimerization initiation site (DIS), downstream of the long terminal repeat within the human immunodeficiency virus type 1 (HIV-1) genome, can form a stem-loop structure (SL1) that has been shown to be involved in the packaging of viral RNA. In order to further determine the role of this region in the virus life cycle, we deleted the 16 nucleotides (nt) at positions +238 to +253 within SL1 to generate a construct termed BH10-LD3 and showed that this virus was impaired in viral RNA packaging, viral gene expression, and viral replication. Long-term culture of these mutated viruses in MT-2 cells, i.e., 18 passages, yielded revertant viruses that possessed infectivities similar to that of the wild type. Cloning and sequencing showed that these viruses retained the original 16-nt deletion but possessed two additional point mutations, which were located within the p2 and NC regions of the Gag coding region, respectively, and which were therefore named MP2 and MNC. Site-directed mutagenesis studies revealed that both of these point mutations were necessary to compensate for the 16-nt deletion in BH10-LD3. A construct with both the 16-nt deletion and the MP2 mutation, i.e., LD3-MP2, produced approximately five times more viral protein than BH10-LD3, while the MNC mutation, i.e., construct LD3-MNC, reversed the defects in viral RNA packaging. We also deleted nt +261 to +274 within the 3′ end of SL1 and showed that the diminished infectivity of the mutated virus, termed BH10-LD4, could also be restored by the MP2 and MNC point mutations. Therefore, compensatory mutations within the p2 and NC proteins, distal from deletions within the DIS region of the HIV genome, can restore HIV replication, viral gene expression, and viral RNA packaging to control levels.
Human immunodeficiency virus type 1 (HIV-1) encapsidates two identical copies of viral genomic RNA that are noncovalently linked at the 5′ end in each virion (7). Specific packaging of viral genomic RNA is dependent on both cis-acting RNA elements and viral structural proteins. These cis-acting RNA elements have largely been mapped to 5′-end noncoding leader sequences near the major splice donor site across a region of approximately 140 nucleotides (nt) (i.e., the E/ψ site), although some RNA sequences in the gag and env genes may also participate in the packaging of viral genomic RNA (1, 11, 21, 22, 27, 30, 32, 37, 41). The E/ψ site exists within specific secondary RNA structures that comprise four stem-loops (i.e., SL1, SL2, SL3, and SL4), as deduced from computer-based and conformational analyses and studies involving nuclease digestion and chemical sensitivity measurements (3, 12, 20). Mutational analysis has shown that SL1, SL3, and SL4 are all important for the packaging of viral genomic RNA (32, 33). Interestingly, viral nucleocapsid protein 7 (NC7), a cleavage product of Gag precursor protein Pr55, is also involved in this process through the activity of the zinc finger motifs and flanking basic amino acids within NC7 (1, 4, 8–10, 15–17, 40). In addition, direct interactions between viral RNA elements, including SL1, SL3, and SL4, and Gag proteins have been reported in cell-free systems (8, 9, 12, 42).
Interestingly, the interstrand binding sites within viral genomic RNA dimers coincide with the location of E/ψ, suggesting that the processes of RNA dimerization and packaging might be mechanistically linked (4, 5, 34). Although the mechanism of dimerization is still unclear, cell-free studies have demonstrated that stem-loop structure SL1 may serve as a dimerization initiation site due to the presence of palindromic loop sequence GCGCGC (2, 14, 24, 31, 36, 48). This has stimulated interest in the activity of this RNA structure in encapsidation and dimerization of viral genomic RNA (6, 13, 25, 35).
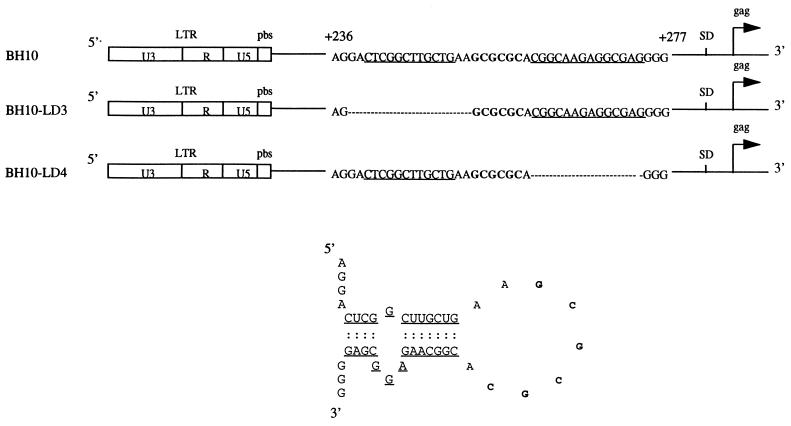
To further investigate the role of SL1 in HIV-1 replication, we have previously generated a mutated HIV-1 proviral DNA clone termed BH10-LD3 in which 16 nt, i.e., nt positions +238 to +253, were deleted in order to disrupt this SL1 region (nt 240 to 274) (29). We now show that the resultant mutated virus, i.e., BH10-LD3, is defective in both packaging of viral genomic RNA and replication capacity. We have also propagated these defective viruses in MT-2 cells for 18 passages and have thereby generated viruses with wild-type infectivity that contain two distinct compensatory point mutations within NC7 and the p2 spacer peptide domain between the capsid (CA) and NC7 regions of Gag. These experiments shed new light on the importance of the SL1 region of viral genomic RNA and the NC7 protein in regard to packaging and, in addition, provide evidence of the remarkable ability of HIV-1 to acquire compensatory mutations that can act to restore wild-type viral replication kinetics, even when such mutations are located distal to the region of the initial deletion or perturbation and distal to each other.
MATERIALS AND METHODS
Mutagenesis of HIV-1 DNA.
Primers used in the mutagenesis studies described here are illustrated in Table 1. The generation of the BH10-LD3 deletion has been previously described (29) (Fig. 1). We also deleted an additional DNA segment termed BH10-LD4, nt positions +261 to +274 (Fig. 1), through use of PCR methodology and the primer pair DIS-R and Apa-A (Table 1) (44). Each primer contained a convenient restriction site to facilitate cloning, and the primer termed DIS-R was designed to have the desired deletion. Point mutation MNC was introduced through the use of primer NC-A, which contained this substitution, and a second primer termed BssH-S (Table 1). Point mutation MP2 was generated with a PCR-Script Amp cloning kit (Stratagene, La Jolla, Calif.) and primers P2-S and P2-A (Table 1). The combination of mutations MP2 and MNC was achieved through the use of primer pair BssH-S/NC-A (Table 1), in order to amplify the BH10-MP2 DNA template that contained the MP2 substitution.
TABLE 1.
Primers utilized in the experiments
| Name | Sequence | Locationa |
|---|---|---|
| Hpa-S | 5′-CTGCAGTTAACTGGAAGGGCTAATTCACTCCC-3′ | −454–−434 |
| pS | 5′-AGACCAGATCTGAGCCTGGGAG-3′ | +14–+35 |
| BssH-S | 5′-CTGAAGCGCGCACGGCAAGAGG-3′ | +249–+270 |
| DIS-R | 5′-GCTGAAGCGCGCAGGGCGGCGACTGGTGAGTACGCC-3′ | +248–+297 |
| GAG1 | 5′-CAGCATTATCAGAAGGAGCC-3′ | +850–+869 |
| pST | 5′-CATTCTGCAGCTTCCTCATTGATGG-3′ | +968–+944 |
| P2-S | 5′-CAAATACAGCTATCATAATGATGC-3′ | +1444–+1467 |
| P2-A | 5′-GCATCATTATGATAGCTGTATTTG-3′ | +1467–+1444 |
| Apa-A | 5′-CCTAGGGGCCCTGCAATTTCTG-3′ | +1559–+1538 |
| NC-A | 5′-CTAGGGGCCCTGCAATTTCTGGCTATGTGCCC-3′ | +1558–+1527 |
Location of the primer refers to the transcription initiation site (+1).
FIG. 1.
Deletion mutation constructs BH10-LD3 and BH10-LD4, in which DNA positions +238 to +253 and +261 to +274, respectively, have been eliminated. The stem-loop RNA structure (SL1) is shown and is formed by RNA sequences (nt +240 to +274). Nucleotides that form the stem are underlined, and the palindromic sequence (GCGCGC) of the loop is in boldface. LTR, long terminal repeat; SD, splice donor site.
HIV-1 transfection and viral infection studies.
MT-2 and COS-7 cells were cultured in RPMI 1640 and Dulbecco’s modified Eagle’s medium, respectively, each supplemented with 10% fetal calf serum. COS-7 cells were transfected with either wild-type viral DNA or mutated HIV-1 proviral DNA in the presence of calcium phosphate as described previously (44). Progeny virus was harvested 48 h after transfection and was assessed by measuring levels of viral p24 (CA) antigen by enzyme-linked immunosorption assay (Abbott Laboratories, Abbott Park, Ill.) (28).
To monitor the efficiency of transfection, cells were also cotransfected with 2 μg of pSV–β-galactosidase, and cell extracts were analyzed with a β-galactosidase enzyme assay system (Promega, Madison, Wis.). Protein concentrations were standardized through use of a Dc assay kit (Bio-Rad Laboratories, Mississauga, Ontario, Canada). Levels of viral p24 (CA) antigen were corrected based on the transfection efficiencies of the various plasmids employed.
For infectivity experiments, similar amounts of virus (i.e., 3 ng of p24 antigen per 106 cells; equivalent to approximately 105 cpm of reverse transcriptase [RT] activity) were used to infect MT-2 cells. After 2 h, cells were washed twice with serum-free RPMI 1640 to remove unbound viruses and were then maintained in serum-supplemented medium. Culture fluids were collected at various times for determinations of RT activity.
DNA sequencing of mutated BH10-LD3 viruses after long-term culture.
MT-2 cells were infected with BH10-LD3 virus derived from transfected COS-7 cells. No cytopathology was initially observed due to the minimal infectiousness of this virus. However, after maintenance of infected cells in culture for 3 months, cytopathology began to appear, and, at this time, culture fluids were collected for subsequent passage. Thereafter, DNA was extracted from infected MT-2 cells by incubating cell pellets at 37°C for 6 h in lysis buffer containing 0.5% sodium dodecyl sulfate (SDS) and 1 mg of protease K per ml. The lysed suspensions were extracted twice with phenol-chloroform (1:1) and precipitated with 2.5 volumes of 95% ethanol. Viral DNA fragments containing nt +18 to +968 were amplified through use of the pS/pST primer pair (Table 1). PCR products were digested with restriction enzymes BglII and PstI and cloned into vector pSP72. Sequencing was performed with a double-stranded DNA cycle sequencing system (GIBCO BRL, Montreal, Quebec, Canada).
Analysis of viral proteins.
Culture fluids (10 ml) collected from COS-7 cells 48 h after transfection were clarified in a Beckman GS-6R centrifuge at 3,000 rpm for 30 min at 4°C. Viral particles were then pelleted through a 20% sucrose cushion at 40,000 rpm for 1 h at 4°C with an SW41 rotor in a Beckman L8-M ultracentrifuge. Viral pellets were suspended in 50 μl of lysis buffer, of which 10 μl were used in Western blots. Protein samples were fractionated on SDS–12% polyacrylamide gels and transferred to nitrocellulose filters. After being blocked with 5% skim milk–0.05% Tween 20–phosphate buffer at 4°C overnight, the filters were incubated with anti-HIV-p24 or anti-HIV-gp41 immunoglobulin G1 (IgG1) monoclonal antibodies (MAbs) (ID Labs Inc., London, Ontario, Canada) at 37°C for 1 h. After extensive washing with 0.05% Tween 20–phosphate buffer, secondary anti-mouse IgG–horseradish peroxidase-conjugated antibody (Amersham Life Sciences, Oakville, Ontario, Canada) was added for 1 h at 37°C. After thorough washing, viral proteins were visualized with an ECL chemiluminescence detection kit (Amersham Life Science, Amersham Place, England).
Viral RNA analysis.
Virus pellets harvested from culture fluids (30 ml) of transfected COS-7 cells were suspended in 100 μl of TN buffer (50 mM Tris-HCl [pH 7.5], 10 mM NaCl). Five microliters of sample was used to measure the amount of p24 antigen. Viral RNA was then extracted from the remaining viral suspension with an Ultraspec TM-II RNA isolation system (Biotecs, Houston, Tex.), dissolved in RNase-free double-distilled water, and then diluted such that each microliter of RNA-containing sample represented 8 ng of the p24 antigen of wild-type virus.
For slot blot assays, 10 μl of viral RNA samples was treated with 10 U of DNase I (RNase-free; GIBCO BRL) at 37°C for 10 min and then heated to 95°C for 10 min to inactivate this enzyme. To ensure that no contaminating DNA was present in these treated RNA samples, 5 μl of material was digested with RNase A as a negative control. Samples were incubated in 40 μl of buffer containing 50% formamide, 17.5% formaldehyde, and 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) and then denatured at 68°C for 15 min, following which they were immobilized onto nylon membranes with a slot blot apparatus and UV irradiated. The membranes were baked at 80°C for 2 h and prehybridized in buffer containing 50% formamide, 0.5% SDS, 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 M EDTA [pH 7.7]), 5× Denhardt’s solution, and 0.1 mg of salmon sperm DNA (GIBCO BRL) at 42°C for 3 h. HIV-1 proviral DNA was employed as a probe and was labeled with a nick translation kit (Boehringer GmbH, Mannheim, Germany). Hybridization was performed at 42°C overnight in buffer containing 50% formamide, 0.5% SDS, 6× SSPE, 0.1 mg of salmon sperm DNA per ml, and the DNA probe (106 cpm/ml). After being extensively washed, the membranes were exposed to X-ray film.
The viral nucleic acid samples, treated by DNase I as described above, were also quantified by RT-PCR. For this purpose, the DNA primer, i.e., pST (Table 1), was annealed onto the viral RNA and extended by avian myeloblastosis virus (AMV) RT (Pharmacia) in 20 μl containing 50 mM Tris-HCl (pH 7.5), 75 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol, 4 U of AMV RT, 400 μM deoxynucleoside triphosphates, and 20 U of RNA guard (Pharmacia). As a control, similar reactions were performed in the absence of RT to check for any potential DNA contamination. Five microliters of the reverse-transcribed products was then used in a 15-cycle PCR with primer pair GAG1/pST (Table 1) to generate a 119-bp DNA fragment. The GAG1 primer used in this reaction was radioactively labeled in order to visualize reaction products. The PCR samples were then fractionated on 5% acrylamide gels and exposed to X-ray film.
RESULTS
Increased infectiousness of mutated BH10-LD3 virus after long-term culture in MT-2 cells.
Mutated BH10-LD3 proviral DNA has a deletion at nt positions +238 to +253 in the noncoding leader sequence (Fig. 1). Accordingly, we transfected COS-7 cells with either mutated (BH10-LD3) or wild-type (BH10) proviral DNA. The results showed, in accordance with previous observations (29), that mutant construct BH10-LD3 yielded four to five times less viral protein than did wild-type BH10, as shown by both Western blots of cell lysates and pelleted virus particles and enzyme-linked immunosorbent assay (data not shown). Consistently, mutated BH10-LD3 viruses derived from these COS-7 transfections were far less replication competent in MT-2 cells than were wild-type BH10 viruses of the same origin (Fig. 2A).
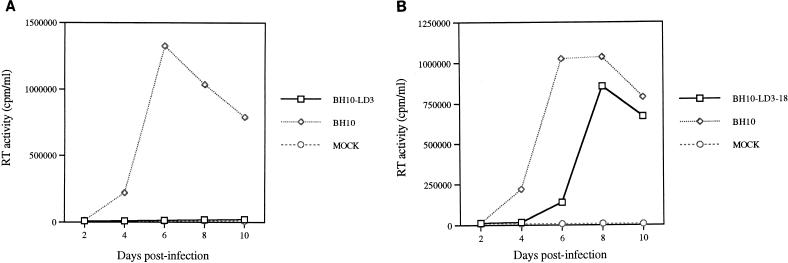
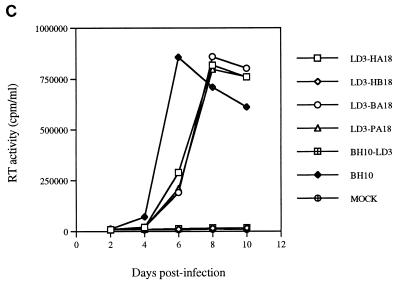
FIG. 2.
Replication capacities of mutated BH10-LD3 and reverted BH10-LD3-18 viruses. (A) Growth curves of wild-type (BH10) and mutated (BH10-LD3) viruses in MT-2 cells infected with viruses harvested from transfected COS-7 cells. MT-2 cells (106 cells) were infected with equivalent amounts of virus based on RT activity (105 cpm per 106 cells), and viral production was monitored by RT assay of culture fluids. Mock infection denotes exposure of cells to heat-inactivated wild-type virus as a negative control. (B) Increased infectiousness of mutated BH10-LD3 virus after long-term culture in MT-2 cells. Reverted virus BH10-LD3-18 was that which was harvested after 18 passages of BH10-LD3 in MT-2 cells. MT-2 cells were infected with equivalent amounts of virus based on RT activity (105 cpm/106 cells). Production of progeny virus was monitored by RT assay. (C) Growth curves of wild-type virus (BH10) and recombinant viruses LD3-HA18, LD3-HB18, LD3-BA18, and LD3-PA18 in MT-2 cells; curves were produced as described above.
Since the BH10-LD3 deletion had disrupted the SL1 RNA structure (Fig. 1), thought to be involved in dimerization, encapsidation, gene expression, and viral replication (2, 6, 13, 14, 24, 25, 31, 35, 36, 48), we were anxious to understand how the deleted sequences (nt +238 to +253) may have impacted these activities. Therefore, MT-2 cells that had been infected with mutated BH10-LD3 virus were passaged for a number of generations to determine whether reverted viruses with increased infectivity might appear and what the mechanism of reversion might be.
Indeed, cytopathology was observed after 18 cell passages, suggesting that a reverted virus might have been generated. To verify whether this was the case, fresh cultures of MT-2 cells were infected with the same amount of either MT-2-derived wild-type BH10 or a potentially reverted virus termed BH10-LD3-18, based on RT activity. The results showed that the BH10-LD3-18 virus grew almost as well as wild-type BH10 (Fig. 2B), suggesting that compensatory mutations might have occurred during long-term passage of the MT-2 cells that had been chronically infected with the mutated BH10-LD3 virus.
Since the most likely region in which such compensatory mutations might have occurred includes the flanking sequences around the original LD3 deletion, i.e., nt +238 to +253, cellular DNA was extracted from MT-2 cells infected by the BH10-LD3-18 virus, amplified by PCR with the pS/pST primer pair, as described above, and cloned into the pSP72 vector. Sequencing showed that the deleted +238 to +253 segment had not reappeared, in whole or in part, in the viral DNA and that no additional mutations could be detected in the flanking sequences. When the amplified fragment (with pS/pST; i.e., nt +18 to +968) was inserted into the equivalent region within wild-type proviral BH10 DNA, we found that the infectious capacity of the resultant recombinant virus was similar to that of the mutated BH10-LD3 virus. These data demonstrate the likelihood that any compensatory mutations responsible for BH10-LD3-18 replication must have been located outside the +18-to-+968 region.
In order to functionally localize such compensatory mutations, a much larger DNA fragment comprising nt −454 to +1548 was amplified from the cellular DNA of MT-2 cells infected by BH10-LD3-18 through use of the Hpa-S/Apa-A primer pair (Table 1) and inserted, as described above, in place of the corresponding region of wild-type BH10 DNA to yield recombinant proviral DNA clone LD3-HA18. Five such clones were selected, and DNA from relevant plasmids was independently transfected into COS-7 cells. Two days later, the culture fluids of these COS-7 cells were used to infect MT-2 cells. Six days thereafter, three of the five cultures studied showed cytopathology and contained high levels of RT activity (Fig. 2C), indicating that the LD3-HA18 viruses were infectious. Sequencing viral DNA derived from these clones confirmed that the BH10-LD3 deletion had occurred. Therefore, the compensatory mutations responsible for replication of BH10-LD3-18 were present within the −454-to-+1548 region.
Of course, 18 passages of MT-2 cells chronically infected by defective BH10-LD3 virus might have yielded a number of spontaneous mutations that could compromise our attempts to identify compensatory mutations. To narrow the region under investigation, we performed subcloning by using the BssHII (i.e., nt +254) restriction site to insert either the DNA fragment from nt −454 to +254 (HpaI-BssHII) or from nt +254 to +1548 (BssHII-ApaI) of LD3-HA18 (i.e., the infectious clone) into proviral BH10-LD3 DNA to yield recombinant proviral clones LD3-HB18 and LD3-BA18, respectively (Fig. 3). Infectivity assays showed that the recombinant LD3-HB18 virus was not viable, while, in contrast, the recombinant LD3-BA18 virus possessed replication capacity similar to that of BH10-HA18 (Fig. 2C). Since, as shown above, the DNA segment (nt +18 to +968) from viral revertant BH10-LD3-18 could not compensate for the LD3 deletion, it follows that the compensatory mutations in question must have been present within the +968-to-+1548 (PstI-ApaI) sequence.
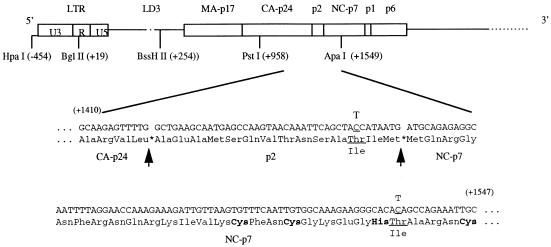
FIG. 3.
Locations of the compensatory mutations MP2 and MNC that restore replication capacity to the mutated BH10-LD3 virus. The amino acid sequences of the spacer peptide p2 and parts of the capsid protein (CA) and nucleocapsid protein 7 (NC7) and the corresponding nucleotide sequences are shown. Arrows illustrate the protease cleavage sites (asterisks) of CA/p2 and p2/NC7. Mutated nucleotides and amino acids are underlined, and the CCHC sequence found within the first zinc finger of NC7 is in boldface. The positions of the restriction sites used in the cloning experiments are also shown. LTR, long terminal repeat.
To further evaluate this subject, the +968-to-+1548 segment of LD3-HA18 was substituted for the equivalent region within BH10-LD3 to generate LD3-PA18. This virus (LD3-pA18) possessed infectiousness similar to that of LD3-HA18 (Fig. 2C). To identify any putative compensatory mutations within this smaller stretch, the +968-to-+1548 region was sequenced. The results revealed two point mutations, one at position +1456 (C→T [Thr→Ile]) within the coding sequence for the p2 spacer peptide between CA and NC7 (termed mutation MP2 [for mutation within p2]) and one at position +1534 (also C→T [Thr→Ile]) within the coding sequence for the first zinc finger motif of NC7 (termed mutation MNC [for mutation within NC]) (Fig. 3).
The MP2 and MNC point mutations are both required to restore the diminished replication capacity of mutated BH10-LD3 virus.
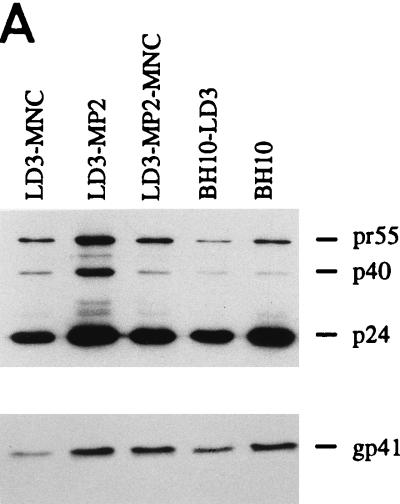
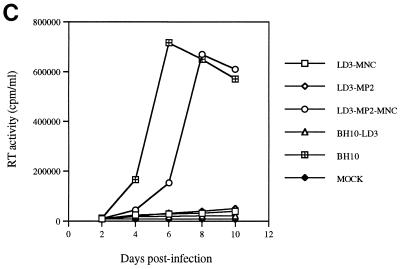
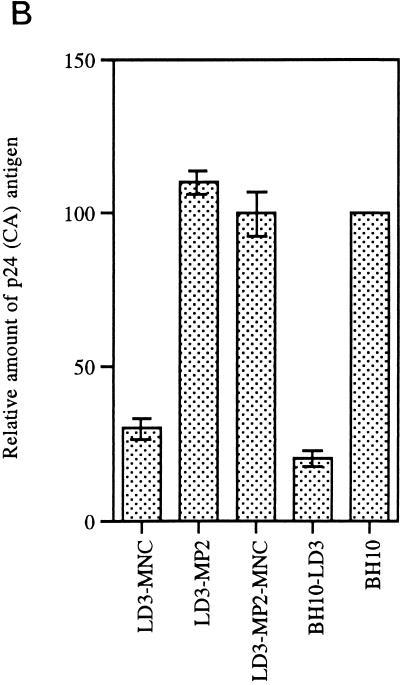
To determine whether the MP2 and MNC point mutations could help restore the reduced infectious capacity of mutated BH10-LD3 virus, site-directed mutagenesis was performed to introduce either or both of these substitutions into BH10-LD3 to yield recombinant clones LD3-MP2, LD3-MNC, and LD3-MP2-MNC. COS-7 cells were transfected with these constructs, and the virus particles in culture fluids were harvested by ultracentrifugation, as described in Materials and Methods. Western blot analysis using MAbs directed against either p24 (CA) or gp41 showed that similar amounts of viral proteins (Gag or Env) were generated in COS-7 cells transfected by the LD3-MP2, LD3-MP2-MNC, and wild-type BH10 constructs; in contrast, both LD3-MNC and BH10-LD3 yielded far less viral protein (Fig. 4A and B). Therefore, the MP2 mutation was seemingly responsible for the increased level of viral protein production associated with the LD3-MP2 recombinant virus.
FIG. 4.
Site-directed mutagenesis to illustrate that point mutations MP2 and MNC can compensate for the LD3 deletion. (A) Viral protein analysis by Western blotting. Constructs LD3-MNC, LD3-MP2, and LD3-MP2-MNC have the BH10-LD3 deletion as well as point mutations MNC, MP2, or both MNC and MP2, respectively. Virus particles harvested from transfected COS-7 cells were subjected to Western blotting with either anti-HIV p24 (CA) IgG1 MAb or anti-HIV gp41 IgG MAb. The molecular masses (in kilodaltons) of viral proteins are shown on the right. (B) Relative amounts of p24 (CA) antigen in culture fluids are shown in the graph, with levels for wild-type virus arbitrarily set at 100. (C) Growth curves of wild-type and mutated viruses in MT-2 cells infected with equivalent amounts of virus based on RT activity, as described in the legend to Fig. 2. The production of progeny virus was monitored by observing levels of RT activity in culture fluids.
We next monitored the infectious capacities of these various viruses in MT-2 cells as described above. The results showed that neither point mutation alone (i.e., LD3-MNC or LD3-MP2) could restore wild-type replication capacity to the BH10-LD3 deletion construct (Fig. 4C). However, both the MP2 and MNC mutations were jointly able to restore viral replication capacity, within the context of construct LD3-MP2-MNC, to a level equivalent to that of the BH10-HA18 virus (Fig. 4C).
We also inserted these two point mutations, MNC and MP2, separately and together, into wild-type BH10 virus to generate recombinant clones BH10-MNC, BH10-MP2, and BH10-MP2-MNC. When these constructs were transfected into COS-7 cells, the viruses thus generated possessed wild-type characteristics in regard to both infectiousness and protein band pattern as determined by Western blotting (data not shown). These data suggest that the amino acid substitutions encoded by both MNC and MP2 (Thr→Ile), involving changes from a hydrophilic to a hydrophobic moiety, were necessary for effective interactions to take place between the altered NC and p2 proteins that were generated and the disrupted SL1 region of BH10-LD3. However, these changes must have been relatively insignificant in the context of interactions between these same altered proteins and a wild-type nondisrupted SL1 structure.
Restoration of diminished viral RNA packaging in mutated BH10-LD3 viruses by compensatory mutations.
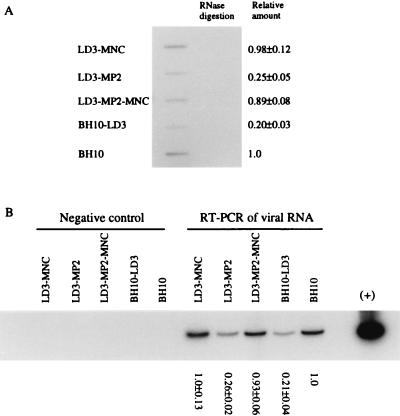
To further investigate the defects caused by the LD3 deletion and restoration of viral replication by the MP2 and MNC point mutations, we first analyzed the packaging of viral genomic RNA into both wild-type BH10 and mutated BH10-LD3 viruses. Viral RNA was extracted from purified viral particles harvested from transfected COS-7 cells, and the amount of virus was standardized on the basis of levels of p24 antigen. A quantity of viral RNA equivalent to 40 ng of p24 antigen was then used in slot blot assays under conditions in which RNA samples were treated with 10 U of DNase I (RNase free) for 10 min at 37°C to eliminate potential DNA contamination. An RNase A digestion control was also performed to ensure that there was no contamination with DNA. The results showed that the amount of viral RNA packaged by the mutated BH10-LD3 viruses was four- to fivefold lower than that packaged by the wild-type BH10 virus (Fig. 5A). To further test this point, we performed RT-PCR to amplify a 119-nt RNA fragment (nt +850 to +968) within the gag region as a reflection of the packaging of full-length viral genomic RNA. We again observed defective packaging of viral RNA by BH10-LD3 virus (Fig. 5B).
FIG. 5.
Viral RNA packaging in wild-type and mutated viruses. (A) Analysis of viral RNA packaging by slot blotting. Viral particles were harvested from culture fluids of transfected COS-7 cells and centrifuged through a 20% sucrose cushion. Viral RNA was extracted and dissolved in double-distilled water to a concentration equivalent to 8 ng of p24 (CA) antigen per μl. After treatment with RNase-free DNase I, RNA samples were subjected to slot blotting. As a control, samples were digested with RNase A to rule out possible DNA contamination. Relative RNA content was quantified by molecular image analysis. Levels of RNA in wild-type HIV-1 were arbitrarily set at 1.0. (B) Analysis of viral RNA packaging by RT-PCR. Reverse transcription was performed by using AMV RT to extend viral RNA that had been annealed to DNA primer pST. Reverse transcription products were amplified in a 15-cycle PCR by using primer pair GAG1/pST to generate a 119-bp DNA fragment. Relative amounts of DNA products were quantified by molecular imaging, with wild-type levels arbitrarily set at 1.0. Reactions run without RT served as a negative control to exclude any potential DNA contamination. As a positive control, the 15-cycle PCR was also performed with 5 ng of proviral BH10 DNA.
The fact that mutations MP2 and MNC had restored infectivity to LD3-mutated viruses dictated that we also evaluate RNA packaging in viruses that contained these compensatory mutations, i.e., LD3-MNC, LD3-MP2, and LD3-MP2-MNC. The results of Fig. 5A show by slot blot that viral RNA packaging in the doubly mutated virus, i.e., LD3-MP2-MNC, was restored to wild-type levels (Fig. 5A). Viruses containing the MNC substitution, i.e., LD3-MNC, also showed significant restoration of packaging (Fig. 5A), while viruses containing the MP2 mutation, i.e., LD3-MP2, were still significantly impaired in this regard. These results were confirmed by RT-PCR experiments (Fig. 5B).
Partial restoration of infectiousness of mutated BH10-LD4 virus by the MP2 and MNC mutations.
The disruption of the SL1 region of viral RNA by the LD3 deletion may potentially have caused the observed impairment of viral replication in a way that permitted the two compensatory mutations, MP2 and MNC, to restore the functional role of this region. To shed further light on this subject, we asked whether deletion of the 3′ end of the SL1 region would also impair viral replication in a manner that might be compensated for by the MP2 and MNC mutations. Accordingly, we deleted the +261-to-+274 segment of viral DNA that encodes the 3′ end of the SL1 region of viral genomic RNA to generate construct BH10-LD4 (Fig. 1). We also inserted both the MP2 and MNC point mutations into BH10-LD4 to generate LD4-MP2-MNC.
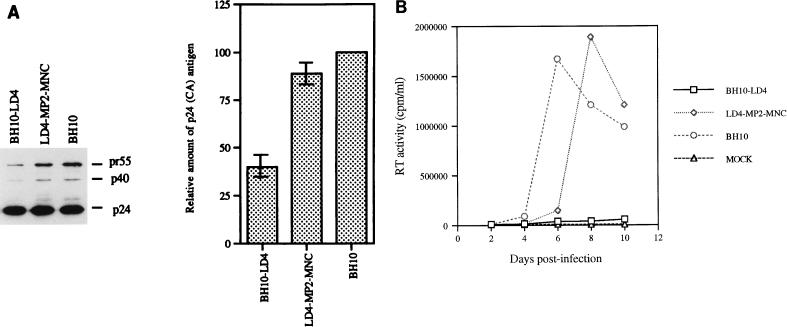
COS-7 cells were transfected with these virus constructs to generate relevant mutated viruses that were then studied in Western blot and infectivity assays. The results showed that the mutated BH10-LD4 and LD4-MP2-MNC viruses possessed viral protein band patterns similar to that of wild-type BH10 virus (Fig. 6A). When equivalent quantities of these viruses, based on p24 levels, were used to infect MT-2 cells, a peak in RT activity was observed after 6 days with the wild-type BH10 virus. In contrast, the mutated BH10-LD4 virus replicated poorly and generated little RT activity, while virus LD4-MP2-MNC showed only a brief 2-day delay in growth compared with wild-type virus (Fig. 6B). Thus, MP2 and MNC mutations also compensated for the defect caused by the BH10-LD4 deletion.
FIG. 6.
Ability of point mutations MP2 and MNC to compensate for the BH10-LD4 deletion. (A) Analysis of viral proteins by Western blotting. Viral particles were harvested from culture fluids of transfected COS-7 cells and subjected to Western blotting with anti-HIV p24 (CA) IgG1 MAb. The molecular masses (in kilodaltons) of viral proteins are indicated on the right. Construct LD4-MP2-MNC has the BH10-LD4 deletion as well as the MP2 and MNC point mutations. Relative amounts of p24 (CA) antigen in culture fluids are shown in the graph, with levels for wild-type virus arbitrarily set at 100. (B) Growth curves of wild-type and mutated viruses in MT-2 cells infected with equivalent amounts of virus as described in the legend to Fig. 2. The production of progeny virus was monitored by RT assay of culture fluids.
DISCUSSION
The encapsidation of viral genomic RNA is dependent on a stretch of cis-acting RNA elements located around the major splice donor site (7). Our results show that nucleotide segment +238 to +253, which constitutes part of the SL1 structure, is involved in the efficient packaging of viral RNA. We have also shown that long-term culture of a virus with this segment deleted, BH10-LD3, led to two compensatory point mutations located within the Gag-coding region that could restore both RNA packaging and replication capacity.
The RNA sequences that flank the major splice donor site in HIV-1 include several RNA stem-loop structures (i.e., SL1 to SL4) (3, 12, 20). It had originally been thought that RNA sequences downstream of the major splice donor were responsible for the specific encapsidation of viral genomic RNA in a manner that would exclude the packaging of spliced viral RNA species. However, RNA sequences upstream of the splice donor site, which include stem-loop structure SL1, are also reported to be involved in the efficient packaging of viral RNA (6, 33, 35). We have now confirmed and extended these observations through analysis of viral deletion mutant BH10-LD3 in which the SL1 structure had been disrupted, thereby diminishing both the efficiency of RNA packaging and viral infectivity.
We have also identified a compensatory mutation termed MNC within the gag gene that can largely restore packaging efficiency. The NC7 protein of HIV-1 has been shown to be involved in viral genomic packaging through studies in which the exchange of NC between HIV-1 and murine leukemia virus resulted in a switch in the specific packaging of the respective viral RNAs (10). Further mutational analysis showed that both the zinc finger motifs of NC and flanking basic amino acids played important roles in this regard (1, 4, 8–10, 15–17, 40), and cell-free experiments have shown that specific interactions between NC7 and the SL1, SL3, and SL4 viral RNA structures represent important signals for packaging (12), a concept supported as well by structural analysis of NC7 bound to the SL3 stem-loop element (19). The compensatory mutation termed MNC is located within the first zinc finger motif of NC7 and represents a Thr→Ile substitution in the primary structure (Fig. 3). Since the MNC mutation can restore wild-type levels of packaging to mutated viruses containing a disruption of the SL1 region, our data suggest that a specific interaction between the first zinc finger of NC7 and SL1 must contribute to viral RNA packaging. Hence, the cis-acting (i.e., RNA SL1) and trans-acting (i.e., NC7) elements involved in this process can genetically complement one another.
Another role of SL1 is its involvement in the dimerization of viral genomic RNA. This region has been proposed to initiate the dimerization process through the activity of palindromic loop sequence GCGCGC, termed the kissing-loop (2, 14, 24, 31, 36, 48). Although in vivo mutational studies of SL1 suggest that this region contributes little to the maturation of RNA dimers (6, 43), others have reported that disruption or deletion of SL1 resulted in abnormalities of dimerization (13, 25). Current work in our laboratory deals with the effects of the BH10-LD3 deletion on dimerization and the role in this regard of the compensatory mutation MNC.
Although the MNC mutation compensated for the reduction in the efficiency of RNA packaging caused by the BH10-LD3 deletion, the impaired infectiousness of BH10-LD3 could only be reversed in the presence of a second compensatory mutation termed MP2. This mutation is located within the 14-amino-acid p2 spacer peptide, found between the capsid (CA) and NC7 proteins within Gag, and represents a Thr→Ile substitution within a stretch of four amino acids at the C terminus of this moiety (Fig. 3). These four amino acids constitute the 5′ end of the first cleavage site recognized by the viral protease during the processing of Gag precursor proteins (18, 39). The role of the p2 peptide in the ordered processing of Gag polyproteins, essential for virion maturation, is still unclear, although deletion of p2 was shown to interfere with virus assembly and to result in fewer infectious particles, and more aberrant morphology, in spite of the presence of the final processed products of Gag (23, 38). The alteration of the cleavage site between p2 and NC7 by MP2 may result in either negative or positive modulation of Gag polyprotein processing.
Cell-free studies have shown that cleavage of the nucleocapsid p15 protein (NC 15) by HIV-1 protease is an RNA-dependent process (45, 46). Hence, the efficient processing of Gag may be dependent on interactions with viral genomic RNA. Since the LD3 deletion reduced the packaging efficiency of viral RNA, it may also have interfered with Gag polyprotein cleavage. While the MNC mutation may have compensated for the encapsidation defect in BH10-LD3, this virus would still contain a disrupted SL1 region that would compromise the quality of the expected interaction with NC7 and potentially affect the processing of Gag precursor proteins. The MP2 mutation may conceivably compensate for this defect by facilitating the cleavage of Gag precursor proteins in the LD3-MP2-MNC virus.
As noted above, the introduction of the MP2 and MNC mutations into wild-type BH10 virus had little effect on either viral protein expression or replication. This suggests that the nature of the Thr→Ile substitutions within the p2 and NC proteins did not result in significant alterations in the abilities of these proteins to interact with viral genomic RNA in the SL1 region in regard to signalling for packaging and encapsidation. In contrast, the presence of these mutations was clearly necessary to restore the effectiveness of such interactions to virus with the 16-nt deletion within the dimerization initiation site i.e., construct BH10-LD3. Current studies involve electron microscopy of cells infected by these various wild-type and mutated viruses to better understand the role of the MP2 and MNC mutations on viral assembly.
In addition to the foregoing, we have also shown that the MP2 mutation leads to increased protein production in mutated LD3-MP2 virus. This result is consistent with recent reports that noncoding sequences in the leader region between the primer binding site and the major splice donor are required for efficient viral gene expression (26, 29). Although the SL1 region may play a posttranscriptional role in the expression of unspliced viral RNA, it is also possible that the LD3 deletion reduced overall HIV gene expression and, hence, resulted in diminished levels of Gag proteins, as shown in previous studies (29). Genomic elements that can inhibit expression of Gag proteins have been identified within the Gag-coding region (46, 47). These “instability sequences” (INS) have been localized by mutagenesis studies to both the MA (p17)- and CA (p24)-encoding regions but not to the p2 peptide (45). It is thought that the instability of HIV-1 RNA may be due to the richness of A’s and T’s in viral genomic RNA, and, indeed, mutations that enhanced viral RNA stability were shown to involve mostly substitutions of either A or T for C or G, which eliminated the INS (45). In contrast, the MP2 mutation involves a C→T change, making it unlikely that this substitution results in increased stability of viral RNA. Therefore, other mechanisms must enable the MP2 mutation to restore wild-type levels of protein production and replication competence to the mutated BH10-LD3 virus.
In summary, this paper documents the selection of two compensatory mutations, both of which are located distal to the LD3 deletion, that resulted in impaired packaging of viral RNA and infectivity. One of these changes, MNC, is located within NC7, while the other is in the spacer p2 peptide between CA and NC7. Our results also provide further evidence for the requirements for both cis-acting RNA elements (i.e., SL1) and trans-acting viral structural proteins (i.e., NC7) in the encapsidation of viral RNA. Further study of the MP2 and MNC mutations in regard to protein-RNA interactions will help to define the roles of p2, NC7, and the SL1 region of viral RNA in viral assembly and infectiousness.
REFERENCES
- 1.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging results in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awang G, Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry. 1993;32:11453–11457. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- 3.Baudin G, Marquet R, Isel C, Darlix J L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 4.Bender W, Chien Y-H, Chattopadhyay S, Vogt P K, Gardner M B, Davidson N. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J Virol. 1978;25:888–896. doi: 10.1128/jvi.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender W, Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976;7:595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 8.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 9.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkowitz R D, Ohagen A, Höglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clavel F, Orenstein J M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clever J L, Wong M L, Parslow T G. Requirement for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 17.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gowda S D, Stein A S, Engleman E G. Identification of protein intermediates in the processing of the p55 HIV-1 gag precursor in cells infected with recombinant vaccinia virus. J Biol Chem. 1989;264:8459–8462. [PubMed] [Google Scholar]
- 19.Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3Ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 20.Harrison G P, Lever A M L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi T, Shioda T, Iwakura Y, Shibuta H. RNA packaging signal of human immunodeficiency virus type 1. Virology. 1992;188:590–599. doi: 10.1016/0042-6822(92)90513-o. [DOI] [PubMed] [Google Scholar]
- 22.Kim H J, Lee K, O’Rear J J. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology. 1994;198:336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 23.Krausslich J G, Facke M, Heuser A M, Konvalinka J, Zentgraf J. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 25.Laughrea M, Jette L, Mak J, Kleiman L, Liang C, Wainberg M A. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J Virol. 1997;71:3397–3406. doi: 10.1128/jvi.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenz C, Scheid A, Schaal H. Exon 1 leader sequences downstream of U5 are important for efficient human immunodeficiency virus type 1 gene expression. J Virol. 1997;71:2757–2764. doi: 10.1128/jvi.71.4.2757-2764.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X G, Mak J, Arts E J, Gu Z X, Kleiman L, Wainberg M A, Parniak M A. Effects of alterations of primer-binding site sequences on human immunodeficiency virus type 1 replication. J Virol. 1994;68:6198–6206. doi: 10.1128/jvi.68.10.6198-6206.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang C, Li X G, Quan Y D, Laughrea M, Kleiman L, Hiscott J, Wainberg M A. Sequence elements downstream of the human immunodeficiency virus type 1 long terminal repeat are required for efficient viral gene transcription. J Mol Biol. 1997;272:167–177. doi: 10.1006/jmbi.1997.1239. [DOI] [PubMed] [Google Scholar]
- 30.Luban J, Goff S P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marquet R, Paillart J C, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22:145–151. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murti K G, Bondurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J Virol. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paillart J-C, Berthoux L, Ottmann M, Darlix J-L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paillart J C, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 37.Parolin C, Dorfman T, Palú G, Göttlinger H, Sodroski J. Analysis in human immunodeficiency virus type 1 vector of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–3895. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The P2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettit S C, Simsic J, Loeb D D, Everitt L, Hutchison III C A, Swanstrom R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J Biol Chem. 1991;266:14539–14547. [PubMed] [Google Scholar]
- 40.Poon D T K, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richardson J H, Child L A, Lever A M L. Packaging of human immunodeficiency virus type 1 RNA requires cis-acting sequences outside the 5′ leader region. J Virol. 1993;67:3997–4005. doi: 10.1128/jvi.67.7.3997-4005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakaguchi K, Zambrano N, Baldwin E T, Shapiro B A, Erickson J W, Omichinski J G, Clore G M, Gronenborn A M, Appella E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc Natl Acad Sci USA. 1993;90:5219–5223. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakuragi J I, Panganiban A T. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J Virol. 1997;71:3250–3254. doi: 10.1128/jvi.71.4.3250-3254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory element allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B K, Pavlakis G N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz S, Felber B K, Pavlakis G N. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]