Abstract.
We encountered a Chinese girl with pseudohypoparathyroidism type 1A (PHP1A) and her mother with pseudopseudohypoparathyroidism (PPHP). Sequencing analysis of GNAS-Gsα revealed a heterozygous c.212+2T>C variant (NM_000516.4) affecting the canonical splice donor site of intron 2 in the girl and her mother. RT-PCR performed on mRNA samples obtained from cycloheximide-treated and cycloheximide-untreated lymphoblastoid cell lines of this girl revealed the utilization of an alternative splice donor site at 33–34 bp from the boundary between exon 2 and intron 2 and the production of an aberrant mRNA with a retention of a 32 bp intronic sequence between exon 2 and exon 3 (p.(Gly72Lysfs*39)), which satisfied the condition for the occurrence of nonsense-mediated mRNA decay, as predicted by SpliceAI. This study revealed the molecular consequences of disruption of the canonical splice donor site and confirmed the clinical utility of SpliceAI.
Keywords: pseudohypoparathyroidism, GNAS-Gsα, splice donor site mutation, nonsense-mediated mRNA decay
Highlights
● We encountered a girl with pseudohypoparathyroidism type 1A and her mother with pseudopseudohypoparathyroidism.
● We identified a novel GNAS-Gsα splice donor site variant that created an alternative splice donor site and produced an aberrant mRNA subject to nonsense-mediated mRNA decay.
● We revealed the molecular consequence of the disruption of the canonical splice donor site.
Introduction
The α subunit of the stimulatory G protein (Gsα), encoded by exons 1–13 of GNAS (hereafter referred to as GNAS-Gsα), plays a critical role in the signal transduction of multiple G-protein-coupled receptors (GPCRs), including PTH-receptor (PTH1R), TSHR, LHCGR, FSHR, and GHRHR (1). While GNAS-Gsα is biallelically expressed in most tissues, including the skeletal system, it is predominantly expressed from the maternal allele in a few tissues, such as the renal proximal tubule, thyroid, gonad, and pituitary, because genomic imprinting is regulated by the methylation pattern of GNAS differentially methylated regions (GNAS-DMRs) (2,3,4). Thus, compromised expression of maternally inherited GNAS-Gsα leads to pseudohypoparathyroidism type 1A (PHP1A) associated with resistance to PTH as well as other hormones such as TSH, LH/FSH, and GHRH, and Albright’s hereditary osteodystrophy (AHO), which is characterized by obesity, round face, short stature, brachydactyly, subcutaneous ossifications, and intellectual disability, whereas compromised expression of paternally transmitted GNAS-Gsα results in pseudopseudohypoparathyroidism (PPHP) with AHO but without hormonal resistance (5). In addition, methylation defects in GNAS-DMRs usually result in pseudohypoparathyroidism type 1B (PHP1B) with hormonal resistance but without AHO (5).
Here, we report a novel GNAS-Gsα splice donor site variant in a girl with PHP1A and her mother with PPHP. Molecular analysis revealed that an alternative splice donor site was utilized to produce an aberrant mRNA subject to nonsense-mediated mRNA decay (NMD).
Clinical Report
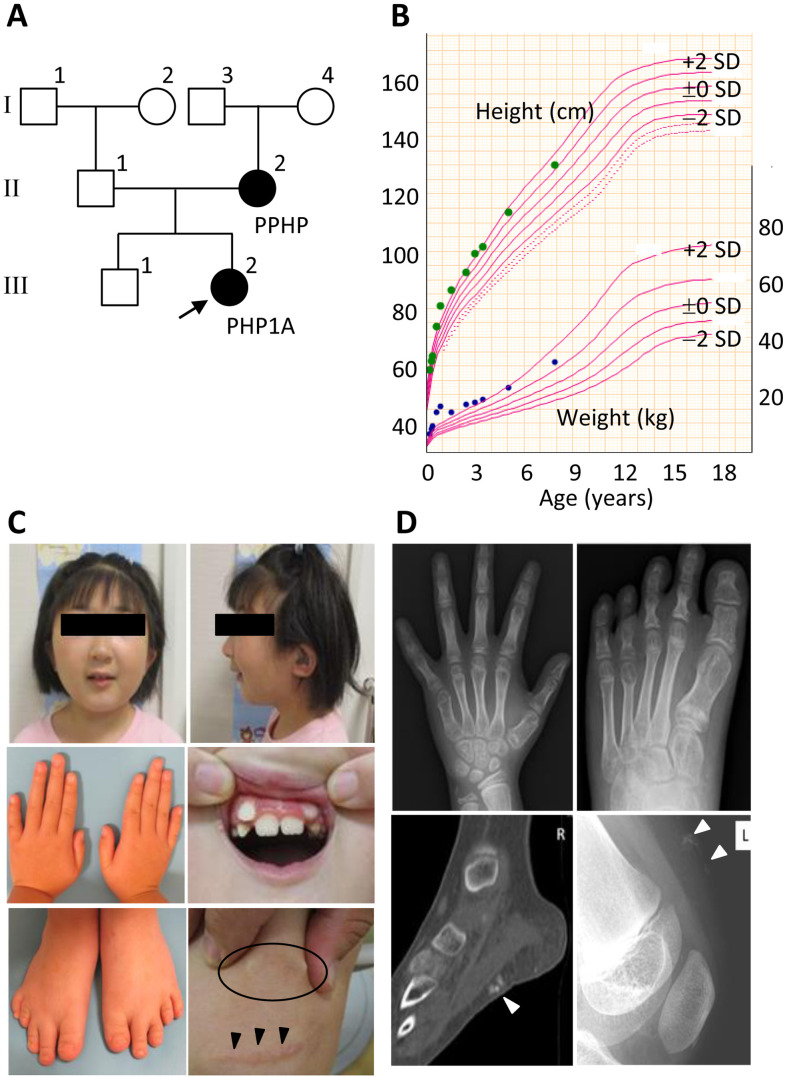
We encountered a Chinese girl with PHP1A (the proband) and her mother with PPHP (Fig. 1A). The girl (III-2) was born as the second child to non-consanguineous parents at 37 wk and 4 d of gestation after an uneventful pregnancy and delivery. At birth, her length was 43.0 cm (–2.1 SD), and her weight was 2,646 g (–0.2 SD). Shortly after birth, she was referred to a local hospital because of an elevated TSH level on newborn screening for congenital hypothyroidism, although she exhibited no clinical features of hypothyroidism such as neonatal jaundice, hypotonia, poor feeding, umbilical hernia, and macroglossia. At that time, her serum TSH was 53.27 mIU/mL (reference range, 0.36–9.75 mIU/mL), and free T4 was 2.10 ng/dL (0.70–2.47 ng/dL). Thus, the patient was diagnosed with subclinical congenital hypothyroidism and treated with l-thyroxine. Furthermore, congenital bilateral sensorineural deafness was identified during the neonatal period. Around 5 mo of age, multiple subcutaneous ossifications were observed on the abdomen, back, and lower extremities. Marked weight gain was also observed up to 12 mo of age, which became inconspicuous afterward (Fig. 1B).
Fig. 1.
Summary of clinical findings. A: Pedigree of the family. The girl (III-2) indicated by an arrow is the proband. B: The growth chart of the girl with PHP1A plotted on the growth curves of Japanese girls. C: Photographs of the girl at the ages of 7 yr and 10 mo. The circle indicates subcutaneous ossification, and the arrowheads indicate the postoperative scar from the subcutaneous osteotomy. D: Roentgenograms of the girl. The arrowheads indicate subcutaneous ossifications.
At 7 yr and 10 mo of age, the patient was referred to our hospital because of AHO-like features. Her height was 130.6 cm (+1.3 SD), and her weight was 31.8 kg (+1.6 SD). Physical examination revealed a round face, low-set ears, misaligned teeth, micrognathia, brachydactyly of the bilateral fifth fingers and fourth toes, and multiple subcutaneous ossifications of the upper and lower limbs (Fig. 1C). Furthermore, several postoperative scars derived from the subcutaneous osteotomy were observed (Fig. 1C). She also had intellectual disability with speech delay. Laboratory tests revealed a low-normal serum calcium level of 8.9 mg/dL (8.8–10.8 mg/dL), a high-normal level of serum inorganic phosphorous concentration of 5.5 mg/dL (3.7–5.6 mg/dL), and an elevated intact PTH level of 125 pg/mL (10–65 pg/mL). Bone survey delineated brachydactyly of the bilateral fifth metacarpal bones, fourth metatarsal bones, and multiple subcutaneous ossifications in the legs (Fig. 1D). Based on these findings, the patient was diagnosed as having PHP1A.
Her mother (II-2) had multiple subcutaneous ossifications and normal serum calcium, inorganic phosphorus, and intact PTH levels. Therefore, she was clinically diagnosed as having PPHP. The father (II-1) and older brother (III-1) were clinically healthy, with no discernible AHO features. The maternal grandfather (I-3) was free from PHP1A or PPHP clinical features, although he was obese and had diabetes mellitus. The maternal grandmother (I-4) exhibited no AHO features.
Molecular Studies
This study was approved by the Institutional Review Board Committee of the Hamamatsu University School of Medicine and was performed after obtaining written informed consent for the publication of clinical and molecular findings, including photographs.
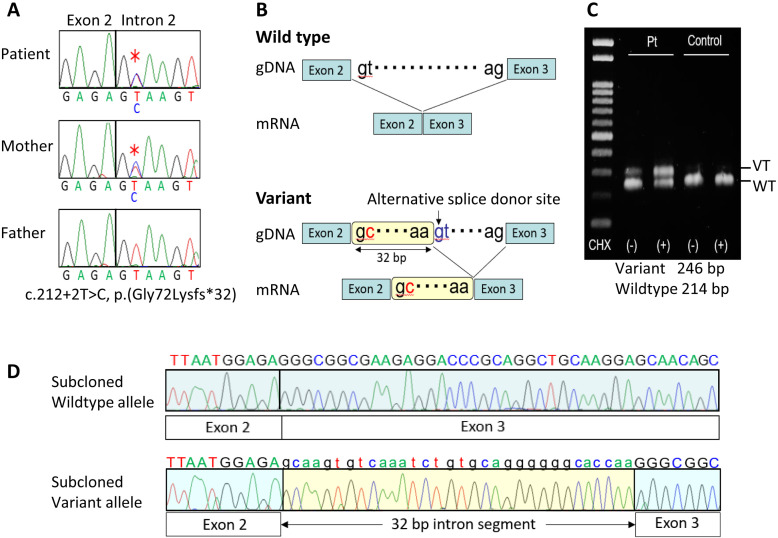
Direct sequencing was performed for exons 1–13 of GNAS-Gsα, using leukocyte genomic DNA samples obtained from the girl and her parents. Consequently, a maternally derived c.212+2T>C variant (NM_000516.4), affecting the canonical splice donor site of intron 2, was identified in this girl (Fig. 2A). This variant was absent from the three representative public databases (gnomAD, http://gnomad.broadinstitute.org/; HGVD, http://www.hgvd.genome.med.kyoto-u.ac.jp; and 38 KJPN, https://jmorp.megabank.tohoku.ac.jp/) and in-house database.
Fig. 2.
Summary of the molecular findings. A: Electrochromatograms showing a maternally derived splice donor site variant (c.212+2T>C) in intron 2 of GNAS-Gsα (marked with multiple asterisks) obtained using a forward primer hybridizing to exon 2 (5′-CTCTGCGTCGAAATGTCAAG-3′) and a reverse primer hybridizing to exon 3 (5′-TGGTTGCCTTCTCACCATC-3′). B: Schematic representation of the utilization of an alternative splice donor site and production of an aberrant mRNA predicted by SpliceAI. C: RT-PCR analysis of mRNA extracted from CHX-untreated and CHX-treated LCLs of the girl and a control subject. The primers utilized were: forward, 5′-GGGTGCTGGAGAATCTGGTA-3′; and reverse, 5′- TGGTTGCCTTCTCACCATC-3′. D: Electrochromatograms of subcloned wildtype and variant mRNA sequences obtained by RT-PCR for CHX-treated LCLs of the girl.
SpliceAI (https://github.com/Illumina/SpliceAI) predicted the utilization of an alternative splice donor site at 33–34 bp from the boundary between exon 2 and intron 2, as well as the production of an aberrant mRNA with a retention of a 32 bp intronic sequence between exon 2 and exon 3 (p.(Gly72Lysfs*39)), thereby satisfying the condition for the occurrence of NMD (Fig. 2B) (6). To examine this possibility, reverse transcription (RT)-PCR was performed on mRNA samples obtained from lymphoblastoid cell lines (LCLs) of this girl and a healthy control, which were incubated for 8 hours with or without the NMD inhibitor cycloheximide (CHX), yielding normal 214 bp products and aberrant 246 bp products which was clearly identified in the CHX-treated LCLs of this girl (Fig. 2C). The RT-PCR products of CHX-treated LCLs of this girl were subcloned using a TOPO TA Cloning Kit (Invitrogen), and direct sequencing of the subcloned RT-PCR products confirmed the presence of the wildtype mRNA and the aberrant mRNA retaining a 32 bp intronic sequence, as predicted by Splice AI (Fig. 2D).
Discussion
We identified a novel GNAS-Gsα splice donor site variant (c.212+2T>C) in a girl with PHP1A and her mother with PPHP. This variant was shown to produce mRNA subject to NMD by utilizing an alternative splice donor site, as predicted by SpliceAI. Although a certain amount of mRNA may have escaped NMD, such mRNA is predicted to yield a drastically truncated protein p.(Gly72Lysfs*39). Thus, this study revealed the molecular consequences of the disruption of the canonical splice donor site. According to the ACMG guidelines (7), this variant is classified as “pathogenic”, because it is positive for PVS1 (disruption of the canonical ± 1 or 2 splice sites), PS3 (confirmation of NMD by RT-PCR), and PM2 (absence from the public databases and in-house database). This variant was of maternal origin in this girl with PHP1A and is predicted to reside on the paternally transmitted allele of the mother with PPHP. In this regard, since the maternal grandfather had no clinical features of PHP1A or PPHP, the GNAS-Gsα variant may have occurred as a de novo event in the mother.
This girl showed an elevated serum TSH level, congenital bilateral sensorineural deafness, multiple subcutaneous ossifications, and excessive weight gain in the neonatal to infantile period, before she was found to have multiple AHO-like features and asymptomatic PTH resistance in childhood. The clinical course was fairly characteristic of PHP1A, because it has been reported that [1] TSH resistance has frequently been identified as the initial hormonal abnormality in PHP1A (8, 9); [2] congenital hearing impairment has been reported in PHP1A with a prevalence of 11% (10); [3] the occurrence of multiple subcutaneous ossifications has been reported in early infancy of PHP1A patients (8, 11, 12); [4] early onset of obesity has also frequently been observed in PHP1A (13); and [5] PTH resistance usually becomes clinically discernible in childhood (5). Notably, early recognition of PHP1A-compatible clinical features would serve to make the diagnosis of PHP1A before the clinical symptoms of PTH resistance, such as hypocalcemic seizures, become discernible. In this girl, several features, such as elevated serum TSH level, sensorineural deafness, subcutaneous ossifications, and weight gain are considered as the characteristic features which permit early recognition of PHP1A. In addition, although a strict genotype–phenotype correlation has not been established for PHP1A, subcutaneous ossifications are more frequent in PHP1A and PPHP patients with amorphic GNAS-Gsα variants than in those with hypomorphic GNAS-Gsα variants (8, 11, 12). Thus, multiple subcutaneous ossifications can be considered an indication of amorphic GNAS-Gsα variants.
Conclusions
We identified a novel GNAS-Gsα splice donor site variant shared by a girl with PHP1A and her mother with PPHP. The molecular studies demonstrated the production of aberrant mRNA subject to NMD, providing strong support for the clinical utility of SpliceAI.
Conflict of interests
The authors have no conflicts of interest to declare.
Acknowledgments
This work was supported by grants from the Japan Agency for Medical Research and Development (AMED) (23ek0109587h), Grants-in-Aid for Scientific Research (JP20K08178 [C]) from the Japan Society for the Promotion of Science, and a Shizuoka Children’s Hospital Research Grant.
References
- 1.Weinstein LS, Yu S, Warner DR, Liu J. Endocrine manifestations of stimulatory G protein alpha-subunit mutations and the role of genomic imprinting. Endocr Rev 2001;22: 675–705. [DOI] [PubMed] [Google Scholar]
- 2.Yu S, Yu D, Lee E, Eckhaus M, Lee R, Corria Z, et al. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein alpha-subunit (Gsalpha) knockout mice is due to tissue-specific imprinting of the gsalpha gene. Proc Natl Acad Sci USA 1998;95: 8715–20. doi: 10.1073/pnas.95.15.8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayward BE, Barlier A, Korbonits M, Grossman AB, Jacquet P, Enjalbert A, et al. Imprinting of the G(s)alpha gene GNAS1 in the pathogenesis of acromegaly. J Clin Invest 2001;107: R31–6. doi: 10.1172/JCI11887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantovani G, Ballare E, Giammona E, Beck-Peccoz P, Spada A. The gsalpha gene: predominant maternal origin of transcription in human thyroid gland and gonads. J Clin Endocrinol Metab 2002;87: 4736–40. doi: 10.1210/jc.2002-020183 [DOI] [PubMed] [Google Scholar]
- 5.Mantovani G, Spada A, Elli FM. Pseudohypoparathyroidism and Gsα-cAMP-linked disorders: current view and open issues. Nat Rev Endocrinol 2016;12: 347–56. doi: 10.1038/nrendo.2016.52 [DOI] [PubMed] [Google Scholar]
- 6.Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med 2006;12: 306–16. doi: 10.1016/j.molmed.2006.05.005 [DOI] [PubMed] [Google Scholar]
- 7.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17: 405–24. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sano S, Nakamura A, Matsubara K, Nagasaki K, Fukami M, Kagami M, et al. (Epi)genotype-phenotype analysis in 69 Japanese patients with pseudohypoparathyroidism type I. J Endocr Soc 2017;2: 9–23. doi: 10.1210/js.2017-00293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinsker JE, Rogers W, McLean S, Schaefer FV, Fenton C. Pseudohypoparathyroidism type 1a with congenital hypothyroidism. J Pediatr Endocrinol Metab 2006;19: 1049–52. doi: 10.1515/JPEM.2006.19.8.1049 [DOI] [PubMed] [Google Scholar]
- 10.Shoemaker AH, Jüppner H. Nonclassic features of pseudohypoparathyroidism type 1A. Curr Opin Endocrinol Diabetes Obes 2017;24: 33–8. doi: 10.1097/MED.0000000000000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiele S, Werner R, Grötzinger J, Brix B, Staedt P, Struve D, et al. A positive genotype-phenotype correlation in a large cohort of patients with pseudohypoparathyroidism type Ia and pseudo-pseudohypoparathyroidism and 33 newly identified mutations in the GNAS gene. Mol Genet Genomic Med 2015;3: 111–20. doi: 10.1002/mgg3.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salemi P, Skalamera Olson JM, Dickson LE, Germain-Lee EL. Ossifications in Albright hereditary osteodystrophy: role of genotype, inheritance, sex, age, hormonal status, and BMI. J Clin Endocrinol Metab 2018;103: 158–68. doi: 10.1210/jc.2017-00860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grüters-Kieslich A, Reyes M, Sharma A, Demirci C, DeClue TJ, Lankes E, et al. Early-onset obesity: unrecognized first evidence for GNAS mutations and methylation changes. J Clin Endocrinol Metab 2017;102: 2670–7. doi: 10.1210/jc.2017-00395 [DOI] [PMC free article] [PubMed] [Google Scholar]