Abstract
HMGA2, a pivotal transcription factor, functions as a versatile regulator implicated in the progression of diverse aggressive malignancies. In this study, mass spectrometry was employed to identify ubiquitin-specific proteases that potentially interact with HMGA2, and USP48 was identified as a deubiquitinating enzyme of HMGA2. The enforced expression of USP48 significantly increased HMGA2 protein levels by inhibiting its degradation, while the deprivation of USP48 promoted HMGA2 degradation, thereby suppressing tumor invasion and metastasis. We discovered that USP48 undergoes SUMOylation at lysine 258, which enhances its binding affinity to HMGA2. Through subsequent phenotypic screening of small molecules, we identified DUB-IN-2 as a remarkably potent pharmacological inhibitor of USP48. Interestingly, the small-molecule inhibitor targeting USP48 induces destabilization of HMGA2. Clinically, upregulation of USP48 or HMGA2 in cancerous tissues is indicative of poor prognosis for patients with colorectal cancer (CRC). Collectively, our study not only elucidates the regulatory mechanism of DUBs involved in HMGA2 stability and validates USP48 as a potential therapeutic target for CRC, but also identifies DUB-IN-2 as a potent inhibitor of USP48 and a promising candidate for CRC treatment.
Key words: Colorectal cancer, Invasion and metastasis, Post-translational modification, Ubiquitination, SUMOylation, USP48, HMGA2, Specific inhibitors
Graphical abstract
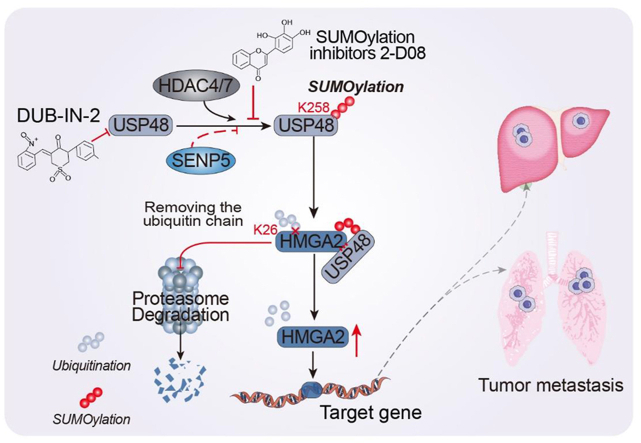
This study not only elucidates the regulatory mechanism of USP48 involved in HMGA2 stability, but also identifies DUB-IN-2 as a potent inhibitor of USP48 and a promising candidate for CRC treatment.
1. Introduction
Colorectal cancer (CRC) is a prevalent malignancy worldwide, with approximately 1.8 million cases diagnosed annually1. The primary cause of mortality in CRC is its metastatic potential, which occurs in 40% of cases2. Despite this, the molecular mechanisms underlying tumor metastasis remain incompletely understood. Therefore, it is crucial to decipher the precise molecular pathways driving CRC metastasis, particularly in high-risk individuals. Therefore, it is imperative to investigate the aggressive characteristics and identify potential molecular targets in CRC. Ubiquitination is a tightly regulated post-translational modification that directs proteins for degradation and modulates protein function. A substantial body of evidence indicates that ubiquitination plays a pivotal role in the pathogenesis of cancer, and targeting this process has emerged as an effective therapeutic strategy for various types of cancers, including CRC3, 4, 5, 6. Like other post-translational modifications, ubiquitin chains can be cleaved from substrate proteins by DUBs7. There are approximately 100 human DUBs that have been identified and classified into six categories: USPs, OTUs, JAMMs, MJDs, UCHs, and MCPIPs8,9. Dysregulation of deubiquitination occurs frequently in tumorigenesis. For instance, the pro-metastatic protein Snail1 is subject to deubiquitination by USP110, USP3711,12, USP9X13, and USP27X14, which enhances Snail1 stability and promotes tumorigenesis. USP2215, USP2816,17, USP3718, and OTUD6A19 directly deubiquitinate c-Myc, increasing tumorigenesis, while USP9X indirectly reduces c-Myc by stabilizing its E3 ligase Fbw7, thereby inhibiting CRC growth20.
HMGA2 is an architectural transcription factor composed of 109 amino acids and three AT-hooks, which are basic DNA-binding domains. By binding to the AT-rich regions of DNA through its AT-hooks, HMGA2 modulates transcription by altering chromatin structure, thereby enabling the transcriptional machinery to approach targets for activating or repressing gene expression in mammalian cells21,22. The expression of HMGA2 is significantly upregulated during tumorigenesis, while it is rarely detected in normal adult tissues23. Patients diagnosed with breast cancer24, CRC25,26, and lung cancer27,28 exhibit a higher likelihood of survival when presenting low levels of HMGA2, thereby indicating the pro-carcinogenic effects of this protein. Moreover, HMGA2 is implicated in DNA repair29, stem cell renewal30, tumor invasion and metastasis31, as well as differentiation32. By regulating multiple target genes, HMGA2 contributes to tumorigenesis. For instance, the binding of HMGA2 to the promoter region of STAT3 triggers its transcriptional activation, subsequently leading to the secretion of CCL2 that facilitates macrophage recruitment and promotes cancer progression25. HMGA2 interacts with MSI2 and Beclin1 to regulate autophagy, thereby inhibiting the growth of malignant peripheral nerve sheath tumors associated with NF133. Moreover, HMGA2 exerts direct activation on multiple pro-metastatic genes such as SNAI1, CXCR4 and SLUG34.
Additionally, the regulatory cascades governing HMGA2 expression during cancer progression have been a subject of interest among researchers. The Lin-28B-let-7-HMGA2 axis, coordinated by STAT3, is responsible for initiating epithelial-to-mesenchymal transition (EMT) in breast cancer cells35. It should be emphasized that HMGA2 function is significantly influenced by post-translational modifications, and phosphorylation at the C-terminus of the protein may exert an impact on its DNA binding ability36. Acetylation of HMGA2 at lysine 26 can augment its DNA-binding affinity34. Treatment with arsenic trioxide has been demonstrated to induce SUMOylation of HMGA2, resulting in the formation of nuclear foci around promyelocytic leukemia protein (PML) nuclear bodies and ultimately promoting PML degradation37. However, the current understanding of post-translational modifications in regulating HMGA2 expression in CRC remains limited.
Accumulating evidence suggests that DUBs play a critical role in maintaining cellular protein homeostasis and regulating cellular processes involved in the initiation and progression of cancer. The cellular processes regulated by USP48, an essential member of the USP subfamily, remain largely unexplored in current literatures. USP48 has been shown to be dysregulated in multiple types of malignancies and exhibits a unique ability to promote or suppress tumorigenesis in an environment-dependent manner. It is noteworthy that the overexpression of USP48 in glioblastoma has been demonstrated to play a crucial role in tumorigenesis, as it stabilizes Gli1 protein and activates Gli-dependent transcriptional regulation, thereby promoting disease progression38. In hepatocellular carcinoma, the expression of USP48 is down-regulated, which is regulated by Mettl14-induced m6A modification and subsequently stabilizes SIRT6 to attenuate HCC glycolysis and malignancy39. However, the precise role of USP48 in CRC biology remains elusive.
In this study, we have demonstrated that USP48 functions as a deubiquitinase for HMGA2, thereby suppressing its degradation and promoting CRC cell migration. Moreover, we have identified lysine 258 (Lys-258) SUMOylation as a critical factor in augmenting the ability of USP48 to modulate HMGA2. Additionally, DUB-IN-2 has been discovered as a promising USP48 inhibitor with great potential as a candidate therapeutic agent for CRC. In conclusion, our findings provide compelling evidence that USP48 plays a significant role in regulating the stability of HMGA2 in CRC, highlighting its potential as a novel target for DUB-based cancer therapy.
2. Methods
2.1. Cell lines and cell culture
SW480 and HEK293T cell lines were procured from the Chinese National Cell Line Resource Infrastructure (Beijing, China), while LOVO, HT29, HCT116, SW620, RKO, DLD1 and HCT8 cell lines were generously donated by Procell Life Science & Technology Co., Ltd. (Wuhan, China). The normal colon immortalized epithelial cell line NCM460 was obtained from INCELL (San Antonio, Texas, USA). The manufacturer's recommended culture conditions were employed for all human cell lines. STR DNA fingerprinting was performed to authenticate all cell lines.
2.2. Patient specimens
Samples of CRC tissue were obtained from the First Affiliated Hospital of the University of Science and Technology of China (USTC). None of the patients had undergone any preoperative chemotherapy or radiotherapy.
2.3. Plasmids
We constructed the HA-USP48 plasmid by cloning human USP48 cDNA into the pcDNA3.1-HA vector. Lentiviral vectors Plvx-Puro-HA expressing USP48 were generated through the cloning of USP48 cDNA, while lentiviral vectors Plvx-Puro-Myc expressing HMGA2 were generated by inserting HMGA2 cDNA into the Plvx-Puro-Myc lentiviral vector. Human HMGA2 cDNA was cloned into pcDNA3.1-Myc to generate the Myc-HMGA2 plasmid. To produce the GST-HMGA2 protein, we utilized pGEX-6P-1-GST and human cDNA. Our research team collaborated with our biotechnological support partner, Shuai Zhou, to synthesize or modify the following constructs: HA-USP48 and Myc-HMGA2, GST-HMGA2, USP48 ΔUSP, USP48 ΔDUSP, USP48 ΔUBL, Myc-HMGA2 D1, Myc-HMGA2 D2, Myc-HMGA2 D3, Myc-HMGA2 D4, USP48 shRNA #1, USP48 shRNA #2, USP48 shRNA #3, HMGA2 shRNA #1, HMGA2 shRNA #2, HMGA2 shRNA#3, His-Ub-WT, His-Ub(K6/K11/K27/K29/K33/K48/K63), V5-Ubc9, His-SUMO1/2/3, Myc-PIAS1/2/3/4, Myc-HDAC4/7, and Flag-SENP1/2/3/4/5/6/7. We performed site-directed mutagenesis to introduce specific point mutations. Myc-HMGA2 was utilized as a template to generate the K26R and K34R mutants, while HA-USP48 served as a template for generating the K258R, K359R, K468R, K898R, and K1003R mutants. The Myc-HMGA2 mutants with lysine substitutions at positions K26, K34, K46, K53, K56, K58, K62, K66, K67, K74, K82, K90 and K91 were procured from General Biol (Chuzhou, China) and all constructs generated in this study were all validated through DNA sequencing.
2.4. Antibodies and other reagents
Anti-HA, anti-DYKDDDDK, anti-Myc, and anti-His antibodies were provided by Cell Signaling Technology (CST, Danvers, MA, USA), while Santa Cruz Biotechnology (Dallas, TX, USA) was the source of the anti-USP48 antibody. The anti-USP48, anti-HMGA2, anti-Tubulin and anti-V5 antibodies were supplied by Proteintech (Wuhan), and the PTM BioLab (Hanzhou, China) provided the anti-Ub antibody. Anti-c-Myc, anti-His, and anti-HA agarose beads were supplied by Medchem Express (MCE) based in NJ, USA. 2-D08, MG132 and CQ were provided by SelleckChem located in Houston, TX, USA.
2.5. Immunoblotting (IB) and immunoprecipitation (IP) assays
We employed an IP lysis buffer (MCE) supplemented with a protease inhibitor cocktail (MCE) to lyse cells at 4 °C for 30 min. The resulting lysates were centrifuged, separated by SDS-PAGE, and subjected to IB analysis. For the detection of protein–protein interactions, cells were lysed in an IP solution. After incubating with specific antibodies 12 h at 4 °C on a revolving shaker, the supernatants were treated with MCE Protein A/G Magnetic Beads for 1 h at room temperature. The immunoprecipitates were washed five times using PBST buffer (PBST: 1 × PBS + 0.5% Tween-20, pH 7.4). Prior to IB analysis, the samples were boiled in 1 × SDS loading buffer and eluted. To detect HMGA2-associated proteins, approximately 4 × 108 SW480 cells overexpressing Myc-HMGA2 or Myc-vector were harvested and then lysed in an IP lysis buffer supplemented with a protease inhibitor cocktail. The cellular lysates were immunoprecipitated with anti-Myc antibody and protein A/G magnetic beads (MCE) at 4 °C for an appropriate duration. The experiment involved both MYC-tagged HMGA2 and Myc-vector control proteins. Following washing of the beads with wash buffers, the binding proteins were eluted for 10 min at room temperature using an elution buffer. After magnetic separation of the beads, residual supernatants containing the target antigen were immediately diluted with a neutralizing buffer (0.1 mol/L NaOH). The supernatants were subsequently heated, separated by SDS-PAGE, and subjected to Coomassie brilliant blue (CBB) staining. Mass spectrometry sequencing and data analysis were conducted following band excision. Approximately 4 × 108 SW480 cells overexpressing HA-USP48 or HA-vector were harvested in IP lysis buffer supplemented with cocktail for USP48-binding protein identification. IP of cell lysates, containing either HA-tagged USP48 or the HA vector control, was performed at 4 °C using anti-HA (CST) and protein A/G magnetic beads (MCE). The remaining steps followed a similar experimental process used to detect HMGA2-related proteins.
2.6. Mass spectrometry analysis
HCT116 cells were transfected with Myc-HMGA2 and subsequently lysed. The resulting lysate was subjected to IP using anti-Myc affinity magnetic beads. The immunoprecipitated proteins were subjected to protein electrophoresis, followed by Coomassie blue staining. The entire bands were then excised for in-gel trypsin digestion and subsequent drying. Mass spectrometry analysis was performed to identify the protein composition of the samples (PTM BioLab). Similarly, HA-USP48 was transfected into HCT116 cells and lysed cells were subjected to IP using anti-HA affinity magnetic beads. The protein composition of the immunoprecipitates was determined by mass spectrometry following the same procedure as mentioned above (APTBIO, Shanghai, China).
2.7. qRT-PCR
Total RNA was extracted using TRIzol reagent (Takara, Dalian, China) and subjected to qRT-PCR. One microgram of RNA was reverse-transcribed using a PrimeScript™ RT reagent kit (Takara). The expression levels of the target genes were normalized to GAPDH. A list of qRT-PCR primers available is provided in Supporting Information Table S1.
2.8. Stable cell lines and lentiviral vectors
Lentiviral vectors were generated by co-transfecting HEK293T cells with helper and lentiviral plasmids. PsPAX2, Rev, and pMD2G were used to create lentiviral vectors for USP48 and HMGA2. After transfection, the virus-containing medium was collected and centrifuged for 6 min at 1.2 × 104 rpm. The collected medium was then filtered using 0.45 μm Millipore filters and collected at 1.2 × 104 rpm. Following concentration, selected cells were infected with the lentiviral particles for 20–24 h and treated with antibiotics and 8 μg/mL polybrene (Biosharp, Hefei, China) for one week. Stable knockdown was achieved by infecting cells with lentiviral particles expressing shRNAs, followed by puromycin selection. The sequences of the shRNA used in the experiment were listed in Supporting Information Table S2.
2.9. Immunofluorescence
Immunofluorescence was performed by growing HEK293T cells and CRC cells transfected with the corresponding plasmids on a confocal plate. The cells were then washed, fixed, permeabilized, and blocked according to the manufacturer's instructions. Following this, appropriate antibodies as identified above were used for staining and incubated with secondary antibodies coupled with fluorescent dyes. DAPI (Sigma–Aldrich, St. Louis, MO, USA) was utilized for counterstaining the nuclei.
2.10. Transwell assay and wound-healing assay
After transfection for 24 h, a total of 5 × 104 CRC cells were collected and seeded into the upper chambers of 24-well Transwells. For invasion assays, the upper chambers were coated with Matrigel (Sigma–Aldrich). The lower chambers were filled with medium containing 15% fetal bovine serum (FBS, Procell). Following incubation at 37 °C for 20 h, the cells adhered to the upper surface of the membrane were gently removed using a cotton swab. The migrated cells were fixed with 4% formaldehyde and subsequently stained using 0.1% crystal violet. Additionally, a wound-healing assay was conducted by seeding transfected CRC cells in 6-well plates and incubating them 24 h. Scratch wounds were generated by scraping the cell layer using a sterile 200 μL pipette tip. After culturing for 48 h in media supplemented with 0.5% FBS, the cells were observed under a microscope.
2.11. Recombinant protein purification
HEK293T cells were transfected with HA-labelled USP48 or its mutant HA-labelled USP48–C98S. Subsequently, cell lysates were added to the culture dishes of the transfected cells at a temperature of 4 °C with an incubation time of 30 min. Sonication disrupts the cell membrane and releases the proteins into solution, then the lysate is centrifuged at 4 °C for approximately 15 min to remove any insoluble debris. Pre-equilibrated MCE anti-HA beads are added to the supernatant and then washed with Equilibration/Wash Buffer to remove impurities. The purified HA-tagged USP48-WT/USP48-C98S protein was eluted from the magnetic beads with elution buffer and washed twice. The purified recombinant proteins were subsequently analysed. To express proteins in bacteria, E. coli strain BL21 cells (General Biol) containing the GST and GST-HMGA2 plasmids were induced with 0.5 mmol/L isopropyl-β-d-1-1-thiogalactopyranoside at 37 °C for 16 h. The cells were lysed and sonicated in a lysis buffer (50 mmol/L Tris-HCl, pH 7.5, 0.5% Triton X-100, 200 mmol/L NaCl, 10% glycerol, 1 mmol/L dithiothreitol (DTT), and 1 mmol/L phenylmethylsulphonyl fluoride). After centrifugation, the lysates were incubated with glutathione sepharose 4B (GE Healthcare) at 4 °C for 4 h. The resin was washed three to five times with lysis buffer containing 300 mmol/L NaCl, followed by two washes with phosphate-buffered saline (PBS). The protein bound to glutathione sepharose beads was confirmed using SDS-PAGE and then aliquoted for long-term storage at −80 °C. The GST-HMGA2 protein was ultimately eluted using a solution containing 50 mmol/L Tris-HCl (pH 8.0), 10% glycerol, and 1 mmol/L DTT supplemented with reduced glutathione.
2.12. GST pull-down assay
Purified GST-HMGA2 or GST-tag was obtained from E. coli strain BL21 using GST agarose beads. HA-USP48, purified from HEK293T cells, was incubated with either GST or GST-HMGA2 coupled to the aforementioned beads at 4 °C for 5 h. IB analysis was performed after washing the beads thrice with PBS.
2.13. Deubiquitination of HMGA2 in vivo and in vitro
The in vivo deubiquitination of HMGA2 was investigated in HEK293T, DLD1, SW480, and HCT116 cells that were transfected with 10 μmol/L of MG132. The cells were lysed using RIPA buffer (Servicebio, Wuhan) containing protease inhibitors and subsequently treated with specified primary antibodies. In vitro deubiquitination assays were conducted using Myc-HMGA2 and His-Ub expressed by transfected HEK293T cells. After 48 h, HMGA2 was enriched using anti-Myc affinity magnetic beads. HA-USP48 and USP48-C98S were expressed and purified with anti-HA affinity magnetic beads in HEK293T cells. The purified proteins were eluted with a concentration of 150 ng/mL of HA peptide, followed by incubation with Myc-HMGA2 in a deubiquitination buffer (50 mmol/L Tris-HCl, pH 7.4, 1 mmol/L MgCl2, and 1 mmol/L DTT) for 2 h at 37 °C. The Myc-bound beads were washed thrice with PBS before being subjected to IB analysis.
2.14. In vitro DUB activity assay
Purified HA-USP48 (0.05 μg/μL) was pre-incubated with either 5 μmol/L DUB-IN-2 or Spautin-1 for 10 min, followed by combination with K48-linked Di-ubiquitin (U2701, KS-V Peptide, Hefei) at a concentration of 0.1 μg/μL in the presence of the compound and incubation in a DUB reaction buffer (50 mmol/L HEPES, pH 7.5, 100 mmol/L NaCl, 2 mmol/L TCEP) at 37 °C for 1.5 h. The reaction was terminated by adding 2 × Tricine sample buffer (Bio-Rad, Hercules, CA, USA) and incubating at 40 °C for 20 min. The samples were subsequently loaded onto a Tris-Tricine gel (Bio-Rad) for immunoblotting analysis using a specific antibody against ubiquitin.
2.15. In vivo SUMOylation analysis
To investigate the SUMOylation of USP48, we employed two distinct denaturation protocols. (1) SDS denaturation method: Cells in 6 cm dishes were lysed using 500 μL SDS-A buffer (100 mmol/L Tris-HCl, pH 6.8, 1% SDS, and 10% glycerol), freshly supplemented with 1 mmol/L PMSF, 2 μg/mL aprotinin and 20 mmol/L NEM. The lysates were then boiled for 10 min before being diluted seven-fold with SDS-B buffer (100 mmol/L Tris-HCl, pH 6.8, and 10% glycerol) containing fresh additions of PMSF (1 mmol/L), aprotinin (2 μg/mL) and NEM (20 mmol/L). Subsequently, the samples underwent sonication and centrifugation (1.5 × 104×g, 15 min, 4 °C) to eliminate debris. The resulting lysates were subjected to immunoprecipitation using either His or HA antibody and Protein A beads. Following this step, the beads were washed thrice with SDS-B buffer before being eluted in 2 × SDS loading buffer. (2) Guanidine/urea denaturation method: The experimental procedure in this section was strictly conducted in accordance with the operating manual of HisPur™ Ni-NTA magnetic beads (Thermo Fisher Scientific). The indicated plasmids were transfected into cells in 10 cm dishes. After 48 h, the cells were lysed using 1 mL of Pierce IP lysis buffer (Thermo Fisher Scientific) to prepare protein extracts. Reconstitute the protein extract by diluting it with an equal volume of equilibration buffer (100 mmol/L Na3PO4, 600 mmol/L NaCl, 6 mol/L guanidine-HCl, 0.05% Tween-20, 30 mmol/L imidazole, pH 8.0). The sample prepared in the preceding step was combined with 2 mg of HisPur™ Ni-NTA magnetic beads that had been washed with equilibration buffer and subsequently subjected to vortexing for 10 s followed by mixing on an end-over-end rotator for a duration of 30 min. Collect the beads and add 400 μL of wash buffer (100 mmol/L Na3PO4, 600 mmol/L NaCl, 6 mol/L guanidine-HCl, 0.05% Tween-20, 50 mmol/L imidazole, pH 8.0) to the tube. Vortex for 10 s to mix and repeat twice. Collect the beads again and add 100 μL of elution buffer (100 mmol/L Na3PO4, 600 mmol/L NaCl, 6 mol/L guanidine-HCl, 250 mmol/L imidazole, pH 8.0). Vortex for another15 s. Incubate the beads on a rotating platform for 15 min, followed by collection of the beads using a magnetic stand. Carefully remove and preserve the supernatant containing the His-tagged protein.
2.16. Immunohistochemistry (IHC)
The CRC tissue microarrays (TMAs) were procured from Shanghai Outdo Biotech Co. Ltd., comprising paired cancerous and adjacent normal tissues. Immunohistochemical staining of HMGA2 and USP48 was carried out on paraffin-embedded tissue blocks utilized for clinical diagnosis, employing the heat-induced epitope retrieval method and following a standardized protocol. Subsequently, a semiquantitative analysis was conducted by assigning scores to both the intensity and proportion of staining. The intensity score ranged from 0 (none) to 3 (strong), while the proportion score ranged from 1 (25%) to 4 (>75%). Two pathologists independently evaluated the IHC staining. The H-Scores of USP48 or HMGA2 were determined by multiplying the intensity and extent scores (0–12) of stained cells. Low expression was defined as H-Scores <6, while high expression was defined as H-Scores ≥6.
2.17. Animal models
BALB/c nude mice for each experimental group were procured from GemPharmatech Co., Ltd. (Nanjing, China). A liver metastasis model was established in 4-week-old BALB/C mice by injecting 1 × 106 cell suspensions, dissolved in 40 μL PBS, into the subcapsular spleen. The mice were euthanized 6 weeks post-injection. Four-week-old BALB/c nude mice were injected intravenously with 2 × 106 cells in 150 μL PBS suspension to establish a lung metastasis model. The mice were sacrificed 6 weeks after tumor cell injection. Excised lungs and livers were fixed in phosphate buffered formalin and then subjected to tomographic examination with HE staining and panoramic scanning. Finally, metastatic nodules in the lung and liver were carefully examined. Experimental procedure in vivo: After 7 days of the injection, the mice were treated intraperitoneally with 2-D08 (4 mg/kg) or DUBs-IN-2 (1 mg/kg) every 2 days for 35 days. The solvents used for 2-D08 were 5% DMSO, 40% PEG300, and 55% saline. The solvents used for DUBs-IN-2 were 50% PEG300 and 50% saline as recommended by the manual.
2.18. Proximity ligation assay
The proximity ligation assay (PLA) analysis was conducted in accordance with previously established protocols40. HEK293T cells were cultured on a Biosharp confocal dish for no less than 18 h before being subjected to two rounds of PBS washing and fixed in 4% formaldehyde PBS solution at room temperature for 15 min. The resulting samples were then rinsed with TBST. The dishes were subsequently blocked with a BSA blocking solution at 37 °C for 1.5 h, followed by 24 h incubation with antibody combinations at 4 °C. After washing with TBST, proximity ligation was performed using Rabbit PLUS and Mouse MINUS Duolink in situ PLA kits (Sigma–Aldrich) according to the manufacturer's instructions. The dishes were subsequently immersed in DAPI solution for nuclear restaining. The images were analyzed using Zeiss confocal software and captured by the Zeiss LSM710 confocal microscope system (Carl Zeiss GmbH, Jena, Germany).
2.19. Statistics
For comparisons between two independent groups, a Student's t-test (two-tailed) was employed unless otherwise specified. The correlation between USP48 and HMGA2 was analyzed statistically using the chi-square test and linear regression analysis. The Kaplan–Meier survival curves were evaluated using a log-rank test. Statistical significance was defined as a P-value of 0.05 or less. The quantification was conducted using GraphPad Prism 8 or Excel 2019. The IB results presented in the figures were obtained from three independent experiments with consistent outcomes, unless otherwise specified in the legends.
2.20. Study approval
The collection and analysis of all tumors were approved by the research ethics committee at the First Affiliated Hospital of USTC (2021-KY-087), and all experiments were conducted in accordance with the Helsinki Declaration of 1975. All animal experiments were performed with approval from The Institutional Animal Care and Use Committee of The First Affiliated Hospital of USTC (202106091541000573097), adhering to the Guide for the Care and Use of Laboratory Animals.
2.21. Data availability
The complete dataset can be accessed through the article, and Supporting Information.
3. Results
3.1. USP48 interacts with HMGA2
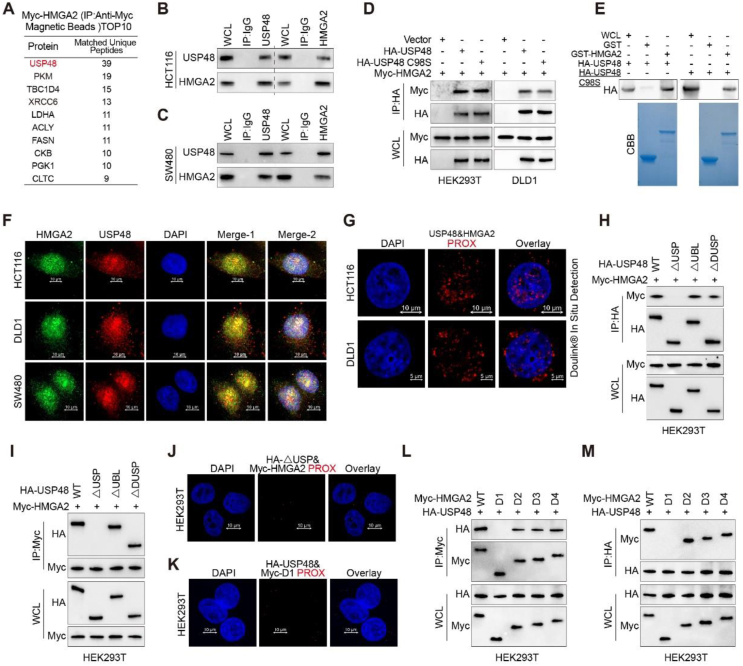
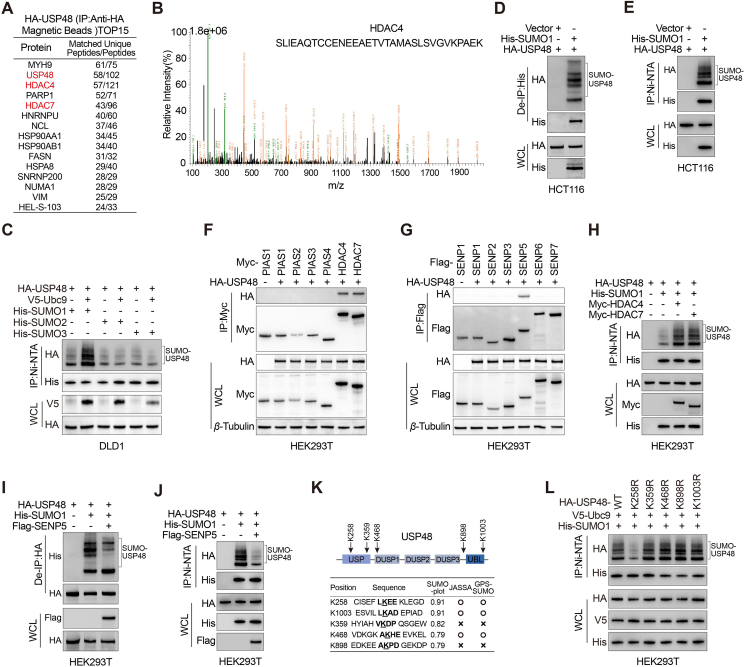
We established a HCT116 cell line stably expressing Myc-HMGA2, and subsequently purified whole-cell extracts from these cells using anti-Myc affinity magnetic beads for mass spectrometry analysis to identify potential deubiquitinating enzymes responsible for stabilizing HMGA2. Mass spectrometry revealed the presence of USP48 in the HMGA2 immunoprecipitates with high peptide coverage (Fig. 1A, Supporting Information Table S3 and Fig. S1A). Co-immunoprecipitation (Co-IP) assays were used to investigate the interaction between USP48 and HMGA2 in vivo. Ectopically expressed Myc-tagged HMGA2 and HA-tagged USP48 were detected in each other's immunoprecipitates (Fig. S1B). Endogenous USP48 was also found to interact with endogenous HMGA2 in SW480, HCT116, and DLD1 cells (Fig. 1B, C and Fig. S1C). Co-IP assays further demonstrated that Myc-tagged HMGA2 could be detected in immunoprecipitates of either HA-USP48 or its catalytically inactive mutant C98S (HA-USP48 C98S) in HEK293T and DLD1 cells (Fig. 1D), indicating that deubiquitinating activity of USP48 is not required for this interaction. Moreover, purified GST-HMGA2 exhibited the ability to directly interact with HA-tagged USP48 under cell-free conditions, as opposed to GST control (Fig. 1E), indicating a direct binding between HMGA2 and USP48. Immunofluorescence staining revealed co-localization of endogenous HMGA2 and USP48 primarily within the nucleus of CRC cells (Fig. 1F). Furthermore, the interaction between endogenous USP48 and HMGA2 was validated through an in-situ proximity ligation assay (PLA) in DLD1, SW480, and HCT116 cells (Fig. 1G and Fig. S1D). In addition, exogenous HA-USP48 and Myc-HMGA2 proteins were also confirmed to interact with each other in HEK293T cells (Fig. S1E and F). Truncating mutation analysis revealed that the USP domain of USP48 is essential for its interaction with HMGA2 (Fig. 1H‒J and Fig. S1G). We also observed that the binding of HMGA2 D1 to USP48 is absent, indicating the indispensability of the N-terminus (amino acids 1–43) for its interaction with USP48 (Fig. 1K‒M). These findings suggest a specific interaction between USP48 and HMGA2, indicating that USP48 directly binds to HMGA2.
Figure 1.
USP48 interacts with HMGA2. (A) HMGA2-interacting proteins were detected by mass spectrometry using anti-Myc affinity magnetic beads, following transfection of Myc-HMGA2 into HCT116 cells for 36 h. (B–C) IP assays were performed on HCT116 and SW480 cell lysates using control IgG, anti-USP48, or anti-HMGA2 antibodies, followed by detection of resulting immunoprecipitates with appropriate antibodies. (D) HEK293T and DLD1 cells were transfected with Myc-HMGA2 alone or in combination with HA-tagged USP48-WT or USP48 CS, and cell lysates were analyzed by IP with anti-HA affinity magnetic beads followed by IB with antibodies against Myc and HA. (E) Purified HA-USP48 or HA-USP48 C98S from HEK293T cells was incubated with purified recombinant GST-HMGA2 or GST, respectively. The retained USP48 or USP48 C98S on sepharose was detected using the HA antibody. (F) HCT116, DLD1 and SW480 cells were stained with USP48 antibody (red) and HMGA2 antibody (green), then analyzed using confocal microscopy. Nuclei were counterstained with DAPI (blue). Scale bar, 10 μm. (G) In situ PLA was used to detect the interaction between endogenous USP48 and HMGA2 in HCT116 and DLD1 cells. Representative merged PLA and nuclei (DAPI) images from the experiments are shown, with upper panel scaled at 10 μm and lower panel at 5 μm. (H) HEK293T cells were co-transfected with Myc-HMGA2 and either full-length HA-tagged USP48 or its deletion mutants, followed by IP using anti-HA magnetic beads and IB with antibodies against HA and Myc. (I) HEK293T cells were co-transfected with Myc-HMGA2 and either full-length HA-tagged USP48 or its deletion mutants, followed by IP using anti-Myc magnetic beads and IB with antibodies against HA and Myc. (J) In situ PLA was employed to investigate the interaction between exogenous Myc-HMGA2 WT and HA-USP48 △USP in HEK293T cells, with a scale bar of 10 μm. (K) In situ PLA was employed to investigate the interaction between exogenous HA-USP48 WT and Myc-HMGA2 D1 in HEK293T cells, with a scale bar of 10 μm. (L) HEK293T cells were co-transfected with HA-USP48 and either full-length Myc-tagged HMGA2 or its deletion mutants, followed by IP using anti-Myc magnetic beads and IB with antibodies against HA and Myc. (M) HEK293T cells were co-transfected with HA-USP48 and either full-length Myc-tagged HMGA2 or its deletion mutants, followed by IP using anti-HA magnetic beads and IB with antibodies against HA and Myc. The presented panels depict representative outcomes from three independent experiments. The panels presented illustrate representative results obtained from three independent experiments.
3.2. USP48 maintains HMGA2 stability
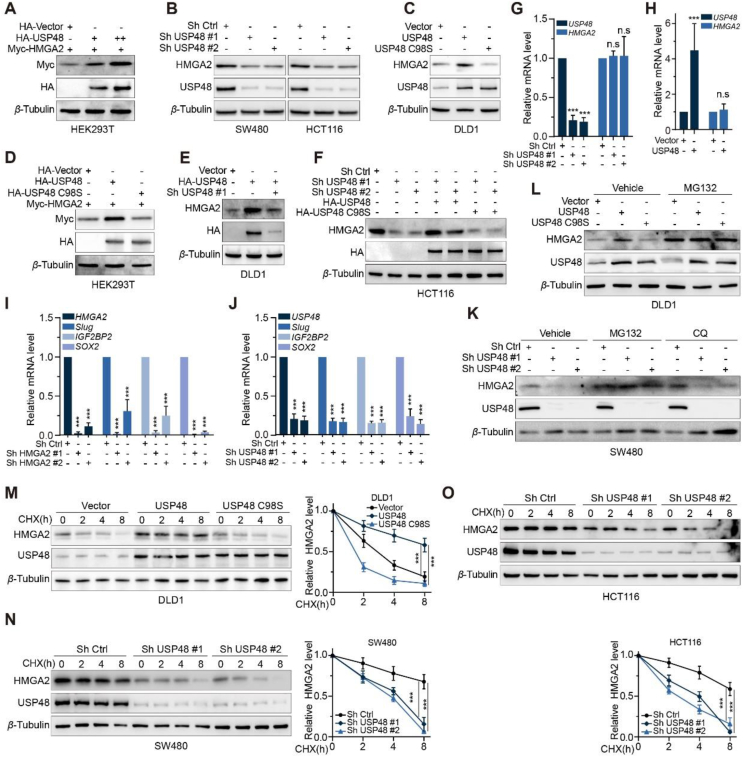
We have identified an interaction between USP48 and HMGA2, and our study further investigated the involvement of USP48 in maintaining the stability of HMGA2. To this end, we transfected HEK293T cells with HA-tagged USP48 and Myc-tagged HMGA2. Our findings indicate that USP48 expression enhances HMGA2 protein expression in a dose-dependent manner (Fig. 2A). Additionally, overexpression of USP48 in DLD1 cells leads to a dose-dependent increase in HMGA2 expression (Supporting Information Fig. S2A). Conversely, depletion of the USP48 gene in CRC cells led to a reduction in HMGA2 protein levels (Fig. 2B and Fig. S2B).
Figure 2.
USP48 maintains HMGA2 stability. (A) Increasing amounts of HA tagged USP48 were transfected into HEK293T cells along with equal amounts of Myc-HMGA2. Cell lysates were analyzed using IB and antibodies against Myc. “+” denotes 2.5 μg plasmid transfection, while “++” indicates 5 μg. (B) Two independent shRNAs were employed to silence USP48 expression in SW480 and HCT116 cells, followed by IB analysis of HMGA2 protein levels. (C) USP48-WT or USP48–C98S was overexpressed in DLD1 cells, followed by analysis of HMGA2 protein levels. (D) Equal amounts of Myc-HMGA2 and either USP48-WT or USP48–C98S were co-transfected into HEK293T cells, followed by IB analysis using Myc antibodies. (E) DLD1 cells were transduced with HA-tagged USP48-WT alone or in combination with USP48 shRNA, and HMGA2 levels were analyzed by IB analysis. (F) HCT116 cells were transduced with USP48 shRNA and either HA-tagged USP48-WT or USP48–C98S, followed by IB analysis to analyze HMGA2 levels. (G) qRT-PCR analysis was conducted to assess the expression of HMGA2 mRNA in SW480 cells with depleted endogenous USP48 via two independent shRNA. (H) The expression of HMGA2 mRNA was assessed by qRT-PCR in DLD1 cells following transduction with either vector control or HA-USP48. (I) qRT-PCR was performed to evaluate the expression levels of Slug, IGF2BP2 and SOX2 in SW480 cells transduced with two distinct HMGA2 shRNA constructs. (J) The expression levels of Slug, IGF2BP2 and SOX2 were quantified by qRT-PCR in SW480 cells that had been transduced with USP48 shRNA. (K) SW480 cells transfected with two independent USP48 shRNA were treated with DMSO, MG132 (10 μmol/L), or CQ (20 mmol/L) for 6 h, followed by IB analysis of USP48 and HMGA2. (L) DLD1 cells transfected with vector control, or HA-USP48 were treated with DMSO or MG132 (10 μmol/L) for 6 h, and then USP48 and HMGA2 were analyzed. (M) DLD1 cells were transfected with vector control, HA-USP48 or USP48-C98S, treated with 50 μg/mL of CHX, collected at the indicated times, and then subjected to IB analysis with antibodies against USP48 and HMGA2. Quantification of HMGA2 levels relative to β-Tubulin is shown. (N) SW480 cells with stable expression of either control shRNA or USP48 shRNA were treated with 50 μg/mL CHX, harvested at indicated time points, and analyzed by IB using antibodies against HMGA2 and USP48. Quantification of HMGA2 levels relative to β-Tubulin is presented as mean ± SD. (O) HCT116 stably expressing control shRNA or USP48 shRNA were treated with 50 μg/mL CHX, harvested at the indicated times, and then subjected to IB with antibodies against HMGA2 and USP48. Quantification of HMGA2 levels relative to β-Tubulin is shown. One-way ANOVA test (G, I, J, M‒O). Unpaired two-tailed Student's t-test (H). All experiments were performed independently at least three times. ∗∗∗P < 0.001, ns indicates no statistical significance.
To investigate whether the deubiquitinating activity of USP48 is responsible for the increased levels of HMGA2 protein, we overexpressed wild-type (WT) and mutant (USP48-C98S) forms of USP48 in DLD1 and HEK293T cells. Our findings demonstrate that only the expression of USP48-WT led to an increase in HMGA2 levels, while USP48-C98S had no effect on protein abundance (Fig. 2C and D). Moreover, the co-expression of USP48 shRNA counteracted the elevated endogenous HMGA2 levels induced by USP48 overexpression in both HEK293T and DLD1 cells (Fig. 2E and Fig. S2C). We observed a significant reduction in HMGA2 protein expression upon USP48 depletion, which could be almost completely restored by overexpressing USP48-WT but not USP48-C98S (Fig. 2F). Interestingly, no alterations in HMGA2 mRNA levels were observed upon either USP48 overexpression or knockdown (Fig. 2G and H). Consistent with our findings, there was a positive correlation between HMGA2 protein expression in CRC cell lines and USP48 protein expression (Fig. S2D and E). We subsequently investigated the expression of three HMGA2 target genes that had been previously reported, and observed a significant downregulation of Slug, IGF2BP2, and SOX2 upon knockdown of HMGA2 in SW480 cells (Fig. 2I and Fig. S2F). Similarly, knockdown of USP48 also resulted in reduced mRNA levels of Slug, IGF2BP2, and SOX2 in SW480 cells (Fig. 2J).
We further investigated the role of USP48 in HMGA2 protein stability by examining two major pathways for cellular protein degradation: the ubiquitin‒proteasome system and autophagy-lysosome pathway41. We observed that the depletion of USP48 resulted in a reduction of HMGA2 expression, which was almost completely restored by the addition of MG132 but not chloroquine (CQ) (Fig. 2K and Fig. S2G). The expression of HMGA2 was significantly diminished in DLD1 cells stably expressing Vector or USP48-C98S, as compared to those expressing USP48-WT. However, this reduction was nearly completely reversed upon treatment with MG132 (Fig. 2L). We employed cycloheximide (CHX) to halt protein synthesis and demonstrated that USP48 modulated the stability of HMGA2. Our findings revealed that enforced expression of USP48-WT, but not USP48-C98S, led to increased stability of HMGA2 protein (Fig. 2M). Conversely, knockdown of USP48 in SW480 and HCT116 cells resulted in destabilization of HMGA2 protein (Fig. 2N and O). Our findings collectively indicate that USP48 specifically regulates the stability of HMGA2 protein, rather than affecting HMGA2 mRNA levels.
3.3. USP48 deubiquitinates HMGA2
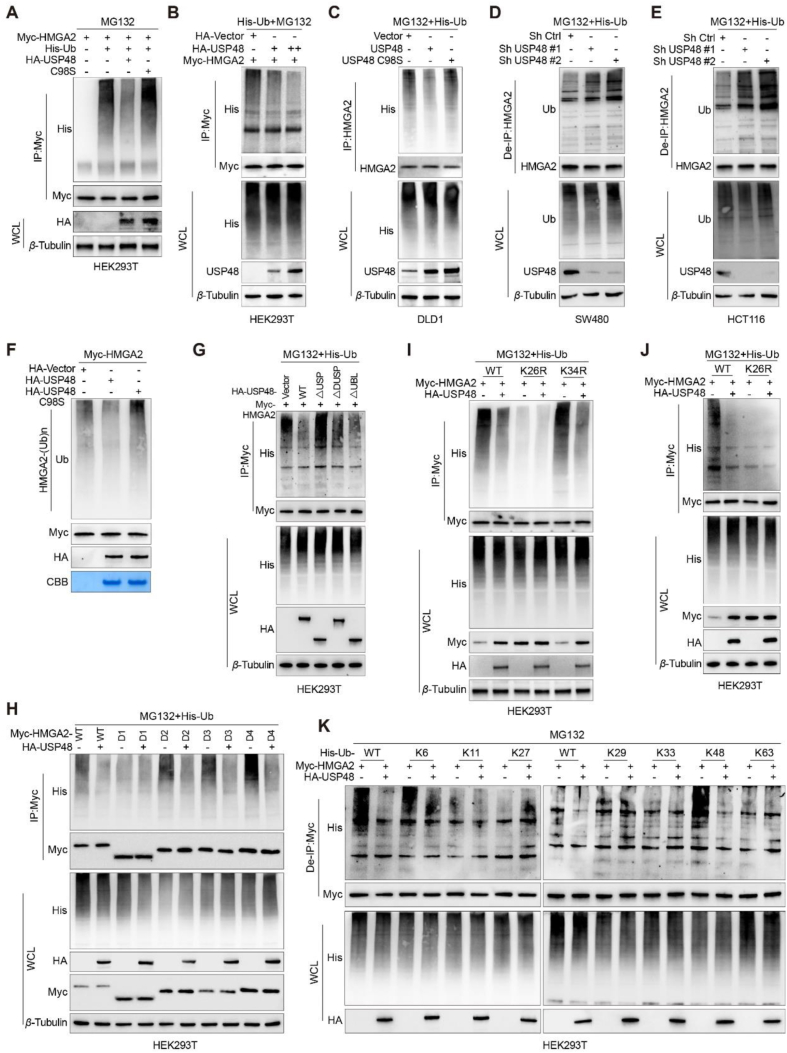
We examined whether USP48 mediates the deubiquitination of HMGA2 by co-transfecting Myc-HMGA2 and His-ubiquitin into HEK293T cells with either wild-type or C98S mutant USP48. Our findings demonstrate that USP48-WT significantly attenuates the ubiquitination of HMGA2, whereas USP48-C98S exerts no effect (Fig. 3A). Furthermore, we observed a dose-dependent impact of USP48-WT on the ubiquitination of HMGA2 in HEK293T cells (Fig. 3B). Moreover, the ectopic expression of USP48-WT resulted in a decrease in endogenous HMGA2 polyubiquitination in DLD1 cells (Fig. 3C), while the downregulation of USP48 led to an increase in HMGA2 polyubiquitination in SW480 and HCT116 cells (Fig. 3D and E). Our in vitro deubiquitination assays have further validated that USP48 specifically removes ubiquitin chains on HMGA2, but not on USP48-C98S mutant (Fig. 3F). Additionally, we have investigated the contribution of both the USP domain and C98 residue to the deubiquitination activity of USP48. Our findings indicate that the USP domain is indispensable for USP48's function as a deubiquitinase, and removal of this domain results in the loss of USP48's ability to regulate HMGA2 ubiquitination (Fig. 3G). Furthermore, the co-expression of HMGA2 truncation mutants and His-ubiquitin with USP48-WT in HEK293T cells has demonstrated that the N-terminal region of HMGA2 harbors potential sites for deubiquitination by USP48 (Fig. 3H). We constructed mutant plasmids that mutated two lysine residues (K26R and K34R) of the N-terminal region of HMGA2 to arginine one by one. At the same time, we also constructed a mutant plasmid with each lysine retained one by one (K26, lysine 26 retained, all other lysines were mutated to arginine), and all other lysines were mutated to arginine. The analysis of site-directed mutagenesis further confirms the critical role of Lys26 in the deubiquitination process of HMGA2 by USP48 (Fig. 3I and Supporting Information Fig. S3A), and the mutated form of HMGA2-K26R was not deubiquitinated by USP48 (Fig. 3J). Protein degradation via the proteasome is primarily initiated by Lys48-linked ubiquitin chains, whereas Lys63-linked ubiquitin chains are predominantly responsible for other cellular processes that do not necessitate proteasome-mediated degradation42. To identify the specific ubiquitination chains targeted by USP48 on HMGA2, we employed all seven lysine-specific mutants of ubiquitin. Our findings demonstrate that USP48 selectively deubiquitinates K6-linked and K48-linked ubiquitination chains attached to K26 residue of the HMGA2 protein (Fig. 3K and Fig. S3B). Our findings demonstrate that USP48 acts as a deubiquitinase and is essential for regulating the ubiquitination process of HMGA2. Specifically, it removes K6 and K48-linked ubiquitin chains from position K26 on the HMGA2 protein.
Figure 3.
USP48 deubiquitinates HMGA2. (A) HEK293T cells were co-transfected with Myc-HMGA2, His-Ub, and either HA-tagged USP48-WT or USP48-C98S. Cell lysates were then subjected to IP using anti-Myc magnetic beads followed by IB analysis with antibodies against His, HA, and Myc. Prior to harvesting, the cells were treated with 10 μmol/L MG132 for 6 h. (B) Increasing amounts of HA-tagged USP48 and equal amounts of Myc-HMGA2 were transfected into the cells, followed by IP with Myc antibodies and IB analysis with anti-His antibodies to detect ubiquitinated HMGA2 after treatment with MG132 for 6 h. (C) DLD1 cells were transfected with USP48-WT or USP48-C98S. After 48 h, the cells were treated with MG132 for 6 h. The whole-cell lysates were subjected to IP with HMGA2 antibodies and IB analysis with anti-Ub antibodies to detect ubiquitinated HMGA2. (D) SW480 cells were transfected with shRNA as indicated, and lysates were subjected to IP using HMGA2 antibody, followed by IB analysis with antibodies against Ub and HMGA2. (E) HCT116 cells were transfected with shRNA as indicated, and lysates were subjected to IP using HMGA2 antibody, followed by IB analysis with antibodies against Ub and HMGA2. Cells were treated with 10 μmol/L MG132 for 6 h prior to harvesting. Before IP, lysates were denatured at 95 °C for 5 min in the presence of 1% SDS, then diluted tenfold with lysis buffer and sonicated. (F) The ubiquitinated Myc-HMGA2 protein was subjected to in vitro deubiquitination assay using USP48-WT or USP48-C98S, and the reaction mixes were analyzed by IB analysis. (G) Myc-HMGA2 was co-transfected with either USP48-WT or USP48 deletion mutants into HEK293T cells. Whole-cell lysates were subjected to IP using Myc antibody, followed by IB analysis with anti-His antibodies after treatment with MG132 for 6 h to detect ubiquitinated HMGA2. (H) De-ubiquitination assay of HMGA2 WT and mutant domains (D1‒D4) was performed in HEK293T cells co-transfected with His-Ub and HA-USP48, followed by treatment with 10 μmol/L MG132 for 6 h. (I) De-ubiquitination assay of HMGA2 in HEK293T cells co-transfected with His-Ub, HA-USP48, Myc-HMGA2-K26R, and Myc-HMGA2-K34R and treated with 10 μmol/L MG132 for 6 h. (J) De-ubiquitination assay of HMGA2 in HEK293T cells co-transfected with His-Ub, HA-USP48, Myc-HMGA2-K26R and treated with 10 μmol/L MG132 for 6 h. (K) Myc-HMGA2, HA-USP48 and His-Ub plasmids with different ubiquitin linkages (K6, K11, K27, K29, K33, K48 or K63) were co-transfected into HEK293T cells to analyze HMGA2 ubiquitination. The data shown in all panels are representative of three independent experiments.
3.4. Ablation of USP48 expression impairs the metastatic potential of CRC cells
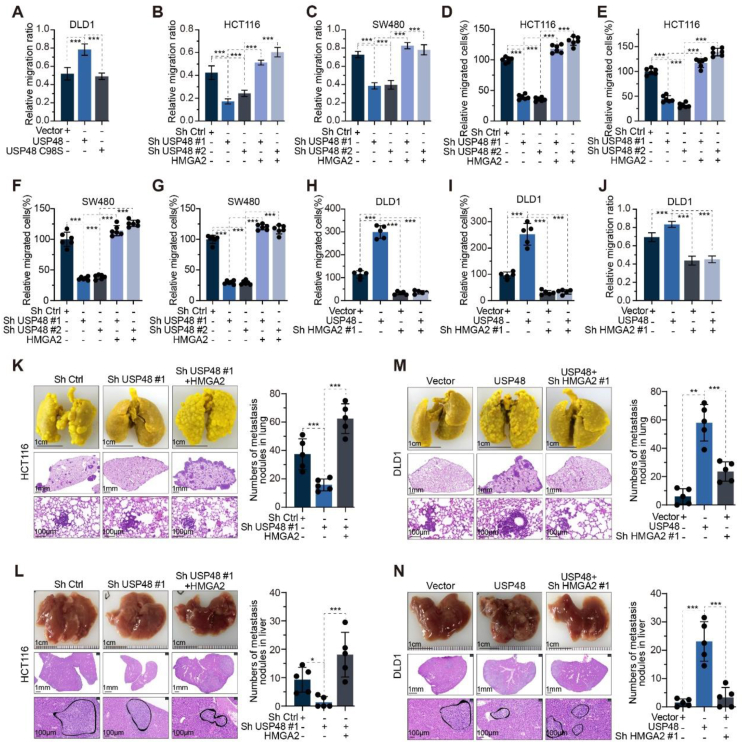
Our research findings prompted us to investigate whether USP48 facilitates invasion and metastasis in CRC. As anticipated, USP48-WT, but not USP48-C98S, augmented the metastatic and invasive potential of CRC cells (Fig. 4A and Supporting Information Fig. S4A and S4B). Previous studies have suggested that HMGA2 plays a pivotal role in the development of CRC25,43. Therefore, we investigated whether USP48 regulates HMGA2 to promote invasion and metastasis of CRC cells. To achieve this goal, we overexpressed HMGA2 in SW480 and HCT116 cells with or without knockdown of USP48. The inhibition of CRC cell migration and invasion was significantly observed upon the knockdown of USP48, while the metastatic and invasive phenotype induced by USP48 knockdown was reversed through re-expression of HMGA2 (Fig. 4B‒G and Fig. S4C‒G). Furthermore, knockdown of HMGA2 in DLD1 cells with and without USP48 overexpression was conducted. The results showed that USP48 overexpression significantly enhanced the migration and invasion abilities of CRC cells, while blocking HMGA2 expression reversed the phenotype caused by USP48 overexpression (Fig. 4H‒J and Fig. S4H‒J). Our in vitro experiments have demonstrated the pivotal role of USP48 in facilitating CRC invasion and migration. Consequently, we proceeded to investigate the function of USP48 in promoting CRC metastasis in vivo.
Figure 4.
Ablation of USP48 expression impairs the metastatic potential of CRC cells. (A) Wound-healing assay of DLD1 cells transfected with USP48-WT, USP48-C98S, or vector control. Migration rates in each group were quantified by measuring four different wound areas. (B–G) USP48 knockdown was performed in HCT116 and SW480 cells, with or without HMGA2 overexpression, followed by quantification of migration rates through measurement of five distinct wound areas (B, C). The relative number of migrated cells in each group was determined by counting the cells in five random fields of view (D–G). (H–J) USP48 was upregulated in DLD1 cells with or without HMGA2 knockdown. The relative number of migrated cells was quantified by counting the cells in five random fields of view (H, I), while migration rates were determined by measuring five different wound areas (J). (K–L) USP48 knockdown was conducted in HCT116 cells, with or without HMGA2 overexpression. Representative images of lung tissue and HE-stained sections are presented in K, while the number of metastatic nodules in each group was calculated (K). Representative images of liver tissue and HE-stained sections are shown in (L), along with the calculation of metastatic nodules for each group (L). (M–N) USP48 was upregulated in DLD1 cells with or without HMGA2 knockdown. Representative images of the lung and HE-staining sections are shown in (M), and the metastasis nodules in each group were calculated (M). Representative images of the liver and HE-staining sections are shown in N, and the metastasis nodules in each group were calculated (N). The Scale bars were shown in (K–N). Data are presented as mean ± SD. One-way ANOVA test (A–N). All in vitro experiments were performed independently at least three times. ∗∗∗P < 0.001.
Our findings consistently demonstrate that knockdown of USP48 in CRC cells significantly suppresses tumor metastasis compared to control cells in liver and lung metastasis models of CRC. Furthermore, the overexpression of HMGA2 counteracts the effect of USP48 silencing and promotes tumor metastasis (Fig. 4K and L). Additionally, the depletion of HMGA2 in CRC cells reversed the promoting effect of USP48 on liver and lung metastasis models (Fig. 4M and N). Collectively, these findings suggest that USP48 enhances tumor metastasis largely by regulating HMGA2 in vitro and in vivo.
3.5. The function of USP48 on HMGA2 is significantly enhanced by SUMOylation at Lys-258
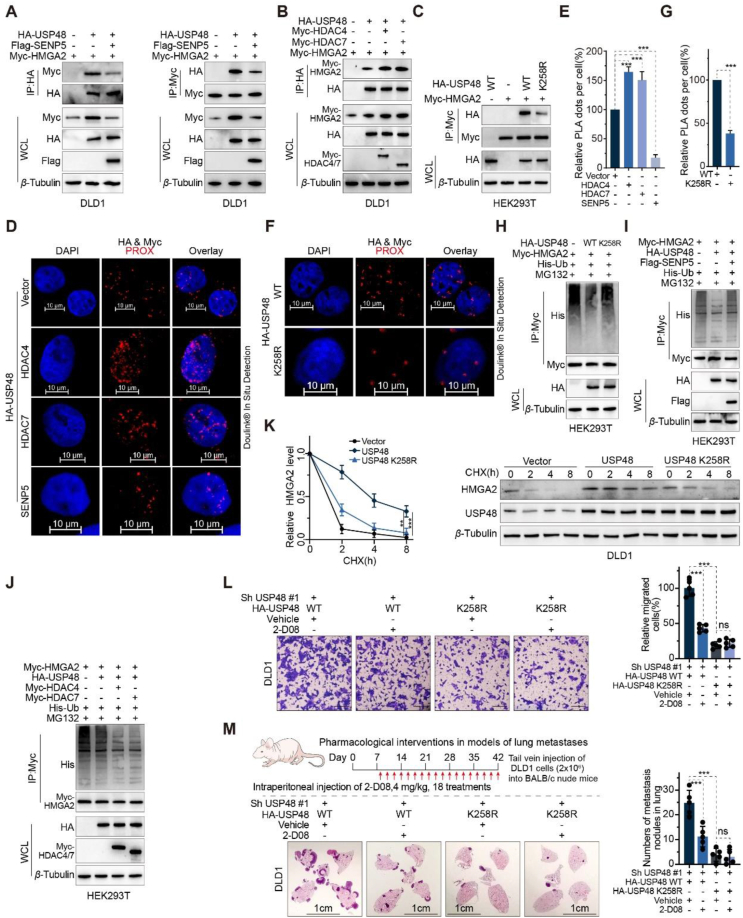
SUMOylation plays a critical role in the survival of eukaryotic cells by modulating the stability, interaction, and activity of hundreds of proteins through various pathways. USP25 interacts with SUMO2/3, resulting in a decrease in its binding affinity towards ubiquitin chains and attenuated DUB activity44. USP5 interacts with and modulates the activity of Cav3.2 calcium channel, yet its binding capacity is impaired by SUMO2/3 modification45. Furthermore, the SUMOylation of OTUB2 at Lys-233 facilitates its interaction with YAP/TAZ and augments its deubiquitinating activity46. USP48 functions as a positive regulator of HMGA2 protein, exerting a pivotal role in the invasion and migration of CRC cells. We aimed to explore the potential regulation of USP48 through other post-translational modifications and their impact on its activity as a deubiquitinating enzyme for HMGA2 protein. The diversity of post-translational modifications on target proteins often depends on their interactions with various protein partners. To characterize the USP48 interactome, we employed mass spectrometry and affinity purification techniques. Through mass spectrometry with high peptide sequence coverage, non-classical E3 SUMO-protein ligase HDAC4 and HDAC7 were identified in USP48-containing immunoprecipitates (Fig. 5A and B, Supporting Information Table S4 and Fig. S5A). Our literature review and USP48-interactome mass spectrometry analysis suggest that there may be SUMOylation occurring at a specific lysine residue of the USP48 protein, which could potentially impact its ability to deubiquitinate the HMGA2 protein. In mammalian cells, there exist five paralogues of SUMO, among which SUMO1, SUMO2 and SUMO3 have been the focus of more extensive research compared to their counterparts, namely SUMO4 and SUMO547. To further explore the role of SUMO-mediated modifications of USP48 in CRC cells, Ni-NTA magnetic bead-pulldowns were performed in DLD1 human CRC cells after cotransfection of HA-USP48 WT and His-SUMO1, His-SUMO2 or His-SUMO3 plasmids. Our results show that USP48 is mostly modified by SUMO1, in a SUMO conjugating enzyme Ubc9 dependent process (Fig. 5C). In CRC cells, it was observed that endogenous USP48 underwent poly-SUMOylation (Fig. 5D and E). According to the USP48-interactome, our findings indicate that USP48 exhibits specific interactions with HDAC4 and HDAC7, both of which are SUMO E3 ligases, as well as SENP5, a SUMO-specific protease (Fig. 5F and G). Co-expression of HDAC4 or HDAC7 facilitated the SUMOylation of USP48 protein (Fig. 5H), while SENP5 inhibited the SUMOylation of USP48 protein (Fig. 5I and J). Three independent computational programs, including SUMOplotTM Analysis Programme (Abgent, https://www.abcepta.com.cn/sumoplot), JASSA48 and GPS-SUMO49, were employed for bioinformatics analysis of USP48 to identify potential SUMOylation sites. The analysis revealed five consensus motifs for SUMO-conjugation with a score greater than 0.5 in Abgent, among which five are evolutionarily conserved from mouse to human (Fig. 5K and Fig. S5B). Additionally, substitution of the putative SUMOylation site Lys258 (USP48-K258R) with arginine resulted in a near-complete abrogation of USP48 protein SUMOylation (Fig. 5L). These findings suggest that HDAC4/7 and SENP5 play a role in the dynamic regulation of USP48 protein SUMOylation, with Lys258 identified as a potential site for this modification. Next, we investigated the impact of SUMOylation on USP48's enzymatic activity as a deubiquitinating enzyme for HMGA2. Our findings demonstrate that the SUMO E3 ligases HDAC4 and HDAC7 enhance the binding ability between HMGA2 and USP48 in CRC cells, while the SUMO protease SENP5 significantly inhibits this interaction (Fig. 6A and B). To investigate the impact of SUMOylation on USP48's enzymatic activity as a deubiquitinating enzyme for HMGA2, we assessed the interaction between HMGA2 and USP48. Our findings indicate that compared to USP48-WT, the interaction between HMGA2 and USP48-K258R mutant was relatively weak (Fig. 6C). We conducted PLA experiments to determine the subcellular localization of SUMOylated USP48. Our findings suggest that the majority of PLA signals were detected in the nucleus, providing compelling evidence for increased binding between SUMOylated USP48 protein and HMGA2 (Fig. 6D‒G). Compared to USP48 WT, the USP48–K258R mutant or cotransfection of USP48 with SENP5 did not significantly reduce HMGA2 ubiquitination (Fig. 6H and I). However, cotransfection of HDAC4 or HDAC7 with USP48 was more effective in reducing HMGA2 ubiquitination (Fig. 6J). Consistent with these findings, the USP48-K258R was less efficient than USP48-WT in stabilizing HMGA2 (Fig. 6K). Cotransfection of USP48 with HDAC4 or HDAC7 synergistically enhances HMGA2 stability, while cotransfection with SENP5 exhibits a weaker effect (Supporting Information Fig. S6A‒C). Next, we investigated the role of USP48 SUMOylation in driving invasive metastasis in CRC. Interestingly, we found that reconstitution of USP48 WT in endogenous USP48-deficient DLD1 cells significantly increased cell migration capacity as well as lung metastasis compared to the USP48 K258R mutant (Fig. 6L and M). In addition, the pharmacological effects of SUMOylation inhibitors 2-D08 significantly reduced the migration and lung metastasis of USP48 WT recombinant cells (Fig. 6L and M). However, no significant effect of 2-D08 was observed in cells reconstituted with the USP48 K258R mutant (Fig. 6L and M). The findings suggest that SUMOylation of USP48 significantly enhances its ability to stabilize HMGA2 and promote CRC invasion and metastasis in a HMGA2-dependent manner.
Figure 5.
USP48 is SUMOylated by SUMO1 at Lys258. (A) Mass spectrometry detection of USP48-interacting proteins (obtained from anti-HA affinity magnetic beads) after HCT116 cells were transfected with HA-USP48 for 36 h. (B) Mass-spectrometry analysis of a HDAC4 peptide in HA-USP48 precipitate. (C) DLD1 cells were co-transfected with His-SUMO1, His-SUMO2, or His-SUMO3, with the HA-USP48 in the presence and absence of ubiquitin-conjugating enzyme 9 (V5-Ubc9), and cell lysates were subjected to Ni-NTA magnetic beads pulldown followed by IB with antibodies against His, HA, and V5. (D–E) IB analysis of total lysates, anti-His immunoprecipitates (D) and Ni-NTA magnetic beads pulldown products (E) of HCT116 cells transfected with control vector or His-SUMO1 as indicated. The intensity of total USP48 band was quantitated and shown. (F) IB analysis of total lysates and anti-HA immunoprecipitates of HEK293T cells transfected with HA-USP48 and Myc-tagged PIAS1, PIAS2, PIAS3, PIAS4, HDAC4, or HDAC7 constructs, as indicated. (G) IB analysis of total lysates and anti-HA immunoprecipitates of HEK293T cells transfected with HA-USP48 and flag-tagged SENP1, SENP2, SENP3, SENP5, SENP6, or SENP7 constructs, as indicated. (H) IB analysis of total lysates and Ni-NTA magnetic beads pulldown products of HEK293T cells transfected with HA-USP48, His-SUMO1 and Myc-HDAC4 or Myc-HDAC7 constructs, as indicated. (I–J) IB analysis of total lysates, anti-HA immunoprecipitates (I) and Ni-NTA magnetic beads pulldown products (J) of HEK293T cells transfected with HA-USP48, His-SUMO1 and Flag-SENP5 constructs, as indicated. (K) A schematic representation of the five predicted SUMO-conjugation motifs (ψKXE) in the USP48 protein. Down-hand side, bioinformatics analysis of USP48 using three independent computational programmes to detect SUMOylation sites (SUMOplotTM Analysis Programme (Abgent), JASSA and GPS-SUMO). (L) IB analysis of total lysates and Ni-NTA magnetic beads pulldown products of HEK293T cells transfected with V5-Ubc9, His-SUMO1, and HA-USP48 WT or USP48 SUMO mutants (USP48 K258R, USP48 K359R, USP48 K468R, USP48 K898R, and USP48 K1003R), as indicated.
Figure 6.
The function of USP48 on HMGA2 is significantly enhanced by SUMOylation at Lys258. (A) IB analysis of total lysates and anti-HA (left panel) or anti-Myc immunoprecipitates (right panel) from DLD1 cells transfected with control vector or Flag-SENP5. (B) IB analysis of total lysates and anti-HA immunoprecipitates derived from DLD1 cells transfected with the control vector, Myc-HDAC4, or Myc-HDAC7, as indicated. (C) IB analysis of total lysates and anti-Myc immunoprecipitates derived from HEK293T cells transfected with HA-USP48 WT or K258R and Myc-HMGA2, as indicated. (D–E) Representative images are shown with merged PLA and DAPI channels from PLA experiments. Scale bar, 10 μm. Each red dot represents the detection of the USP48–HMGA2 interaction complex, and the graphs representing mean ± SD are shown in (E). (F–G) HEK293T cells were transfected with HA-USP48 WT or K258R. After fixation, in situ PLA for USP48-HMGA2 was performed with anti-HA and anti-Myc antibodies. Scale bar, 10 μm. Each red dot represents the detection of the USP48-HMGA2 interaction complex, and the graphs representing mean ± SD are shown in (G). (H) IB analysis of total lysates and anti-Myc immunoprecipitates of HEK293T cells transfected with His-Ub, HA-USP48 WT/K258R and Myc-HMGA2 and treated with MG132 (10 μmol/L for 6 h), as indicated. (I–J) IB analysis of total lysates and anti-Myc immunoprecipitates of HEK293T cells transfected with Myc-HMGA2, His-Ub, HA-USP48, and control vector, Flag-SENP5 (I), Myc-HDAC4 (J), or Myc-HDAC7 (J) and treated with MG132 (10 μmol/L for 6 h), as indicated. (K) DLD1 cells were transfected with specific plasmids, as indicated and treated with CHX (50 μg/mL) for the indicated time points. Quantification of HMGA2 levels relative to β-Tubulin is shown. (L) DLD1 cells stably expressing USP48 shRNA were transfected with indicated plasmids and treated with 200 μmol/L 2-D08 or DMSO for 24 h. Graphic representation of the migration ability from cells described above was examined by transwell migration assay. Scale bar, 200 μm. (M) Representative images of lung sections were stained with HE, and metastatic nodules were calculated in each group. Four-week-old BALB/c nude mice were injected intravenously with 2 × 106 cells as in (I). 2-D08 (4 mg/kg) or vehicle was injected intraperitoneally on Day 8 and every other day for 5 weeks. Scale bar, 1 cm. Data are presented as mean ± SD. One-way ANOVA test (E). Unpaired two-tailed Student's t-test (G). All experiments were performed independently at least three times. ∗∗∗P < 0.001, ns indicates no statistical significance.
3.6. DUB-IN-2 hinders the deubiquitinating activity of USP48 and induces the degradation of HMGA2
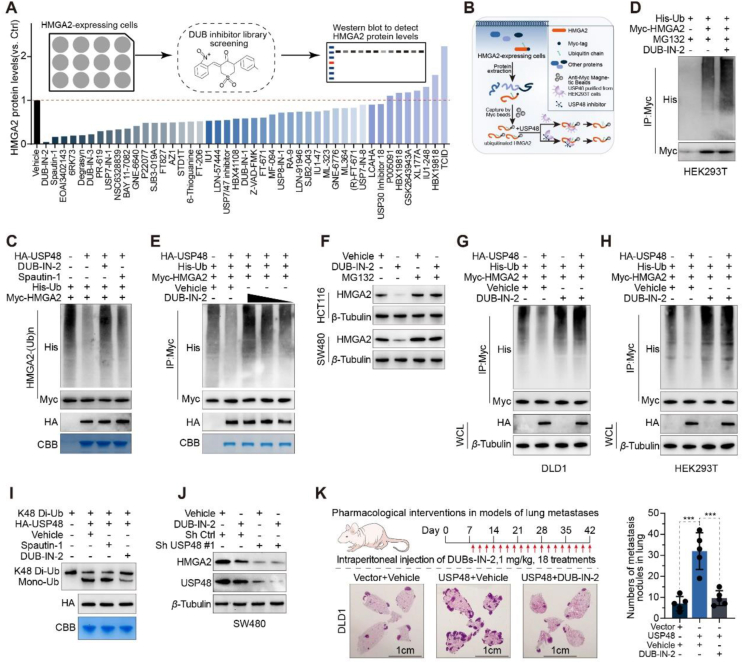
Our research has demonstrated that targeting USP48 leads to the enzyme-mediated degradation of HMGA2, which has prompted us to investigate the potential of USP48 inhibitors in CRC therapy. Unfortunately, there is currently a lack of reports on small molecule inhibitors targeting USP48. In order to assess the potential of small molecule inhibitors in degrading HMGA2, we conducted a screening based on levels of the HMGA2 protein. We treated SW480 cells with high expression of HMGA2 with a library of 45 previously reported DUB inhibitors individually and assessed their impact on HMGA2 protein levels (Fig. 7A, Supporting Information Fig. S7A and B). We have identified two inhibitors, namely DUB-IN-2 and Spautin-1, which significantly reduced the levels of HMGA2 protein. Among them, DUB-IN-2 exhibited the most substantial effect. To determine whether the decline in HMGA2 induced by the inhibitor was dependent on USP48, we conducted in vitro deubiquitination experiments. Initially, USP48 that had been purified from HEK293T cells overexpressing HA-USP48 underwent pre-treatment using either a DUB inhibitor or DMSO before being incubated with ubiquitinated HMGA2. Subsequently, the degree of HMGA2 ubiquitination was assessed (Fig. 7B). Our results indicated that DUB-IN-2 effectively disrupted the USP48-mediated reduction in HMGA2 ubiquitination, suggesting a near-complete inhibition of USP48's deubiquitinating activity (Fig. 7C). In contrast, Spautin-1 did not significantly impact the deubiquitination of HMGA2 by USP48, implying alternative mechanisms for its effect on HMGA2 stability (Fig. 7C). We aimed to investigate the regulatory role of DUB-IN-2 in HMGA2 ubiquitination. Our findings demonstrated that treatment with DUB-IN-2 significantly enhances dose-dependent ubiquitination of HMGA2 (Fig. 7D and E). Furthermore, we evaluated the reliance of DUB-IN-2-induced downregulation of HMGA2 on USP48 and the ubiquitin‒proteasome system. Treatment with MG132 reversed the downregulation of HMGA2 induced by DUB-IN-2, indicating that DUB-IN-2 facilitated the proteasomal degradation of HMGA2 (Fig. 7F). In vivo deubiquitination experiments also demonstrated that DUB-IN-2 significantly disrupted the reduction in USP48-induced ubiquitination level of HMGA2 (Fig. 7G and H). Additionally, through in vitro deubiquitination experiments utilizing ubiquitin chains as substrates, we have discovered that USP48 has the ability to cleave K48-linked Di-ubiquitin. This activity was inhibited by DUB-IN-2 but not by Spautin-1 (Fig. 7I). The treatment of DUB-IN-2 significantly attenuated the maintenance of HMGA2 stability by USP48 (Fig. S7C). Moreover, our observations suggest that DUB-IN-2 destabilizes HMGA2 via USP48 in SW480 cells transfected with control shRNA. However, this effect was not observed in SW480 cells transfected with USP48 shRNA, indicating that DUB-IN-2 induces HMGA2 degradation by inhibiting the deubiquitination activity of USP48 (Fig. 7J). Next, we investigated whether DUB-IN-2 also exerted sustained pharmacological inhibition of USP48 in vivo. It was found that significantly fewer lung metastases occurred in mice treated with DUB-IN-2 for 5 weeks compared to the vehicle group (Fig. 7K). Taken together, these results demonstrate the efficacy of DUB-IN-2 in inhibiting USP48 enzyme activity and antagonizing USP48-HMGA2 signaling during CRC metastasis.
Figure 7.
DUB-IN-2 hinders the deubiquitinating activity of USP48 and induces the degradation of HMGA2. (A) Relative protein level of HMGA2 in the screening of DUBs inhibitors. The SW480 cells were treated with the library of 45 reported DUB inhibitors at a concentration of 2.5 μmol/L for 12 h as indicated, and then, the protein levels of HMGA2 were assessed by Western blotting. (B) Schematic diagram of USP48 activity detection after treatment with compounds in vitro. (C) Ubiquitinated HMGA2 was immunoprecipitated from HEK293T cells transfected with Myc-HMGA2 and His-Ub plasmids with after 10 μmol/L MG132 treatment. Purified USP48 from HEK293T cells overexpressing HA-USP48 was pretreated with DUB inhibitors for 1 h and subsequently incubated with ubiquitinated HMGA2 for 6 h. Then, the ubiquitylation level of HMGA2 was evaluated. (D) HEK293T cells were co-transfected with MYC-HMGA2 and His-ubiquitin, and then treated with 2.5 μmol/L DUB-IN-2 24 h. After pretreatment with MG132 (10 μmol/L) for 6 h, anti-Myc beads were used for IP, and anti-Myc and His antibodies were used for IB analysis. (E) The deubiquitinating effect of USP48 on HMGA2 upon treatment with different concentrations of DUB-IN-2. HA-USP48 was exposed to 5, 2.5 and 1 μmol/L DUB-IN-2 and then incubated with ubiquitinated HMGA2. The ubiquitylation level of HMGA2 was measured. (F) IB analysis of HMGA2 and β-Tubulin in HCT116 and SW480 cells pretreated with 2.5 μmol/L DUB-IN-2 24 h and then treated with MG132 (10 μmol/L) for 6 h. (G, H) DLD1 and HEK293T cells were co-transfected with MYC-HMGA2, HA-USP48 and His-ubiquitin, and then treated with 2.5 μmol/L DUB-IN-2 24 h. After pretreatment with MG132 (10 μmol/L) for 6 h, anti-Myc beads were used for IP, and anti-Myc, anti-HA and His antibodies were used for IB analysis. (I) Purified HA-USP48 was pretreated with Spautin-1 (2.5 μmol/L) or DUB-IN-2 (2.5 μmol/L) for 10 min and then incubated with K48-linked Di-ubiquitin in the presence of the compound at 37 °C for 1.5 h. Samples were then analyzed by IB analysis with a ubiquitin-specific antibody. (J) IB analysis of HMGA2 and β-Tubulin in control and USP48-knockdown SW480 cells treated with 2.5 μmol/L DUB-IN-2 24 h. (K) Representative images of lung sections were stained with HE, and metastatic nodules were calculated in each group. Four-week-old BALB/c nude mice were injected intravenously with 2 × 106 DLD1 cells with high USP48 expression. DUB-IN-2 (1 mg/kg) or vehicle was injected intraperitoneally on Day 8 and every other day for 5 weeks. Scale bar, 1 cm ∗∗∗P < 0.001.
3.7. Elevated levels of USP48 are positively associated with the upregulation of HMGA2 and serve as a predictor for an unfavorable prognosis in patients with CRC
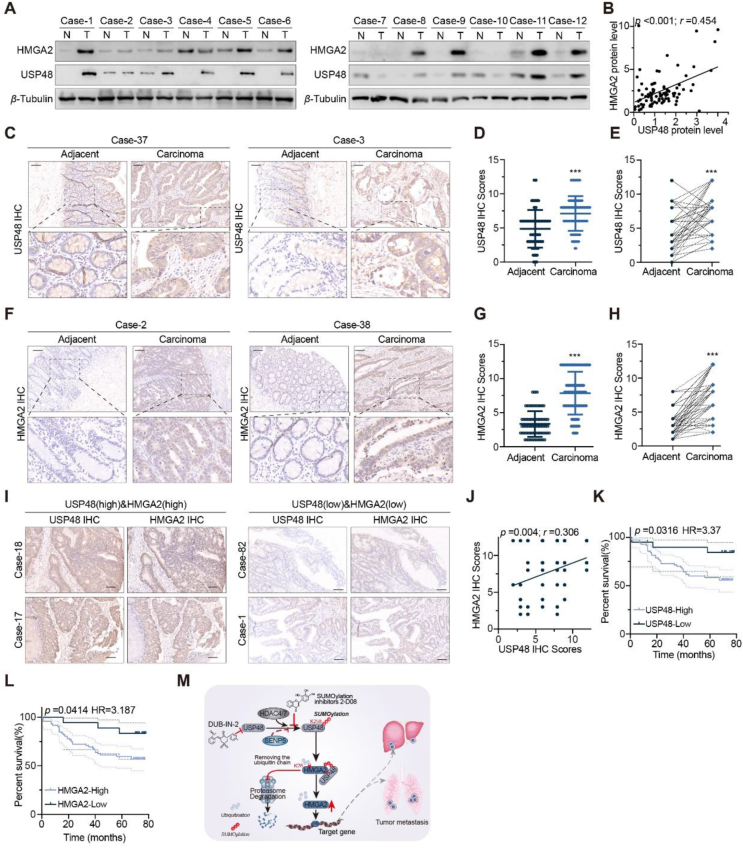
Recent studies have revealed a strong correlation between the upregulation of HMGA2 protein in CRC and malignant biological phenotypes, including epithelial–mesenchymal transition, invasion, metastasis, and chemotherapy resistance. Our hypothesis posits that USP48 upregulation leads to deubiquitination and stabilization of HMGA2 protein, thereby impacting CRC cell invasion and metastasis. To validate our hypothesis, we investigated the correlation between USP48 and HMGA2 protein expression in human CRC specimens. Our findings, as shown in Fig. 8A and B, demonstrated a significant positive correlation between USP48 and HMGA2 protein levels across 42 pairs of CRC specimens (P < 0.001, Pearson r = 0.454). To further assess the clinical significance of the USP48-HMGA2 axis and establish their correlation in CRC, we performed IHC staining to examine the expression of these proteins in consecutive sections of tissue microarrays (TMAs) consisting of 90 CRC tissues and 86 adjacent normal tissues. The IHC results revealed a significant upregulation of USP48 and HMGA2 expression levels in CRC tissues compared to adjacent or matched adjacent tissues (Fig. 8C‒H). Furthermore, our data also indicates a positive correlation between the IHC scores of USP48 and HMGA2 in these tumor samples (Fig. 8I and J). Notably, patients with high levels of USP48 or HMGA2 exhibited shorter overall survival than those with low levels of these proteins (Fig. 8K and L). Taken together, these human CRC data strongly support our experimental findings regarding USP48-mediated HMGA2 stabilization (Fig. 8M).
Figure 8.
Elevated levels of USP48 are positively associated with the upregulation of HMGA2 and serve as a predictor for an unfavorable prognosis in patients with CRC. (A) Cell lysates from 42 pairs CRC tissues and matched adjacent tissues were blotted with USP48 and HMGA2 antibodies. (B) Correlation analysis of USP48 and HMGA2 in CRC tissues and matched adjacent tissues. Statistical analyses were performed using the chi-square test. Pearson r indicates the correlation coefficient. (C–E) Representative images of USP48 highly expressed in CRC tissues and IHC score of USP48 in CRC and matched adjacent tissue, respectively. Data are presented as mean ± SD. The comparison between cancer tissues and adjacent or matched adjacent tissues can be conducted using either a Wilcoxon unpaired test or paired t-test. (F–H) Representative images of HMGA2 highly expressed in CRC tissues and IHC score of HMGA2 in CRC and matched adjacent tissue, respectively. Data are presented as mean ± SD. The comparison between cancer tissues and adjacent or matched adjacent tissues can be conducted using either a Wilcoxon unpaired test or paired t-test. (I) Representative images showing the simultaneous high or low expression of both USP48 and HMGA2 in CRC tissues with a scale bar of 100 μm. (J) USP48 protein scores (X axis) in CRC positively correlate with HMGA2 protein scores (Y axis) in individual patients. The P-value was calculated from a linear regression analysis. r is the correlation coefficient. (K–L) Kaplan–Meier curves of the overall survival of CRC patients (90 patients), stratified by USP48 (K) or HMGA2 (L) protein levels. The P-value was calculated from a log-rank test. HR, hazard ratio. (M) Working model of SUMOylated USP48-mediated HMGA2 deubiquitination. ∗∗∗P < 0.001.
4. Discussion
The modulation of transcriptional activity by HMGA2 has been linked to uncontrolled cancer progression, rapid metastasis, and poor overall survival across a range of malignancies. In their study on chemotherapy-resistant gastric cancer, Fan et al. found evidence supporting the role of the HMGA2‒FOXL2 axis in regulating both EMT and metastatic potential50. Furthermore, prior research has demonstrated that HBXIP facilitates the acetylation of HMGA2 at Lys-26 through activation of the AKT signaling pathway34. Subsequently, AKT activation leads to phosphorylation and activation of PCAF in esophageal squamous cell carcinoma34. By inhibiting the ubiquitination and proteasomal degradation of PCAF, HMGA2 accumulates and contributes to carcinogenesis. However, the mechanisms underlying the regulation of HMGA2 protein activity and stability in CRC remain elusive. In this study, USP48 was identified as a novel DUB that can stabilize HMGA2 and prevent its degradation in CRC. Importantly, enhanced USP48 expression was positively associated with elevated HMGA2 levels in CRC tissue and predicted unfavorable survival outcomes for patients afflicted with this disease.
DUBs have garnered significant attention in cancer progression due to their ability to stabilize oncoproteins. Numerous studies have demonstrated the critical roles played by USPs in tumor invasion and metastasis. For example, USP1 promotes breast cancer metastasis by deubiquitinating KPNA251. Additionally, USP3 accelerates gastric cancer development by stabilizing COL9A3/COL6A5 through deubiquitination52. USP11 stabilizes PPP1CA and promotes the growth and metastasis of CRC by activating the ERK/MAPK pathway, indicating its potential as a prognostic marker53. As a member of the deubiquitinating enzyme family, the diverse roles of USP48 across different tumor types can be ascribed to a multitude of factors, encompassing the intricacies of cancer biology, the context-specific nature of tissues, and the intricate interplay among various molecular pathways. USP48 specifically interacts with Gli1 to remove ubiquitin and regulate the Hedgehog signaling pathway, thereby promoting glioblastoma tumorigenesis38. In addition, USP48 plays a pivotal role in the development of numerous aggressive cancers due to its ability to deubiquitinate β-catenin, which is known to initiate non-small cell lung cancer54. USP48 bound GSDME and removed K48-linked ubiquitination at positions K120 and K189 to promote pyroptosis in cancer55. Furthermore, USP48 plays an oncogene role in hepatocellular carcinoma, stabilising SIRT6 to attenuate HCC glycolysis and malignancy39. This study has identified that USP48 directly interacts with HMGA2, and overexpression of USP48 significantly increases the protein expression of HMGA2 while decreasing its ubiquitination. Conversely, knockdown of USP48 enhances HMGA2 degradation. Furthermore, our findings suggest that USP48 stabilizes HMGA2 through K6- and K48-linked deubiquitination. We have determined that Lys26 in the N-terminal region of HMGA2 plays a critical role in the deubiquitination of HMGA2 by USP48. Interestingly, we have observed a positive correlation between the expression levels of HMGA2 and USP48 in both CRC patient samples and cell lines. Our findings suggest that USP48 partially regulates HMGA2 to promote the malignant phenotype of CRC. USP48 promoted the migration and invasion of CRC cells by upregulating HMGA2 expression. The pro-metastatic effect induced by USP48 overexpression was reversed upon depletion of HMGA2. Interestingly, we discovered that SUMOylation on Lys-258 significantly enhanced USP48-mediated deubiquitination of HMGA2. Our findings demonstrated that HDAC4 and HDAC7 promoted the SUMOylation of USP48, while SENP5 cleaved it. The lysine residue 258 in USP48 is evolutionarily conserved, and its SUMOylation plays a critical role in stabilizing HMGA2. Notably, the introduction of K258R mutation into USP48 abolished its SUMOylation and compromised its ability to stabilize HMGA2. Although we failed to identify a conserved SIM motif in HMGA2, our findings demonstrated that the SUMOylated USP48 could still strongly interact with HMGA2, thereby enhancing the affinity of USP48 for ubiquitin chains and augmenting its DUB activity. We hypothesize that the SUMOylation of USP48 may interact with a non-classically conserved SIM motif (V-V-Q-K) in the HMGA2 protein, or induce conformational changes in USP48 to better accommodate the HMGA2 protein44. The precise mechanism underlying the enhancement of USP48-mediated deubiquitination of HMGA2 protein by SUMOylation remains incompletely elucidated in this study, but it represents a topic for our future investigations. The protein expression levels of USP48 and HMGA2 were significantly elevated in CRC tissues compared to adjacent normal tissues, and a positive correlation was observed between the expressions of USP48 and HMGA2 in CRC samples. Our study identifies USP48 as a novel deubiquitinating enzyme for HMGA2, providing insights into the regulatory mechanism of HMGA2 in CRC. In vivo and in vitro functional investigations have demonstrated that overexpression of USP48 promotes the proliferation of CRC cells. The signaling pathway mediated by the USP48–HMGA2 axis plays a critical role in promoting invasion and metastasis of CRC cells. Patients with relatively high levels of both USP48 and HMGA2 exhibit poor overall survival rates. Therefore, targeting USP48 may represent a promising therapeutic strategy for CRC. We have established a screening model based on HMGA2 stability and identified DUB-IN-2 as a potent inhibitor of USP48. Mechanistically, DUB-IN-2 promotes the ubiquitination and proteasomal degradation of HMGA2, in part through its inhibition of USP48. Although DUB-IN-2 may not exhibit drug-like properties in humans, our findings demonstrate the feasibility of small molecule inhibition of USP48's HMGA2 deubiquitinating activity, providing a promising starting point for developing USP48 inhibitors.
5. Conclusions
In summary, our studies provide new insights into the function of USP48 in CRC cell invasion and metastasis through deubiquitination and stabilization of HMGA2 oncoprotein. We then demonstrate that inhibition of USP48 by shRNA or its newly discovered inhibitor DUB-IN-2 triggers HMGA2 degradation, thereby inhibiting CRC cell metastasis in vitro and in vivo. Furthermore, SUMOylation of USP48 significantly enhanced its ability to stabilize HMGA2 and promote CRC cell invasion and metastasis in an HMGA2-dependent manner. The pharmacological effects of SUMOylation inhibitor 2-D08 significantly reduced the pro-metastatic effects of USP48 in CRC. In summary, our study not only reveals the mechanism of USP48 regulation of HMGA2 ubiquitination degradation, but also validates USP48 as a potential therapeutic target for CRC, and suggests that DUB-IN-2 is promising as a USP48 inhibitor for the treatment of CRC.
Author contributions
Liang Wu, Can Cheng and Hanhui Yao conceived and planned the project, conducted data analysis, and drafted the manuscript. Liang Wu, Can Cheng, and Liang Wu designed and executed experiments while Yang Wu, Liang Zhu, Jingwen Liu, Heng Li and Zhengyi Liu. Evaluated experimental data through visual structuring techniques. The study was supervised by Liang Wu, Zhengdong Fang, Hanhui Yao, and Hejie Hu who also secured funding.
Conflicts of interest
The authors have declared that no conflict of interest exists.
Acknowledgments
We extend our sincere appreciation to Mingyang Gao, Shuai Zhou, and Wenxiang Fang for their exceptional technical assistance. This research was financially supported by the National Natural Science Foundation of China (No. 82103543), the Fundamental Research Funds for the Central Universities (No. WK9110000159, China), Natural Science Foundation of Anhui Province (No. 2108085QH340, China), and the Postdoctoral Science Foundation of China (No. 2021M693082).
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2024.01.006.
Contributor Information
Hejie Hu, Email: huhejie@163.com.
Zhengdong Fang, Email: fangzhengdong@126.com.
Liang Wu, Email: wu8722@ustc.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Keum N., Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in globocan 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Harrigan J.A., Jacq X., Martin N.M., Jackson S.P. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoeller D., Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 5.Senft D., Qi J., Ronai Z.A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18:69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wertz I.E., Wang X. From discovery to bedside: targeting the ubiquitin system. Cell Chem Biol. 2019;26:156–177. doi: 10.1016/j.chembiol.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Komander D., Clague M.J., Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 8.Abdul Rehman S.A., Kristariyanto Y.A., Choi S.Y., Nkosi P.J., Weidlich S., Labib K., et al. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell. 2016;63:146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Sonego M., Pellarin I., Costa A., Vinciguerra G.L.R., Coan M., Kraut A., et al. USP1 links platinum resistance to cancer cell dissemination by regulating Snail stability. Sci Adv. 2019;5 doi: 10.1126/sciadv.aav3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L., Zhao N., Zhou Z., Chen J., Han S., Zhang X., et al. PLAGL2 promotes the proliferation and migration of gastric cancer cells via USP37-mediated deubiquitination of Snail1. Theranostics. 2021;11:700–714. doi: 10.7150/thno.47800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai J., Li M., Wang X., Li L., Li Q., Hou Z., et al. USP37 promotes lung cancer cell migration by stabilizing Snail protein via deubiquitination. Front Genet. 2019;10:1324. doi: 10.3389/fgene.2019.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan T., Yang X., Liang H., Chen J., Chen Y., Zhu Y., et al. Deubiquitinating enzyme USP9X regulates metastasis and chemoresistance in triple-negative breast cancer by stabilizing Snail1. J Cell Physiol. 2022;237:2992–3000. doi: 10.1002/jcp.30763. [DOI] [PubMed] [Google Scholar]
- 14.Lambies G., Miceli M., Martinez-Guillamon C., Olivera-Salguero R., Pena R., Frias C.P., et al. TGFβ-activated USP27X deubiquitinase regulates cell migration and chemoresistance via stabilization of Snail1. Cancer Res. 2019;79:33–46. doi: 10.1158/0008-5472.CAN-18-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D., Hong A., Park H.I., Shin W.H., Yoo L., Jeon S.J., et al. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J Cell Physiol. 2017;232:3664–3676. doi: 10.1002/jcp.25841. [DOI] [PubMed] [Google Scholar]
- 16.Ruiz E.J., Pinto-Fernandez A., Turnbull A.P., Lan L., Charlton T.M., Scott H.C., et al. USP28 deletion and small-molecule inhibition destabilizes c-Myc and elicits regression of squamous cell lung carcinoma. Elife. 2021;10 doi: 10.7554/eLife.71596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui B., Luo Y., Tian P., Peng F., Lu J., Yang Y., et al. Stress-induced epinephrine enhances lactate dehydrogenase a and promotes breast cancer stem-like cells. J Clin Invest. 2019;129:1030–1046. doi: 10.1172/JCI121685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan J., Deng Q., Jiang C., Wang X., Niu T., Li H., et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene. 2015;34:3957–3967. doi: 10.1038/onc.2014.327. [DOI] [PubMed] [Google Scholar]
- 19.Peng Y., Liu J., Wang Z., Cui C., Zhang T., Zhang S., et al. Prostate-specific oncogene OTUD6A promotes prostatic tumorigenesis via deubiquitinating and stabilizing c-Myc. Cell Death Differ. 2022;29:1730–1743. doi: 10.1038/s41418-022-00960-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan O.M., Carvalho J., Spencer-Dene B., Mitter R., Frith D., Snijders A.P., et al. The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J Clin Invest. 2018;128:1326–1337. doi: 10.1172/JCI97325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garabedian A., Jeanne Dit Fouque K., Chapagain P.P., Leng F., Fernandez-Lima F. AT-hook peptides bind the major and minor groove of AT-rich DNA duplexes. Nucleic Acids Res. 2022;50:2431–2439. doi: 10.1093/nar/gkac115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sgarra R., Rustighi A., Tessari M.A., Di Bernardo J., Altamura S., Fusco A., et al. Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. FEBS Lett. 2004;574:1–8. doi: 10.1016/j.febslet.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Fedele M., Visone R., De Martino I., Troncone G., Palmieri D., Battista S., et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–471. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 24.Sgarra R., Pegoraro S., Ros G., Penzo C., Chiefari E., Foti D., et al. High mobility group a (HMGA) proteins: molecular instigators of breast cancer onset and progression. Biochim Biophys Acta Rev Cancer. 2018;1869:216–229. doi: 10.1016/j.bbcan.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Wang X., Wang J., Zhao J., Wang H., Chen J., Wu J. HMGA2 facilitates colorectal cancer progression via STAT3-mediated tumor-associated macrophage recruitment. Theranostics. 2022;12:963–975. doi: 10.7150/thno.65411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen R.X., Chen X., Xia L.P., Zhang J.X., Pan Z.Z., Ma X.D., et al. N(6)-Methyladenosine modification of circ NSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu L., Ma Y., Zhang H., Lu Q.J., Yang L., Jiang G.N., et al. HMGA2 regulates circular RNA ASPH to promote tumor growth in lung adenocarcinoma. Cell Death Dis. 2020;11:593. doi: 10.1038/s41419-020-2726-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai F.Q., Li C.R., Fan X.Q., Tan L., Wang R.T., Jin H. Mir-150-5p inhibits non-small-cell lung cancer metastasis and recurrence by targeting HMGA2 and β-Catenin signaling. Mol Ther Nucleic Acids. 2019;16:675–685. doi: 10.1016/j.omtn.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobersch S., Rubio K., Singh I., Gunther S., Graumann J., Cordero J., et al. Positioning of nucleosomes containing γ-H2AX precedes active DNA demethylation and transcription initiation. Nat Commun. 2021;12:1072. doi: 10.1038/s41467-021-21227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai J., Yokomizo-Nakano T., Kubota S., Sun Y., Kanai A., Iimori M., et al. Overexpression of HMGA2 activates IGF2BP2 and remodels transcriptional program of Tet2-deficient stem cells in myeloid transformation. Oncogene. 2021;40:1531–1541. doi: 10.1038/s41388-020-01629-w. [DOI] [PubMed] [Google Scholar]
- 31.Chen X., Zeng K., Xu M., Liu X., Hu X., Xu T., et al. P53-induced mir-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA and HMGA2. Cell Death Dis. 2019;10:131. doi: 10.1038/s41419-018-1188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan L., Xu H., Chen G., Wei X., Yu B., Ye J., et al. Silencing of HMGA2 reverses retardance of cell differentiation in human myeloid leukaemia. Br J Cancer. 2018;118:405–415. doi: 10.1038/bjc.2017.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang K., Guo W., Ren T., Huang Y., Han Y., Zhang H., et al. Knockdown of HMGA2 regulates the level of autophagy via interactions between MSI2 and Beclin1 to inhibit NF1-associated malignant peripheral nerve sheath tumour growth. J Exp Clin Cancer Res. 2019;38:185. doi: 10.1186/s13046-019-1183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y., Wang X., Xu F., Zhang L., Wang T., Fu X., et al. The regulation of acetylation and stability of HMGA2 via the HBXIP -activated AKT-PCAF pathway in promotion of esophageal squamous cell carcinoma growth. Nucleic Acids Res. 2020;48:4858–4876. doi: 10.1093/nar/gkaa232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo L., Cheng X., Chen H., Chen C., Xie S., Zhao M., et al. Induction of breast cancer stem cells by M1 macrophages through lin-28b-let-7-HMGA2 axis. Cancer Lett. 2019;452:213–225. doi: 10.1016/j.canlet.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 36.Sgarra R., Maurizio E., Zammitti S., Lo Sardo A., Giancotti V., Manfioletti G. Macroscopic differences in HMGA oncoproteins post-translational modifications: C-terminal phosphorylation of HMGA2 affects its DNA binding properties. J Proteome Res. 2009;8:2978–2989. doi: 10.1021/pr900087r. [DOI] [PubMed] [Google Scholar]
- 37.Stabell M., Saether T., Rohr A.K., Gabrielsen O.S., Myklebost O. Methylation-dependent sumoylation of the architectural transcription factor HMGA2. Biochem Biophys Res Commun. 2021;552:91–97. doi: 10.1016/j.bbrc.2021.02.099. [DOI] [PubMed] [Google Scholar]
- 38.Zhou A., Lin K., Zhang S., Ma L., Xue J., Morris S.A., et al. Gli1-induced deubiquitinase USP48 aids glioblastoma tumorigenesis by stabilizing Gli1. EMBO Rep. 2017;18:1318–1330. doi: 10.15252/embr.201643124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du L., Li Y., Kang M., Feng M., Ren Y., Dai H., et al. USP48 is upregulated by Mettl14 to attenuate hepatocellular carcinoma via regulating SIRT6 stabilization. Cancer Res. 2021;81:3822–3834. doi: 10.1158/0008-5472.CAN-20-4163. [DOI] [PubMed] [Google Scholar]
- 40.Soderberg O., Gullberg M., Jarvius M., Ridderstrale K., Leuchowius K.J., Jarvius J., et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 41.Tai H.C., Schuman E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 42.Zheng N., Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Wang J., Wu J. Emerging roles for HMGA2 in colorectal cancer. Transl Oncol. 2021;14 doi: 10.1016/j.tranon.2020.100894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meulmeester E., Kunze M., Hsiao H.H., Urlaub H., Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 45.Garcia-Caballero A., Zhang F.X., Chen L., M'Dahoma S., Huang J., Zamponi G.W. Sumoylation regulates USP5-Cav3.2 calcium channel interactions. Mol Brain. 2019;12:73. doi: 10.1186/s13041-019-0493-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z., Du J., Wang S., Shao L., Jin K., Li F., et al. OTUB2 promotes cancer metastasis via hippo-independent activation of YAP and TAZ. Mol Cell. 2019;73 doi: 10.1016/j.molcel.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 47.Zubiete-Franco I., Garcia-Rodriguez J.L., Lopitz-Otsoa F., Serrano-Macia M., Simon J., Fernandez-Tussy P., et al. Sumoylation regulates LKB1 localization and its oncogenic activity in liver cancer. EBioMedicine. 2019;40:406–421. doi: 10.1016/j.ebiom.2018.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beauclair G., Bridier-Nahmias A., Zagury J.F., Saib A., Zamborlini A. Jassa: a comprehensive tool for prediction of sumoylation sites and sims. Bioinformatics. 2015;31:3483–3491. doi: 10.1093/bioinformatics/btv403. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., et al. Gps-sumo: a tool for the prediction of sumoylation sites and sumo-interaction motifs. Nucleic Acids Res. 2014;42:W325–W330. doi: 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong J., Wang R., Ren G., Li X., Wang J., Sun Y., et al. HMGA2-FOXL2 axis regulates metastases and epithelial-to-mesenchymal transition of chemoresistant gastric cancer. Clin Cancer Res. 2017;23:3461–3473. doi: 10.1158/1078-0432.CCR-16-2180. [DOI] [PubMed] [Google Scholar]
- 51.Ma A., Tang M., Zhang L., Wang B., Yang Z., Liu Y., et al. USP1 inhibition destabilizes KPNA2 and suppresses breast cancer metastasis. Oncogene. 2019;38:2405–2419. doi: 10.1038/s41388-018-0590-8. [DOI] [PubMed] [Google Scholar]
- 52.Wu X., Wang H., Zhu D., Chai Y., Wang J., Dai W., et al. USP3 promotes gastric cancer progression and metastasis by deubiquitination-dependent COL9A3/COL6A5 stabilisation. Cell Death Dis. 2021;13:10. doi: 10.1038/s41419-021-04460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun H., Ou B., Zhao S., Liu X., Song L., Liu X., et al. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine. 2019;48:236–247. doi: 10.1016/j.ebiom.2019.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang P., Li L., Wang B., Ran X., Yang S., Luo Y., et al. Mir-489-3p promotes malignant progression of non-small cell lung cancer through the inactivation of Wnt/β-Catenin signaling pathway via regulating USP48. Respir Res. 2022;23:93. doi: 10.1186/s12931-022-01988-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren Y., Feng M., Hao X., Liu X., Li J., Li P., et al. USP48 stabilizes gasdermin e to promote pyroptosis in cancer. Cancer Res. 2023;83:1074–1093. doi: 10.1158/0008-5472.CAN-22-1812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete dataset can be accessed through the article, and Supporting Information.