Abstract
We have constructed a series of vectors based on simian foamy virus type 1 (SFV-1) to define the minimum cis-acting elements required for gene transfer. To characterize these vectors, we inserted the coding sequence of the bacterial lacZ gene linked to the cytomegalovirus immediate-early gene promoter. Introduction of a deletion mutation in the leader region between the 5′ long terminal repeat and the start of the gag gene at position 1659 to 1694 completely abrogated gene transfer by the SFV-1 vector. Deletion of 39 nucleotides from position 1692 to 1731 in the leader region resulted in a significant reduction in the transducing-particle titer. Furthermore, we have identified a second cis-acting element located at the 3′ end of the pol gene between position 6486 and 6975 to be critical for SFV-1 vector transduction. These results identify the two important cis-acting elements required for SFV-1 vector construction, and the finding of a cis-acting element in the pol gene is unique among retroviruses.
Foamy viruses belong to a distinct genus of retroviruses. These viruses share many features with other retroviruses, including the structural genes of the gag, pro, pol, and env genes, which encode core proteins, protease, reverse transcriptase, integrase, and envelope protein, respectively. Similar to retroviruses with complex genome organization, foamy viruses possess additional genes modulating viral replication (11, 13, 17, 22, 28, 30, 38). Foamy viruses have unique features that distinguish them from other retroviruses. The Gag protein of foamy virus lacks the Cys-His motif present in the nucleocapsid (NC) domains of other retroviruses (11, 13, 17, 22, 28). Three glycine-arginine-rich motifs are found in the corresponding region of the foamy virus NC domain. One of these glycine-arginine-rich motifs is required for nucleic acid binding and virus replication, suggesting that the Gag protein of foamy viruses may be more analogous to the core protein of hepatitis B virus (40). Retroviruses use either a minus ribosomal frameshift or a suppression of stop codons to generate the Gag-Pro-Pol precursor polyprotein, which is subsequently processed by the viral protease to produce Gag, Pro, and Pol cleavage products (7). Unlike that of other retroviruses, the pro-pol gene product of foamy viruses is translated from a subgenomic message which lacks the Gag domain (5, 39). Translation of the protease-polymerase products is initiated from an ATG initiator codon located at the 5′ end of the protease gene (9, 19). Foamy viruses transcribe an RNA pregenome as a late event in replication, and large amounts of infectious foamy virus particles were shown to contain double-stranded DNA, suggesting that foamy viruses have a distinct replication pathway with features of both retroviruses and hepadnaviruses (8, 39).
Foamy viruses are found in many mammalian species. However, these viruses appear to be nonpathogenic, although the virus is widely distributed within the natural host and can be recovered from several organs and tissues, including the brain and peripheral blood leukocytes (15). Foamy viruses also have a broad host range in cell culture with respect to cell type and species in which they can be propagated; host cell types include fibroblasts and epithelial, lymphoid, and neural cells (21, 25). A foamy virus isolate from one species can also infect other mammalian species (15, 32). Thus, foamy viruses offer unique opportunities for developing viral vector systems to deliver genes into several cell types of many species. The size of the genome of foamy virus (13 kb) suggests that more heterologous DNA can be accommodated in foamy virus-based vectors than in murine and avian retroviral vectors (8 kb). Recently, it was demonstrated that heterologous genes can be transduced by foamy viruses (24, 31, 34). Furthermore, the efficiency of transduction in primate hematopoietic cells by foamy virus vectors compared favorably with those obtained for murine leukemia virus vectors (14). Although the molecular mechanism of foamy virus replication has been characterized to a great extent, several important features necessary for vector construction remain to be determined. Among them, signals important for packaging the foamy virus RNA genome into virion particles have not been identified. Here we defined two cis-acting regions required for simian foamy virus type 1 (SFV-1)-based vectors to mobilize heterologous genes.
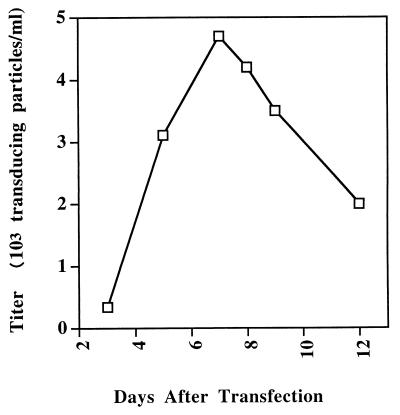
SFV-1 vectors were constructed by using lacZ as a reporter gene. Cloning procedures were done according to standard protocols (33). The coding sequence of the lacZ gene was placed downstream of the cytomegalovirus (CMV) immediate-early gene promoter for expression. All plasmids were derived from an SFV-1 infectious proviral DNA clone, pSFV-1. The construction of pSFV-1 was described previously (24). An SFV-1 vector with a marker gene (CMV-lacZ) was constructed by replacing the region from position 7077 to 10750, which encompasses the envelope and accessory genes located between env and the 5′ long terminal repeat (pV7-9) (Fig. 1). Placing the lacZ gene under the control of the constitutively expressing heterologous CMV promoter avoids the requirement for the foamy transactivator (tas) gene to express the reporter gene. The pV7-9 construct was transfected into 293 (human fibroblast) cells along with helper plasmid pSFV-1. As a control, the pV7-9 plasmid was transfected into 293 cells in the absence of the helper plasmid. Transfections were performed by a liposome-mediated method with the reagent Lipofectamine (Life Technologies, Inc., Gaithersburg, Md.). For each transfection experiment, duplicate cell cultures were transfected with 1.5 μg of vector plasmid and 1.5 μg of helper plasmid or carrier plasmid. Efficiency of transfection was determined by staining cells for β-galactosidase (β-Gal) expression. In situ staining for β-Gal activity was performed by fixing cells with 0.25% glutaraldehyde and staining with 0.2% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Blue-stained positive cells were viewed under a microscope and scored. The β-Gal staining showed that 60 to 70% of cells were expressing the reporter gene, indicating high transfection efficiency. Cell culture media from these transfected cells were collected at different days and filtered through a 0.45-μm-pore-size membrane filter. Supernatants containing virus particles were used to infect fresh 293 cells. Three days after infection, infected cells were stained for β-Gal expression, and blue-stained positive cells were scored. Figure 1 shows transduction efficiency of transducing particles harvested from vector- and helper-transfected cells at different times. The pV7-9 vector construct produced titers ranging from 0.34 × 103 to 4.7 × 103 transducing particles/ml, with higher titers obtained from samples harvested at days 5 to 9 after transfection. None of the cells infected with supernatant harvested from cells transfected with pV7-9 alone were positive for β-Gal staining. These titers obtained with the SFV-1 vector were comparable to that described for human foamy virus vectors (31).
FIG. 1.
Transduction efficiency of SFV-1 vector pV7-9. The genome organization of vector pV7-9 is shown in Fig. 2. Cells of the human fibroblast line 293 were cotransfected with pV7-9 and pSFV-1. The cell culture medium was changed 24 h after transfection. The medium was harvested at intervals and filtered and used to infect fresh 293 cells. The vector titer was measured by staining for β-Gal.
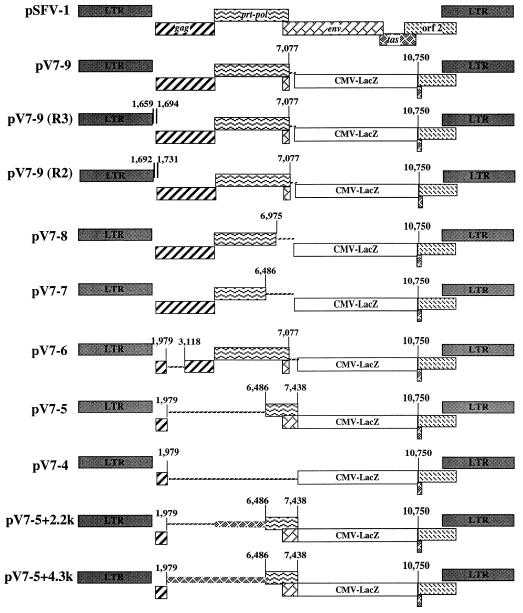
In order to define the cis-acting regions required for the SFV-1 vector, we introduced deletions in the gag and pol regions of pV7-9 as detailed in Fig. 2. cis-acting elements required for packaging the retroviral genome are located in the leader sequence at the 5′ end downstream from the splice donor site (18, 20, 35, 37). Sequences in the gag gene of retroviral genomes have also been implicated as contributing to efficient encapsidation of the viral RNA (1, 3, 4). The 5′ cis-acting elements contain stem-loop structures that are implicated in packaging and dimerization of retroviral genomes (2, 12, 16). The splice donor site for the foamy viruses is located in the R region 51 bases downstream from the transcription initiation site (26). Computer sequence analysis of the R-U5 region revealed a stable secondary RNA structure (23, 29). Similar analysis of sequences of the region between the beginning of R and the start of the gag gene also revealed a stable structure (20a). This region is highly conserved among foamy viruses with similar predicted RNA secondary structures. To determine the minimum sequence required for the SFV-1 vector, we removed the region from position 1979 to 7077 containing the entire pol gene and a portion of gag (pV7-4). This construct, therefore, included the leader sequence at the 5′ end and the first 249 nucleotides of the gag gene. pV7-4 was cotransfected along with helper pSFV-1 into 293 cells. Interestingly, supernatant harvested from the transfected cells had no transducing particles (Table 1). For other retroviruses, although less efficient, the packaging signal located in the leader sequence is sufficient for packaging the viral genome (1, 3, 4). To determine if more gag sequences are required for the SFV-1 vector, pV7-6 was constructed by deletion of sequences extending from position 1979 to 3118 within the gag gene of the parental vector, pV7-9. Supernatant harvested from pV7-6-transfected cells showed a transduction efficiency with titers equivalent to that obtained with the parental vector, pV7-9, 1.2 × 103 transducing particles/ml (Table 1), suggesting that there is no cis-acting element in the middle portion of gag. Since foamy virus replication has unique features distinct from replication of other retroviruses, we tested whether the leader sequence is critical for the SFV-1 vector. Introduction of a small deletion between the primer binding site and the gag gene at position 1692 to 1731 (pV7-9-R2) resulted in substantial reduction of the titer (33-fold). When the region from position 1659 to 1694 (pV7-9-R3) was deleted, no transduction of the reporter gene was observed. This deleted region includes the conserved palindrome that was implicated in human foamy virus RNA dimerization (10). Therefore, similar to the case for other retroviruses, the region between the primer binding site and the gag gene is critical for the foamy virus vector, implying the presence of a cis-acting element responsible for viral genome packaging.
FIG. 2.
SFV-1 vectors. Shown is the genome organization of pSFV-1 and vector derivatives. The thin hatched lines represent the deleted regions. The thick hatched lines are heterologous DNA fragments. CMV-lacZ is the coding sequence of the lacZ gene linked to the CMV immediate-early gene promoter. The numbers represent sequence positions in the proviral genome of SFV-1.
TABLE 1.
Transduction of the 293 cell line by SFV-1 vectorsa
| Vector | Titer (transducing particles/ml) |
|---|---|
| pV7-9 | 2.9 × 103 ± 285 |
| pV7-8 | 5.6 × 103 ± 367 |
| pV7-7 | 0 ± 0 |
| pV7-6 | 1.2 × 103 ± 244 |
| pV7-5 | 0 ± 0 |
| pV7-4 | 0 ± 0 |
| pV7-9 (R3) | 0 ± 0 |
| pV7-9 (R2) | 90 ± 7 |
| pV7-5+4.3k | 3.3 × 103 ± 244 |
| pV7-5+2.2k | 30 ± 8 |
Human 293 fibroblasts were infected with the indicated SFV-1 vectors. Transducing particles for infection were harvested at day 7 from cells cotransfected with the vectors and pSFV-1. Titers were measured in replicates from independent transfections by staining cells for β-Gal.
The fact that transduction was not observed with the construct pV7-4 led us to conclude that a second cis-acting element may be located either in the gag gene downstream from position 3118 or in the pol gene. To identify a second cis-acting element, we generated deletion mutations in the pol gene that removed the regions from positions 6975 to 7077 (pV7-8) and 6486 to 7077 (pV7-7). The pV7-8 vector gave a transduction efficiency with a titer (5.6 × 103 transducing particles/ml) equivalent to that of the parental vector, pV7-9 (Table 1). When the region between position 6486 and 7077 (pV7-7) was removed, however, none of the cells infected with supernatant harvested from transfected cultures were positive for β-Gal expression. This observation implied that a second cis-acting element was located in the pol gene between position 6486 and 6975. In all the transfected cells, levels of cytopathology were observed that were equivalent to that of the cell culture transfected with helper virus DNA alone. Furthermore, supernatant harvested from cells transfected with the different vector constructs and the helper pSFV-1 induced the same level of cytopathology, indicating similar virus titers in all the samples. Therefore, the packaging efficiency of the helper was not affected.
To further elucidate whether two cis-acting elements were critical for the SFV-1 vector, we constructed a vector containing the leader sequence, including the first 249 nucleotides of the gag gene, and a second region located in the pol gene (pV7-5). Interestingly, transduction of the marker gene was not observed with the pV7-5 vector. This construct was substantially smaller than the SFV-1 genome, which may have affected its packaging efficiency. To determine if size reduction of the vector affected gene transfer, a 4,300-nucleotide-long heterologous DNA fragment (restriction enzyme HindIII-digested fragment of the lambda phage genome) was cloned into the pV7-5 plasmid (pV7-5+4.3k) (Fig. 2). The pV7-5+4.3k vector had a titer (3.3 × 103 transducing particles/ml) equivalent to that of the pV7-9 vector. When a 2,200-nucleotide heterologous DNA fragment (restriction enzyme HindIII-digested fragment of the lambda phage genome) was inserted into the pV7-5 plasmid (pV7-5+2.2k) (Fig. 2), the titer was reduced substantially (97-fold). pV7-6, which had a deletion of 1,120 nucleotides in the gag gene, however, had a titer that was equivalent (1.2 × 103 transducing particles/ml) to that of pV7-9. In all transfected cell cultures, the titer of the helper virus was equivalent to that of the cell culture transfected with pSFV-1 alone. Taken together, these results suggested that two cis-acting elements were necessary for the SFV-1 vector, one located at the leader region and a second one in the pol gene between position 6486 and 6975. Furthermore, either a minimum size or a minimum distance between the two cis-acting elements has to be maintained for the SFV-1 vector.
The absolute requirement of a second cis-acting element located in the pol gene for the SFV-1 vector is unique among retroviruses. The principal signal for the major retroviral vector used in gene delivery, murine leukemia virus as well as avian spleen necrosis virus, is located in the 5′ leader sequence of the viral RNA between the major splice donor site and the gag ATG (20). Murine leukemia virus vectors containing the 5′ leader sequence are capable of generating viral particles at significant titers (4). The efficiency of packaging is further enhanced by the 5′ gag sequences by at least 1 order of magnitude. For SFV-1, however, the titer was reduced to zero in the absence of the second cis-acting region. It remains to be determined whether this region contains a packaging signal sequence. Sequences in these regions are highly conserved and show more than 83% homology among the primate foamy viruses (22). Interestingly, a second polypurine tract (PPT), most likely used as a second site of initiation of plus-strand DNA synthesis to produce a gapped unintegrated DNA intermediate, is located within this cis-acting region (27, 36). This PPT is conserved in all foamy viruses, including the bovine and feline foamy virus isolates. For human immunodeficiency virus, the central PPT is required for optimal replication (6). It is not known whether the second PPT is necessary for foamy virus replication to the extent that it has to be included in vector construction.
Acknowledgments
This research was supported by a National Institutes of Health grant (AI39126) to A. Mergia.
REFERENCES
- 1.Adam M A, Miller A D. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J Virol. 1988;62:3802–3806. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alford R L, Honda S, Lawrence C B, Belmont J W. RNA secondary structure analysis of the packaging signal for Moloney murine leukemia virus. Virology. 1991;183:611–619. doi: 10.1016/0042-6822(91)90990-s. [DOI] [PubMed] [Google Scholar]
- 3.Armentano D, Yu S-F, Kantoff P W, von Ruden T, Anderson W F, Gilboa E. Effect of internal viral sequences on the utility of retroviral vectors. J Virol. 1987;61:1647–1650. doi: 10.1128/jvi.61.5.1647-1650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender M A, Palmer T D, Gelinas R E, Miller A D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987;61:1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodem J, Löchelt M, Winkler I, Flower R P, Delius H, Flügel R M. Characterization of the spliced pol transcript of feline foamy virus: the splice acceptor site of the pol transcript is located in gag of foamy viruses. J Virol. 1996;70:9024–9027. doi: 10.1128/jvi.70.12.9024-9027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charneau P, Alizon M, Clavel F. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J Virol. 1992;66:2814–2820. doi: 10.1128/jvi.66.5.2814-2820.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin J. Retroviridae and their replication. In: Fields B N, editor. Fields virology. New York, N.Y: Raven Press; 1990. pp. 1437–1500. [Google Scholar]
- 8.Enssle J, Fischer N, Moebes A, Mauer B, Smola U, Rethwilm A. Carboxy-terminal cleavage of the human foamy virus Gag precursor molecule is an essential step in the viral life cycle. J Virol. 1997;71:7312–7317. doi: 10.1128/jvi.71.10.7312-7317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enssle J, Jordan I, Mauer B, Rethwilm A. Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc Natl Acad Sci USA. 1996;93:4137–4141. doi: 10.1073/pnas.93.9.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erlwein O, Cain D, Fischer N, Rethwilm A, McClure M O. Identification of sites that act together to direct dimerization of human foamy virus RNA in vitro. Virology. 1997;229:251–258. doi: 10.1006/viro.1997.8438. [DOI] [PubMed] [Google Scholar]
- 11.Flugel R M, Rethwilm A, Maurer B, Darai G. Nucleotide sequence analysis of the env gene and its flanking regions of the human spumaretrovirus reveals two novel genes. EMBO J. 1987;6:2077–2084. doi: 10.1002/j.1460-2075.1987.tb02473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison G P, Lever A M L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herchenroder O, Renne R, Loncar D, Cobb E K, Murthy K K, Schneider J, Mergia A, Luciw P A. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV) Virology. 1994;201:187–199. doi: 10.1006/viro.1994.1285. [DOI] [PubMed] [Google Scholar]
- 14.Hirata R K, Miller A D, Andrews R G, Russell D W. Transduction of hematopoietic cells by foamy virus vectors. Blood. 1996;88:3654–3661. [PubMed] [Google Scholar]
- 15.Hooks J J, Detrick-Hooks B. Spumavirinae: foamy virus group infections. Comparative aspects and diagnosis. In: Kurstak E, Kurstak C, editors. Comparative diagnosis of viral disease. New York, N.Y: Academic Press, Inc.; 1981. pp. 599–618. [Google Scholar]
- 16.Knight J B, Si Z H, Stoltzfus C M. A base-paired structure in the avian sarcoma virus 5′ leader is required for efficient encapsidation of RNA. J Virol. 1994;68:4493–4502. doi: 10.1128/jvi.68.7.4493-4502.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kupiec J, Kay A, Hayat M, Ravier R, Peries J, Galibert F. Sequence analysis of the simian foamy virus type 1 genome. Gene. 1991;101:185–194. doi: 10.1016/0378-1119(91)90410-d. [DOI] [PubMed] [Google Scholar]
- 18.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Löchelt M, Flügel R M. The human foamy virus pol gene is expressed as a Pro-Pol polyprotein and not as a Gag-Pol fusion protein. J Virol. 1996;70:1033–1040. doi: 10.1128/jvi.70.2.1033-1040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mann R, Mulligan R C, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 20a.Mergia, A. Unpublished data.
- 21.Mergia A, Leung N J, Blackwell J. Cell tropism of the simian foamy virus type 1 (SFV-1) J Med Primatol. 1996;25:2–7. doi: 10.1111/j.1600-0684.1996.tb00185.x. [DOI] [PubMed] [Google Scholar]
- 22.Mergia A, Luciw P A. Replication and regulation of primate foamy viruses. Virology. 1991;184:475–482. doi: 10.1016/0042-6822(91)90417-a. [DOI] [PubMed] [Google Scholar]
- 23.Mergia A, Pratt-Lowe E, Shaw K E S, Renshaw-Gegg L W, Luciw P A. cis-acting regulatory regions in the long terminal repeat of simian foamy virus type 1. J Virol. 1992;66:251–257. doi: 10.1128/jvi.66.1.251-257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mergia A, Wu M. Characterization of provirus clones of simian foamy virus type 1. J Virol. 1998;72:817–822. doi: 10.1128/jvi.72.1.817-822.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikovits J A, Hoffman P M, Rethwilm A, Ruscetti F W. In vitro infection of primary and retrovirus-infected human leukocytes by human foamy virus. J Virol. 1996;70:2774–2780. doi: 10.1128/jvi.70.5.2774-2780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muranyi W, Flügel R M. Analysis of splicing patterns of human spumaretrovirus by polymerase chain reaction reveals complex RNA structures. J Virol. 1991;65:727–735. doi: 10.1128/jvi.65.2.727-735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann-Haefelin D, Schweizer M, Corsten B, Matz B. Detection and characterization of infectious DNA intermediates of a primate foamy virus. J Gen Virol. 1986;67:1993–1999. doi: 10.1099/0022-1317-67-9-1993. [DOI] [PubMed] [Google Scholar]
- 28.Renne R, Freidl E, Schweizer M, Fleps U, Turek R, Neumann-Haefelin D. Genomic organization and expression of simian foamy virus type 3 (SFV-3) Virology. 1992;186:597–608. doi: 10.1016/0042-6822(92)90026-l. [DOI] [PubMed] [Google Scholar]
- 29.Renne R, Mergia A, Renshaw-Gegg L W, Neumann-Haefelin D, Luciw P A. Regulatory elements in the long terminal repeat (LTR) of simian foamy virus type 3. Virology. 1993;192:365–369. doi: 10.1006/viro.1993.1045. [DOI] [PubMed] [Google Scholar]
- 30.Renshaw R W, Casey J W. Transcriptional mapping of the 3′ end of the bovine syncytial virus genome. J Virol. 1994;68:1021–1028. doi: 10.1128/jvi.68.2.1021-1028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell D W, Miller A D. Foamy virus vectors. J Virol. 1996;70:217–222. doi: 10.1128/jvi.70.1.217-222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saib A, Neves M, Giron M-L, Guillemin M-C, Valla J, Peries J, Canivet M. Long-term persistent infection of domestic rabbits by the human foamy virus. Virology. 1997;228:263–268. doi: 10.1006/viro.1996.8383. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Schmidt M, Rethwilm A. Replicating foamy virus-based vectors directing high level expression of foreign genes. Virology. 1995;210:167–178. doi: 10.1006/viro.1995.1328. [DOI] [PubMed] [Google Scholar]
- 35.Sorge J, Ricci W, Hughes S H. cis-acting RNA packaging locus in the 115-nucleotide direct repeat of Rous sarcoma virus. J Virol. 1983;48:667–675. doi: 10.1128/jvi.48.3.667-675.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tobaly-Tapiero J, Kupiec J-J, Santillana-Hayat M, Canivet M, Peries J, Emanoil-Ravier R. Further characterization of the gapped DNA intermediates of human spumavirus: evidence for a dual initiation of plus-stranded DNA synthesis. J Gen Virol. 1991;72:605–608. doi: 10.1099/0022-1317-72-3-605. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe S, Temin H M. Encapsidation sequences for spleen necrosis virus, an avian retrovirus, are between the 5′ long terminal repeat and the start of the gag gene. Proc Natl Acad Sci USA. 1982;79:5986–5990. doi: 10.1073/pnas.79.19.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler I, Bodem J, Hass L, Zemba M, Delius H, Flower R, Flügel R M, Löchelt M. Characterization of the genome of feline foamy virus and its proteins shows distinct features different from those of primate spumaviruses. J Virol. 1997;71:6727–6741. doi: 10.1128/jvi.71.9.6727-6741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 40.Yu S F, Edelmann K, Strong R K, Moebes A, Rethwilm A, Linial M L. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J Virol. 1996;70:8255–8262. doi: 10.1128/jvi.70.12.8255-8262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]