Abstract
Parotid mucoepidermoid carcinoma (P-MEC) is a significant histopathological subtype of salivary gland cancer with inherent heterogeneity and complexity. Existing clinical models inadequately offer personalized treatment options for patients. In response, we assessed the efficacy of four machine learning algorithms vis-à-vis traditional analysis in forecasting the overall survival (OS) of P-MEC patients. Using the SEER database, we analyzed data from 882 postoperative P-MEC patients (stages I–IVA). Single-factor Cox regression and four machine learning techniques (random forest, LASSO, XGBoost, best subset regression) were employed for variable selection. The optimal model was derived via stepwise backward regression, Akaike Information Criterion (AIC), and Area Under the Curve (AUC). Bootstrap resampling facilitated internal validation, while prediction accuracy was gauged through C-index, time-dependent ROC curve, and calibration curve. The model’s clinical relevance was ascertained using decision curve analysis (DCA). The study found 3-, 5-, and 10-year OS rates of 0.887, 0.841, and 0.753, respectively. XGBoost, BSR, and LASSO stood out in predictive efficacy, identifying seven key prognostic factors including age, pathological grade, T stage, N stage, radiation therapy, chemotherapy, and marital status. A subsequent nomogram revealed a C-index of 0.8499 (3-year), 0.8557 (5-year), and 0.8375 (10-year) and AUC values of 0.8670, 0.8879, and 0.8767, respectively. The model also highlighted the clinical significance of postoperative radiotherapy across varying risk levels. Our prognostic model, grounded in machine learning, surpasses traditional models in prediction and offer superior visualization of variable importance.
Keywords: Machine learning, Parotid mucoepidermoid carcinoma, Nomogram, Postoperative radiotherapy, SEER
Subject terms: Head and neck cancer, Risk factors
Introduction
Parotid gland carcinoma represents an uncommon, markedly heterogeneous malignancy of the head and neck region1. As per the World Health Organization (WHO) classification, over 20 histological subtypes exist, encompassing adenoid cystic carcinoma, mucoepidermoid carcinoma (MEC), secretory carcinoma, among others, with MEC being the predominant tissue type2. MEC is characterized as a malignant glandular epithelial neoplasm, characterized by the presence of mucinous, intermediate, and epidermoid cells exhibiting columnar, clear cell, and cancer-like features. Based on invasiveness and differentiation, MEC is stratified into low, intermediate, and high-grade malignant neoplasms3–5.
The tumor’s pathological grade is an important factor in determining the prognosis for MEC patients, and it often guides treatment approaches6–9. However, the TNM system and pathological grade fall short in estimating survival or informing adjuvant therapy-related decisions10. Previous research has shown that high-grade tumors and those with positive margins are at a higher risk of recurrence and spread. Post-operative radiation therapy can help to prevent the recurrence of the tumor and improve overall outcomes for these patients11–13. Therefore, postoperative management is crucial for the long-term prognosis of parotid mucoepidermoid carcinoma (P-MEC) patients. Additionally, the postoperative stage is also a critical period for disease monitoring and making clinical decisions, such as the development of postoperative radiation therapy plans. Hence, a comprehensive and accurate postoperative survival prediction model is needed to integrate multiple clinical pathological features to guide treatment decision-making and disease monitoring.
Although the preoperative phase and initial treatment strategies are undoubtedly important in the management of P-MEC, the majority of patients undergo surgery as the primary treatment modality. Therefore, the objective of this study is to establish a personalized, clinically valuable, and effective predictive model specifically for postoperative P-MEC. Prior research has been limited by small sample sizes, brief study durations, and single-center designs. In contrast, the Surveillance, Epidemiology, and End Results (SEER) database serves as a comprehensive trove of clinical data collected from a diverse array of cancer patients across multiple registries spanning the vast expanse of the United States. The database’s wide-reaching coverage imbues it with the capacity to offer a representative sample of cancer incidence and survival rates across the entire US populace, thereby enhancing the credibility and robustness of research studies10. Machine learning methods have been extensively employed in other tumor prediction models, with their analytical and visual capabilities serving as potent tools for clinical forecasting. Machine learning approaches consider a broader range of variable relationships compared to conventional statistical methodologies. To date, no studies have explored machine learning-based predictive models for postoperative P-MEC14,15.
A nomogram integrating visual representations with hazard probability scores for each prognostic factor to construct a proportional hazards regression model was developed and validated using a large SEER population database and screening suitable variables via traditional and machine learning methods. Moreover, risk stratification based on the nomogram scores substantially enhances clinical applicability, offering clinicians tailored treatment strategies for patients with postoperative P-MEC.
Method
Study population: SEER data
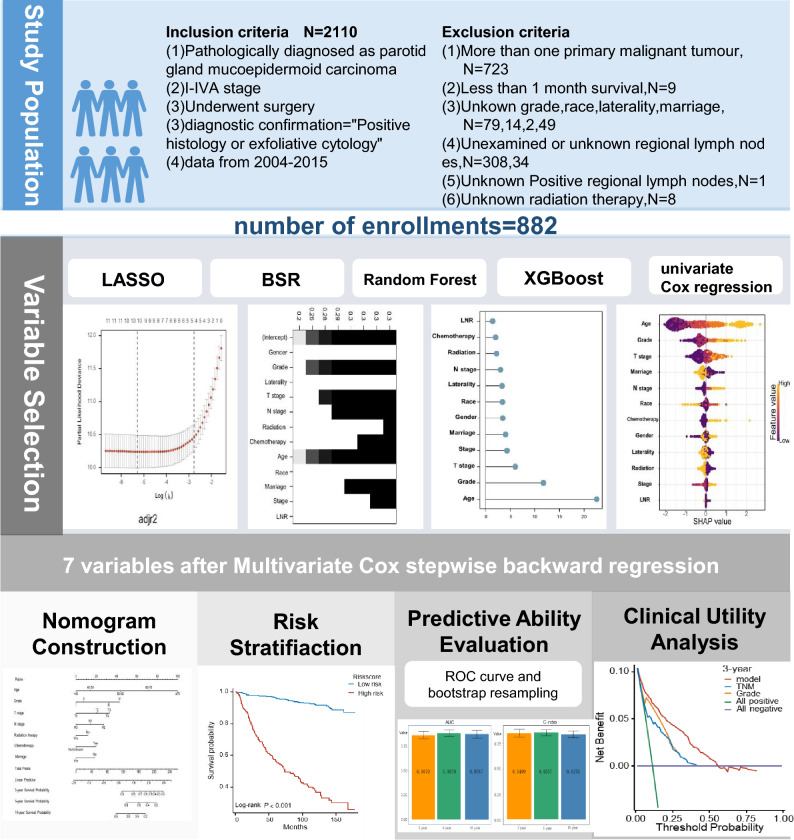
For this study, SEER*STAT software (version 8.4.0) was employed to extract clinical data of patients with P-MEC between 2004 and 2015.The criterias for the inclusion of P-MEC patients were as follows: (1) patients with a primary site of “parotid gland: 07.9”; (2) patients with pathologically confirmed stage I-IVA P-MEC; (3) post-surgical patients; (4) patients with diagnostic confirmation of “Positive histology or exfoliative cytology”. Additionally, patients who were not primary cases, lived less than 1 month, had unclear basic information, were not examined for regional lymph nodes, or had inaccurate positive lymph node results were excluded. The screening process is illustrated in Fig. 1.
Figure 1.
Analysis flow for the development and evaluation of models for parotid mucoepidermoid carcinoma(P-MEC).
Variable selection
To identify the clinical variables that significantly impact the overall survival (OS) of patients, various statistical methods were employed, including extreme gradient boosting (XGBoost), least absolute shrinkage and selection operator (LASSO), best subsets regression (BSR), random forest (RF), and Cox proportional hazard regression. The LNR, representing the ratio of regional nodes testing positive among those examined, was calculated, and its cut-off point was determined using X-tile software. Subsequently, the clinical variables were screened using stepwise backward regression with the “MASS” package. Model fitting was evaluated by the Akaike Information Criterion (AIC), and the area under the receiver operating characteristic curve (AUC) was used to evaluate the model’s accuracy. The ‘survival’ package was used for proportional hazards hypothesis testing and Cox regression analysis, the ‘caret’ package for RF analysis with default settings and 20-fold cross-validation, the ‘shapviz’ and ‘xgboost’ packages for XGBoost analysis, the ‘glmnet’ package for LASSO analysis, and the ‘leaps’ package for BSR analysis.
Nomogram construction
The nomogram, an effective visual tool for predicting survival outcomes, was constructed using the “rms,” “timeROC,” and “nomogramFormula” packages, based on the variables included in the Cox proportional hazards regression model. The nomogram values were assigned to each patient, and the ROC curve analysis was utilized to determine the optimal cutoff value for risk stratification.
Model validation
The calibration curves were used to assess the agreement between the predicted probabilities and observed outcomes, and the ROC curve and AUC were utilized to evaluate the model’s discrimination ability. The internal validation was performed using the bootstrapping method, where 1000 resampling was performed to validate the model, using the “riskRegression” and “pec” packages16. Moreover, to identify the clinical utility of the model, the decision curve analysis (DCA) through the “stdca.R” package was employed17. Additionally, the Kaplan–Meier (K–M) method was used to compare the OS of various groups, and the log-rank test was used to evaluate its significance with a P-value threshold of < 0.05. The statistical analyses were performed using R software (R version 4.2.1), and the seed was set to 1234 for all the tests.
Results
Screening and characteristics of the patients
This study examined 882 patients with stage I–IVA P-MEC, who met the inclusion–exclusion criteria, from the SEER database between 2004 and 2015. Figure 1 illustrates the patient selection process, while Table 1 summarizes patients’ demographic and clinicopathological characteristics. The lymph node ratio (LNR) cut-off was determined using X-tile analysis, with a resultant cut-off of 1.15%. The median (95% CI) follow-up time was 99 (92–105) months, and the median (IQR) age at diagnosis was 52 (37–66) years. A majority of the patients were white (661, 74.9%), with most tumors being grade II (396, 44.9%), stage I (353, 40%), T1-stage (381, 43.2%), N0-stage (685, 77.7%), and LNR0 (686, 77.8%) according to the AJCC 6th stage. All variables, except for chemotherapy (94.2% vs 5.8%), had proportions exceeding 10%. The study encompassed 12 variables, including age, gender, grade, stage, tumor (T) stage, node (N) stage, radiation, chemotherapy, laterality, marriage, and LNR. Nine factors—age, gender, grade, stage, T stage, N stage, radiation, chemotherapy, and LNR—were selected based on univariate Cox regression. Multivariate Cox regression revealed that four factors (age, grade, T stage, and chemotherapy) were independent risk factors, each with P-values less than 0.05. In the multivariate analysis, individuals aged 60–70 years (HR = 5.936, 95% CI = 3.016–11.681, P < 0.001), those over 70 years old (HR = 11.962, 95% CI = 6.303–22.703, P < 0.001), Grade III (HR = 2.324, 95% CI = 1.235–4.375, P = 0.009), Grade IV (HR = 3.148, 95% CI = 1.710–5.795, P < 0.001), T2 (HR = 3.162, 95% CI = 1.059–9.440, P = 0.039), T3 (HR = 4.300, 95% CI = 1.501–12.316, P = 0.007), T4 (HR = 4.414, 95% CI = 1.439–13.535, P = 0.009), and chemotherapy (HR = 1.721, 95% CI = 1.096–2.703, P = 0.018) emerged as independent risk factors for overall survival (OS). Nevertheless, radiation(HR = 0.750, 95% CI = 0.525–1.072, P = 0.114), LNR (HR = 0.868, 95% CI = 0.114–6.602, P = 0.891), and other variables demonstrated no prognostic value (Table 2).
Table 1.
Demographic and clinical characteristics of patients with P-MEC.
| Characteristics | n (%) |
|---|---|
| Age | |
| Median (IQR) | 52.00 [37.00, 66.00] |
| < 40 | 254 (28.8) |
| 40–50 | 146 (16.6) |
| 50–60 | 173 (19.6) |
| 60–70 | 138 (15.6) |
| ≥ 70 | 171 (19.4) |
| Gender | |
| Female | 445 (50.5) |
| Male | 437 (49.5) |
| Race | |
| White | 669 (74.9) |
| Black | 120 (13.6) |
| Other | 101 (11.5) |
| Grade | |
| I | 234 (26.5) |
| II | 396 (44.9) |
| III | 120 (13.6) |
| IV | 132 (15) |
| Stage | |
| I | 353 (40) |
| II | 202 (22.8) |
| III | 170 (18.9) |
| IVA | 161 (18.3) |
| Tumor (T) stage | |
| T1 | 381 (43.2) |
| T2 | 257 (29.1) |
| T3 | 145 (16.4) |
| T4 | 99 (11.2) |
| Node (N) stage | |
| N0 | 685 (77.7) |
| N1 | 110 (12.5) |
| N2 | 87 (9.9) |
| Radiation | |
| No | 450 (51) |
| Yes | 432 (49) |
| Chemotherapy | |
| No/unknown | 831 (94.2) |
| Yes | 51 ( 5.8) |
| Laterality | |
| Left | 413 (46.8) |
| Right | 469 (53.2) |
| Marriage | |
| No | 390 (44.2) |
| Yes | 492 (55.8) |
| Lymph node ratio (%) | |
| Median (IQR) | 0.00 [0.00, 0.00] |
| LNR0 | 686 (77.8) |
| LNR1 | 196 (22.2) |
Grade is codefined as follows: Grade I, well differentiated, Grade II, moderately differentiated; Grade III, poorly differentiated, Grade IV, undifferentiated; anaplastic.
Lymph node ratio, the number of positive lymph nodes divided by the number of neck lym-ph nodes dissected. LNR0, LNR(%) < 1.15. LNR1, LNR(%) ≥ 1.15.
IQR, inter quartile range.
Table 2.
Univariate and multivariate analyses for OS in patients with P-MEC.
| Characteristics | Univariable analysis | Multivariable analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age | < 0.001 | |||
| < 40 | Reference | Reference | ||
| 40–50 | 1.453 (0.628–3.363) | 0.383 | 1.305 (0.561–3.036) | 0.536 |
| 50–60 | 3.282 (1.656–6.505) | < 0.001 | 2.594 (1.286–5.234) | 0.008 |
| 60–70 | 7.195 (3.766–13.746) | < 0.001 | 5.936 (3.016—11.681) | < 0.001 |
| ≥ 70 | 18.925 (10.379—34.508) | < 0.001 | 11.962 (6.303—22.703) | < 0.001 |
| Gender | < 0.001 | |||
| Female | Reference | Reference | ||
| Male | 2.112 (1.563–2.855) | < 0.001 | 1.101 (0.797–1.522) | 0.560 |
| Race | < 0.001 | |||
| White | Reference | Reference | ||
| Black | 0.533 (0.319–0.891) | 0.016 | 1.444 (0.840–2.483) | 0.184 |
| Other | 0.437 (0.237–0.806) | 0.008 | 0.839 (0.449–1.570) | 0.584 |
| Grade | < 0.001 | |||
| I | Reference | Reference | ||
| II | 1.668 (0.975–2.856) | 0.062 | 1.346 (0.773–2.343) | 0.294 |
| III | 6.837 (3.981–11.741) | < 0.001 | 2.324 (1.235–4.375) | 0.009 |
| IV | 9.505 (5.654–15.978) | < 0.001 | 3.148 (1.710–5.795) | < 0.001 |
| Stage | < 0.001 | |||
| I | Reference | Reference | ||
| II | 1.668 (1.029–2.706) | 0.038 | 0.481 (0.145–1.596) | 0.232 |
| III | 3.353 (2.173–5.175) | < 0.001 | 0.480 (0.149–1.544) | 0.218 |
| IVA | 5.950 (3.970–8.917) | < 0.001 | 0.462 (0.130–1.638) | 0.232 |
| Tumor (T) stage | < 0.001 | |||
| T1 | Reference | Reference | ||
| T2 | 2.199 (1.454–3.325) | < 0.001 | 3.162 (1.059–9.440) | 0.039 |
| T3 | 4.210 (2.773–6.390) | < 0.001 | 4.300 (1.501–12.316) | 0.007 |
| T4 | 5.440 (3.534–8.376) | < 0.001 | 4.414 (1.439- 13.535) | 0.009 |
| Node (N) stage | < 0.001 | |||
| N0 | Reference | Reference | ||
| N1 | 2.972 (2.067–4.274) | < 0.001 | 1.807 (0.226–14.438) | 0.577 |
| N2 | 4.849 (3.400–6.916) | < 0.001 | 2.517 (0.300–21.156) | 0.395 |
| Radiation | < 0.001 | |||
| No | Reference | Reference | ||
| Yes | 2.074 (1.531–2.810) | < 0.001 | 0.750 (0.525–1.072) | 0.114 |
| Chemotherapy | < 0.001 | |||
| No/unknown | Reference | Reference | ||
| Yes | 3.871 (2.569–5.832) | < 0.001 | 1.721 (1.096–2.703) | 0.018 |
| Laterality | 0.648 | |||
| Left | Reference | |||
| Right | 0.935 (0.702–1.246) | 0.648 | ||
| Marriage | 0.327 | |||
| No | Reference | |||
| Yes | 0.866 (0.649–1.154) | 0.326 | ||
| Lymph node ratio 9%) | < 0.001 | |||
| LNR0 | Reference | Reference | ||
| LNR1 | 3.647 (2.731–4.869) | < 0.001 | 0.868 (0.114–6.602) | 0.891 |
Statistical significance indicated by P < 0.05 has been denoted in bold.
Variable selection
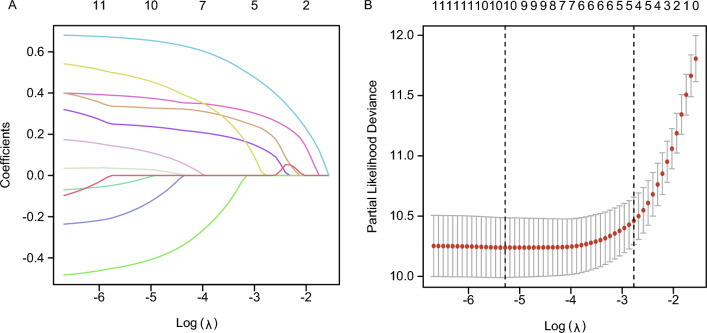
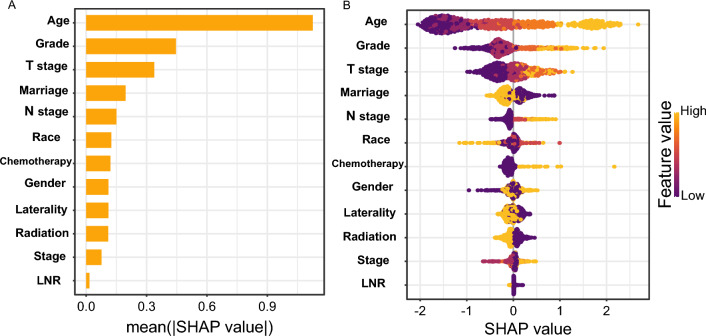
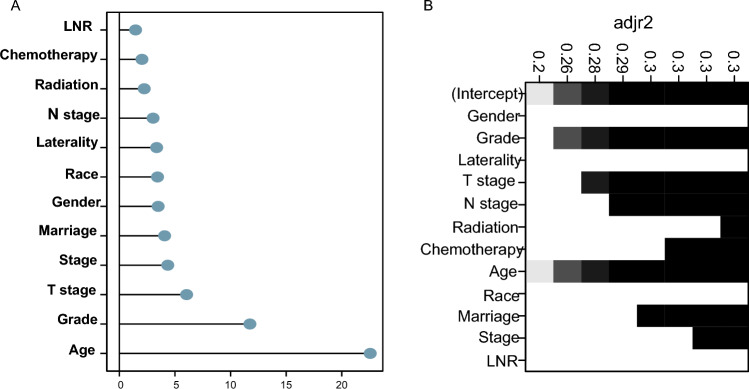
Figure 2A displays the relationship between the LASSO coefficients and the regularization parameter, lambda (λ), and demonstrates the variable selection process and the effect of λ on the coefficients. The “lambda.min” value, which represents the lambda value corresponding to the minimum likelihood deviation or the highest C-index, was utilized for selecting tuning parameters in LASSO regression. Another vertical line was “lambda.1se,” which corresponds to the most regularized model within one standard error of the minimum (Fig. 2B). The “λ.min” (λ = 0.0050724) was chosen for the best predictive performance. A ten-fold cross-validation was employed. Ten variables were chosen through the LASSO regression algorithm, including age, gender, grade, T stage, N stage, radiation, chemotherapy, laterality, marriage, and LNR. Employing the adjusted R-squared maximum of the BSR, we selected eight variables: age, grade, stage, T stage, N stage, radiation, chemotherapy, and marriage(Fig. 3). In the RF model and XGBoost, we independently extracted the top 10 variables, excluding laterality, radiation (RF), and LNR (XGBoost) (Fig. 4). We assessed the key performance of machine learning and traditional statistics using AUC and AIC. Multivariate Cox stepwise backward regression reconfirmation identified LASSO, BSR, and XGBoost as the best of the five screening methods based on both AUC (AUC = 88.4) and AIC (AIC = 2118.9) criteria (Table 3).
Figure 2.
Predictor Screening: the least absolute shrinkage and selection operator (LASSO) regression and fivefold cross-validation.
Figure 3.
Predictor Screening: A SHAP plot and a feature importance plot are visualizations used to interpret XGBoost model results.
Figure 4.
Predictor Screening: (A) Random Forest importance plot; (B) Best Subset Regression (BSR), it selected the best subset of predictor variables to accurately model a response variable.
Table 3.
Screening results of five methods for identifying predictors.
| Characteristics | Univariate Cox regression | LASSO regression | Besubset regression | Random forest | XGboost | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Preliminary | Final | Preliminary | Final | Preliminary | Final | Preliminary | Final | Preliminary | Final | |
| Age | + | + | + | + | + | + | + | + | + | + |
| Gender | + | + | + | + | ||||||
| Race | + | + | + | |||||||
| Grade | + | + | + | + | + | + | + | + | + | + |
| Stage | + | + | + | + | ||||||
| T stage | + | + | + | + | + | + | + | + | + | + |
| N stage | + | + | + | + | + | + | + | + | + | |
| Radiation | + | + | + | + | + | + | + | + | ||
| Chemotherapy | + | + | + | + | + | + | + | + | + | + |
| Laterality | + | + | ||||||||
| Marriage | + | + | + | + | + | + | + | + | ||
| Lymph node ratio(%) | + | + | + | |||||||
| Total number | 10 | 6 | 10 | 7 | 8 | 7 | 10 | 6 | 10 | 7 |
| AUC(5 years) | 87.7 | 88.4 | 88.4 | 88.0 | 88.4 | |||||
| AIC | 2131.3 | 2122.3 | 2123.9 | 2118.9 | 2122.6 | 2118.9 | 2129.0 | 2120.0 | 2124.2 | 2118.9 |
Final, the results obtained after the model undergone Multivariate Cox stepwise backward regression.
Statistical significance indicated by P < 0.05 has been denoted in bold.
Nomogram construction
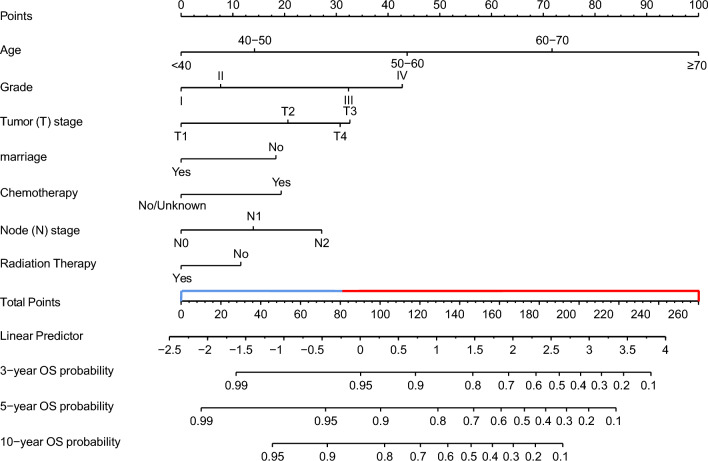
Consequently, we constructed a nomogram with seven variables from the three algorithms (LASSO, BSR, and XGBoost), including age, grade, tumor stage, node stage, chemotherapy, radiation, and marriage. We developed an OS-nomogram capable of predicting a patient’s 3-, 5-, and 10-year OS rates using these variables (Fig. 5). By converting clinical, pathological, and therapeutic factors into points, the nomogram accurately predicted OS. The total risk point score, calculated by summing all points, significantly correlated with 3-, 5-, and 10-year OS. We utilized a 5-year ROC curve to determine the optimum risk score cut-off point. Kaplan–Meier curves revealed that low-risk group patients (risk score < 80.29) had better survival prognosis compared to high-risk group patients (risk score ≥ 80.29, log-rank test, P < 0.001) (Fig. S1).
Figure 5.
A survival nomogram for predicting overall survival (OS) for patients with P-MEC. (1) When using the nomogram, seven predictors were quantified as “point” based on patient-specific factors and then the sum of the “point” corresponded to the “total point” below, which corresponded to the 3, 5, 10 year OS ; (2) The optimal cut-off total point was 80.29 (the median of patients’ point), which divided the patients into high-risk group and low-risk group.
Predictive ability evaluation
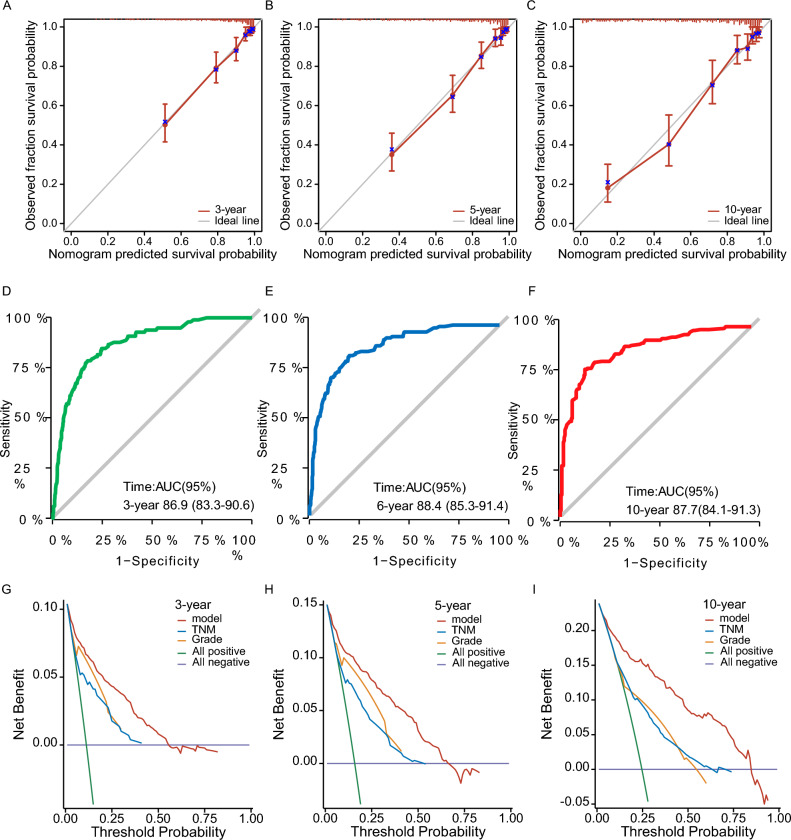
We evaluated the predictive ability of our nomogram by constructing time-dependent receiver operating characteristic (ROC) curves at 3, 5, and 10 years. The ROC curves demonstrated excellent discriminative capacity of our model, with areas under the curves (AUCs) of 86.9 (95% CI = 83.3–90.6), 88.4 (95% CI = 83.5–91.4), and 87.7 (95% CI = 84.1–91.3) (Fig. 6). This indicates that our model has high accuracy in predicting overall survival in parotid MEC patients.
Figure 6.
(A–C) The calibration curves. The calibration curves of the nomogram predicting (A) 3-years, (B) 5-years, and (C) 10-years OS. (D–F) Time dependent ROC curve. (D) ROC curves for 3-year, (E) 5-year, and (F) 10-year overall survival rates. (G–I) Decision curve analysis (DCA) plot. (G) DCA plot for 3-year, (H) 5-year, and (I) 10-year overall survival rates.
We also performed 1000 bootstrap resampling analyses on the dataset and generate calibration plots for the prediction model. The calibration plots showed that the curves closely aligned with the 45-degree line, indicating a well-calibrated model in practical use (Fig. 6). Furthermore, the 1000 bootstrap resamplings indicated good concordance between actual and predicted values in both the training and validation datasets, as evidenced by C-index (3-year, 0.8499, 0.775–0.914; 5-year 0.8557, 0.793–0.911; 10-year, 0.8375, 0.772–0.897) and AUC (3-year, 0.8670, 95 CI% = 0.787–0.935; 5-year, 0.8879, 95 CI% = 0.82–0.945; 10-year, 0.8767, 95 CI% = 0.792–0.947). (Fig. 7). These results further support the reliability and accuracy of our prediction model.
Figure 7.
This figure presents a bootstrap analysis of a dataset, displaying the 3-year and 5-year AUC and C-index values. The analysis was performed using 1000 bootstrap replicates. The figure demonstrates the accuracy and predictive power of the model for the specified time intervals.
Clinical utility analysis
To determine the clinical utility of our prediction model, we utilized the decision curve analysis (DCA) plot. The DCA plot illustrates the net benefit of the prediction model across a spectrum of threshold probabilities. Our model demonstrates clinical utility, as evidenced by its net benefit curve lies above both two lines across the range of threshold probabilities (Fig. 6). This suggests that our prediction model is more effective than TNM stage or grade and can aid in making clinical decisions for P-MEC patients.
In summary, our nomogram exhibited excellent predictive ability and calibration, as well as clinical utility, indicating its potential usefulness in clinical practice.
Discussion
In clinical research, traditional linear statistical methods, such as multivariate regression and correlation analysis, often encounter challenges due to the complexity of biological variables.These variables frequently display nonlinear relationships and exhibit conditional dependencies. The fundamental assumption of these methods is that variables are independent, and there exist linear relationships among them. However, these assumptions often fall short in practice. Machine learning offers a robust solution to these problems. ML techniques can capture nonlinear relationships and conditional dependencies in data, demonstrating high accuracy and flexibility when handling clinical research data. To date, several ML-based predictive models have been developed, including models based on conditional survival forests and random survival forests for the study of advanced salivary gland cancer and primary salivary gland cancer, respectively18,19.The determination of prognostic factors exhibits considerable variation among patients with P-MEC, largely due to differences in pathological grading and disease stage. Furthermore, the establishment of reliable prognostic factors remains a challenging task due to limitations such as patient size and subjective grading. However, our research incorporates an expanded set of ML algorithms. Among the algorithms employed in this study, XGBoost, BSR, and LASSO screening displayed excellent overall predictive performance.
Our study identified seven independent prognostic factors and designed Kaplan–Meier plots for risk stratification, potentially providing valuable insights for clinical practice. Age, grade, T stage, N stage, radiotherapy, chemotherapy, and marital status have been determined as independent prognostic factors for the OS of patients with P-MEC.
Age, grade, T stage, and N stage are factors that have been extensively explored and validated across 18 studies published between 1968 and 20208. Beyond these parameters, our study incorporates additional variables such as radiotherapy, chemotherapy, and marital status. Subgroup analyses were conducted to further understand the study population. The role of radiotherapy in treating mucoepidermoid carcinoma of the salivary glands has not been thoroughly examined in prior research. Our in-depth analysis of patients who underwent postoperative radiotherapy suggests that individuals in low-risk categories may not derive significant benefits from this treatment20–23. Additionally, our findings regarding LNR diverge from those of previous studies, which will be elaborated upon in subsequent sections of this paper.
A review of the National Cancer Database and a meta-analysis further confirmed the importance of age as a prognostic factor24. This meta-analysis of survival factors in P-MEC patients showed that in 33% of search studies, older age was associated with worse survival outcomes (HR, 1.02–6.86)8. A 30-year retrospective study on a predominantly MEC affected sample also demonstrated a more favorable prognosis for children compared to adults (95% ± 1.5% compared with 59% ± 0.5% (P < 0.001))25.
Our study found that the prognosis of P-MEC patients is grade-dependent, with poor prognoses observed for patients with high grade, consistent with prior research8. The inclusion of marital status in the model was supported by previous studies, suggesting that married patients may experience better prognoses due to factors such as “spousal protection”26,27.
Currently, the treatment of primary mucoepidermoid carcinoma of the parotid gland is based on the overall treatment strategy of parotid gland cancer, according to the National Comprehensive Cancer Network (NCCN) guidelines27. The role of radiotherapy as a protective prognostic factor for parotid cancer patients has been a subject of controversy20–23. For patients with advanced, high-grade (poorly differentiated or undifferentiated), nerve or vascular invasion, insufficient or positive margins, extraparotid invasion, or lymph node involvement, postoperative radiotherapy is recommended. Our study further analyzed the clinical value of postoperative radiotherapy at different risk levels. Compared with surgery alone, postoperative radiotherapy did not significantly improve the survival of patients in the low-risk group (P = 0.973). In contrast, there was a significant therapeutic advantage in survival performance for patients in the high-risk group (P = 0.002).This suggests that the low-risk population may not benefit from postoperative radiation therapy in cases of postoperative P-MEC, potentially preventing overtreatment. Previous studies have reported successful postoperative radiotherapy for low-grade pT2N0 P-MEC and cases where postoperative radiotherapy significantly improved survival in advanced, high-grade patients. Our results attempt to analyze the optimal population that can benefit from postoperative radiotherapy by integrating pathologic and demographic characteristics, providing limited evidence for the choice of treatment options28–30. Further multicenter, large sample retrospective or prospective studies are needed to validate these findings.
During surgery for parotid gland cancer, lymph node dissection and biopsy are performed. Generally, the association between LNR and OS in parotid gland cancer is worth affirming. However, due to differences in research endpoints, subjects, and statistical methods, our study suggests that LNR is not an independent prognostic factor for P-MEC patients. Q. Fang et al. reviewed more than 20 years of postoperative patients with parotid cancer and confirmed our viewpoint. Furthermore, the study believed that more than two positive parotid lymph nodes reduced the control of recurrence-free survival (RFS)31. A German retrospective cohort study after parotid cancer surgery also found that LNR was not the best prognostic factor32. Due to the failure of LNR to consider the condition of lymph nodes in the parotid gland, the study suggests that the evaluation of lymph nodes in the parotid gland should be included. Consequently, more analysis is required to determine the effect of LNR on the prognosis of P-MEC patients.
This study is subject to several limitations. Firstly, its retrospective design may lead to selection bias. Secondly, the absence of data on nerve and vascular invasion could compromise the accuracy of the findings. Additionally, the reliance on cancer data primarily from North America restricts the generalizability of the results to other regions, particularly Asia. Furthermore, both the development and validation cohorts were sourced from the same database, which might affect the robustness of the conclusions. Although the nomogram and risk stratification systems underwent comprehensive internal validation, the limited number of P-MEC patients prevented external validation, potentially limiting the external applicability of the findings. Moreover, the study did not account for all possible factors influencing patient survival, such as more detailed treatment protocols (e.g., radiation dosage, chemotherapy drugs) and more comprehensive information on individual heterogeneity (e.g., molecular markers). Despite these limitations, our clinical prediction model shows substantial potential in predicting the survival rate of P-MEC patients based on SEER database, offering reliable results for clinical decision-making.
Conclusion
In conclusion, this study developed a machine learning-based nomogram for predicting overall survival in postoperative P-MEC patients, incorporating age, pathological grade, T stage, N stage, radiation therapy, chemotherapy, and marital status as independent prognostic factors.The feature selection capabilities of machine learning not only significantly enhance model-building process but also offer improved predictive performance and enable effective visualization of variable importance. The nomogram has the potential to aid clinicians in making personalized treatment recommendations and clinical management decisions for P-MEC patients, thereby improving patient outcomes.
Supplementary Information
Acknowledgements
We thank the National Cancer Institute for providing the SEER data set.
Abbreviations
- P-MEC
Parotid mucoepidermoid carcinoma
- OS
Overall survival
- AIC
Akaike information criterion
- AUC
Area under the curve
- DCA
Decision curve analysis
- WHO
World Health Organization
- MEC
Mucoepidermoid carcinoma
- SEER
Surveillance, Epidemiology, and End Results
- XGBoost
Extreme gradient boosting
- LASSO
Least absolute shrinkage and selection operator
- BSR
Best subsets regression
- RF
Random forest
- K–M
Kaplan–Meier
- IQR
Interquartile range
- ROC
Receiver operating characteristic
- NCCN
National Comprehensive Cancer Network
- LNR
Lymph node ratio
- RFS
Recurrence-free survival
Author contributions
Zongwei Huang and Zihan Chen contributed equally to this work, including conducting the majority of the experiments, analyzing the data, and drafting the manuscript. Ying Li and Ting Lin were involved in data collection and contributed to the design of the experiments. Sunqin Cai and Wenxi Wu contributed to the data analysis and interpretation. Lishui Wu and Siqi Xu provided the materials and analysis tools and were also involved in revising the manuscript critically for important intellectual content. Jun Lu and Sufang Qiu, the co-corresponding authors, made substantial contributions to the conception and design of the work, and they also revised the manuscript and gave final approval of the version to be published. All authors read and approved the final manuscript.
Funding
This work was supported by the grants of Science and Technology Program of Fujian Province, China (2018Y2003); Fujian Provincial Clinical Research Center for Cancer Radiotherapy and Immunotherapy (2020Y2012); Supported by the National Clinical Key Specialty Construction Program (2021); Fujian Clinical Research Center for Radiation and Therapy of Digestive, Respiratory and Genitourinary Malignancies; National Natural Science Foundation of China (82072986); Major Research Projects for Young and Middle-aged Researchers of Fujian Provincial Health Commission (2021ZQNZD010); Science and Technology Pilot Program of Fujian Province, China (2021Y0053); Wu Jieping Medical Foundation (320.6750.2021-01-27); Joint Funds for the Innovation of Science and Technology, Fujian province (2021Y9196); High-level Talent Training Program of Fujian Cancer Hospital (2022YNG07); Innovative Medicine Subject of Fujian Provincial Health Commission, China (2021CXA029); and Natural Science Foundation of Fujian province (2023J01121763).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zongwei Huang and Zihan Chen.
Contributor Information
Jun Lu, Email: lujvip@126.com.
Sufang Qiu, Email: sufangqiu@fjmu.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-58329-8.
References
- 1.Guzzo M, et al. Major and minor salivary gland tumors. Crit. Rev. Oncol. Hematol. 2010;74:134–148. doi: 10.1016/j.critrevonc.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Skálová A, Hyrcza MD, Leivo I. Update from the 5th edition of the World Health Organization classification of head and neck tumors: Salivary glands. Head Neck Pathol. 2022;16:40–53. doi: 10.1007/s12105-022-01420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Healey WV, Perzin KH, Smith L. Mucoepidermoid carcinoma of salivary gland origin. Classification, clinical-pathologic correlation, and results of treatment. Cancer. 1970;26:368–388. doi: 10.1002/1097-0142(197008)26:2<368::AID-CNCR2820260219>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 4.Nishida H, Kusaba T, Kawamura K, Oyama Y, Daa T. Histopathological aspects of the prognostic factors for salivary gland cancers. Cancers. 2023;15:1236. doi: 10.3390/cancers15041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peraza A, Gómez R, Beltran J, Amarista F. Mucoepidermoid carcinoma An update and review of the literature. J. Stomatol. Oral Maxillofac. Surg. 2020;121:713–720. doi: 10.1016/j.jormas.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Niu X, Fang Q, Liu F. Role of intraparotid node metastasis in mucoepidermoid carcinoma of the parotid gland. BMC Cancer. 2019;19:1–5. doi: 10.1186/s12885-019-5637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goode RK, Auclair PL, Ellis GL. Mucoepidermoid carcinoma of the major salivary glands: Clinical and histopathologic analysis of 234 cases with evaluation of grading criteria. Cancer Interdiscip. Int. J. Am. Cancer Soc. 1998;82:1217–1224. doi: 10.1002/(sici)1097-0142(19980401)82:7<1217::aid-cncr2>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 8.Cheng EY, et al. Clinicopathological predictors of survival for parotid mucoepidermoid carcinoma: A systematic review. Otolaryngol. Head Neck Surg. 2023;168:611–618. doi: 10.1177/01945998221086845. [DOI] [PubMed] [Google Scholar]
- 9.Lin X, Lin Q, Chen X, Wang D. Number of positive lymph nodes combined with the logarithmic ratio of positive lymph nodes predicts long-term survival for patients with node-positive parotid gland carcinoma after surgery: A SEER population-based study. Eur. Arch. Oto Rhino Laryngol. 2023 doi: 10.1007/s00405-023-07848-7. [DOI] [PubMed] [Google Scholar]
- 10.Shafique K, et al. Pathologic grading of mucoepidermoid carcinomas of the salivary gland and its effect on clinicopathologic follow-up: An institutional experience. Human Pathol. 2020;98:89–97. doi: 10.1016/j.humpath.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Park G, Lee S-W. Postoperative radiotherapy for mucoepidermoid carcinoma of the major salivary glands: Long-term results of a single-institution experience. Radiat. Oncol. J. 2018;36:317. doi: 10.3857/roj.2018.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh-Laskar S, et al. Mucoepidermoid carcinoma of the parotid gland: Factors affecting outcome. Head neck. 2011;33:497–503. doi: 10.1002/hed.21477. [DOI] [PubMed] [Google Scholar]
- 13.Taylor ZC, et al. Overall and cause-specific survival for mucoepidermoid carcinoma of the major salivary glands: Analysis of 2210 patients. World J. Clin. Oncol. 2020;11:1029. doi: 10.5306/wjco.v11.i12.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan X, et al. A survival prediction model via interpretable machine learning for patients with oropharyngeal cancer following radiotherapy. J. Cancer Res. Clin. Oncol. 2023;149(10):6813–6825. doi: 10.1007/s00432-023-04644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangil T, et al. Predicting clinical outcomes of radiotherapy for head and neck squamous cell carcinoma patients using machine learning algorithms. J. Big Data. 2022;9:1–19. doi: 10.1186/s40537-022-00578-3. [DOI] [Google Scholar]
- 16.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. CRC Press; 1994. [Google Scholar]
- 17.Vickers AJ, Elkin EB. Decision curve analysis: A novel method for evaluating prediction models. Med. Decis. Mak. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, et al. Prognostic risk factor of major salivary gland carcinomas and survival prediction model based on random survival forests. Cancer Med. 2023 doi: 10.1002/cam4.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TH, Eun Y-G, Lee YC. A machine learning based survival prediction model for advanced parotid gland mucoepidermoid carcinoma. Oral Oncol. 2021;124:105664–105664. doi: 10.1016/j.oraloncology.2021.105664. [DOI] [PubMed] [Google Scholar]
- 20.Jiang W-M, et al. Prediction of long-term survival outcome by lymph node ratio in patients of parotid gland cancer: A retrospective study. Front. Surg. 2022 doi: 10.3389/fsurg.2022.903576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer M, et al. Prediction of outcome by lymph node ratio in patients with parotid gland cancer. Clin. Otolaryngol. 2017;42:98–103. doi: 10.1111/coa.12672. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, et al. Nomogram to predict the prognosis of parotid gland mucoepidermoid carcinoma: A population-based study of 1306 cases. PeerJ. 2019;7:e7237. doi: 10.7717/peerj.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carta F, et al. Survival in patients with primary parotid gland carcinoma after surgery—Results of a single-centre study. Curr. Oncol. 2023;30:2702–2714. doi: 10.3390/curroncol30030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajasekaran K, et al. Mucoepidermoid carcinoma of the parotid gland: A National Cancer Database study. Am. J. Otolaryngol. 2018;39:321–326. doi: 10.1016/j.amjoto.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 25.Sultan I, et al. Salivary gland carcinomas in children and adolescents: A population-based study, with comparison to adult cases. Head Neck. 2011;33:1476–1481. doi: 10.1002/hed.21629. [DOI] [PubMed] [Google Scholar]
- 26.Rendall MS, Weden MM, Favreault MM, Waldron H. The protective effect of marriage for survival: A review and update. Demography. 2011;48:481–506. doi: 10.1007/s13524-011-0032-5. [DOI] [PubMed] [Google Scholar]
- 27.Olarte LS, Megwalu UC. The impact of demographic and socioeconomic factors on major salivary gland cancer survival. Otolaryngol.Head Neck Surg. 2014;150:991–998. doi: 10.1177/0194599814526556. [DOI] [PubMed] [Google Scholar]
- 28.Terhaard C, et al. The role of radiotherapy in the treatment of malignant salivary gland tumors. Int. J. Radiat. Oncol. Biol. Phys. 2005;61:103–111. doi: 10.1016/j.ijrobp.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Olsen MP, Mitchell AO, Miles EF. Postoperative radiation therapy for parotid mucoepidermoid carcinoma. Case Rep. Oncol. Med. 2014 doi: 10.1155/2014/345128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, et al. Role of postoperative radiotherapy in nonmetastatic head and neck adenoid cystic carcinoma. J. Natl. Compr. Cancer Netw. 2020;18:1476–1484. doi: 10.6004/jnccn.2020.7593. [DOI] [PubMed] [Google Scholar]
- 31.Fang Q, Zhang X, Dai L, Luo R, Yuan J. Association between factor of parotid lymph node and prognosis in parotid cancer. Eur. J. Surg. Oncol. 2023 doi: 10.1016/j.ejso.2023.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Kouka M, et al. Role of intraparotid and neck lymph node metastasis in primary parotid cancer surgery: A population-based analysis. Cancers. 2022;14:2822. doi: 10.3390/cancers14122822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.