Abstract
The conserved miR-183/96/182 cluster (miR-183C) is expressed in both corneal resident myeloid cells (CRMCs) and sensory nerves (CSN) and modulates corneal immune/inflammatory responses. To uncover cell type-specific roles of miR-183C in CRMC and CSN and their contributions to corneal physiology, myeloid-specific miR-183C conditional knockout (MS-CKO), and sensory nerve-specific CKO (SNS-CKO) mice were produced and characterized in comparison to the conventional miR-183C KO. Immunofluorescence and confocal microscopy of flatmount corneas, corneal sensitivity, and tear volume assays were performed in young adult naïve mice; 3′ RNA sequencing (Seq) and proteomics in the trigeminal ganglion (TG), cornea and CRMCs. Our results showed that, similar to conventional KO mice, the numbers of CRMCs were increased in both MS-CKO and SNS-CKO vs age- and sex-matched WT control littermates, suggesting intrinsic and extrinsic regulations of miR-183C on CRMCs. The number of CRMCs was increased in male vs female MS-CKO mice, suggesting sex-dependent regulation of miR-183C on CRMCs. In the miR-183C KO and SNS-CKO, but not the MS-CKO mice, CSN density was decreased in the epithelial layer of the cornea, but not the stromal layer. Functionally, corneal sensitivity and basal tear volume were reduced in the KO and SNS-CKO, but not the MS-CKO mice. Tear volume in males is consistently higher than female WT mice. Bioinformatic analyses of the transcriptomes revealed a series of cell-type specific target genes of miR-183C in TG sensory neurons and CRMCs. Our data elucidate that miR-183C imposes intrinsic and extrinsic regulation on the establishment and function of CSN and CRMCs by cell-specific target genes. miR-183C modulates corneal sensitivity and tear production through its regulation of corneal sensory innervation.
Subject terms: Cell biology, Computational biology and bioinformatics, Immunology, Molecular biology, Neuroscience, Systems biology, Experimental models of disease, Corneal diseases
Introduction
The cornea is an avascular, transparent tissue between the eye and the external environment. It provides visual clarity and two thirds of the refractive power of the eye1–3. It also serves as first-line defense against microbial infection and other insults1–3. Multiple cell types of different origins, including corneal epithelium, stromal keratocytes, endothelial cells, resident immune cells (CRICs) and nerves1–4, work in sync to confer the unique architecture and functionalities of the cornea1–4. Although rare (1.2–5% in mouse5–7), CRICs are a diverse population and have essential roles in almost every aspect of the development and functions of the cornea under both physiological and pathological conditions8–16. The cornea is the most densely sensory-innerved tissue in the body17–19. Sensory innervation of the cornea not only provides bodily sensation to various stimuli but also plays important roles in the homeostasis of the cornea as well as the pathogenesis of corneal diseases through neuroimmune interactions18–33.
microRNAs (miRNAs) are small, non-coding RNAs and are post-transcriptional regulators of gene expression34–37. They play an important role in human diseases38–45 and are viable therapeutic targets46–49. However, their roles in the neuroimmune interaction in health and diseases of the cornea are still largely unknown. The evolutionarily-conserved, paralogous miRNA cluster, miR-183/96/182 cluster (referred to as the miR-183C from here on) is highly, specifically expressed in and required for the normal development and functions of sensory neurons50–52. Inactivation of miR-183C in conventional knockout (KO) mouse models results in multisensory defects51,53–55. Point mutations in the seed sequence of miR-96 results in non-syndromic hearing loss in both mouse56 and human57. In addition, this miRNA cluster is also expressed and plays important roles in both innate4,7,58–62 and adaptive immune cells63–70.
In the cornea, we demonstrated that inactivation of miR-183C in a conventional KO mouse model results in decreased nerve density and reduced expression of capsaicin receptor TRPV1 and pro-inflammatory neuropeptide substance P (sP) precursor gene Tac160. Inactivation or knockdown of miR-183C in innate immune cells, e.g. macrophages (Mϕ) and neutrophils, reduces their production of pro-inflammatory cytokines, however, enhances their phagocytosis and intracellular bacterial killing capacity60,61. These result in a reduced inflammatory response to bacterial infection, e.g. Pseudomonas aeruginosa (PA) and contribute to a decreased severity of PA keratitis60,61,71. In addition to its effect on peripheral Mϕ and neutrophils, we showed that inactivation of the miR-183C results in an increased number of steady-state CRMCs in naïve mice4,7 and myeloid cell infiltration into the cornea in the event of PA infection7. Collectively, these findings suggest a pivotal regulatory role of miR-183C in fine-tuning neuroimmune interactions in the cornea.
However, since in the conventional KO mice, miR-183C is simultaneously inactivated in all miR-183C-expressing cells, including both CSN and CRMCs7,60, the phenotypes observed reflect a composite effect of loss-of-function of miR-183C in both cell types. This precludes distinguishing whether the changes observed in CSN and CRMCs are intrinsic functions of miR-183C in sensory nerves and myeloid cells, respectively, or an extrinsic effect through neuroimmune interaction. To resolve this conundrum, we produced sensory neuron-specific (SNS) and myeloid cell-specific (MS) conditional knockout mice (CKO). Here we report the first characterization of these CKO mice in comparison to their corresponding wild type (WT) littermate control mice at their steady state. Our results reveal sensory neuron- and myeloid cell-specific functions of miR-183C and their impact on the homeostasis of the cornea.
Methods
Mice
All experiments and procedures involving animals and their care were pre-reviewed and approved by the Wayne State University Institutional Animal Care and Use Committee and carried out in accordance with National Institute of Health and Association for Research in Vision and Ophthalmology (ARVO) guidelines (Approved protocol number: IACUC-22-05-4618). The study is reported in accordance with ARRIVE guidelines. Euthanasia was performed by cervical dislocation under anesthesia with isoflurane followed by thoracotomy.
The miR-183C KO—the miR-183CGT/GT mice, are on a 129S2/C57BL/6-mixed background51 and were originally derived from a gene-trap (GT) embryonic stem cell clone51,72,73. Csf1r-EGFP or MacGreen mice74 were purchased from the Jackson Laboratory (Stock number. 018549). In this strain, the EGFP transgene is under the control of the 7.2-kb mouse colony stimulating factor 1 receptor (Csf1r) promoter, allowing specific expression of EGFP in the mononuclear phagocyte system (MPS) myeloid cells, including monocytes (MCs), Mϕ and dendritic cells (DCs)74,75. The Csf1r-EGFP mice were bred with miR-183CGT/+ to produce miR-183C KO [Csf1r-EGFP(+);miR-183CGT/GT] and WT mice [Csf1r-EGFP( +);miR-183C+/+] on the background of Csf1r-EGFP as described previously7.
Mice with miR-183C CKO allele, the miR-183Cf/f, were provided by Dr. Patrick Ernfors, Karolinska Institutet, Sweden through the European Mouse Mutant Archive (EMMA. ID: EM12387). The miR183Cf allele has two loxP sites flanking the 5′ and 3′ ends of the miR-183C for robust Cre-mediated miR-183C CKO76. The myeloid-specific, LysM-Cre mice77 were purchased from The Jackson Laboratory (Stock number: 004781). The LysM-Cre knock-in/knock-out allele has a nuclear-localized (NLS)-Cre recombinase inserted into the first coding ATG of the lysozyme 2 gene (Lyz2), both abolishing endogenous Lyz2 gene function and placing NLS-Cre expression under the control of the endogenous Lyz2 promoter/enhancer elements. Therefore, the LysM-Cre allows myeloid-specific expression of Cre recombinase. The sensory nerve specific Nav1.8-Cre mice78 were kindly provided by Dr. John N. Wood, University College London, through Dr. Theodore J. Price, University of Texas Dallas. This strain is now available at the Jackson Laboratory (Stock Number: 036564). The voltage-gated sodium channel Nav1.8 (encoded by the Scn10a gene) is one of the signature genes of the majority of nociceptive sensory neurons in the TG and dorsal root ganglia (DRG)33,79–81. Nav1.8 promoter-driven Cre recombinase (Nav1.8-Cre) is expressed in nearly all corneal nociceptive sensory nerves33,78,82. The reporter strain, R26LSL-RFP(+/+) mice83, also known as Ai14 (Stock number: 007914, the Jackson Laboratory) has a loxP-flanked STOP cassette (LSL) in front of a tdTomato RFP cassette, all of which are inserted into the ROSA26 locus83. The LSL prevents the transcription of tdTomato RFP; however, when Cre recombinase is present, the LSL cassette will be excised to allow the expression of tdTomato RFP.
miR-183C MS-CKO mice [LysM-Cre(+/−);miR-183Cf/f;Csf1r-EGFP(+/+)], in which MPS myeloid cells are labeled with EGFP, were produced by breeding of the LysM-Cre(+/−), miR-183Cf/f and Csf1r-EGF(+/−) mice. Their littermates [LysM-Cre(−/−);miR-183Cf/f;Csf1r-EGFP(+/+)] are used as WT controls. SNS-CKO mice [Nav1.8-Cre(+/−);miR-183Cf/f;R26LSL-RFP(+/+);Csf1r-EGFP(+/+)] and their WT littermates [Nav1.8-Cre(+/−);miR-183C+/+;R26LSL-RFP(+/+);Csf1r-EGFP(+/+)] were produced by breeding of Nav1.8-Cre (+/−), miR-183Cf/f, R26LSL-RFP and Csf1r-EGF(+/−) mice. All breeding showed a normal mendelian inheritance pattern.
Sex is considered as a biological variance in all studies. 8–12 weeks old, male and female mice were used as separate groups in all experiments. The age, sex and number of the mice in each experiment are specified in the figure legends and/or the text.
Hematoxylin and eosin (H&E) staining of paraffin sections of mouse cornea
Mouse eyes were enucleated and fixed in 4% formalin. The embedding, sectioning and H&E staining were performed by Excalibur Pathology Inc (Oklahoma City, OK). Comparable sagittal sections through the optic nerves were imaged under a DM4000b brightfield microscope (Leica).
Fluorescence-activated cell sorting (FACS)
8–12 weeks old, naïve MS-CKO and KO mice and age- and sex matched WT littermate controls were used to isolate Csf1r-EGFP + myeloid cells from the cornea and spleen for DNA and RNA preparations (see below). For corneal samples, corneas anterior to the limbi from 6–7 mice/genotype were carefully dissected and pooled for single cell preparation as described before4,7. For spleen cells, mononuclear cells were isolated following a standard protocol84. FACS was conducted on a Sony SY3200 cell sorter at the Microscopy, Imaging and Cytometry Resources Core (MICR), Wayne State University (WSU) to isolate Csf1r-EGFP + myeloid cells as we described previously4,7. EGFP- corneal and spleen mononuclear cells were used as negative controls to optimize the gating.
DNA and genotyping PCR
Genomic DNA was prepared from mouse tail, cornea, TG, FACS-sorted Csf1-EGFP + myeloid cells using the gMax DNA mini kit (IBI Scientific) following manufacturer’s instruction as described previously85. Subsequently, DNA concentration and quality were assayed on the Nanodrop 2000 (ThermoFisher Scientific). 100 ng (from mouse cornea, TG and tails) or 4 ng (from FACS-sorted myeloid cells of the cornea and spleen) genomic DNA was used for genotyping PCR to detect tissue and/or cell type-specific recombination at genomic DNA level as we described previously51,85. PCR of 18 s rRNA was amplified and used as a loading control. The genomic structure and primers are illustrated in Fig. S1. The sequences of the primers are: 7Fintron1: 5′-TACCCTGAGTGTGTCTCAATC-3′; CKO-R: 5′-GCAGAGTCACAAACATGTGTAGC-3′; 18 s rRNA Forward: 5′-GTAACCCGTTGAACCCCATT-3′; 18 s rRNA Reverse: 5′-CCATCCAATCGGTAGTAG CG-3′.
RNA preparation and qRT-PCR
Total RNA was prepared using the miRVana miRNA isolation kit (Life Technologies, Foster City, CA, USA) or the RNeasy (Qiagen, Frederick, MD, USA) for miRNA or mRNA studies, respectively, as described previously50,86,87. Quantitative (q)RT-PCR for miRNAs was performed using Taqman miRNA primers and RT-PCR kit (Life Technologies) with snRNA U6 as an endogenous control as described before50,87. For protein-coding genes, qRT-PCR was performed using a QuantiFast SYBR Green RT-PCR kit and QuantiTect primers (Qiagen) with 18 s rRNA as endogenous controls50,51,87.
Tear volume measurement
Tear volume was measured by the Zone-Quick Phenol Red Thread (PRT) test (Yokota Co. Ltd). Manufacturer’s instruction was followed with modifications. Briefly, under light anesthesia by isoflurane, the lower eyelid was pulled down slightly; the PRT is placed between the palpebral conjunctiva of the lower eyelid and the eyeball at a point approximately 1/3 of the distance from the lateral canthus for 15 s. The length of the thread which turned red was measured.
Corneal sensitivity test
Corneal sensitivity to mechanical stimuli was measured by a blink threshold test using a Cochet and Bonnet aesthesiometer (Western Ophthalmics). Briefly, the tip of a nylon filament, starting from 6 cm, was applied perpendicularly to the central cornea. The length at which the mouse blinks was registered as the blink threshold.
Corneal flatmount, immunofluorescence (IF), confocal microscopy, and quantification of Csf1r-EGFP + corneal resident MPS cells and corneal nerve density
Corneal flatmount and IF was performed as described previously7,71. Briefly, mice were euthanized; eyes were enucleated and transferred to cold phosphate buffered saline (PBS). Under a dissecting scope (VWR International, Radnor, PA), the cornea anterior to the limbus was carefully dissected out. The corneas were transferred to cold 1% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB), pH 7.4 for 1 hour (h) at 4 °C. For direct confocal microscopy, the cornea was flattened by six evenly spaced cuts from the periphery toward the center and mounted in Vectashield media with DAPI (Vector Laboratories, Burlingame, CA) on Superfrost Plus slides (Fisherbrand). For IF, after fixation, the corneas were incubated in a blocking buffer with 2% normal goat serum (NGS) (Vector labs) in PBS for 30 min at room temperature (RT); then, permeabilized with 0.1% Triton X-100 in the blocking buffer for 30 min at RT. Subsequently, corneal tissues were incubated with mouse anti-β-Tubulin III (1/600 dilution. Cat. No. 801201, BioLegend, San Diego, CA, USA) antibodies in the blocking buffer for 72 h at 4 °C. After washes with PBS, the corneas were incubated with AF546–conjugated goat anti-mouse IgG (1/1000 dilution. Cat. No. A-11003, Thermo Fisher Scientific) for overnight at 4 °C. After washing, the corneas were flattened and mounted on slides. Negative controls were treated similarly with omission of the primary antibody. All slides were imaged using a TCS SP8 laser confocal microscope (Leica Microsystems Inc. Buffalo Grove, IL). To capture the images of the entire cornea, a series of Z-stacked images under 10 × objective were taken across the entire cornea and stitched together. These stitched Z-stacked images were merged/flattened for cell counting or nerve density quantification using Adobe Photoshop CS6 (64 bit) and ImageJ 1.52p software (http://imagej.nih.gov/ij. NIH, Bethesda, MD, USA) as described previously7. Briefly, the raw image was first converted to the 16-bit grayscale; then the black and white binary image is optimized for the threshold to faithfully represent the original image and the cell density. The EGFP + cells in the entire cornea were counted. To quantify nerve density, the mean value of pixels in a 500-μm2 square covering the center of the whorl-like subbasal plexus was quantified as the nerve density of the center; the ones in three 500-μm2 squares randomly placed in the periphery of the cornea were measured, the average of which was recorded as nerve density of the peripheral cornea. Corneas from at least n = 3 mice/sex/genotype were quantified.
3′ RNA sequencing and data analysis
mRNAs were isolated from TG, corneas or FACS-sorted Csf1r-EGFP + myeloid cells of young adult (8–12 weeks old) male mice. For miR-183C KO strain, n = 3 for both KO and WT controls; for SNS-CKO strain, n = 3 for SNS-CKO mice, n = 4 for WT controls; for MS-CKO strain, n = 5 for both MS-CKO and WT controls were used. To isolate Csf1r-EGFP + myeloid cells, 12 corneas of 6 KO and 14 corneas of 7 WT control mice or 20 corneas of 10 MS-CKO or 10 WT control mice were pooled for FACS isolation. Subsequently, 3′ mRNA-Seq libraries were prepared by the Genome Sciences Core (GSC), WSU using the QuantSeq 3′ mRNA-Seq Library Prep kit FWD (Lexogen, Greenland, NH)88. High sensitivity D1000 ScreenTape peak range between 100 and 700 bp were in accordance with Lexogen guidelines. The libraries were sequenced on NovaSeq sequencer (Illumina). Reads were aligned to the mouse genome (Build mm10) using a public available software STAR89 and tabulated for each gene region using HTSeq90. Differential expressed genes (DEGs) between KO or CKO vs WT controls were identified by edgeR91. miR-183C predicted target genes were identified by TargetScan algorithm (Targetscan.org)92–96 in the upregulated genes in CKO or KO vs WT control and are recognized as miR-183C target genes in these tissues. Statistical significance of enrichment of miR-183C targets in upregulated genes was analyzed by Chi-square with Yates’ correction using the Analyze a 2 × 2 Contingency Table (GraphPad. https://www.graphpad.com/quickcalcs/contingency1/). Functional annotation analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID)97 as described before51.
ELISA assay of CX3CL1 in the TG and cornea
Mouse CX3CL1/Fractalkine Quantikine ELISA kit (Cat. No. MCX310. R&D Systems) was used for analysis of protein levels of CX3CL1 in the TG and the cornea of miR-183C KO mice. Sensitivity of this kit is 0.08–0.32 ng/ml. 12 weeks old, male, naïve KO and age- and sex-matched WT littermate control mice were used for this assay. n = 5/genotype. ELISA assay was performed following the manufacturer’s instruction as described before60,71.
Proteomics in TG and corneas
TG and corneas of adult, male miR-183C KO mice and age- and sex-matched WT littermate controls mice (n = 3/genotype) were harvested, snap-frozen in liquid nitrogen and subjected to proteomics in the WSU Proteomics Core as described before98. Briefly, tissue samples were homogenized in 100 ul of 2% Lithium dodecyl sulfate, 40 mM triethylammonium bicarbonate buffer (TEAB, Honeywell Fluka Cat. No. 60-044-974). Then, the samples were reduced and alkylated by incubating with 5 mM DL-Dithiothretol (DTT, Sigma Cat. No. D5545) for 1 h at 37 °C followed by addition of 15 mM Iodoacetamide (IAA, Sigma Cat. No. I1149) for 30 min (min) at RT in the dark. After IAA quenching with 5 mM DTT, samples were acidified by addition of 2 μl of 12% phosphoric acid. Then, proteins were precipitated by adding 700 μl 90% MeOH, 100 mM TEAB and incubating for 1 h at 37 °C and overnight at − 20 °C. Precipitates were pelleted by centrifugation for 1 min at 16,000×g, washed with 90% MeOH, 10 mM TEAB, and dried on the bench before resuspended in 25 μl of 50 mM HEPES (pH 8.2). Then, 0.25 ug of Trypsin (Promega, Cat. No. V5113) was added to each sample and incubated at 47 °C for 1 h and at 37 °C, overnight to complete the digestion. Subsequently, samples were labeled with Tandem Mass Tag 6plex (TMT-6plex) reagents by adding the selected reagent to each sample and 2-h incubation at RT. 140 µg of each TMT-6plex reagent (ThermoFisher, USA) was used for each 20 μg sample. All samples were then pooled and speed-vac’ed to dryness. Fractionation was achieved using high pH reversed phase spin cartridges (Pierce, Cat. No. 84868). A step gradient of increasing ACN concentrations was applied to elute bound peptides in nine fractions. Each fraction was dried in a vacuum centrifuge and stored until analysis by mass spectrometry. The mass spectrometry analysis was performed using a Vanquish-Neo chromatography system with an Acclaim PepMap 100 trap column (100 µm × 2 cm, C18, 5 µm, 100 Å), and an Easy-Spray PepMap RSLC C18 75 μm × 25 cm column (Thermo scientific). LC–MS/MS was performed on an Orbitrap Eclipse MS system operated with Real Time Search (RTS) active. Subsequently, mass spectrometry data were processed by Proteome Discoverer version 2.4 using the Sequest HT algorithm with Percolator and the mus musculus Uniprot FASTA database (downloaded March 8, 2021, 17,035 entries). False discovery rate (FDR) was calculated by enabling the peptide sequence analysis using a decoy database, and a cut-off of 1% was used for identifications. Quantitative values were normalized using the total Master Protein signal in each TMT channel.
Statistical analysis
When the comparison was made among more than 2 conditions, one-way ANOVA with Bonferroni’s multiple comparison test was employed (GraphPad Prism); adjusted p < 0.05 was considered significant. Otherwise, a two-tailed Student’s t test was used to determine the significance; p < 0.05 was considered significant. Each experiment was repeated at least once to ensure reproducibility and data from a representative experiment are shown. Quantitative data is expressed as the mean ± SEM.
Results
Sensory nerve- and myeloid cell-specific knockout of miR-183C results in no major morphological changes of the cornea
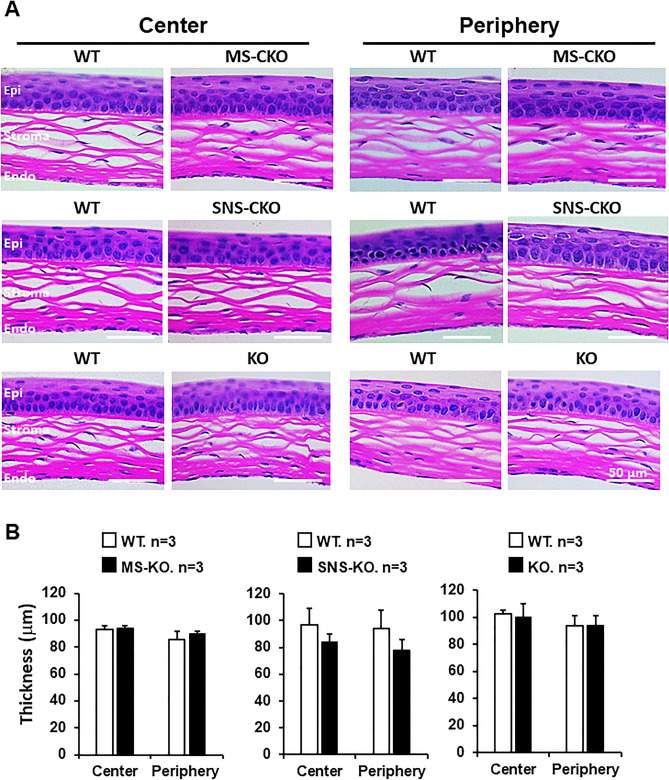
To uncover tissue-specific functions of the miR-183C, we produced sensory neuron- and myeloid cell-specific miR-183C CKO mice (Fig. S1). To detect whether inactivation of miR-183C in sensory nerves and/or myeloid cells causes major histological and morphological changes in the cornea, we performed H&E staining in cross-sections of the corneas of SNS-CKO, MS-CKO and KO mice and their WT control littermates. No gross morphological changes were observed in the cornea of miR-183C SNS-CKO, MS-CKO and conventional KO mice, when compared to their WT controls (Fig. 1). The total thickness (Fig. 1) and thickness of the epithelial and stromal layers (not shown) had no significant differences between the CKOs and KO vs their age- and sex-matched WT controls.
Figure 1.
Inactivation of miR-183C has no significant impact on the gross histological architecture of the cornea. (A) H&E staining of cross-sections of the corneas of MS-CKO, SNS-CKO and miR-183C conventional KO and age- and sex-matched WT control littermates. (B) Measurement of the thickness of the corneas. Epi epithelium, Endo endothelium.
Inactivation of miR-183C in either sensory neurons or myeloid cells results in increased number of CRMCs
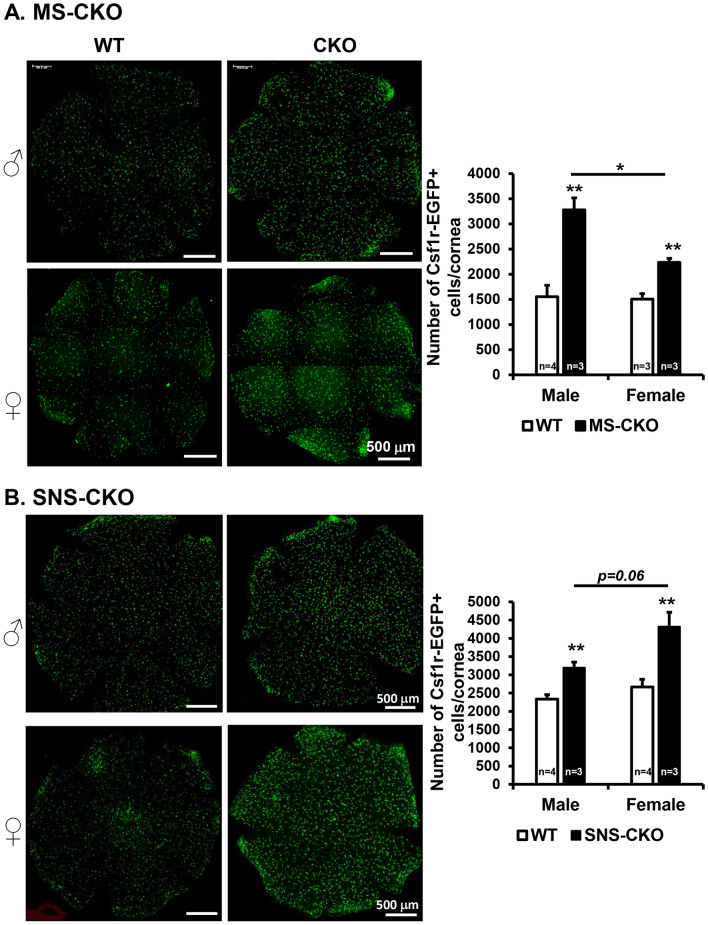
Quantification of Csf1r-GFP + CRMCs of young adult, naïve mice showed that the numbers of Csf1r-EGFP + resident myeloid cells in MS-miR-183C CKO mice [3278 ± 268/cornea (n = 3) in male; 2234 ± 80/cornea (n = 3) in female] were significantly increased in both male and female mice, when compared to age-matched WT controls [1697 ± 225/cornea (n = 3) in male, 1507 ± 107/cornea (n = 3) in female] (Fig. 2A), suggesting miR-183C intrinsically regulates the number of CRMCs under homeostatic condition. In WT controls, male and female mice showed no difference in the number of CRMCs, however, in MS-CKO, the male vs female mice showed increased number of CRMCs (Fig. 2A), suggesting a potential sex-related modulation.
Figure 2.
Inactivation of miR-183C in either sensory neurons or myeloid cells results in increased number of corneal resident myeloid cells (CRMCs). Compressed confocal images of flatmount cornea of young adult MS-CKO (A) and SNS-CKO sand their age- and sex-matched WT control littermates (B). **p < 0.01.
To test whether miR-183C in CSN has an extrinsic regulation on the CRMC population, we quantified the number of CRMCs in the SNS-CKO mice. Intriguingly, sensory nerve-specific inactivation of miR-183C also resulted in an increased number of CRMCs when compared to age- and sex-matched WT controls (Fig. 2B). In male, it was increased by ~ 35% [3197 ± 151/cornea in SNS-CKO (n = 3) vs 2335 ± 121/cornea in WT controls (n = 4)]; while in the female by ~ 56% in female [3197 ± 151/cornea in SNS-CKO (n = 3) vs 2772 ± 298/cornea in WT controls (n = 4)] (Fig. 2B). The numbers of CRMCs showed no significant differences between male and female in WT, however, an increased trend (p = 0.06) in the female vs male in the SNS-CKO mice.
Inactivation of miR-183C in sensory neurons, but not in CRMCs, results in decreased sensory nerve density in the epithelial layer
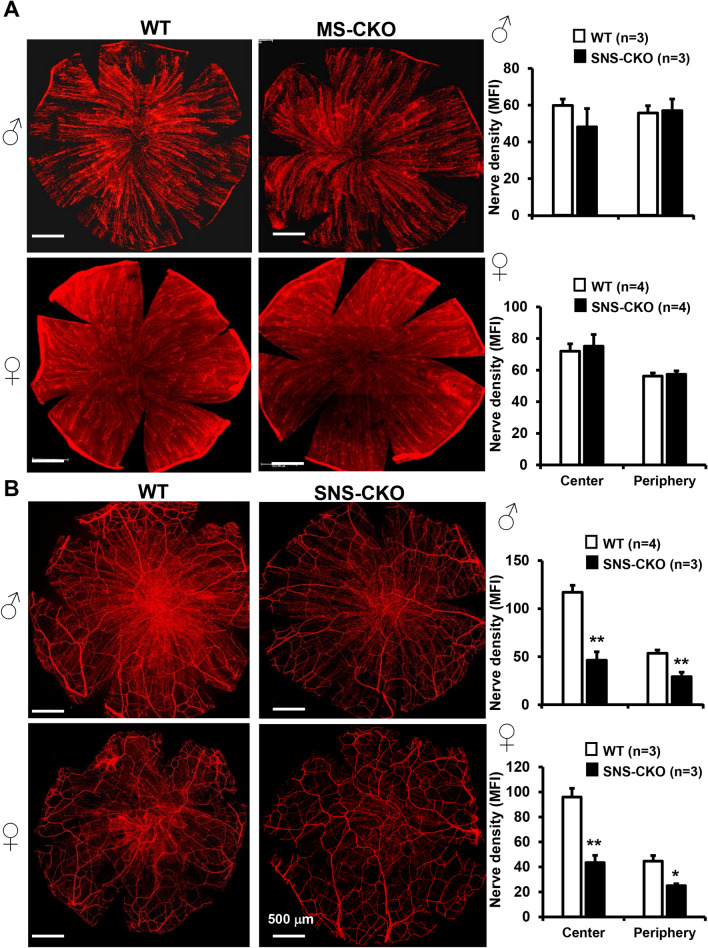
Previously, we showed that corneal nerve density is significantly decreased in the miR-183C KO mice60. To dissect the contribution of miR-183C in corneal sensory nerves or resident myeloid cells to this phenotype, we quantified corneal nerve density in the SNS-CKO and MS-CKO mice. β-III tubulin IF of flatmount cornea showed no difference in corneal nerve density of the MS-CKO vs their WT littermate control mice in both male and female (Fig. 3A), suggesting loss of miR-183C in CRMCs had no contribution to the decreased corneal nerve density in the miR-183C KO mice60.
Figure 3.
Inactivation of miR-183C in sensory neurons, but not in the CRMCs, results in decreased sensory nerve density. Compressed confocal images of flatmount corneas of young adult MS-CKO (A) and SNS-CKO sand their age- and sex-matched WT control littermates (B). *p < 0.05; **p < 0.01. MFI mean fluorescence intensity.
However, the corneal nerve density was significantly decreased in the SNS-CKO vs WT littermate control mice in both male and female (Fig. 3B), in both the whorl center and peripheral regions of the cornea, suggesting that decreased nerve density in the miR-183C conventional KO mice60 is a result of loss of miR-183C in corneal sensory nerves; miR-183C imposes an intrinsic regulation of corneal sensory innervation.
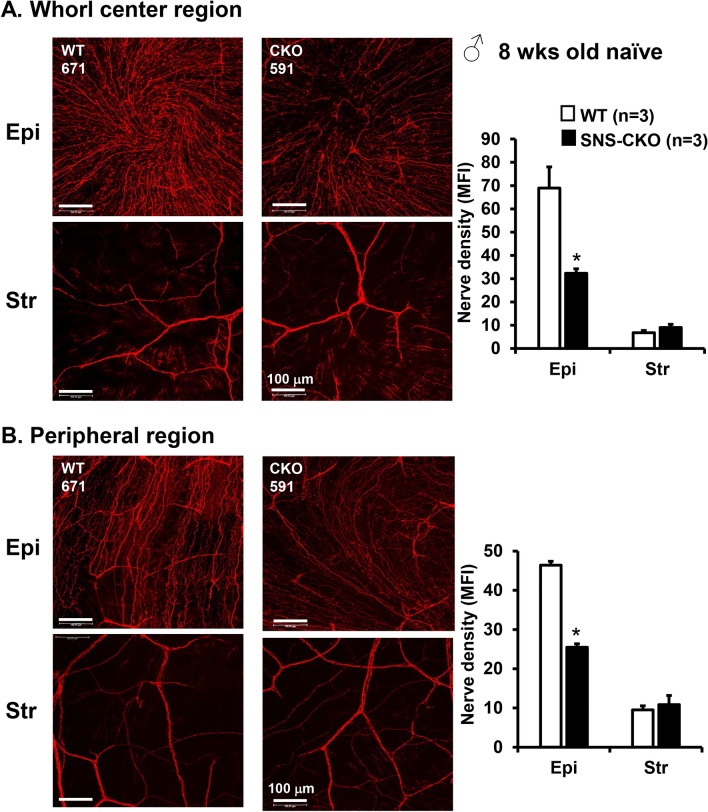
To further probe into the reduction of sensory nerve density, we quantified the sensory nerve density in the epithelial and stromal layers separately (Fig. 4). The layer-specific analyses showed that, in both the whorl center and peripheral regions of male mice, it is the density of the fine terminal nerves in the epithelial layer that is significantly decreased, but not the large nerves in the stromal layer (Fig. 4), suggesting that miR-183C regulates the terminal differentiation and/or extension of sensory nerves in the epithelial layer of the cornea, but has no significant impact on the establishment of large sensory nerves in the stromal layer. Similar effect was observed in female mice (Fig. S2).
Figure 4.
Reduction of corneal sensory nerve density in the SNS-CKO is caused by decreased sensory nerve density in the epithelial layer but not the stromal layer. Compressed confocal images of corneal sensory nerves of the epithelial layers (Epi) and stromal layers (Str) of the whorl center areas (A) and peripheral regions (B) of flatmount corneas of young adult, male SNS-CKO sand their age- and sex-matched WT control littermates. *p < 0.05.
Inactivation of miR-183C in sensory nerves results in decreased corneal sensitivity to mechanical stimuli
Since inactivation of miR-183C in sensory neurons resulted in decreased sensory nerve density (Figs. 3, 4), we hypothesize that the functions of the sensory nerve are compromised in the cornea. To test this hypothesis, we first tested corneal sensitivity to mechanical stimuli using a Cochet and Bonnet aesthesiometer. Our result showed in both miR-183C KO (Fig. 5A) and SNS-CKO mice (Fig. 5B), corneal sensitivity was significantly decreased, when compared to their WT littermate. In the conventional KO, corneal sensitivity was decreased by ~ 23% and 33% in male and female mice, respectively. In the SNS-CKO, corneal sensitivity was significantly reduced by ~ 10 and 11% in male and female mice, respectively. However, in MS-CKO mice, inactivation of miR-183C showed no impact on corneal sensitivity. These data suggest that miR-183C in corneal sensory nerves intrinsically regulate its function. Decreased corneal sensitivity in miR-183C KO is a result of loss of miR-183C in the sensory nerve.
Figure 5.
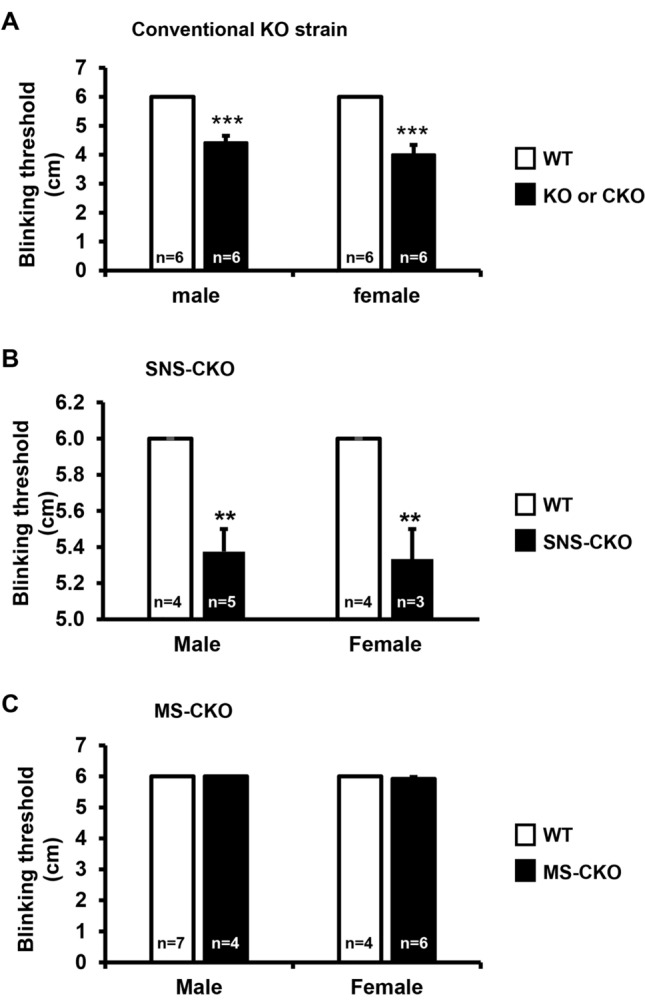
Inactivation of miR-183C in sensory nerves, but not in the myeloid cells, results in decreased corneal sensitivity to mechanical stimuli. Sensitivity test using a Cochet and Bonnet aesthesiometer in young adult (8–12 weeks old) miR-183C conventional KO (A), SNS-CKO (B) and MS-CKO mice and their age- and sex-matched WT control littermates (C). **p < 0.01; ***p < 0.001.
Inactivation of miR-183C in sensory nerves results in decreased basal tear production
It is known that corneal sensory innervation plays important role in regulating tear production27,99. Since inactivation of miR-183C in sensory neurons resulted in decreased sensory nerve density and sensitivity to mechanical stimuli (Figs. 3, 4, 5), we further hypothesize that miR-183C may play a role in tear production through its regulation of corneal sensory innervation. To test this hypothesis, we measured tear volumes using the PRT test. Our result showed that in both the miR-183C KO and SNS-CKO mice, tear volume was significantly decreased when compared to their WT littermates (Fig. 6A,B). In male mice, the tear volume was decreased by ~ 81% in the miR-183C KO and ~ 71% in the SNS-CKO mice; while in female, it was decreased by ~ 55% and 59% in the conventional KO and SNS-CKO, respectively (Fig. 6A,B). However, tear volume of MS-CKO mice showed no difference compared to their WT littermate controls (Fig. 6C). These data suggest that miR-183C modulates basal tear production through regulation of corneal sensory innervation.
Figure 6.
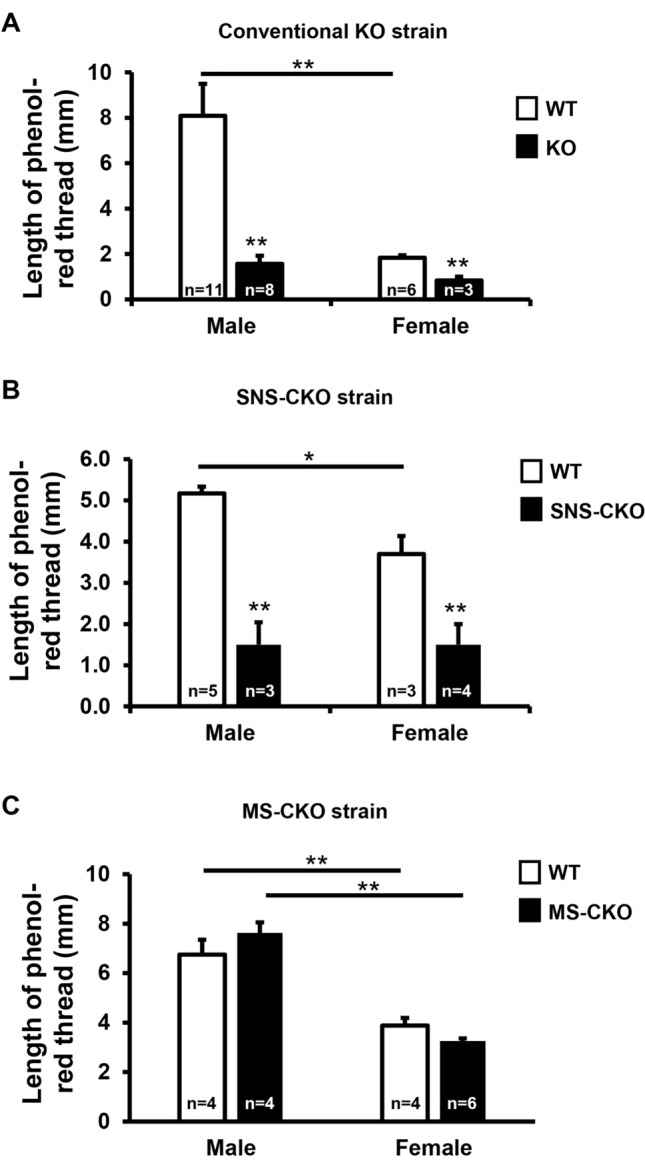
Inactivation of miR-183C in sensory nerves, but not in myeloid-cells, results in decreased basal tear volume. Phenol-red thread assays in young adult (8–12 weeks old) miR-183C conventional KO (A), SNS-CKO (B) and MS-CKO mice and their age- and sex-matched WT control littermates (C). *p < 0.05; **p < 0.01.
In addition, our data showed that the tear volume in female vs male WT mice was consistently reduced in all three strains, by ~ 77%, 28%, and 43% in the conventional KO, SNS-CKO, and MS-CKO mice, respectively, suggesting a sex-dependent difference of tear production (Fig. 6A–C).
miR-183C modulates the functions of sensory neurons and myeloid cells through its regulation of cell type-specific target genes
To start to understand the molecular mechanisms underlying miR-183C’s regulation of corneal functions, we performed 3′ mRNA sequencing88,100 in the TG and corneas of young adult, naïve SNS-CKO, MS-CKO and miR-183C KO mice and their corresponding WT littermate controls. Comparison of the transcriptomes identified a series of DEGs in the TG and corneas of SNS-CKO, MS-CKO and miR-183C KO mice vs their WT controls (Fig. S3; Tables S1–S6). Since miRNAs negatively regulate gene expression35, to identify tissue-specific targets of miR-183C, we searched predicted target genes of miR-183C among the upregulated genes in KO or CKO vs their WT controls in both TG and cornea. This analysis yielded a series of tissue-specific target genes of miR-183C (Tables S7–S12). Statistical analysis showed that miR-183C predicted target genes are significantly enriched among the upregulated genes in the TG and cornea of SNS-CKO or KO mice vs their corresponding WT littermate controls. However, no enrichment of miR-183C predicted target genes was detected in the upregulated genes of either TG or cornea of miR-183C MS-CKO mice, possibly a result of the rarity of the CRMCs in the TG or cornea4,7.
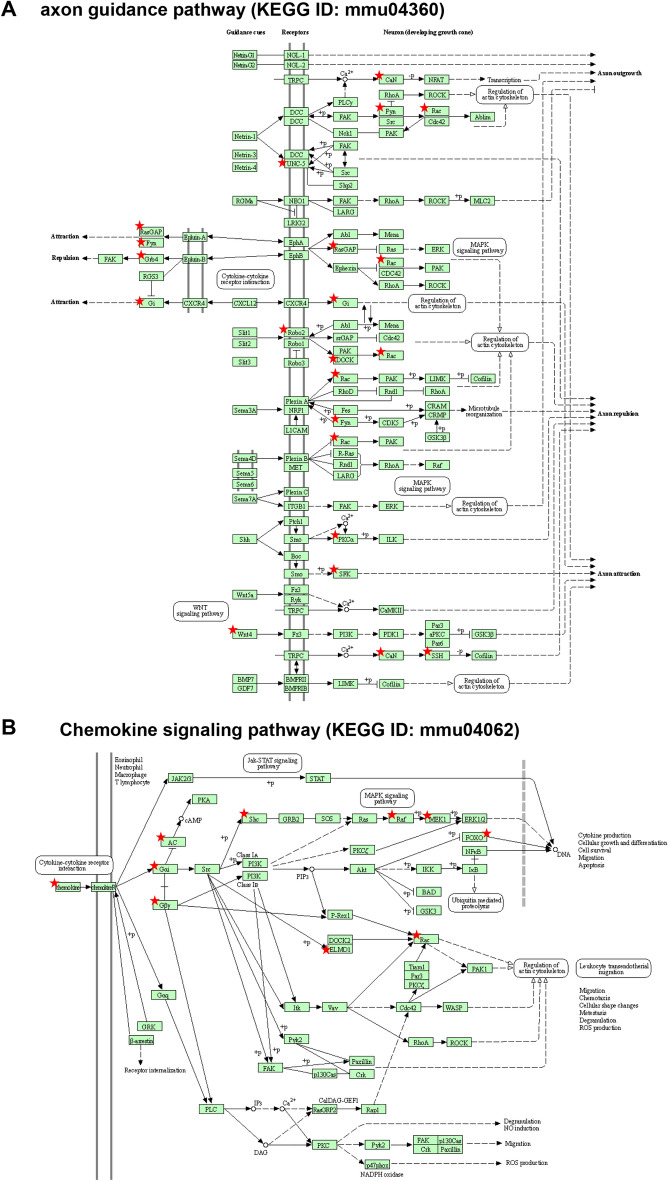
Functional annotation analyses of the tissue-specific target genes identified in the TG and cornea of SNS-CKO and KO mice showed similar overlapping results (Tables S13–S20). Since miR-183C is predominantly expressed in the TG sensory neurons when compared to myeloid cells (> 50 folds; Fig. S4) and that CRMCs are a rare population in the cornea (1–3% of total corneal cells)4,7, we reasoned that gene expression changes and miR-183C targets identified in the TG and cornea of SNS-CKO and KO mice represents TG sensory neuron-specific changes and miR-183C targets. Gene ontology (GO) analysis of the tissue-specific target genes identified in the TG of SNS-CKO and KO mice (Tables S13, S17, S21) showed significant enrichment in neuronal axon projection-, immune response- and epithelial cell proliferation and migration-related biological processes (Table 1). Consistent with the GO analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed enrichment in axon guidance (Fig. 7A), synaptic function pathways (Table 2; Tables S14, S18, S22). In the cornea, miR-183C target genes identified in the SNS-CKO and miR-183C KO mice (Tables S15, S16, S19, S20, S23, S24) are prominently enriched in regulation of synaptic functions, in addition to axonogenesis and axon projection (Tables 3 and 4). These data collectively suggest that miR-183C regulates sensory neuronal projection and synaptic function through directly targeting genes involved in these biological processes. Besides neuronal related functions, miR-183C targets in TG sensory neurons are also enriched in epithelial cell proliferation and migration-, cell–cell adhesion-, fibroblast and endothelial cell migration-related biological processes, suggesting indirect regulation of miR-183C on other cell types in the cornea through its modulation of corneal sensory innervation (Tables 1, 2, 3, 4). Furthermore, genes involved in chemokine signaling pathway, e.g. CX3CL1, GNG5, GNG12, GNAI3, ADCY6, SHC1, BRAF, ELMO1, RAC1, MAP2K1, FOXO3 (Fig. 7B), leukocyte trans-endothelial migration, e.g. GNAI3, MSN, PRKCA, RAC1, and FcγR-mediated phagocytosis pathways, e.g. MAP2K1, PRKCE, PRKCA, RAC1, are also enriched in miR-183C target genes of TG sensory neurons (Tables 2, 4), suggesting potential roles in neuropeptide production and neuroimmune interactions.
Table 1.
Neuronal, immune and epithelial-related GO terms enriched in miR-183C targets in TG sensory neurons.
| GO ID | Terms | P | Genes | Fold enrichment |
|---|---|---|---|---|
| Neuronal related | ||||
| 0010976 | Positive regulation of neuron projection development | 0.0023 | EHD1, RGS2, ANKRD27, ITGA3, STMN2, FYN, CX3CL1 | 5.17 |
| 1902667 | Regulation of axon guidance | 0.0428 | NOVA2, MYCBP2 | 45.59 |
| 0031175 | Neuron projection development | 0.0431 | EHD1, GDNF, SHC1, STMN2, FYN | 3.78 |
| 0007411 | Axon guidance | 0.0508 | KLF7, GAP43, MYCBP2, PAX6, ANK3 | 3.58 |
| Immune related | ||||
| 0050729 | Positive regulation of inflammatory response | 0.0355 | CEBPA, CD47, LDLR, CX3CL1 | 5.53 |
| 0071560 | Cellular response to transforming growth factor beta stimulus | 0.0003 | ZEB1, NOX4, FYN, WNT2, TGFBR1, WNT4 | 10.39 |
| 0036120 | Cellular response to platelet-derived growth factor stimulus | 0.0152 | PRKCE, RASA1, FYN | 15.78 |
| Epithelial related | ||||
| 0010634 | Positive regulation of epithelial cell migration | 0.0373 | ITGA3, PRKCE, PPM1F | 9.77 |
| 0050680 | Negative regulation of epithelial cell proliferation | 0.0279 | ZEB1, CELF1, PAX6, MTSS1 | 6.08 |
Figure 7.
miR-183C target genes in TG sensory neurons are enriched in axon guidance (A) chemokine signaling pathways (B).
Modified from www.genome.jp/kegg/pathway.html. Star-labeled molecules or complexes are or contain target genes of miR-183C.
Table 2.
Neuronal and immune-related KEGG pathways enriched in miR-183C targets in TG sensory neurons.
| KEGG ID | Term | P | Genes | Fold enrichment |
|---|---|---|---|---|
| Neuronal related | ||||
| mmu04725 | Cholinergic synapse | 0.0124 | GNA11, KCNJ14, FYN, GNG12, ADCY6 | 5.48 |
| mmu04724 | Glutamatergic synapse | 0.0132 | GRIA1, PPP3R1, SLC1A1, GNG12, ADCY6 | 5.39 |
| mmu04722 | Neurotrophin signaling pathway | 0.0161 | MAP3K3, SHC1, PSEN2, BRAF, FOXO3 | 5.08 |
| mmu04360 | Axon guidance | 0.0574 | PPP3R1, RASA1, NCK2, FYN, WNT4 | 3.39 |
| Immune related | ||||
| mmu04062 | Chemokine signaling pathway | 0.0189 | SHC1, BRAF, FOXO3, GNG12, CX3CL1, ADCY6 | 3.84 |
Table 3.
Neuronal, cellular adhesion and migration-related GO terms enriched in miR-183C targets in the cornea of SNS-CKO and KO mice.
| GO ID | Term | P | Genes | Fold Enrichment |
|---|---|---|---|---|
| Neuronal function related | ||||
| 0010976 | Positive regulation of neuron projection development | 3.21E-04 | ANKRD27, FUT9, AVIL, DAB2IP, CNTN1, FYN, PRKD1, CX3CL1 | 6.16 |
| 2000300 | Regulation of synaptic vesicle exocytosis | 0.002954 | NRN1, SV2C, PRKCE, PRKCA, CACNA1E | 8.38 |
| 0001764 | Neuron migration | 0.004744 | MEF2C, TRIM46, FYN, FAT3, RAC1, CELSR3 | 5.48 |
| 0048813 | Dendrite morphogenesis | 0.005398 | KLF7, ELAVL4, FYN, RAC1 | 11.18 |
| 0030030 | Cell projection organization | 0.014212 | UBXN10, RAB34, GPR22, TSC1, FAM149B, TAPT1 | 4.19 |
| 0030517 | Negative regulation of axon extension | 0.0173 | TRIM46, AATK, RTN4 | 14.75 |
| 0048812 | Neuron projection morphogenesis | 0.022254 | MAP2K1, ANKRD27, DAB2IP, RAC1 | 6.63 |
| 0010977 | Negative regulation of neuron projection development | 0.024328 | TSC1, HES1, RTN4, PTPRG | 6.41 |
| 0048870 | Cell motility | 0.030182 | MAP2K1, ELMO1, RAC1 | 10.96 |
| 0050772 | Positive regulation of axonogenesis | 0.033101 | ROBO2, ZEB2, MAP2K1 | 10.43 |
| 0007411 | Axon guidance | 0.044784 | ROBO2, KLF7, CNTN1, UNC5D, RAC1 | 3.73 |
| Cellular adhesion and migration related | ||||
| 0007155 | Cell adhesion | 0.001191 | CD164, PRKCE, FIBCD1, PRKCA, CX3CL1, PCDH17, CELSR3, CNTN1, HES1, FAT3, TGFBI, RAC1, DGCR2 | 3.03 |
| 0010595 | Positive regulation of endothelial cell migration | 9.78E-04 | ANXA3, PDCD6, PRKCA, PRKD1, RAC1 | 11.31 |
| 0010764 | Negative regulation of fibroblast migration | 0.002543 | ZEB2, RAC1, CYGB | 38.87 |
| 0022409 | Positive regulation of cell–cell adhesion | 0.01618 | SOX2, MAGI1, PDE4D | 15.27 |
Table 4.
Neuronal and immune-related KEGG pathways enriched in miR-183C targets in the cornea of SNS-CKO and KO mice.
| KEGG ID | Term | P | Genes | Fold enrichment |
|---|---|---|---|---|
| Neuronal and synaptic function related | ||||
| mmu04360 | Axon guidance | 0.0011 | ROBO2, GNAI3, FYN, PRKCA, UNC5D, RAC1, SSH2 | 5.84 |
| mmu04725 | Cholinergic synapse | 0.0060 | MAP2K1, GNG5, GNAI3, FYN, PRKCA | 6.74 |
| mmu04726 | Serotonergic synapse | 0.0103 | MAP2K1, GNG5, DUSP1, GNAI3, PRKCA | 5.76 |
| mmu04921 | Oxytocin signaling pathway | 0.0174 | MAP2K1, MEF2C, CACNA2D1, GNAI3, PRKCA | 4.94 |
| mmu04727 | GABAergic synapse | 0.0201 | GABARAPL2, GNG5, GNAI3, PRKCA | 6.79 |
| mmu04713 | Circadian entrainment | 0.0258 | GNG5, GNAI3, PRKCA, GRIA3 | 6.16 |
| mmu04724 | Glutamatergic synapse | 0.0380 | GNG5, GNAI3, PRKCA, GRIA3 | 5.30 |
| Immune function related | ||||
| mmu04062 | Chemokine signaling pathway | 0.0081 | MAP2K1, GNG5, ELMO1, GNAI3, RAC1, CX3CL1 | 4.72 |
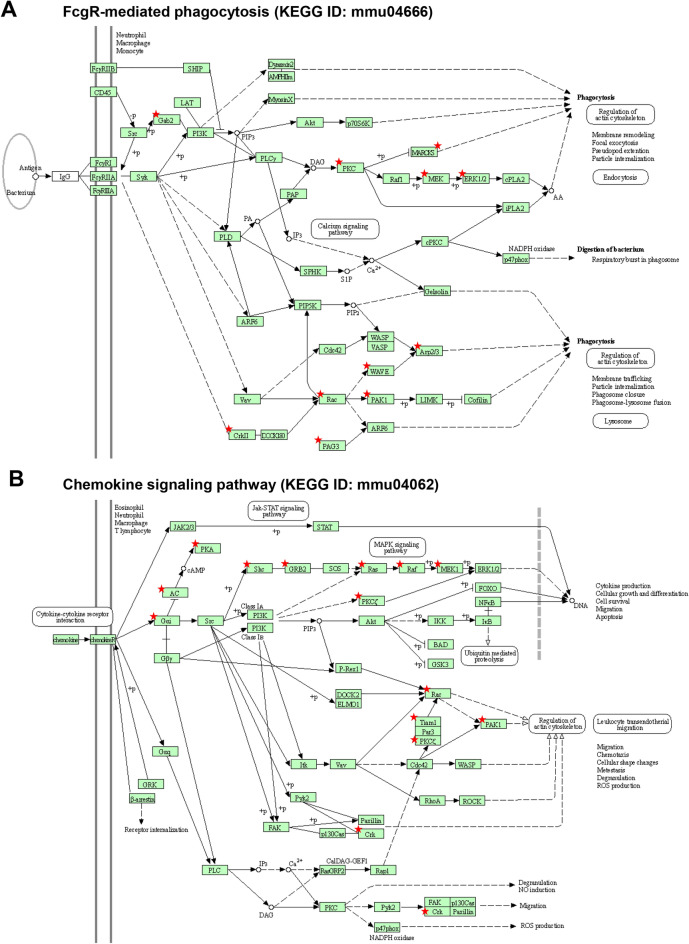
| mmu04666 | Fc gamma R-mediated phagocytosis | 0.0225 | MAP2K1, PRKCE, PRKCA, RAC1 | 6.50 |
| mmu04670 | Leukocyte transendothelial migration | 0.0414 | GNAI3, MSN, PRKCA, RAC1 | 5.12 |
To further identify myeloid cell-specific target genes of miR-183C, we isolated the Csf1r-EGFP + CRMCs from young adult miR-183C MS-CKO and conventional KO mice and their age- and sex-matched WT control mice. KEGG pathway analyses of miR-183C targets identified in CRMCs of both MS-CKO and conventional KO (Tables S25 and S26) are consistently enriched in multiple immune/inflammation-related pathways (Table 5; Table S27), including FcγR-mediated phagocytosis (Fig. 8A), chemokine signaling (Fig. 8B), FcεRI signaling, TGFβ signaling pathway, TNF signaling pathways, and several microbial infection pathways (Table 5; Table S27). These data suggest that miR-183C regulates the functions of myeloid cells through simultaneously targeting multiple genes in different pathways. Intriguingly, miR-183C targets are also enriched in neuronal function, especially synaptic function-related pathways (Table S28), and cell–cell interaction pathways (Table S29), suggesting potential indirect modulation of neuronal innervation and other cell types of the cornea.
Table 5.
Immune/infection-related KEGG pathways enriched in miR-183C targets of CRMCs of KO and MS-CKO mice.
| KEGG ID | Term | P | Genes | Fold enrichment |
|---|---|---|---|---|
| mmu05100 | Bacterial invasion of epithelial cells | 2.98E-04 | ITGB1, ACTR2, CTTN, SHC1, CAV1, ARPC1B, RAC1, WASL, CRK, WASF2, CD2AP | 4.12 |
| mmu04666 | Fc gamma R-mediated phagocytosis | 3.87E-04 | ACTR2, MAP2K1, MARCKS, PAK1, PRKCE, ARPC1B, MAPK1, ASAP1, GAB2, RAC1, CRK, WASF2 | 3.67 |
| mmu04664 | Fc epsilon RI signaling pathway | 0.002041 | MAP2K3, MAPK9, MAP2K1, MAPK1, GRB2, FYN, KRAS, GAB2, RAC1 | 3.88 |
| mmu05135 | Yersinia infection | 0.002728 | ITGB1, MAP2K3, ACTR2, GIT2, MAP2K1, ARHGEF12, ARPC1B, WASL, MAPK9, MAPK1, RAC1, CRK, WASF2 | 2.74 |
| mmu04062 | Chemokine signaling pathway | 0.003045 | MAP2K1, SHC1, GNAI3, BRAF, GNG12, PRKCZ, ADCY6, TIAM1, PAK1, GNG5, MAPK1, GRB2, KRAS, RAC1, CRK, PRKACB | 2.37 |
| mmu05166 | Human T-cell leukemia virus 1 infection | 0.006333 | ATF2, MAP3K3, EGR1, MAP2K1, CRTC1, XIAP, TGFBR1, ADCY6, PPP3CA, MAPK9, ZFP36, CCND2, CREB3L1, CREB3L2, MAPK1, EP300, KRAS, PRKACB | 2.06 |
| mmu05132 | Salmonella infection | 0.007681 | MAP2K3, ACTR2, RALA, MAP2K1, ARPC1B, WASL, PIK3C2A, RHOB, MAPK9, SNX18, PAK1, BCL2, EXOC4, MAPK1, RAC1, PFN1, KPNA3, PFN2 | 2.02 |
| mmu05163 | Human cytomegalovirus infection | 0.007979 | ATF2, MAP2K1, ARHGEF12, GNAI3, TSC1, GNG12, ADCY6, PPP3CA, GNG5, GNA11, CREB3L1, CREB3L2, MAPK1, GRB2, KRAS, RAC1, CRK, PRKACB | 2.02 |
| mmu05165 | Human papillomavirus infection | 0.015215 | MAGI1, ITGB1, PRKCI, MAP2K1, ITGA3, TSC1, PPP2R5C, LAMC1, PRKCZ, PPP2CA, PPP2CB, CCND2, CREB3L1, CREB3L2, MAPK1, EP300, GRB2, HES1, KRAS, WNT2, PRKACB, WNT4 | 1.74 |
| mmu04650 | Natural killer cell mediated cytotoxicity | 0.020466 | PPP3CA, MAP2K1, PAK1, SHC1, MAPK1, GRB2, FYN, BRAF, KRAS, RAC1 | 2.45 |
| mmu05145 | Toxoplasmosis | 0.037313 | ITGB1, MAP2K3, MAPK9, GNAI3, BCL2, XIAP, MAPK1, LAMC1, LDLR | 2.35 |
| mmu04350 | TGF-beta signaling pathway | 0.039069 | ACVR1, PPP2CA, SMAD1, PPP2CB, EP300, MAPK1, SKIL, TGFBR1, SMAD7 | 2.33 |
| mmu05170 | Human immunodeficiency virus 1 infection | 0.040389 | MAP2K3, MAP2K1, FBXW11, GNAI3, GNG12, PPP3CA, MAPK9, PAK1, GNG5, GNA11, BCL2, MAPK1, KRAS, RAC1, CRK | 1.79 |
| mmu04668 | TNF signaling pathway | 0.048672 | MAP2K3, CYLD, MAPK9, ATF2, MAP2K1, CREB3L1, CREB3L2, XIAP, MAPK1 | 2.23 |
Figure 8.
miR-183C target genes in CRMCs are enriched in FcγR-mediated phagocytosis (A) and chemokine signaling pathways (B).
Modified from www.genome.jp/kegg/pathway.html. Star-labeled molecules or complexes are or contain target genes of miR-183C.
Cx3cl1 and a dozen of other genes were validated as miR-183C targets in TG sensory neurons.
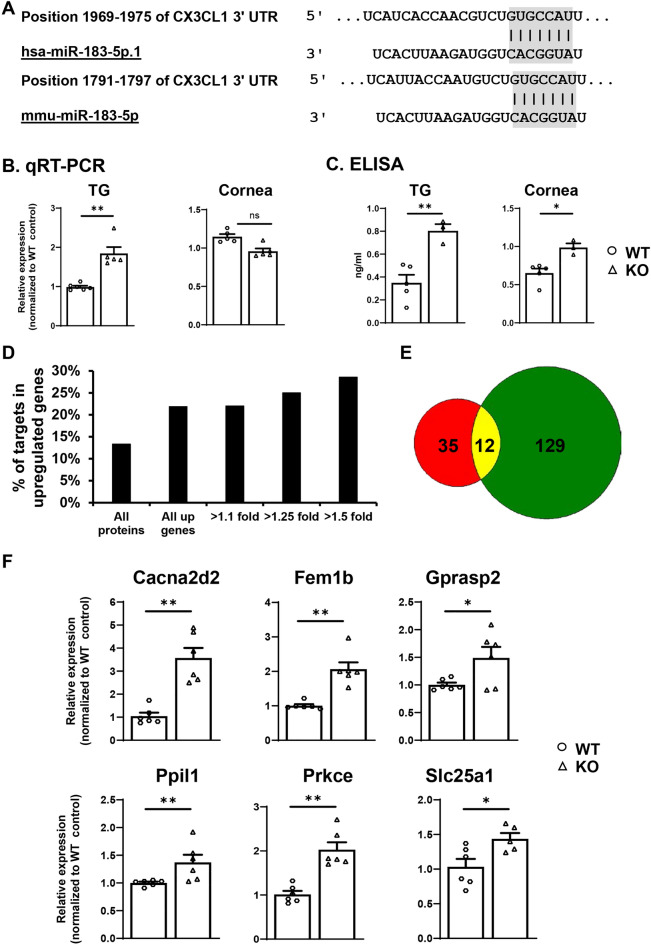
Cx3cl1, also called fractalkine or neurotactin, is a type I transmembrane (TM) chemokine and is the only member of the CX3C or delta chemokine subfamily101–103. It is known to promote chemotactic migration of microglia in the central nervous system (CNS)104,105, mediate the homing of resident myeloid cells in the cornea106 and recruitment of Mϕ in other tissues107. RNA seq data revealed that Cx3cl1 was a potential target gene of miR-183C. It is significantly increased in the TG of miR-183C KO (Table S4) and the cornea of SNS-CKO mice (Table S1) by ~ 1.89 and 1.42 folds, respectively, when compared to their WT controls. Target prediction showed that Cx3cl1 held a conserved target site for miR-183 in its transcript of both human and mouse (Fig. 9A). To validate these findings, we performed qRT-PCR in the TG and corneas of the miR-183C KO and age- and sex-matched WT control mice. Our results confirmed significant upregulation of Cx3cl1 in the TG of the KO vs WT control mice, although no significant change of Cx3cl1 was detected in the cornea (Fig. 9B).
Figure 9.
Validation of miR-183C target genes by qRT-PCR, ELISA assay and proteomics. (A) Sequence alignment of miR-183C with its target site in in 3′ untranslated region (UTR) of human (upper) and mouse Cx3cl1 transcript (lower panel). (B) qRT-PCR; (C) ELISA assay on Cx3cl1 in the TG and cornea of miR-183C KO and WT control mice. (D,E) Proteomics study in the TG of KO and WT control mice. (D) Percentage of miR-183C target genes in upregulated proteins of the TG of KO vs WT control mice; (E) Venn diagram of miR-183C target genes detected by proteomics (Red) and 3′ RNA seq (Green). 12 target genes (yellow) were detected by both methods; 6 of which were further validated by qRT-PCR (F).
miRNAs regulate downstream genes by base-pairing to the target sites in their transcripts to induce the breakdown and/or translation inhibition of the mRNAs35. This regulation ultimately results in decreased expression at protein level35. To further validate the observation of Cx3cl1 at the protein level, we performed ELISA assay in the TG and corneas of miR-183C KO and age- and sex-matched WT controls. Our results showed that CX3CL1 was significantly upregulated in both TG and cornea in the KO vs WT control mice at its protein level (Fig. 9C), supporting that miR-183C targets Cx3cl1 in TG sensory neurons in vivo.
To systemically validate the 3′ RNA seq-identified miR-183C targets at the protein level, we performed proteomics in the TG and corneas of the miR-183C KO mice and age- and sex-matched WT littermate controls. Our results showed that predicted target genes of miR-183C (Table S31) were significantly enriched (p = 0.0006) in the upregulated proteins in the TG of the KO vs WT control mice (Table S30), suggesting specific detection of miR-183C target genes. Consistently, the percentage of miR-183C target genes among the upregulated proteins increased in correlation with the extent of upregulation (Fig. 9D). Twelve (25.5%) of the 47 target genes detected by proteomics (Table S31), including Cacna2d2, Eea1, Femb1, Gprasp2, Hook3, Nedd4l, Ppil1, Prkce, Rbm25, Slc25a1, were also detected by 3′ RNA sequencing (Fig. 9E). Therefore, these 12 target genes of miR-183C are validated at the protein level. To further validate these target genes at RNA level, we performed qRT-PCR on six of the 12 targets validated by proteomics, including Cacna2d2, Fem1b, Gprasp2, Ppil1, Prkce, and Slc25a1. All 6 were confirmed to be significantly upregulated in the TG of the KO vs age- and sex-matched WT control mice (Fig. 9F). Among these, Gprasp2 and Prkce were also upregulated in the cornea of the KO (Fig. S5A) and in the TG of SNS-CKO vs their corresponding age- and sex-matched WT control mice (Fig. S5B), respectively.
For the proteomics in the cornea, similar with the RNA seq data, predicted miR-183C target genes (Table S33) showed no enrichment in the upregulated proteins (Table S32), possibly a result of the low representation of proteins from corneal sensory nerves and CRMCs in total corneal proteins4,7.
Discussion
To delineate the cell type-specific functions of miR-183C in corneal sensory nerves and innate immune cells, we created and characterized a SNS- and a MS-CKO mouse models, in comparison to the conventional KO mice. Our data showed that inactivation of miR-183C in corneal nerves, but not in innate immune cells, resulted in decreased corneal sensory nerve density and reduced sensitivity to mechanical stimuli, suggesting intrinsic regulation of miR-183C on corneal sensory innervation. Our data showed that, in the SNS-CKO mice, the reduction of sensory nerve density occurs specifically in the fine, terminal neurites in the epithelial layer, including the subbasal plexus, while the large-diameter nerves in the stromal layer remain unaffected, suggesting a critical role of miR-183C in the terminal differentiation and projection of corneal sensory nerves. This is consistent with its functions in the other sensory organs, e.g. the photoreceptors of the retina51,54 and the inner ear hair cells108,109, as well as myeloid cells59, suggesting a common feature of miR-183C regulating terminal functional differentiation of primary sensory neurons and innate immune cells. Consistent with this observation, our RNA seq data showed that target genes of miR-183C in the TG were significantly enriched with genes involved in axonal guidance and neuronal projection regulation as well as synaptic functions, suggesting that miR-183C regulates corneal sensory nerve functions by directly targeting neuronal projection and synaptic release-related genes. Multiple genes in the same pathway are simultaneously targeted. This exemplifies the mode of action of miRNAs—quantitatively regulating multiple genes in the same pathway. Their effect on each individual gene may not cause a major phenotypic change; however, working in concert, they impose significant functional consequence7,110–113. In this case, the simultaneous regulation of multiple genes in the neuronal projection and synaptic functional pathways contributes to the decreased sensory nerve density and levels of neuropeptides in the cornea of the SNS-CKO and miR-183C KO mice, which affects the neuroimmune response of the cornea to bacterial infection60.
Comparing to the conventional miR-183C KO mice, the decrease of the corneal sensitivity to mechanical stimuli was modest (~ 10% vs 23–33% in conventional KO mice). This is most possibly a result of the incomplete inactivation of miR-183C in the SNS-CKO mice. In the conventional miR-183C KO model, the miR-183C is inactivated in all cells that normally express miR-183C, including all TG sensory neurons51. However, the SNS-CKO is based on Nav1.8-promoter driven Cre recombinase-induced deletion of miR-183C76,78,82. Although it is expressed in nearly all small-diameter nociceptors, Nav1.8-Cre is expressed in less than a third of the large-diameter mechanoreceptive and proprioceptive neurons78,114–116. Therefore, miR-183C is possibly intact in two thirds of the mechanoreceptors in the SNS-CKO mice, resulting in only modest decrease of corneal sensitivity to mechanical stimuli measured by the Cochet and Bonnet aesthesiometer in this study.
MS-CKO of miR-183C resulted in increased CRMCs, suggesting intrinsic regulation of miR-183C in CRMCs. Our comparative study of the transcriptomes of CRMCs of miR-183C KO and MS-CKO mice vs their WT controls revealed myeloid-specific target genes of miR-183C, which are significantly enriched in a range of immune/infection related pathways, including chemokine signaling and phagocytosis pathways. These data provide new insight into the mechanisms of intrinsic regulation of miR-183C on the functions of innate myeloid cells, e.g. cytokine production and phagocytosis and intracellular killing capacity that we reported previously7,60,61.
Intriguingly, SNS-CKO of miR-183C also led to increased number of CRMCs in naïve mouse cornea, suggesting that normal function of miR-183C in sensory nerves has an extrinsic regulation on the establishment of CRMCs, indicative of neuroimmune interaction in the maintenance of the homeostasis of the cornea. KEGG pathway analysis of miR-183C target genes in TG sensory neurons showed significant enrichment of genes involved in chemokine signaling pathway, including Cx3cl1. Inactivation of miR-183C-resulted disinhibition/upregulation of Cx3cl1 was further validated by qRT-PCR and ELISA assays. Neuron-produced chemokine Cx3cl1, also called fractalkine, is known to promote chemotactic migration of microglia in the CNS104,105, mediate the homing of CRMCs106 and recruitment of Mϕ in other tissues107. These observations support a hypothesis that inactivation of miR-183C disinhibits Cx3cl1 in the TG sensory neurons and resulted in increased expression of Cx3cl1 in the cornea of the SNS-CKO mice, which enhances the recruitment of myeloid cells to the cornea, resulting in an increased number of the CRMCs. Additional studies are warranted to further test this hypothesis. If proven, it will uncover a new mechanism underlying the extrinsic regulation of miR-183C in TG neurons on the establishment of the CRMCs in the cornea.
Furthermore, our RNA seq data showed that miR-183C target genes in TG sensory neurons are also enriched in pathways regulating other corneal cell types, e.g. epithelial, endothelial cell and fibroblast migration; while target genes in CRMCs are also enriched in neuronal synaptic functions and cell–cell interaction and migration pathway. These data suggest that, in addition to the intrinsic regulation of sensory innervation and CRMCs, miR-183C exerts extrinsic regulation on neuro-immune-epithelial-stromal-endothelial interactions. This is consistent with our recent report of a single-cell transcriptome study that miR-183C serves as a checkpoint of corneal resident immune cells and shapes the cellular landscape of the cornea4. Different cell types form the microenvironment or niche of the cornea and work in concert to endow the cornea with its unique architecture and functionalities and maintain homeostasis1–4. Perturbation of the functions of one cell type imposes global impact on other cell types and the overall function of the cornea in both physiological and various pathological conditions4.
In addition to increased numbers of CRMCs in both MS-CKO and SNS-CKO vs their corresponding WT control mice, we identified sex-dependent differences of the number of CRMCs in both MS-CKO and SNS-CKO. Myeloid-specific inactivation of miR-183C resulted in an increased number of CRMCs in male vs female MS-CKO mice, while sensory neuron-specific knockout of miR-183C led to a decreased trend of CRMCs in the male vs female SNS-CKO mice, although there were no differences of CRMC numbers between male and female WT mice of both MS-CKO and SNS-CKO strains. Sex-dependent differences in immune cell composition and immune responses are affected by sex chromosome-linked genes, sex hormones and variations in levels of sex hormones and their receptors overtime118. Although sex differences in both innate and adaptive immunity have been well documented119,120, insights into sexual dimorphism of tissue resident immunity remains sparse118,121–123 and completely unknown in CRICs. Based on our observations, we predict that miR-183C regulates sex chromosome-linked genes, and/or sex hormones and/or their receptors, and/or other genes involved in the establishment of CRICs. Inactivation of miR-183C alters the expression levels of these genes, leading to changes of the numbers of CRMCs through intrinsic and/or extrinsic mechanisms, in which neuroimmune interaction may play an important role. The fact that tissue-specific inactivation of miR-183C in myeloid cells or sensory neurons resulted in opposite trends of the numbers of CRMCs suggests tissue-specific mechanisms of miR-183C in TG sensory neurons and myeloid cells regulating the establishment of CRMCs. Major efforts in the future are warranted to test these hypotheses.
For the first time, here we discovered that both naïve miR-183C KO and SNS-CKO, but not the MS-CKO mice, had significantly decreased tear volume, when compared to their age- and sex-matched WT controls. These data suggest miR-183C enhances basal tear production through its regulation of corneal sensory innervation. It is known that the primary afferent sensory neurons innervating the cornea play important roles in the basal tear production and secretion evoked by noxious stimulation of the cornea through the brainstem tear reflex arc27. Corneal sensory neurons sense dry condition and other environmental stressors, send primary afferent projections to activate the neurons in the spinal trigeminal nucleus (Vsp) and regulates lacrimation through the efferent autonomic arm of the reflex arc, which include the preganglionic parasympathetic neurons in/around the superior and inferior salivatory nuclei (SSN and ISN) and postganglionic parasympathetic neurons in the pterygopalatine ganglion (PG)27. The autonomic reflex promotes the production of the watery component of the tears by the lacrimal glands; mucin component of the tears by the conjunctival goblet cells, and lipid content by the meibomian glands27. We speculate that decreased corneal sensory innervation caused by loss of miR-183C in sensory neurons imposes a negative impact on proper lacrimation through the cornea-brainstem tear reflex arc. In addition, tear producing tissues, including lacrimal glands (Fig. S6), meibomian glands and conjunctival goblet cells are subjected to sensory innervation themselves124–131. Based on its effect in corneal sensory innervation, we predict that inactivation of miR-183C in sensory neurons innervating these tissues may result in defects in sensory nerve density in them. Currently, little is known about whether and how sensory innervation of lacrimal and meibomian glands contributes to tear volume128,132. Nevertheless, our data suggest that miR-183C may play a role in dry eye disease (DED). Additional studies dedicated to these topics are warranted to confirm these hypotheses and uncover the underlying mechanisms.
To our surprise, we observed significant decrease of basal tear volume in female vs male mice in all three strains used in this study, which are in a mixed genetic background of C57BL/6 and129S51. It is well known that DED is more prevalent in women compared to men133–143, especially in the autoimmune DED, Sjögren’s syndrome144. Over 90% of all patients with Sjögren’s syndrome are women144. The female gender is considered a risk factor for the development of DED133. Gender and sex steroids are known to have significant and multitudes of impact on the morphology and functions of tear producing tissues including the lacrimal gland141–143,145–148, meibomian glands128,131 and conjunctival goblet cells149. The acinar area of the lacrimal glands of males are larger than that of females in multiple species including human146. Sex hormones modulates tear volume, secretory components, protein contents and lipid profiles of the tears128,141–143,146–148. Androgen promotes lipogenesis in the meibomian gland; androgen deficiency leads to meibomian gland dysfunction (MGD) and evaporative dry eye, while estrogen reduces lipid synthesis in the meibomian gland and promote both MGD and evaporative dry eye128. Androgen receptors are only detected in male but not female goblet cells149. Human male and female goblet cells have different responses to pro-inflammatory and pro-resolving mediators149. In spite of these observations, sex-related difference in basal tear volumes has not been reported in either humans or animals141,145,148. Our observation of decreased tear volume in female mice appears to be in-line with the fact that female gender has been considered a risk factor for the development of DED133. The discrepancy between our observation and others145,150 could come from different species and/or strains of animals as well as different methodology of measurement151. Nevertheless, our data suggest a sex-dependent difference in basal tear volume and further studies are warranted to uncover the underlying mechanisms.
To our knowledge, this is the first example that a miRNA cluster plays significant, functional roles in the corneal homeostasis through direct regulations of both sensory innervation and CRMCs and modulation of neuroimmune interaction. The SNS-CKO and MS-CKO mouse models developed in this report provide critical tools to fully uncover the roles of miR-183C in sensory nerves or myeloid cells under different pathological conditions, e.g., bacterial keratitis and DED. Such knowledge will be essential to develop cell type-specific strategies targeting miRNAs for the treatment of corneal diseases.
Supplementary Information
Acknowledgements
This work is supported by grants from the National Eye Institute, National Institutes of Health (R01 EY026059 to SX; R01 EY016058, R01 EY035231, and P30 EY004068 to LDH); a Research to Prevent Blindness unrestricted grant to the Department of Ophthalmology, Visual and Anatomical Science, Wayne State University School of Medicine; and the Bridge fund from the Office of Vice President for Research (OVPR) of the Wayne State University. The WSU Proteomics Core is supported through NIH grants P30ES020957, P30CA022453 and S10OD030484 (PS).
Author contributions
N.G., M.S., K.G., G.L., H.Z., S.M., A.P., M.S., M.F.A.S. conducted experiments and data collection. N.G., G.L. S.X. performed data analyses. P.S. and R.R. performed proteomics experiment and analyses. L.D.H participated in experimental design and manuscript revision. S.X. conceived, designed and directed the entire study, conducted experiments, data collection, wrote, revised and submitted the manuscript.
Data availability
The datasets of 3′ RNA sequencing generated during the current study are available in the Gene Expression Omnibus (GEO) repository (accession number: GSE250213). The proteomics data have been submitted to the repository Proteomics IDEntifications Database (PRIDE. Accession number: PXD05033). All data and protocols generated in this study are available upon request from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-58403-1.
References
- 1.Lwigale PY. Corneal development: Different cells from a common progenitor. Prog. Mol. Biol. Transl. Sci. 2015;134:43–59. doi: 10.1016/bs.pmbts.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Cvekl A, Tamm ER. Anterior eye development and ocular mesenchyme: New insights from mouse models and human diseases. Bioessays. 2004;26:374–386. doi: 10.1002/bies.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DelMonte DW, Kim T. Anatomy and physiology of the cornea. J. Cataract. Refract. Surg. 2011;37:588–598. doi: 10.1016/j.jcrs.2010.12.037. [DOI] [PubMed] [Google Scholar]
- 4.Li W, et al. The miR-183/96/182 cluster is a checkpoint for resident immune cells and shapes the cellular landscape of the cornea. Ocular Surf. 2023;30:17–41. doi: 10.1016/j.jtos.2023.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sosnova M, Bradl M, Forrester JV. CD34+ corneal stromal cells are bone marrow-derived and express hemopoietic stem cell markers. Stem Cells. 2005;23:507–515. doi: 10.1634/stemcells.2004-0291. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, et al. CCR2(-) and CCR2(+) corneal macrophages exhibit distinct characteristics and balance inflammatory responses after epithelial abrasion. Mucosal. Immunol. 2017;10:1145–1159. doi: 10.1038/mi.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coku A, et al. The miR-183/96/182 cluster regulates the functions of corneal resident macrophages. Immunohorizons. 2020;4:729–744. doi: 10.4049/immunohorizons.2000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niederkorn JY. Cornea: Window to ocular immunology. Curr. Immunol. Rev. 2011;7:328–335. doi: 10.2174/157339511796196593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barabino S, Chen Y, Chauhan S, Dana R. Ocular surface immunity: Homeostatic mechanisms and their disruption in dry eye disease. Prog. Retin. Eye Res. 2012;31:271–285. doi: 10.1016/j.preteyeres.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Burns AR, Rumbaut RE, Smith CW. gamma delta T cells are necessary for platelet and neutrophil accumulation in limbal vessels and efficient epithelial repair after corneal abrasion. Am. J. Pathol. 2007;171:838–845. doi: 10.2353/ajpath.2007.070008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Burns AR, Han L, Rumbaut RE, Smith CW. IL-17 and VEGF are necessary for efficient corneal nerve regeneration. Am. J. Pathol. 2011;178:1106–1116. doi: 10.1016/j.ajpath.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byeseda SE, et al. ICAM-1 is necessary for epithelial recruitment of gammadelta T cells and efficient corneal wound healing. Am. J. Pathol. 2009;175:571–579. doi: 10.2353/ajpath.2009.090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Burns AR, Miller SB, Smith CW. CCL20, gammadelta T cells, and IL-22 in corneal epithelial healing. FASEB J. 2011;25:2659–2668. doi: 10.1096/fj.11-184804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skelsey ME, Mellon J, Niederkorn JY. Gamma delta T cells are needed for ocular immune privilege and corneal graft survival. J. Immunol. 2001;166:4327–4333. doi: 10.4049/jimmunol.166.7.4327. [DOI] [PubMed] [Google Scholar]
- 15.McClellan SA, Huang X, Barrett RP, van Rooijen N, Hazlett LD. Macrophages restrict Pseudomonas aeruginosa growth, regulate polymorphonuclear neutrophil influx, and balance pro- and anti-inflammatory cytokines in BALB/c mice. J. Immunol. 2003;170:5219–5227. doi: 10.4049/jimmunol.170.10.5219. [DOI] [PubMed] [Google Scholar]
- 16.Sarkar J, et al. CD11b+GR1+ myeloid cells secrete NGF and promote trigeminal ganglion neurite growth: implications for corneal nerve regeneration. Invest. Ophthalmol. Vis. Sci. 2013;54:5920–5936. doi: 10.1167/iovs.13-12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marfurt CF, Cox J, Deek S, Dvorscak L. Anatomy of the human corneal innervation. Exp. Eye Res. 2010;90:478–492. doi: 10.1016/j.exer.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Muller LJ, Marfurt CF, Kruse F, Tervo TM. Corneal nerves: Structure, contents and function. Exp. Eye Res. 2003;76:521–542. doi: 10.1016/S0014-4835(03)00050-2. [DOI] [PubMed] [Google Scholar]
- 19.Guerrero-Moreno A, Baudouin C, Melik Parsadaniantz S, Reaux-Le Goazigo A. Morphological and functional changes of corneal nerves and their contribution to peripheral and central sensory abnormalities. Front. Cell Neurosci. 2020;14:610342. doi: 10.3389/fncel.2020.610342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaheen BS, Bakir M, Jain S. Corneal nerves in health and disease. Surv. Ophthalmol. 2014;59:263–285. doi: 10.1016/j.survophthal.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Streilein JW, Okamoto S, Sano Y, Taylor AW. Neural control of ocular immune privilege. Ann. N Y Acad. Sci. 2000;917:297–306. doi: 10.1111/j.1749-6632.2000.tb05396.x. [DOI] [PubMed] [Google Scholar]
- 22.Blanco T, Saban DR. The cornea has "the nerve" to encourage immune rejection. Am. J. Transplant. 2015;15:1453–1454. doi: 10.1111/ajt.13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerrero-Moreno A, Fakih D, Parsadaniantz SM, Reaux-Le Goazigo A. How does chronic dry eye shape peripheral and central nociceptive systems? Neural. Regen. Res. 2021;16:306–307. doi: 10.4103/1673-5374.290895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belmonte C, et al. TFOS DEWS II pain and sensation report. Ocular Surf. 2017;15:404–437. doi: 10.1016/j.jtos.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantelli F, Micera A, Sacchetti M, Bonini S. Neurogenic inflammation of the ocular surface. Curr. Opin. Allergy Clin. Immunol. 2010;10:498–504. doi: 10.1097/ACI.0b013e32833e16cc. [DOI] [PubMed] [Google Scholar]
- 26.Hamrah P, Seyed-Razavi Y, Yamaguchi T. Translational immunoimaging and neuroimaging demonstrate corneal neuroimmune crosstalk. Cornea. 2016;35(Suppl 1):S20–S24. doi: 10.1097/ICO.0000000000001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng ID, Kurose M. The role of corneal afferent neurons in regulating tears under normal and dry eye conditions. Exp. Eye Res. 2013;117:79–87. doi: 10.1016/j.exer.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, et al. Substance P delays apoptosis, enhancing keratitis after Pseudomonas aeruginosa infection. Invest. Ophthalmol. Vis. Sci. 2008;49:4458–4467. doi: 10.1167/iovs.08-1906. [DOI] [PubMed] [Google Scholar]
- 29.Lighvani S, Huang X, Trivedi PP, Swanborg RH, Hazlett LD. Substance P regulates natural killer cell interferon-gamma production and resistance to Pseudomonas aeruginosa infection. Eur. J. Immunol. 2005;35:1567–1575. doi: 10.1002/eji.200425902. [DOI] [PubMed] [Google Scholar]
- 30.Hazlett LD, et al. Spantide I decreases type I cytokines, enhances IL-10, and reduces corneal perforation in susceptible mice after Pseudomonas aeruginosa infection. Invest. Ophthalmol. Vis. Sci. 2007;48:797–807. doi: 10.1167/iovs.06-0882. [DOI] [PubMed] [Google Scholar]
- 31.McClellan SA, Zhang Y, Barrett RP, Hazlett LD. Substance P promotes susceptibility to Pseudomonas aeruginosa keratitis in resistant mice: Anti-inflammatory mediators downregulated. Invest. Ophthalmol. Vis. Sci. 2008;49:1502–1511. doi: 10.1167/iovs.07-1369. [DOI] [PubMed] [Google Scholar]
- 32.Foldenauer ME, McClellan SA, Barrett RP, Zhang Y, Hazlett LD. Substance P affects growth factors in Pseudomonas aeruginosa-infected mouse cornea. Cornea. 2012;31:1176–1188. doi: 10.1097/ICO.0b013e31824d6ffd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin T, et al. Pseudomonas aeruginosa-induced nociceptor activation increases susceptibility to infection. PLoS Pathog. 2021;17:e1009557. doi: 10.1371/journal.ppat.1009557. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 35.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 36.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 37.Lee RC, Feinbaum RL, Ambros VT. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 38.Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu. Rev. Genom. Hum. Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 39.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 40.Lee YS, Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mencia, A. et al. in 6th Molecular Biology of Hearing and Hearing Deafness Conference.
- 42.Hughes AE, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am. J. Hum. Genet. 2011;89:628–633. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iliff BW, Riazuddin SA, Gottsch JD. A single-base substitution in the seed region of miR-184 causes EDICT syndrome. Invest. Ophthalmol. Vis. Sci. 2012;53:348–353. doi: 10.1167/iovs.11-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conte I, et al. MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc. Natl. Acad. Sci. U S A. 2015 doi: 10.1073/pnas.1401464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu S. microRNAs and inherited retinal dystrophies. Proc. Natl. Acad. Sci. U S A. 2015;112:8805–8806. doi: 10.1073/pnas.1511019112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeyaseelan K, Herath WB, Armugam A. MicroRNAs as therapeutic targets in human diseases. Expert Opin. Ther. Targets. 2007;11:1119–1129. doi: 10.1517/14728222.11.8.1119. [DOI] [PubMed] [Google Scholar]
- 47.van Rooij E, Olson EN. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J. Clin. Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang B, Farwell MA. microRNAs: A new emerging class of players for disease diagnostics and gene therapy. J. Cell Mol. Med. 2008;12:3–21. doi: 10.1111/j.1582-4934.2007.00196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stenvang J, Kauppinen S. MicroRNAs as targets for antisense-based therapeutics. Expert. Opin. Biol. Ther. 2008;8:59–81. doi: 10.1517/14712598.8.1.59. [DOI] [PubMed] [Google Scholar]
- 50.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 51.Lumayag S, et al. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc. Natl. Acad. Sci. USA. 2013;110:E507–E516. doi: 10.1073/pnas.1212655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 53.Fan J, et al. Maturation arrest in early postnatal sensory receptors by deletion of the miR-183/96/182 cluster in mouse. Proc. Natl. Acad. Sci. U S A. 2017;114:E4271–E4280. doi: 10.1073/pnas.1619442114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang L, et al. miR-183/96 plays a pivotal regulatory role in mouse photoreceptor maturation and maintenance. Proc. Natl. Acad. Sci. U S A. 2017;114:6376–6381. doi: 10.1073/pnas.1618757114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang CJ, et al. Ablation of mature miR-183 leads to retinal dysfunction in mice. Invest. Ophthalmol. Vis. Sci. 2020;61:12. doi: 10.1167/iovs.61.3.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lewis, M. et al. in 6th Molecular Biology of Hearing and Deafness Conference.
- 57.Mencia A, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 58.Donatelli SS, et al. TGF-beta-inducible microRNA-183 silences tumor-associated natural killer cells. Proc. Natl. Acad. Sci. USA. 2014;111:4203–4208. doi: 10.1073/pnas.1319269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wurm AA, et al. Disruption of the C/EBPalpha-miR-182 balance impairs granulocytic differentiation. Nat. Commun. 2017;8:46. doi: 10.1038/s41467-017-00032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muraleedharan CK, et al. Inactivation of the miR-183/96/182 cluster decreases the severity of pseudomonas aeruginosa-induced keratitis. Invest. Ophthalmol. Vis. Sci. 2016;57:1506–1517. doi: 10.1167/iovs.16-19134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muraleedharan, C. K. et al. The miR-183/96/182 cluster regulates macrophage functions in response to pseudomonas aeruginosa. J. Innate Immun. 1–12. 10.1159/000495472 (2019). [DOI] [PMC free article] [PubMed]
- 62.Wang J, et al. miR-183-96-182 cluster is involved in invariant NKT cell development, maturation, and effector function. J. Immunol. 2019 doi: 10.4049/jimmunol.1900695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ichiyama K, et al. The MicroRNA-183-96-182 cluster promotes T helper 17 cell pathogenicity by negatively regulating transcription factor foxo1 expression. Immunity. 2016;44:1284–1298. doi: 10.1016/j.immuni.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stittrich AB, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat. Immunol. 2010;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 65.Kelada S, et al. miR-182 and miR-10a are key regulators of Treg specialisation and stability during Schistosome and Leishmania-associated inflammation. PLoS Pathog. 2013;9:e1003451. doi: 10.1371/journal.ppat.1003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li YF, et al. Loss of miR-182 affects B-cell extrafollicular antibody response. Immunology. 2016;148:140–149. doi: 10.1111/imm.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wan C, et al. MicroRNA 182 inhibits CD4(+)CD25(+)Foxp3(+) Treg differentiation in experimental autoimmune encephalomyelitis. Clin. Immunol. 2016;173:109–116. doi: 10.1016/j.clim.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 68.Wang Z, et al. Deletion of microRNA-183-96-182 cluster in lymphocytes suppresses anti-DsDNA autoantibody production and IgG deposition in the kidneys in C57BL/6-Fas(lpr/lpr) mice. Front. Genet. 2022;13:840060. doi: 10.3389/fgene.2022.840060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ichiyama K, Dong C. The role of miR-183 cluster in immunity. Cancer Lett. 2019;443:108–114. doi: 10.1016/j.canlet.2018.11.035. [DOI] [PubMed] [Google Scholar]
- 70.Yu H, Liu Y, Bai L, Kijlstra A, Yang P. Predisposition to Behcet's disease and VKH syndrome by genetic variants of miR-182. J. Mol. Med. 2014;92:961–967. doi: 10.1007/s00109-014-1159-9. [DOI] [PubMed] [Google Scholar]
- 71.McClellan S, et al. Prophylactic knockdown of the miR-183/96/182 cluster ameliorates pseudomonas aeruginosa-induced keratitis. Invest. Ophthalmol. Vis. Sci. 2021;62:14. doi: 10.1167/iovs.62.15.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hansen J, et al. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc. Natl. Acad. Sci. USA. 2003;100:9918–9922. doi: 10.1073/pnas.1633296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schnutgen F, et al. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc. Natl. Acad. Sci. USA. 2005;102:7221–7226. doi: 10.1073/pnas.0502273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sasmono RT, et al. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood. 2003;101:1155–1163. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- 75.MacDonald KP, et al. The colony-stimulating factor 1 receptor is expressed on dendritic cells during differentiation and regulates their expansion. J. Immunol. 2005;175:1399–1405. doi: 10.4049/jimmunol.175.3.1399. [DOI] [PubMed] [Google Scholar]
- 76.Peng C, et al. miR-183 cluster scales mechanical pain sensitivity by regulating basal and neuropathic pain genes. Science. 2017;356:1168–1171. doi: 10.1126/science.aam7671. [DOI] [PubMed] [Google Scholar]
- 77.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- 78.Stirling LC, et al. Nociceptor-specific gene deletion using heterozygous NaV1.8-Cre recombinase mice. Pain. 2005;113:27–36. doi: 10.1016/j.pain.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 79.Kozak CA, Sangameswaran L. Genetic mapping of the peripheral sodium channel genes, Scn9a and Scn10a, in the mouse. Mamm. Genome. 1996;7:787–788. doi: 10.1007/s003359900235. [DOI] [PubMed] [Google Scholar]
- 80.Souslova VA, Fox M, Wood JN, Akopian AN. Cloning and characterization of a mouse sensory neuron tetrodotoxin-resistant voltage-gated sodium channel gene, Scn10a. Genomics. 1997;41:201–209. doi: 10.1006/geno.1997.4669. [DOI] [PubMed] [Google Scholar]
- 81.Abrahamsen B, et al. The cell and molecular basis of mechanical, cold, and inflammatory pain. Science. 2008;321:702–705. doi: 10.1126/science.1156916. [DOI] [PubMed] [Google Scholar]
- 82.Agarwal N, Offermanns S, Kuner R. Conditional gene deletion in primary nociceptive neurons of trigeminal ganglia and dorsal root ganglia. Genesis. 2004;38:122–129. doi: 10.1002/gene.20010. [DOI] [PubMed] [Google Scholar]
- 83.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kruisbeek AM. Isolation of mouse mononuclear cells. Curr. Protoc. Immunol. 2001;3:1. doi: 10.1002/0471142735.im0301s39. [DOI] [PubMed] [Google Scholar]
- 85.Xu S, et al. Mutation screening in the miR-183/96/182 cluster in patients with inherited retinal dystrophy. Front. Cell. Dev. Biol. 2020;8:619641. doi: 10.3389/fcell.2020.619641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li C, McClellan SA, Barrett R, Hazlett LD. Interleukin 17 regulates Mer tyrosine kinase-positive cells in pseudomonas aeruginosa keratitis. Invest. Ophthalmol. Vis. Sci. 2014;55:6886–6900. doi: 10.1167/iovs.14-14522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kovacs B, Lumayag S, Cowan C, Xu S. microRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Invest. Ophthalmol. Vis. Sci. 2011;52:4402–4409. doi: 10.1167/iovs.10-6879. [DOI] [PubMed] [Google Scholar]
- 88.Moll P, Ante M, Seitz A, Reda T. QuantSeq 3′ mRNA sequencing for RNA quantification. Nat. Methods. 2014;11:441. doi: 10.1038/nmeth.f.376. [DOI] [Google Scholar]
- 89.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:1. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garcia DM, et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 97.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 98.Tan Z, Yi X, Carruthers NJ, Stemmer PM, Lubman DM. Single amino acid variant discovery in small numbers of cells. J. Proteome Res. 2019;18:417–425. doi: 10.1021/acs.jproteome.8b00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vereertbrugghen A, Galletti JG. Corneal nerves and their role in dry eye pathophysiology. Exp. Eye Res. 2022;222:109191. doi: 10.1016/j.exer.2022.109191. [DOI] [PubMed] [Google Scholar]
- 100.Ma F, et al. A comparison between whole transcript and 3' RNA sequencing methods using Kapa and Lexogen library preparation methods. BMC Genomics. 2019;20:9. doi: 10.1186/s12864-018-5393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zlotnik A, Yoshie O. Chemokines: A new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 102.Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: Basic chemokinese grammar for immune cells. Annu. Rev. Immunol. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 103.Stievano L, Piovan E, Amadori A. C and CX3C chemokines: Cell sources and physiopathological implications. Crit. Rev. Immunol. 2004;24:205–228. doi: 10.1615/critrevimmunol.v24.i3.40. [DOI] [PubMed] [Google Scholar]
- 104.Harrison JK, et al. Role for neuronally derived fractalkine in mediating interactions between neurons and CX3CR1-expressing microglia. Proc. Natl. Acad. Sci. USA. 1998;95:10896–10901. doi: 10.1073/pnas.95.18.10896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pawelec P, Ziemka-Nalecz M, Sypecka J, Zalewska T. The impact of the CX3CL1/CX3CR1 axis in neurological disorders. Cells. 2020;9:1. doi: 10.3390/cells9102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chinnery HR, et al. The chemokine receptor CX3CR1 mediates homing of MHC class II-positive cells to the normal mouse corneal epithelium. Invest. Ophthalmol. Vis. Sci. 2007;48:1568–1574. doi: 10.1167/iovs.06-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ishida Y, Gao JL, Murphy PM. Chemokine receptor CX3CR1 mediates skin wound healing by promoting macrophage and fibroblast accumulation and function. J. Immunol. 2008;180:569–579. doi: 10.4049/jimmunol.180.1.569. [DOI] [PubMed] [Google Scholar]
- 108.Kuhn S, et al. miR-96 regulates the progression of differentiation in mammalian cochlear inner and outer hair cells. Proc. Natl. Acad. Sci. USA. 2011;108:2355–2360. doi: 10.1073/pnas.1016646108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Geng R, et al. The microRNA-183/96/182 cluster is essential for stereociliary bundle formation and function of cochlear sensory hair cells. Sci. Rep. 2018;8:18022. doi: 10.1038/s41598-018-36894-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 112.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nowakowski TJ, et al. Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat. Neurosci. 2018;21:1784–1792. doi: 10.1038/s41593-018-0265-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Patil MJ, Hovhannisyan AH, Akopian AN. Characteristics of sensory neuronal groups in CGRP-cre-ER reporter mice: Comparison to Nav18-cre, TRPV1-cre and TRPV1-GFP mouse lines. PLoS One. 2018;13:e0198601. doi: 10.1371/journal.pone.0198601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Akopian AN, Sivilotti L, Wood JN. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature. 1996;379:257–262. doi: 10.1038/379257a0. [DOI] [PubMed] [Google Scholar]
- 116.Djouhri L, et al. The TTX-resistant sodium channel Nav1.8 (SNS/PN3): expression and correlation with membrane properties in rat nociceptive primary afferent neurons. J. Physiol. 2003;550:739–752. doi: 10.1113/jphysiol.2003.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lauro C, et al. The chemokine CX3CL1 reduces migration and increases adhesion of neurons with mechanisms dependent on the beta1 integrin subunit. J. Immunol. 2006;177:7599–7606. doi: 10.4049/jimmunol.177.11.7599. [DOI] [PubMed] [Google Scholar]
- 118.Forsyth KS, Jiwrajka N, Lovell CD, Toothacre NE, Anguera MC. The conneXion between sex and immune responses. Nat. Rev. Immunol. 2024 doi: 10.1038/s41577-024-00996-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Klein SL, Flanagan KL. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 120.Wilkinson NM, Chen HC, Lechner MG, Su MA. Sex differences in immunity. Annu. Rev. Immunol. 2022;40:75–94. doi: 10.1146/annurev-immunol-101320-125133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zazara DE, Belios I, Lucke J, Zhang T, Giannou AD. Tissue-resident immunity in the lung: A first-line defense at the environmental interface. Semin. Immunopathol. 2022;44:827–854. doi: 10.1007/s00281-022-00964-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Han J, Fan Y, Zhou K, Blomgren K, Harris RA. Uncovering sex differences of rodent microglia. J. Neuroinflamm. 2021;18:74. doi: 10.1186/s12974-021-02124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Scotland RS, Stables MJ, Madalli S, Watson P, Gilroy DW. Sex differences in resident immune cell phenotype underlie more efficient acute inflammatory responses in female mice. Blood. 2011;118:5918–5927. doi: 10.1182/blood-2011-03-340281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zoukhri D, Hodges RR, Dartt DA. Lacrimal gland innervation is not altered with the onset and progression of disease in a murine model of Sjogren's syndrome. Clin. Immunol. Immunopathol. 1998;89:126–133. doi: 10.1006/clin.1998.4597. [DOI] [PubMed] [Google Scholar]
- 125.Brundl M, Garreis F, Schicht M, Dietrich J, Paulsen F. Characterization of the innervation of the meibomian glands in humans, rats and mice. Ann. Anat. 2021;233:151609. doi: 10.1016/j.aanat.2020.151609. [DOI] [PubMed] [Google Scholar]
- 126.Luhtala J, Palkama A, Uusitalo H. Calcitonin gene-related peptide immunoreactive nerve fibers in the rat conjunctiva. Invest. Ophthalmol. Vis. Sci. 1991;32:640–645. [PubMed] [Google Scholar]
- 127.Luhtala J, Uusitalo H. The distribution and origin of substance P immunoreactive nerve fibres in the rat conjunctiva. Exp. Eye Res. 1991;53:641–646. doi: 10.1016/0014-4835(91)90224-3. [DOI] [PubMed] [Google Scholar]
- 128.Knop E, Knop N, Millar T, Obata H, Sullivan DA. The international workshop on meibomian gland dysfunction: Report of the subcommittee on anatomy, physiology, and pathophysiology of the meibomian gland. Invest. Ophthalmol. Vis. Sci. 2011;52:1938–1978. doi: 10.1167/iovs.10-6997c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kimura M, et al. A nerve-goblet cell association promotes allergic conjunctivitis through rapid antigen passage. JCI Insight. 2023;8:1. doi: 10.1172/jci.insight.168596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Swamynathan SK, Wells A. Conjunctival goblet cells: Ocular surface functions, disorders that affect them, and the potential for their regeneration. Ocular Surf. 2020;18:19–26. doi: 10.1016/j.jtos.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Thody AJ, Shuster S. Control and function of sebaceous glands. Physiol. Rev. 1989;69:383–416. doi: 10.1152/physrev.1989.69.2.383. [DOI] [PubMed] [Google Scholar]
- 132.Dartt DA. Neural regulation of lacrimal gland secretory processes: Relevance in dry eye diseases. Prog. Retin. Eye Res. 2009;28:155–177. doi: 10.1016/j.preteyeres.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Caffery, B. E. et al. CANDEES. The Canadian dry eye epidemiology study. Adv. Exp. Med. Biol.438, 805–806 (1998). [PubMed]
- 134.Ahn JM, et al. Prevalence of and risk factors associated with dry eye: The Korea National Health and Nutrition Examination Survey 2010–2011. Am. J. Ophthalmol. 2014;158:1205–1214. doi: 10.1016/j.ajo.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 135.Schaumberg DA, Sullivan DA, Buring JE, Dana MR. Prevalence of dry eye syndrome among US women. Am. J. Ophthalmol. 2003;136:318–326. doi: 10.1016/s0002-9394(03)00218-6. [DOI] [PubMed] [Google Scholar]
- 136.Uchino M, et al. Prevalence of dry eye disease among Japanese visual display terminal users. Ophthalmology. 2008;115:1982–1988. doi: 10.1016/j.ophtha.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 137.Schaumberg DA, Dana R, Buring JE, Sullivan DA. Prevalence of dry eye disease among US men: Estimates from the Physicians' Health Studies. Arch. Ophthalmol. 2009;127:763–768. doi: 10.1001/archophthalmol.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Uchino M, et al. Prevalence and risk factors of dry eye disease in Japan: Koumi study. Ophthalmology. 2011;118:2361–2367. doi: 10.1016/j.ophtha.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 139.Um SB, Kim NH, Lee HK, Song JS, Kim HC. Spatial epidemiology of dry eye disease: Findings from South Korea. Int. J. Health Geogr. 2014;13:31. doi: 10.1186/1476-072X-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang Y, Chen H, Wu X. Prevalence and risk factors associated with dry eye syndrome among senior high school students in a county of Shandong Province China. Ophthal. Epidemiol. 2012;19:226–230. doi: 10.3109/09286586.2012.670742. [DOI] [PubMed] [Google Scholar]
- 141.Sullivan DA, et al. TFOS DEWS II sex, gender, and hormones report. Ocular Surf. 2017;15:284–333. doi: 10.1016/j.jtos.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 142.Wagner H, Fink BA, Zadnik K. Sex- and gender-based differences in healthy and diseased eyes. Optometry. 2008;79:636–652. doi: 10.1016/j.optm.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 143.Truong S, Cole N, Stapleton F, Golebiowski B. Sex hormones and the dry eye. Clin. Exp. Opt. 2014;97:324–336. doi: 10.1111/cxo.12147. [DOI] [PubMed] [Google Scholar]
- 144.Homma, M., Sugai, S., Tojo, T., Miyasaka, N. & Akizuki, M. Sjoren's syndrome: State of the Art. (Kugler Press, 1994).
- 145.Sullivan DA, Hann LE, Yee L, Allansmith MR. Age- and gender-related influence on the lacrimal gland and tears. Acta Ophthalmol. (Copenh) 1990;68:188–194. doi: 10.1111/j.1755-3768.1990.tb01902.x. [DOI] [PubMed] [Google Scholar]
- 146.Cornell-Bell AH, Sullivan DA, Allansmith MR. Gender-related differences in the morphology of the lacrimal gland. Invest. Ophthalmol. Vis. Sci. 1985;26:1170–1175. [PubMed] [Google Scholar]
- 147.Sullivan DA, Bloch KJ, Allansmith MR. Hormonal influence on the secretory immune system of the eye: Androgen regulation of secretory component levels in rat tears. J. Immunol. 1984;132:1130–1135. doi: 10.4049/jimmunol.132.3.1130. [DOI] [PubMed] [Google Scholar]
- 148.Sullivan DA, Allansmith MR. Hormonal modulation of tear volume in the rat. Exp. Eye Res. 1986;42:131–139. doi: 10.1016/0014-4835(86)90037-0. [DOI] [PubMed] [Google Scholar]
- 149.Yang M, et al. Sex-based differences in conjunctival goblet cell responses to pro-inflammatory and pro-resolving mediators. Sci. Rep. 2022;12:16305. doi: 10.1038/s41598-022-20177-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sullivan DA, Bloch KJ, Allansmith MR. Hormonal influence on the secretory immune system of the eye: androgen control of secretory component production by the rat exorbital gland. Immunology. 1984;52:239–246. [PMC free article] [PubMed] [Google Scholar]
- 151.Pieczynski J, Szulc U, Harazna J, Szulc A, Kiewisz J. Tear fluid collection methods: Review of current techniques. Eur. J. Ophthalmol. 2021;31:2245–2251. doi: 10.1177/1120672121998922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets of 3′ RNA sequencing generated during the current study are available in the Gene Expression Omnibus (GEO) repository (accession number: GSE250213). The proteomics data have been submitted to the repository Proteomics IDEntifications Database (PRIDE. Accession number: PXD05033). All data and protocols generated in this study are available upon request from the corresponding author.