Abstract
Bacterial biofilm-associated infection was one of the most serious threats to human health. However, effective drugs for drug-resistance bacteria or biofilms remain rarely reported. Here, we propose an innovative strategy to develop a multifunctional antimicrobial agent with broad-spectrum antibacterial activity by coupling photosensitizers (PSs) with antimicrobial peptides (AMPs). This strategy capitalizes on the ability of PSs to generate reactive oxygen species (ROS) and the membrane-targeting property of AMPs (KRWWKWIRW, a peptide screened by an artificial neural network), synergistically enhancing the antimicrobial activity. In addition, unlike conventional aggregation-caused quenching (ACQ) photosensitizers, aggregation-induced emission (AIE) PSs show stronger fluorescence emission in the aggregated state to help visualize the antibacterial mechanism. In vitro antibacterial experiments demonstrated the excellent killing effects of the developed agent against both Gram-positive (G+) and Gram-negative (G–) bacteria. The bacterial-aggregations induced ability enhanced the photoactivatable antibacterial activity against G– bacteria. Notably, it exhibited a significant effect on destroying MRSA biofilms. Moreover, it also showed remarkable efficacy in treating wound infections in mice in vivo. This multifunctional antimicrobial agent holds significant potential in addressing the challenges posed by bacterial biofilm-associated infections and drug-resistant bacteria.
Key words: Photodynamic therapy, Antimicrobial peptides, Multidrug-resistant bacteria, Aggregation-induced emission, Anti-biofilm
Graphical abstract
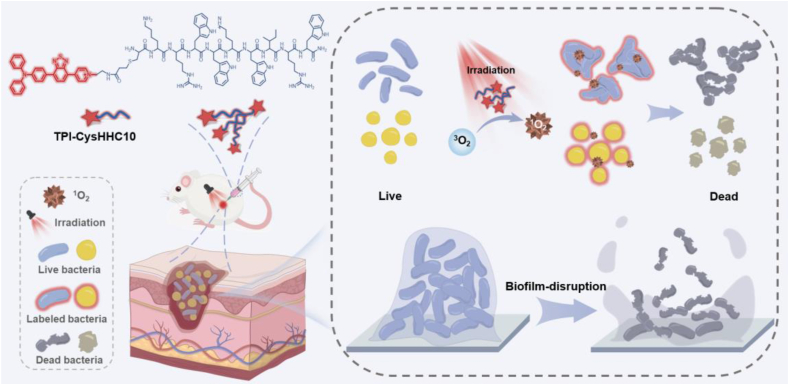
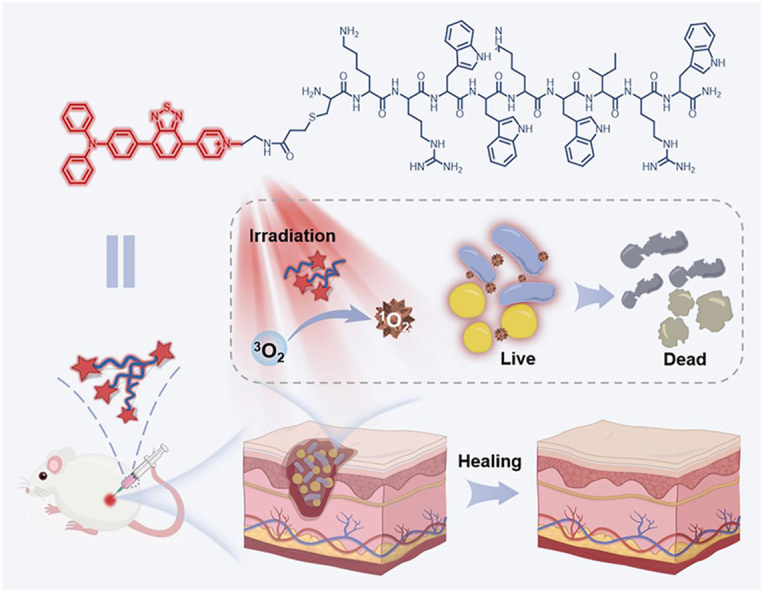
Schematic illustration of TPI-CysHHC10 comprising AMPs and AIE photosensitizer as a photoactivatable, broad-spectrum, antibacterial agent for combating biofilms.
1. Introduction
Bacterial infection was one of the most serious threats to human health, killing millions of people worldwide each year1. However, due to the emergence of bacterial resistance, many traditional antibiotics had weakened or even almost lost their antimicrobial activity against diseases caused by many species of bacteria2, 3, 4, 5, 6, 7. In addition, the formation of bacterial biofilms, a population of microorganisms encapsulated in an extracellular matrix whose main constituents were proteins and polysaccharides, was undoubtedly exacerbating the current unoptimistic treatment status. Therefore, the research and development of new efficient antibacterial drugs and novel antibacterial strategies were of great significance for mankind to break out of this dilemma. Fortunately, there were many potential antibacterial agents under study, such as cationic photosensitizers8, metabolic probes9, metal nanomaterials10,11, antimicrobial peptides (AMPs)12, 13, 14, 15, and cationic polymers16, 17, 18.
As a large category of new antibacterial materials, AMPs have many advantages. Antimicrobial peptides, as an important element of the innate immune system of many animals and plants in the ecological environment12,19, exhibit excellent antibacterial activity against both gram-negative (G–) and gram-positive bacteria (G+)20. Its most widespread mode of action was to increase the permeability or disrupt bacterial membranes leading to extravasation of bacterial contents, resulting in bacterial death21. Because of this, it was nearly impossible for bacteria to develop resistance to AMPs due to their different modes of action compared to traditional antibiotics. HHC10 (KRWWKWIRW) is a peptide screened by an artificial neural network with certain antibacterial activity against both G– and G+ bacteria, its antimicrobial activity is mainly derived from the destruction of bacterial membranes22,23. The broad-spectrum antibacterial activity of AMPs and the characteristics of not easily developing bacterial resistance made them one of the promising antibacterial agents that have been widely explored in recent years.
However, for AMPs, there is still the possibility of drug resistance in bacteria. It is reported that bacteria could change the permeability and fluidity of the membrane by changing the composition and structure of the membrane, thus producing resistance to prevent interaction with AMPs24. On the other hand, the real-time monitoring and visualization of the interaction process between AMPs and bacteria is of great significance to reveal the antibacterial mechanism of AMPs. However, the use of conventional imaging techniques, such as scanning electron microscope (SEM), transmission electron microscope (TEM), etc., often requires complex sample preparation and instrument operation procedures, which can't achieve real-time monitoring. On the contrary, fluorescence imaging technology can be used for dynamic and real-time observation with high sensitivity and simple operation25,26. Furthermore, many fluorescent molecules not only have excellent luminescent properties but also can be used as photosensitizers (PSs) to induce the production of ROS and promote their application in photodynamic therapy.
Photodynamic antibacterial therapy was on the rise as an effective sterilization strategy21,27. The main principle is to use the ROS produced by PSs after light irradiation to disrupt the bacterial membrane or cause irreversible damage to the proteins and nucleic acids inside the bacteria, to achieve the purpose of antibacterial28,29. In addition, PSs have also been extensively studied in the field of anti-tumor30. Due to the indiscriminate destruction of membrane structures or biological macromolecules antibacterial effects while preventing bacteria from developing drug resistance. In addition, unlike conventional aggregation-caused quenching (ACQ) photosensitizers, aggregation-induced emission (AIE) PSs show stronger fluorescence emission and higher ROS generation efficiency in the aggregated state31.
Herein, we propose a novel strategy to combine PSs with AMPs to form a new class of antibacterial conjugates with broad-spectrum antibacterial and antibiofilm effects (Scheme 1). We developed the TPI-CysHHC10 by conjugating the fluorophore with AMPs via a thiol-ene reaction. The excellent fluorescence properties of the fluorophores have helped to reveal visualizations of the interaction patterns between TPI-CysHHC10 and bacteria, and its photodynamic antibacterial activity was enhanced by the excellent singlet oxygen generation ability of the fluorophores. On the other hand, TPI-CysHHC10 could not only kill G+ bacteria but also help to destroy the outer membrane, an important barrier of G– bacteria. In addition, TPI-CysHHC10 was found to induce aggregation of G– bacteria, including E. coli and P. aeruginosa. The formation of bacterial aggregations helped to overcome the limited diffusion range of ROS and improve its overall antibacterial efficiency. In this way, it can play a better role in the photodynamic antibacterial effect. The disruption function of AMPs to the outer membrane of bacteria played a crucial role in the antibacterial activity of TPI-CysHHC10 against G– bacteria. Compared with the reported AMP-functionalized PS, TPI-CysHHC10 has the advantages of broad spectrum, high photosensitive activity, and good biocompatibility32, 33, 34, 35. This approach of coupling antimicrobial peptides with a membrane-destroying function to AIE PSs will provide a novel strategy for the treatment of bacterial infections.
Scheme 1.
Schematic illustration of TPI-CysHHC10 comprising AMPs and AIE photosensitizer as a photoactivatable, broad-spectrum, antibacterial agent for combating biofilms. TPI-CysHHC10 was initially attached to the bacterial surface and inserted into the bacterial membrane structure by means of the positivity of molecules and the affinity of antimicrobial peptides to the bacterial membrane. Subsequently, under light irradiation, the photodynamic antibacterial activity of TPI-CysHHC10 was triggered. In addition, TPI-CysHHC10 could induce the aggregation of G– bacteria and produce ROS in the site, which was helpful in maximizing the antibacterial effect of ROS in their effective range. On the other hand, the small molecular size and excellent water solubility of TPI-CysHHC10 helped to penetrate the biofilms to achieve an efficient photodynamic anti-biofilm effect inside.
2. Materials and methods
2.1. Materials and instruments
All required materials and instruments were included in Supporting Information. The synthesis and characterization of TPI-CysHHC10 can also be obtained from Supporting Information.
2.2. ROS detection
Using ABDA as an indicator of 1O2 production efficiency, its decomposition under singlet oxygen reduces the absorbance between 350 and 420 nm. ABDA was mixed with TPI-CysHHC10 solution (containing ABDA: 50 μmol/L, TPI-CysHHC10: 10 μmol/L white light irradiation (60 mW/cm2) for different times (0, 10, 20, 30, 40, 50, 60, 70, 80, 90 and 100 s), the UV absorption spectra (300–700 nm) of each sample were measured. The 1O2 generation efficiency was estimated by comparing the degree of absorbance reduction at 378 nm.
2.3. Bacteria imaging
Bacteria were collected by centrifugation at 4000 rpm for 5 min and washed with sterilized PBS three times. Then, the bacterial precipitate was resuspended with 300 μL PBS, and 1.5 μL/3 μL TPI-CysHHC10 (1 mmol/L) solution was added and incubated for a different time. Then 10 μg/mL Hoechst 33342 was added and incubated for another 30 min. All procedures were maintained at room temperature. After incubation, bacteria were collected and washed with sterilized PBS and then placed on a microscope glass slide, and the fluorescence images were obtained by CLSM.
2.4. In vitro antibacterial activity
The antibacterial activity assays of TPI-CysHHC10, TPI-PA, and HHC10 were evaluated by the traditional plate counting method. Bacteria were harvested by centrifuging at 4000 rpm for 5 min and then washed with sterilized PBS three times. Then, the bacteria precipitate obtained from the broth was resuspended with PBS to obtain a bacterial solution with a designed concentration (OD600 = 1.0). To prepare the samples for antibacterial determinations, the bacterial solution was diluted 1000-fold by PBS and then mixed with equal volumes of TPI-CysHHC10, TPI-PA, HHC10, or polymyxin(G–)/vancomycin(G+) at various concentrations. After that, for the group of polymyxin(G–)/vancomycin(G+), HHC10, TPI-PA, and TPI-CysHHC10 in the darkness, the samples were incubated at 37 °C for 2 h. For the group of TPI-CysHHC10 and TPI-PA under irradiation, the samples were incubated at 37 °C for 30 min and illuminated with white light for 15 min, and incubated for another 75 min. Then, 100 μL of each sample was dropped onto the plate and spread evenly. After incubation for 24 h at 37 °C, the surviving number of colonies forming units (CFU) on each plate was then enumerated by viable counting.
2.5. Cytotoxicity assay
HeLa cells were cultured using a DMEM medium containing 10% fetal bovine serum, and the flask was placed in an incubator containing 5% CO2 at 37 °C. Cells were seeded into 96-well plates (about 5 103 cells per well), and placed in the above incubator for 24 h. After removing the medium, 100 μL of various concentrations of TPI-CysHHC10 dissolved in the medium were added to each well. Cell viability was subsequently assessed by the MTT method. Briefly, 100 μL fresh medium containing 10 μL MTT stock solution (5 mg/mL) was added into each well and incubated at 37 °C for 4 h. After the liquid was removed from each well, 100 μL DMSO was added to dissolve crystal violet, and the absorption value at 570 nm was measured using a microplate reader. The cell viability in each group was compared to the control group without treatment.
2.6. Hemolysis experiment
Mice arterial blood (blood from the heart) was harvested and heparin was added for anticoagulation. The anticoagulant blood was centrifuged at 1500 rpm for 5 min and red blood cells were collected. The obtained red blood cells were further washed twice with PBS and diluted. 500 μL of TPI-CysHHC10 at different concentrations (128, 64, 32, 16, 8, 4, 2, 1, and 0.5 μmol/L), with saline as the negative control and H2O as the positive control, were prepared and mixed with 500 μL red blood cell suspension. The mixture was incubated at 37 °C for 2 h. The mixture was then centrifuged and imaged and the absorbance at 540 nm of the supernatant was determined. The hemolysis rate was calculated concerning Eq. (1):
| (1) |
2.7. In vivo antibacterial activity
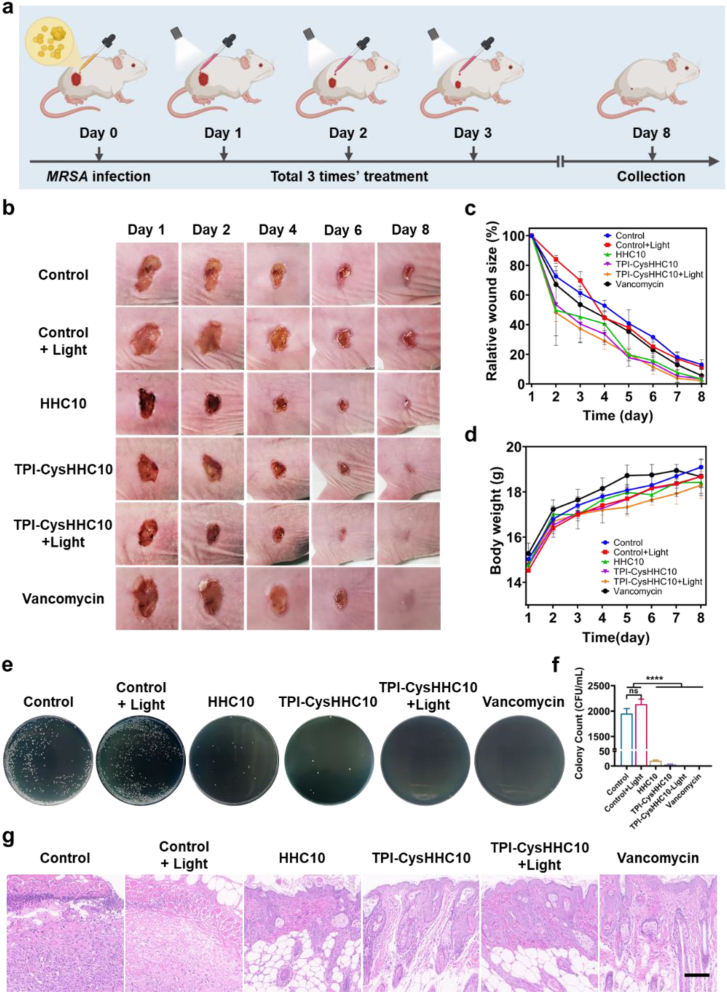
All procedures of animal experiments were approved by the Animal Care and Use Committee of Central South University and complied with all relevant ethical regulations. Female BALB/c mice (6 weeks) were purchased from Hunan SJA Laboratory Animal Co., Ltd. (Hunan, China). BALB/c mice were randomly divided into six groups: (1) bacteria-infected group without any treatment (Control); (2) bacteria-infected group with white light irradiation (Light); (3) bacteria-infected group with HHC10 treatment alone (HHC10); (4) bacteria-infected group with TPI-CysHHC10 alone treatment (TPI-CysHHC10); (5) bacteria-infected group with TPI-CysHHC10 treatment and white light irradiation (TPI-CysHHC10 + Light); (6) bacteria-infected group with Vancomycin alone treatment (Vancomycin) (n = 4 in each group). The mice were anesthetized by injection of 1.5% pentobarbital sodium. Then two full-thickness injuries (∼8 mm diameter) on each mouse were created. The bacteria-infected wound models were established by inoculating with 20 μL bacteria suspension (1.0 × 109 CFU/mL of MRSA) on the surface of wounds. After 24 h post-infection, 20 μL of physiological saline, TPI-CysHHC10, HHC10, and vancomycin (10 μmol/L) was added to the wound and kept for 30 min. Subsequently, the wounds were irradiated by white light (60 mW/cm2) for 10 min or kept under darkness. The treatment of drugs and with or without white light irradiation was performed on the first day and continued for another two days with a total of three times. The wound healing processes were dynamically monitored through measurements of the wound size, at the same time the wound was photographed. On Day 8, tissues from the infected sites of all mice were collected for further evaluation. The agar plate counting method was used to evaluate the bacterial content in the infected site.
2.8. Immunohistochemical analysis and H&E staining
The extracted tissues were fixed in 4% paraformaldehyde, and then paraffin-embedded, and sliced using standard protocols for hematoxylin-eosin (H&E) staining. And they were finally examined using an inverted microscope.
3. Results and discussion
3.1. Synthesis and characterization of TPI-CysHHC10
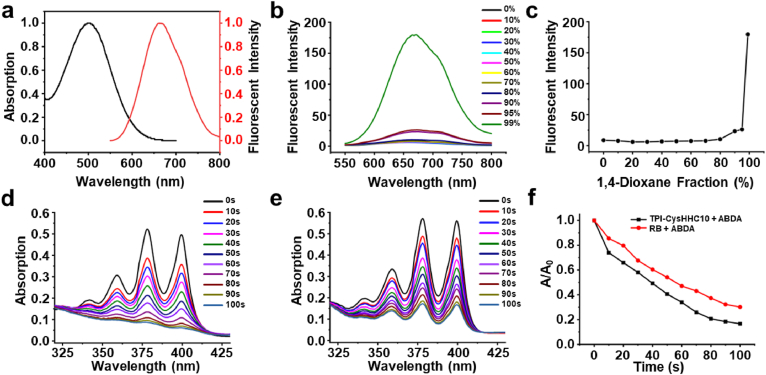
The synthesis route was shown in Supporting Information Scheme S1. TPI-CysHHC10 was successfully synthesized and the fluorophore and peptide were conjugated by the thiol-ene reaction. The structure of TPI-CysHHC10 was confirmed by 1H NMR and HRMS. The results were shown in Supporting Information Figs. S9–S11. 1As shown in Fig. S9, we could find the multi-charge peaks ([M–Br+H]2+: 1051.0133, [M–Br+2H]3+: 701.0117, [M–Br+3H]4+: 526.0113, [M–Br+4H]5+: 421.0105) corresponding to the charge numbers of 2, 3, 4, and 5, respectively. The peptide could disrupt the cell membrane of bacteria, the fluorophore contributed to the singlet oxygen generation that synergistically destroyed bacterial structure with antimicrobial peptides to achieve excellent antibacterial activity. TPI-CysHHC10 had obvious UV absorption in the wavelength range of 400 to 600 nm, with a maximum absorption wavelength of 500 nm (Fig. 1a). The maximum emission of TPI-CysHHC10 in aggregation state was located at 670 nm, indicating its long-wavelength emission characteristics and large Stokes shifts (170 nm). As shown in Fig. 1b–c, as the ratio of the poor solvent 1,4-dioxane increased, the fluorescence intensity of TPI-CysHHC10 also increased. When the ratio of 1,4-dioxane reached 99%, the fluorescence intensity increased sharply, and TPI-CysHHC10 was in an aggregated state, revealing the obvious AIE property of TPI-CysHHC10. PSs with AIE properties also had higher ROS generation ability in the aggregated state36,37, which was of great benefit to the efficiency of PSs aggregated on the bacterial outer membrane and generated ROS after exposure to white light radiation.
Figure 1.
Characterization of TPI-CysHHC10. (a) Normalized UV–vis spectra and fluorescent spectra of TPI-CysHHC10. (b) Fluorescent spectra curve and (c) plotting of the emission maximum of TPI-CysHHC10 in different fractions of H2O/1,4-dioxane mixture. Photodegradation curve of ABDA with (d) TPI-CysHHC10 (in PBS) and (e) Rose Bengal (in PBS) under white light irradiation (60 mW/cm2). cTPI-CysHHC10 = cRose Bengal = 10 μmol/L, cABDA = 50 μmol/L. (f) Decomposition rates of ABDA under white light irradiation at 378 nm absorption.
The exceptional light absorption characteristics of TPI-CysHHC10 across a broad range of wavelengths within the visible light spectrum have rendered it highly suitable for inducing ROS generation when subjected to white light irradiation, thereby facilitating the effective photodynamic eradication of bacteria. We used Rose Bengal (RB) as a reference and ABDA as an indicator to evaluate the singlet oxygen-generating capacity of TPI-CysHHC1038. The 1O2 generation was judged by the decrease of ABDA absorption at 378 nm. In Fig. 1d–f, in the presence of a combined solution of ABDA and TPI-CysHHC10, the absorption intensity of ABDA at 378 nm exhibited a gradual reduction with increasing illumination duration. Notably, the observed efficiency of TPI-CysHHC10 in this context surpassed that of RB.
3.2. In vitro antimicrobial activity
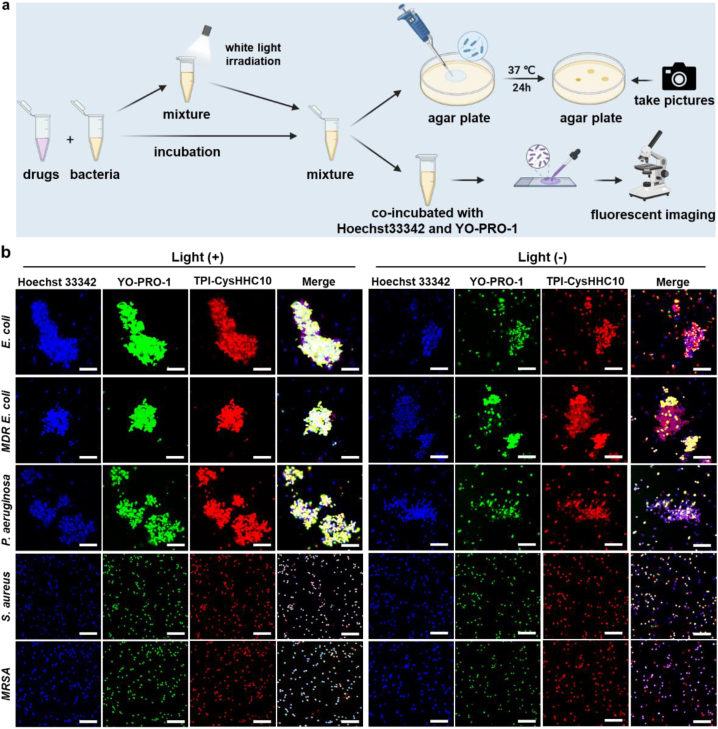
To explore the influence of AMPs on the antibacterial activity of fluorophores, we used the traditional plate counting method to evaluate the bactericidal activity of TPI-CysHHC10 and compared it with antibiotics vancomycin and polymyxin B. Escherichia coli (E. coli), multidrug-resistant Escherichia coli (MDR E. coli), Acinetobacter baumannii (A. baumannii), Klebsiella pneumoniae (K. pneumoniae), and Pseudomonas aeruginosa (P. aeruginosa) were chosen as the representative of G– bacteria, Staphylococcus aureus (S. aureus), Streptococcus pneumoniae (S. pneumoniae), and methicillin-resistant Staphylococcus aureus (MRSA) were chosen as the representative of G+. As shown in Fig. 2a, drugs with different concentrations were incubated with bacteria at a density of 5 × 105 colony-forming units per milliliter (CFU/mL), and then divided into light groups and non-light groups. In the non-light group, the mixture of bacteria and drugs was incubated at 37 °C for 2 h. In the light group, the mixture of bacteria and drugs was incubated at 37 °C for 30 min, then exposed to white light (60 mW/cm2) for 15 min, and then incubated for 75 min. Following the incubation period, 100 μL aliquots from each group of the mixture were dispensed onto agar plates and allowed to incubate for a duration of 24 h at 37 °C. The TPI-CysHHC10 groups with or without white light irradiation were stained by Hoechst 33342 and YO-PRO-1, and then placed on a microscope glass slide, and fluorescence images were acquired by confocal laser scanning microscope (CLSM). As shown in Fig. 2b and Supporting Information Fig. S14, for non-light groups, only a small number of bacteria showed green fluorescence (signal of YO-PRO-1, which stained the dead bacteria). On the contrary, for the groups with white light irradiation, most bacteria showed green fluorescence, indicating that the ROS produced under light irradiation played a crucial role in improving the antibacterial activity of TPI-CysHHC10.
Figure 2.
Dead-alive staining assays. (a) Schematic illustration of drugs incubated with bacteria for agar plate experiments, and dead-alive staining assays. (b) CLSM images of E. coli, MDR E. coli, P. aeruginosa, S. aureus, and MRSA after incubation with TPI-CysHHC10 (10 μmol/L) (red fluorescence) and then without or with light irradiation for 15 min. Then co-staining with Hoechst 33342 (blue fluorescence, a nucleic acid dye for all bacteria) and YO-PRO-1 (green fluorescence, a dye for dead bacteria). Light intensity: 60 mW/cm2. Scale bar: 10 μm.
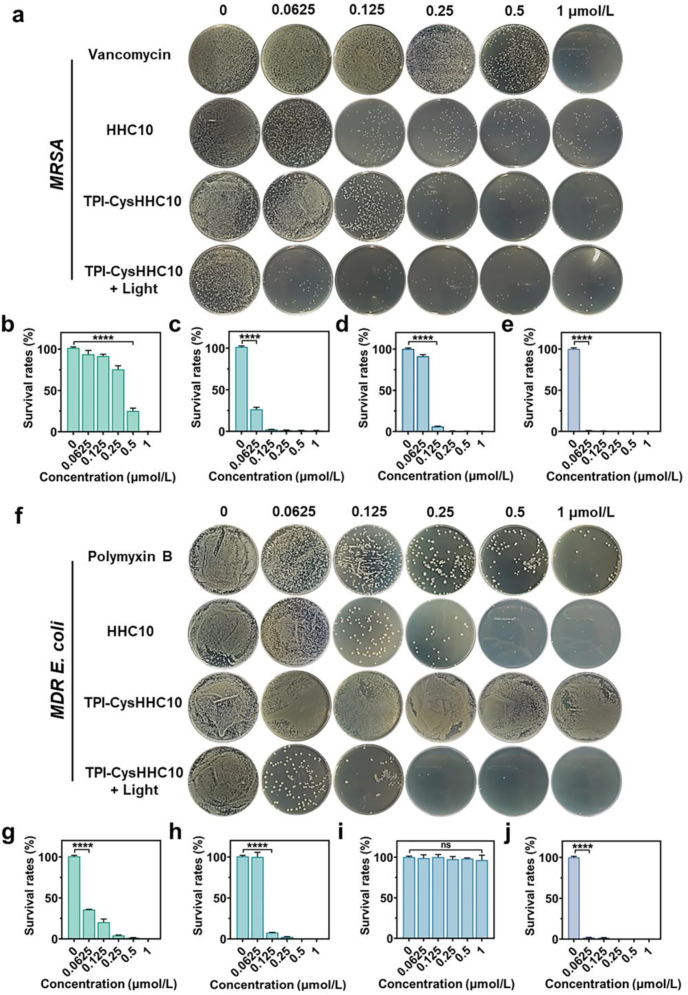
Then, the results of the colony-counting assays also confirm the above results. As shown in Fig. 3 and Supporting Information Figs. S15–S25, the minimum inhibitory concentration (MIC) of TPI-CysHHC10 under white-light irradiation was determined against E. coli, MDR E. coli, P. aeruginosa, S. aureus, MRSA, A. baumannii, K. pneumoniae, and S. pneumoniae, yielding MIC values of 125, 62.5, 125, 62.5, 62.5, 125, 62.5, and 62.5 nmol/L, respectively. Notably, TPI-CysHHC10 exhibited superior antibacterial activity compared to conventional antibiotics such as vancomycin or polymyxin B against these bacterial strains. This observation was further supported by the growth curves depicted in Supporting Information Figs. S26–S30.
Figure 3.
Agar plate experiments. (a) Plate photographs of MRSA LB plates treated with vancomycin, HHC10, TPI-CysHHC10, and TPI-CysHHC10 + Light at different concentrations. (b–e) Quantized survival rates of MRSA via colony counting on LB agar plates treated with vancomycin (b), HHC10 (c), TPI-CysHHC10 (d), and TPI-CysHHC10 + Light (e). (f) Plate photographs of MDR E. coli LB plates treated with vancomycin, HHC10, TPI-CysHHC10, and TPI-CysHHC10 + Light at different concentrations. (g–j) Quantized survival rates of MDR E. coli via colony counting on LB agar plates treated with polymyxin B (g), HHC10 (h), TPI-CysHHC10 (i), and TPI-CysHHC10 + Light (j). Data are mean SD (n = 3). ∗∗∗∗P < 0.0001, ns: no significance. Light irradiation: 60 mW/cm2, 15 min.
However, it should be noted that the antibacterial activity of TPI-CysHHC10 in the absence of light irradiation was found to be comparable to or even lower than that of AMPs alone. This discrepancy suggests that the incorporation of the fluorophore in TPI-CysHHC10 may influence the underlying mechanism of action of AMPs, thereby affecting its antibacterial efficacy. The antimicrobial efficacy of AMPs can be enhanced through their covalent linkage with AIE fluorophore, followed by the induction of a photodynamic effect using white light irradiation. In this study, the synthesized compound TPI-CysHHC10 demonstrated remarkable bactericidal activity at remarkably low concentrations against a wide range of bacteria, including both G– and G+ strains. Notably, even drug-resistant bacteria were susceptible to the photodynamic killing effect mediated by TPI-CysHHC10. This shows that the utilization of a combination of fluorophore-conjugated photosensitizers (PSs) and antimicrobial peptides (AMPs) is mutually advantageous. G– bacteria possess a thicker lipid outer membrane compared with G+ bacteria39,40. Nevertheless, the lifetime of ROS is very short41, therefore, small molecular PSs cannot cause obvious damage to the bilayer membrane structure of G– bacteria only by classical adsorption. However, through the combination of AMPs and PSs, PSs could bind closely to the surface of bacteria with the assistance of antimicrobial peptides. Subsequently, upon light irradiation, ROS generated by the PSs can effectively destroy the outer membrane of bacteria in the range of their lifespan. Therefore, TPI-CysHHC10 had a broad-spectrum bactericidal effect on G+ and G– bacteria.
3.3. Antibacterial mechanism
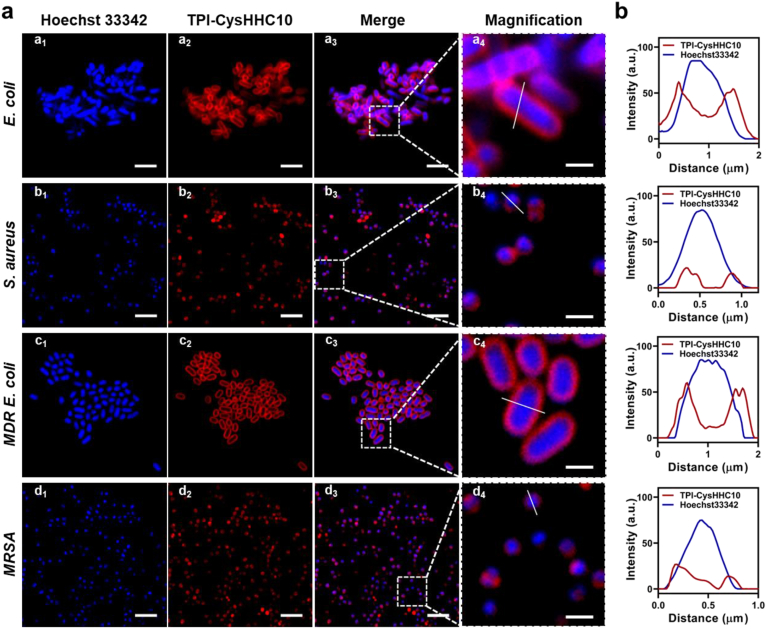
To explore the antibacterial mechanism of TPI-CysHHC10, we conducted bacterial co-localization experiments and SEM experiments. We used TPI-CysHHC10 to stain the bacteria and co-incubated them with Hoechst 33342. As shown in Fig. 4a, the bacteria after co-incubation with TPI-CysHHC10 and nucleic acid dye could be observed under CLSM and all the bacterial cells showed blue fluorescence inside, which indicated that the nucleic acids inside the bacteria are successfully stained and could be used to determine the location of the bacteria. In contrast, the red fluorescence (TPI-CysHHC10) appears only on the bacterial wall, which indicated that TPI-CysHHC10 does not enter the interior of the bacteria, but only binds to the bacterial surface to exert its antibacterial effect, suggesting that TPI-CysHHC10 acted as a bactericide through antibacterial peptides with ROS. The fluorescence intensity profiles confirmed that the sample interacts with the cell membrane of bacteria, as indicated by the anticoincidence of the blue and red fluorescence (Fig. 4b).
Figure 4.
Bacteria staining assays. (a) CLSM images of E. coli, S. aureus, MDR E. coli, and MRSA after incubation with 10 μmol/L of TPI-CysHHC10 and 10 μg/mL Hoechst 33342 for 10 min. The red and blue channels were merged and magnified to check the colocalization of the two channels. (b) Corresponding intensity profiles of (a4–d4). Scale bars: 4 μm (a1–d1, a2–d2, and a3–d3), 1 μm (a4–d4).
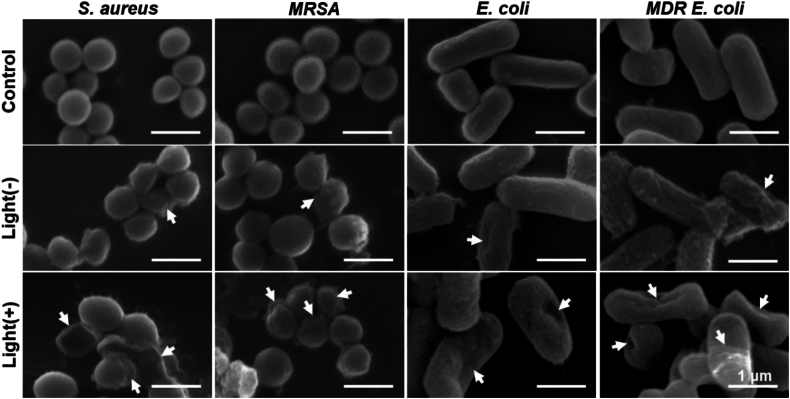
To validate the membrane disruption effect of TPI-CysHHC10, we further performed SEM experiments. E. coli and S. aureus were selected as representatives of G– and G+ bacteria, respectively. As shown in Fig. 5, the surfaces of the untreated bacteria appeared intact, smooth, and regular. However, after treatment with TPI-CysHHC10 followed by a 2-h dark incubation, the majority of the bacteria displayed rough surfaces, with some exhibiting wrinkles and depressions. Subsequent treatment with TPI-CysHHC10 followed by light irradiation resulted in surface depressions in most bacteria, and some bacteria were completely denatured, leading to the observation of only fragmented bacterial remnants under electron microscopy. These results indicated that TPI-CysHHC10 tends to aggregate on the membrane of bacteria to cause the flowing out of inner nucleic acids or proteins by increasing membrane permeability or disrupting membrane structure through the combined action of ROS and AMPs.
Figure 5.
SEM experiments. SEM images of S. aureus, MRSA (Gram-positive bacteria), and E. coli and MDR E. coli (Gram-negative bacteria) with different treatments (PBS, TPI-CysHHC10, and TPI-CysHHC10 + Light). The arrows indicated the wrinkles and depressions of the bacterial membrane. Light irradiation: 60 mW/cm2, 15 min. Scale bars: 1 μm.
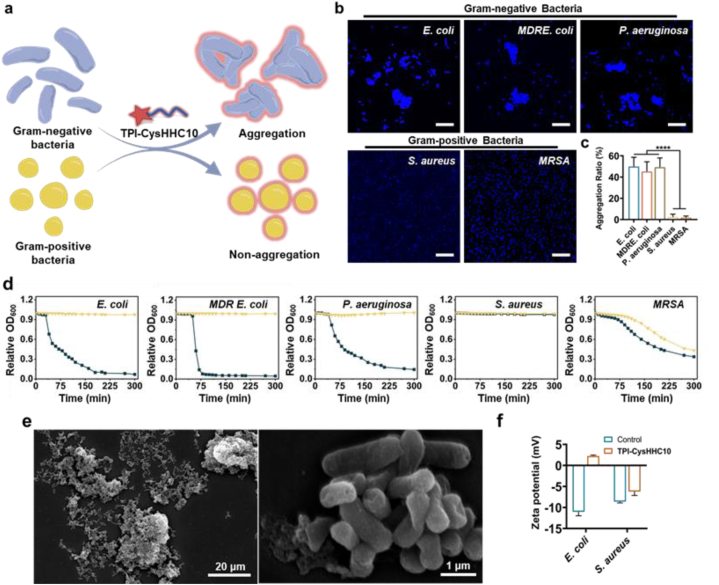
In addition, an interesting phenomenon was found during the experiment that TPI-CysHHC10 exhibits distinct modes of action against G– and G+ bacteria. In the presence of TPI-CysHHC10, aggregations of G– bacteria were observed, while not observed for G+ bacteria (Fig. 6a). This phenomenon was confirmed through bacterial staining and CLSM imaging, as depicted in Fig. 6b. The images showed bacterial aggregation in E. coli, P. aeruginosa, and MDR E. coli, whereas no aggregation was observed in S. aureus and MRSA. The bacterial aggregation degree was quantified by determining the proportion of aggregations larger than the size of four bacteria. The results revealed that more than 40% of G– bacteria formed aggregations within 30 min of TPI-CysHHC10 treatment, whereas G+ bacteria did not show such aggregation (Fig. 6c). Additionally, the optical density at 600 nm (OD600) of the supernatant from various bacterial suspensions treated with TPI-CysHHC10 was measured using the turbidimetric method. As depicted in Fig. 6d, the OD600 of the supernatant from G– bacteria (E. coli, P. aeruginosa, and MDR E. coli) decreased over time upon treatment with TPI-CysHHC10, and the formation of flocculent precipitation was observed. In contrast, the OD600 of the control group and the groups treated with HHC10 or TPI-PA did not exhibit significant changes (Supporting Information Fig. S31). This indicates that TPI-CysHHC10 induced the formation of bacterial aggregations, leading to enhanced sedimentation. SEM results further confirmed the formation of bacterial aggregations (Fig. 6e). However, both the TPI-CysHHC10 treatment group and the control group of MRSA showed a decrease in OD600, suggesting that the sedimentation observed in MRSA was likely due to inherent bacterial characteristics rather than the action of TPI-CysHHC10. However, S. aureus did not precipitate, indicating that TPI-CysHHC10 could not induce aggregations in G+ bacteria.
Figure 6.
Bacteria aggregation. (a) Illustration of G– bacterial aggregation induced by TPI-CysHHC10. (b) CLSM images of E. coli, MDR E. coli, P. aeruginosa, S. aureus, and MRSA after incubation with 10 μmol/L of TPI-CysHHC10 and stained by 10 μg/mL Hoechst 33342 for 10 min. Scale bar: 20 μm. (c) The aggregation ratio of bacteria treated by TPI-CysHHC10. Error bars: mean SD (n = 20). ∗∗∗∗P < 0.0001. (d) Aggregation as measured by turbidimetry for E. coli, MDR E. coli, P. aeruginosa, S. aureus, and MRSA in the absence or presence of TPI-CysHHC10 (20 μmol/L). (e) SEM images of E. coli aggregations treated with TPI-CysHHC10 (10 μmol/L). Scale bar: 20 μm (left); 1 μm (right). (f) Zeta potential of E. coli and S. aureus treated with 10 μmol/L of TPI-CysHHC10.
To explore the difference, E. coli, and S. aureus were selected as the representative G– bacteria and G+ bacteria, respectively, to assess the change in bacterial zeta potential before and after TPI-CysHHC10 treatment (Fig. 6f). The results showed that zeta potential of E. coli and S. aureus increased. However, the distinctive feature was observed in E. coli, where the zeta potential underwent a reversal from negative to positive after TPI-CysHHC10 treatment. We hypothesize that this potential reversal is responsible for the aggregation of G– bacteria. Following the potential reversal, the bacterial surface became positively charged, generating electrostatic attraction with other bacteria carrying negative potentials, thereby promoting aggregation. Once bacterial aggregations form, TPI-CysHHC10 becomes embedded within them, facilitating the effective antibacterial action of short-lived reactive oxygen species within their range. This mechanism provides effective assistance against G– bacteria with their resilient outer membranes, contributing to the unique advantage of TPI-CysHHC10 over other photosensitizers in targeting G– bacteria.
3.4. Anti-biofilm
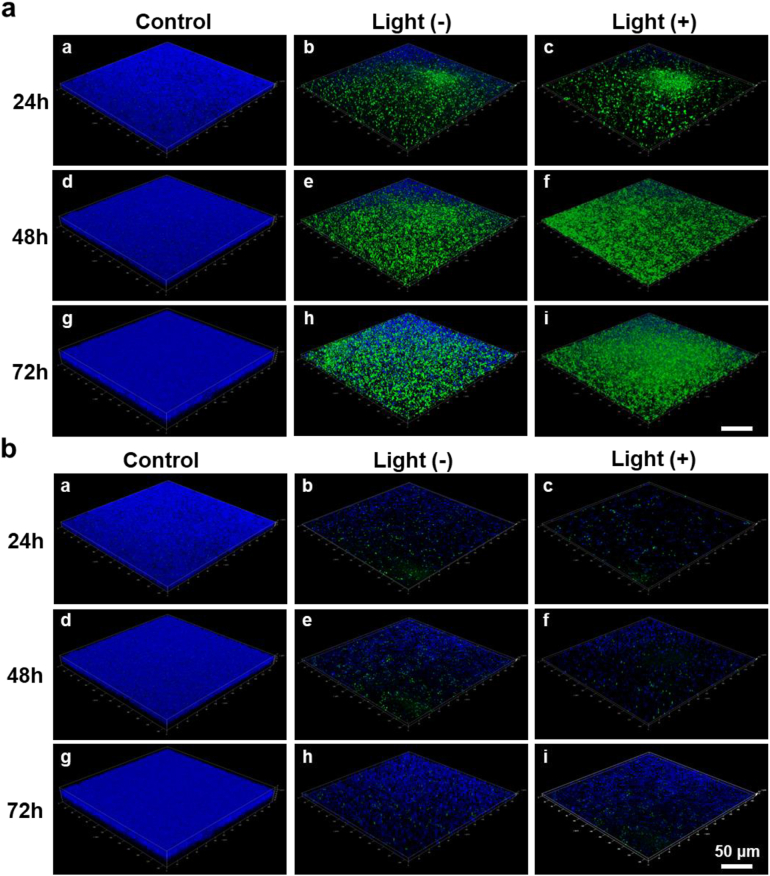
Biofilm infections are usually associated with chronic infections and are rarely cured42, 43, 44. Biofilms exist in structured microbial communities, which typically contain a variety of microorganisms embedded in protective polymers that hinder the penetration of antimicrobial agents45, 46, 47. To investigate the impact of TPI-CysHHC10 on biofilms formation and eradication of mature biofilms, we utilized CLSM to visualize the biofilms at different time points (24, 48, 72 h). The biofilms were imaged in a layer-by-layer manner using stack scanning to generate three-dimensional (3D) images. Fig. 7 showed a blue fluorescent biofilm in the control group, indicating the formation of a biofilm filled with living bacteria. After incubation with TPI-CysHHC10, only a few dead bacteria were observed in the CLSM images, indicated by several green fluorescence markers. This confirms the effectiveness of TPI-CysHHC10 in preventing the formation of MRSA biofilms. In addition to the practical importance of inhibiting biofilm growth for the application of antibiotics, the scavenging effect of mature biofilms is also important for the treatment of bacterial infections. Hence, we also investigated the ability of TPI-CysHHC10 to scavenge the mature MRSA biofilms. Biofilms at different stages of maturation (24, 48, 72 h) were incubated with TPI-CysHHC10 for 1 h, followed by a 15-min light illumination. The biofilms were then with Hoechst 33342 and YO-PRO-1 and examined using CLSM. Interestingly, the CLSM images revealed that the bacterial biofilms were partially disrupted even without light treatment. However, after 15 min of illumination, more pronounced destruction of the biofilms was observed, accompanied by scattered green fluorescence (Fig. 7).
Figure 7.
Anti-biofilm assays. (a) CLSM 3D imaging of MRSA-biofilms co-incubation with PBS or TPI-CysHHC10 (10 μmol/L) at different maturation stages (24, 48, and 72 h). (b) CLSM 3D imaging of MRSA-biofilm co-incubation with PBS or TPI-CysHHC10 (10 μmol/L) for 24, 48, and 72 h. The samples were co-staining with Hoechst 33342 (blue fluorescence) and YO-PRO-1 (green fluorescence) before CLSM imaging. Light irradiation: 60 mW/cm2, 15 min. Scale bars: 50 μm.
The anti-biofilm activity was further quantified by crystal violet staining (Supporting Information Figs. S32–S34). Compared with the control group, the biofilm biomass of MRSA in the non-light group was 10.7%, 17.3%, and 29.2%, and that in the light group was 8%, 11.7%, and 15.4% after 24, 48, and 72 h of incubation with TPI-CysHHC10, respectively. This also confirms the growth inhibitory effect of TPI-CysHHC10 on biofilms. On the other hand, the biomass of mature biofilms for 24, 48, and 72 h treated with TPI-CysHHC10 in the non-light group was 11.3%, 15.6%, and 30.1%, and that in the light group was 9.2%, 10.5%, and 11.2%, respectively. These results showed that most of the biofilms were destroyed and the bacteria died, indicating that TPI-CysHHC10 was effective in eradicating mature biofilms.
3.5. In vivo antibacterial activity
The in vivo efficacy of TPI-CysHHC10 in terms of its antibacterial properties and wound-healing acceleration was assessed using a mouse model with full-thickness wounds infected by MRSA. The infected wounds, with an 8 mm diameter, were treated with various substances including physiological saline solution, HHC10, TPI-CysHHC10, TPI-CysHHC10+Light, and vancomycin. Photographic images of the wound skin were captured on Days 1, 2, 4, 6, and 8, as shown in Fig. 8b. Throughout the experiment, all groups showed a trend of wound shrinkage. The wound area analysis in Fig. 8c demonstrated a steady decrease in the wound area for each treatment group. By the eighth day, the wounds in all drug treatment groups were almost healed, with the TPI-CysHHC10 + Light group displaying the smallest relative wound area of only 2.2%, the lowest among all groups. The control group and control + Light group exhibited wound areas of 13.1% and 11.4%, respectively. At the same time, the body weight of the mice in each group increased steadily over time (Fig. 8d). To evaluate the presence of residual bacteria in the wounds, the wound tissue from each group on the eighth day was homogenized, diluted, and spread on LB agar plates. Fig. 8e and f and Supporting Information Fig. S35 illustrated that only the control group exhibited bacterial residue in the wound, while no bacterial residue was observed in the wounds of the TPI-CysHHC10 + Light group. To evaluate the healing effect, the skin tissue at the wound site on Day 8 was analyzed by H&E staining (Fig. 8g), which showed that there was still significant skin inflammation in the control group with more macrophage infiltration, in contrast, little inflammation was found in the TPI-CysHHC10 photodynamic treatment group. These results demonstrate that TPI-CysHHC10 is an excellent anti-infective and wound-healing agent for photodynamic therapy of wound infection, without affecting the growth of mice.
Figure 8.
In vivo antibacterial activity. (a) Schematic illustration of the in vivo treatment process. (b) Photographs of the wound during treatment with the different drugs on Days 1, 2, 4, 6, and 8. (c) Relative wound size of the infected area. (d) The body weights of the mice. Error bars: mean ± SD (n = 4). (e) Plate photographs of bacterial plates collected from the wound site on mice with different treatments. (f) Quantification of the bacterial colonies on LB agar plates. Error bars: mean ± SD (n = 3). ∗∗∗∗P < 0.0001, ns: no significance. (g) H&E staining imaging of skin wounds with different treatments on Day 8. Scale bar: 100 μm.
Furthermore, in vitro cytotoxicity test, hemolysis test, and in vivo H&E analysis of the main organic (heart, liver, spleen, lung, kidney) showed excellent biocompatibility of TPI-CysHHC10 and HHC10. The cytotoxicity of TPI-CysHHC10 and HHC10 in vitro was determined by methyl thiazolyl tetrazolium (MTT) assays and HeLa cells. As shown in Supporting Information Figs. S36–S38, the cell viability of HeLa remained above 90% when the concentration of TPI-CysHHC10 was up to 64 μmol/L after co-cultured for 24 h, while the HHC10 also remained above 80%. The hemolysis test results indicated its safety for in vivo therapy (Supporting Information Figs. S39–S40). Merely no hemolytic activity was observed at concentrations below 32 μmol/L of TPI-CysHHC10 and HHC10, and low hemolytic activity was observed at concentrations as high as 64 μmol/L (hemolysis rate: 7.9% and 12.3%). Afterward, in vivo biocompatibility of TPI-CysHHC10 and HHC10 is investigated. The heart, liver, spleen, lung, and kidney of mice in each group on Day 8 of treatment were collected for biopsy section analysis. The results showed that the organs of mice in each group showed normal morphology (Supporting Information Fig. S41), which indicated that TPI-CysHHC10 had no significant toxicity in vivo.
4. Conclusions
In conclusion, we have proposed a novel strategy for the development of antibacterial conjugates by combining PSs with AMPs, resulting in a new class of broad-spectrum antibacterial agents. The excellent fluorescence properties of the fluorophores have enabled the visualizations of the interaction patterns between TPI-CysHHC10 and bacteria. Additionally, the excellent singlet oxygen generation ability of the fluorophores has enhanced the photodynamic antibacterial activity of TPI-CysHHC10. On the other hand, the disruption function of AMPs towards the bacterial outer membrane has played a crucial role in the antibacterial activity of TPI-CysHHC10 against G– bacteria. TPI-CysHHC10 not only exhibits bactericidal effects against G+ bacteria but also aids in the destruction of the outer membrane, an important barrier of G– bacteria. The aggregation-inducing effect of TPI-CysHHC10 on G– bacteria facilitates the effective utilization of ROS generated by the conjugates, which are encapsulated within bacterial aggregates. This localized action of ROS within a small range enhances the photodynamic antibacterial effect of TPI-CysHHC10. While killing planktonic bacteria, TPI-CysHHC10 has also demonstrated promising efficacy in inhibiting the formation of MRSA biofilms and destroying mature MRSA biofilms. In the treatment of MRSA-infected wounds, TPI-CysHHC10 has also exhibited excellent photodynamic antibacterial effects and favorable biocompatibility. The coupling of antimicrobial peptides with membrane-disrupting properties to photosensitizers represents a novel and effective strategy for the treatment of bacterial infections. This multifunctional antimicrobial agent holds significant potential in addressing the challenges posed by bacterial biofilm-associated infections and drug-resistant bacteria. Further research and development of this agent could lead to substantial advancements in the field of bacterial infection treatment.
Author contributions
Duoyang Fan: Conceptualization, Formal analysis, Data curation, Investigation, Writing-original draft. Xiaohui Liu: Formal analysis, Investigation, Data curation, Validation. Yueming Ren: Formal analysis, Data curation. Ziheng Luo: Formal analysis. Yanbing Li: Formal analysis. Jie Dong: Writing-review & editing. Seraphine V. Wegner: Writing-review & editing. Fei Chen: Conceptualization, Supervision, Funding acquisition, Writing-review & editing. Wenbin Zeng: Conceptualization, Supervision, Resources, Project administration, Funding acquisition, Writing-review & editing.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
The authors gratefully acknowledge the financial support from the National Natural Science Foundation of China (grant Nos. 82272067, 81971678, 22107123, and M-0696) Natural Science Foundation of Hunan Province (grant Nos. 2022JJ80052, 2022JJ40656, 20231120077, China), Scientific Research Fund of Hunan Provincial Education Department (22B0009, China), and the Central South University Innovation-Driven Research Program (2023CXQD004, China).
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.12.016.
Contributor Information
Fei Chen, Email: fechen@csu.edu.cn.
Wenbin Zeng, Email: wbzeng@hotmail.com.
Appendix A. Supporting information
The following is the Supporting data to this article.
References
- 1.Looke D.F., Gottlieb T., Jones C.A. The global challenges of infectious diseases. Med J Aust. 2015;202:225–227. doi: 10.5694/mja15.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson D.J., Jenkins T.C., Evans S.R., Harris A.D., Weinstein R.A., Tamma P.D., et al. The role of stewardship in addressing antibacterial resistance: stewardship and infection control committee of the antibacterial resistance leadership group. Clin Infect Dis. 2017;64:S36–S40. doi: 10.1093/cid/ciw830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catalano A., Iacopetta D., Ceramella J., Scumaci D., Giuzio F., Saturnino C., et al. Multidrug resistance (MDR): a widespread phenomenon in pharmacological therapies. Molecules. 2022;27:616. doi: 10.3390/molecules27030616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huh A.J., Kwon Y.J. "Nanoantibiotics": a new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J Control Release. 2011;156:128–145. doi: 10.1016/j.jconrel.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Levy S.B., Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 6.Pelgrift R.Y., Friedman A.J. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv Drug Deliv Rev. 2013;65:1803–1815. doi: 10.1016/j.addr.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q., Lv Y., Pang J., Li X., Lu X., Wang X., et al. In vitro and in vivo activity of d-serine in combination with β-lactam antibiotics against methicillin-resistant Staphylococcus aureus. Acta Pharm Sin B. 2019;9:496–504. doi: 10.1016/j.apsb.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Li C., Wu Q., Wen H., Sun T., Xie Z. A cationic BODIPY photosensitizer decorated with quaternary ammonium for high-efficiency photodynamic inhibition of bacterial growth. J Mater Chem B. 2022;10:4967–4973. doi: 10.1039/d2tb00778a. [DOI] [PubMed] [Google Scholar]
- 9.Wang W., Lin L., Du Y., Song Y., Peng X., Chen X., et al. Assessing the viability of transplanted gut microbiota by sequential tagging with d-amino acid-based metabolic probes. Nat Commun. 2019;10:1317. doi: 10.1038/s41467-019-09267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Y., Zheng W., Tran K., Kamilar E., Bariwal J., Ma H., et al. Hydrophilic nanoparticles that kill bacteria while sparing mammalian cells reveal the antibiotic role of nanostructures. Nat Commun. 2022;13:197. doi: 10.1038/s41467-021-27193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rai M.K., Deshmukh S.D., Ingle A.P., Gade A.K. Silver nanoparticles: the powerful nanoweapon against multidrug-resistant bacteria. J Appl Microbiol. 2012;112:841–852. doi: 10.1111/j.1365-2672.2012.05253.x. [DOI] [PubMed] [Google Scholar]
- 12.Deo S., Turton K.L., Kainth T., Kumar A., Wieden H.J. Strategies for improving antimicrobial peptide production. Biotechnol Adv. 2022;59 doi: 10.1016/j.biotechadv.2022.107968. [DOI] [PubMed] [Google Scholar]
- 13.Gan B.H., Gaynord J., Rowe S.M., Deingruber T., Spring D.R. The multifaceted nature of antimicrobial peptides: current synthetic chemistry approaches and future directions. Chem Soc Rev. 2021;50:7820–7880. doi: 10.1039/d0cs00729c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan D., Liu X., Ren Y., Bai S., Li Y., Luo Z., et al. Functional insights to the development of bioactive material for combating bacterial infections. Front Bioeng Biotechnol. 2023;11 doi: 10.3389/fbioe.2023.1186637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Splith K., Neundorf I. Antimicrobial peptides with cell-penetrating peptide properties and vice versa. Eur Biophys J. 2011;40:387–397. doi: 10.1007/s00249-011-0682-7. [DOI] [PubMed] [Google Scholar]
- 16.Khoerunnisa, Mazrad Z.A.I., In I., Park S.Y. pH-switchable bacteria detection using zwitterionic fluorescent polymer. Biosens Bioelectron. 2017;90:394–402. doi: 10.1016/j.bios.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Sun J., Ma X., Li R., Lin M., Shu L., Chen X. Antimicrobial nanostructured assemblies with extremely low toxicity and potent activity to eradicate Staphylococcus aureus biofilms. Small. 2022;19 doi: 10.1002/smll.202204039. [DOI] [PubMed] [Google Scholar]
- 18.Zhang T., An W., Sun J., Duan F., Shao Z., Zhang F., et al. Terminal lysozyme conjugation to a cationic polymer enhances antimicrobial activity and overcomes antimicrobial resistance. Nano Lett. 2022;22:8294–8303. doi: 10.1021/acs.nanolett.2c03160. [DOI] [PubMed] [Google Scholar]
- 19.Otvos L., Jr. Antibacterial peptides isolated from insects. J Pept Sci. 2000;6:597. doi: 10.1002/1099-1387(200010)6:10<497::AID-PSC277>3.0.CO;2-W. 11. [DOI] [PubMed] [Google Scholar]
- 20.Li W., Separovic F., O'Brien-Simpson N.M., Wade J.D. Chemically modified and conjugated antimicrobial peptides against superbugs. Chem Soc Rev. 2021;50:4932–4973. doi: 10.1039/d0cs01026j. [DOI] [PubMed] [Google Scholar]
- 21.Jia Q., Song Q., Li P., Huang W. Rejuvenated photodynamic therapy for bacterial infections. Adv Healthcare Mater. 2019;8 doi: 10.1002/adhm.201900608. [DOI] [PubMed] [Google Scholar]
- 22.Cherkasov A., Hilpert K., Jenssen H., Fjell C.D., Waldbrook M., Mullaly S.C., et al. Use of artificial intelligence in the design of small peptide antibiotics effective against a broad spectrum of highly antibiotic-resistant superbugs. ACS Chem Biol. 2009;4:65–74. doi: 10.1021/cb800240j. [DOI] [PubMed] [Google Scholar]
- 23.Fjell C.D., Jenssen H., Hilpert K., Cheung W.A., Panté N., Hancock R.E.W., et al. Identification of novel antibacterial peptides by chemoinformatics and machine learning. J Med Chem. 2009;52:2006–2015. doi: 10.1021/jm8015365. [DOI] [PubMed] [Google Scholar]
- 24.Maria-Neto S., de Almeida K.C., Macedo M.L., Franco O.L. Understanding bacterial resistance to antimicrobial peptides: from the surface to deep inside. Biochim Biophys Acta. 2015;1848:3078–3088. doi: 10.1016/j.bbamem.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Yoon J. Turning an FDA-approved therapeutic into an AIEgen for imaging live bacteria and for bacterial detection. Aggregate. 2021;2:e47. [Google Scholar]
- 26.Sun J., He X. AIE-based drug/gene delivery system: evolution from fluorescence monitoring alone to augmented therapeutics. Aggregate. 2022;3:e282. [Google Scholar]
- 27.Tian S., Bai H., Li S., Xiao Y., Cui X., Li X., et al. Water-soluble organic nanoparticles with programable intermolecular charge transfer for NIR-II photothermal anti-bacterial therapy. Angew Chem Int Ed. 2021;60:11758–11762. doi: 10.1002/anie.202101406. [DOI] [PubMed] [Google Scholar]
- 28.Li X., Lee S., Yoon J. Supramolecular photosensitizers rejuvenate photodynamic therapy. Chem Soc Rev. 2018;47:1174–1188. doi: 10.1039/c7cs00594f. [DOI] [PubMed] [Google Scholar]
- 29.Li T., Xu H., Xu Y., Yu T., Tang J., Li K., et al. Efficient delivery of chlorin e6 by polyglycerol-coated iron oxide nanoparticles with conjugated doxorubicin for enhanced photodynamic therapy of melanoma. Mol Pharm. 2021;18:3601–3615. doi: 10.1021/acs.molpharmaceut.1c00510. [DOI] [PubMed] [Google Scholar]
- 30.Lin L., Chi J., Yan Y., Luo R., Feng X., Zheng Y., et al. Membrane-disruptive peptides/peptidomimetics-based therapeutics: promising systems to combat bacteria and cancer in the drug-resistant era. Acta Pharm Sin B. 2021;11:2609–2644. doi: 10.1016/j.apsb.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y., Zhao D., Wang G., Wang Y., Cao L., Sun J., et al. Recent progress of hypoxia-modulated multifunctional nanomedicines to enhance photodynamic therapy: opportunities, challenges, and future development. Acta Pharm Sin B. 2020;10:1382–1396. doi: 10.1016/j.apsb.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng G., Yuan Y., Fang H., Zhang R., Xing B., Zhang G., et al. A light-up probe with aggregation-induced emission characteristics (AIE) for selective imaging, naked-eye detection and photodynamic killing of Gram-positive bacteria. Chem Commun. 2015;51:12490–12493. doi: 10.1039/c5cc03807c. [DOI] [PubMed] [Google Scholar]
- 33.Bao P., Li C., Ou H., Ji S., Chen Y., Gao J., et al. A peptide-based aggregation-induced emission bioprobe for selective detection and photodynamic killing of Gram-negative bacteria. Biomater Sci. 2021;9:437–442. doi: 10.1039/d0bm01330g. [DOI] [PubMed] [Google Scholar]
- 34.Lei X., Qiu L., Lan M., Du X., Zhou S., Cui P., et al. Antibacterial photodynamic peptides for staphylococcal skin infection. Biomater Sci. 2020;8:6695–6702. doi: 10.1039/d0bm01467b. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y., Zhou W., Xiao L., Lan Q., Li M., Liu Y., et al. Bacitracin-engineered BSA/ICG nanocomplex with enhanced photothermal and photodynamic antibacterial activity. ACS Omega. 2022;7:33821–33829. doi: 10.1021/acsomega.2c02470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu F., Xu S., Liu B. Photosensitizers with aggregation-induced emission: materials and biomedical applications. Adv Mater. 2018;30 doi: 10.1002/adma.201801350. [DOI] [PubMed] [Google Scholar]
- 37.Wang C., Wang J., Xue K., Xiao M., Sun Z., Zhu C. A receptor-targeting AIE photosensitizer for selective bacterial killing and real-time monitoring of photodynamic therapy outcome. Chem Commun. 2022;58:7058–7061. doi: 10.1039/d2cc02230c. [DOI] [PubMed] [Google Scholar]
- 38.Lambert C.R., Kochevar I.E. Does rose bengal triplet generate superoxide anion? J Am Chem Soc. 1996;118:3297–3298. [Google Scholar]
- 39.Domalaon R., Idowu T., Zhanel George G., Schweizer F. Antibiotic hybrids: the next generation of agents and adjuvants against Gram-negative pathogens? Clin Microbiol Rev. 2018;31 doi: 10.1128/CMR.00077-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Telhig S., Ben Said L., Zirah S., Fliss I., Rebuffat S. Bacteriocins to thwart bacterial resistance in Gram negative bacteria. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.586433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharman W.M., Allen C.M., van Lier J.E. In: Methods in enzymology. Lester P., Helmut S., editors. Academic Press; Amsterdam: 2000. Role of activated oxygen species in photodynamic therapy; pp. 376–400. [DOI] [PubMed] [Google Scholar]
- 42.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 43.Lakemeyer M., Zhao W., Mandl F.A., Hammann P., Sieber S.A. Thinking outside the box—novel antibacterials to tackle the resistance crisis. Angew Chem Int Ed. 2018;57:14440–14475. doi: 10.1002/anie.201804971. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y., Li Y., Shi L. Controlled drug delivery systems in eradicating bacterial biofilm-associated infections. J. Control Release. 2021;329:1102–1116. doi: 10.1016/j.jconrel.2020.10.038. [DOI] [PubMed] [Google Scholar]
- 45.Lencova S., Svarcova V., Stiborova H., Demnerova K., Jencova V., Hozdova K., et al. Bacterial biofilms on polyamide nanofibers: factors influencing biofilm formation and evaluation. ACS Appl Mater Interfaces. 2021;13:2277–2288. doi: 10.1021/acsami.0c19016. [DOI] [PubMed] [Google Scholar]
- 46.Darvishi S., Tavakoli S., Kharaziha M., Girault H.H., Kaminski C.F., Mela I. Advances in the sensing and treatment of wound biofilms. Angew Chem Int Ed. 2022;61 doi: 10.1002/anie.202112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sauer K., Stoodley P., Goeres D.M., Hall-Stoodley L., Burmolle M., Stewart P.S., et al. The biofilm life cycle: expanding the conceptual model of biofilm formation. Nat Rev Microbiol. 2022;20:608–620. doi: 10.1038/s41579-022-00767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.