Abstract
Small molecule drugs play a pivotal role in the arsenal of anticancer pharmacological agents. Nonetheless, their small size poses a challenge when directly visualizing their localization, distribution, mechanism of action (MOA), and target engagement at the subcellular level in real time. We propose a strategy for developing triple-functioning drug beacons that seamlessly integrate therapeutically relevant bioactivity, precise subcellular localization, and direct visualization capabilities within a single molecular entity. As a proof of concept, we have meticulously designed and constructed a boronic acid fluorescence drug beacon using coumarin–hemicyanine (CHB). Our CHB design includes three pivotal features: a boronic acid moiety that binds both adenosine triphosphate (ATP) and adenosine diphosphate (ADP), thus depleting their levels and disrupting the energy supply within mitochondria; a positively charged component that targets the drug beacon to mitochondria; and a sizeable conjugated luminophore that emits fluorescence, facilitating the application of structured illumination microscopy (SIM). Our study indicates the exceptional responsiveness of our proof-of-concept drug beacon to ADP and ATP, its efficacy in inhibiting tumor growth, and its ability to facilitate the tracking of ADP and ATP distribution around the mitochondrial cristae. Furthermore, our investigation reveals that the micro-dynamics of CHB induce mitochondrial dysfunction by causing damage to the mitochondrial cristae and mitochondrial DNA. Altogether, our findings highlight the potential of SIM in conjunction with visual drug design as a potent tool for monitoring the in situ MOA of small molecule anticancer compounds. This approach represents a crucial advancement in addressing a current challenge within the field of small molecule drug discovery and validation.
Key words: Mitochondrial, Drug beacon, Sub-cellular, Small molecule, Energy supply, Anti-tumor
Graphical abstract
Drug beacon CHB inhibits energy production by binding to ATP/ADP in mitochondria, disrupting the structure and function of mitochondria, exerting anti-tumor effects.
1. Introduction
Cancer drug discovery and development represent monumental and resource-intensive endeavors1, 2, 3, 4. However, given the projected increase in the global cancer burden over the next decade, the urgency for new cancer drugs remains paramount. While recent years have witnessed a dramatic surge in biologics development, such as antibody- and cell-based therapies for cancer, small molecule drugs, characterized by molecular weights typically below 900 Da, continue to constitute a significant class of anticancer therapeutics5, 6, 7. Within this context, the development of targeted drugs that exert their effects by engaging a single target has received immense interest within the scientific community. Consequently, a pivotal step in the targeted drug development process involves the direct validation of an on-target mechanism of action (MOA)4,8. Despite the several methods available to investigate the MOA of small molecule drugs, achieving direct in-cell and in vivo visualization remains a formidable challenge.
In recent years, fluorescence imaging has emerged as a viable strategy for tracking intracellular drug activity, visualizing drug dynamics, elucidating off-target activity, assessing MOA, and aiding in disease diagnosis9, 10, 11, 12, 13, 14, 15, 16, 17, 18. Furthermore, fluorescence labeling can be seamlessly integrated with super-resolution microscopic techniques to provide insights at the nanometer scale15,18, 19, 20, 21, 22, 23, 24. For instance, super-resolution fluorescence imaging has revealed novel transport pathways of platinum complexes (Pt2L) within organelles10, time-dependent distribution patterns of dextran within mitochondria and lysosomes12, and the ability of a natural product, magnoflorine, to target ClO– within mitochondria11. Nonetheless, a significant drawback in these efforts is that traditional strategies for drug visualization often necessitate the use of fluorescent protein sensors, which present challenges in terms of intracellular delivery, or drug-fluorophore conjugates, which may unpredictably affect drug efficacy, off-target activity, and MOA25. Furthermore, directly fluorescing drug-like molecules tend to be disfavored in drug development due to their propensity to generate false positive signals in large screening campaigns reliant on fluorescent signal read-outs11,26, 27, 28. Altogether, there is a need for improved strategies for the direct visualization of targeted therapeutics to enhance our capacity to assess drug effects in real time.
In response to these challenges, we have introduced a novel molecular concept known as the drug beacon. Drug beacons are small molecule drug-like compounds designed to include three key components: (1) a pharmacophore critical for bioactivity; (2) a sizeable conjugated luminophore that emits fluorescence, enabling the application of advanced microscopy and imaging techniques, such as structured illumination microscopy (SIM)29, 30, 31; and (3) a subcellular localization anchor that acts as a homing device, directing the drug beacon to a specific organelle. In our proof-of-concept study, we developed a drug beacon designed to disrupt mitochondrial energy metabolism, a well-validated target for anticancer drugs32,33. In particular, we designed a drug beacon that employs a boronic acid-based pharmacophore to scavenge both ATP and ADP, a coumarin- and hemocyanin-based luminophore, and a positively charged group to facilitate mitochondrial localization. We validated that this coumarin–hemocyanin-based boronic acid fluorescence drug beacon (CHB) compound exhibits high-affinity binding to both ATP and ADP, both in vitro and in vivo, and that this binding is pivotal for its potent anticancer activity. Moreover, owing to its fluorescence properties, CHB allowed us to visualize the distribution and function of this drug beacon within mitochondria, resulting in the discovery that ATP and ADP depletion results in damage to mitochondrial membrane potential (MMP) and mitochondrial cristae, subsequently triggering mitochondrial dysfunction (Scheme 1). Therefore, along with confirming the feasibility of the drug beacon concept for tracking drug micro-dynamics, we provide evidence that targeting the structural integrity of mitochondrial cristae may represent a novel avenue for cancer drug discovery and development.
Scheme 1.
The chemical structure of drug beacon CHB and the schematic representation of the anti-tumor effect of CHB blocking mitochondrial energy metabolism. The boronic acid group bonded to the hydroxyl group of ATP/ADP to form a CHB-ATP/ADP complex, which depleted the ATP/ADP pool, resulting in the disorders of mitochondrial H+ homeostasis, mitochondrial depolarization, and damage to the mitochondrial cristae, ultimately, resulting in mitochondrial dysfunction.
2. Materials and methods
2.1. Proof-of-concept design and initial validation of a drug beacon targeting mitochondrial energy metabolism
All materials were procured from commercial sources. CTP and GTP were acquired from MedChemExpress (Monmouth Junction, NJ, USA). Culture bottles and 15-, 50-mL plastic conical centrifuge tubes were obtained from Guangzhou Jet Bio-Filtration Co., Ltd. NMR spectra were recorded using a Bruker Advance 600 MHz spectrometer (Germany), with shifts referenced relative to the internal solvent signals. ESI-MS spectra were recorded on a Thermo Finnigan LCQ DECa XP spectrometer (USA), with the quoted m/z values representing the significant peaks in the isotopic distribution.
2.2. Synthesis and characterization of CHB
Compound Hc (0.15 g, 1 mmol) and 4-(bromomethyl) phenylboronic acid (0.42 g, 2 mmol) were dissolved in acetonitrile (20 mL) and then refluxed overnight. The mixture was purified by gel chromatography with the eluent as MeOH/DCM (1:8) eluent after the removal of the solvent in vacuo, and a pale-yellow solid was obtained after drying (0.12 g, 0.5 mmol, 50.0%). Subsequently, a mixture of the previous product (0.11 g, 0.37 mmol) and 7-(diethylaMino)-2-oxo-2H-chroMene-3-carbaldehyde (0.09 g, 0.37 mmol) was refluxed for 6 h in ethanol (10 mL) (Supporting Information Scheme S1). The crude compound was obtained after the removal of the solvent in vacuo and then purified by gel chromatography with the eluent as MeOH/DCM (1:6). Dark blue solid (CHB) was obtained after drying (0.104 g, 0.20 mmol, 58.0%). 1H NMR (600 MHz, DMSO-d6, δ, ppm) (Supporting Information Fig. S5): δ 8.89 (s, 1 H), 8.38 (d, J = 15.6 Hz, 1 H), 8.10 (s, 2 H), 7.97 (d, J = 15.6 Hz, 1 H), 7.89 (dd, J = 5.4, 3.1 Hz, 1 H), 7.82 (t, J = 7.4 Hz, 3 H), 7.57 (dd, J = 10.0, 6.2 Hz, 3 H), 7.34 (d, J = 7.9 Hz, 2 H), 6.96–6.89 (m, 1 H), 6.69 (s, 1 H), 5.78 (s, 2 H), 3.57 (d, J = 7.0 Hz, 4 H), 1.84 (s, 6 H), 1.18 (t, J = 7.0 Hz, 6 H). 13C NMR (151 MHz, DMSO-d6, Supporting Information Fig. S6): δ 181.98, 172.47, 159.70, 158.29, 154.81, 151.60, 151.23, 143.56, 141.66, 135.63, 135.36, 133.14, 129.59, 129.04, 126.46, 123.56, 115.09, 112.69, 112.02, 110.22, 109.85, 97.08, 52.01, 49.66, 45.43, 26.90, 21.54, and 12.98. HRMS (positive mode, m/z (Supporting Information Fig. S7): Calcd. 521.2606, Found 521.1707 for [M]+.
2.3. Cell culture and staining
Cells were cultured in complete Dulbecco's modified Eagle medium (DMEM) (VivaCell, Shanghai, China) supplemented with 10% fetal bovine serum (FBS; VivaCell) and penicillin–streptomycin (100 units/mL) in a 5% CO2 humidified incubator at 37 °C. The CHB stock solution (5.0 × 10−3 mol/L) was prepared in dimethyl sulfoxide (DMSO) and diluted with the working concentration of DMEM. The solution of the required concentration was added to cells for incubation in a 5% CO2 humidified incubator at 37 °C.
2.4. Cytotoxicity assay
The cytotoxicity assay of CHB was determined using the Cell Counting Kit-8 (CCK-8) Assay (Suzhou, China). HeLa cells were seeded with complete DMEM in 96-well microplates, reaching a cell density of 6000 cells/well, and then cultured in an atmosphere containing 5% CO2 for 24 h at 37 °C. The culture medium was replaced with 100 μL of CHB at different concentrations (0.0, 0.1, 0.5, 1.0, 3.0, 5.0, 10.0, 15.0, 20.0, and 25.0 μmol/L) in a complete medium and incubated for 24 h at 37 °C. Subsequently, cells were incubated with 10 μL of CCK-8 solution for 2 h at 37 °C, and the absorption wavelength was measured using an enzyme labeling apparatus (Synergy HT) at 450 nm.
2.5. OMX 3D-SIM imaging and analysis
Cells were seeded on a 35-mm glass-bottom microwell dish and cultured in complete DMEM supplemented with 10% FBS. After 24 h of incubation, cells were stained with CHB for various durations, washed with phosphate-buffered saline (PBS) (VivaCell) five times, and free DMEM three times. Finally, the cells were cultured in DMEM and imaged using an OMX 3D-SIM microscope (Delta Vision, Inc.) with a 60 × /1.42 numerical aperture oil-immersion objective lens and solid-state lasers. Images were acquired at 512 × 512 resolution using z-stacks with a step size of 0.125 μm, and CHB was excited at 561 nm and emitted at 655–705 nm.
2.6. Flow cytometry analysis
HeLa cells were seeded in 6-well plates at a density of 1.0 × 106 cells/well in complete DMEM at 37 °C for 24 h. The culture medium was then replaced with 1 mL of fresh medium containing CHB at concentrations of 0.0, 0.1, 0.5, 1.0, 5.0, 10.0, and 20.0 μmol/L. After a 1-h treatment with CHB, the cells were washed with PBS twice, digested with trypsin, rewashed with precooled PBS twice, and resuspended in 500 μL of PBS. The samples were then analyzed using flow cytometry (CytoFLEX).
2.7. Cell transfection
For cell transfection, 2500 ng of DNa was combined with 8 μL of Turbofect transfection solution in 250 μL of free DMEM to create the transfection mixtures. After incubating the mixtures at room temperature for 20 min, cells in 35-mm dishes were replaced with 750 μL of complete DMEM, and the mixtures were added to the cells. After 3 h of transfection, the transfection medium was replaced with 1 mL of complete DMEM containing penicillin–streptomycin solution. Cells were incubated at 37 °C and stained with CHB (0.5 μmol/L, 1 h) for a colocalization assay.
2.8. Colocalization assay
To observe the localization of CHB in living cells, imaging was conducted using the OMX 3D-SIM microscope. Cells were cultured in 35-mm glass-bottom culture dishes at 37 °C for 24 h. Subsequently, cells were stained with CHB at different concentrations (0.5 and 1.0 μmol/L) for varying durations (1 and 3 h). The cells were washed twice with PBS and incubated with Mito-tracker Green (100 nmol/L, λex = 488 nm) for 0.5 h to label mitochondria. Finally, the cells were rewashed and imaged using SIM.
2.9. In vitro endocytic pathways
HeLa cells were divided into four groups: those pretreated with chlorpromazine (CPZ, 20.0 μmol/L, 1 h, an inhibitor of endocytosis), those incubated at a lower temperature (4 °C, 1 h, an energy inhibitor), and the control group (37 °C, 1 h). After washing the cells with PBS twice, they were stained with CHB (0.5 μmol/L) for 1 h at 37 °C. Subsequently, the cells were washed with PBS five times and completed DMEM thrice before visualization under the OMX 3D-SIM microscope with 561-nm excitation.
2.10. Seahorse XF test
HeLa cells were seeded at a density of 1.0 × 104 cells/well in DMEM supplemented with 10% FBS in XFe96 cell culture plates (Agilent Technologies, USA). After 24 h of incubation, DMEM was replaced with XF DMEM containing 1.0 mmol/L sodium pyruvate, 10.0 mmol/L glucose, and 2.0 mmol/L l-glutamine at pH 7.4. All cells were treated with 1.0 μmol/L oligomycin A, 1.0 μmol/L FCCP, and 500.0 nmol/L rotenone/antimycin A. The oxygen consumption rates (OCRs) of the cells were assessed using the XF Cell Mito Stress Test Kit (Agilent Technologies, USA), and the Seahorse XF96 analyzer (Agilent Technologies, USA) was used for OCR measurement.
2.11. Tumor model establishment
All animal study procedures were executed according to the protocols approved by Shandong First Medical University Animal Care and Use Committee. Female BALB/c nude Crli mice (4 weeks old) were housed under standard conditions. HeLa cells were suspended in a 50-mL centrifugal tube and subcutaneously injected in a volume of 200 μL into the left rear thigh of the mice at a density of 4.0 × 107 cells/mL. Drug treatment commenced when the tumor volumes reached approximately 150 mm3. Tumor volume was calculated according to Eq. (1):
| Volume (V) = Width × Length × Height × π/6 | (1) |
2.12. In vivo antitumor therapy
When the tumor volume reached 150 mm3, the tumor-bearing mice were randomly divided into five groups (n = 5) and injected in situ with Control, oligomycin A (5.0 μmol/L), CHB (5.0 μmol/L, 1:999, DMSO: PBS), CHB (15.0 μmol/L, 3:997, DMSO: PBS), and CHB (25.0 μmol/L, 5:995, DMSO: PBS) according to a previously published animal experiment scheme1 and our cell viability results. After 1 day, the mice in all five groups received another intratumoral injection with the same drug. Tumor volume and body weights were recorded over the 14-day treatment period. Hematoxylin and eosin staining of the tumor tissues and normal tissues (heart, liver, spleen, lung, and kidney) from each group were also conducted.
2.13. Preparation of CHB/oligomycin A-loaded folate-targeted liposomes
Folate receptor α (FRα) is an appealing target for anticancer drug delivery2. Liposomes were prepared using the film hydration method with the following lipid composition: egg yolk lecithin, cholesterol, DSPE-Mpeg 2000, and FA-PEG-DSPE in a ratio of 50:50:1:1.5. Subsequently, 400 μL of CHB/oligomycin A (0.5 mg/mL) was added. The solvent was then evaporated using a rotary evaporator, forming a dried thin film at the bottom of the flask. The lipid film was rehydrated with 1 mL of phosphate-buffered saline (pH 7.4) and subjected to sonication for 10 min. The hydrated lipid film was transferred to a hydrated dialysis bag and placed in a magnetic stirrer for mixing. The drug-loaded liposomes and non-excluded drugs were separated from each other by preequilibrating in a PBS solution (pH 7.4).
2.14. Statistical analyses
Statistical analysis was performed using Prism 8 (GraphPad). Normality and lognormality tests were conducted to assess the distribution of data. In cases of normal distribution, statistical comparisons were performed using a Student's t-test. For non-normally distributed data, statistical comparisons were made using a Mann–Whitney test. Significance levels were set at n.s. (no significant difference), ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001. Data are presented as mean ± SEM, and the analyses were based on data obtained from three replicates. The corresponding figure legends indicate the statistical significance and sample sizes in all graphs.
3. Results and discussion
3.1. Proof-of-concept design and initial validation of a drug beacon that blocks mitochondrial energy metabolism
In pursuing a robust proof-of-concept drug beacon, our design prioritized the integration of a high-performance fluorophore capable of producing a bright and stable signal for drug beacon detection. Additionally, we sought to incorporate a pharmacophore with high bioactivity within the drug beacon molecule and a homing functionality for effective delivery to a specific organelle. For our initial design, we opted for a coumarin- and hemocyanin-based luminophore (CH) (Scheme 1), given its excellent fluorescence properties under SIM conditions, as supported by previous studies conducted by us20 and others30,34,35. We coupled the CH luminophore with phenylboronate to create the resulting CHB drug beacon. This design featured boronic acid as the pharmacophore, anticipated to bind to the ortho hydroxyl groups of ATP and ADP (Scheme 1), based on previous research findings36,37. Our drug beacon design also incorporated a phenyl ring as a linker, which contributed to a more extensive conjugated system aimed at enhancing fluorescence properties. Finally, the CHB molecule included a positive charge as a mitochondria-targeting feature (Scheme 1)20. The synthesis of the CHB drug beacon molecule and its structural confirmation are described in the Methods section (Supporting Information Figs. S1–S7).
To characterize CHB, we examined its fluorescence properties in a DMSO-PBS solution (2:998, v/v, pH = 7.4). To align with the laser parameters of our SIM device (Supporting Information Table S1), we assessed the fluorescence spectrum of CHB at excitation wavelengths of λex = 405, 488, 561, and 640 nm, revealing strong fluorescence emission from CHB (λex = 561 nm) (Supporting Information Fig. S8). To determine whether CHB binds to ATP and ADP and how this interaction influences its fluorescence properties, we conducted titration studies to monitor changes in the fluorescence intensity of CHB at 673.8 nm upon the addition of ATP or ADP (Fig. 1A and B; Supporting Information Fig. S9). The fluorescence intensity at 673.8 nm increased progressively with rising concentrations of ATP (0.0–10.0 mmol/L) or ADP (0.0–0.5 mmol/L). We calculated the limit of detection (LOD) of CHB for ATP/ADP as 218.7 and 14.0 μmol/L, respectively, according to Eq. (2):
| (2) |
where σ represents the standard deviation of the blank, and k signifies the slope between the fluorescence intensity and ATP/ADP concentration. This indicates that CHB is sufficiently sensitive to bind mitochondrial ATP and ADP. Furthermore, we calculated the binding affinities as shown in Eqs. (3), (4):
| (3) |
| (4) |
where a and A0 indicate the ultraviolet absorptions of CHB at 673.8 nm with and without ATP/ADP (1.0 mmol/L), respectively. Amax represents the ultraviolet absorption of CHB in excess ATP/ADP at 673.8 nm, whereas [ATP/ADP] is the concentration of the ATP or ADP added at A0. Based on the above formula, the association constant Ka-ATP was 2.14 × 104 mol/L and Ka-ADP was 2.08 × 104 mol/L.
Figure 1.
The characterization of the ATP and ADP-binding properties. (A, B) Fluorescence intensity alteration of CHB (10.0 μmol/L) with the gradual addition of ATP (A, 0.0–10.0 mmol/L) and ADP (B, 0.0–0.5 mmol/L) in DMSO-PBS (2:998, v/v, pH = 7.40). λex = 561 nm, λem = 655–705 nm. Slit widths: λex = 2.5 nm, λem = 2.5 nm. (C) Selected comparison of CHB (10.0 μmol/L) between glucose and ATP/ADP (5.0 mmol/L) in DMSO-PBS (2:998, v/v, pH = 7.40). Slit widths: λex = 5 nm, λem = 5 nm. (D–H) Binding modes of ATP-synthase (1BMF.pdb) with substrates ADP/ATP/CHB/CHB-ADP/CHB-ATP. The substrates were represented by the ball and stick models. 1BMF residues were represented by the stick models. The C, N, O, and P atoms were colored in green (substrate)/white (1BMF.pdb), blue, red, and orange, respectively. The green/dark orange/pink dash lines represented the H-bond/electrostatic/hydrophobic interactions. (I) The presentation of CHB depleting ATP/ADP.
To evaluate the selectivity of CHB for ATP/ADP, we investigated CHB's response to various molecular species commonly found in biological systems, including biological diols, NMPs, NDPs, and NTPs. Our observations revealed that the fluorescence intensity of CHB significantly changed in the presence of ATP and ADP; however, it remained unaffected by other analytes (Fig. 1C and Supporting Information Fig. S10). We also demonstrated that the binding of CHB to ATP and ADP remained unaltered even in the presence of high concentrations of carbohydrates or proteins such as bovine serum albumin and human serum albumin (Supporting Information Figs. S11 and S12). It is known that the pH value may affect the binding of CHB with ATP and ADP; thus, we investigated the stability of CHB in various pH solutions (Supporting Information Fig. S13). Our findings indicated that after the addition of ATP or ADP, the fluorescence intensity of CHB (λem = 673.8 nm) remained consistently high in buffers with pH values ranging from 3.0 to 9.0, suggesting that the drug beacon should maintain stable performance under most physiological pH conditions.
We also explored the possibility of CHB directly binding to ATP synthase and inhibiting ATP production. To investigate this, we conducted molecular docking studies involving CHB, ATP, ADP, as well as CHB-ATP and CHB-ADP complexes, with ATP synthase (1BMF.pdb) as the target. Our results indicated that only ATP and ADP formed stable complexes with ATP synthase, as indicated by the calculated binding energies (Fig. 1D–I). This suggests that CHB is unlikely to exert its effects through mechanisms other than binding to and sequestering ATP and ADP.
Altogether, these results suggest that CHB is well-suited for the dual-binding of ATP and ADP under physiological conditions, offering characteristics such as high fluorescence signal intensity, high selectivity, and remarkable stability.
3.2. Tracking the distribution and consumption of ADP and ATP with CHB at the subcellular level
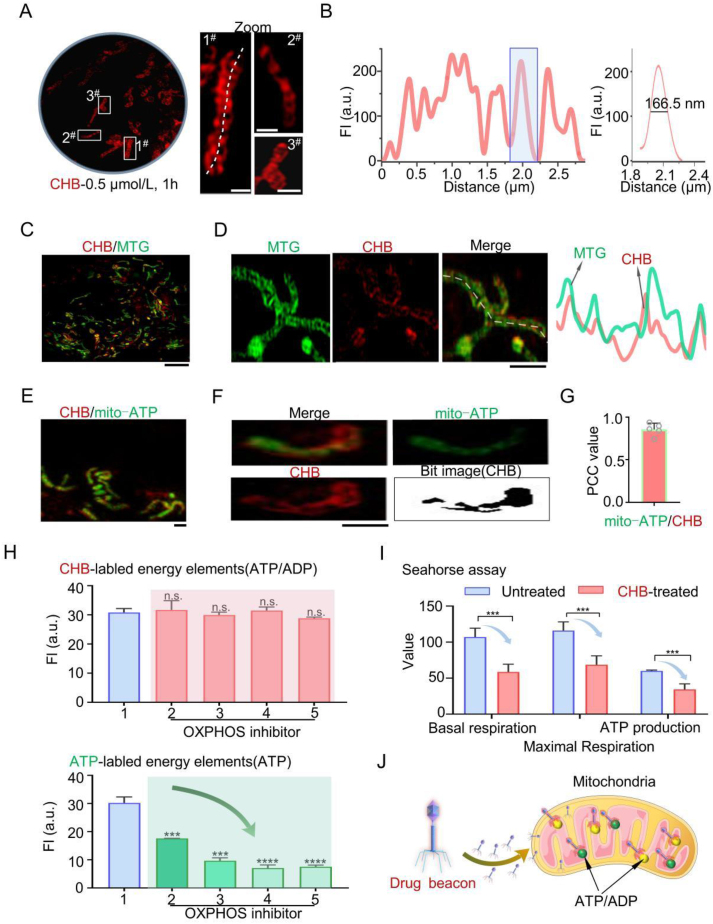
Given our intention for CHB to function as a visualization drug, we evaluated its capacity to visualize its MOA at the intended targets within mitochondria. Initially, we established that CHB entered cells primarily through energy-dependent endocytosis (Supporting Information Figs. S14 and S15) and determined the appropriate tracking concentration to be 0.5 μmol/L CHB based on cytotoxicity assessments (Supporting Information Fig. S16). Next, we employed confocal microscopy and SIM to characterize the subcellular location of CHB in live cells. Conventional confocal microscopy struggled to visualize organelles stained with CHB (Supporting Information Fig. S17). In contrast, SIM successfully revealed the fiber-like morphology of mitochondria stained with CHB (Fig. 3A and B, red color), thus confirming the targeting of CHB within mitochondria, as intended. To further verify mitochondrial localization, HeLa cells were co-stained with a mitochondrial membrane tracker (Mito Tracker Green [MTG], 100.0 nmol/L, 30 min)19 and CHB (0.5 μmol/L, 1 h). The results showed red fiber-shaped structures within the green-labeled mitochondria, with a high Pearson's correlation coefficient (PCC) of 0.89 (Fig. 2C and Supporting Information S18).
Figure 3.
CHB consumes ATP-ADP around cristae, resulting in mitochondrial morphological damage. (A) Mitochondrial cristae damage of HeLa cells under CHB treatment. (B) Quantitative analysis of the damage to mitochondrial cristae without or with CHB treatment. (n = 3 cells, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001) (C) Green/red ratio of HeLa stained with JC-1 probe after CHB-treatment. (D) The related distribution of fluorescence intensity on white solid lines in fluorescence imaging. (E) Damaged mitochondria in the tumor tissues are shown in transmission electron microscopy (upper panel) and the representative images of single mitochondria morphology in each group (below panel). Damaged mitochondrial cristae were observed in the treated groups and the damaged level in the CHB-treated group was higher than that in the oligomycin A-treated group. (F) The cristae length changes of the tumor tissues in all groups. Data is expressed as the mean ± SEM (n = 10 mitochondria). (G) The cristae number changes of the mitochondria in the tumor tissues. Data is expressed as the mean ± SEM (n = 6 mitochondria). (H) Schematic diagram illustrating CHB binding of ATP and ADP to damage the mitochondria. CHB channel: λex = 561 nm, λem = 655–705 nm; MTG channel: λex = 488 nm, λem = 500–550 nm.
Figure 2.
CHB tracks ADP/ATP distribution and the consumption of ATP in the mitochondria. (A) SIM images of HeLa cells stained with CHB (0.5 μmol/L) for 1 h. Scale bar = 1 μm. (B) Distribution of the fluorescence intensity on the solid lines across mitochondria cristae in untreated HeLa cells stained with CHB (0.5 μmol/L) for 1 h. (C) SIM images of HeLa cells stained with CHB (0.5 μmol/L, 1 h) and MTG (100.0 nmol/L). Scale bar = 5 μm. (D) Fluorescence intensity profiles of mitochondrial cristae (right panel) co-stained with CHB and MTG (left panel). (E–G) Mitochondria co-stained with CHB (0.5 μmol/L, 1 h) and the commercial mitochondria ATP probe pCMV-Mito-AT1.03 (mito-ATP) in HeLa cells and its PCC value. Scale bar = 1 μm. (H) Fluorescence intensity changes of CHB and pCMV-Mito-AT1.03 in HeLa cells treated with OXPHOS inhibitors. 1. Untreated; 2. Oligomycin A; 3. Antimycin A; 4. Carboxin; 5. Rotenone. Data are expressed as the mean ± SEM (n = 3 cells, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001). (I) Mito Stress Test Kit results of Seahorse cell energy metabolism analysis system in HeLa cells without or with CHB (0.5 μmol/L) treatment for 1 h. (J) Schematic diagram illustrating CHB binding with ATP and ADP. CHB channel: λex = 561 nm, λem = 655–705 nm; MTG channel: λex = 488 nm, λem = 500–550 nm; mitochondrial ATP commercial probe pCMV-Mito-AT1.03 channel: λex = 488 nm, λem = 500–550 nm.
Within mitochondria, ATP production primarily occurs in the cristae. Further analysis of CHB distribution within mitochondria revealed that the drug beacon co-localized with the cristae (Fig. 2D). To confirm CHB's binding to ADP and ATP, HeLa cells were co-stained with CHB (0.5 μmol/L, 1 h) and a commercial dye for mitochondrial ATP, pCMV-Mito-AT1.03 (mito-ATP) (Fig. 2E and F and Supporting Information S19), as well as a control dye for cellular matrix-localized ATP, pCMV-AT1.03 (cellular matrix-ATP) (Supporting Information Fig. S20). The results demonstrated that CHB co-localized with mito-ATP, with a PCC value of up to 0.86 (Fig. 3G), whereas minimal overlap was observed between CHB and cellular matrix-ATP (Fig. S20). These findings further confirmed that CHB binds ATP in mitochondria while simultaneously facilitating ATP visualization using SIM.
To affirm that CHB also binds mitochondrial ADP, we inhibited ATP synthesis using various oxidative phosphorylation (OXPHOS) inhibitors (OXPHOS is an enzyme that synthesizing ATP from ADP)38. HeLa cells were pretreated with OXPHOS inhibitors (Supporting Information Fig. S21). Despite a significant reduction in the mitochondrial ATP production, we observed no significant change in the fluorescence intensity of CHB labeling (Fig. 2H). These results suggested that CHB also binds mitochondrial ADP, aligning with our design of CHB as a dual-binding drug beacon. Moreover, we assessed whether CHB inhibited ATP production using the Mito Stress Test Kit and found that basal respiration and ATP production decreased when cells were exposed to CHB at 0.5 μmol/L for 1 h (Fig. 2I). Furthermore, it was confirmed that CHB binds mitochondrial ADP and, in conjunction with its ability to bind and decrease ATP levels, disrupts the mitochondrial respiratory chain and energy metabolism. Additionally, CHB enabled clear visualization of mitochondrial ATP and ADP distribution and levels, allowing us to demonstrate, for the first time, that mitochondrial cristae are surrounded by ATP and ADP in living cells (Fig. 2J).
3.3. Visualizing CHB-induced mitochondrial morphology damage
Exploiting the intrinsic fluorescence of CHB and its compatibility with SIM imaging, we visualized the effects of the drug beacon on mitochondrial morphology to gain a more granular understanding of its MOA. HeLa cells were treated with CHB (0.5 and 1.0 μmol/L) for 1 and 3 h and co-stained with an MMP-independent probe, MitoTracker® GreenFM, before SIM imaging. The results revealed that as the CHB concentration and treatment time increased, green filamentous mitochondria transitioned into rod- and round-shaped structures (Fig. 3A and B and Supporting Information Fig. S22). Quantitative analysis of mitochondrial morphology further showed an increase in round mitochondria (Fig. S22), with a significant difference observed between treated and untreated HeLa cells (Fig. 3B). Normal MMP is crucial for maintaining a functional mitochondrial energy source and normal physiological functions39,40. To demonstrate that CHB disrupted the mitochondrial function of HeLa cells, we examined changes in MMP with and without treatment. As the CHB concentration and exposure time increased, the green fluorescence intensity became stronger, indicating the depolarization of MMP and impaired mitochondrial function (Fig. 3C). This aligns with the proposed MOA of CHB, where it depletes ATP and ADP around mitochondrial cristae, triggering morphological damage. Similar results were observed in HepG2 cells (Supporting Information Fig. S23). Moreover, under mild CHB treatment, CHB-labeled ADP/ATP localized to the cristae, as revealed by the high overlap of fluorescence curves (Fig. 3D). However, with continued depletion of ATP/ADP by CHB, mitochondrial morphology was damaged, as indicated by the low overlap of fluorescence curves (Fig. 3D), accompanied by mitochondrial (green) fission and the release of ATP (red color) from damaged mitochondria. Finally, transmission electron microscopy confirmed the underlying mechanism of drug-induced mitochondrial morphological damage in tumor tissues from mice subjected to different treatments (Fig. 3E). We observed damage and reduced mitochondrial cristae in the CHB-treated groups (Fig. 3E–G). These findings suggest that CHB interacts with the ADP/ATP pool around mitochondrial cristae, sequestering these molecules and profoundly perturbing mitochondrial energetics. This leads to the observed damage to mitochondrial morphology and mitochondrial destruction.
To further confirm that the interactions between CHB and ATP/ADP are critical for mitochondrial damage, we synthesized CHE, a CHB analog lacking the essential boronic acid moiety required for ATP/ADP binding (see Supporting Information Scheme S2, Figs. S24–S28 for the synthesis route and CHE characterization). We demonstrated that CHE exhibited negligible cytotoxicity to HeLa cells (Supporting Information Fig. S29) and caused no mitochondrial damage (Supporting Information Fig. S30). These studies support our conclusion that the damage to mitochondrial morphology was caused by the direct, boronic acid-mediated binding of CHB to ATP/ADP.
3.4. Mitochondrial dysfunction triggered by CHB-induced ATP/ADP pool depletion results in cell death
Under pathological conditions, damage to the mitochondrial respiratory chain leads to an increase in reactive oxygen species (ROS), which directly contributes to the damage of mitochondrial cristae, ultimately resulting in mitochondrial destruction, cellular damage, and cell death41, 42, 43. Herein, we confirmed that MitoTracker® Red FM (MTRFM, Invitrogen) labels mitochondria in an MMP-dependent manner, allowing it to remain associated with mitochondria when MMP collapses (Supporting Information Fig. S31). To investigate whether CHB-induced depletion of ATP and ADP levels triggers changes in mitochondrial morphology by affecting ROS levels, we stained cells with MTRFM and then exposed them to CHB (0.5 μmol/L, 1 h and 1.0 μmol/L, 3 h) before labeling with the commercial ROS probe DCFH-Da for 0.5 h, and compared this to untreated cells to detect ROS levels (Fig. 4A). We found that the ROS levels in CHB (0.5 μmol/L, 1 h)-treated cells were similar to those in untreated cells. However, treatment with higher CHB concentrations and longer durations (1.0 μmol/L, 3 h) resulted in a significant increase in ROS levels (Fig. 4B). Therefore, the observed increase in ROS likely directly contributes to the destruction of mitochondrial cristae and, consequently, the mitochondria themselves.
Figure 4.
CHB-induced ATP/ADP depletion triggers mitochondrial dysfunction. (A) Colocalization of the ROS probe (stained with DCFH-DA), CHB, and Mito Tracker® Red FM (MTRFM). (B) ROS level without or with CHB treatment. The ROS level in the CHB (1.0 μmol/L, 3 h, damaged mitochondria) treatment group was higher than that in the CHB (0.5 μmol/L, 1 h, normal mitochondria) treatment and control groups. Data are expressed as the mean ± SEM. n = 3 cells, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. (C) The distribution of mtDNA in the mitochondria was revealed by co-staining with MitoTracker® Red FM (MTRFM) and CHB (0.5 μmol/L, 1 h, normal mitochondria and 1.0 μmol/L, 3 h, destroyed mitochondria), with the white lines indicating the region of interest for fluorescence measurement (D). (E) The illustration of CHB upregulates the ROS levels to damage the mitochondria by consuming ATP/ADP around the cristae. The schematic representation of higher ROS levels induces mtDNA escape from damaged mitochondria. (F) The entire process of visualizing the excellent anti-tumor properties of drug beacon-CHB in mitochondria from the action targets to mitochondrial cristae damage to mitochondrial dysfunction. CHB is located in the mitochondria and consumes ATP‒ADP to destroy mitochondria respiration, inducing mitochondrial cristae and function damage. Decreased ATP production cannot provide adequate energy for cell growth, thus causing cell death. ROS commercial probe DCFH-Da channel: λex = 488 nm, λem = 500–550 nm; mtDNA commercial probe pigcogreen channel: λex = 488 nm, λem = 500–550 nm; CHB channel: λex = 561 nm, λem = 673.8 nm; MitoTracker® Red FM channel: λex = 561 nm, λem = 580–627 nm.
Previous studies have shown that elevated levels of mitochondrial ROS can also damage to mitochondrial DNA (mtDNA) damage44. We observed that mtDNA co-localized with CHB (0.5 μmol/L, 1 h, in the absence of mitochondrial damage; Fig. 4C). However, upon induction of mitochondrial damage (1.0 μmol/L CHB, 3 h), we noted an increase in the size of mtDNA (Fig. 5C and Supporting Information Fig. S32; mtDNA stained with PicoGreen, and mitochondria stained with MTRFM). This change in size likely indicates the occurrence of a recently described phenomenon known as mtDNA gel-like phase transition45,46. At this stage, mtDNA and CHB localization no longer overlapped, aligning with CHB-mediated destruction of cristae structure, which initiates mitochondrial morphological damage and the release of mtDNA (Fig. 4D and E). These experiments further support the proposed MOA of CHB, which involves mitochondrial ADP and ATP depletion, leading to the inhibition of mitochondrial respiration, increased ROS levels, morphological damage starting with mitochondrial depolarization and cristae structure destruction, and eventual release of mtDNA.
Figure 5.
CHB exhibits an inhibition effect on tumor growth. (A) The schema of the treatment process for HeLa tumor-bearing mice. (B) Representative photographs of tumors in mice were taken on the 14th day after starting the treatments. (C) The digital photographs of tumors dissected from mice in different treated groups. (D, E) Relative tumor volumes and body weight after treatment with different therapies. P < 0.05 is considered significant. Data is expressed as the mean ± SEM (n = 5 mice in control and oligomycin A-treated group; n = 6 mice in CHB-treated group, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001). (F) Representative hematoxylin and eosin staining of tumors after different treatments.
3.5. Characterization of the biological activity of CHB
Given that CHB strongly binds both ATP and ADP, it was predicted that this molecule would exhibit potent bioactivity, particularly in the context of cancer cells that are highly sensitive to the inhibition of mitochondrial energy metabolism. Accordingly, the antitumor effects of CHB were evaluated and compared to oligomycin A15, a commercially available ATP inhibitor, in nude mice transplanted with HeLa cells. When the tumor volume reached approximately 150 mm3, different formulations were injected into the tumors in situ2 selected based on toxicity studies conducted in HeLa cells (Fig. 5A and Supporting Information Fig. S16). Following the completion of the treatment process, oligomycin demonstrated some inhibitory effect on tumor growth compared to the untreated group, consistent with previous reports15. The inhibitory effects of CHB were significantly more potent (Fig. 5B and C). Tumor volume decreased in the CHB-treated groups (Fig. 5D). In contrast, body weight remained unchanged (Fig. 5E). Moreover, the distribution of CHB in the tumor tissues in situ was recorded, and CHB was found to be concentrated within the tumor tissues (Supporting Information Fig. S33). These results confirmed that the antitumor effect of CHB was significantly higher than that of oligomycin A under the same conditions, supporting the superior ability of dual-binding compounds in tumor growth inhibition.
Furthermore, the histological analysis of tumor tissues from animals treated with intratumoral injection revealed noticeable changes in the morphology and structure of tumor cells compared to the control group (Fig. 5F). No significant changes were observed in other tissues under these conditions (Supporting Information Figs. S34 and S35). Similar results were obtained with intravenous injection (Supporting Information Figs. S36–S38). Collectively, these data demonstrate that CHB exhibits strong tumor-suppressive effects without affecting healthy tissues in a short time frame.
4. Conclusions
In this study, we developed a strategy for enhancing the monitoring of subcellular drug dynamics through in situ super-resolution imaging visualization. Our proof-of-concept drug beacon molecule, CHB, was designed to integrate a dual-binding pharmacophore (boronic acid) that can bind both ADP and ATP, with mitochondrial targeting and fluorescent features. CHB was demonstrated to be a robust imaging agent with a strong fluorescence signal. Additionally, it was shown that CHB binds both ATP and ADP levels within mitochondria. CHB allowed for the direct visualization of its distribution and effects in live cells, enabling the identification of mitochondrial cristae. Mechanistically, CHB was found to interact with mitochondrial ATP and ADP, leading to ATP consumption, damage to the mitochondrial respiratory chain and cristae, ROS production, and disruption of normal mitochondrial morphology. This cascade of events prevented sufficient ATP production to support normal cellular activities and functions (Supporting Information Movie S1).
The following is the supplementary data related to this article:
Additionally, this work introduced the concept of drug beacon design, where the luminophore and pharmacophore are integrated into a single molecule. This approach offers advantages over previous tools for drug visualization that use modified drug molecules, which can introduce uncertainty regarding their MOA and off-target effects. The fluorescent signal of CHB could trace the entire process of drug action in mitochondria, from localization to changes in mitochondrial morphology. It also provided insights into the cellular uptake and subcellular localization of the drug beacon. Overall, this study paves the way for developing additional drug beacons for in situ monitoring of drug MOA and micro-dynamics. This approach has the potential to accelerate the drug validation process, minimize off-target toxicity, and reveal additional targets for anticancer drug development.
Author contributions
Huimin Chen and Youxiao Ren cultured cell. Guiqian Fang, Youxiao Ren and Huimin Chen synthesized and characterized CHB. Huimin Chen performed confocal laser scanning microscopy. Weiwei Zou performed Mito Stress Test Kit. Huimin Chen and Youxiao Ren conducted animal experiment. Kang Ying and Zhiwei Yang conducted calculated work. Qixin Chen conceived the project, designed the experiments, and wrote the manuscript with the help of all authors.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This work was financially supported by the Shandong Province Key R&D Program (Major Technological Innovation Project) (2021CXGC010501, China), National Natural Science Foundation of China (Nos. 22107059, 22007060), Natural Science Foundation of Shandong Province (ZR2021QH057, ZR2022QH304, ZR2020QB166, China), Innovation Team of Shandong Higher School Youth Innovation Technology Program (2021KJ035, China), Taishan Scholars Program (TSQN202211221, China), Shandong Science Fund for Excellent Young Scholars (ZR2022YQ66, China), Young Elite Scientists Sponsorship Program of China Association for Science and Technology (CACM-2023-QNRC1-02), Academic Promotion Programme of Shandong First Medical University (No. 2019LJ003, China). We thank Translational Medicine Core Facility of Shandong University and Laboratory of Prof. Shuping Zhang of Shandong First Medical University for consultation and instrument support. Thanks are due to Prof. Yixuan Tang of Shandong First Medical University for helpful discussions and assistance with the experiments.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.11.022.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Gao M., Yu F.B., Lv C.J., Choo J.B., Chen L.X. Fluorescent chemical probes for accurate tumor diagnosis and targeting therapy. Chem Soc Rev. 2017;46:2237–2271. doi: 10.1039/c6cs00908e. [DOI] [PubMed] [Google Scholar]
- 2.Ding X.L., Liu M.D., Cheng Q., Guo W.H., Niu M.T., Huang Q.X., et al. Multifunctional liquid metal-based nanoparticles with glycolysis and mitochondrial metabolism inhibition for tumor photothermal therapy. Biomaterials. 2022;281 doi: 10.1016/j.biomaterials.2022.121369. [DOI] [PubMed] [Google Scholar]
- 3.Zhao R.Y., Fu J.H., Zhu L.J., Chen Y., Liu B. Designing strategies of small-molecule compounds for modulating non-coding RNAs in cancer therapy. J Hematol Oncol. 2022;15:14. doi: 10.1186/s13045-022-01230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu C., Yu L.F., Miao Y.X., Liu X.L., Yu Z.J., Wei M.J. Peptide-drug conjugates (PDCs): a novel trend of research and development on targeted therapy, hype or hope? Acta Pharm Sin B. 2023;13:498–516. doi: 10.1016/j.apsb.2022.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tu L., Liao Z.H., Luo Z., Wu Y.L., Herrmann A., Huo S.D. Ultrasound-controlled drug release and drug activation for cancer therapy. Explorations. 2021;1 doi: 10.1002/EXP.20210023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Q.Q., Yang W., Zhong M., Lin Z.X., Gray N.E., Xian Y.F. Animal models of Alzheimer's disease: preclinical insights and challenges. Acta Materia Medica. 2023;2:192–215. [Google Scholar]
- 7.Fu M.H., Han X., Chen B., Guo L., Zhong L., Hu P., et al. Cancer treatment: from traditional Chinese herbal medicine to the liposome delivery system. Acta Materia Medica. 2022;1:486–506. [Google Scholar]
- 8.Yu S.J., Zhao R.R., Zhang B.C., Lai C.M., Li L.Y., Shen J.W., et al. Research progress and application of the CRISPR/Cas9 gene-editing technology based on hepatocellular carcinoma. Asian J Pharm Sci. 2023;18 doi: 10.1016/j.ajps.2023.100828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z.P., Zhang L.J., Song Y., He J.Y., Wu L., Zhao C., et al. Hierarchical targeted hepatocyte mitochondrial multifunctional chitosan nanoparticles for anticancer drug delivery. Biomaterials. 2015;52:240–250. doi: 10.1016/j.biomaterials.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Liu L.Y., Fang H., Chen Q., Chan M.H., Ng M., Wang K.N., et al. Multiple-color platinum complex with super-large Stokes shift for super-resolution imaging of autolysosome escape. Angew Chem Int Ed Engl. 2020;59:19229–19236. doi: 10.1002/anie.202007878. [DOI] [PubMed] [Google Scholar]
- 11.Wei Y.C., Kong L.X., Chen H.M., Liu Y.Y., Xu Y.F., Wang H., et al. Super-resolution image-based tracking of drug distribution in mitochondria of a label-free naturally derived drug molecules. Chem Eng J. 2021;429 [Google Scholar]
- 12.Chen H.M., Wang H., Wei Y.C., Hu M.M., Dong B., Fang H.B., et al. Super-resolution imaging reveals the subcellular distribution of dextran at the nanoscale in living cells. Chin Chem Lett. 2022;33:1865–1869. [Google Scholar]
- 13.Wang K.N., Liu L.Y., Qi G., Chao X.J., Ma W., Yu Z., et al. Light-driven cascade mitochondria-to-nucleus photosensitization in cancer cell ablation. Adv Sci. 2021;8 doi: 10.1002/advs.202004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y.Y., Zhang C.Y., Wei Y.C., Chen Q.X., Wang Y.F. De novo-designed landmine warfare strategy luminophore for super-resolution imaging reveal ONOO- evolution in living cells. Chem Eng J. 2021;422 [Google Scholar]
- 15.Zhou H., Zeng X.D., Li A.G., Zhou W.Y., Tang L., Hu W.B., et al. Upconversion NIR-II fluorophores for mitochondria-targeted cancer imaging and photothermal therapy. Nat Commun. 2020;11:6183. doi: 10.1038/s41467-020-19945-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y., Zeng X.D., Xiao Y.L., Liu C.H., Zhu H., Zhou H., et al. Novel dual-function near-infrared II fluorescence and PET probe for tumor delineation and image-guided surgery. Chem Sci. 2018;9:2092–2097. doi: 10.1039/c7sc04774f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F.R., Zhang X.L., Jing J., Zhang X.L. A Golgi-targeted fluorescent probe for imaging H2O2 and releasing H2S during Golgi stress. Dyes Pigments. 2023;219 [Google Scholar]
- 18.Liu F.R., Jing J., Zhang X.L. Research progress of fluorescent probes for cysteine targeting cellular organelles. Chin J Org Chem. 2023;43:2053–2067. [Google Scholar]
- 19.Chen Q.X., Shao X.T., Tian Z.Q., Chen Y., Mondal P., Liu F., et al. Nanoscale monitoring of mitochondria and lysosome interactions for drug screening and discovery. Nano Res. 2019;12:1009–1015. [Google Scholar]
- 20.Chen Q.X., Fang H.B., Shao X.T., Tian Z.Q., Geng S.S., Zhang Y.M., et al. A dual-labeling probe to track functional mitochondria-lysosome interactions in live cells. Nat Commun. 2020;11:6290. doi: 10.1038/s41467-020-20067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Q.X., Shao X.T., Hao M.G., Fang H.B., Guan R.L., Tian Z.Q., et al. Quantitative analysis of interactive behavior of mitochondria and lysosomes using structured illumination microscopy. Biomaterials. 2020;250 doi: 10.1016/j.biomaterials.2020.120059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C.Y., Liu W., Wang S.F., Li H.J., Lv Z.L., Zhang F., et al. Super-resolution nanoscopy by coherent control on nanoparticle emission. Sci Adv. 2020;6 doi: 10.1126/sciadv.aaw6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao X.T., Fang Z., Li M.L., Zou W.W., Wang Q.C., Zhang C.Y., et al. Graphene oxide nanosheets induce mitochondrial fragmentation by cutting through membrane. ACS Mater Lett. 2023;5:2308–2316. [Google Scholar]
- 24.Li P.L., Shang X.Y., Jiao Q.L., Mi Q., Zhu M.Q., Ren Y.D., et al. Alteration of chromatin high-order conformation associated with oxaliplatin resistance acquisition in colorectal cancer cells. Explorations. 2023;3 doi: 10.1002/EXP.20220136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang W.J., Yuan C.N., Zhu L.Q., Du S.B., Qian L.H., Ge J.Y., et al. Recent advances in construction of small molecule-based fluorophore-drug conjugates. J Pharm Anal. 2020;10:434–443. doi: 10.1016/j.jpha.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baell J.B., Nissink J.W.M. Seven year itch: pan-assay interference compounds (PAINS) in 2017-utility and limitations. ACS Chem Biol. 2018;13:36–44. doi: 10.1021/acschembio.7b00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng X.D., Xiao Y.L., Lin J.C., Li S.S., Zhou H., Nong J.X., et al. Near-infrared II dye-protein complex for biomedical imaging and imaging-guided photothermal therapy. Adv Healthcare Mater. 2018;7 doi: 10.1002/adhm.201800589. [DOI] [PubMed] [Google Scholar]
- 28.Shao X.T., Meng C.C., Song W.J., Zhang T., Chen Q.X. Subcellular visualization: organelle-specific targeted drug delivery and discovery. Adv Drug Deliv Rev. 2023;199 doi: 10.1016/j.addr.2023.114977. [DOI] [PubMed] [Google Scholar]
- 29.Zeng Z.L., Liew S.S., Wei X., Pu K.Y. Hemicyanine-based near-infrared activatable probes for imaging and diagnosis of diseases. Angew Chem Int Ed Engl. 2021;60:26454–26475. doi: 10.1002/anie.202107877. [DOI] [PubMed] [Google Scholar]
- 30.Lan J.S., Liu L., Zeng R.F., Qin Y.H., Liu Y., Jiang X.Y., et al. Rational modulation of coumarin-hemicyanine platform based on OH substitution for higher selective detection of hypochlorite. Chem Commun. 2020;56:1219–1222. doi: 10.1039/c9cc06477j. [DOI] [PubMed] [Google Scholar]
- 31.Shu W., Wu Y.L., Zang S.P., Su S., Kang H., Jing J., et al. A mitochondria-targeting highly specific fluorescent probe for fast sensing of endogenous peroxynitrite in living cells. Sens Actuators B. 2020;303 [Google Scholar]
- 32.Shchepina L.A., Pletjushkina O.Y., Avetisyan A.V., Bakeeva L.E., Fetisova E.K., Izyumov D.S., et al. Oligomycin, inhibitor of the F0 part of H+-ATP-synthase, suppresses the TNF-induced apoptosis. Oncogene. 2002;21:8149–8157. doi: 10.1038/sj.onc.1206053. [DOI] [PubMed] [Google Scholar]
- 33.Sosa V., Moliné T., Somoza R., Paciucci R., Kondoh H., Me L.L. Oxidative stress and cancer: an overview. Ageing Res Rev. 2013;12:376–390. doi: 10.1016/j.arr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X., Kwon Y.H., Kim G.M., Ryu J.H., Yoon J.Y. A ratiometric fluorescent probe based on a coumarin-hemicyanine scaffold for sensitive and selective detection of endogenous peroxynitrite. Biosens Bioelectron. 2015;64:285–291. doi: 10.1016/j.bios.2014.08.089. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y.C., Zhu C.C., Yang Z.H., Chen J.J., He Y.F., Jiao Y., et al. A ratiometric fluorescent probe for rapid detection of hydrogen sulfide in mitochondria. Angew Chem Int Ed Engl. 2013;52:1688–1691. doi: 10.1002/anie.201207701. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X.G., Wang Y.X., Zheng C., Li C.X. Phenylboronic acid-functionalized glycopolymeric nanoparticles for biomacromolecules delivery across nasal respiratory. Eur J Pharm Biopharm. 2012;82:76–84. doi: 10.1016/j.ejpb.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Kim D.H., Hee S.Q., Norris A.J., Faull K.F., Eckhert C.D. Boric acid inhibits adenosine diphosphate-ribosyl cyclase non-competitively. J Chromatogr A. 2006;1115:246–252. doi: 10.1016/j.chroma.2006.02.066. [DOI] [PubMed] [Google Scholar]
- 38.Capaldi R.A., Aggeler R., Turina P., Wilkens S. Coupling between catalytic sites and the proton channel in F1F0-type ATPases. Trends Biochem Sci. 1994;19:284–289. doi: 10.1016/0968-0004(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 39.Cui L.Y., Gouw A.M., LaGory E.L., Guo S.H., Attarwala N., Tang Y., et al. Mitochondrial copper depletion suppresses triple-negative breast cancer in mice. Nat Biotechnol. 2021;39:357–367. doi: 10.1038/s41587-020-0707-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y.Q., Zhang X.J., Zhou M.J., Nan X.Y., Chen X.F., Zhang X.H. Mitochondrial-targeting lonidamine-doxorubicin nanoparticles for synergistic chemotherapy to conquer drug resistance. ACS Appl Mater Interfaces. 2017;9:43498–43507. doi: 10.1021/acsami.7b14577. [DOI] [PubMed] [Google Scholar]
- 41.Chen C., Chen Y., Guan M.X. A peep into mitochondrial disorder: multifaceted from mitochondrial DNA mutations to nuclear gene modulation. Protein Cell. 2015;6:862–870. doi: 10.1007/s13238-015-0175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou T.T., Zhang R.F., Jian C.S., Ding W.Q., Wang Y.R., Ling S.K., et al. NDUFAB1 confers cardio-protection by enhancing mitochondrial bioenergetics through coordination of respiratory complex and supercomplex assembly. Cell Res. 2019;29:754–766. doi: 10.1038/s41422-019-0208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleele T., Rey T., Winter J., Zaganelli S., Mahecic D., Perreten Lambert H., et al. Distinct fission signatures predict mitochondrial degradation or biogenesis. Nature. 2021;593:435–439. doi: 10.1038/s41586-021-03510-6. [DOI] [PubMed] [Google Scholar]
- 44.Caielli S., Athale S., Domic B., Murat E., Chandra M., Banchereau R., et al. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J Exp Med. 2016;213:697–713. doi: 10.1084/jem.20151876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long Q., Zhou Y.S., Wu H., Du S.W., Hu M.L., Qi J.T., et al. Phase separation drives the self-assembly of mitochondrial nucleoids for transcriptional modulation. Nat Struct Mol Biol. 2021;28:900–908. doi: 10.1038/s41594-021-00671-w. [DOI] [PubMed] [Google Scholar]
- 46.Feric M., Demarest T.G., Tian J., Croteau D.L., Bohr V.A., Misteli T. Self-assembly of multi-component mitochondrial nucleoids via phase separation. EMBO J. 2021;40 doi: 10.15252/embj.2020107165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.