Abstract
Antimicrobial resistance (AMR) has become a global health crisis in need of novel solutions. To this end, antibiotic combination therapies, which combine multiple antibiotics for treatment, have attracted significant attention as a potential approach for combating AMR. To facilitate advances in antibiotic combination therapies, most notably in investigating antibiotic interactions and identifying synergistic antibiotic combinations however, there remains a need for automated high-throughput platforms that can create and examine antibiotic combinations on-demand, at scale, and with minimal reagent consumption. To address these challenges, we have developed a Robotic-Printed Combinatorial Droplet (RoboDrop) platform by integrating a programmable droplet microfluidic device that generates antibiotic combinations in nanoliter droplets in automation, a robotic arm that arranges the droplets in an array, and a camera that images the array of thousands of droplets in parallel. We further implement a resazurin-based bacterial viability assay to accelerate our antibiotic combination testing. As a demonstration, we use RoboDrop to corroborate two pairs of antibiotics with known interactions and subsequently identify a new synergistic combination of cefsulodin, penicillin, and oxacillin against a model E. coli strain. We therefore envision RoboDrop becoming a useful tool to efficiently identify new synergistic antibiotic combinations toward combating AMR.
Key words: Droplet microfluidics, Antimicrobial resistance, Antibiotic combinatorial screening, Contact printing, Robotics, Pharmaceutical analysis, High-throughput, Antibiotic interactions
Graphical abstract
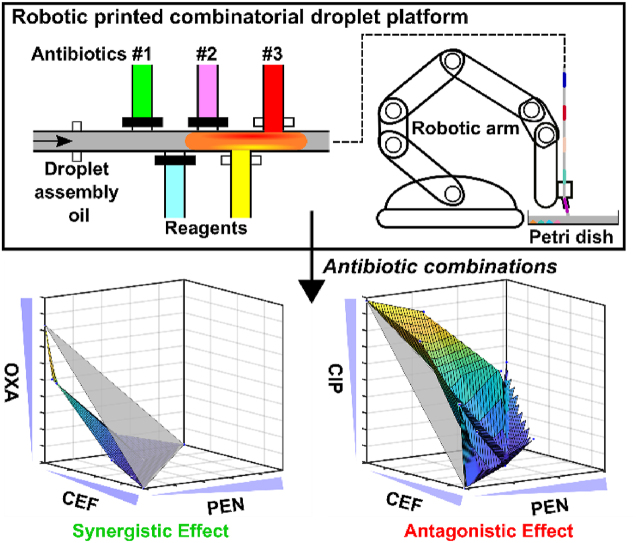
We report the robotic-printed combinatorial droplet platform as a potential solution to automate and miniaturize the screening of antibiotic combinations to identify synergistic combinations for combating antimicrobial resistance.
1. Introduction
Antimicrobial resistance (AMR) has become a severe public health concern. The US Centers for Disease Control and Prevention (CDC) reports that over 2.8 million AMR infections occur in the United States each year, resulting in more than 35,000 deaths1. The incidence of bacterial resistance to commonly used antibiotics and the emergence of multidrug-resistant bacteria continue to rise due to the absence of new antibiotics and overuse of broad-spectrum antibiotics2, 3, 4, 5. Several non-traditional antimicrobial drugs such as quorum sensing inhibitors6,7, antimicrobial peptides8,9, and bacteriophage9,10 have been investigated to combat AMR, relieving the burden of discovering new antibiotics. However, their lengthy discovery pipelines restrict their timely implementation. Rapid diagnostics and antimicrobial susceptible test (ID/AST) tools have been developed to deliver diagnostic results within hours. Compared to the lengthy turnaround diagnostic time, typically days, in traditional clinical microbiology lab, these tools allow the prescription of antibiotics based on evidence, reduce the use of broad-spectrum antibiotic, and thereby retard the development of AMR11, 12, 13, 14, 15, 16, 17, 18, 19, 20. However, ID/AST tools cannot serve as a treatment for patients with AMR, emphasizing the necessity for alternative therapies that do not require the lengthy discovery of new antibiotics.
Antibiotic combination therapy, wherein two or more antibiotics are combined as a single treatment regime, offers a promising strategy for accomplishing effective treatment using existing antibiotics while bypassing the lengthy discovery of new antibiotics21, 22, 23. By targeting multiple pathways and mechanisms, antibiotic combinations can increase the likelihood in combating multidrug resistant bacteria. For instance, combinatorial therapies have become essential for treating Mycobacterium tuberculosis (a highly infectious bacteria typically in the respiratory system)24,25, and multidrug-resistant Gram-negative bacteria such as Acinetobacter spp.26, 27, 28 and carbapenemase-producing Enterobacteriaceae29,30. To further proceed with the practice for other diseases, there is a critical need to screen combinations using various antibiotic candidates and determine whether the combined effect is greater than or less than the expected effects from the individual antibiotics, classified as synergism and antagonism, respectively31. In particular, synergistic antibiotic combinations have the potential to match or even outperform individual antibiotics in efficacy—potentially shortening treatment duration—while using lower drug doses and thus reducing side effects32,33. Current antibiotic combination studies have predominantly focused on pairwise interactions, with relatively limited exploration of higher-order combinations23,34, 35, 36, 37 Nonetheless, the potential of higher-order combinations remains substantial because their broader spectrum of activity create more challenges for bacteria to develop resistance38. However, practical realization of higher-order combinations faces challenges posed by exponential increase in the number of combinations and intricate analysis for interpreting the interactions22,39,40. As a result, there is a need for antibiotic combinatorial screening system to enable both pairwise and higher-order combinations to investigate new synergistic antibiotic combinations.
Currently, the most widely used antibiotic combinatorial screening systems are based on microtiter plates, which handle the liquids in a microliter scale. However, as the number of antibiotic combinations increases, this approach can lead to large reagent consumption and lab-intensive process. Microfluidics have been leveraged to reduce reagent consumption while screening different drug combinations41, 42, 43, 44, 45, 46, 47, 48, 49. Some of the systems showed the ability to automate the screening process and alleviate the manual pipetting, such as combining inkjet printing43, contact printing50, electrowetting-on-dielectric (EWOD)45, and Actuate-to-Open Valve47. While these technologies significantly reduce reagent consumption and automate the screening process, the full capacity of throughput and scalability has yet to be demonstrated as only tens of conditions have been demonstrated. On the other hand, recent advancements in droplet microfluidic systems have brought significant improvements35,36,51, 52, 53, 54, 55 but still exhibit various limitations concerning reagent/sample consumption, automation, throughput, scalability, and on-demand control of antibiotic combinations. For instance, a previously demonstrated nanoliter droplet array platform, which relied on random mixing of pre-generated droplets, has shown potential in reducing reagent/sample consumption, achieving high throughput and scalability36. However, it involves several manual steps and lacks the ability to generate antibiotic combinations on-demand. Conversely, another approach, the combinatorial nanodroplet platform, has successfully generated combinatorial droplets using pneumatic valves, addressing issues of on-demand control of droplet compositions, and retaining low reagent/sample consumption35. Nevertheless, its automation, scalability and throughput are limited due to the finite droplet storage capacity within tubing, manual operation requirements, and the necessity of an additional detection chip. Consequently, there remains a pressing need for a system that can effectively address these limitations.
In response, we present the Robotic-Printed Combinatorial Droplet (RoboDrop) system for automated and miniaturized screening of antibiotic combinations. The system incorporates a microfluidic device with pneumatic valve-based droplet control system to programmatically produce droplets with reagents/samples in the nanoliter range. The resulting droplets are then transported into a tubing fixed to a robotic arm, which automatically prints the ejected droplets into an array for subsequent analysis. To demonstrate the capacity and programmability of RoboDrop, we generated and printed an array of thousands of droplets on a substrate composed of green and blue food dyes alternatively using the microfluidic device. We then incorporated resazurin-based bacterial viability assay in RoboDrop, which allowed us to examine antibiotic combinations within hours. We verified the utility of our platform with previously reported synergistic and antagonistic antibiotic pairs. As an entry point for higher-order combinations, we used RoboDrop to investigate three-antibiotic combinations, from which we identified a new synergistic combination. These results demonstrate the potential of RoboDrop for screening pairwise and higher-order antibiotic combinations.
2. Materials and methods
2.1. Microfluidic device design
The microfluidic device is composed of two polydimethylsiloxane (PDMS) layers with one top fluidic layer for droplet generation and one bottom valve layer for controlling the actuation of the microvalves. The top fluidic layer is composed of one oil inlet port, 13 sample inlet ports, and one droplet outlet port. The droplet assembly is taken place at a channel with a width of 200 μm to help squeeze the droplets and ensure better merging of different samples and reagents. Immediately downstream of the droplet assembly channel, the outlet of the device is designed to have three steps with gradually increased height to ensure smooth droplet transportation into PTFE tubing. The bottom valve layer is composed of 14 ports that control the actuation of the corresponding microvalves, including one port that controls the oil inlet and 13 ports that control the injection of samples and reagents.
2.2. Master mold fabrication
The photomasks for both the fluidic layer and valve layer were designed using AutoCAD 2021 (Autodesk, San Rafael, CA, USA) and subsequently printed onto high precision transparencies at 20,000 dpi by CAD/Art Services Inc. (Bandon, OR, USA). The master molds for both layers were microfabricated using standard photolithography on separate 4-inch silicon wafers (Polishing Corporation of America, Santa Clara, CA, USA). To fabricate the master mold for the fluidic layer, SPR-220-7 positive photoresist (Microchem Corp., Newton, MA, USA) was first spin-coated onto the wafer with a height of 35 μm, which serves as the segments to interact with the bottom valve layer. Due to the rounded structure of the SPR-220-7 segments after hard baking, the device allowed a push-up valve architecture, where the bottom valve layer can collapse into the SPR-220-7 patterns on the top fluidic layer to ensure tight sealing. The remaining fluidic layers were fabricated by spin-coating 4 additional layers of SU-8 3050 (negative photoresist; Microchem Corp., Newton, MA, USA) with gradually increased height onto the silicon wafer. Predesigned alignment markers were used and referenced during the alignment of the SPR-220-7 segments and different SU-8 3050 fluidics channels. To fabricate the master mold for the valve layer, a single layer of SU8-3025 (negative photoresist; Microchem Corp., Newton, MA, USA) was spin-coated onto a silicon wafer with a height of 30 μm via standard photolithography.
2.3. Microfluidic device fabrication
The microfluidic device was fabricated with PDMS using multilayer soft lithography. Both master molds were firstly silanized with chlorotrimethylsilane (Sigma–Aldrich, St. Louis, MO, USA) in a desiccator for 10 min to reduce adhesion between PDMS and the master molds. The thin valve layer was obtained by spin-coating PDMS (SYLGARD 184 Silicone Elastomer Kit, Dow Corning, Midland, MI, USA) with 15:1 base to curing agent ratio on the valve layer master mold at 1000 rpm. The spin-coated mold was immediately baked at 80 °C for 20 min. The thick fluidic PDMS layer was prepared by directly pouring 51.7 g of PDMS with 10:1 base to curing agent ratio onto the fluidic layer master mold, and baked at 80 °C for 20 min. After baking, cured PDMS was peeled off from the fluidic layer master mold and the cured PDMS on the valve master mold remained attached. Both peeled fluidic PDMS layer and valve PDMS layer on the master mold was treated with an oxygen plasma machine (42 W, 500 mTorr, 45 s), and then manually aligned and temporarily bonded under Stemi DV4 Microscopy (Carl Zeiss AG, Oberkochen, Germany). After an additional bake at 80 °C for 10 min, both PDMS layers became a permanently bonded double layer, which was then peeled off from the valve layer master mold before holes were punched through all ports. The double-layer PDMS was then permanently bonded with a 64 mm × 50 mm cover glass (Ted Pella Inc., Redding, CA, USA) upon oxygen plasma treatment (42 W, 500 mTorr, 45 s) and bake (80 °C, 10 min), completing the fabrication. The fabricated devices were stored at 80 °C oven until use.
2.4. Microfluidic device operation and characterization
In our microfluidic device, nanoliter droplets were programmatically assembled by actuating the microvalves that were interacted with a set of solenoid valves controlled by a custom code written in MATLAB (MathWorks, Natick, MA, USA). Water-filled Tygon microbore tubing (0.02-inch ID and 0.06-inch OD; Cole-Parmer, Vernon Hills, IL, USA) was used as the interface between the microvalves and the solenoid valves. Each tubing was connected to a 23-gauge blunt needle (MCMaster-Carr, Elmhurst, IL, USA), which was inserted at its designated valve inlet port. The pressure was set to 30 psi to ensure fully closed microvalves during the operation and avoid cross-talk from the samples and reagents. The pressure was set to be 5 psi for the oil inlet and 2.5 psi for all sample and reagent inlets. By controlling the opening and closing time of each microvalve, nanoliter droplets can be assembled with the desired compositions and volume. The oil we used to push the generated droplets was fluorinated oil FC-40 (3M, Two Harbors, MN, USA).
Before the operation, the microfluidic device was continuously flushed with Rain-X for 15 min and flashed with air. The surface-treated microfluidic device was then baked at 80 °C for 30 min before usage. This surface treatment was to form a hydrophobic channel surface to avoid aqueous droplets attaching to the channel wall, which could cause residues and cross-contamination. During the operation, a pre-designed injection instruction was created to programmatically and automatically control the droplet assembly with desired compositions. The generated droplets were then sequentially flowed into the PTFE tubing (Cole-Parmer, Vernon Hills, IL, USA) from the device outlet, where the tubing was directly inserted into the microfluidic device outlet port.
The microvalve opening time and the droplet volume were correlated to precisely control the droplet composition and volume. With pressure set to 2.5 psi, triplicate droplets composed of fluorescence dyes were generated with different microvalve opening time ranging from 0.1 to 0.6 s with a 0.1 s increment. The droplets were flowed through a simple one-channel microfluidic device and detected using our custom laser-induced fluorescence (LIF) detector. The droplet image with 0.6 s microvalve opening time was taken under a fluorescence microscope (Olympus IX71, Shinjuku, Tokyo, Japan) as the reference to calculate the droplet flow rate, shown in Eq. (1):
| (1) |
where A is droplet area, h is channel height, and t is the flowing time obtained from LIF detector. The droplet volume with other microvalve opening time were then calculated by multiplexing the corresponding flowing time and the calculated droplet flow rate.
2.5. Robotic arm mount and operation
The mount that interfaces the robotic arm, tubing fixer (fingertight one-piece fitting, Cole–Parmer, Vernon Hills, IL, USA), and the PTFE tubing was designed in SolidWorks and 3D printed using high temp resin on a Form 2 printer (Formlabs, Somerville, MA, USA). The 3D-printed mount was designed to have a hole to insert the tubing from the top and a hollow pillar at the bottom to tighten the tubing fixer. During the operation, the PTFE tubing was inserted from the top of the mount, through the hollow pillar, and all the way to the tubing fixer, which holds the tubing tightly. The hollow pillar was designed with an angle of 70° to help the smooth printing of the droplets onto the substrate.
The uArm Swift Pro robotic arm was chosen and purchased from Ufactory (Guangdong, China). The operation of the arm was automatically controlled using a custom program in Python. Once the program started, the arm was first controlled to move in the x-y-z axis to the designated starting point on the substrate, with the PTFE tubing tip touching onto the substrate surface. Then the program asked for the size of the petri dish as the input information to calculate 90% of the largest square inside the petri dish as the designated printing area. After the calibration steps, the robotic arm will automatically move the tubing around the designated printing area in a serpentine shape. The flowing droplets from the tubing will be printed onto the downstream substrate individually and orderly.
2.6. Petri dish preparation and array characterization
The petri dish (Fisher Scientific, Waltham, MA, USA) was firstly treated with 20 mL of 2% (3-aminopropyl) triethoxysilane (APTES) (Sigma–Aldrich, St. Louis, MO, USA) in ethanol for 30 min on 50 °C hot plate. The heated petri dish was then rinsed with ethanol, dried with N2 gas, and placed in a 65 °C oven for 30 min. After 30 min, the surface-treated petri dish was kept at room temperature before immediate usage. During the use, the petri dish was firstly filled with the droplet carrier oil, FC-40 oil (3M, Two Harbors, MN, USA), to reduce the droplet evaporation issues during the printing.
The printed array density was characterized by controlling the oil injection time in between the droplets. More specifically, food dye was used to continuously generate and print 75-nL droplets on a 10-cm diameter petri dish with different oil injection time of 5 and 15 s. With the robotic arm set to move at the minimum speed, the array density increased with shorter oil injection time. The contact printing of the droplet array was further tested to its maximum capability on a 15-cm diameter petri dish with around 2700 75-nL droplets composed of alternating blue and green food dyes.
2.7. Resazurin-based cell viability assay and on-chip characterization
The resazurin powder was purchased from Sigma–Aldrich (St. Louis, MO, USA) and dissolved to 50 mmol/L stock in nuclease and protease free water purchased from Quality Biological (Gaithersburg, MD, USA), and stored at 4 °C. All drugs, cefsulodin (CEF), penicillin (PEN), ciprofloxacin (CIP), and oxacillin (OXA) were purchased from Sigma–Aldrich (St. Louis, MO, USA) and dissolved to 10 mg/mL stock in nuclease and protease free water for immediate use. The bacteria, E. coli (ATCC 25922), were cultured to 8 × 108 CFU/mL in MH II medium in a shaking incubator at 37 °C, washed three times with fresh MH II medium, aliquoted and stored at −80 °C with glycerol until use. The minimum inhibitory concentrations (MIC) of each antibiotic were tested using the standard microdilution method recommended by the Clinical and Laboratory Standards Institute (CLSI) guidelines, where 5 × 105 CFU/mL of bacteria were cultured with antibiotics, titrating concentrations in 2-fold increments at 37 °C for 16–20 h. The optical density at 600 nm was measured using a plate reader (Synergy H1, BioTek) to determine the MIC based on the bacteria growth. A typical resazurin-based cell viability assay system is composed of the bacteria, resazurin, and drug with various concentrations in Mueller Hinton II (MH II) medium (Sigma–Aldrich, St. Louis, MO, USA).
The on-chip optimization of the assay was achieved by screening different resazurin concentrations at 100, 300, and 500 μmol/L, and bacteria concentrations at 5 × 106, 5 × 107, and 5 × 108 CFU/mL with and without MIC concentration of CEF. The generated droplets were printed onto a 10-cm diameter petri dish and incubated at 37 °C for 6 h before further analysis in MATLAB. During the incubation, the droplet images at multiple time points, 0, 2, 4, and 6 h, were captured with a Sony Alpha a7 II camera for later multi-timepoint analysis in Excel. The data was later used to optimize the colorimetric analysis.
2.8. On-chip drug combinations
The on-chip drug combinations were first verified with two known two-drug combinations, CEF and PEN, and PEN and CIP, where the former one was reported with a synergistic effect and the latter one in bulk showed an antagonistic effect. In brief, the bulk experiment was designed to test 16 different compositions across 0, MIC/4, MIC/2, and MIC of each drug with 5 × 105 CFU/mL bacteria and incubated at 37 °C for 16–20 h and the optical density at 600 nm were measured using a plate reader (Synergy H1, BioTek) to determine the MIC based on the bacteria growth. For on-chip drug combinations, a predesigned injection instruction was designed to automate the droplet microfluidic system to generate droplets that contain contents with the desired combinations. For the two-drug combinations, droplets with 16 different compositions across 0, MIC/4, MIC/2, and MIC of each drug were generated in triplicate along with final concentrations of 500 μmol/L resazurin and 5 × 106 CFU/mL bacteria. The generated droplets were subsequently transported via PTFE tubing and printed onto a surface-treated oil-filled petri dish. The droplet array on the petri dish was then incubated at 37 °C for 6 h before image capture with a Sony Alpha a7 II camera (Tokyo, Japan).
Three-drug combinations were similarly achieved by designing a queue that automates the droplet microfluidics system to generate 64 different compositions across 0, MIC/4, MIC/2, and MIC of each drug. The final concentration for resazurin and bacteria were also 500 μmol/L and 5 × 106 bacteria, respectively. The droplet array on the petri dish was then incubated at 37 °C for 6 h before image capture with a Sony Alpha a7 II camera. The three-drug combinations were replicated three times in separate experiments.
2.9. Data acquisition and analysis
Colorimetric analysis was characterized using the droplet images in the on-chip assay optimization experiment. The example droplet picture represents droplets composed of 5 × 106 CFU/mL and 500 μmol/L resazurin with and without MIC concentration of CEF. During the colorimetric analysis in ImageJ, ROI tool was used to select each droplet before splitting the picture into red, green, and blue channels. In the RGB color model, a higher signal means a higher intensity value in the corresponding channel. The intensity values in each channel range from 0 to 255, where 0 represents no intensity (completely dark) and 255 represents full intensity (completely bright). The signal intensity of each droplet in each channel was measured and normalized to the initial stage and the signal-to-background ratio of the droplets in each channel was later analyzed in Excel. Since the red channel showed the highest SNR among the other channels, red channel intensity was consistently used for colorimetric analysis of all droplet images.
For on-chip drug combinations experiments, dose-response curve model as shown in Eq. (2):
| (2) |
where E is the magnitude of the response, A is the drug concentration, EC50 is the drug concentration that produces a 50% maximal response, and n is the Hill coefficient that describes the cooperativity of the biological interactions. The dose-response curve model was utilized to analyze the IC80 values (i.e., drug concentration required to inhibit 80% of bacterial growth) of the growth curve in OriginLab (OriginLab Corporation), where the drug concentrations were transformed into log scale and the colorimetric signal was normalized between the signal of no-drug condition and highest concentration drug condition in Excel. The IC80 values were later plotted in the x-y plot or x-y-z plane for two-drug or three-drug combinations, respectively, to determine the combinatorial effects. A definition of normalized total distance with direction (NTDD) was proposed to quantitively classify the concave and convex shape of the line or plane, with positive value indicating concave shape and negative value indicating convex shape.
For 2-drug combinations or plot in the x-y axis, the NTDD was determined by first finding the equation of the reference line, connected lines of the IC80 values of the two individual drugs, in the form of Eq. (3):
| (3) |
where A, B, and C are real constants. The distance equation of Eq. (4):
| (4) |
was used to calculate the distance between each IC80 value in coordinates of (x0, y0) to the reference line. The sum of the distance was then normalized by the number of the IC80 values, where the positivity and negativity determine the overall direction of the IC80 values with respect to the IC80 values of the individual drugs.
For 3-drug combinations or plot in the x-y-z axis, the NTDD was determined by first find the equation of the reference plane, connected plane of the IC80 values of the three individual drugs, in the form of Eq. (5):
| (5) |
where A, B, C, and D are real constants. The distance equation of Eq. (6):
| (6) |
was used to calculate the distance between each IC80 value in coordinates of (x0, y0, z0) to the reference plane. The sum of the distance was then normalized by the number of the IC80 values, where the positivity and negativity determine the overall direction of the IC80 values with respect to the IC80 values of the individual drugs. All plots were plotted using OriginLab (OriginLab Corporation) and later organized with Inkscape 1.2.2 (Software Freedom Conservancy, NY, USA).
3. Results and discussion
3.1. Overview of RoboDrop
RoboDrop consists of three main components, a droplet microfluidic device for droplet generation, a tubing segment for droplet transportation, and a robotic arm for droplet printing onto a petri dish (Fig. 1 and Supporting Information Fig. S1). The droplet microfluidic device injects reagents from various microchannels, individually tuned via pneumatic valves to control the open/close status and the opening duration (Fig. 1A). These reagents coalesce into an individual droplet upon contact, which is subsequently carried downstream by carrier oil. This droplet generation cycle is conducted in an automatic means via a custom MATLAB program allowing the generation of numerous droplets, analogous to a droplet assembly pipeline. Colorimetric assays are implemented in the droplets and the droplets then flow into a tubing, which is interfaced with a customized 3D-printed mount fixed to the robotic arm (Fig. 1B). This robotic arm moves the tubing on a petri dish with a designated pattern and speed, allowing for the droplets to be printed sequentially onto a petri dish. The sequence and spatial location barcode the different drug combinations in each droplet. Finally, the assay results in droplets are captured using a commercial camera and colorimetric analysis is performed to quantify the bacterial growth treated with different antibiotic combinations (Fig. 1C).
Figure 1.
Schematic of RoboDrop. RoboDrop is composed of three mean components for (A) droplet generation, (B) droplet transportation, and (C) droplet printing. (A) A pneumatic valve-based nanodroplet microfluidic device was used to programmatically actuate the valves and generate a nanoliter droplet composed of bacteria and resazurin with varying drug combinations. (B) The generated droplets were later pushed by oil into a tubing (PTFE tubing) that is interfaced and fixed with a robotic arm. (C) The robotic arm automatically moves the tubing with programmed movements, where the flowing droplets are printed onto a cover oil-filled petri dish with spatial barcoding. After incubation of the droplets, colorimetric images were taken by a commercial camera for later analysis.
3.2. Programmable control of droplet microfluidics
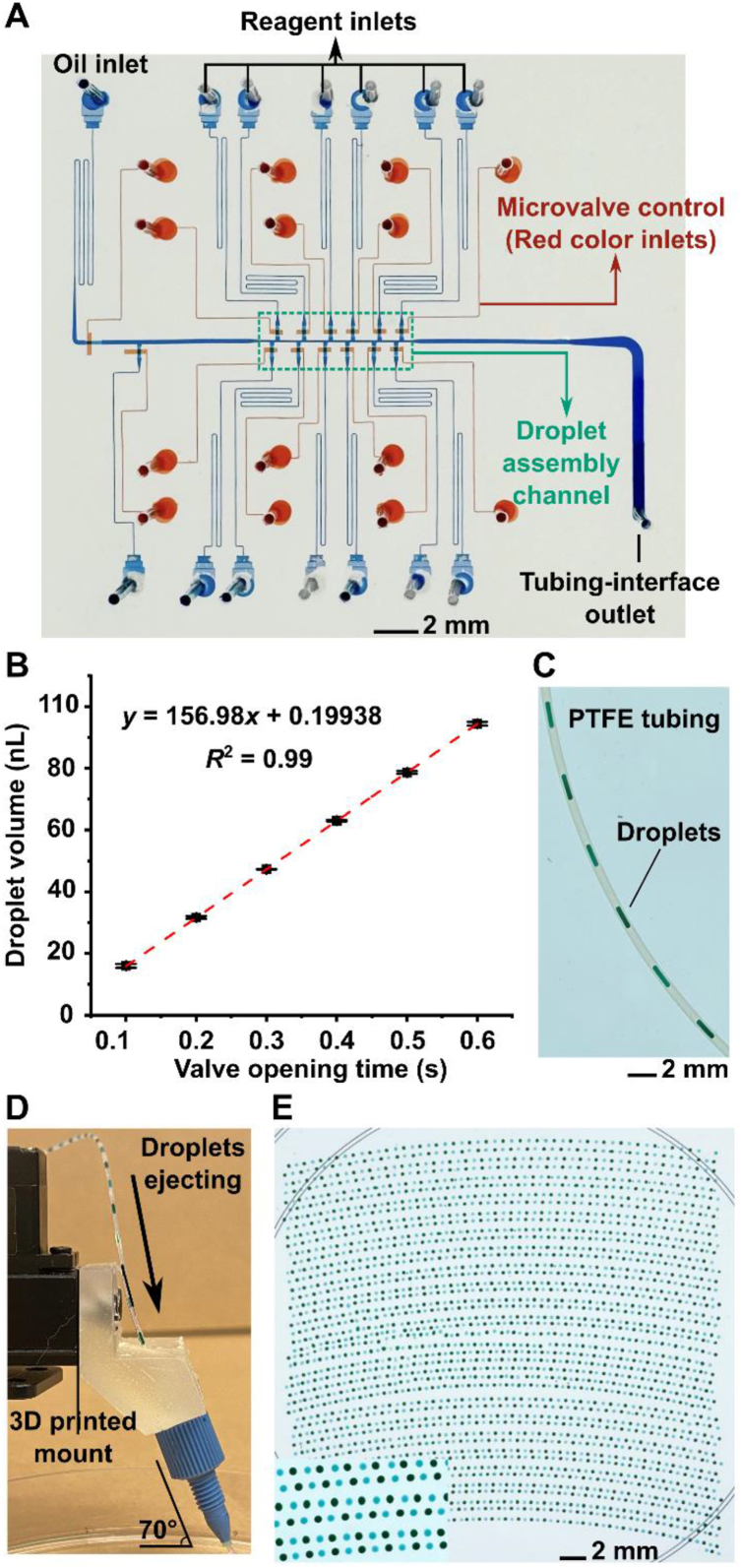
We first designed the pneumatic valve-based microfluidic device and calibrated the valve opening time and the droplet volume to ensure precise control of the droplet composition. Similar to our previous devices16,56, 57, 58, 59, this microfluidic device adopts two-layer architecture, where the bottom valve layer consists of a set of microvalves to control the fluid flow in the top fluidic layer (Fig. 2A). More specifically, the microvalve of the microfluidic device contains a thin PDMS membrane controlled by an external pressure source. When pressure is turned on, the microvalve is closed such that the thin membrane is deflected upward to seal the fluid flow; when pressure is turned off, the microvalve is opened such that the thin membrane is relaxed to allow the fluid flow in the top fluidic layer (Supporting Information Fig. S2). By turning on and off the microvalve, droplet will be assembled in the 100-μm droplet assembly channel, where an upstream oil inlet pushes the droplet to the tubing-interface outlet (Fig. 2A).
Figure 2.
Characterization of RoboDrop. (A) The pneumatic valve-based nanodroplet microfluidic device in RoboDrop is composed of two PDMS layers: the top fluidic layer where droplets are generated (filled with blue dye for visualization) and the bottom valve layer where microvalves are actuated to regulate droplet generation and flow (filled with red dye for visualization). Droplet assembly is conducted in the 100-μm wide droplet assembly channel and an upstream oil inlet introduces the oil phase to push the generated droplets downstream into the tubing-interface outlet. (B) In order to precisely control the volume of each component in the droplet with high precision, the device has been first calibrated by correlating the microvalve opening time and the droplet volume. (C) Upon droplet assembly, the droplets were transported into a PTFE tubing, visualized with droplets composed of food dye. (D) In RoboDop, the PTFE tubing was interfaced with the robotic arm and fixed by a 3D-printed mount with a tubing fixer. The 70° angle of the tubing fixer and the tubing is to ensure smooth transportation of the droplets. (E) Automatic printing of around 2700 droplets composed of green and blue food dyes alternately onto a single petri dish. Data presented in (A) as mean ± SD, n = 3.
To demonstrate the programmable control of the microfluidic device, we utilized water and two distinct food dyes as reagents to form droplets with different compositions. The composition of each droplet was controlled by regulating the number of food dye injections into the water (Supporting Information Video 1). Furthermore, we quantitively analyzed the relationship between the droplet volume and the opening time of pneumatic valves (Fig. 2B). In particular, the droplets were generated using a fluorescence dye and the pneumatic valve opening time was 0.1–0.6 s with a 0.1 s increment, which is the duration of our particular interest as appropriate droplet volumes could be achieved for subsequent drug screening experiments. The droplets were detected by a laser-induced fluorescence (LIF) detection system60 and the volume of the droplets was measured using a reference droplet. The results suggested a great linearity (R2 = 1) between the droplet volume and the valve opening time, demonstrating the precise control of individual reagents and thus the composition of reagents in droplets. Based on this result, a total of 0.48 s of opening time was consistently applied to generate 75-nL droplets in the following experiments if not otherwise mentioned.
The following is the supplementary data related to this article.
2
3.3. Droplet transportation and robotic arm control
The generated droplets from the microfluidic device were sequentially transported into a PTFE tubing that is interfaced and fixed with a custom 3D mount on the robotic arm for automated operation. PTFE tubing was chosen as the droplet transportation media due to its transparency for visualization and high chemical resistance. The droplets composed of food dye were generated to visualize the robust droplet transportation in the tubing (Fig. 2C), where we have previously demonstrated the reliable droplet transition from device to tubing35. Then, the outlet of the tubing was fixed on an affordable robotic arm using a 3D-printed customized mount. In particular, the 3D mount was designed with holes to screw onto the robotic arm, with a top hole to insert the tubing, and a hollow pillar to tighten a commercial tubing fixer (fingertight one-piece fitting) that can fix the PTFE tubing in place (Supporting Information Fig. S3A). The hollow pillar was designed with a 70° angle, resulting in the same angle for the tubing fixer and the PTFE tubing. Such design not only allowed direct contact between the tubing and the substrate but also accommodated spacing for droplet transition, which thereby ensured the smooth transportation of the droplets (Fig. 2D and Fig. S3B).
The robotic arm was utilized to automate the movement of the PTFE tubing for sequential droplet printing onto a petri dish. A Python program was designed to control the robotic arm's movement based on the input of the petri dish size and initial positional calibration. This program allowed for programmable printing of droplets into an array on the petri dish. Once the program started, the robotic arm continuously moved the tubing on the designated printing area in a serpentine shape and constant speed, where the ejected droplets are printed onto the petri dish individually and orderly. RoboDrop utilizes spatial barcoding and location information to accurately retrieve the specific antibiotic combination within each droplet (Supporting Information Fig. S4).
3.4. Contact printing array
The choice of petri dish as the substrate in RoboDrop is based on several advantages such as low cost, commercial availability, and compatibility with biological assay. To ensure the printed droplets stay in place and avoid evaporation, the petri dish was hydrophilized and filled with cover oil prior to the printing. Compared to arrays in physically predefined microreactors (e.g., microwells36,61,62), RoboDrop directly prints the droplets onto a flat petri dish surface without any additional complex substrate design and sophisticated microfabrication steps. Moreover, this design eliminates active feedback control needed to synchronize the droplet generation and robotic arm movement and vertical movement of the robotic arm between different droplet spots, further simplifying the overall system. In addition, to prevent droplet evaporation for assays that require long incubation times or high temperature, the petri dish was filled with FC-40 cover oil, the same oil as the droplet carrier oil. FC-40 oil was chosen for its high oxygen solubility and diffusion coefficient, enabling the unrestricted diffusion of oxygen into the droplets during the bacterial incubation63, 64, 65.
RoboDrop's customizability and flexibility were demonstrated by controlling the droplet array density through adjusting the oil injection time between the droplets and the ability to print thousands of droplets on a single petri dish. By setting the robotic arm to move at its minimum moving speed, we generated two groups of droplets composed of food dye with different oil injection time, 5 and 15 s, and showed the droplet array density decreased with oil injection time (Supporting Information Fig. S5). To demonstrate the high array density required for high-throughput screening, we utilized 3-s oil injection time between adjacent droplets and showed the capacity of automatically printing 2700 droplets composed of two colors of food dyes on a single petri dish (Fig. 2E and Supporting Information Video 2). In addition, the alternative colors of the droplets suggested the programmability of droplet compositions using RoboDrop. The curvature of the printed array indicates the precision limit of the chosen affordable robotic arm, which can be lifted with a higher-precision arm based on user needs.
RoboDrop offers the advantage of volume reduction and scalability, making it suitable for high-throughput screening of a large number of conditions. For instance, if we assume 2700 droplets were generated via RoboDrop on a single petri dish, this is equivalent to twenty-nine 96-well plates or eight 384-well plates. The volume requirement of the droplets can be reduced from 200 to 25 μL to 75 nL, resulting in a 2667- and 333-fold reduction, respectively (Supporting Information Table S1). To screen an even greater number of conditions, the petri dish can simply be replaced every 2700 droplets, allowing for continuous operation without the footprint limitations of a traditional static array.
3.5. Resazurin cell viability assay and on-chip optimization
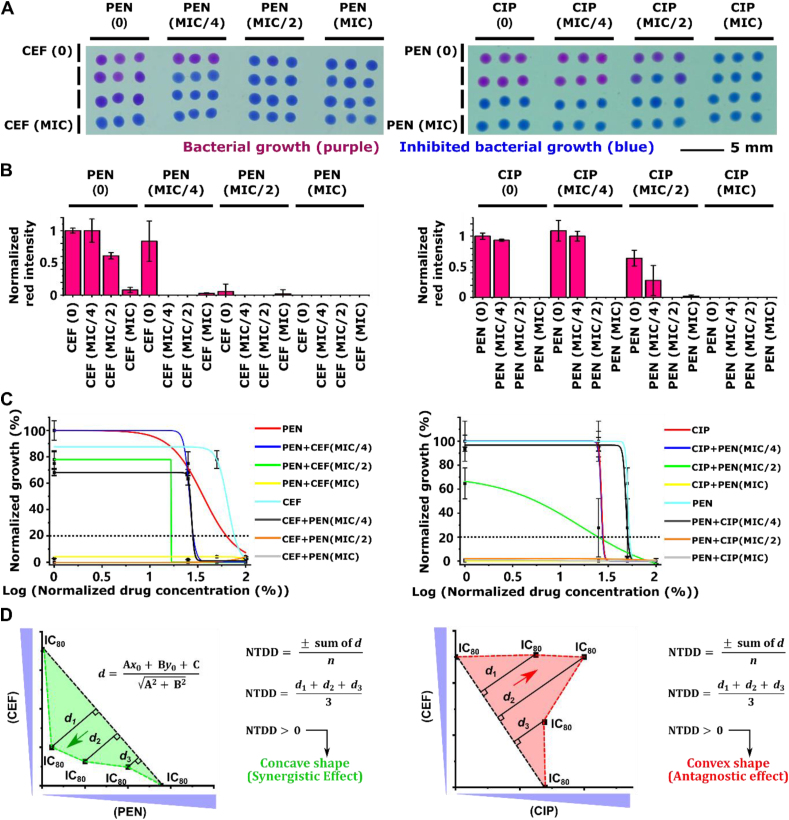
A resazurin-based cell viability assay has been implemented within RoboDrop to screen antibiotic combinations. The mechanism of this assay is that the resazurin can be reduced by NAHD secreted from viable bacterial cells to resorufin, leading to a colorimetric change from blue to purple (Fig. 3A). As the initial demonstration, we generated droplets containing the reference strain of E. coli (ATCC 25922) either with CEF (at its minimum inhibitory concentration, MIC) or without CEF and then detected the E. coli viability based on the colorimetric change between two groups of droplets, with purple indicating bacterial growth and blue indicating inhibited bacterial growth (Fig. 3B, left). For quantitative analysis, the droplet images were split into red, green, and blue channels, where the red channel showed the highest signal-to-noise ratio (SNR) and used throughout this work (Fig. 3B, right; Supporting Information Fig. S6).
Figure 3.
Resazurin-based cell viability assay and on-chip optimization. (A) The mechanism of resazurin-based cell viability assay is that the blue resazurin can be reduced by NADH secreted from viable bacteria cells, E. coli (ATCC 25922) in this study, into purple resorufin. (B) The color change can be used as the criteria to measure bacterial growth, where blue color indicates inhibited growth. (C) The colorimetric analysis of two groups of droplets with and without CEF at its MIC concentration split the raw image into red, green, and blue channels. Red channel among them showed the highest signal-to-noise ratio (SNR) (n = 3). The bacteria concentrations, 5 × 106, 5 × 107, and 5 × 108 CFU/mL, and resazurin concentration, 100, 300, 500 μmol/L, has been screened (n = 3), and 5 × 106 CFU/mL of bacteria with 500 μmol/L of resazurin showed the highest SNR and were used as the final concentration throughout the experiment. Data presented in (B) and (C) as mean ± SD, n = 3.
The resazurin and bacteria concentration has also been optimized and screened with the highest signal-to-noise ratio (SNR). More specifically, we screened three bacteria concentrations, including 5 × 106, 5 × 107, and 5 × 108 CFU/mL and three resazurin concentrations, including 100, 300 and 500 μmol/L, with and without CEF at its MIC concentration, incubated at 37 °C for 6 h as the standard condition. Based on the red channel's intensity change at 6 h, the combination of 5 × 106 CFU/mL of E. coli bacteria and 500 μmol/L resazurin showed the highest SNR (Fig. 3C). To better understand the colorimetric changes in the early stage of the assay, we further collected multi-timepoint measurements at every 2 h. This was done to account for the possibility that a high bacterial concentration might plateau the red channel signal earlier than 6 h. Our results showed that 5 × 106 CFU/mL of E. coli bacteria and 500 μmol/L resazurin consistently showed the highest SNR throughout the entire 6 h of incubation (Supporting Information Fig. S7). The time-lapse measurement of our droplet array allowed us to dynamically optimize and verify the assay condition. This approach is challenging for traditional continuous flow droplet chips66, 67, 68, highlighting the advantages of our system.
3.6. Pairwise drug verification via RoboDrop
We first verified that RoboDrop can correctly identify known synergistic and antagonistic pairs against model E. coli. The synergistic pair of CEF and PEN23,35,69 has been previously reported and the antagonistic pair of PEN and CIP has been verified in house (Supporting Information Fig. S8). Among the three antibiotics, PEN and CEF bind the beta-lactam to inhibit cell wall formation, whereas CIP inhibits DNA replication by targeting the bacterial DNA topoisomerase and DNA-gyrase70, 71, 72. In this experiment, 16 combinations among 0, MIC/4, MIC/2, and MIC of each drug were screened for both drug pairs. Each combination was generated in triplicate droplets, which were printed onto the petri dish manually and subsequently incubated (Fig. 4A). We plotted the normalized intensity of each combination for both drug pairs (Fig. 4B). The signal was normalized between the droplets with no drug and droplets with drug combinations at MIC.
Figure 4.
Pairwise drug verification via RoboDrop. (A) For both synergistic drug pair (CEF and PEN, left column) and antagonistic drug pair (CEF and CIP, right column), 16 different compositions among 0, MIC/4, MIC/2, and MIC have been screened against E. coli (ATCC 25922), with triplicate droplets for each composition. (B) During the colorimetric analysis, the red channel intensity was plotted and normalized to the intensity of the droplets with no drug and the droplets with drug combinations at MIC. (C) Subgroups of the droplet signal were fitted using dose-response curve model to analyze the IC80 values (i.e. the cross-points between the dot line and the fitted curves), where each subgroup fixes one antibiotic dose and vary the other antibiotics' dose. (D) The IC80 values of the subgroups in each drug pair (n = 3) are plotted on the x-y axis. The IC80 values of individual drugs are connected to as the reference line to represent the cutoff for synergistic or antagonistic effect (black dotted line) and the colored dotted line connects all IC80 values. Based on the normalized total distance with direction (NTDD, see Methods for calculation) calculation, positive value of NTDD represents concave shape of the line indicating synergistic interaction and negative value of NTDD represents convex shape of the line indicating antagonistic interactions. Both result matches with the previously reported interactions35 and bulk results (Fig. S6), respectively. Data presented in (C) as mean, n = 3.
For both synergistic and antagonistic drug pairs, 8 subgroups of the droplet colorimetric signal have been fitted using the dose-response curve model to analyze the IC80 values (i.e., the cross point between the dotted line and the fitted curves; drug concentration required to inhibit 80% of bacterial growth) and evaluate the combinatorial effect, where the dose of one antibiotic was fixed and the other was analyzed in each subgroup (Fig. 4C). IC80 values were chosen to evaluate the drug combination's effectiveness in inhibiting bacterial growth, as it is a commonly used metric in drug screening studies35,73, 74, 75. There were 8 subgroups in total as we tested two antibiotics and each antibiotic could be paired with the other, resulting in a total of 4 subgroups per antibiotics. By plotting the IC80 values of the subgroups in the x-y axis, we connected the IC80 values of independent drugs as the individual IC80 values reference line representing the cutoff for synergistic or antagonistic effects (referred hereafter as “reference line”, depicted as the black dotted line in Fig. 4D) and all IC80 values with colored dotted line. The shape of the connected IC80 values line was used to evaluate the combinatorial effects, where concave shape indicates synergistic effects and convex shape indicates antagonistic effects. More specifically, a concave shape means that for the same amount of inhibition, the concentration required for two synergistic drugs is lower than the required concentration of each single drug, and the opposite for a convex shape. A definition of normalized total distance with direction (NTDD, see Methods for calculation) was proposed for each IC80 value with respect to the reference line to quantitively classify the two interactions. The positivity or negativity of the NTDD dictates the overall direction of the points with respect to the reference line, with positive value indicating concave shape and negative value indicating convex shape (Fig. 4D). These results demonstrated that RoboDrop can correctly identify synergistic and antagonistic drug pairs.
3.7. Screening of three-drug combinations via RoboDrop
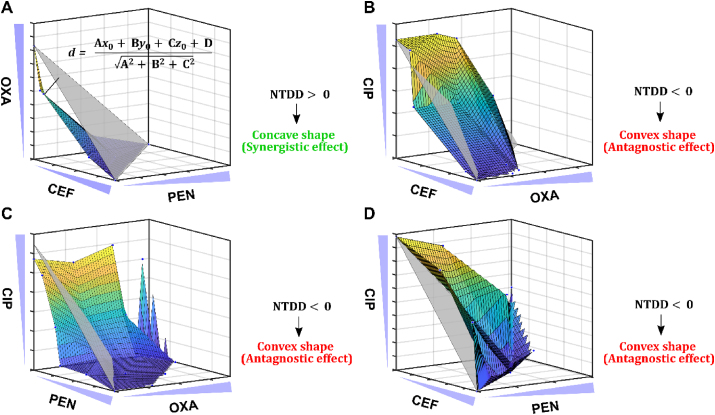
We employed RoboDrop to screen three-drug combinations among four antibiotics (PEN, CEF, OXA, and CIP) to treat a model E. coli strain (ATCC 25922). The choice of PEN, CEF, and OXA is based on their classification as beta-lactam antibiotics, which were selected based on the previously reported hypothesis that antibiotics with similar target of interest tended to show synergistic effects23,76,77. The addition of CIP, fluoroquinolone antibiotic, allowed us to explore the possibility of synergistic effects when an antibiotic from a different category was introduced. Using the four antibiotics, four unique three-drug combinations, OXA + CEF + PEN, CIP + CEF + OXA, CIP + PEN + OXA, CIP + PEN + CEF, were screened with 64 combinations among 0, MIC/4, MIC/2, and MIC of each drug (Supporting Information Fig. S9). For each three-drug combination, 64 droplets with different compositions were printed onto the petri dish and incubated and normalized red channel intensity was plotted on the x-y axis (Supporting Information Fig. S10). Based on the normalized colorimetric signal, 48 subgroups of the droplet signal have been fitted using the dose-response curve model to analyze the IC80 values to evaluate the combinatorial effect, where two antibiotic doses were fixed and the remaining one was analyzed in each subgroup. There were 48 subgroups because each antibiotic could be paired with two other antibiotics, resulting in a total of 16 subgroups per antibiotics, and we tested three antibiotics in total.
After plotting the IC80 values on the x-y-z axis, the IC80 values of the individual drugs were connected as the individual IC80 values reference plane representing the cutoff for synergistic or antagonistic effects (referred hereafter as “reference plane”, depicted as the grey plane in Fig. 5), and all IC80 values were connected into a colored surface. Similarly, we used NTDD to quantitively evaluate the shape of the colored surface and classify the combinatorial effect, with positive NTDD represent concave shape indicating synergistic effects and negative NTDD represents convex shape indicating antagonistic effects. Among the four unique drug combinations, OXA + CEF + PEN was the only combination that showed synergistic effects with a concave surface, indicating the lower dosage requirement with the three synergistic drugs compared with the dosage requirement of each drug to have the same efficacy of bacterial inhibition (Fig. 5A). The rest three drug combinations, CIP + CEF + OXA, CIP + PEN + OXA, CIP + PEN + CEF, all showed antagonistic effects with convex surface (Fig. 5B–D). To better illustrate our finding, we further analyzed all the sub-two-drug combinations among the four unique three-drug combinations. We found that CIP showed antagonistic effects with all other three drugs, most likely because CIP is in a different category from the others. This finding is consistent with previously reported hypothesis23,76,77 where the drugs from the same category are more likely to show synergistic effects but drugs in different categories are more likely antagonistic. Similar phenomenon applies to the three-drug combinations, as CIP is in all antagonistic three-drug combinations. The matching sub-two-drug combinations effects (same-color mark) further supports the robustness and replicability of our RoboDrop (Supporting Information Fig. S11). We currently focus on the reference strain of bacteria for concept demonstration, but recent studies have shown promising results in the discovery of antibiotic combinations to combat drug-resistant strains of bacteria78,79. We envision that our methodology can be extended to identify synergistic antibiotic combinations targeting drug-resistant strains.
Figure 5.
Screening of three-drug combinations via RoboDrop. The IC80 values of all four unique three-drug combinations (n = 3), (A) OXA + CEF + PEN, (B) CIP + CEF + OXA, (C) CIP + PEN + OXA, (D) CIP + PEN + CEF, are plotted in x-y-z axis. The reference plane (grey plane) connects the IC80 values of individual drugs, and the colored surface plane connects all IC80 values of each three-drug combination. Based on the NTDD calculation, the positive NTDD indicates a concave shape (A) therefore a synergistic effect and the negative NTDD indicates a convex shape (B, C, D) therefore an antagonistic effect. Data presented as mean, n = 3.
4. Conclusions
In this work, we developed Robotic-Printed Combinatorial Droplet (RoboDrop) system that achieved screening of antibiotic combinations with small sample/reagent consumption, automation, high-throughput, and high scalability. RoboDrop utilizes a pneumatic valve-based droplet system to programmatically generate nanoliter droplets with desired compositions, which are then transported into PTFE tubing. The tubing is fixed on an affordable robotic arm that can move the tubing and print the droplets on a designated area of a petri dish. The printed droplets on the petri dish were captured by a commercial camera and colorimetric analysis was utilized to analyze the raw images. Then, we demonstrated RoboDrop by integrating it with resazurin-based bacterial viability assay for combinatorial screening, where we verified two known antibiotics combinations and found a new three-drug combination to treat a model E. coli (ATCC 25922) strain, collectively suggesting the robustness and reliability of RoboDrop.
The integration of the resazurin-based cell viability assay with RoboDrop has demonstrated the feasibility for screening of antibiotic combinations. In this work, we chose colorimetric readout with a commercial camera for simplicity, but we foresee that fluorescence detection of resazurin-based assay35,80,81 can be readily achieved by adding the appropriate excitation source and emission camera filter to RoboDrop. Furthermore, RoboDrop can also be utilized with other types of staining-based antibiotic combinatorial screening assay36,41,46,49,82, and other readouts83 such as luminescent and plasmonic readouts. Moreover, the ability of RoboDrop to perform time-lapse measurements enabled us to dynamically optimize and verify the assay conditions, thus demonstrating a superior performance compared to traditional continuous flow droplet chips66, 67, 68. This feature may be particularly advantageous for applications that necessitate continuous monitoring of droplets, which provides accurate and comprehensive understanding of the assay dynamics.
In conclusion, RoboDrop offers a promising platform for screening new antibiotic combinations, and we foresee some additional technical enhancements. To achieve higher throughput and array density, the choice and precision of the robotic arm must be improved, and large-scale droplet printing should be automated to eliminate the need for manual petri dish replacement. Additionally, exploring higher-order antibiotic combinations, such as four or five-drug combination, could provide a more comprehensive understanding of the antibiotic interactions. Finally, a previously reported vacuum-based sample loading system56,58 or autosampler84, 85, 86 can be integrated with RoboDrop's upstream droplet microfluidic device to further scale up the number of antibiotics that RoboDrop can screen. These technical advancements will allow RoboDrop to become a valuable tool for identifying new antibiotic combinations for antibiotic combination therapy to address the critical global health challenge posed by AMR.
Author contributions
Fangchi Shao: Conceptualization, Methodology, Software, Formal Analysis, Investigation, Visualization, Writing-Original Draft. Hui Li: Conceptualization, Methodology, Supervision, Writing-Review & Editing. Kuangwen Hsieh: Conceptualization, Methodology, Supervision, Writing-Review & Editing. Pengfei Zhang: Methodology, Software, Writing-Review & Editing. Sixuan Li: Software, Writing-Review & Editing. Tza-Huei Wang: Conceptualization, Methodology, Writing-Review & Editing, Funding acquisition.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
Research reported in this publication was financially supported by the National Institutes of Health (R01AI117032, R01AI137272, and R01AI138978, USA).
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.11.027.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Center for Disease Control and Prevention. National infection & death estimates for antimicrobial resistance. Available from: https://www.cdc.gov/drugresistance/national-estimates.html.
- 2.Aslam B., Wang W., Arshad M.I., Khurshid M., Muzammil S., Rasool M.H., et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pourmand A., Mazer Amirshahi M., Jasani G., May L. Emerging trends in antibiotic resistance: implications for emergency medicine. Am J Emerg Med. 2017;35:1172–1176. doi: 10.1016/j.ajem.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Frieri M., Kumar K., Boutin A. Antibiotic resistance. J Infect Public Health. 2017;10:369–378. doi: 10.1016/j.jiph.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 6.Tay S.B., Yew W.S. Development of quorum-based anti-virulence therapeutics targeting Gram-negative bacterial pathogens. Int J Mol Sci. 2013;14:16570–16599. doi: 10.3390/ijms140816570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LaSarre B., Federle M.J. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev. 2013;77:73–111. doi: 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilhelmelli F., Vilela N., Albuquerque P., Derengowski L., Silva-Pereira I., Kyaw C. Antibiotic development challenges: the various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front Microbiol. 2013;4:353. doi: 10.3389/fmicb.2013.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joerger R. Alternatives to antibiotics: bacteriocins, antimicrobial peptides and bacteriophages. Poultry Sci. 2003;82:640–647. doi: 10.1093/ps/82.4.640. [DOI] [PubMed] [Google Scholar]
- 10.Ghannad M.S., Mohammadi A. Bacteriophage: time to re-evaluate the potential of phage therapy as a promising agent to control multidrug-resistant bacteria. Iran J Basic Med Sci. 2012;15:693. [PMC free article] [PubMed] [Google Scholar]
- 11.Scherer B., Surrette C., Li H., Torab P., Kvam E., Galligan C., et al. Digital electrical impedance analysis for single bacterium sensing and antimicrobial susceptibility testing. Lab Chip. 2021;21:1073–1083. doi: 10.1039/d0lc00937g. [DOI] [PubMed] [Google Scholar]
- 12.Kaushik A.M., Hsieh K., Mach K.E., Lewis S., Puleo C.M., Carroll K.C., et al. Droplet-based single-cell measurements of 16S rRNA enable integrated bacteria identification and pheno-molecular antimicrobial susceptibility testing from clinical samples in 30 min. Adv Sci. 2021;8 doi: 10.1002/advs.202003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y., Gupta K., Ekinci K.L. All-electrical monitoring of bacterial antibiotic susceptibility in a microfluidic device. Proc Natl Acad Sci USA. 2020;117:10639–10644. doi: 10.1073/pnas.1922172117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Torab P., Wong P.K. Detection of bacterial infection via a fidget spinner. Nat Biomed Eng. 2020;4:577–578. doi: 10.1038/s41551-020-0571-4. [DOI] [PubMed] [Google Scholar]
- 15.Li H., Torab P., Mach K.E., Surrette C., England M.R., Craft D.W., et al. Adaptable microfluidic system for single-cell pathogen classification and antimicrobial susceptibility testing. Proc Natl Acad Sci U S A. 2019;116:10270–10279. doi: 10.1073/pnas.1819569116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P., Kaushik A.M., Hsieh K., Li S., Lewis S., Mach K.E., et al. A cascaded droplet microfluidic platform enables high-throughput single cell antibiotic susceptibility testing at scale. Small Methods. 2022;6 doi: 10.1002/smtd.202101254. [DOI] [PubMed] [Google Scholar]
- 17.Park K.S., Huang C.H., Lee K., Yoo Y.E., Castro C.M., Weissleder R., et al. Rapid identification of health care–associated infections with an integrated fluorescence anisotropy system. Sci Adv. 2016;2 doi: 10.1126/sciadv.1600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaushik A.M., Hsieh K., Chen L., Shin D.J., Liao J.C., Wang T.H. Accelerating bacterial growth detection and antimicrobial susceptibility assessment in integrated picoliter droplet platform. Biosens Bioelectron. 2017;97:260–266. doi: 10.1016/j.bios.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Athamanolap P., Hsieh K., O'Keefe C.M., Zhang Y., Yang S., Wang T.H. Nanoarray digital polymerase chain reaction with high-resolution melt for enabling broad bacteria identification and pheno–molecular antimicrobial susceptibility test. Anal Chem. 2019;91:12784–12792. doi: 10.1021/acs.analchem.9b02344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trick A.Y., Melendez J.H., Chen F.E., Chen L., Onzia A., Zawedde A., et al. A portable magnetofluidic platform for detecting sexually transmitted infections and antimicrobial susceptibility. Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abf6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehta K.C., Dargad R.R., Borade D.M., Swami O.C. Burden of antibiotic resistance in common infectious diseases: role of antibiotic combination therapy. J Clin Diagn Res. 2014;8:ME05. doi: 10.7860/JCDR/2014/8778.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tekin E., White C., Kang T.M., Singh N., Cruz Loya M., Damoiseaux R., et al. Prevalence and patterns of higher-order drug interactions in Escherichia coli. NPJ Syst Biol Appl. 2018;4:1–10. doi: 10.1038/s41540-018-0069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brochado A.R., Telzerow A., Bobonis J., Banzhaf M., Mateus A., Selkrig J., et al. Species-specific activity of antibacterial drug combinations. Nature. 2018;559:259–263. doi: 10.1038/s41586-018-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cottarel G., Wierzbowski J. Combination drugs, an emerging option for antibacterial therapy. Trends Biotechnol. 2007;25:547–555. doi: 10.1016/j.tibtech.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Goldberg D.E., Siliciano R.F., Jacobs W.R., Jr. Outwitting evolution: fighting drug-resistant TB, malaria, and HIV. Cell. 2012;148:1271–1283. doi: 10.1016/j.cell.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo L.C., Lai C.C., Liao C.H., Hsu C.K., Chang Y.L., Chang C.Y., et al. Multidrug-resistant Acinetobacter baumannii bacteraemia: clinical features, antimicrobial therapy and outcome. Clin Microbiol Infect. 2007;13:196–198. doi: 10.1111/j.1469-0691.2006.01601.x. [DOI] [PubMed] [Google Scholar]
- 27.Motaouakkil S., Charra B., Hachimi A., Nejmi H., Benslama A., Elmdaghri N., et al. Colistin and rifampicin in the treatment of nosocomial infections from multiresistant Acinetobacter baumannii. J Infect. 2006;53:274–278. doi: 10.1016/j.jinf.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Tängdén T. Combination antibiotic therapy for multidrug-resistant Gram-negative bacteria. Ups J Med Sci. 2014;119:149–153. doi: 10.3109/03009734.2014.899279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirsch E.B., Tam V.H. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J Antimicrob Chemother. 2010;65:1119–1125. doi: 10.1093/jac/dkq108. [DOI] [PubMed] [Google Scholar]
- 30.Qureshi Z.A., Paterson D.L., Potoski B.A., Kilayko M.C., Sandovsky G., Sordillo E., et al. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob Agents Chemother. 2012;56:2108–2113. doi: 10.1128/AAC.06268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh P.J., Hegreness M.J., Aiden A.P., Kishony R. Drug interactions and the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7:460–466. doi: 10.1038/nrmicro2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cokol M. Drugs and their interactions. Curr Drug Discov Technol. 2013;10:106–113. doi: 10.2174/1570163811310020003. [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann G.R., Lehar J., Keith C.T. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today. 2007;12:34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen C., Zhou A., Khan A., Miller J.H., Yeh P. Pairwise antibiotic interactions in Escherichia coli: triclosan, rifampicin and aztreonam with nine other classes of antibiotics. J Antibiot. 2016;69:791–797. doi: 10.1038/ja.2016.26. [DOI] [PubMed] [Google Scholar]
- 35.Li H., Zhang P., Hsieh K., Wang T.H. Combinatorial nanodroplet platform for screening antibiotic combinations. Lab Chip. 2022;22:621–631. doi: 10.1039/d1lc00865j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulesa A., Kehe J., Hurtado J.E., Tawde P., Blainey P.C. Combinatorial drug discovery in nanoliter droplets. Proc Natl Acad Sci U S A. 2018;115:6685–6690. doi: 10.1073/pnas.1802233115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Y., Huang H.L., Ye X.Q., Cai D.T., Fang J.T., Sun J., et al. Synergistic potential of antimicrobial combinations against methicillin-resistant Staphylococcus aureus. Front Microbiol. 2020;11:1919. doi: 10.3389/fmicb.2020.01919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu M., Tse M.W., Weller J., Chen J., Blainey P.C. The future of antibiotics begins with discovering new combinations. Ann N Y Acad Sci. 2021;1496:82–96. doi: 10.1111/nyas.14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood K.B. Pairwise interactions and the battle against combinatorics in multidrug therapies. Proc Natl Acad Sci U S A. 2016;113:10231–10233. doi: 10.1073/pnas.1612365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foucquier J., Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3 doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J., Taylor D., Agrawal N., Wang H., Kim H., Han A., et al. A programmable microfluidic cell array for combinatorial drug screening. Lab Chip. 2012;12:1813–1822. doi: 10.1039/c2lc21202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Opalski A.S., Ruszczak A., Promovych Y., Horka M., Derzsi L., Garstecki P. Combinatorial antimicrobial susceptibility testing enabled by non-contact printing. Micromachines. 2020;11:142. doi: 10.3390/mi11020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brennan Krohn T., Truelson K.A., Smith K.P., Kirby J.E. Screening for synergistic activity of antimicrobial combinations against carbapenem-resistant Enterobacteriaceae using inkjet printer-based technology. J Antimicrob Chemother. 2017;72:2775–2781. doi: 10.1093/jac/dkx241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu L., Chen H., Canales M., Ciric L. Use of synthesized double-stranded gene fragments as qPCR standards for the quantification of antibiotic resistance genes. J Microbiol Methods. 2019;164 doi: 10.1016/j.mimet.2019.105670. [DOI] [PubMed] [Google Scholar]
- 45.Zhai J., Li C., Li H., Yi S., Yang N., Miao K., et al. Cancer drug screening with an on-chip multi-drug dispenser in digital microfluidics. Lab Chip. 2021;21:4749–4759. doi: 10.1039/d1lc00895a. [DOI] [PubMed] [Google Scholar]
- 46.Li J., Tan W., Xiao W., Carney R.P., Men Y., Li Y., et al. A plug-and-play, drug-on-pillar platform for combination drug screening implemented by microfluidic adaptive printing. Anal Chem. 2018;90:13969–13977. doi: 10.1021/acs.analchem.8b03456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schudel B.R., Choi C.J., Cunningham B.T., Kenis P.J. Microfluidic chip for combinatorial mixing and screening of assays. Lab Chip. 2009;9:1676–1680. doi: 10.1039/b901999e. [DOI] [PubMed] [Google Scholar]
- 48.Shao F., Lee P.W., Li H., Hsieh K., Wang T.H. Emerging platforms for high-throughput enzymatic bioassays. Trends Biotechnol. 2023;41:120–133. doi: 10.1016/j.tibtech.2022.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuster B., Junkin M., Kashaf S.S., Romero-Calvo I., Kirby K., Matthews J., et al. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-020-19058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Z., Li W., Choi G., Yang X., Miao J., Cui L., et al. Arbitrarily accessible 3D microfluidic device for combinatorial high-throughput drug screening. Sensors. 2016;16:1616. doi: 10.3390/s16101616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svensson C.M., Shvydkiv O., Dietrich S., Mahler L., Weber T., Choudhary M., et al. Coding of experimental conditions in microfluidic droplet assays using colored beads and machine learning supported image analysis. Small. 2019;15 doi: 10.1002/smll.201802384. [DOI] [PubMed] [Google Scholar]
- 52.Kehe J., Kulesa A., Ortiz A., Ackerman C.M., Thakku S.G., Sellers D., et al. Massively parallel screening of synthetic microbial communities. Proc Natl Acad Sci U S A. 2019;116:12804–12809. doi: 10.1073/pnas.1900102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Churski K., Kaminski T.S., Jakiela S., Kamysz W., Baranska Rybak W., Weibel D.B., et al. Rapid screening of antibiotic toxicity in an automated microdroplet system. Lab Chip. 2012;12:1629–1637. doi: 10.1039/c2lc21284f. [DOI] [PubMed] [Google Scholar]
- 54.Boedicker J.Q., Li L., Kline T.R., Ismagilov R.F. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip. 2008;8:1265–1272. doi: 10.1039/b804911d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eduati F., Utharala R., Madhavan D., Neumann U.P., Longerich T., Cramer T., et al. A microfluidics platform for combinatorial drug screening on cancer biopsies. Nat Commun. 2018;9:1–13. doi: 10.1038/s41467-018-04919-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zec H.C., Zheng T., Liu L., Hsieh K., Rane T.D., Pederson T., et al. Programmable microfluidic genotyping of plant DNA samples for marker-assisted selection. Microsyst Nanoeng. 2018;4:1–10. [Google Scholar]
- 57.Rane T.D., Zec H.C., Wang T.H. A barcode-free combinatorial screening platform for matrix metalloproteinase screening. Anal Chem. 2015;87:1950–1956. doi: 10.1021/ac504330x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao F., Hsieh K., Zhang P., Kaushik A.M., Wang T.H. Facile and scalable tubing-free sample loading for droplet microfluidics. Sci Rep. 2022;12:1–12. doi: 10.1038/s41598-022-17352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang P., Kaushik A., Hsieh K., Wang T.H. Customizing droplet contents and dynamic ranges via integrated programmable picodroplet assembler. Microsyst Nanoeng. 2019;5:1–12. doi: 10.1038/s41378-019-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rane T.D., Zec H.C., Puleo C., Lee A.P., Wang T.H. Droplet microfluidics for amplification-free genetic detection of single cells. Lab Chip. 2012;12:3341–3347. doi: 10.1039/c2lc40537g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei Y., Zhu Y., Fang Q. Nanoliter quantitative high-throughput screening with large-scale tunable gradients based on a microfluidic droplet robot under unilateral dispersion mode. Anal Chem. 2019;91:4995–5003. doi: 10.1021/acs.analchem.8b04564. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y., Wang T.H. Rapid generation of chemical combinations on a magnetic digital microfluidic array. RSC Adv. 2019;9:21741–21747. doi: 10.1039/c9ra03469b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sklodowska K., Jakiela S. Enhancement of bacterial growth with the help of immiscible oxygenated oils. RSC Adv. 2017;7:40990–40995. [Google Scholar]
- 64.Soitu C., Feuerborn A., Tan A.N., Walker H., Walsh P.A., Castrejón-Pita A.A., et al. Microfluidic chambers using fluid walls for cell biology. Proc Natl Acad Sci U S A. 2018;115:E5926–E5933. doi: 10.1073/pnas.1805449115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Junker B.H., Hatton T.A., Wang D.I. Oxygen transfer enhancement in aqueous/perfluorocarbon fermentation systems: I. Experimental observations. Biotechnol Bioeng. 1990;35:578–585. doi: 10.1002/bit.260350605. [DOI] [PubMed] [Google Scholar]
- 66.Shang L., Cheng Y., Zhao Y. Emerging droplet microfluidics. Chem Rev. 2017;117:7964–8040. doi: 10.1021/acs.chemrev.6b00848. [DOI] [PubMed] [Google Scholar]
- 67.Kulkarni M.B., Goel S. Advances in continuous-flow based microfluidic PCR devices—a review. Eng Res Express. 2020;2 [Google Scholar]
- 68.Zhang Y., Jiang H.R. A review on continuous-flow microfluidic PCR in droplets: advances, challenges and future. Anal Chim Acta. 2016;914:7–16. doi: 10.1016/j.aca.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 69.Chandrasekaran S., Cokol Cakmak M., Sahin N., Yilancioglu K., Kazan H., Collins J.J., et al. Chemogenomics and orthology-based design of antibiotic combination therapies. Mol Syst Biol. 2016;12:872. doi: 10.15252/msb.20156777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tomasz A. The mechanism of the irreversible antimicrobial effects of penicillins: how the beta-lactam antibiotics kill and lyse bacteria. Annu Rev Microbiol. 1979;33:113–137. doi: 10.1146/annurev.mi.33.100179.000553. [DOI] [PubMed] [Google Scholar]
- 71.LeBel M. Ciprofloxacin: chemistry, mechanism of action, resistance, antimicrobial spectrum, pharmacokinetics, clinical trials, and adverse reactions. Pharmacotherapy. 1988;8:3–30. doi: 10.1002/j.1875-9114.1988.tb04058.x. [DOI] [PubMed] [Google Scholar]
- 72.Wright D.B. Cefsulodin. Drug Intell Clin Pharm. 1986;20:845–849. doi: 10.1177/106002808602001104. [DOI] [PubMed] [Google Scholar]
- 73.Russ D., Kishony R. Additivity of inhibitory effects in multidrug combinations. Nat Microbiol. 2018;3:1339–1345. doi: 10.1038/s41564-018-0252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Houdkova M., Chaure A., Doskocil I., Havlik J., Kokoska L. New broth macrodilution volatilization method for antibacterial susceptibility testing of volatile agents and evaluation of their toxicity using modified MTT assay in vitro. Molecules. 2021;26:4179. doi: 10.3390/molecules26144179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kudera T., Doskocil I., Salmonova H., Petrtyl M., Skrivanova E., Kokoska L. In vitro selective growth-inhibitory activities of phytochemicals, synthetic phytochemical analogs, and antibiotics against diarrheagenic/probiotic bacteria and cancer/normal intestinal cells. Pharmaceuticals. 2020;13:233. doi: 10.3390/ph13090233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng F., Kovács I.A., Barabási A.-L. Network-based prediction of drug combinations. Nat Commun. 2019;10:1–11. doi: 10.1038/s41467-019-09186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin N., Ma W., Pei J., Ouyang Q., Tang C., Lai L. Synergistic and antagonistic drug combinations depend on network topology. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singh N., Yeh P.J. Suppressive drug combinations and their potential to combat antibiotic resistance. J Antibiot. 2017;70:1033–1042. doi: 10.1038/ja.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Si Z., Pethe K., Chan Park M.B. Chemical basis of combination therapy to combat antibiotic resistance. JACS Au. 2023;3:276–292. doi: 10.1021/jacsau.2c00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hsieh K., Zec H.C., Chen L., Kaushik A.M., Mach K.E., Liao J.C., et al. Simple and precise counting of viable bacteria by resazurin-amplified picoarray detection. Anal Chem. 2018;90:9449–9456. doi: 10.1021/acs.analchem.8b02096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen F.-E., Kaushik A., Hsieh K., Chang E., Chen L., Zhang P., et al. Toward decentralizing antibiotic susceptibility testing via ready-to-use microwell array and resazurin-aided colorimetric readout. Anal Chem. 2020;93:1260–1265. doi: 10.1021/acs.analchem.0c04095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kilinc D., Schwab J., Rampini S., Ikpekha O.W., Thampi A., Blasiak A., et al. A microfluidic dual gradient generator for conducting cell-based drug combination assays. Integr Biol. 2016;8:39–49. doi: 10.1039/c5ib00209e. [DOI] [PubMed] [Google Scholar]
- 83.Chen W., Shao F., Xianyu Y. Microfluidics-implemented biochemical assays: from the perspective of readout. Small. 2020;16 doi: 10.1002/smll.201903388. [DOI] [PubMed] [Google Scholar]
- 84.Rane T.D., Zec H.C., Wang T.H. A serial sample loading system: interfacing multiwell plates with microfluidic devices. J Lab Autom. 2012;17:370–377. doi: 10.1177/2211068212455169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Menezes R., Dramé Maigné A., Taly V., Rondelez Y., Gines G. Streamlined digital bioassays with a 3D printed sample changer. Analyst. 2020;145:572–581. doi: 10.1039/c9an01744e. [DOI] [PubMed] [Google Scholar]
- 86.Kaminski T.S., Jakiela S., Czekalska M.A., Postek W., Garstecki P. Automated generation of libraries of nL droplets. Lab Chip. 2012;12:3995–4002. doi: 10.1039/c2lc40540g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2