Summary
Recent advancements in image-based pooled CRISPR screening have facilitated the mapping of diverse genotype-phenotype associations within mammalian cells. However, the rapid enrichment of cells based on morphological information continues to pose a challenge, constraining the capacity for large-scale gene perturbation screening across diverse high-content cellular phenotypes. In this study, we demonstrate the applicability of multimodal ghost cytometry-based cell sorting, including both fluorescent and label-free high-content phenotypes, for rapid pooled CRISPR screening within vast cell populations. Using the high-content cell sorter operating in fluorescence mode, we successfully executed kinase-specific CRISPR screening targeting genes influencing the nuclear translocation of RelA. Furthermore, using the multiparametric, label-free mode, we performed large-scale screening to identify genes involved in macrophage polarization. Notably, the label-free platform can enrich target phenotypes without requiring invasive staining, preserving untouched cells for downstream assays and expanding the potential for screening cellular phenotypes even when suitable markers are absent.
Keywords: pooled CRISPR screening, image-based cell sorter, cellular phenotyping, label-free cell analysis, machine learning, high-content cell analysis, flow cytometry
Graphical abstract

Highlights
-
•
High-content phenotyping of large-scale pooled CRISPR screens with ghost cytometry
-
•
Platform enables both label-free and fluorescence-based cell phenotyping
-
•
Kinase-targeting CRISPR screening demonstrated with fluorescence mode
-
•
Label-free screening identifies key genes in macrophage polarization
Motivation
Pooled CRISPR screening based on image information has great potential to map complex genotype-phenotype associations in mammalian cells. However, barriers to efficient cell enrichment based on diverse morphological information remain, limiting large-scale gene perturbation screening across diverse cell phenotypes. Further complications arise when appropriate biomarkers and staining techniques are not available, making phenotypic evaluation under label-free conditions a challenge. To address these challenges, this study aims to increase the feasibility and reach of pooled CRISPR screening by analyzing and sorting both fluorescence and label-free high-content cell phenotypes using a machine vision-based approach.
Tsubouchi et al. present multimodal ghost cytometry for pooled CRISPR screening. Their approach, combining both fluorescence and label-free phenotyping, allows rapid, large-scale gene perturbation analysis. This method successfully identifies genes involved in kinase signaling and macrophage polarization and significantly expands the possibilities for designing perturbation screens.
Introduction
CRISPR-based pooled screening offers several advantages, including increased throughput, reduced cost, and reduced well-to-well batch effects, over conventional array-based approaches for perturbation screening.1,2 In pooled phenotypic screening, cells and intracellular molecules have been labeled with fluorescent dyes, reporters, or immunofluorescent antibodies. Cell phenotyping typically requires quantifying explicitly defined features; thus, fluorescence-based labeling provides distinct advantages owing to its high specificity and sensitivity to the molecule of interest.3 For example, representative values such as total fluorescence are measured from temporal signals obtained in fluorescence-activated cell sorting (FACS), or more detailed features, such as molecular localization and morphologic parameters, are evaluated from optical microscopic images.4,5,6,7,8,9 However, when suitable biomarkers or staining methods are unavailable, and cell phenotypes can only be assessed without labeling, image analysis based on human-recognizable features can become challenging. To address this challenge, machine learning (ML)-based analysis of label-free high-content cell phenotypes emerges as a promising alternative.10,11 In this study, we present a versatile approach for large-scale pooled CRISPR screening, including both fluorescent and label-free high-content cell phenotypes, utilizing a cell sorter based on fluorescence and label-free ghost cytometry (GC) technologies.11,12
Central to GC is an ML-based direct and integrative analysis of cellular morphological information without image reconstruction. In GC, as depicted in Figure 1A (left), fluorescence and stain-free high-content information from cells is compressively converted into temporal signals, which we call ghost motion imaging (GMI) waveforms. This conversion occurs as cells pass through static structured light illumination in a microchannel, and optical interactions are measured using single-pixel detectors. With configurations to observe the optical interactions through different angles and paths, we simultaneously detect multiple different temporal GMI waveforms using multiple detectors. Specifically, fluorescence GMI (flGMI) waveforms excited by a 488-nm laser, as well as forward scattering GMI (fsGMI), backscattering GMI (bsGMI), diffractive GMI (dGMI), and bright-field GMI (bfGMI) waveforms generated by a 405-nm laser, are measured as analogs to their corresponding microscopic images (Figure 1A, left; STAR Methods). To develop a classifier model based on a support vector machine (SVM), we first used a training cell sample to measure multiple GMI waveforms along with ground truth labels created using molecular staining, including surface markers and functional evaluations. Using this training dataset, comprising pairs of waveforms and true labels, we defined the target high-content phenotypes and trained the SVM model (Figure 1A, center). After training, the classifier can predict the labels directly from the multiple GMI waveforms using SVM-based scoring. These SVM scores enable users to estimate sorting performance through metrics such as the precision-recall (PR) curve and the area under the receiver operating characteristic-area under the curve (ROC-AUC) score before conducting any sorting experiments (Figure 1A, right; STAR Methods). During cell sorting, the trained classifier was implemented on a field-programmable gate array (FPGA), which can judge cells in real time using only the generated GMI waveforms, facilitating subsequent cell sorting based on this assessment. This direct analysis of GMI waveforms, devoid of computationally intensive image reconstruction, enhances the speed and accuracy of high-content cell sorting during large-scale screening.
Figure 1.
The ghost cytometry (GC)-based cell sorter employs multimodal GC enabled for fast-pooled CRISPR screening of high-content fluorescent and label-free cell phenotypes
(A) Schematic illustration of the simultaneous acquisition of various cellular morphological and structural information as different ghost motion imaging (GMI) waveforms, such as flGMI, fsGMI, bsGMI, dGMI, and bfGMI, for each cell. These datasets are used to develop machine learning (ML)-based classifiers for high-content cellular phenotypes. GMI waveforms are analogs to microscopic images, enabling subcellular-resolution cell phenotyping.
(B) The workflow of pooled high-content CRISPR screening by the GC-based cell sorter. A gene knockout cell library prepared by using the CRISPR-Cas9 system is treated with compounds or reagents to induce phenotypic changes. The GC-based cell sorter, equipped with a pre-trained ML model, selectively enriches cells displaying the target high-content phenotype for downstream analysis. For gene analysis, the sgRNA regions within the isolated genomic DNAs are amplified and sequenced to identify enriched or depleted genes in response to the treatment. When live cells are used, transcriptomic analysis and cell-based functional assays are widely applicable.
Figure 1B illustrates the workflow of CRISPR-based pooled screening employing the trained classifier in the GC. Initially, cells expressing the Cas9 protein were transduced with pooled CRISPR lentiviral libraries for loss-of-function gene sets and selected for stable viral integration. Subsequently, the pooled knockout cell library was treated with compounds or reagents, manifesting diverse phenotypes. Additional assays, such as immunostaining, could be conducted if necessary. In the GC-based cell sorter, a pre-trained ML model was then applied to selectively enrich cells exhibiting the desired high-content phenotypes. Finally, the sorted cells can be subject to various biological assays, including gene analyses such as genome sequencing, protein assays, and cell-based functional analyses. In the case of standard CRISPR perturbation screening, genomic DNA is extracted from sorted cells, and regions of single guide RNAs (sgRNAs) are amplified by polymerase chain reaction (PCR), pooled, and read using commercially available next-generation sequencing (NGS) platforms to identify genes inducing the target phenotype. When live cells are sorted, transcriptomics analyses using single-cell RNA sequencing and cell-based functional assays are widely applicable.
Results
Pooled fluorescence high-content cellular phenotyping
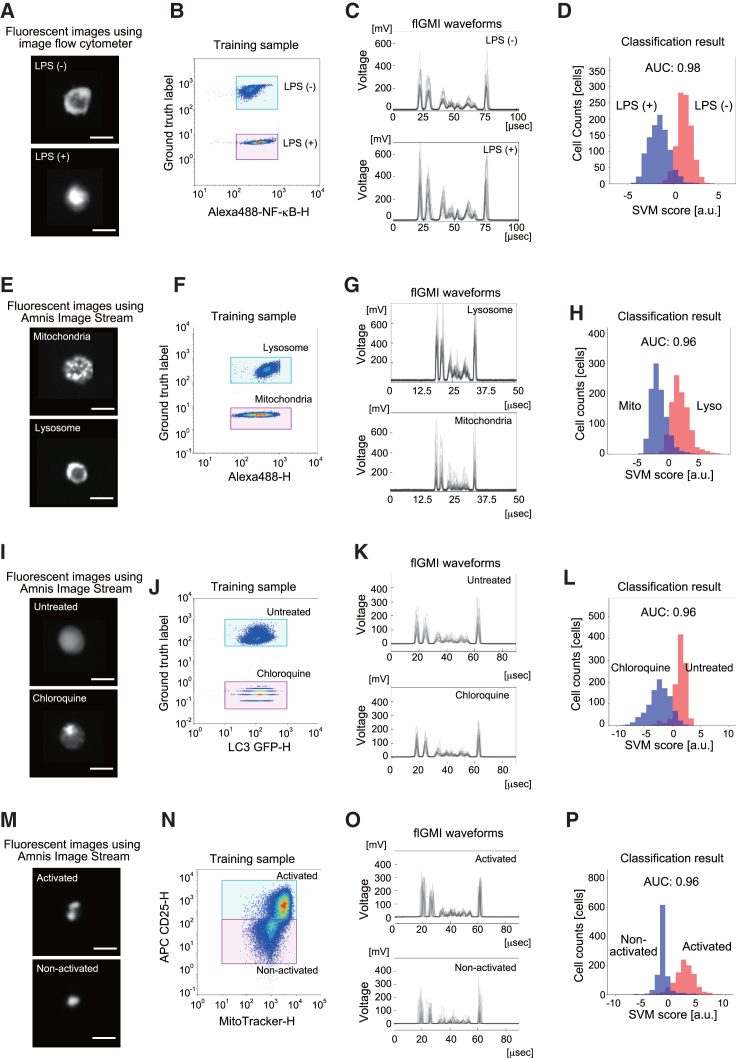
We first investigated the capability of GC to evaluate a wide range of fluorescent high-content cellular phenotypes (Figure 2). Figures 2A–2D illustrate sample images of an intracellular phenotype, specifically the nuclear translocation of RelA protein in THP-1 cells, and outlines the procedure for developing and assessing this classifier in the GC. To train the classifier for nuclear translocation, a cell mixture consisting of lipopolysaccharide (LPS)-stimulated and unstimulated THP-1 cells was prepared. These cells were stained with a combination of anti-RelA primary antibody and Alexa Fluor 488-conjugated secondary antibody. Notably, only the unstimulated cells were stained with a ground-truth marker dye before mixing.13 Note that the total fluorescence intensity for RelA staining in the training sample was similar between LPS-stimulated and unstimulated cells, rendering them challenging to distinguish using conventional FACS (Figure 2B). Consequently, an SVM-based classifier for flGMI waveforms representing nuclear translocation phenotypes was trained, using 1,250 cells for each ground-truth label. When applied to a test dataset comprising 1,000 waveforms, the histogram of returned scores exhibited bimodal peaks colored according to ground-truth labels, demonstrating robust and high performance, as indicated by an AUC score of 0.98 (Figures 2C, 2D, and S1A–S1C). Similarly, we assessed the capacity of GC to distinguish various high-content fluorescence cellular phenotypes. One such instance involves distinguishing between lysosomes and mitochondria in adherent HEK293 cells (Figures 2E–2H and S1D–S1F). Individual cells were immunostained with anti-Lamp-1 primary antibody for lysosomes and anti-complex III-core1 primary antibody for mitochondria. Subsequently, they were stained with Alexa Fluor 488-conjugated secondary antibody, with only the anti-Lamp1-stained cells being additionally marked with a ground-truth marker dye before their mixing. In all cases, the models exhibited excellent performance, achieving AUC scores of 0.96.
Figure 2.
High-content cell phenotypes distinguishable by fluorescence GC
(A–P) Fluorescence GC effectively distinguishes (A)–(D) nuclear translocation of RelA proteins in THP-1 cell suspension, (E)–(H) lysosomes (Lamp1) and mitochondria (COX III) in adherent HEK293 cells, (I)–(L) autophagosome translocation of LC3-GFP proteins in adherent HeLa cells, and (M)–(P) mitochondrial morphological changes in activated and non-activated human primary T cells.
(A), (E), (I), and (M) Representative cell images obtained using Amnis Image Stream. Scale bars, 10 μm.
(B), (F), (J), and (N) Conventional FACS scatterplots depicting the fluorescence intensity of labeling for cellular phenotyping versus that of a ground-truth marker.
(B) Fluorescence intensity of Alexa 488-labeled RelA versus that of fixable far-red dye, which exclusively labeled LPS-unstimulated cells as the ground truth.
(F) Fluorescence intensity of Alexa 488-labeled Lamp1 (lysosome) and COX III (mitochondria) proteins versus that of fixable far-red dye, which labeled only cells with Lamp1 proteins stained as ground truth.
(J) Fluorescence intensity of LC3-GFP proteins versus that of fixable far-red dye, which labeled only untreated cells as ground truth.
(N) Fluorescence intensity of MitoTracker Green versus that of CD25/CD69 double-positive cells as the ground truth.
(C), (G), (K), and (O) Representative fluorescence GMI (flGMI) waveforms of 20 randomly selected cells for each condition.
(D), (H), (L), and (P) Classification of fluorescent cell phenotypes. The ML-based classification performance is displayed as histograms of SVM scores, where red and blue colors correspond to ground-truth labels and area under the ROC curve (AUC) scores, respectively.
Additionally, we investigated LC3-GFP dynamics in adherent HeLa cells in response to autophagy induction (Figures 2I–2L and S1G–S1I). Under normal conditions, LC3 protein is evenly distributed in the cell cytoplasm and nuclei, but during autophagy induction, it aggregates into autophagosomes, forming distinct foci in the cell cytoplasm.14 Treatment with 10 μM chloroquine prevents autophagosome-lysosome fusion and degradation, leading to the accumulation of autophagosomes in the cell cytoplasm.15 To classify LC3-GFP localization, both induced and uninduced HeLa cells expressing LC3-GFP were individually prepared, and autophagosome accumulation in the uninduced cells was stained with CellTracker DeepRed dye as a ground-truth label. For classification, an equal concentration of each cell type was mixed, and 2,000 cells were randomly selected (without overlap), evenly distributed between induced and uninduced cells, and used as training data. The models consistently achieved high performance with AUC scores of 0.96.
Furthermore, we examined whether GC could differentiate between mitochondrial morphological changes in activated and non-activated human primary T cells, given the established connection between T cell fate and mitochondrial dynamics.16,17 Activated and non-activated T cells were separately prepared and stained with MitoTracker Green and the T cell activation markers CD25 and CD69. To classify activated and non-activated cells, each cell type was mixed at an equal concentration. CD25/CD69 double-positive cells represented activated cells, whereas CD25/CD69 double-negative cells represented non-activated T cell populations. For training, 2,000 cells were randomly selected (without overlap) with an equal number of induced and uninduced cells. Consistently, the models demonstrated robust performance, achieving AUC scores of 0.96 (Figures 2M–2P and S1J–S1L). These results confirm the ability of GC to classify a diverse range of high-content fluorescence phenotypes.
Pooled CRISPR screening of fluorescence high-content phenotypes
Subsequently, we focused on nuclear translocation as the target fluorescence high-content phenotype for pooled CRISPR screening. RelA translocates to the nucleus downstream of the Toll-like receptor 4 (TLR4) pathway upon activation by LPS stimulation (Figure 3A).13 In the initial evaluation, the trained classifier in the GC-based cell sorter was applied to a small-scale pooled cell library, where 60 sgRNAs, including those downstream of the TLR4 pathway, were perturbed using the CRISPR-based system. Specifically, 40 sgRNAs targeted 10 genes downstream of the TLR4 signaling pathway as positive controls, whereas 20 non-targeting sgRNAs outside of the pathway served as negative controls (Table S1). Cells exhibiting suppressed RelA nuclear translocation were sorted and subjected to deep sequencing analysis (coverage, 1,166). The results indicated an enrichment of cells containing sgRNAs downstream of the TLR4 pathway in the sorted samples compared with the input (Figures S2).
Figure 3.
High-throughput pooled CRISPR screening of fluorescent high-content phenotypes
(A) Genes downstream of the TLR4 signaling pathway were targeted for knockout using a small-scale library.
(B) Volcano plot visualization of statistical significance (y axis) and magnitude of the change (x axis) before and after cell sorting, with p values calculated using the Mann-Whitney U test. Dashed lines represent the cutoff for hit genes (false discovery rate [FDR] = 0.01).
(C) Fluorescent images display RelA (green) co-localized with nuclei (magenta) inside MYD88, MAP3K7, IRAK4, and TNFRSF CRISPR knockout cells and LPS (−) and LPS (+) cells as controls. Scale bars, 10 μm.
(D) The correlation coefficient between SVM-based prediction probabilities in GC and similarity scores obtained using Amnis Image Stream was 0.914. N = 3 biological replicates.
See also Table S4.
Subsequently, we performed large-scale screening with 7,290 sgRNAs targeting 729 kinase genes (approximately 82.3 coverage), where we did not apply any staining as ground-truth labeling to the cells. This comprehensive screening analyzed 6,000,000 cells and successfully sorted target cells within 2 h (Figures S3). Analysis of the deep sequencing data revealed that the enriched cells contained sgRNAs targeting genes, including MAP3K7, IRAK4, IKBKB, and IKBKG, that are downstream of the TLR4 pathway (Figures 2B and S4). Therefore, we demonstrated the large-scale applicability of GC-based pooled CRISPR screening to fluorescent high-content phenotypes.
Furthermore, we assessed whether the degree of the cellular phenotype could be quantified using fluorescent GC to the same extent as image analysis performed with a commercial image flow cytometer (Amnis Image Stream). Specifically, we compared SVM-based analysis of GMI waveforms and nuclear translocation scores obtained from the analysis of images captured using the Amnis Image Stream. Specifically, we examined CRISPR knockout cell lines targeting MYD88, MAP3K7, IRAK4, and TNFRSF individually. SVM-based prediction probabilities were calculated as values ranging from 0–1 using the trained SVM classifier (STAR Methods), and nuclear translocation scores were determined as the degree of overlap between two images (RelA and nuclear images) using analysis software equipped with the Amnis Image Stream.18,19 The correlation coefficient between these scores was notably high (R = 0.914), indicating that GC-based phenotypic screening was quantitatively comparable with that of high-content microscopy image analysis (Figures 3C, 3D, and S5).
Pooled label-free high-content cellular phenotyping
We investigated the capacity of GC to assess a range of label-free high-content cellular phenotypes, including cell polarization, differentiation, and exhaustion (Figure 4). Figures 4A and 4B present microscopy images and scattering properties associated with label-free morphological phenotypes linked to THP-1 cell polarization from M0 to M1 macrophages. Figures 4C and 4D depict the process for developing and evaluating morphological classifiers within GC. To train a classifier, we separately prepared M0 and M1 macrophages, subsequently combining them as the training sample (Figure S6A–S6D).20 In this study, we employed a combination of forward scatter (FSC), back scatter (BSC), and label-free fsGMI and bfGMI waveforms to train SVM-based classifiers, utilizing 1,000 cells for each ground-truth label. Upon applying the trained model to a test dataset of 1,000 waveforms, the histogram of returned scores exhibited bimodal peaks color coded based on validation labels, resulting in high performance, as indicated by an AUC score of 0.89. This facilitated the classification of M0 and M1 macrophage high-content phenotypes even in the absence of surface markers. While distinguishing M0 from M1 polarized macrophages appears challenging through bright-field microscopy or FACS (Figures 4A and 4B), these results underscore the robust and accurate classification capabilities of GC.
Figure 4.
High-content cell phenotypes distinguishable using label-free GC
(A–J) Label-free GC classified (A)–(D) THP-1-derived M0 and M1 macrophages, (E)–(G) THP-1 monocytes and THP-1-derived macrophages, and (H)–(J) exhausted (LAG3/PD-1 double-positive) and non-exhausted (LAG3/PD-1 double-negative) human primary T cells.
(A), (E), and (H) Representative bright-field cell images on a dish were obtained using a microscope and Amnis Image Stream. Scale bars, 30 μm and 10 μm, respectively.
(B) Conventional fluorescence-activated cell sorting (FACS) scatterplots of forward scatter (FSC) and side scatter (SSC).
(C), (F), and (I) Representative label-free GMI waveforms of 20 randomly selected cells for each condition.
(D), (G), and (J) Classification results for label-free cell phenotypes. The ML-based classification performance is displayed as histograms of SVM scores, where red and blue colors indicate ground-truth labels and AUC scores.
Similarly, we evaluated the capability of classifying other label-free high-content phenotypes, such as THP-1 monocytes and THP-1-derived macrophages (cell differentiation) (Figures 4E–4G and S6E–S6G) as well as exhausted (LAG3/PD-1 double-positive) and non-exhausted (LAG3/PD-1 double-negative) human primary T cells (Figures 4H–4J and S6H–S6J).20,21 In each case, the models exhibited high performance with AUC scores of 0.94 and 0.92, respectively (Figures 4G and 4J). These results suggest that GC can classify various types of label-free high-content phenotypes.
Pooled CRISPR screening of label-free high-content phenotypes
We subsequently focused on morphological changes related to polarization, specifically from inactivated macrophages (M0 macrophages) to classical pro-inflammatory macrophages (M1 macrophages), serving as the target label-free phenotype for pooled CRISPR screening to identify associated genes. Macrophages are pivotal in the innate immune system, involved in critical processes such as tissue repair, inflammation, and cancer. These cells exhibit polarization into various subtypes, each with distinct functions, including cytokine secretion and response to injury or pathogenic threats.22,23 Defining their biological functions using only a few surface markers is often challenging, rendering the isolation of live cell populations based on their functions challenging. Thus, we hypothesized that macrophage polarization correlates with their morphology, prompting us to assess whether our system could identify relevant genes based on changes in label-free high-content cell morphological information without relying on surface markers. Accordingly, we employed the kinase CRISPR library, including various signaling pathways associated with macrophage polarization, particularly within the downstream pathway containing kinase genes.24 To train a classifier in the GC, we separately prepared M0 and M1 macrophages before merging them as the training sample. Only M0 macrophages were stained as the ground-truth label before mixing with the M1 macrophages (Figures S6A–S6D). Using the trained classifier and kinase library and the cells without ground truth labeling, we conducted a large-scale pooled CRISPR screening (approximately 68.6 coverage), sorting cells that exhibited suppression of the M1 polarization phenotype and subjecting them to deep sequencing (Figures 4A–4D, S7, and S8).
Analyzing sgRNA enrichment after sorting revealed several genes with the potential to induce macrophage polarization (Figures 5A and S9; Tables S2 and S3). Notably, interferon γ (IFNγ), a primary cytokine associated with M1 activation, was prominent. JAK2, a major mediator of IFNγ-induced signaling, was among the hit genes.25 Other candidate genes linked to macrophage polarization were also identified. For example, it is reported that DLG2 is altered in response to inflammation and can activate the formation of NLRP3 inflammasomes.26 Considering the function of cellular membrane trafficking, STK16 localizes to the Golgi complex and is involved in transporting secretory vesicles, which may indirectly contribute to cytokine production for pro-inflammatory responses.27 The top-hit gene, BRD2, is reportedly essential for pro-inflammatory cytokine production in macrophages.20,28 Concordantly, reduced expression of the M1 marker and secretion of pro-inflammatory cytokines, such as IFNγ and tumor necrosis factor alpha (TNF-α), in BRD2 CRISPR knockout M1 cells validated the hypothesis that BRD2 plays a pivotal role as a modulator of the M1 inducer gene (Figures 5B, 5C, and S10).
Figure 5.
High-throughput pooled CRISPR screening of label-free high-content phenotypes
(A) Volcano plot displaying statistical significance (y axis) and magnitude of the change (x axis) before and after cell sorting, with p values calculated using the Mann-Whitney U test. Dashed lines represent the cutoff for hit genes (FDR = 0.01).
(B) Expression of macrophage surface markers in control M0, control M1, and BRD2 CRISPR knockout (KO) M1 cells, using CD11b as a pan-macrophage marker and CD38 as an M1-specific marker. N = 3 biological replicates.
(C) Cytokine (IFNγ and TNF-α) release profiling of control M0, control M1, and BRD2 CRISPR KO M1 cells. Supernatants were collected from three independent experiments. Data are presented as mean ± SD; Welch’s t test: ∗p < 0.05, ∗∗∗p < 0.001. N = 3 biological replicates.
See also Table S4.
Collectively, we demonstrated that large-scale GC-based pooled CRISPR screening is applicable for label-free high-content phenotypes, which can be challenging to distinguish with conventional image-feature-based analyses using standard FACS and possibly even conventional microscopes.
Unsupervised representation of GMI waveforms
ML models trained within the GC framework exhibit versatility in addressing various high-content cell phenotypes, depending on the characteristics of the cell population and the classification objective. In this study, we systematically trained models for each pair of cellular phenotypes, specifically defined through ground truth markers, thereby establishing the robust performance of these classification models. This methodological approach can be extended to scenarios where only one phenotype can be defined during the screening process. It can be achieved by leveraging anomaly detection techniques, such as the one-class SVM, which can be readily implemented within the existing GC framework. However, practical situations often arise where suitable molecular markers or staining methods are unavailable for defining a particular cell phenotype. Moreover, the cells of interest within a pooled population may exhibit morphological heterogeneity. In such instances, defining and sorting the desired phenotype solely based on the GMI waveforms becomes imperative.
To explore the potential capabilities of GC in this context, we employed uniform manifold approximation and projection (UMAP) to visualize the GMI waveforms, as depicted in Figures 2C and 4C. Dimensionality reduction through principal-component analysis29 preceded the UMAP analysis (Figure 6). In Figure 6A, the UMAP of flGMI waveforms, obtained from a mixture of LPS-stimulated and unstimulated cells labeled with fluorescently tagged RelA molecules, revealed two distinct clusters. These clusters were subsequently validated as representing LPS-stimulated and unstimulated populations, respectively. The clear separation of these clusters suggests the potential to train classification models based on populations delineated within the UMAP space, similar to those defined using molecular markers.
Figure 6.
Uniform manifold approximation and projection (UMAP) of GMI waveforms
(A) UMAP of flGMI waveforms obtained for LPS-stimulated (orange) and unstimulated (blue) THP-1 cells.
(B) The same UMAP plot as presented in (A) is colored according to SVM scores obtained in the classification of RelA flGMI waveforms for the LPS-treated cells (stimulated versus unstimulated).
(C) UMAP of label-free GMI waveforms obtained for THP-1-derived M0 (blue) and M1 (orange) macrophages.
(D) The UMAP plot presented in (C) is colored according to SVM scores obtained in the classification of label-free multimodal GMI waveforms for the cells at different polarization states (M0 versus M1).
Furthermore, Figure 6B illustrates that cells distinguished within the UMAP space exhibit differential SVM scores, as quantified in Figure 2D. These results indicate that the morphological differences identified through supervised learning algorithms can also be captured via unsupervised learning algorithms, including UMAP and other dimensionality reduction methodologies.
In Figure 6C, similar to the fluorescence scenario, we projected label-free GMI waveforms derived from a mixture of THP-1-derived M0 and M1 macrophages onto the UMAP space. Colors were assigned based on ground-truth labels. While M0 and M1 macrophages did not present as distinct clusters within the UMAP, Figure 6D demonstrates that their distributions correlated with the SVM scores obtained from the classification model in Figure 4D. These findings strongly suggest that GMI waveforms alone hold the potential to define target high-content cellular phenotypes, provided that discernible morphological differences exist in an unsupervised context.
Discussion
An important technical advantage of CRISPR screening using the GC-based cell sorter, compared with conventional FACS-based screening, lies in its ability to analyze and integrate intricate cellular information, leading to more precise cell selection. In cases involving fluorescence-based cell phenotyping, this approach has demonstrated its superiority, especially when distinguishing between cell phenotypes, such as variations in subcellular protein localization or alterations in organelles morphologies, with similar total fluorescence intensity (Figure 2). The advantage of utilizing high-dimensional GMI waveforms for label-free cellular phenotyping is also evident. This is highlighted by the fact that conventional FSCs and side scatters (SSCs) (Figure 4B) were insufficient in distinguishing the polarized state of macrophages, even when utilizing the SVM method (Figure S6D). In addition, the results presented in Figure S6D show that the combination of average characteristics from FSC and SSC, along with detailed GMI waveforms, can synergistically enhance classification accuracy.
We anticipate the potential benefits of combining surface marker-based cell definition with label-free high-content cellular phenotyping within the GC. This combination is particularly valuable for exploring subtle cell differences among immune cell subtypes and various cell states, including activation, exhaustion, and differentiation. Moreover, the unique capability of GC to discern both fluorescent and label-free high-content cellular phenotypes holds potential for discovering novel targets, including gene perturbations and compounds. The fluorescence mode proves effective, particularly when the molecules or subcellular features of interest are pre-defined, enabling screening based on changes in their spatial distribution. Examples include the aggregation or degradation of proteins and alterations in intracellular organelles, serving as indicators of the underlying mechanisms of action.30,31 The label-free GMI waveforms uniquely capture complex morphological changes as a phenotypic response within each entire cell. In future implementations, simultaneous use of fluorescence and label-free GMI waveforms holds the potential for cell-based screening through the combinatorial analysis of changes in fluorescently labeled target proteins and detailed holistic high-content cellular phenotypes.32,33,34
From the viewpoint of applying ML methods in GC, we envision the following cases for supervised and unsupervised methods, respectively. Supervised learning approaches will be powerful when a labeled dataset is available for training classification models. This includes instances where the presence or absence of specific phenotypes can be defined by surface marker expression or scenarios when separate cell preparations with and without the desired phenotypes can be prepared. When accessible, leveraging class label information in supervised learning tends to achieve superior separation of phenotypes compared with unsupervised methods. On the other hand, unsupervised learning methods stand out for their potential in exploratory scenarios, especially in the absence of a labeled training dataset. These include experiments where the expected phenotypes are uncertain or when the target phenotypes cannot be defined only by surface markers. Such situations can typically arise in primary samples during disease progression, infection, or differentiation processes.
Image-based pooled CRISPR screening has witnessed rapid advancements. A noteworthy strategy involves fluorescence microscopy for both fluorescence image-based cellular phenotyping and subsequent reading of DNA barcodes assigned to each cell.6 The use of fluorescence markers is essential to delineate target image features and phenotypes. However, analyzing explicitly discernible image features becomes intricate when suitable biomarkers are unavailable, and cell phenotypes must be evaluated without labeling. Nevertheless, optical microscopy offers the unique advantage of characterizing cellular phenotypes in adherent states, coupled with the potential for high-resolution time-lapse observations, albeit at the cost of throughput.
Another recently introduced screening method relies on the enrichment of fluorescence image phenotypes using a fluorescence image-activated cell sorter (ICS),7 which has demonstrated even higher throughput than that of the current GC-based cell sorter. In ICS, a distinct set of image features is quantified by analyzing the reconstructed images, whereas the GC approach analyzes GMI modalities that are beyond visual comprehension. We acknowledge that the absence of images during GC-based sorting may be a limitation for those who prioritize visual verification. Nonetheless, in an era where machines frequently outperform human capabilities, we believe in the potential of utilizing modalities that are machine suitable. We have also demonstrated that GC can perform phenotyping quantitatively, resembling a 2D image-based analysis of image features (Figures 3C and 3D).
Moreover, in the context of label-free cell phenotyping, whether a limited set of image features computed from non-fluorescent images can sufficiently support optimal analysis remains uncertain. In contrast, GC classifies multimodal GMI waveforms, which include holistic, multiparametric morphological data of cells without requiring image reconstruction or feature extraction. Consequently, our ML-driven, less biased approach has showcased its advantages in detecting both simple molecular phenotypes (e.g., nuclear translocation of fluorescently labeled RelA) and more intricate, holistic phenotypes (e.g., macrophage polarization without labels).
Last, we discuss the similarities and differences in the screening results when compared with recent reports. In one report,7 the authors used HeLa cells and stimulated them with TNF-α to induce nuclear translocation of RelA. They performed a CRISPR screen using their custom nuclear factor κB (NF-κB) pathway-focused library targeting 1,068 genes and obtained MAP3K7, IKBKB, IKBKG, MAP3K7, IRAK4, IKBKB, and IKBKG, which are genes relevant to NF-κB signaling and were also hit in our screens, as statistically predominant hits. In another report,6 the authors used HeLa cells and stimulated with interleukin-1β (IL-1β) and TNF-α for induction of RelA nuclear translocation. They performed a screen using their custom library targeting 963 genes and obtained MAP3K7, IKBKB, IKBKG, MAP3K7, IRAK1, IRAK4, IKBKB, and IKBKG, which are genes relevant to NF-κB signaling and were also hit in our screens, as statistically predominant hits. In this study, we used THP-1 cells and stimulated them with LPS to induce RelA nuclear translocation. We first used a small-scale library targeting pathways containing NF-κB signaling and obtained TRAF6, MAP3K7, MYD88, IKBKB, IRAK1, IRAK4, and TIRAP as statistically predominant hits. We then used kinase library targeting 729 kinase genes and detected five genes, RelA, MAP3K7, IRAK4, IKBKB, and IKBKG, as statistically predominant hits relevant to NF-κB signaling. The large-scale library we used targeted only kinase genes and thus did not include non-kinase genes such as MYD88, which we included in the small-scale library. TAB1 and NFKB1A genes were hit in a report by Feldman et al.6 but not in another by Schraivogel et al.7 or in our screen. As the cell lines and stimulation conditions were different, and the genes in each library were not completely the same, it is difficult to compare the sensitivity directly. Nevertheless, many of the hit genes overlap with the previous two reports.
Collectively, we successfully developed a high-throughput and large-scale pooled CRISPR screening method, facilitated by rapid and selective cell sorting based on machine vision of high-content cellular phenotypes. We anticipate widespread utilization of GC-based cell sorters for screening critical cellular phenotypes by integrating various available ML methods and existing biomarkers with high-content analysis capabilities. When combined with single-cell sequencing techniques,35,36,37,38 this method seamlessly aligns with the pooled screening of various DNA-tagged perturbations, including antibodies,39 compounds,40 short hairpin RNAs (shRNAs),41 and peptides.42 Notably, as it is not confined to fluorescence-based cellular phenotypes, our ML -based, label-free, high-content cell analysis enables the enrichment of target phenotypes without invasive staining, preserving “untouched” cells for downstream functional assays, thereby expanding its applicability across a wide spectrum of biological studies.
Limitations of the study
To perform CRISPR screening using GC-based cell sorting, it is necessary to determine what to screen as the target positive phenotype and train the machine classifier using training samples. The design and preparation of positive and negative phenotypes in the step of training classifiers are the most important tasks for successful screening of genes that induce target image cell phenotypes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-UQCRC1 antibody | Invitrogen | 459140, RRID: AB_2532227 |
| anti-Lamp1 antibody | CST | 9091, RRID: AB_2687579 |
| anti-RelA p65 antibody | CST | 8242, RRID: AB_10859369 |
| Alexa Fluor 488-conjugated secondary antibody | ThermoFisher | A32723, RRID: AB_2633275 |
| Alexa Fluor 488-conjugated secondary antibody | ThermoFisher | A11008, RRID: AB_143165 |
| PE anti-human LAG3 (11C3C65) | BioLegend | 369305, RRID: AB_2629591 |
| APC anti-human PD-1 (EH12.2H7) | BioLegend | 329908, RRID: AB_950475 |
| PE-anti-human CD11b (ICRF4) | BioLegend | 301306, RRID: AB_314158 |
| Alexa Fluor 488 anti-human CD38 (HIT2) | BioLegend | 303512, RRID: AB_493088 |
| APC anti-human CD25 (BC96) | BioLegend | 302610, RRID: AB_314280 |
| BV421 anti-human CD69 (FN50) | BioLegend | 310930, RRID: AB_2561909 |
| Bacterial and virus strains | ||
| LentiBrite GFP-LC3 lentiviral biosensor | MERCK | 17–10193 |
| One Shot Stbl3 Chemically Competent E. coli | ThermoFisher | C737303 |
| Chemicals, peptides, and recombinant proteins | ||
| Chloroquine | Sigma-Aldrich | C6628-25G |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | P8139-1MG |
| LPS | Sigma-Aldrich | L4391-1MG |
| IFNγ | R&D Systems | 300–02 |
| rhIL-2 | Pepro Tech | 200-02-50ug |
| Critical commercial assays | ||
| Human Pro-inflammatory Cytokine Multiplex ELISA Kit | Arigo Biolaboratories Corp | ARG82862 |
| KAPA library quantification kit Illumina platform | Roche | KK4824 |
| QIAamp DNA FFPE Tissue Kit | QIAGEN | 56404 |
| Gel/PCR Extraction Kit | Nippon Genesis | FG-91012 |
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | G7571 |
| TruSeq rapid SBS kit (2 x 151 bp paired-end) | Illumina | FC-402-4023 |
| MiSeq v2 kit (2 x 301 bp paired-end) | Illumina | MS-102-2002 |
| Deposited data | ||
| Raw and analyzed data - Zenodo | This paper |
https://doi.org/10.5281/zenodo.7701145, https://doi.org/10.5281/zenodo.7703670, https://doi.org/10.5281/zenodo.7709846 https://doi.org/10.5281/zenodo.10472989 |
| Sequencing data | This paper | DDBJ submission DRA017748 |
| Experimental models: Cell lines | ||
| HEK293T cells | Applied Biological Materials | T3327 |
| HeLa cells | Japanese Collection of Research Bioresources | JCRB9004 |
| THP-1 cells | ATCC | TIB-202 |
| Recombinant DNA | ||
| pLentiGuide-Puro | Addgene | 52963 |
| pLenti-Cas9-Blast | Addgene | 52962 |
| pMD2.G | Addgene | 12259 |
| psPAX2 | Addgene | 12260 |
| Software and algorithms | ||
| Python scripts | This paper | https://doi.org/10.5281/zenodo.10472989 |
| FlowJo | BD Biosciences | Version 10.9.0 |
| IDEAS | Luminex | Version 6.2 |
| python | python.org | 3.7.12 |
| scikit-learn | scikit-learn.org | 1.0.2 |
| pandas | pandas.pydata.org | 1.3.5 |
| numpy | numpy.org | 1.21.6 |
| optuna | optuna.org | 2.10.1 |
| matplotlib | matplotlib.org | 3.5.3 |
| GCApp | ThinkCyte | v1.2.9.82 |
| Other | ||
| Dulbecco’s Modified Eagle’s Medium (DMEM) | FUJIFILM Wako | 044–29765 |
| RPMI-1640 | FUJIFILM Wako | 189–02025 |
| X-VIVO 15 | Lonza | BEBP04-744Q |
| Penicillin-streptomycin solution | FUJIFILM Wako | 168–23191 |
| Fetal bovine serum (FBS) | HyClone | SH30396.03 |
| 0.25w/v% Trypsin-1mmol/L EDTA・4Na Solution with Phenol Red | FUJIFILM Wako | 209–16941 |
| Earle’s Balanced Salt Solution (EBSS) | ThermoFisher | 14155063 |
| StemSure 2-mercaptoethanol solution | FUJIFILM Wako | 198–15781 |
| Glutamax supplement | ThermoFisher | 35050061 |
| CD3/CD28 Dynabeads | ThermoFisher | 11132D |
Resource availability
Lead contact
Requests for further information and resources relating to this paper should be directed to Sadao Ota (sadaota@solab.rcast.u-tokyo.ac.jp).
Materials availability
Materials used in this study are commercially available.
Data and code availability
-
•All data are reported in the paper or deposited at Zenodo: https://doi.org/10.5281/zenodo.7701145, https://doi.org/10.5281/zenodo.7703670, https://doi.org/10.5281/zenodo.7709846, https://doi.org/10.5281/zenodo.10472989.
-
•NGS fastq files of NFkB screening (replicate 1&2 of pilot screening, related to Figures S2A and S2K) are available in Zenodo: https://doi.org/10.5281/zenodo.7701145.
-
•NGS fastq files of NFkB screening (replicate 1&2 kinase library screening, related to Figures 3B, and S4A–S4C) are available in Zenodo: https://doi.org/10.5281/zenodo.7701145.
-
•NGS fastq files of macrophage label-free screening (replicate 2&3, related Figures S9D–S9G; Table S3) are available in Zenodo: https://doi.org/10.5281/zenodo.7701145.
- •
-
•Text files of similarity score taken by Amnis image flow cytometer (Figure 3D) are available in Zenodo: https://doi.org/10.5281/zenodo.7701145.
-
•NGS fastq files of macrophage label-free screening (replicate 1, related to Figure 5A) are available in Zenodo: https://doi.org/10.5281/zenodo.7703670.
-
•Csv files of ML classifier training and test data (NFkB nuclear translocation, related to Figures 2D and 6) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (mitochondria/lysosome, related to Figure 2H) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (LC3 aggregation, related to Figure 2L) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (T cell mitochondria, related to Figure 2P) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (Macrophage M0/M1, related to Figure 4D) are available in Zenodo https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (monocyte/macrophage, related to Figure 4G) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (T cell exhaustion, related to Figure 4J) are available in Zeonodo: https://doi.org/10.5281/zenodo.7709846.
-
•Text files of SVM-based prediction probabilities in GC (Figures 3D and S5) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•NGS sequencing raw data (fastq files) were also deposited at DNA DataBank of Japan (DDBJ)
-
•Submission: DDBJ: DRA017748 (sadaotalab1-0001_Submission)
-
•BioProject: DDBJ: PRJDB17362 (PSUB022209)
-
•BioSample: DDBJ: SAMD00732055-SAMD00732060 (SSUB028198)
-
•Experiment: DDBJ: DRX509723-DRX509728 (sadaotalab1-0001_Experiment_0001–0024)
-
•Run: DDBJ: DRR525841-DRR525847 (sadaotalab1-0001_Run_0001–0020)
-
•
-
•
-
•Data analysis was performed using code written in Python, which is available in Zenodo. To analyze the data, csv files are imported and run Python code.
-
•Python code file (svm.py) for ML classification and drawing confusion matrix, SVM histogram, and PR curve, and calculation of probabilities from SVM score (Figures 3D, S1L, and S6J) is available in Zenodo: https://doi.org/10.5281/zenodo.10472989.
-
•Python code file (umap_plot.py) for UMAP plot (Figure 6) is available in Zenodo: https://doi.org/10.5281/zenodo.10472989.
-
•Instruction file (how_to_use.txt) is also available in Zenodo: https://doi.org/10.5281/zenodo.10472989.
-
•
-
•
Any additional information needed to re-analyze the data reported in this paper are available from the lead contact upon request.
Experimental model and study participant details
Cell culture
HEK293T cells (gender: female) were purchased from Applied Biological Materials (abm) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (FUJIFILM Wako), supplemented with 10% fetal bovine serum (FBS) (HyClone) and 1% penicillin-streptomycin solution (FUJIFILM Wako) at 37°C with 5% CO2. Routine testing for mycoplasma contamination was conducted using nested PCR with culture medium as the template.
HeLa cells were obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank and cultured in DMEM (FUJIFILM Wako), supplemented with 10% FBS (HyClone) and 1% penicillin-streptomycin solution (FUJIFILM Wako) at 37°C with 5% CO2.
HeLa cells (gender: female) expressing LC3-GFP were transduced by adding LentiBrite GFP-LC3 Lentiviral Biosensor (Millipore) at a multiplicity of infection (MOI) of 20 for 24 h, following the manufacturer’s instructions. After 48 h post-infection, cells were passaged into complete culture medium. Routine testing for mycoplasma contamination was conducted using nested PCR with culture medium as the template. Cells were either maintained in complete media or incubated for 4 h in Earle’s Balanced Salt Solution (EBSS) (ThermoFisher) containing 20 μM chloroquine (Sigma-Aldrich) to induce autophagosome accumulation.
THP-1 human monocytic leukemia cells (gender: male) were obtained from ATCC and cultured in RPMI-1640 (FUJIFILM Wako), supplemented with 10% FBS (HyClone), 1% penicillin-streptomycin (FUJIFILM Wako), and 1% StemSure 2-mercaptoethanol solution (FUJIFILM Wako) at 37°C with 5% CO2. Routine testing for mycoplasma contamination was conducted using nested PCR with culture medium as the template. To activate TLR4 signaling, THP-1 cells were stimulated with lipopolysaccharide (LPS) (Sigma-Aldrich) at a concentration of 300 ng/mL for 1 h at 37°C with 5% CO2. For macrophage differentiation and polarization from THP-1 cells, THP-1 monocytes were differentiated into resting macrophages (M0) using 100 nM phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich) for 72 h, followed by 24 h in PMA-free medium. For M1 polarization, M0 macrophages were further cultured in M1-polarization medium containing 100 ng/mL LPS (Sigma-Aldrich) and 20 ng/mL IFNγ (R&D Systems) for 24 h, starting on the third day of PMA treatment [20].
For primary T cells, Pan-T cells were acquired from Precision for Medicine and cultured in X-VIVO 15 (Lonza), supplemented with 10% FBS (HyClone), 1% Glutamax supplement (Thermo Fisher), 1% penicillin-streptomycin (FUJIFILM Wako), and 1% StemSure 2-mercaptoethanol solution (FUJIFILM Wako) at 37°C with 5% CO2. For activation, Pan-T cells were cultured in the presence of CD3/CD28 Dynabeads (Thermo Fisher) and 25 U/mL rhIL-2 (Pepro Tech) for 4 days. For non-activated cells, Pan-T cells were cultured with only 25 U/mL rhIL-2 for 4 days. To mimic transient stimulation for the T cell exhaustion experiment, cells were cultured with CD3/CD28 Dynabeads and 25 U/mL rhIL-2 for 3 days, followed by incubation in the presence of only rhIL-2 for the remaining 11 days of culture [21].
Method details
Ghost cytometry (GC)-based cell sorter
In the optical setup of the GC-based cell sorter prototype, diffractive optical elements (DOEs) are employed to generate structured illumination of light for acquiring ghost motion imaging (GMI) waveforms. The structured light pattern generated using a 488 laser is utilized to measure fluorescence GMI (flGMI) waveforms, whereas that produced using a 405 laser is employed for measuring label-free GMI (fsGMI, bsGMI, dGMI, and bfGMI) waveforms. In the fluidic setup, a sample volume is controlled using a syringe pump, and a sheath flow is driven by a pressure pump. Within the microfluidic device, cells are initially hydrodynamically focused to form a tight stream. Subsequently, they undergo structured illumination by light and are then sorted using a piezo-based microfluidic sorting device, based on the analysis using a support vector machine (SVM) model implemented i.n a field programmable gate array (FPGA), similar to a method reported previously.12 During large-scale CRISPR screenings, cells flowed at an input rate of approximately 1,000 cells/s. This prototype allowed us to flow cells, whose size ranged from 4 to 40 μm, at a flow rate up to 40 μL/min, at a maximum rate of 3,000 events/s, with an optical resolution of approximately 0.7 μm, which was estimated by the single spot size of the structured illumination. The updated information on machines is available on Thinkcyte.43
Construction of small-scale CRISPR sgRNA expression plasmids
gRNA spacer inserts were prepared through a single-pot reaction to phosphorylate and anneal ssDNA pairs. To generate each spacer fragment (see Table S1), a T4 polynucleotide kinase reaction sample was prepared with two ssDNAs in accordance with the manufacturer’s protocol (Takara) and subjected to the following thermal cycling conditions: 37°C for 30 min; 95°C for 5 min; 70 cycles of 12 s starting at 95°C and decreasing by 1°C per cycle, followed by incubation at 25°C. The annealed spacer inserts were subsequently ligated into a pLentiGuide-Puro vector (Addgene) via Golden Gate Assembly using BsmBI (NEB) and T4 DNA ligase (NEB). The assembly process followed these thermal cycling conditions: 15 cycles of 37°C for 5 min and 20°C for 5 min, 55°C for 30 min, and then maintenance at 4°C.44 pLenti-Cas9-Blast plasmids were obtained from Addgene. Plasmid sequences were validated using Sanger sequencing.
Amplicon sequencing
Genomic DNA from fixed cells was extracted using the QIAamp DNA FFPE Tissue Kit (QIAGEN) or the hotshot method.37 For each sample, target regions were amplified using the extracted genomic DNA as the PCR template with the corresponding first HTS primer pair (see Table S1). The PCR was conducted following the previously established protocol [43]. The PCR product was purified using the FastGene Gel/PCR Extraction Kit (Nippon Genetics) and re-amplified with custom Illumina index primers (see Table S1). Each indexed library was electrophoresed in a 2% agarose gel, and the expected band was purified using the FastGene Gel/PCR Extraction Kit (Nippon Genetics). The sequencing libraries were quantified using qPCR with the KAPA Library Quantification Kit Illumina (KAPA Biosystems) for multiplexing. The multiplexed libraries were quantified using the same qPCR protocol and sequenced with 30–40% PhiX control using Illumina HiSeq2500 (HiSeq Rapid SBS kit; 2 x 151 bp paired-end) or MiSeq (MiSeq v2 kit; 2 x 301 bp paired-end).
Lentiviral production and transduction
HEK293T cells were seeded onto a 10-cm plate at a density of 100,000 cells/cm2. After 20 h, cells were transfected with pMD2.G (0.75 μg) (Addgene), psPAX2 (2.25 μg) (Addgene), and a lentiviral transfer plasmid (3.6 μg) using polyethylenimine (PEI) MAX (Cosmobio).44 The media was changed after 24 h. Viral supernatant was harvested 48 h post-transfection and filtered through 0.45 μm cellulose acetate filters. Virus titer was determined using CellTiter-Glo (Promega), following the manufacturer’s protocol. THP-1 cells expressing Cas9 protein were transduced in bulk with the library at a low MOI (0.15 viral particles per cell) by adding viral supernatant supplemented with polybrene (8 μg/mL) and centrifuging at 510 g for 1 h at 37°C. After 24 h post-infection, cells were passaged into media containing selection antibiotics, 0.5 μg/mL puromycin (Sigma-Aldrich) and 10 μg/mL blasticidin (Sigma-Aldrich).
Immunostaining
HEK293T cells were washed once with PBS and treated with 0.25 w/v% Trypsin-EDTA solution (FUJIFILM Wako) for 5 min at 37°C to detach them from the culture dish. Cells were fixed with 4% formaldehyde in PBS (Thermo Fisher) for 20 min at room temperature. To visualize mitochondria, cells in PBS were incubated at 95°C for 5 min, blocked with 5% FBS and 0.3% Triton X-100 (Sigma-Aldrich, #T8787-100 mL) in PBS solution for 30 min at room temperature, and incubated with a 1/200 diluted anti-UQCRC1 antibody for Complex III-core1 (Invitrogen, #459140) in the blocking buffer overnight at 4°C. After washing with wash buffer (0.5% FBS in PBS), cells were incubated with Alexa Fluor 488-conjugated secondary antibody (Thermo Fisher, #A32723) for 1 h at room temperature. To visualize lysosomes, cells were permeabilized with 0.2% Triton X-100 in PBS for 3 min at room temperature after fixation. After washing with 0.5% BSA in PBS, cells were blocked with 5% BSA in PBS for 1 h at room temperature and incubated with a 1/200 diluted anti-Lamp1 antibody (CST, #9091) in 1% BSA in PBS overnight at 4°C. After washing with 0.5% BSA in PBS, cells were incubated with Alexa Fluor 488-conjugated secondary antibody (Thermo Fisher, #A11008) for 1 h at room temperature.
THP-1 cells were fixed with 4% formaldehyde in PBS (Thermo Fisher, #R37814) for 20 min at room temperature. After washing with PBS once, cells were incubated with ice-cold methanol for 5 min at −20°C. After washing with 0.5% BSA in PBS, cells were blocked with 5% BSA in PBS for 1 h at room temperature and incubated with anti-RelA p65 antibody (CST, #8242) in 1% BSA in PBS overnight at 4°C. After washing with 0.5% BSA in PBS, cells were incubated with Alexa Fluor 488-conjugated secondary antibody (Thermo Fisher, #A11008) for 1 h at room temperature.
Flow cytometry
For flow cytometry and ghost cytometry, the following monoclonal antibodies were employed: PE anti-human LAG3 (11C3C65), APC anti-human PD-1 (EH12.2H7), PE anti-human CD11b (ICRF4), Alexa Fluor 488 anti-human CD38 (HIT2), APC anti-human CD25 (BC96), and BV421 anti-human CD69 (FN50), along with their corresponding isotype controls (all from BioLegend). Cell viability was assessed using 7-AAD (BD Biosciences) or Zombie NIR staining (BioLegend). Nuclear staining was performed using Hoechst 33342 (Thermo Fisher, #H3570). Cells were suspended in a staining buffer (2% FSB with 1 mM EDTA in PBS) containing human FcR blocking reagent (Miltenyi Biotec, #130-059-901) and incubated with monoclonal antibodies for 30 min at 4°C. Flow cytometry data were acquired using the Attune Flow Cytometer (Thermo Fisher) or JSAN (Bay Bioscience). Fluorescence was compared to corresponding isotype-stained controls. FlowJo software (Tree Star Inc.) was used for data analysis. Cell images in Amnis Image Stream were captured using the FlowSight system, and black or gray areas were added to the surroundings to ensure uniformly sized images for improved visualization in figures. Similarity scores for RelA and Hoechst were obtained using IDEAS 6.2 (Luminex).
ELISA
THP-1 cells were polarized as described earlier. The supernatant was collected from three independent experiments, pooled, and subjected to the Human Pro-inflammatory Cytokine Multiplex ELISA Kit (Arigo Biolaboratories Corp) following the manufacturer’s protocol. Optical density (OD) was read with a microplate reader (Speark, Tecan) at 450 nm, and average absorbance values were calculated for each set of standards and samples. The experiments were performed as three independent replicates.
Experimental conditions for fluorescence and label-free GC
Electronic settings were detailed previously.11,12 Briefly, all photomultiplier tubes (PMTs) were purchased from Hamamatsu Photonics Inc. PMTs with a frequency of 10 MHz and built-in amplifiers were used for detecting the flGMI, fsGMI, bsGMI, and dGMI, whereas PMTs with frequencies of 200 kHz, 1 MHz, or 10 MHz detected fluorescence and BSC signals. FSC signals were obtained using either a photodetector or a 200 kHz PMT. Multi-pixel photon counters from Hamamatsu Photonics Inc. were used for detecting bfGMI. DC signals for flGMI, fsGMI, bsGMI, bfGMI, dGMI, and FSC were filtered using an electronic high-pass filter. PMT signals were recorded with electronic filters using a digitizer or an FPGA development board with a custom analog-to-digital converter. The digitizer and/or FPGA continually collected fixed-length signal segments from each color channel simultaneously, with a fixed trigger condition applied to the FSC signals.
Cells were passed through either a quartz flow cell (Hamamatsu) or a polydimethylsiloxane (PDMS)-based microfluidic device using a custom pressure pump and/or syringe pump (KD Scientific). The quartz flow cell had a channel cross-section dimension of 150 × 150 μm2 at the measurement position, with the sheath fluid (IsoFlow, Beckman Coulter) driven at a pressure of 305 kPa for cell analysis. The sample fluid was driven at a flow rate between 10 and 40 μL/min. For cell sorting, custom handmade sorting chips were used. The PDMS device had a channel with a cross-section dimension of 32 × 80 μm2 at the measurement position. The sheath flow was driven at a pressure of about 180–270 kPa, and the sample fluid was driven at a flow rate of 20 μm/min during cell sorting. For sorting, a piezoelectric (PZT) actuator implemented on the sorting chip was driven by an input voltage, displacing the fluid containing target cells toward a collection channel.
Machine learning details were described previously.11,12 For binary cell classification, a support vector machine (SVM) algorithm with a radial basis function (rbf) kernel from the scikit-learn library was used. All model training and validations were performed using equal sample amounts for each class label. We avoided the leakage interference of the signals from ground truth markers with the imaging signal by carefully choosing dyes and designing the configrations of optical illuminations. The number of cells for training and testing SVM is described in subsequent sections. After training, probability or decision functions were computed using the predict_proba method or the decision_function method from the scikit-learn library, serving as SVM scores. Score distributions for each sample were visualized as histograms using the matplotlib library. We evaluated trained machine learning models using accuracy, receiver operating characteristic (ROC) curves, and area under the ROC curve (AUC or ROC-AUC). ROC curves were plotted using the matplotlib library after calculating the true positive rate (tpr) and false positive rate (fpr) with the scikit-learn library. Precision-recall pairs for different SVM score thresholds were computed with the scikit-learn library, and precision-recall (PR) curves were plotted using the matplotlib library. Hyperparameters (regularization and kernel parameters in SVM) were optimized through 3-fold cross-validation of the AUC score.
Classification of lysosomes and mitochondria
HEK293T cells were immunostained with anti-Lamp1 and anti-UQCRC1 antibodies as described above. As a ground truth label, anti-Lamp1 antibody-stained cells were incubated with the LIVE/DEAD Fixable Far-Red (FFR) Dead Cell staining kit (Thermo Fisher Scientific). To classify the lysosomal and mitochondrial organelle fluorescence distribution patterns, each cell type was mixed at an equal concentration. The mixed suspension was allowed to flow through the fluorescence GC system. Cells were gated using FSC/SSC scatterplots to remove doublets and debris from the training samples. flGMI waveforms were used as the classification modality. Within the data, 2,500 and 1,000 cells were randomly selected (without overlap), with an equal number of anti-Lamp1-and anti-UQCRC1-stained cells used as training and testing data, respectively.
Classification of autophagosome localization of LC3-GFP proteins in HeLa cells
Autophagosome-induced and -uninduced HeLa cells expressing LC3-GFP proteins were prepared individually, as described earlier. After washing once with PBS, cells were incubated with 0.25 w/v% Trypsin-EDTA solution for 5 min at 37°C to detach them gently from the culture dish. As a ground truth label, the accumulation of autophagosome-uninduced cells was stained with CellTracker DeepRed Dye (Thermo Fisher Scientific). To classify LC3-GFP autophagosome localization, each cell line was mixed at equal concentrations. The mixed suspension was allowed to flow through the fluorescence GC system. Cells were gated using FSC/SSC scatterplots to remove doublets and debris from the training samples. flGMI waveforms were used as the classification modality. Within the data, 2,000 and 1,000 randomly selected cells (without overlap) with equal numbers of induced and uninduced cells were used as training and testing data, respectively.
Classification of mitochondrial morphological changes in activated and non-activated human primary T cells
Activated and non-activated T cells cultured for 4 days were individually stained with MitoTracker Green according to the manufacturer’s instructions. Following PBS washing, cells were further labeled with cell surface markers and the viability dye mentioned previously. To classify activated and non-activated cells, the cell populations were mixed equally and passed through the fluorescence GC system. FSC/SSC scatterplot gating was applied to remove doublets and debris, and live cells were selected. CD25/CD69 double-positive cells were designated as activated, while CD25/CD69 double-negative cells were identified as non-activated T cell populations. Classification was based on flGMI waveforms. In the dataset, 2,000 and 1,000 cells from each category (activated and non-activated, respectively) were randomly selected without overlap for training and testing.
Classification of THP-1-derived monocytes and macrophages
THP-1-derived monocytes and macrophages were stained with cell surface markers and the viability dye described earlier. To classify these cells, FSC/SSC scatterplot gating was used to eliminate doublets and debris. After gating for live cells, CD11b-positive cells were identified as macrophages, whereas CD11b-negative cells were categorized as monocytes. Classification relied on fsGMI, bsGMI, and dGMI waveforms. In the dataset, 1,000 and 500 cells from each category (monocytes and macrophages) were randomly selected without overlap for training and testing, respectively.
Classification of exhausted and non-exhausted cells
Transiently-stimulated T cells were stained with cell surface markers and the viability dye mentioned earlier. To classify exhausted and non-exhausted cells, FSC/SSC scatterplot gating was used to remove doublets and debris, and live cells were selected. LAG3/PD-1 double-positive cells were classified as exhausted, whereas LAG3/PD-1 double-negative cells were considered non-exhausted T cell populations. Classification relied on fsGMI, bsGMI, and dGMI waveforms. In the dataset, 400 and 400 cells from each category (exhausted and non-exhausted) were randomly selected without overlap for training and testing, respectively.
Classification and sorting of CRISPR library using fluorescence GC
THP-1 cells were immunostained with the previously described anti-RelA antibody. Cells unstimulated with LPS were labeled with the LIVE/DEAD FFR dead cell staining kit (Thermo Fisher Scientific) as a reference label. Classification of LPS-stimulated and -unstimulated cell populations was performed with equal concentrations of each cell type passing through the fluorescence GC system. FSC/SSC scatterplot gating was applied to exclude doublets and debris from the training dataset. Classification was based on flGMI waveforms. In the dataset, 1,250 and 1,000 cells from each category (LPS-stimulated and unstimulated) were randomly selected without overlap for training and testing, respectively.
The pooled CRISPR library cells were stimulated with LPS and stained with anti-RelA antibody described earlier. The trained classifier was implemented on an FPGA, allowing real-time cell judgment using GMI signals, enabling subsequent cell sorting. The time for judgment of a single cell with the FPGA was 6.0 μs. For small-scale library sorting, 100,000 cells were processed, with 70,000 cells predicted as positive and sorted using the trained model (coverage 1,166). The coverage is the cell number of after sorting divided by the number of gRNAs. The experiments were performed as two independent replicates. The cells expressing 40 out of 60 pooled sgRNAs were labeled with a ground truth marker to validate the sorting. Sorting purity was validated, with 90.8% of cells in the sorted sample labeled with the ground truth marker (Figure S2C). The result closely matched the predicted sorting purity of 92.6% and a recovery of 96.3% (Figure S2B). Reproducibility of the trained model was assessed, with an average AUC score of 0.98 ± 0.02 (n = 7) and a high Zʹ-factor value of 0.83 (Figure S2D).45 For sorting of the kinase library, 6,000,000 cells (approximately 82.3 coverage) were processed, with approximately 600,000 cells predicted as positive and sorted using the trained model within 2 h (Figure S2J). The experiments were performed as four independent replicates.
Classification and sorting of the CRISPR kinase library using label-free GC
THP-1-derived M0 and M1 macrophages were individually prepared as previously described. Following a single PBS wash, the cells were incubated at 37°C with a 0.25% w/v Trypsin-EDTA solution (FUJIFILM Wako) for 5 min and gently scraped using a Cell Lifter (Corning) to detach them from the culture dish. As a ground truth label, M0 macrophages were stained with CellTracker Green CMFDA Dye (Thermo Fisher). For the classification of M0 and M1 macrophages, an equal concentration of each cell type was mixed, and this mixture was passed through the fluorescence GC system. FSC/SSC scatterplot gating was employed to eliminate doublets and debris from the training dataset. Classification was based on fsGMI and bfGMI waveforms. In this dataset, 1,000 cells from each category (M0 and M1 macrophages) were randomly selected without overlap for training, with an additional 1,000 cells for testing. The reproducibility of creating a trained model was evaluated (the average AUC score was 0.85 ± 0.06, n = 7), and a high Zʹ-factor value from the SVM score (0.75, n = 7) was obtained (Figure S7C).45 For validation of differentiation and polarization, M0 and M1 macrophages in the training sample were stained with known M0 and M1 CD markers individually (Figures S7D–S7G).20
The pooled CRISPR kinase library cells were differentiated and incubated with an M1-polarization medium. For sorting the kinase library, 6,000,000 cells (approximately 68.6 coverage) were processed by the sorter, and approximately 500,000 cells predicted as positive were sorted within 2 h using a classifier trained and implemented on an FPGA. To enhance purity, a threshold of SVM scores >1 was set for cell sorting. In cases where the percentage of target cells in the pre-sorting sample was 8.3%, the predicted purity was 37.5%, and the predicted recovery was 16.7% in the sorted sample (Figures S7A and S7B). These experiments were performed as three independent replicates.
Quantification and statistical analysis
Adapter trimming and demultipexing were performed using Ultraplex.46 The large-scale screening data were analyzed according to the published bioinformatics pipeline (kampmannlab.ucsf.edu/mageck-inc).47,48 Briefly, raw sequencing reads from next-generation sequencing were cropped and aligned to the reference using Bowtie2 to determine sgRNA counts in each sample. Subsequently, counts files from two samples for comparison were input into MAGeCK, where log2 fold changes (LFCs) and p values were calculated for each sgRNA. Gene-level knockout phenotypic scores were determined by averaging LFCs from the top 3 sgRNAs targeting the gene with the most significant p values.
Statistical significance for each gene was determined by comparing the set of p values for sgRNAs targeting it with the set of p values for non-targeting control sgRNAs, employing the Mann–Whitney U test. A cutoff value was selected based on the distribution of all products to ensure a false discovery rate (FDR) of < 0.01.
Acknowledgments
We thank all members of ThinkCyte Inc. for extending experimental support and their discussions, with special recognition to Keisuke Toda, Yasuhiro Kajiwara, Hikari Morita, and Keiji Nakagawa for their roles in instrument development. We also acknowledge Hiroaki Adachi for his support during data analysis, Hiroaki Suita and Kaoru Komoriya for their assistance with biological experiments, and Keisuke Wagatsuma for their insightful scientific discussions. We would like to express our appreciation to Greg Schneider, Andy Wu, and Willem Westra for reviewing the manuscript review and to Kaori Shiina from The University of Tokyo for her assistance with sequencing data analysis. This work was supported by the Product Commercialization Alliance (PCA) program funded by the New Energy and Industrial Technology Development Organization (NEDO; 20001038-0).
Author contributions
A.T. and S.O. conceived the study, designed the experiments, interpreted the results, and wrote the manuscript. S.O. supervised the study. A.T. developed the experimental tools and performed most of the experiments, including construction of the CRISPR library, flow cytometry analysis, cell culture, cell staining, GC data analysis, and NGS library preparation. Y.A. performed the experiments and analyzed the data using GC and NGS. Y.K. designed and fabricated the microfluidic sorting chips, integrated the GC-based cell sorter, and performed all GC-based cell sorting experiments. Y.M. performed all GC-based cell sorting experiments. H.N. wrote the manuscript. Y.Y. performed the computational analysis of the NGS data. K.T. performed the GC analysis. S.I., H.A., and N.Y. provided conceptual input for the pooled CRISPR screening, CRISPR library design, library cloning strategy, and technical inputs. All authors contributed to manuscript revision.
Declaration of interests
S.O. is a co-founder and a board member of ThinkCyte Inc., a company engaged in the development of GC-based cell sorting. A.T., Y.A., Y.K., Y.Y., H.N., Y.M., and K.T. are employees of ThinkCyte, Inc. A.T., Y.A., Y.K., Y.M., Y.Y., H.N., and K.T. have shares of stock options from ThinkCyte, Inc. S.O., A.T., Y.A., Y.K., Y.M., Y.Y., H.N., and T.K. have filed patent applications for this study.
Published: March 25, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2024.100737.
Supplemental information
References
- 1.Joung J., Konermann S., Gootenberg J.S., Abudayyeh O.O., Platt R.J., Brigham M.D., Sanjana N.E., Zhang F. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 2017;12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346 doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 3.Bray M.A., Singh S., Han H., Davis C.T., Borgeson B., Hartland C., Kost-Alimova M., Gustafsdottir S.M., Gibson C.C., Carpenter A.E. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 2016;11:1757–1774. doi: 10.1038/nprot.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panganiban R.A., Park H.R., Sun M., Shumyatcher M., Himes B.E., Lu Q. Genome-wide CRISPR screen identifies suppressors of endoplasmic reticulum stress-induced apoptosis. Proc. Natl. Acad. Sci. USA. 2019;116:13384–13393. doi: 10.1073/pnas.1906275116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler E.C., Vu A.Q., Einstein J.M., DiSalvo M., Ahmed N., Van Nostrand E.L., Shishkin A.A., Jin W., Allbritton N.L., Yeo G.W. Pooled CRISPR screens with imaging on microraft arrays reveals stress granule-regulatory factors. Nat. Methods. 2020;17:636–642. doi: 10.1038/s41592-020-0826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldman D., Singh A., Schmid-Burgk J.L., Carlson R.J., Mezger A., Garrity A.J., Zhang F., Blainey P.C. Optical Pooled Screens in Human Cells. Cell. 2019;179:787–799.e17. doi: 10.1016/j.cell.2019.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schraivogel D., Kuhn T.M., Rauscher B., Rodríguez-Martínez M., Paulsen M., Owsley K., Middlebrook A., Tischer C., Ramasz B., Ordoñez-Rueda D., et al. High-speed fluorescence image-enabled cell sorting. Science. 2022;375:315–320. doi: 10.1126/science.abj3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan X., Stuurman N., Ribeiro S.A., Tanenbaum M.E., Horlbeck M.A., Liem C.R., Jost M., Weissman J.S., Vale R.D. High-content imaging-based pooled CRISPR screens in mammalian cells. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202008158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C., Lu T., Emanuel G., Babcock H.P., Zhuang X. Imaging-based pooled CRISPR screening reveals regulators of lncRNA localization. Proc. Natl. Acad. Sci. USA. 2019;116:10842–10851. doi: 10.1073/pnas.1903808116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christiansen E.M., Yang S.J., Ando D.M., Javaherian A., Skibinski G., Lipnick S., Mount E., O'Neil A., Shah K., Lee A.K., et al. In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images. Cell. 2018;173:792–803.e719. doi: 10.1016/j.cell.2018.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ugawa M., Kawamura Y., Toda K., Teranishi K., Morita H., Adachi H., Tamoto R., Nomaru H., Nakagawa K., Sugimoto K., et al. In Silico-Labeled Ghost Cytometry. Elife. 2021;10 doi: 10.7554/eLife.67660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ota S., Horisaki R., Kawamura Y., Ugawa M., Sato I., Hashimoto K., Kamesawa R., Setoyama K., Yamaguchi S., Fujiu K., et al. Ghost cytometry. Science. 2018;360:1246–1251. doi: 10.1126/science.aan0096. [DOI] [PubMed] [Google Scholar]
- 13.Barton G.M., Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 14.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami N., Oyama F., Gu Y., McLennan I.S., Nonaka I., Ihara Y. Accumulation of tau in autophagic vacuoles in chloroquine myopathy. J. Neuropathol. Exp. Neurol. 1998;57:664–673. doi: 10.1097/00005072-199807000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Buck M.D., O'Sullivan D., Klein Geltink R.I., Curtis J.D., Chang C.H., Sanin D.E., Qiu J., Kretz O., Braas D., van der Windt G.J.W., et al. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rambold A.S., Pearce E.L. Mitochondrial Dynamics at the Interface of Immune Cell Metabolism and Function. Trends Immunol. 2018;39:6–18. doi: 10.1016/j.it.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Maguire O., Collins C., O'Loughlin K., Miecznikowski J., Minderman H. Quantifying nuclear p65 as a parameter for NF-kappaB activation: Correlation between ImageStream cytometry, microscopy, and Western blot. Cytometry A. 2011;79:461–469. doi: 10.1002/cyto.a.21068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander J., Smola P.B., Schoelkopf B., Dale S. A brandford Book; 1999. Advances in Large-Margin Classifiers. [Google Scholar]
- 20.Surdziel E., Clay I., Nigsch F., Thiemeyer A., Allard C., Hoffman G., Reece-Hoyes J.S., Phadke T., Gambert R., Keller C.G., et al. Multidimensional pooled shRNA screens in human THP-1 cells identify candidate modulators of macrophage polarization. PLoS One. 2017;12 doi: 10.1371/journal.pone.0183679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corselli M., Saksena S., Nakamoto M., Lomas W.E., 3rd, Taylor I., Chattopadhyay P.K. Single cell multiomic analysis of T cell exhaustion in vitro. Cytometry A. 2022;101:27–44. doi: 10.1002/cyto.a.24496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Y., Xu X.H., Jin L. Macrophage Polarization in Physiological and Pathological Pregnancy. Front. Immunol. 2019;10:792. doi: 10.3389/fimmu.2019.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerneur C., Cano C.E., Olive D. Major pathways involved in macrophage polarization in cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1026954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He L., Jhong J.H., Chen Q., Huang K.Y., Strittmatter K., Kreuzer J., DeRan M., Wu X., Lee T.Y., Slavov N., et al. Global characterization of macrophage polarization mechanisms and identification of M2-type polarization inhibitors. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keane S., Herring M., Rolny P., Wettergren Y., Ejeskär K. Inflammation suppresses DLG2 expression decreasing inflammasome formation. J. Cancer Res. Clin. Oncol. 2022;148:2295–2311. doi: 10.1007/s00432-022-04029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J., Yang X., Li B., Wang J., Wang W., Liu J., Liu Q., Zhang X. STK16 regulates actin dynamics to control Golgi organization and cell cycle. Sci. Rep. 2017;7 doi: 10.1038/srep44607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belkina A.C., Nikolajczyk B.S., Denis G.V. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J. Immunol. 2013;190:3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leland McInnes J.H., James M. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv. 2020 doi: 10.48550/arXiv.1802.03426. Preprint at. [DOI] [Google Scholar]
- 30.Wagner B.K., Schreiber S.L. The Power of Sophisticated Phenotypic Screening and Modern Mechanism-of-Action Methods. Cell Chem. Biol. 2016;23:3–9. doi: 10.1016/j.chembiol.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An S., Fu L. Small-molecule PROTACs: An emerging and promising approach for the development of targeted therapy drugs. EBioMedicine. 2018;36:553–562. doi: 10.1016/j.ebiom.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandrasekaran S.N., Ceulemans H., Boyd J.D., Carpenter A.E. Image-based profiling for drug discovery: due for a machine-learning upgrade? Nat. Rev. Drug Discov. 2021;20:145–159. doi: 10.1038/s41573-020-00117-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moffat J.G., Vincent F., Lee J.A., Eder J., Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- 34.Mullard A. The phenotypic screening pendulum swings. Nat. Rev. Drug Discov. 2015;14:807–809. doi: 10.1038/nrd4783. [DOI] [PubMed] [Google Scholar]
- 35.Replogle J.M., Norman T.M., Xu A., Hussmann J.A., Chen J., Cogan J.Z., Meer E.J., Terry J.M., Riordan D.P., Srinivas N., et al. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat. Biotechnol. 2020;38:954–961. doi: 10.1038/s41587-020-0470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., Marjanovic N.D., Dionne D., Burks T., Raychowdhury R., et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaitin D.A., Weiner A., Yofe I., Lara-Astiaso D., Keren-Shaul H., David E., Salame T.M., Tanay A., van Oudenaarden A., Amit I. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167:1883–1896.e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 38.Adamson B., Norman T.M., Jost M., Cho M.Y., Nuñez J.K., Chen Y., Villalta J.E., Gilbert L.A., Horlbeck M.A., Hein M.Y., et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867–1882.e21. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGinnis C.S., Patterson D.M., Winkler J., Conrad D.N., Hein M.Y., Srivastava V., Hu J.L., Murrow L.M., Weissman J.S., Werb Z., et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat. Methods. 2019;16:619–626. doi: 10.1038/s41592-019-0433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sims D., Mendes-Pereira A.M., Frankum J., Burgess D., Cerone M.A., Lombardelli C., Mitsopoulos C., Hakas J., Murugaesu N., Isacke C.M., et al. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome Biol. 2011;12:R104. doi: 10.1186/gb-2011-12-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nim S., Jeon J., Corbi-Verge C., Seo M.H., Ivarsson Y., Moffat J., Tarasova N., Kim P.M. Pooled screening for antiproliferative inhibitors of protein-protein interactions. Nat. Chem. Biol. 2016;12:275–281. doi: 10.1038/nchembio.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.ThinkCyte VisionSort Technical Specifications. https://thinkcyte.com/download/visionsort-technical-specifications/
- 44.Sakata R.C., Ishiguro S., Mori H., Tanaka M., Tatsuno K., Ueda H., Yamamoto S., Seki M., Masuyama N., Nishida K., et al. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 2020;38:865–869. doi: 10.1038/s41587-020-0509-0. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J.H., Chung T.D., Oldenburg K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 46.Wilkins O.G., Capitanchik C., Luscombe N.M., Ule J. Ultraplex: A rapid, flexible, all-in-one fastq demultiplexer. Wellcome Open Res. 2021;6:141. doi: 10.12688/wellcomeopenres.16791.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., Liu X.S. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian R., Gachechiladze M.A., Ludwig C.H., Laurie M.T., Hong J.Y., Nathaniel D., Prabhu A.V., Fernandopulle M.S., Patel R., Abshari M., et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron. 2019;104:239–255.e12.e212. doi: 10.1016/j.neuron.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•All data are reported in the paper or deposited at Zenodo: https://doi.org/10.5281/zenodo.7701145, https://doi.org/10.5281/zenodo.7703670, https://doi.org/10.5281/zenodo.7709846, https://doi.org/10.5281/zenodo.10472989.
-
•NGS fastq files of NFkB screening (replicate 1&2 of pilot screening, related to Figures S2A and S2K) are available in Zenodo: https://doi.org/10.5281/zenodo.7701145.
-
•NGS fastq files of NFkB screening (replicate 1&2 kinase library screening, related to Figures 3B, and S4A–S4C) are available in Zenodo: https://doi.org/10.5281/zenodo.7701145.
-
•NGS fastq files of macrophage label-free screening (replicate 2&3, related Figures S9D–S9G; Table S3) are available in Zenodo: https://doi.org/10.5281/zenodo.7701145.
- •
-
•Text files of similarity score taken by Amnis image flow cytometer (Figure 3D) are available in Zenodo: https://doi.org/10.5281/zenodo.7701145.
-
•NGS fastq files of macrophage label-free screening (replicate 1, related to Figure 5A) are available in Zenodo: https://doi.org/10.5281/zenodo.7703670.
-
•Csv files of ML classifier training and test data (NFkB nuclear translocation, related to Figures 2D and 6) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (mitochondria/lysosome, related to Figure 2H) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (LC3 aggregation, related to Figure 2L) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (T cell mitochondria, related to Figure 2P) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (Macrophage M0/M1, related to Figure 4D) are available in Zenodo https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (monocyte/macrophage, related to Figure 4G) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•Csv files of ML classifier training and test data (T cell exhaustion, related to Figure 4J) are available in Zeonodo: https://doi.org/10.5281/zenodo.7709846.
-
•Text files of SVM-based prediction probabilities in GC (Figures 3D and S5) are available in Zenodo: https://doi.org/10.5281/zenodo.7709846.
-
•NGS sequencing raw data (fastq files) were also deposited at DNA DataBank of Japan (DDBJ)
-
•Submission: DDBJ: DRA017748 (sadaotalab1-0001_Submission)
-
•BioProject: DDBJ: PRJDB17362 (PSUB022209)
-
•BioSample: DDBJ: SAMD00732055-SAMD00732060 (SSUB028198)
-
•Experiment: DDBJ: DRX509723-DRX509728 (sadaotalab1-0001_Experiment_0001–0024)
-
•Run: DDBJ: DRR525841-DRR525847 (sadaotalab1-0001_Run_0001–0020)
-
•
-
•
-
•Data analysis was performed using code written in Python, which is available in Zenodo. To analyze the data, csv files are imported and run Python code.
-
•Python code file (svm.py) for ML classification and drawing confusion matrix, SVM histogram, and PR curve, and calculation of probabilities from SVM score (Figures 3D, S1L, and S6J) is available in Zenodo: https://doi.org/10.5281/zenodo.10472989.
-
•Python code file (umap_plot.py) for UMAP plot (Figure 6) is available in Zenodo: https://doi.org/10.5281/zenodo.10472989.
-
•Instruction file (how_to_use.txt) is also available in Zenodo: https://doi.org/10.5281/zenodo.10472989.
-
•
-
•
Any additional information needed to re-analyze the data reported in this paper are available from the lead contact upon request.