Abstract
Breast cancer ranks first in incidence and fifth in cancer-related deaths among all types of cancer globally. Among breast cancer, triple-negative breast cancer (TNBC) has few known therapeutic targets and a poor prognosis. Therefore, new therapeutic targets and strategies against TNBC are required. We found that androgen-induced basic leucine zipper (AIbZIP), also known as cyclic AMP–responsive element-binding protein 3-like protein 4 (CREB3L4), which is encoded by Creb3l4, is highly upregulated in a particular subtype of TNBC, luminal androgen receptor (LAR) subtype. We analyzed the function of AIbZIP through depletion of AIbZIP by siRNA knockdown in LAR subtype TNBC cell lines, MFM223 and MDAMB453. In AIbZIP-depleted cells, the proliferation ratios of cells were greatly suppressed. Moreover, G1–S transition was inhibited in AIbZIP-depleted cells. We comprehensively analyzed the expression levels of proteins that regulate G1–S transition and found that p27 was specifically upregulated in AIbZIP-depleted cells. Furthermore, we identified that this p27 downregulation was caused by protein degradation modulated by the ubiquitin–proteasome system via F-box protein S-phase kinase-associated protein 2 (SKP2) upregulation. Our findings demonstrate that AIbZIP is a novel p27–SKP2 pathway-regulating factor and a potential molecule that contributes to LAR subtype TNBC progression.
Implications:
This research shows a new mechanism for the proliferation of LAR subtype TNBC regulated by AIbZIP, that may provide novel insight into the LAR subtype TNBC progression and the molecular mechanisms involved in cell proliferation.
Introduction
Breast cancer ranks first in incidence and fifth highest in cancer-related deaths among all cancer types globally. Global cancer statistics from 2020 showed that approximately 2.3 million new cases and 685,000 deaths were associated with this disease (1). Breast cancer is divided into three types defined mainly by receptor and protein expression: luminal breast cancer; HER2-positive breast cancer; and triple-negative breast cancer (TNBC). Luminal breast cancer is characterized by the expression of either estrogen receptor or progesterone receptor, and inhibitors targeting these are very effective for treatment of this type of breast cancer. HER2-positive breast cancer is characterized by the expression of HER2, and targeted mAbs have greatly improved the prognosis of this cancer type. TNBC is defined by the absence of estrogen receptor, progesterone receptor, and HER2, and exhibits complicated and diverse characteristics compared with luminal breast cancer and HER2-positive breast cancer. Thus, the prognosis of TNBC is poor (2, 3). Several treatments for TNBC were recently developed, including chemotherapy, pembrolizumab, sacituzumab-govitecan, and targeted therapies such as olaparib for BRCA-mutated TNBC. While these treatments are promising, their efficacy is not ideal. The effectiveness of pembrolizumab against TNBC was demonstrated by several clinical trials; however, resistance to pembrolizumab remains a challenge (4, 5). Sacituzumab-govitecan greatly improved the prognosis of refractory metastatic TNBC; however, the response rate was only 33%. Thus, the identification of new therapeutic targets in TNBC is required for the development of more effective treatments (4–6).
It has been further subdivided TNBC into several subtypes according to gene expression. As one example, Lehmann and colleagues categorized 587 cases of TNBC into six subtypes on the basis of gene expression: basal-like 1 (BL1); basal-like 2 (BL2); immunomodulatory (IM); mesenchymal-like (M); mesenchymal stem-like (MSL); and luminal androgen receptor (LAR) subtype (7). This classification system is widely accepted and has contributed to the identification of several effective therapeutic targets that were overshadowed in the overall TNBC analysis (6, 8).
LAR subtype TNBC represents 15% to 20% of all TNBC cases and exhibits the poorest prognosis and lowest response to chemotherapy compared with other TNBC subtypes. Moreover, this TNBC subtype has a low complete remission rate and short-term relapse-free survival (RFS; refs. 8, 9). In molecular aspects, LAR subtype TNBC has high expression of androgen receptor (AR) and genes associated with androgenic hormonally-regulated pathways (7). Consequently, there have been many attempts to develop new strategies to treat LAR subtype TNBC with drugs that inhibit AR activity. However, their therapeutic value is limited, and effective treatment for LAR subtype TNBC has not yet been established (8). Therefore, the identification of new targets to inhibit proliferation, invasion, and metastasis of LAR subtype TNBC is required.
Old astrocyte specifically induced substance (OASIS) family proteins are composed of OASIS [also known as cyclic AMP–responsive element-binding protein 3-like protein 1 (CREB3L1)], BBF2 human homolog on chromosome 7 (BBF2H7; also known as CREB3L2), Luman (also known as CREB3), CREB-H (also known as CREB3), and androgen-induced basic leucine zipper (AIbZIP; also known as CREB3L4; ref. 10). These proteins localize to the endoplasmic reticulum (ER) membrane and function as transcription factors when cleaved by S1P and S2P protease (10, 11). We previously demonstrated that OASIS family proteins function in many cellular processes such as stress response, cell differentiation, cell survival, cell proliferation, tissue formation, and diseases (12–19). In addition, recent studies have suggested that these proteins are involved in the onset and progression of malignant tumors (18–21). For example, the OASIS is downregulated in patients with glioma, breast cancer, and bladder cancer. Low expression of OASIS in these cancers causes high metastatic ability and unfavorable prognosis (22–24). In contrast, the expression of OASIS is upregulated in thyroid cancer, resulting in the acquisition of stem cell–like undifferentiated status and high proliferative potential (25, 26). BBF2H7, another OASIS family protein, is induced in several types of glioblastoma and accelerates cancer cell proliferation and malignancy (27, 28). In low-grade fibromyxoid sarcoma (LGFMS), gene translocation of the genes encoding these OASIS family proteins leads to fusion events with the fused in sarcoma (FUS) gene (29, 30). The chimera protein FUS/BBF2H7 is a common characteristic of LGFMS, and rare cases of the chimeric FUS/CREB3L1 have also been reported. While the precise functions of these chimeric proteins in the development of LGFMS are still unclear, these fusion proteins may upregulate several genes including CD24 and forkhead box L1 genes (30). Together, these studies have indicated a robust link between OASIS family members and tumorigenesis. However, the overall mechanisms of the regulation for tumorigenesis by these proteins have been still unclear.
AIbZIP was originally identified as a gene that is upregulated by androgen treatment in prostate cancer cell line (31). As well as the other members of OASIS family, AIbZIP may be associated with the pathology of cancer. Indeed, several studies suggested that AIbZIP contributes to cancer progression in several cancer types (32–36). The demethylation of the Aibzip promoter region and following increased transcription of Aibzip mRNA promote the onset of prostate cancer (32). The high level of Aibzip mRNA expression is involved in short-term RFS in breast cancer (37). These reports suggest that AIbZIP may be associated with the pathogenesis of cancers like as the other OASIS family members. However, the molecular basis for regulating tumorigenesis by AIbZIP remains to be largely unelucidated.
To examine the potential role and mechanism of AIbZIP in malignant tumor, we performed this study. In silico analysis revealed that AIbZIP was highly expressed in LAR, a subtype of TNBC with very poor prognosis. We investigated the role of AIbZIP in LAR and discovered that AIbZIP plays a significant role in cell cycle regulation. Our data further demonstrated that the p27-F-box protein S-phase kinase-associated protein 2 (SKP2) pathway is involved in this cell cycle regulation. These findings are significant in that they reveal an aspect of the molecular mechanism of LAR subtype TNBC progression.
Material and Methods
Cell culture, reagents, and siRNA
MDAMB453 (HTB-131, RRID:CVCL_0418) and MDAMB468 (HTB-132, RRID:CVCL_0419) cells were purchased from ATCC. MFM223 (98050130, RRID:CVCL_1408) cells were purchased from European Collection of Authenticated Cell Cultures. MDAMB453 and MDAMB468 cells were maintained in L-15 medium (Gibco) supplemented with 10% FBS (Biosera) and 3 g/L NaHCO3 at 37°C in 5% CO2. MFM223 cells were maintained in minimum essential medium eagle (Wako) supplemented with 10% FBS at 37°C in 5% CO2. A total of 0.07 g/L penicillin and 0.1 g/L streptomycin were added into all mediums. All cell lines were obtained within 3 years and passaged in our laboratory for fewer than 6 months after receipt. Characterization was performed by each cell bank. Mycoplasma contamination testing was performed by examining the absence of cytoplasmic dotting patterns in DNA staining (last date was November 2023). Cells were not kept in culture for more than 25 passages. For cell treatments, 10 μmol/L MG132 (Wako) and 100 μg/mL cycloheximide (CHX; Wako) were used. Plasmids were transfected using Everyday Transfection (EZ Biosystems) according to the manufacturer's protocol. To introduce AIbZIP, myc-tagged-AIbZIP plasmid, which was described previously (18), was used. Stable cell lines were established using transposon. Plasmids for Sleepingbeauty based transposon vectors were obtained from addgene. Donor plasmids (pSBTet-Pur: #60507; ref. 38) were transfected together with the expression plasmid for a mutant form of transposase, SB100X [pCMV(CAT)T7-SB100: #34879; ref. 38]. cDNA of human Skp2 was obtained by reverse transcription using ReverTra Ace (TOYOBO) and RNA purified from MDAMB453 cells. Skp2 was cloned by PCR with primers containing Flag coding sequence.
(Forward) 5′-CTCGAAAGGCCTCTGCCACCATGGACTACAAGGACGACGATGACAAGATGCACAGGAAGCACCTCCAG-3′
(Reverse) 5′-ATGGAAGCTTGGCCTCTATCATAGACAACTGGGCTTTTGC-3′
Integrated cells were selected using puromycin (2 μg/mL).
RNAi
For knockdown of Aibzip and Skp2, siRNA targeting human Aibzip (we previously described; ref. 18), Aibzip#2 (sense; 5′-GAACCAAGAAUUACAGAAA-3′, antisense; 5′-UUUCUGUAAUUCUUGGUUC-3′), Aibzip#3 (sense; 5′-CAGAAAAUCUGGAGACCCA-3′, antisense; 5′-UGGGUCUCCAGAUUUUCUG-3′) and Skp2 (s12892; Ambion) and control siRNA (AM4611; Ambion) were used. siRNAs were transfected using RNAiMax (Thermo Fisher Scientific) as described in the manufactures protocol.
Protein preparation and Western blotting
Proteins were extracted from cell lines using cell lysis buffer containing 50 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 1% Triton X-100, and Protease Inhibitor Cocktail Set V (Wako) at 4°C. The lysates were incubated on ice for 30 minutes and sonicate lysates 10% duty cycle 20 times. After centrifugation at 15,000 × g for 10 minutes, the protein concentrations of the supernatants were determined using a bicinchoninic acid assay kit (Thermo Fisher Scientific). Equal amounts of protein were applied to SDS-PAGE. For immunoblotting, the following antibodies were used; anti-β-actin (1:10,000; MAB1501, RRID:AB_2223041, Merk Millipore), anti-p27 (1:1,000; #2552, RRID:AB_10693314, Cell Signaling Technology), anti-p21 (1:1,000; #2947, RRID:AB_823586, Cell Signaling Technology), anti-AKT (1:1,000, #4691, RRID:AB_915783, Cell Signaling Technology), anti-phospho-AKT (1:1,000; #4060, RRID:AB_2315049, Cell Signaling Technology), anti-p16 (1:500; sc-1661, RRID:AB_628067, Santa Cruz Biotechnology), anti-CyclinE (1:1,000; SC-247, RRID:AB_627357, Santa Cruz Biotechnology), anti-CyclinD1 (1:1,000, sc-8396, RRID:AB_627344, Santa Cruz Biotechnology), anti-RB (1:1,000, sc-102, RRID:AB_628209, Santa Cruz Biotechnology), anti-phospho-RB (1:500; SC-271930, RRID:AB_10655683, Santa Cruz Biotechnology), anti-Src (1:1,000; SC-8056, RRID:AB_627306, Santa Cruz Biotechnology), anti-phospho-Src (1:500, sc-81521, RRID:AB_1125729, Santa Cruz Biotechnology), anti-CDK2 (1:500; sc-6248, RRID:AB_627238, Santa Cruz Biotechnology), anti-SKP2 (1:1,000, sc-74477, RRID:AB_2187653, Santa Cruz Biotechnology), anti-AIbZIP (1:2,000; H00148327-AP51, RRID:AB_10551536, Abnova). The band intensity was quantified with CSAnalyzer4 (ATTO).
Immunofluorescence staining and DNA staining
The cells transfected with Aibzip targeting siRNA and control siRNA were cultured on coverslip (Matsunami Glass) for 72 hours and fixed in 4% paraformaldehyde (PFA) for 30 minutes. After three times wash with PBS, the cells were permeabilized in 0.1% Triton-X 100 for 5 minutes followed by blocking with 1% BSA. Cells were incubated with anti-p27 (1:250; SC-528, RRID:AB_632129, Santa Cruz Biotechnology) antibody at 4°C overnight. For secondary antibodies, goat anti-rabbit IgG F(ab′)2 fragments conjugated with Alexa 568 were used. For DNA staining, DRAQ5 (1:3,000, BioStatus Limited) were used. Confocal fluorescent images were acquired using FV1000D (Evident). Procession and analysis of images was performed with ImageJ (NIH, Rockville, MD; ref. 39). Nuclear areas were defined with DRAQ5, and binary images were generated using “Threshold (Li, auto)” and whole cell areas were defined with p27 signal and the binary images were generated using (Triangle, auto) in ImageJ. Nuclear p27 intensity and whole cell p27 intensity were analyzed by “analyze particles” in ImageJ. To quantify the intensity of p27 in cytosol, subtract the sum of nuclear p27 intensity from whole cell p27 intensity.
RNA isolation and quantitative PCR
Total RNA was isolated from cells using ISOGEN (Wako) according to the manufacturer's protocol. First-strand cDNA was synthesized from 1 μg RNA in a 20 μL reaction volume, using a random primer (Takara), ReverTra Ace (TOYOBO). Quantitative PCR was performed using KAPA SYBR FAST (Kapa Biosystems). The reverse-transcribed cDNA was measured by real-time PCR assay using a LightCycler 480 Instrument II (Roche). The following primer are used; 5′-TGCTAAGCAGCTGTTCCAGA-3′ (Skp2 forward), 5′-AAGATTCAGCTGGGTGATGG-3′ (Skp2 reverse), 5′-AGTCCATTTGATCAGCGGAGACTCG-3′ (Cdkn1b forward), 5′-TCGCACGTTTGACATCTTTCTCCC-3′ (Cdkn1b reverse), 5′-TCCTCCCTGGAGAAGAGCTAC-3′ (Actb forward), 5′-TCCTGCTTGCTGATCCACAT-3′ (Actb reverse).
Cell proliferation assay
Cells were seeded at 1 × 105 cells/well and cultured for 2–8 days. Suspended cells were mixed with 0.4% trypan blue, and the cell number was determined using a Countess 3 (Invitrogen). For the bromodeoxyuridine (BrdU) incorporation assay, cells were seeded at 1 × 105 cells/well and treated with 100 μmol/L BrdU (Sigma-Aldrich) for 24 hours. After fixation by 4% PFA, cells were treated with 1N HCl for 15 minutes. Anti-BrdU antibody (1:250; #5292, RRID:AB_10548898, Cell Signaling Technology) and DRAQ5 (1:3,000), respectively, were used to detect BrdU-positive cell and nucleus. Cells were visualized under a confocal microscope (FV1000D). For flow cytometric analysis, cells (1 × 106 cells) were fixed in 70% ethanol for 30 minutes at real temperature. After centrifugation, cell pellets were resuspended with staining buffer containing 12.5 μg/mL propidium iodide (Sigma-Aldrich) 0.2 mg/mL RNase A (Nippon Gene), and 0.1% Triton X-100 for 30 minutes. An LSR Fortessea X-20 (BD Biosciences) was used for analysis.
Luciferase assay
MFM223 and MDAMB453 cells were plated at 5 × 104 cells/dish and cultured for 2 days. Cells were transfected with 0.2 μg of the pGL3 basic reporter plasmid (Promega) carrying the firefly luciferase gene (SKP2-Luc) driven by the human Skp2 promoter or mutant reporter constructs harboring a Skp2 promoter mutant lacking the CRE-binding site (ΔCRE-Luc) or the Skp2 promoter in which the CRE-binding site was mutated (mut-CRE-Luc) together with 0.02 μg of the pRL-SV40 co-reporter plasmid carrying the Renilla luciferase gene (Promega). Transfections were performed using Avalanche-Everyday Transfection Reagent (EZ Biosystems). The Skp2 promoter was identified using the University of California, Santa Cruz genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway; ref. 40). SKP2-Luc reporter constructs were generated by PCR; the ΔCRE-Luc and mut-CRE-Luc constructs were generated by inverse PCR using SKP2-Luc as a template.
To enhance cleavage of AIbZIP, cells were treated with 1 μmol/L brefeldin A (Sigma-Aldrich) for 24 hours. After 36 hours, luciferase activities were measured using a Dual-Luciferase Reporter Assay System (Promega) and a GloMax Multi+ Detection System (Promega) following the manufacturer's protocol. Relative luciferase activities were determined as the ratio between firefly and Renilla luciferase activities.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP Assay kit (Sigma-Aldrich) following the manufacturer's protocol as described previously (14). Briefly, MFM223 and MDAMB453 cells were plated at 5 × 105 cells/dish and cultured for 2 days. To promote the cleavage of AIbZIP, cells were treated with 1 μmol/L brefeldin A (Sigma-Aldrich) for 24 hours. Cells were cross-linked using 1% formaldehyde for 15 minutes at 37°C, followed by inactivation with 0.15 mol/L glycine for 5 minutes at room temperature. The cells were then lysed with SDS lysis buffer and sonicated (20 × 10 seconds sonication pulses at 1 minute intervals; Sonifier 250, Branson). Equal amounts of chromatin from each sample were incubated overnight at 4°C with anti-AIbZIP (Abnova) or anti-Histone H3 (sc-517576, RRID:AB_2848194, Santa Cruz Biotechnology) antibodies or mouse IgG (#5415, RRID:AB_10829607, Cell Signaling Technology). Cross-linking was reversed by incubating for 6 hours at 65°C, and DNA was purified by phenol-chloroform extraction and ethanol precipitation. Purified DNA was used for PCR analysis. The primers used to detect the human Skp2 promoter were 5′-CTGGAATATGTCACGCTCCCTACTC-3′ (forward) and 5′-GACAGGTCTCATAAATAGCAACATG-3′ (reverse).
Data collection and analysis
To compare the expression level of Aibzip mRNA between normal and cancer in pan cancer view, UALCAN (http://ualcan.path.uab.edu/index.html) was used (41). To compare the expression level of Aibzip mRNA between breast cancer subtypes, The Cancer Genome Atlas (TCGA) database was interrogated with using cBioPortal (cBioPortal, https://www.cbioportal.org/; ref. 42) and UCSC Xena Browser (UCSC Xena, https://xenabrowser.net; ref. 43). For subtyping breast cancer, previous study (44) was referred. ER type was classified as luminal type. Other types were classified as named. The validity of TCGA-BRCA database was certificated by following reports (45–47). We added supplementary table showing patient ID and subtype of breast cancer we applied (Supplementary Table S1). To create volcano plot, ggVolcanoR was used (48). In analysis of mRNA sequence, genes with zero in numerator or denominator in fold change calculation and genes with duplicate names with lower fold change were removed to conform the standard of ggVolcanoR. Androgen response genes were referred gene set enrichment analysis of Human Gene Set: HALLMARK_ANDROGEN_RESPONSE (49). GSE167213 dataset was obtained from Gene Expression Omnibus of NCBI (50). The validity and details are referred by the previous article (51).
Statistical analysis
Statical comparisons were made using unpaired Student t test. Differences were considered statically significant at P < 0.05. P value of less than 0.05, 0.01, and 0.001 are indicated as *, **, and ***, respectively. n.s. means not significant. To construct the boxplot, Microsoft Excel was used.
Data availability
The data generated in this study are available upon request from the corresponding author.
Results
AIbZIP is upregulated in LAR subtype TNBC
We initially performed a bioinformatic analysis to investigate the relationship between AIbZIP and cancer. We compared Aibzip mRNA expression levels between tumorous and normal tissues in TCGA database (Fig. 1A). Aibzip mRNA levels tended to be higher in many cancer types compared with those in normal tissues. In particular, Aibzip mRNA levels were second highest in breast cancer next to prostate cancer among all cancer types and were much higher than those of normal tissue (Fig. 1A). To assess the significance of AIbZIP expression in breast cancer, mRNA expression profiles were compared between normal tissue and tumor. Aibzip mRNA was increased about three times higher in tumor tissue (P-value 1.02 × 10−20), and this fold change ranks in the top 12% (2,210/19,905 genes) and P-value ranks top 15% (2,927/19,905 genes; Fig. 1B; Supplementary Table S2). Subsequently, we investigated which types of breast cancer express high levels of AIbZIP. Aibzip mRNA expression was approximately three times higher in luminal, HER2, and LAR subtypes than in normal tissue. However, other subtypes of TNBC, BL1, BL2, M, IM, MSL, and unclassified (UNC), exhibited almost the same levels of Aibzip mRNA expression as normal tissues (Fig. 1C). In addition, we analyzed another database of TNBC mRNA expression data (GSE167213). The expression of Aibzip mRNA was also higher in the LAR subtype compared with other subtypes of TNBC in this database (Supplementary Fig. S1A), indicating that AIbZIP may have an impact on the cancerous characteristics of luminal type, HER2 type, and LAR type breast cancer. Among them, the therapeutic target for the luminal type and HER2 type is clarified (2, 3). That of LAR subtype TNBC is inhibition of AR activity. However, this treatment has only limited effect (8). Thus, we focused on the role of AIbZIP in LAR subtype TNBC due to the necessary to develop new therapeutic strategies.
Figure 1.
AIbZIP is upregulated in LAR subtype TNBC. A, Comparison of Aibzip mRNA expression in cancerous and normal tissues in a pan-cancer view obtained by UALCAN. The vertical axis indicates mRNA expression levels [transcripts per million (TPM) +1], which was converted to a logarithmic function with base 2. BLCA, bladder urothelial carcinoma; BRCA, breast carcinoma; CESC, cervical squamous cell carcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin melanoma; THCA, thyroid carcinoma; THYM, thymoma; STAD, stomach adenocarcinoma; UCEC, uterine corpus endometrial carcinoma. *, **, and *** means P < 0.05, P < 0.01, and P < 0.001 versus normal tissue. B, Volcano plot of all gene expression profiling from normal tissue versus tumor. Each point represents the difference in expression (log fold-change) and statistical significance (P-value) between the two groups. Aibzip (also called Creb3l4) is showed by red colored point. The X-axis represents log fold change value and Y-axis represents −log10P-value. C,Aibzip mRNA expression levels are shown by a box plot. Midline means median. Number of samples: normal = 114, luminal = 710, HER2 = 158, LAR = 17, BL1 = 34, BL2 = 17, M = 40, IM = 36, MSL = 22, UNC = 13. *** means P < 0.001 versus normal tissue. D, Volcano plot of all gene expression profiling from all TNBC versus LAR subtype TNBC. Aibzip (also called Creb3l4) is showed by red colored point. E, Volcano plot of androgen response gene expression profiling from all TNBC versus LAR subtype TNBC. Aibzip (also called Creb3l4) is showed by red colored point.
To examine the significance of AIbZIP expression in LAR subtype TNBC, we compared the mRNA expression profiles of all genes and androgen response–related genes in LAR with normal tissue and all TNBC. AIbZIP was upregulated about 3.2 times higher in the LAR subtype TNBC compared with normal tissue (P-value 9.52 × 10−21). This fold change value ranks in the top 12% of all examined genes (2,155/19,445 genes) and the P-value ranks in the top 2.7% (519 out 19,445 genes; Supplementary Fig. S2A; Supplementary Table S2). Among the genes regulated by the androgen response pathway, the fold change value of AIbZIP ranks in the top 10% genes (10/102 genes) and the P-value ranks in the top 3% (3/102 genes; Supplementary Fig. S2B; Supplementary Table S2). AIbZIP was upregulated about 2.4 times higher in the LAR subtype TNBC compared with all TNBC (P-value is 2.11 × 10−8). The fold change value ranks in the top 5.5% of genes (1,064/19,526 genes) and the P-value ranks in the top 0.32% (62/19,526 genes; Fig. 1D; Supplementary Table S2). Among the genes regulated by androgen response pathway, the fold change value of AIbZIP ranks in the top 13% (13/102 genes) and the P-value ranks in the top 2% (2/102 genes; Fig. 1E; Supplementary Table S2). These findings indicate that AIbZIP is significantly upregulated in the LAR subtype compared with other subtypes of TNBC and normal tissue. Among the genes associated with the androgen response pathway, AIbZIP is upregulated in LAR subtype cells, suggesting the importance of AIbZIP in the development of LAR subtype TNBC.
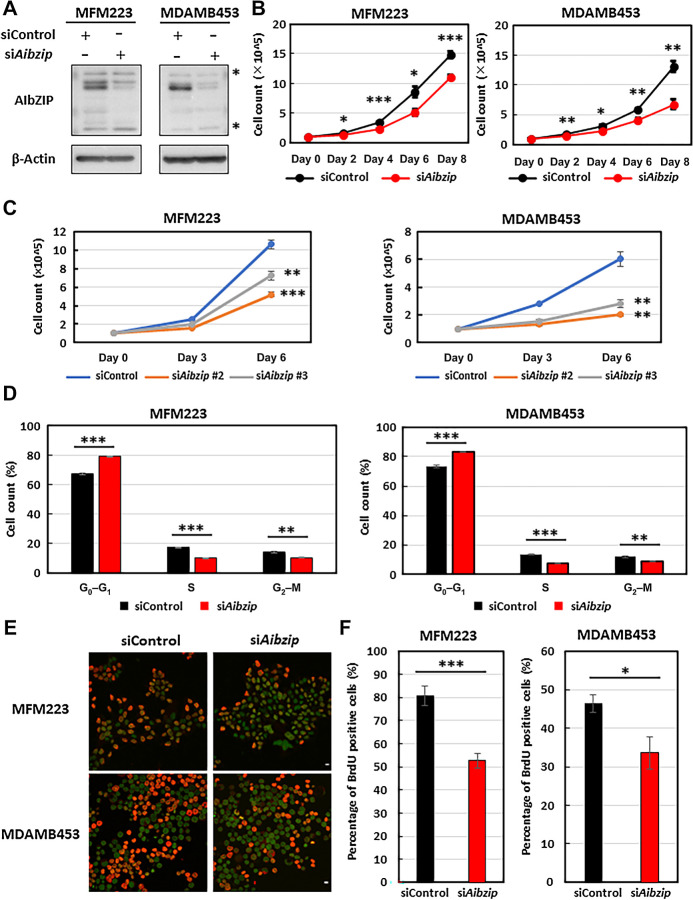
AIbZIP regulates cell proliferation in LAR subtype TNBC cells
To verify the impact of cell proliferation of AIbZIP on LAR subtype TNBC cells, we transfected AIbZIP-targeted siRNA (Aibzip siRNA) into MFM223 and MDAMB453 cell lines, which are typical models of LAR subtype TNBC (7). Western blotting showed that Aibzip knockdown effectively suppressed AIbZIP expression (Fig. 2A). Initially, the cell proliferation rate of these cells was investigated, and we found that that of Aibzip siRNA-transfected cells was significantly suppressed compared with control siRNA-transfected cells (Fig. 2B). Transfection of two siRNAs targeting AIbZIP that recognize distinct sequences also suppressed cell proliferation (Fig. 2C). Notably, Aibzip siRNA transfection in the non-LAR subtype TNBC model cell line MDAMB468 (7), which expresses very low levels of AIbZIP (Supplementary Fig. S3A), did not affect the cell proliferation (Supplementary Fig. S3B). Subsequently, the effects of AIbZIP depletion on cell cycle progression in MFM223 and MDAMB453 cells were investigated by cytometric analysis with propidium iodide staining. The proportion of cells in G0-G1–phase was approximately 10% higher, and that of cells in S-phase reduced approximately to half in Aibzip siRNA-transfected cells compared with control siRNA-transfected cells (Fig. 2D). These results are similar to those of MFM223 and MDAMB453 cells that were transfected with siRNA targeting prolactin-induced protein (PIP; ref. 52), which is considered essential for cell cycle progression (53), suggesting that AIbZIP has a significant impact on the transition from G1- to S-phase, as well as PIP. To provide additional evidence that supports the hypothesis that AIbZIP is involved in S-phase entry, BrdU assays were performed to determine the frequency of cells undergoing DNA synthesis. This assay revealed that incorporation of BrdU was decreased in Aibzip siRNA-transfected cells (Fig. 2E and F; Supplementary Fig. S4A and S4B). Taken together, we concluded that AIbZIP is involved in G1–S transition in the cell lines of LAR subtype TNBC.
Figure 2.
AIbZIP depletion reduces cell proliferation and S-phase entry of cell cycle. A, Western blot analysis of AIbZIP in MFM223 and MDAMB453 cells transfected with Aibzip siRNA. Asterisk; nonspecific bands. B, Cell numbers of MFM223 and MDAMB453 cells transfected with Aibzip siRNA or control siRNA (mean ± SD, n = 5). C, Cell numbers of MFM223 and MDAMB453 cells transfected with Aibzip siRNA #2, Aibzip siRNA #3, or control siRNA (mean ± SD, n = 3). D, Flow cytometric analysis of MFM223 and MDAMB453 cells transfected with Aibzip siRNA (mean ± SD, n = 3). E, BrdU incorporation assay in MFM223 and MDAMB453 cells transfected with Aibzip siRNA (BrdU: red, DRAQ5: green). Bar: 10 μm. F, Percentages of BrdU-positive cells among DRAQ5-positive cells in E (mean ± SD, n = 5).
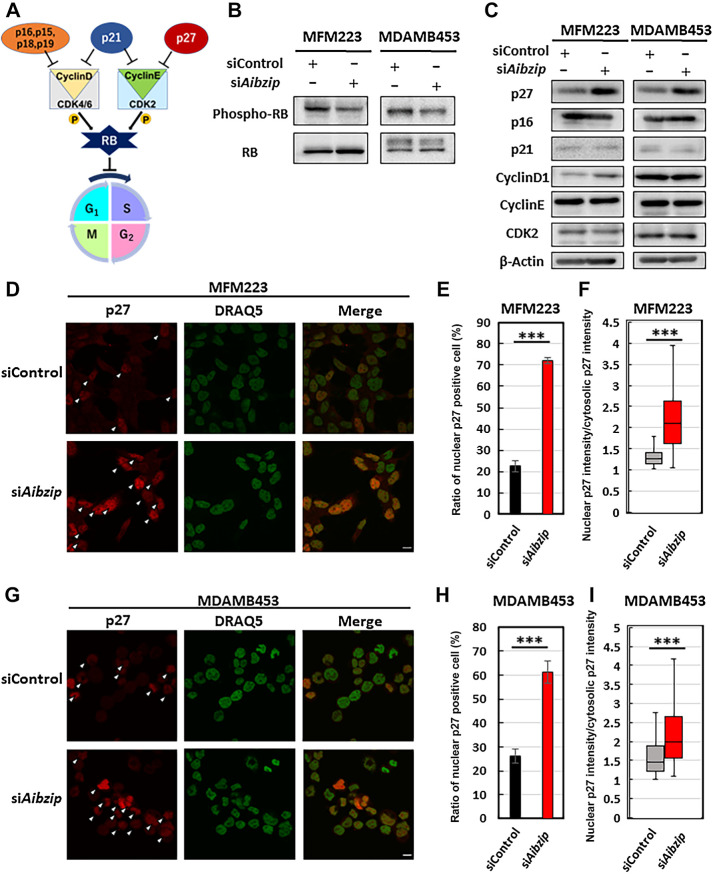
Upregulation of p27 by AIbZIP
G1–S transition is rigidly monitored. The central regulator of G1–S transition is RB–E2F complex (Fig. 3A), which downregulates many genes that encode critical cell cycle regulators to inhibit cell cycle progression. Phosphorylation of RB and subsequent divergence of RB–E2F complex is required to advance the cell cycle into S-phase (54). To investigate whether AIbZIP governs this system, we quantified the phosphorylation status of RB in MFM223 and MDAMB453 by Western blotting. As expected, RB phosphorylation levels were suppressed in Aibzip-siRNA transfected cells (Fig. 3B; Supplementary Fig. S5A). The phosphorylation of RB is regulated by multiple factors. Cyclin D–cyclin-dependent kinase (CDK) 4/6 complex and cyclin E–CDK2 complex directly phosphorylate RB. In addition, p21, p16, p27, and other CDK inhibitors suppress these complexes (Fig. 3A). Retention of the exquisite balance of these proteins contributes to accurate cell cycle progression (55). To analyze which factors AIbZIP mediates for RB phosphorylation, the expression levels of these factors were compared by Western blotting and p27 was only identified as the protein that was about doubly altered in both cell lines (Fig. 3C; Supplementary Figs. S6A, S6B, and S7A). In MDAMB468 cells, Aibzip siRNA transfection had no impact on p27 expression level (Supplementary Fig. S3C). Next, the subcellular localization and expression intensity of p27 in Aibzip siRNA-transfected cells were investigated by immunofluorescence staining with anti-p27 antibody (Fig. 3D–I). In MFM223 cells, control siRNA-transfected cells exhibited only 23% nuclear p27-positive cells, while Aibzip siRNA-transfected cells exhibited 72% (Fig. 3E). In MDAMB453 cells, nuclear p27-positive cells were observed in 26% of control siRNA-transfected cells and 61% of Aibzip siRNA-transfected cells (Fig. 3H). Moreover, the intensity of p27 nuclear signals was higher in Aibzip siRNA-transfected nuclear p27-positive cells than in control siRNA-transfected nuclear p27-positive cells (Fig. 3F and I), indicating that p27 was upregulated and accumulated in the nucleus by Aibzip siRNA transfection. These findings indicate AIbZIP-mediated p27 regulation and we focused on the detailed mechanisms of this regulation. Taken together, we concluded that AIbZIP suppresses the expression of p27, which is a CDK inhibitor, and promotes cell proliferation.
Figure 3.
Depletion of AIbZIP promotes p27 upregulation and its translocation into the nucleus. A, Schema of important factors associated with G1-S–phase transition of cell cycle. B, Western blot analysis of RB and its phosphorylated form (Thr821/Thr826) in MFM223 and MDAMB453 cells transfected with Aibzip siRNA. C, Western blot analysis of p27, p16, p21, Cyclin D1, Cyclin E, and CDK2 in MFM223 and MDAMB453 cells transfected with Aibzip siRNA. D, Immunofluorescence staining of p27 in MFM223 cells transfected with Aibzip siRNA (p27: red, DRAQ5: green). Arrowheads indicate the cells which p27 was localized to the nucleus. Bar: 10 μm. E, Percentages of cells in which p27 was localized to the nucleus. (mean ± SD, n = 4). F, Ratio of nuclear p27 intensity divided by cytosolic p27 intensity (mean ± SD, siControl; n = 33 cells, siAibzip; n = 86 cells). Midline means median. G, Immunofluorescence staining of p27 in MDAMB453 cells transfected with Aibzip siRNA (p27: red, DRAQ5: green). Arrowheads indicate the cells with p27 localized to the nucleus. Bar: 10 μm. H, Percentages of cells in which p27 was localized to the nucleus (mean ± SD, n = 4). I, Ratio of nuclear p27 intensity divided by cytosolic p27 intensity (mean ± SD, siControl; n = 60 cells, siAibzip; n = 39 cells). Midline means median.
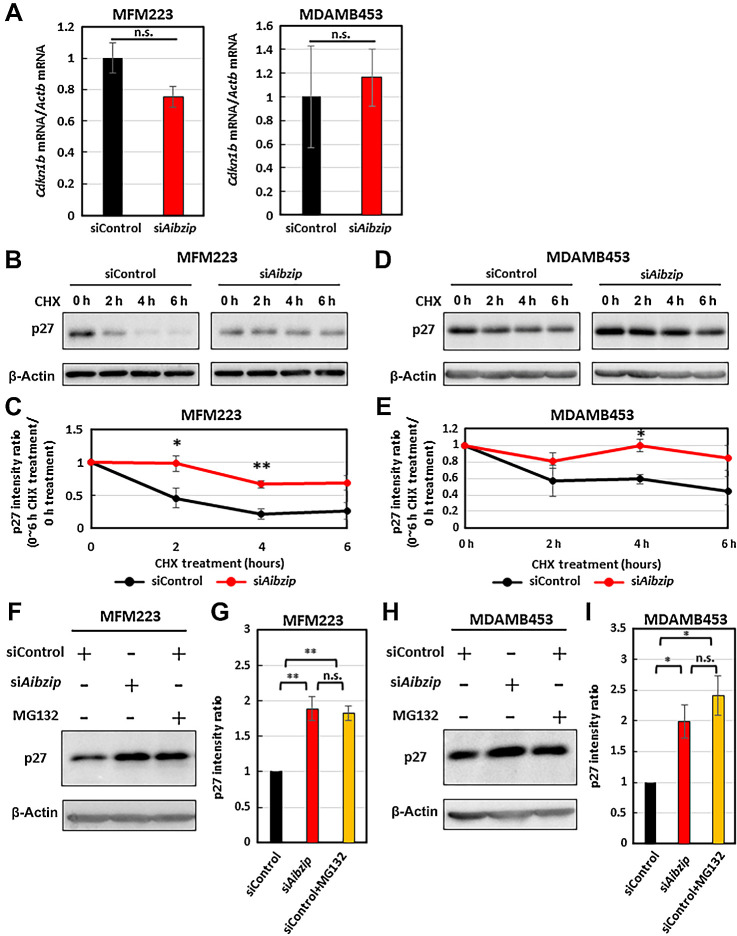
AIbZIP promotes p27 degradation
The next question to address was how AIbZIP inhibits the expression and nuclear accumulation of p27. Some evidence suggests that p27 expression is regulated at the mRNA level by various transcription factors (56), while other evidence indicates that p27 is also regulated by protein degradation via the ubiquitin–proteasome pathway (57, 58). We first checked the mRNA levels of p27 (Cdkn1b) in Aibzip siRNA-transfected cells by quantitative RT-PCR. However, contrary to protein levels, Cdkn1b mRNA levels were not upregulated as much in Aibzip siRNA-transfected cells (Fig. 4A). Therefore, AIbZIP-dependent p27 downregulation may not occur at the transcriptional level, but rather at the posttranslational level. We next evaluated p27 protein stability in Aibzip siRNA-transfected cells by blocking new protein synthesis with CHX treatment. As shown in Fig. 4B–E, p27 protein levels were reduced by 78.4% in MFM223 cells and 40.4% in MDAMB453 cells in control siRNA-transfected cells following treatment with 100 μg/mL CHX for 4 hours. However, in Aibzip siRNA-transfected cells, p27 protein levels were reduced by only 33.7% and 0.055% in MFM223 and MDAMB453 cells, respectively. It is known that p27 is degraded via the ubiquitin–proteasome pathway. Therefore, p27 protein levels were analyzed in MFM223 and MDAMB453 cells treated with proteasome inhibitor MG132 by Western blotting. In control siRNA-transfected cells, MG132 treatment upregulated p27 protein levels to the same level as in the Aibzip siRNA-transfected cells (Fig. 4F–I). These results indicate that AIbZIP degrades p27 via the ubiquitin–proteasome system.
Figure 4.
Knockdown of Aibzip inhibits p27 degradation. A, Quantitative PCR analysis of Cdkn1b mRNA in MFM223 and MDAMB453 cells transfected with Aibzip siRNA. mRNA levels were normalized by Actb. (mean ± SD, n = 4). Western blot analysis of p27 in MFM223 cells (B) and MDAMB453 cells (D) transfected with Aibzip siRNA. Cells were treated with 100 μg/mL CHX for the indicated times. C and E, Quantification of p27 protein expression levels in B and D (mean ± SD, n = 3). Western blot analysis of p27 in MFM223 (F) and MDAMB453 (H) cells transfected with Aibzip siRNA. Cells were treated with 10 μmol/L MG132 for 6 hours. G and I, Quantification of p27 protein expression levels in F and H (mean ± SD, n = 3).
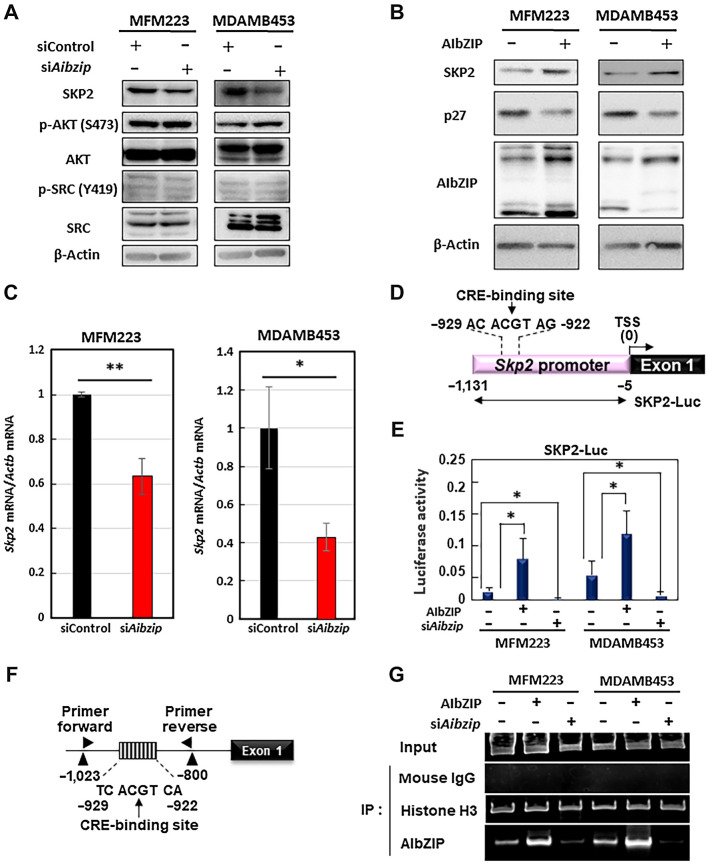
AIbZIP directly upregulates Skp2 transcription
It has been reported that the stability of p27 protein is regulated by several proteins such as SKP2, AKT, and SRC. We quantified the expression levels of SKP2, AKT, and SRC and found that SKP2, which is an E3 ubiquitin ligase that recognizes and ubiquitinates p27 (59), was downregulated in Aibzip siRNA-transfected cells (Fig. 5A; Supplementary Figs. S7A, S8A, and S8B). In MDAMB468 cells, SKP2 protein expression level was not affected by Aibzip siRNA transfection (Supplementary Fig. S3C). Conversely, phosphorylated AKT and SRC, which are phosphate kinases that phosphorylate p27 and lead to its degradation (58), were not changed in Aibzip siRNA-transfected cells (Fig. 5A; Supplementary Fig. S8A and S8B). To make sure the relationship between AIbZIP and SKP2, we transiently overexpressed AIbZIP in MFM223 and MDAMB453 cells, and found that SKP2 expression was upregulated in AIbZIP-introduced cells (Fig. 5B). On the contrary, p27 expression was downregulated in these cells, indicating that AIbZIP does indeed upregulates SKP2. Furthermore, Skp2 mRNA expression levels were evaluated by quantitative RT-PCR and they were also decreased in Aibzip siRNA-transfected cells (Fig. 5C). Luciferase assays were performed using a reporter construct carrying a 1.0-kb promoter region of Skp2 (SKP2-Luc; Fig. 5D) and reporter activity was significantly induced by expression of AIbZIP (Fig. 5E). By contrast, knockdown of Aibzip reduced reporter activity (Fig. 5E). These results suggest that SKP2 is regulated by AIbZIP at the transcription level. A putative AIbZIP consensus binding site (10, 31), the cAMP response element (CRE)-binding site, was identified in the Skp2 promoter region (−929 to −922 bp from the Skp2 transcription start site; Fig. 5D; Supplementary Fig. S9A). Induction of reporter activity was reduced in MFM223 and MDAMB453 cells transfected with the ΔCRE-Luc reporter (lacking the CRE-binding site) or mut-CRE-Luc reporter (replacing a mutated CRE-binding site) under non-treatment and AIbZIP-introduced conditions (Supplementary Fig. S9B and S9C). Chromatin immunoprecipitation assay detected the binding of AIbZIP to the endogenous Skp2 promoter in these cells (Fig. 5F and G). Binding to the promoter was increased by the introduction of AIbZIP. High level of AIbZIP binding was diminished by knockdown of Aibzip. We also observed an increase of reporter activity after the treatment with brefeldin A (BFA), which was previously demonstrated to cleave endogenous AIbZIP (ref. 18; Supplementary Fig. S10A and S10B). This induction was suppressed in cells with Aibzip knockdown or when ΔCRE-Luc or mut-CRE-Luc were used. The increased amount of AIbZIP binding to the Skp2 promoter by BFA was also canceled by the knockdown of Aibzip (Supplementary Fig. S10C). These findings suggest that AIbZIP upregulates SKP2 by binding directly to CRE-binding site within the Skp2 promoter and facilitates Skp2 transcription in MFM223 and MDAMB453 cells.
Figure 5.
Skp2 is a target of AIbZIP. A, Western blot analysis for SKP2, AKT and its phosphorylated form (S473), SRC and its phosphorylated form (Y419) in MFM223 and MDAMB453 cells transfected with Aibzip siRNA. B, Western blot analysis of p27 and SKP2 in MFM223 and MDAMB453 cells under non-treatment and AIbZIP-introduced conditions. C, Quantitative PCR analysis for Skp2 mRNA in MFM223 and MDAMB453 cells transfected with Aibzip siRNA. mRNA levels were normalized by Actb (mean ± SD, n = 4). D, Schematic of the Skp2 promoter region and reporter constructs. Numbers indicate the distance from the transcriptional start site. Luc, luciferase gene, TSS, transcriptional start site. E, Luciferase assays of MFM223 and MDAMB453 cells introduced AIbZIP or transfected with Aibzip siRNA (mean ± SD, n = 4). F, Schematic of the Skp2 promoter and the annealing sites of the primer set used in the chromatin immunoprecipitation assay. Numbers indicate the distance from the transcriptional start site. G, The results of PCR amplification of the Skp2 promoter containing the CRE-binding site after immunoprecipitation by the indicated antibodies. Mouse IgG and Histone H3 were used as negative and positive control, respectively.
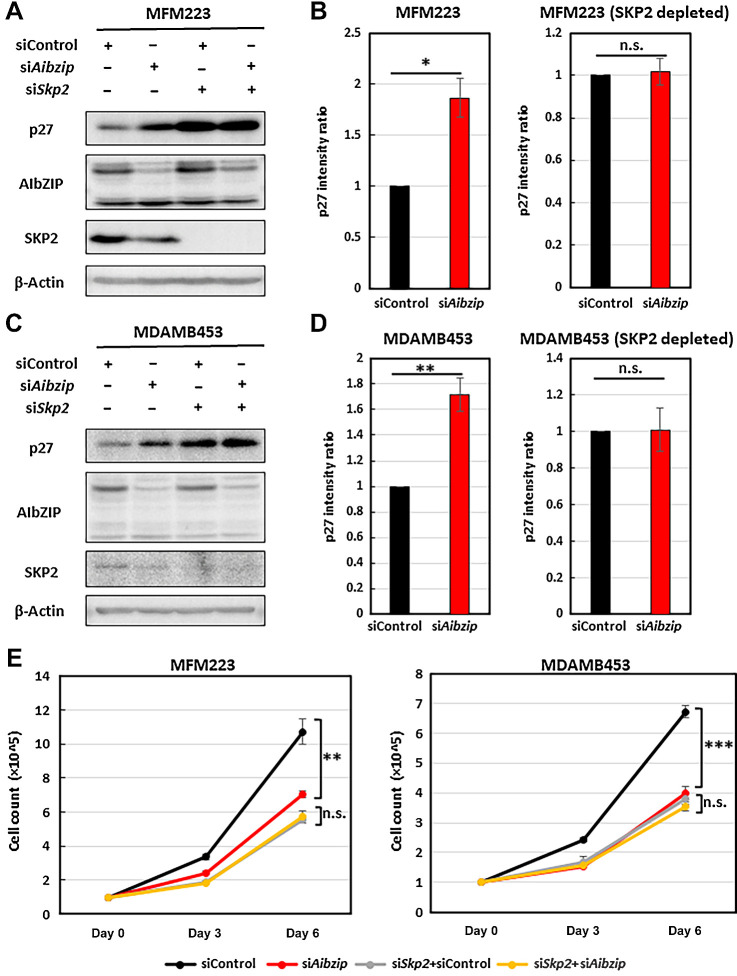
Accelerated cell proliferation and the downregulation of p27 by AIbZIP mediates the SKP2
To confirm that the increase in p27 and the suppression of cell proliferation induced by Aibzip siRNA transfection was entirely due to the SKP2-mediated pathway, we depleted Skp2 by siRNA knockdown. In control cells, Aibzip knockdown alone increased p27 protein (Figs. 3C and 6A–D); however, in SKP2-depleted cells, Aibzip knockdown did not affect the protein levels of p27 (Fig. 6A–D). Moreover, in control cells, Aibzip knockdown alone suppressed cell proliferation (Figs. 2B and 6E); however, suppression of cell proliferation by Aibzip knockdown was not affected in SKP2-depleted cells. To determine whether overexpression of SKP2 compensates for the effect of Aibzip knockdown, we established a MDAMB453 cell line that expresses Flag-tagged-SKP2 in the presence of doxycycline (tet-Flag-SKP2-MDAMB453). Knockdown of Aibzip suppressed cell proliferation with the upregulation of p27 in this cell line in the absence of doxycycline. After the addition of doxycycline, in spite of Aibzip knockdown, cell growth was not inhibited and the expression level of p27 was not also changed (Supplementary Fig. S11A and S11B). These results suggest that AIbZIP regulates cell proliferation via downregulation of p27 mediated by SKP2 upregulation.
Figure 6.
Aibzip knockdown-induced p27 upregulation and cell proliferation suppression are inhibited by Skp2 depletion. Western blot analysis of p27, AIbZIP, and SKP2 in MFM223 cells (A) and MDAMB453 cells (C) transfected with Aibzip siRNA and/or Skp2 siRNA. B and D, Quantification of p27 protein expression levels in A and C (mean ± SD, n = 3). E, Numbers of MFM223 cells and MDAMB453 cells transfected with Aibzip siRNA and/or Skp2 siRNA (mean ± SD, n = 5).
Discussion
In this study, we demonstrated that AIbZIP induces LAR subtype TNBC cell proliferation via acceleration of the cell cycle with following evidence; depletion of AIbZIP by siRNA; (i) suppressed cell proliferation; (ii) increased the population of cells in G1-phase and reduced those in S-phase; (iii) reduced BrdU incorporation; and (iv) reduced the phosphorylation levels of RB. Moreover, we demonstrated that AIbZIP degrades p27 protein via upregulation of SKP2 at the transcriptional level by the following evidence: (i) depletion of AIbZIP induced p27 upregulation; (ii) depletion of AIbZIP inhibited p27 degradation in CHX chase assays; (iii) AIbZIP directly bound to the Skp2 promotor region and facilitated Skp2 transcription; and (iv) AIbZIP depletion–induced p27 upregulation was inhibited by SKP2 depletion and overexpression.
Several studies reported that AIbZIP is highly upregulated and can promote malignant behavior in several cancer types. For example, it was reported that AIbZIP is upregulated in lung adenocarcinoma. Furthermore, AIbZIP upregulates the TGFβ pathway via induction of ADCK5 and contributes to cancer cell proliferation (36). Other research demonstrated that AIbZIP binds to the Pcna promoter region, suggesting that it leads to cancer cell proliferation in non-LAR subtype breast cancer cells (35). We also previously reported that AIbZIP modulates cell proliferation by downregulating p21 via inhibition of OASIS activation in a prostate cancer cell line (18). These findings indicate that the role of AIbZIP in cell proliferation is diverse and may differ between cell types. We identified a novel mechanism by which AIbZIP regulates cancer cell proliferation via the SKP2–p27 pathway in LAR subtype TNBC, and further studies are required to verify whether this regulation of cell cycle progression is conserved in other cell types. Indeed, in many cancers including luminal and HER2-positive breast cancer, AIbZIP is highly upregulated, as shown in Fig. 1A and C. Therefore, this novel pathway may contribute to cancer progression in other cancer types.
Previous studies reported that the expression of SKP2 and following p27 degradation are necessary for cancer progression and survival in many types of cancer cells (58, 60), and SKP2 expression is correlated with low prognosis in almost all cancers including breast cancer (61, 62). However, cancer treatment strategies targeting SKP2 have not yet been widely used in the clinical stages. Therefore, many studies explored the mechanisms of SKP2 regulation. Several proto-oncoproteins such as E2f1, Mycn, Foxm1, Elk1, Nfkb1, Sp1, and Gabp1 have been reported to upregulate Skp2 at the transcriptional level (63). Furthermore, other studies reported that MYC and FOXM1 are highly upregulated and contribute to Skp2 transcription and cancer progression in basal subtype TNBC, which is a major TNBC subtype other than LAR subtype (64, 65). However, until now, it has not been determined how SKP2 is regulated in LAR subtype TNBC. Our findings that AIbZIP directly binds to the Skp2 promotor region and facilitates its transcription help elucidate the mechanism of SKP2 regulation in LAR subtype TNBC and suggest that AIbZIP plays an equivalent role to other SKP2-regulating proto-oncoproteins such as MYC or FOXM1.
The most significant aspect of LAR subtype TNBC is the expression of AR. However, whether AR promotes or suppresses cell proliferation and tumor progression in TNBC remains controversial and no strategies targeting AR have been established (66–68). Therefore, identifying genes regulated by AR and that are involved in tumor growth may lead to the establishment of novel targets for therapy against AR-positive TNBC. These studies will also deepen understanding of the functions and roles of AR in TNBC. In this research, we analyzed the impact of AIbZIP on cell proliferation in two LAR subtype cell line models, MFM223 and MDAMB453 cells. These cell lines exhibit different responses after the androgen stimuli. The proliferation of MFM223 cells is inhibited by androgen stimulation (69) and that of MDAMB453 cells is promoted in response to the stimulation (70). Our current study showed that the depletion of AIbZIP suppresses the proliferations of these cells. Thus, at least in these cancer model cell lines, it is certain that AIbZIP plays a significant role in cell proliferation. However, we have not determined how AIbZIP acts on cancer cell growth in vivo. Future studies using in vivo models such as xenograft or patient-derived xenograft models are essential to investigate whether AR-mediated AIbZIP induction contributes to LAR subtype TNBC cell proliferation.
Taken together, our study identified AIbZIP as a molecule that promote cancer cell proliferation via SKP2-p27 axis in LAR subtype TNBC model cell lines. Our findings will provide new insights into the LAR subtype TNBC progression and the molecular mechanisms involved in cell proliferation.
Supplementary Material
Supplementary Table 2
Supplementary figure 1. AIbZIP is upregulated in LAR subtype TNBC.
Supplementary figure 2. AIbZIP is upregulated in LAR subtype TNBC compared to normal tissue.
Supplementary figure 3. Aibzip siRNA transfection does not affect cell proliferation and p27 and SKP2 expression in a non-LAR subtype TNBC cell line.
Supplementary figure 4. BrdU incorporation was decreased in AIbZIP depleted cells.
Supplementary figure 5. Protein expression levels of phosphorylated RB.
Supplementary figure 6. Protein expression levels of G1-S transition–associated proteins.
Supplementary figure 7. Aibzip siRNA transfection upregulates p27 and downregulates SKP2.
Supplementary figure 8. Protein expression levels of p27 post-translation regulation–associated proteins.
Supplementary figure 9. AIbZIP induced luciferase reporter activity is depending on CRE-binding site of Skp2.
Supplementary figure 10. AIbZIP directly induces the expression of SKP2 by binding to CRE-binding site in the Skp2 promoter after the treatment with brefeldin A.
Supplementary figure 11. SKP2 overexpression compensates for the suppression of cell proliferation and p27 upregulation induced by AIbZIP siRNA transfection.
Supplementary Figure Legends 1-11
Supplementary Table 1
Acknowledgments
We thank H. Nikki March, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this article. We thank Eric Kowarz and Zsuzsanna Izsvak for providing plasmids. This work was partly supported by grants from the JSPS KAKENHI (23K17413), Takeda Science Foundation, MSD Life Science Foundation, The UBE Foundation, the Program of the Network-type Joint Usage/Research Center for Radiation Disaster Medical Science, Analysis Center of Life Science, the Natural Science Center for Basic Research and Development, Hiroshima University.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
A. Saito reports grants from JSPS KAKENHI (23K17413), Takeda Science Foundation, MSD Life Science Foundation, The UBE Foundation; non-financial support from the Program of the Network-type Joint Usage/Research Center for Radiation Disaster Medical Science and the Natural Science Center for Basic Research and Development, Hiroshima University outside the submitted work. Y. Kamikawa reports non-financial support from the Program of the Network-type Joint Usage/Research Center for Radiation Disaster Medical Science, the Natural Science Center for Basic Research and Development, Hiroshima University; grants from JSPS KAKENHI (22K06129), The Asahi Glass Foundation, and Radiation Effects Association outside the submitted work. K. Imaizumi reports grants from JSPS KAKENHI (23K17413) outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
T. Ito: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft. A. Saito: Conceptualization, data curation, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing. Y. Kamikawa: Data curation, formal analysis, investigation, methodology. N. Nakazawa: Data curation, formal analysis, validation. K. Imaizumi: Conceptualization, data curation, supervision, funding acquisition, validation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Lei S, Zheng R, Zhang S, Wang S, Chen R, Sun K, et al. Global patterns of breast cancer incidence and mortality: a population-based cancer registry data analysis from 2000 to 2020. Cancer Commun 2021;41:1183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Howlader N, Cronin KA, Kurian AW, Andridge R. Differences in breast cancer survival by molecular subtypes in the United States. Cancer Epidemiol Biomark Prev 2018;27:619–26. [DOI] [PubMed] [Google Scholar]
- 3. Prat A, Pineda E, Adamo B, Galván P, Fernández A, Gaba L, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015;24:S26–35. [DOI] [PubMed] [Google Scholar]
- 4. Lu L, Niu Z, Chao Z, Fu C, Chen K, Shi Y. Exploring the therapeutic potential of ADC combination for triple-negative breast cancer. Cell Mol Life Sci 2023;80:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim H, Choi J-M, Lee K. Immune checkpoint blockades in triple-negative breast cancer: current state and molecular mechanisms of resistance. Biomedicines 2022;10:1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yin L, Duan J-J, Bian X-W, Yu S-C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res 2020;22:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marra A, Trapani D, Viale G, Criscitiello C, Curigliano G. Practical classification of triple-negative breast cancer: intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. NPJ Breast Cancer 2020;6:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thompson KJ, Leon-Ferre RA, Sinnwell JP, Zahrieh DM, Suman VJ, Metzger FO, et al. Luminal androgen receptor breast cancer subtype and investigation of the microenvironment and neoadjuvant chemotherapy response. NAR Cancer 2022;4:zcac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Asada R, Kanemoto S, Kondo S, Saito A, Imaizumi K. The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. J Biochem 2011;149:507–18. [DOI] [PubMed] [Google Scholar]
- 11. Sampieri L, Di Giusto P, Alvarez C. CREB3 transcription factors: ER-golgi stress transducers as hubs for cellular homeostasis. Front Cell Dev Biol 2019;7:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kondo S, Murakami T, Tatsumi K, Ogata M, Kanemoto S, Otori K, et al. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat Cell Biol 2005;7:186–94. [DOI] [PubMed] [Google Scholar]
- 13. Kondo S, Saito A, Hino S-I, Murakami T, Ogata M, Kanemoto S, et al. BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol Cell Biol 2007;27:1716–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saito A, Hino S, Murakami T, Kanemoto S, Kondo S, Saitoh M, et al. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat Cell Biol 2009;11:1197–204. [DOI] [PubMed] [Google Scholar]
- 15. Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, et al. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol 2009;11:1205–11. [DOI] [PubMed] [Google Scholar]
- 16. Saito A, Kanemoto S, Kawasaki N, Asada R, Iwamoto H, Oki M, et al. Unfolded protein response, activated by OASIS family transcription factors, promotes astrocyte differentiation. Nat Commun 2012;3:967. [DOI] [PubMed] [Google Scholar]
- 17. Saito A, Kanemoto S, Zhang Y, Asada R, Hino K, Imaizumi K. Chondrocyte proliferation regulated by secreted luminal domain of ER stress transducer BBF2H7/CREB3L2. Mol Cell 2014;53:127–39. [DOI] [PubMed] [Google Scholar]
- 18. Cui X, Cui M, Asada R, Kanemoto S, Saito A, Matsuhisa K, et al. The androgen-induced protein AIbZIP facilitates proliferation of prostate cancer cells through downregulation of p21 expression. Sci Rep 2016;6:37310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saito A, Kamikawa Y, Ito T, Matsuhisa K, Kaneko M, Okamoto T, et al. p53-independent tumor suppression by cell-cycle arrest via CREB/ATF transcription factor OASIS. Cell Rep 2023;42:112479. [DOI] [PubMed] [Google Scholar]
- 20. Howley BV, Link LA, Grelet S, El-Sabban M, Howe PH. A CREB3-regulated ER-Golgi trafficking signature promotes metastatic progression in breast cancer. Oncogene 2018;37:1308–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yuxiong W, Faping L, Bin L, Yanghe Z, Yao L, Yunkuo L, et al. Regulatory mechanisms of the cAMP-responsive element binding protein 3 (CREB3) family in cancers. Biomed Pharmacother 2023;166:115335. [DOI] [PubMed] [Google Scholar]
- 22. Rose M, Schubert C, Dierichs L, Gaisa NT, Heer M, Heidenreich A, et al. OASIS/CREB3L1 is epigenetically silenced in human bladder cancer facilitating tumor cell spreading and migration in vitro. Epigenetics 2014;9:1626–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ward AK, Mellor P, Smith SE, Kendall S, Just NA, Vizeacoumar FS, et al. Epigenetic silencing of CREB3L1 by DNA methylation is associated with high-grade metastatic breast cancers with poor prognosis and is prevalent in triple negative breast cancers. Breast Cancer Res 2016;18:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu L-Q, Feng L-F, Nan C-R, Zhao Z-M. CREB3L1 and PTN expressions correlate with prognosis of brain glioma patients. Biosci Rep 2018;38:BSR20170100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo H, Xia X, Kim GD, Liu Y, Xue Z, Zhang L, et al. Characterizing dedifferentiation of thyroid cancer by integrated analysis. Sci Adv 2021;7:eabf3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pan Z, Li L, Fang Q, Qian Y, Zhang Y, Zhu J, et al. Integrated bioinformatics analysis of master regulators in anaplastic thyroid carcinoma. Biomed Res Int 2019;2019:9734576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sheng Z, Li L, Zhu LJ, Smith TW, Demers A, Ross AH, et al. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat Med 2010;16:671–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iwamoto H, Matsuhisa K, Saito A, Kanemoto S, Asada R, Hino K, et al. Promotion of cancer cell proliferation by cleaved and secreted luminal domains of ER stress transducer BBF2H7. PLoS One 2015;10:e0125982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mertens F, Fletcher CDM, Antonescu CR, Coindre J-M, Colecchia M, Domanski HA, et al. Clinicopathologic and molecular genetic characterization of low-grade fibromyxoid sarcoma, and cloning of a novel FUS/CREB3L1 fusion gene. Lab Invest 2005;85:408–15. [DOI] [PubMed] [Google Scholar]
- 30. Möller E, Hornick JL, Magnusson L, Veerla S, Domanski HA, Mertens F. FUS-CREB3L2/L1-positive sarcomas show a specific gene expression profile with upregulation of CD24 and FOXL1. Clin Cancer Res 2011;17:2646–56. [DOI] [PubMed] [Google Scholar]
- 31. Qi H, Fillion C, Labrie Y, Grenier J, Fournier A, Berger L, et al. AIbZIP, a novel bZIP gene located on chromosome 1q21.3 that is highly expressed in prostate tumors and of which the expression is up-regulated by androgens in LNCaP human prostate cancer cells. Cancer Res 2002;62:721–33. [PubMed] [Google Scholar]
- 32. Cheong A, Zhang X, Cheung Y-Y, Tang W-Y, Chen J, Ye S-H, et al. DNA methylome changes by estradiol benzoate and bisphenol A links early-life environmental exposures to prostate cancer risk. Epigenetics 2016;11:674–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim T-H, Park J-M, Kim M-Y, Ahn Y-H. The role of CREB3L4 in the proliferation of prostate cancer cells. Sci Rep 2017;7:45300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Inagaki Y, Yasui K, Endo M, Nakajima T, Zen K, Tsuji K, et al. CREB3L4, INTS3, and SNAPAP are targets for the 1q21 amplicon frequently detected in hepatocellular carcinoma. Cancer Genet Cytogenet 2008;180:30–6. [DOI] [PubMed] [Google Scholar]
- 35. Pu Q, Lu L, Dong K, Geng W, Lv Y, Gao H. The novel transcription factor CREB3L4 contributes to the progression of human breast carcinoma. J Mammary Gland Biol Neoplasia 2020;25:37–50. [DOI] [PubMed] [Google Scholar]
- 36. Ai C, Rong T, Chen Z, Shen W, Huang K, Li Q, et al. Cyclic AMP responsive element binding protein 3-like 4/AarF domain containing kinase 5 axis facilitates proliferation, migration and invasion of lung adenocarcinoma cells by modulating the TGFβ pathway. Biotechnol Bioprocess Eng 2023;28:192–202. [Google Scholar]
- 37. Jing X, Liang H, Hao C, Yang X, Cui X. Overexpression of MUC1 predicts poor prognosis in patients with breast cancer. Oncol Rep 2019;41:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kowarz E, Löscher D, Marschalek R. Optimized sleeping beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J 2015;10:647–53. [DOI] [PubMed] [Google Scholar]
- 39. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020;38:675–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS One 2016;11:e0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015;163:506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, et al. A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell 2018;33:690–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mullan KA, Bramberger LM, Munday PR, Goncalves G, Revote J, Mifsud NA, et al. ggVolcanoR: a Shiny app for customizable visualization of differential expression datasets. Comput Struct Biotechnol J 2021;19:5735–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hartung C, Porsch M, Stückrath K, Kaufhold S, Staege MS, Hanf V, et al. Identifying high-risk triple-negative breast cancer patients by molecular subtyping. Breast Care 2021;16:637–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Naderi A, Vanneste M. Prolactin-induced protein is required for cell cycle progression in breast cancer. Neoplasia 2014;16:329–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Debily M-A, Marhomy SE, Boulanger V, Eveno E, Mariage-Samson R, Camarca A, et al. A functional and regulatory network associated with PIP expression in human breast cancer. PLoS One 2009;4:e4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Engeland K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ 2022;29:946–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009;9:153–66. [DOI] [PubMed] [Google Scholar]
- 56. Reynisdóttir I, Massagué J. The subcellular locations of p15(Ink4b) and p27(Kip1) coordinate their inhibitory interactions with cdk4 and cdk2. Genes Dev 1997;11:492–503. [DOI] [PubMed] [Google Scholar]
- 57. Hnit SST, Xie C, Yao M, Holst J, Bensoussan A, De Souza P, et al. p27(Kip1) signaling: transcriptional and post-translational regulation. Int J Biochem Cell Biol 2015;68:9–14. [DOI] [PubMed] [Google Scholar]
- 58. Vervoorts J, Lüscher B. Post-translational regulation of the tumor suppressor p27(KIP1). Cell Mol Life Sci 2008;65:3255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 2006;6:369–81. [DOI] [PubMed] [Google Scholar]
- 60. Lee Y, Lim H-S. Skp2 inhibitors: novel anticancer strategies. Curr Med Chem 2016;23:2363–79. [DOI] [PubMed] [Google Scholar]
- 61. Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer 2008;112:1415–24. [DOI] [PubMed] [Google Scholar]
- 62. Wang Z, Fukushima H, Inuzuka H, Wan L, Liu P, Gao D, et al. Skp2 is a promising therapeutic target in breast cancer. Front Oncol 2012;1:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cai Z, Moten A, Peng D, Hsu C-C, Pan B-S, Manne R, et al. The Skp2 pathway: a critical target for cancer therapy. Semin Cancer Biol 2020;67:16–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Duffy MJ, O'Grady S, Tang M, Crown J. MYC as a target for cancer treatment. Cancer Treat Rev 2021;94:102154. [DOI] [PubMed] [Google Scholar]
- 65. Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett 2013;328:198–206. [DOI] [PubMed] [Google Scholar]
- 66. Gerratana L, Basile D, Buono G, De Placido S, Giuliano M, Minichillo S, et al. Androgen receptor in triple negative breast cancer: a potential target for the targetless subtype. Cancer Treat Rev 2018;68:102–10. [DOI] [PubMed] [Google Scholar]
- 67. Ravaioli S, Maltoni R, Pasculli B, Parrella P, Giudetti AM, Vergara D, et al. Androgen receptor in breast cancer: the “5W” questions. Front Endocrinol 2022;13:977331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kolyvas EA, Caldas C, Kelly K, Ahmad SS. Androgen receptor function and targeted therapeutics across breast cancer subtypes. Breast Cancer Res 2022;24:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hackenberg R, Lüttchens S, Hofmann J, Kunzmann R, Hölzel F, Schulz KD. Androgen sensitivity of the new human breast cancer cell line MFM-223. Cancer Res 1991;51:5722–7. [PubMed] [Google Scholar]
- 70. Hall RE, Birrell SN, Tilley WD, Sutherland RL. MDA-MB-453, an androgen-responsive human breast carcinoma cell line with high level androgen receptor expression. Eur J Cancer 1994;30A:484–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 2
Supplementary figure 1. AIbZIP is upregulated in LAR subtype TNBC.
Supplementary figure 2. AIbZIP is upregulated in LAR subtype TNBC compared to normal tissue.
Supplementary figure 3. Aibzip siRNA transfection does not affect cell proliferation and p27 and SKP2 expression in a non-LAR subtype TNBC cell line.
Supplementary figure 4. BrdU incorporation was decreased in AIbZIP depleted cells.
Supplementary figure 5. Protein expression levels of phosphorylated RB.
Supplementary figure 6. Protein expression levels of G1-S transition–associated proteins.
Supplementary figure 7. Aibzip siRNA transfection upregulates p27 and downregulates SKP2.
Supplementary figure 8. Protein expression levels of p27 post-translation regulation–associated proteins.
Supplementary figure 9. AIbZIP induced luciferase reporter activity is depending on CRE-binding site of Skp2.
Supplementary figure 10. AIbZIP directly induces the expression of SKP2 by binding to CRE-binding site in the Skp2 promoter after the treatment with brefeldin A.
Supplementary figure 11. SKP2 overexpression compensates for the suppression of cell proliferation and p27 upregulation induced by AIbZIP siRNA transfection.
Supplementary Figure Legends 1-11
Supplementary Table 1
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.



![Figure 1. AIbZIP is upregulated in LAR subtype TNBC. A, Comparison of Aibzip mRNA expression in cancerous and normal tissues in a pan-cancer view obtained by UALCAN. The vertical axis indicates mRNA expression levels [transcripts per million (TPM) +1], which was converted to a logarithmic function with base 2. BLCA, bladder urothelial carcinoma; BRCA, breast carcinoma; CESC, cervical squamous cell carcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin melanoma; THCA, thyroid carcinoma; THYM, thymoma; STAD, stomach adenocarcinoma; UCEC, uterine corpus endometrial carcinoma. *, **, and *** means P < 0.05, P < 0.01, and P < 0.001 versus normal tissue. B, Volcano plot of all gene expression profiling from normal tissue versus tumor. Each point represents the difference in expression (log fold-change) and statistical significance (P-value) between the two groups. Aibzip (also called Creb3l4) is showed by red colored point. The X-axis represents log fold change value and Y-axis represents −log10 P-value. C, Aibzip mRNA expression levels are shown by a box plot. Midline means median. Number of samples: normal = 114, luminal = 710, HER2 = 158, LAR = 17, BL1 = 34, BL2 = 17, M = 40, IM = 36, MSL = 22, UNC = 13. *** means P < 0.001 versus normal tissue. D, Volcano plot of all gene expression profiling from all TNBC versus LAR subtype TNBC. Aibzip (also called Creb3l4) is showed by red colored point. E, Volcano plot of androgen response gene expression profiling from all TNBC versus LAR subtype TNBC. Aibzip (also called Creb3l4) is showed by red colored point.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/d702/10985479/b7f531debb9f/373fig1.jpg)