Abstract
Background:
Allogeneic bone marrow transplant (alloBMT) in people living with HIV (PLWH) can lead to the undetectable levels of HIV reservoirs in blood, even using highly sensitive assays. However, with antiretroviral therapy (ART) interruption, rebound of HIV viremia occurs. The source of this rebound viremia is of interest in HIV cure strategies.
Methods:
Within a trial of alloBMT in individuals with hematologic malignancies and HIV (ClinicalTrials.gov, NCT01836068), one recipient self-interrupted ART after achieving >99.5% host cell replacement in peripheral blood by day 147 and developed severe acute retroviral syndrome with meningoencephalitis at 156 days post-alloBMT. We isolated replication-competent HIV using a quantitative viral outgrowth assay at −100 and −25 days pre-alloBMT and from the same time points pre-alloBMT for HIV DNA and cell-associated RNA from peripheral blood mononuclear cells and resting memory CD4+ T-cells. We isolated HIV RNA in plasma and cerebrospinal fluid (CSF) at viral rebound. We sequenced the RT-region of pol and performed neighbor-joining phylogenetic reconstruction.
Results:
Phylogenetic analysis revealed an identical viral sequence at both pre-alloBMT timepoints accounting for 9/34 sequences (26%) of the sampled HIV reservoir. This sequence population grouped with viral rebound sequences from plasma and CSF with high sequence homology.
Discussion:
Despite >99.5% replacement of host cells in peripheral blood, ART interruption led to HIV viral rebound in plasma and CSF. Further, the rebound virus matched replication competent virus from resting memory CD4+ T-cells pre-alloBMT. This case underscores that HIV-infected recipient cells can persist after alloBMT, and that latent replication-competent virus can re-establish infection.
Keywords: HIV viral rebound, ART interruption, HIV latent reservoir, allogeneic stem cell transplant
INTRODUCTION
Antiretroviral therapy (ART) reduces HIV viremia to undetectable levels in people living with HIV (PLWH), however the persistence of a latent viral reservoir (LVR) within resting memory CD4+ T-cells remains a barrier to cure.1–3 Two individuals have been cured following allogeneic bone marrow transplant (alloBMT) for where donor cells were homozygous for the CCR5delta32 mutation which is associated with resistance to R5-tropic HIV.4, 5 There has been interest in exploring whether alloBMT with CCR5 wild-type donors can lead to elimination of HIV reservoirs.6–11
The initial study of this approach included the two ‘Boston patients’ who underwent alloBMT and subsequently were reported to have undetectable HIV RNA and cell-associated HIV DNA in peripheral blood and tissues. Following analytical ART interruption, HIV viral rebound occurred in both participants despite <0.0010% residual host cells.12, 13 Several studies have reported on alloBMT recipients with HIV and viral rebound without a clear origin of rebound virus.8, 10, 12 Here, we detail another case where a PLWH was nonadherent to ART post-alloBMT and developed acute retroviral syndrome, including severe meningoencephalitis. Through sequence analysis, we studied rebound virus in peripheral blood and cerebrospinal fluid (CSF), comparing it to pre-alloBMT replication-competent virus in resting memory CD4+ T-cells.
METHODS
Study design
This was a sub-study of a case within a clinical trial investigating the safety and efficacy of alloBMT for PLWH with continuous optimized ART (NCT01836068).8 Participants provided informed consent.
Measurement of donor cell chimerism
PCR of short tandem repeats was used to determine the relative proportion of donor and recipient DNA after alloBMT. Applied Biosystems PCR (Fisher Scientific) or AmpflSTR Identifier PCR Amplification Kit (Thermo Fisher) was used on unsorted peripheral blood mononuclear cells (PBMCs) and sorted CD3+ T-cells to determine the degree of donor replacement. The limit of detection of this assay was determined by the number of input cells and ranged from 0.5–1% at different timepoints for this participant.
HIV viral outgrowth from resting CD4+ T-cells
The HIV LVR was measured in resting memory CD4+ T-cells (rCD4+ T-cells) prior to alloBMT using a quantitative viral outgrowth assay (qVOA) as previously described.2, 14 Briefly, rCD4+ T-cells were isolated from whole blood and plated at limiting-dilution with γ-irradiated PBMCs from HIV-uninfected donors. Cells underwent ex vivo activation by the T-cell mitogen, phytohaemagglutinin. Viral outgrowth was amplified by the addition of the MOLT-4/CCR5+ cell-line on day 2 of the culture and detected by ELISA for HIV p24 antigen in the supernatant by day 21. Post-alloBMT, the participant missed study visits and later opted out of further follow-up, therefore qVOA samples were not obtained post-alloBMT.
Isolation of HIV nucleic acid
Viral RNA from supernatant of p24-positive viral outgrowth wells which represents inducible replication-competent virus were isolated using the Zymo Research ZR Viral RNA Kit. TRIzol Reagent was used according to manufacturer’s instructions for both cell-associated RNA (CA-RNA) and DNA isolation from viably frozen 10×106 PBMCs and pelleted 2×106 rCD4+ T-cells at 100- and 25-days pre-alloBMT, which were collected in the same blood draw as for the qVOA. Viral RNA was isolated from peripheral blood plasma and CSF at the time of viral rebound 156-days post-alloBMT using QIAmp Viral RNA kit (Qiagen). RNA was treated with DNase (DNase I, amplification grade, ThermoFisher Scientific) and DNA with RNaseA (PureLink™ RNaseA, ThermoFisher Scientific).
PCR amplification of HIV RNA and DNA
A One-Step first-round PCR for cDNA synthesis was done using SuperScript-III One-Step RT-PCR with Platinum Taq High-Fidelity DNA polymerase (Invitrogen). Primers targeted the RT-region of pol, HIV_RTpol_F-(5’-CAG-GAG-CAG-ATA-CAG-TAT-AGA-AG-3’) and HIV_RTpol_R-(5’-AGT-CTT-TCC-CCA-TAT-TAC-TAT-GCT-TTC-3’). PCR conditions were: 50°C/60min, 94°C/2min, 30 cycles of 94°C/25sec, 55°C/30sec and 68°C/1.5min, with a final step of 68°C/5min. Nested second-round PCR was carried-out with Platinum PCR SuperMix (Invitrogen) using first-round product, and primers nestedHIV_RT_F-(5’-AAT-TGG-GCC-TGA-AAA-TCC-ATA-3’) and nestedHIV_RT_R-(5’-AGT-TCA-TAA-CCC-ATC-CAA-AG-3’). PCR conditions were: 94°C/2min, 35 cycles of 94°C/30sec, 55°C/30sec and 72°C/1.5min, followed by a final step of 72°C/10min.
For HIV DNA, first-round PCR for HIV DNA was done using Platinum PCR SuperMix (Invitrogen) with the HIV_RTpolF and HIVRTpolR primers as stated above under the following PCR conditions: 94°C/2min, 35 cycles of 94°C/25sec, 55°C/30sec and 68°C/1.5min, with a final step of 68°C/5min. Nested second-round PCR were carried out under the same conditions as above. PCR products were run on 2% agarose gel and positive bands were excised and purified by the QIAquick Gel Extraction Kit (Qiagen). Viral sequencing by Sanger method using nested second-round primers.
Viral Sequencing
The HIV RT-region of pol was sequenced using the nested second-round PCR primers. Rebound HIV from peripheral blood and CSF-RNA, as well as CA-RNA and DNA from PBMCs and rCD4+ T-cells was sequenced using a 454 next-generation sequencing (NGS) platform.15 In the case for 454 NGS, bulk PCR was performed for each sample type and, consensus sequences, defined as unique sequence reads over the region of interest in that sample with >0.2% prevalence were generated. Any reads that were not the full target length were excluded. Positive viral outgrowth supernatant RNA was sequenced using standard Sanger sequencing; sequences with multiple peaks in the electropherograms were eliminated. NGS based sequencing was performed in a separate laboratory from Sanger based sequencing to eliminate possible cross-contamination.
Sequence and Phylogenetic analysis
All chromatograms obtained by Sanger sequencing were manually inspected for multiple peaks and all had a Q-score >30. Consensus sequences from deep sequencing were generated with a cut-off of >0.2% of total sequence reads. Using Multiple Alignment using Fast Fourier Transform (MAFFT) with the FFT-NS-1 200PAM/k=2 algorithm a 497-bp alignment (HXB2-coordinates:2717–3204) was generated. A neighbor-joining phylogeny was reconstructed, and viral genetic diversity was calculated as average pairwise distance (APD) in MEGAX. Maximum-likelihood using PhyML confirmed the pairwise-distance approach topology that Neighbor-joining based methods inferred. ART drug resistance mutations non-nucleoside reverse-transcriptase inhibitors (NNRTI) and nucleoside reverse-transcriptase inhibitors (NRTI) were predicted using the HIV Drug Resistance Database (Stanford University). Phylogenetic tree topology was consistent with and without the inclusion of drug resistant mutated sequences. HyperMut was used to detect APOBEC-induced hypermutation (p<0.05 for significance) (https://www.hiv.lanl.gov/). Trees visualized in FigTreev1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). Subtype B reference sequences were included for phylogenetic analyses: B.NL.00.671_00T36.AY423387, B.TH.90.BK132.AY173951, and B.US.98.1058_11.AY331295.
RESULTS
Case presentation
Participant 3 received a reduced-intensity conditioning match-related donor alloBMT for treatment of multiply relapsed acute myeloid leukemia. HIV had been diagnosed 9 years prior. Baseline HIV plasma-RNA was 42,000 copies/mL and absolute CD4 count nadir of 70 cells/μL. He was treated with multiple ART regimens over time and had challenges with ART adherence prior to alloBMT. Tropism testing demonstrated R5-tropic HIV (Monogram Biosciences), and the most recent HIV genotype (~4 years pre-alloBMT) revealed the K103N mutation in RT with additional clinical records indicating the presence of the M184V mutation (Celera Diagnostics, ViroSeq v2.8 assay). At day −25 prior to alloBMT, the participant was on abacavir, lamivudine, raltegravir, and ritonavir-boosted darunavir with plasma HIV plasma-RNA of <20 copies/mL and CD4 of 409 cells/μL. Maraviroc replaced ritonavir to minimize drug interactions during transplant.
Per protocol, when high dose cyclophosphamide was given days 3 and 4 post-transplant for graft-versus-host-disease prophylaxis, enfuvirtide was also administered to ensure continuous ART should nausea and vomiting interrupt oral ART. At 28 days post-alloBMT, chimerism (i.e., the percentage of donor vs. host cells) was measured using short-tandem repeats and >99% donor cell chimerism was achieved.
By 84-days post-alloBMT, HIV plasma-RNA was <20 copies/mL. The participant began missing clinic and study visits. At 100-days post-alloBMT, the patient developed fevers, and was hospitalized at 146-days post-alloBMT, with confusion which progressed to obtundation. A lumbar puncture showed 28 white blood cells/μL (normal range 0–3), protein 150 mg/dL (normal range 50–80), and glucose 50 mg/dL (normal range 50–80) indicating meningoencephalitis. All bacteria, mycobacteria, and fungal cultures were negative. CSF was negative for EBV, CMV, JC, VZV, HSV, and WNV by PCR; cryptococcal antigen was not detected. HIV plasma-RNA was 25,518 copies/mL and HIV CSF-RNA was 17,000 copies/mL (Abbott Laboratories, IL). Subcutaneous enfuvirtide was initiated since oral ART not possible. After 3 days, mental status improved, and oral ART was resumed. After 10 days, the participant was discharged and by 190-days post-alloBMT HIV plasma-RNA was 23 copies/mL.8
Pre-alloBMT measurement and genetic analysis
The HIV LVR day-100 pre-alloBMT was measured to be 6.29 (2.20–17.97 95% CI) Infectious Units per Million cells (IUPM) and at day-25 pre-alloBMT was 0.76 (0.43–1.36 95% CI) IUPM by qVOA. The HIV LVR was measured at 100- and 25-days pre-alloBMT with 22/32 and 12/32 wells (34 total wells) found to be p24 positive, respectively. The viral genetic diversity of the replication-competent HIV LVR was 1.1% APD 100-days and 1.2% APD 25-days pre-alloBMT with an overall diversity of 1.1% APD (Figure 1A). There were identical sequences (6/34 total sequences; 17.6%) sampled from both pre-alloBMT timepoints that did not possess predicted drug resistance mutations that represented the HIV LVR, these sequences represented within the 11.5% overall clonal population of the HIV LVR sampled (3/26 unique viral sequences) (Figure 1A). Major ART resistance mutations K103N and/or M184V were found in 12/34 outgrowth sequences at both pre-alloBMT time points.
Figure 1. Neighbor-joining phylogeny of the RT-pol region of participant 3.
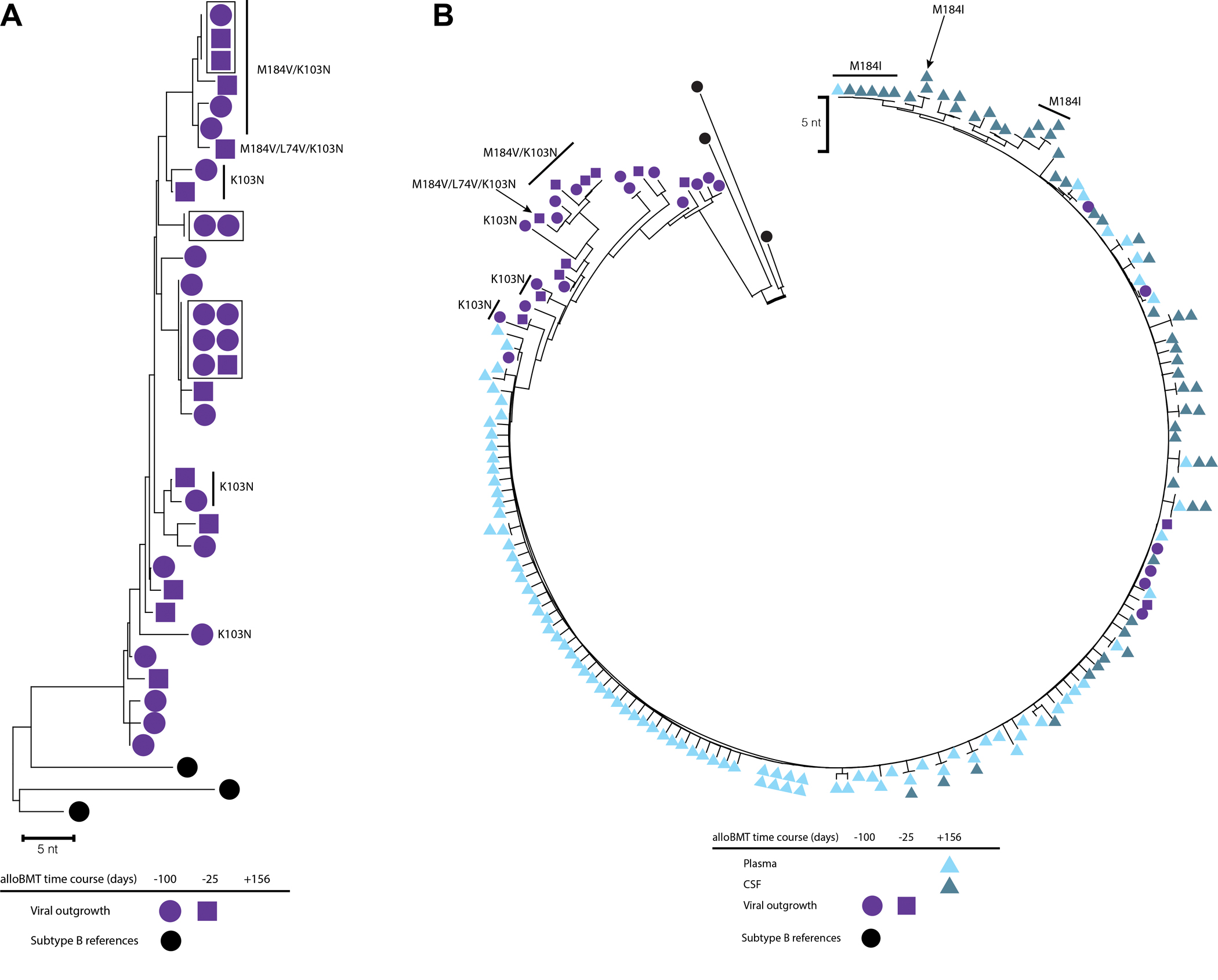
(A) Phylogenetic reconstruction of replication-competent viral outgrowth 25- and 100-days pre-alloBMT. Identical sequences are boxed. (B) Phylogenetic reconstruction of replication-competent viral outgrowth pre-alloBMT and rebound viremia from plasma and CSF 156-days post-alloBMT following self-interruption of ART. Major ART drug resistance mutations are labeled. Colors indicate sample isolate and shape for time point. The scale bar is set to 5 nucleotides.
Post-alloBMT viral rebound genetic analysis
The RT-pol region of HIV in plasma and CSF at rebound were sequenced. Consensus sequences from rebound plasma-RNA were 92 sequences and from CSF-RNA, 55 sequences. There was almost complete sequence homology between rebound peripheral blood plasma virus (0.4% APD) and CSF (0.4% APD) with 0.4% APD between compartments (Figure 1B). The majority of rebound HIV quasi-species did not have predicted drug resistance. A sub-branch of several CSF sequences with one plasma consensus sequence were predicted to have the M184I mutation (Figure 1B). Rebound viral populations were compared to sequences from the HIV LVR pre-alloBMT, where 9/34 (26%) of the total sequences matched rebound quasi-species. Pre-alloBMT compared to viral rebound diversity differed by 0.04% APD (1.07% pre-alloBMT vs. plasma, and 1.11% pre-alloBMT vs. CSF).
Cellular HIV DNA and CA-RNA analysis
Consensus sequences from HIV DNA and CA-RNA from PBMCs and rCD4+ T-cells pre-alloBMT were obtained. At 100-days pre-alloBMT, the number of sequences isolated from PBMC CA-RNA were 72 sequences, rCD4+ T-cell DNA (n=79) and CA-RNA (n=86); and 25-days pre-alloBMT from PBMC DNA (n=69) and CA-RNA (n=74), rCD4+ T-cell DNA (n=73) and CA-RNA (n=87). HIV DNA sequences from PBMCs 100-days pre-alloBMT could not be obtained. Since only a small percentage of integrated proviruses are intact, it is likely that the majority of DNA sequences obtained had defects that were not captured through sequencing. Because of this, it is likely that only a few select cells with integrated inducible intact provirus could possibly give rise to rebound virus. These inducible intact replication-competent proviruses from rCD4+ T-cells that were isolated from the qVOA were interspersed in the phylogeny (Figure 2). There was an overall mean APD of 1.3%. Major ART drug resistance was common with M184V and/or K103N mutations (Figure 1A, 11/34 qVOA sequences from pre-alloBMT). Additionally, E138K and Y188D, and L74V were also observed in cellular-derived sequences. While the M184I mutation was detected in a sub-population of rebound CSF and plasma viral sequences, it was also observed in PBMC CA-RNA 25-days pre-alloBMT (n=9) (Figure 2). However, PBMC CA-RNA from 25-days pre-alloBMT and rebound quasi-species with the M184I mutation were genetically distinct (1.6% APD). In addition, some of the CA-RNA sequences from rCD4+ T-cells that were sampled 25-days pre-alloBMT were most genetically similar to the rebound virus. APOBEC hypermutated sequences were detected in PBMC HIV DNA (n=4) and in CA-RNA (n=2) at 25-days pre-alloBMT.
Figure 2. Neighbor-joining phylogeny of the RT-pol region of participant 3 including cellular sequences.
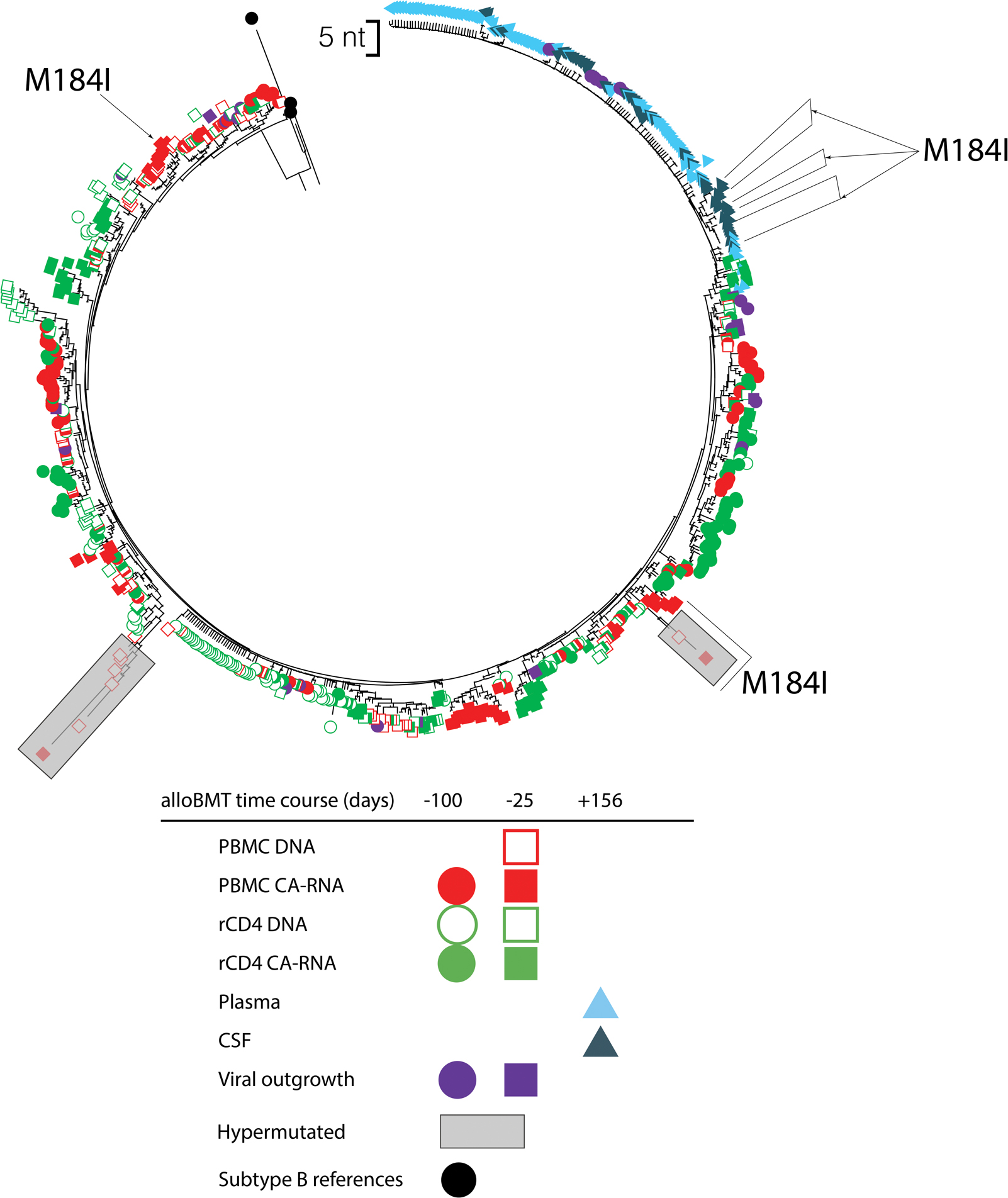
Phylogenetic reconstruction of replication-competent viral outgrowth 25 and 100 days pre-alloBMT with corresponding HIV-1 DNA and cell-associated RNA from PBMCs and rCD4+ T-cells. In addition, rebound viremia from plasma and CSF 156-days post-alloBMT following self-interruption of ART are included. The sequences that had the M184I mutation are labeled. Shaded regions indicate predicted APOBEC hypermutated sequences using HyperMut (p<0.05). Colors indicate sample isolate and shape for time point. The scale bar is set to 5 nucleotides.
DISCUSSION
There are a few reports of acute retroviral syndrome including meningoencephalitis following alloBMT, either due to unplanned16 or analytic treatment interruption (ATI).12, 13, 17 In primary HIV infection, the incidence of meningoencephalitis associated with acute retroviral syndrome is 9–55%.18, 19 In the circumstance surrounding the two Boston Patients’, ‘Patient A’ had clinical evidence of meningoencephalitis following ATI with symptoms of acute retroviral syndrome.12 CSF sampling revealed low-grade lymphocytic pleocytosis consistent with HIV meningitis.12
Cases of meningoencephalitis with HIV rebound have raised questions with regard to the role of the CNS as an HIV reservoir. In the case of the Boston patients’, HIV sequence analysis of CSF was not possible. HIV may persist deep in tissues including frontal motor cortex of the brain as demonstrated in post-mortem specimens.20 Similarly, there is CNS persistence in SIV-infected pigtail macaques after long-term suppressive ART.21 Furthermore, persistent replicating HIV in the CNS during 3 years of ART has been reported in one individual.22 Evidence of a CNS reservoir, not withstanding, in our case, the rebound virus matched virus from the reservoir of resting memory CD4+ T-cells isolated pre-BMT from cell culture. Interestingly, other studies have not observed a direct clonal connection between rebound virus after analytic treatment interruptions and virus isolated from the latent reservoir using cell culture techniques.23, 24 However, these trials investigated the impact of broadly neutralizing antibody therapies which ultimately had little effect on the size of the HIV reservoir, in contrast to hematopoietic stem cell transplantation.
Severe meningoencephalitis may reflect the lack of a donor CD8+ T-cell response against HIV rather than evidence of an anatomic sanctuary or reservoir in CNS cells. Recently, the IciStem Consortium demonstrated incomplete T-cell reconstitution in PLWH following alloBMT, where there could be small yet persistent HIV-specific CD8+ T-cell responses in some individuals.25 The extent on the potency of these small pools of CD8+ T-cells and their ability to prevent serious clinical complications (e.g., meningoencephalitis) remain to be investigated. In the Boston patients, there were negligible HIV-specific T-cell responses measured after alloBMT nor during the analytic treatment interruption.12, 13 These cases, including the one we report here, demonstrate a unique and serious clinical risk associated with interrupting ART after alloBMT. Possible mechanisms to employ a more effective CD8+ T-cell response following alloBMT include the use of engineered HIV-specific T-cells that are capable of secreting broadly neutralizing antibodies against HIV envelope to elicit antibody-dependent cellular cytotoxicity26 or the use of donor derived engineered HIV-specific T cells.27–29 The latter is being investigated in an ongoing clinical trials for PLWH who require alloBMT for clinical reasons (NCT04248192).
Some limitations of the study include the use of bulk PCR rather than limiting dilution PCR on samples that contain genetically diverse templates. This allowed for the possibility of in vitro recombinants to occur which may not represent true within-host diversity. Thus, due to this approach, we were unable to quantify the number of unique viruses as well as being able to reduce PCR recombination and sequencing error. At the time our specific interests were broad differences within the viral populations of this individual. Additionally, we only examined a subgenomic region of HIV. A more highly diverse region such as full-length env or near full-length sequencing would have increased our ability to detect genetic differences.
In summary, we found that interruption of ART post-alloBMT can lead to a serious clinical acute retroviral syndrome including meningoencephalitis which may be due to the lack of HIV immunity in naïve donor cells. This unique case suggests that recipient cells persist early after alloBMT and that a single predominant viral population latent in resting memory CD4+ T-cells can re-establish infection.
Acknowledgments:
We thank the study staff and the Participants for their generous willingness to be included in the study. All participants provided written informed consent for the study. We would also like to thank Dr. Susanna Lamers of BioInfoExperts for her assistance with the sequence submission process.
Funding Information:
This work was supported by amfAR (108707-54-RKRL), the Johns Hopkins University Center for AIDS Research (P30AI094189), the National Cancer Institute (grant numbers K23CA177321-01A1, P01CA225618-01A1, P01CA015396, and P30CA006973), and was supported in part by the Division of Intramural Research, NIAID, NIH.
Footnotes
Declaration of interests: CMD reports grants from Abbvie, grants from GlaxoSmithKine, and from Gilead Sciences, outside the submitted work. All other authors have nothing to disclose.
Sequence Data: All sequence data can be access in Genbank accessions MZ890863-MZ891581.
References
- 1.Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. May 8 1997;387(6629):183–8. doi: 10.1038/387183a0 [DOI] [PubMed] [Google Scholar]
- 2.Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. Nov 14 1997;278(5341):1295–300. doi: 10.1126/science.278.5341.1295 [DOI] [PubMed] [Google Scholar]
- 3.Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. Jun 2003;9(6):727–8. doi: 10.1038/nm880 [DOI] [PubMed] [Google Scholar]
- 4.Hütter G, Nowak D, Mossner M, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. Feb 12 2009;360(7):692–8. doi: 10.1056/NEJMoa0802905 [DOI] [PubMed] [Google Scholar]
- 5.Gupta RK, Abdul-Jawad S, McCoy LE, et al. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature. Apr 2019;568(7751):244–248. doi: 10.1038/s41586-019-1027-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambinder RF, Capoferri AA, Durand CM. Haemopoietic cell transplantation in patients living with HIV. Lancet HIV. Sep 2020;7(9):e652–e660. doi: 10.1016/s2352-3018(20)30117-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambinder RF, Wu J, Logan B, et al. Allogeneic Hematopoietic Cell Transplant for HIV Patients with Hematologic Malignancies: The BMT CTN-0903/AMC-080 Trial. Biol Blood Marrow Transplant. Nov 2019;25(11):2160–2166. doi: 10.1016/j.bbmt.2019.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand CM, Capoferri AA, Redd AD, et al. Allogeneic bone marrow transplantation with post-transplant cyclophosphamide for patients with HIV and haematological malignancies: a feasibility study. Lancet HIV. Sep 2020;7(9):e602–e610. doi: 10.1016/s2352-3018(20)30073-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston C, Harrington R, Jain R, Schiffer J, Kiem HP, Woolfrey A. Safety and Efficacy of Combination Antiretroviral Therapy in Human Immunodeficiency Virus-Infected Adults Undergoing Autologous or Allogeneic Hematopoietic Cell Transplantation for Hematologic Malignancies. Biol Blood Marrow Transplant. Jan 2016;22(1):149–56. doi: 10.1016/j.bbmt.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koelsch KK, Rasmussen TA, Hey-Nguyen WJ, et al. Impact of Allogeneic Hematopoietic Stem Cell Transplantation on the HIV Reservoir and Immune Response in 3 HIV-Infected Individuals. J Acquir Immune Defic Syndr. Jul 1 2017;75(3):328–337. doi: 10.1097/qai.0000000000001381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salgado M, Kwon M, Gálvez C, et al. Mechanisms That Contribute to a Profound Reduction of the HIV-1 Reservoir After Allogeneic Stem Cell Transplant. Ann Intern Med. Nov 20 2018;169(10):674–683. doi: 10.7326/m18-0759 [DOI] [PubMed] [Google Scholar]
- 12.Henrich TJ, Hanhauser E, Marty FM, et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med. Sep 2 2014;161(5):319–27. doi: 10.7326/m14-1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henrich TJ, Hu Z, Li JZ, et al. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis. Jun 1 2013;207(11):1694–702. doi: 10.1093/infdis/jit086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laird GM, Rosenbloom DI, Lai J, Siliciano RF, Siliciano JD. Measuring the Frequency of Latent HIV-1 in Resting CD4⁺ T Cells Using a Limiting Dilution Coculture Assay. Methods Mol Biol. 2016;1354:239–53. doi: 10.1007/978-1-4939-3046-3_16 [DOI] [PubMed] [Google Scholar]
- 15.Redd AD, Collinson-Streng A, Martens C, et al. Identification of HIV superinfection in seroconcordant couples in Rakai, Uganda, by use of next-generation deep sequencing. J Clin Microbiol. Aug 2011;49(8):2859–67. doi: 10.1128/jcm.00804-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf T, Rickerts V, Staszewski S, et al. First case of successful allogeneic stem cell transplantation in an HIV-patient who acquired severe aplastic anemia. Haematologica. Apr 2007;92(4):e56–8. doi: 10.3324/haematol.11394 [DOI] [PubMed] [Google Scholar]
- 17.Kang EM, de Witte M, Malech H, et al. Nonmyeloablative conditioning followed by transplantation of genetically modified HLA-matched peripheral blood progenitor cells for hematologic malignancies in patients with acquired immunodeficiency syndrome. Blood. Jan 15 2002;99(2):698–701. doi: 10.1182/blood.v99.2.698 [DOI] [PubMed] [Google Scholar]
- 18.Newton PJ, Newsholme W, Brink NS, Manji H, Williams IG, Miller RF. Acute meningoencephalitis and meningitis due to primary HIV infection. BMJ. 2002;325(7374):1225–1227. [PMC free article] [PubMed] [Google Scholar]
- 19.Wendel KA, McArthur JC. Acute Meningoencephalitis in Chronic Human Immunodeficiency Virus (HIV) Infection: Putative Central Nervous System Escape of HIV Replication. Clinical Infectious Diseases. 2003;37(8):1107–1111. doi: 10.1086/378300 [DOI] [PubMed] [Google Scholar]
- 20.Chaillon A, Gianella S, Dellicour S, et al. HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J Clin Invest. Apr 1 2020;130(4):1699–1712. doi: 10.1172/jci134815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gama L, Abreu CM, Shirk EN, et al. Reactivation of simian immunodeficiency virus reservoirs in the brain of virally suppressed macaques. AIDS. 2017;31(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joseph SB, Kincer LP, Bowman NM, et al. Human Immunodeficiency Virus Type 1 RNA Detected in the Central Nervous System (CNS) After Years of Suppressive Antiretroviral Therapy Can Originate from a Replicating CNS Reservoir or Clonally Expanded Cells. Clin Infect Dis. Sep 27 2019;69(8):1345–1352. doi: 10.1093/cid/ciy1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salantes DB, Zheng Y, Mampe F, et al. HIV-1 latent reservoir size and diversity are stable following brief treatment interruption. The Journal of Clinical Investigation. 07/February/ 2018;128(7):3102–3115. doi: 10.1172/JCI120194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen YZ, Lorenzi JCC, Krassnig L, et al. Relationship between latent and rebound viruses in a clinical trial of anti–HIV-1 antibody 3BNC117. Journal of Experimental Medicine. 2018;215(9):2311–2324. doi: 10.1084/jem.20180936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eberhard JM, Angin M, Passaes C, et al. Vulnerability to reservoir reseeding due to high immune activation after allogeneic hematopoietic stem cell transplantation in individuals with HIV-1. Sci Transl Med. May 6 2020;12(542)doi: 10.1126/scitranslmed.aay9355 [DOI] [PubMed] [Google Scholar]
- 26.Powell AB, Ren Y, Korom M, et al. Engineered Antigen-Specific T Cells Secreting Broadly Neutralizing Antibodies: Combining Innate and Adaptive Immune Response against HIV. Mol Ther Methods Clin Dev. Dec 11 2020;19:78–88. doi: 10.1016/j.omtm.2020.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel S, Hanajiri R, Grant M, et al. HIV-Specific T Cells Can Be Generated against Non-escaped T Cell Epitopes with a GMP-Compliant Manufacturing Platform. Mol Ther Methods Clin Dev. Mar 13 2020;16:11–20. doi: 10.1016/j.omtm.2019.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sung JA, Patel S, Clohosey ML, et al. HIV-Specific, Ex Vivo Expanded T Cell Therapy: Feasibility, Safety, and Efficacy in ART-Suppressed HIV-Infected Individuals. Mol Ther. Oct 3 2018;26(10):2496–2506. doi: 10.1016/j.ymthe.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel S, Lam S, Cruz CR, et al. Functionally Active HIV-Specific T Cells that Target Gag and Nef Can Be Expanded from Virus-Naïve Donors and Target a Range of Viral Epitopes: Implications for a Cure Strategy after Allogeneic Hematopoietic Stem Cell Transplantation. Biol Blood Marrow Transplant. Mar 2016;22(3):536–41. doi: 10.1016/j.bbmt.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]


