Abstract
Immature dentate granule cells (DGCs) generated in the hippocampus during adulthood are believed to play a unique role in dentate gyrus (DG) function. Although immature DGCs have hyperexcitable membrane properties in vitro, the consequences of this hyperexcitability in vivo remain unclear. In particular, the relationship between experiences that activate the DG, such as exploration of a novel environment (NE), and downstream molecular processes that modify DG circuitry in response to cellular activation is unknown in this cell population. We first performed quantification of immediate early gene (IEG) proteins in immature (5-week-old) and mature (13-week-old) DGCs from mice exposed to a NE. Paradoxically, we observed lower IEG protein expression in hyperexcitable immature DGCs. We then isolated nuclei from active and inactive immature DGCs and performed single-nuclei RNA-Sequencing. Compared to mature nuclei collected from the same animal, immature DGC nuclei showed less activity-induced transcriptional change, even though they were classified as active based on expression of ARC protein. These results demonstrate that the coupling of spatial exploration, cellular activation, and transcriptional change differs between immature and mature DGCs, with blunted activity-induced changes in immature cells.
Keywords: adult neurogenesis, Arc, dentate gyrus, hippocampus, immediate early gene, single-nuclei RNA-seq, Smart-Seq2
1 |. INTRODUCTION
The dentate gyrus (DG) of the mammalian hippocampus continues to produce new neurons even in adulthood (Kempermann et al., 2015). The precise contribution of adult-born neurons to the function of the hippocampus remains a subject of debate and may differ depending on these neurons’ developmental stage. Within a few months of their birth in the rodent hippocampus, adult-born dentate granule cells (DGCs) reach maturity and attain morphological and physiological features that are generally considered indistinguishable from their developmentally generated counterparts (Laplagne et al., 2006; van Praag et al., 2002). During their maturation process, however, immature DGCs possess distinct electrophysiological properties.
Immature DGCs under 6–7 weeks of age show an intrinsic electrophysiological hyperexcitability. They are characterized by high input resistance, high efficacy of firing to current injection, and increased activation in response to perforant path stimulation (Dieni et al., 2016; Marín-Burgin et al., 2012; Mongiat et al., 2009; Yang et al., 2015). These immature cells also show enhanced long-term potentiation (LTP), with a reduced threshold for induction and increased amplitude (Ge et al., 2007; Schmidt-Hieber et al., 2004). Reduced synaptic connectivity may counterbalance cellular hyperexcitability, at least in part. Immature DGCs are more limited by excitatory drive than mature cells, and stimulation of the entorhinal cortex has been reported to poorly recruit immature DGCs (Dieni et al., 2013, 2016). While in vivo data on the firing patterns of immature cells are limited, immature DGCs have been shown to generate more calcium transients than mature cells in one study using head-fixed imaging (Danielson et al., 2016). Combined, these results have led to considerable speculation about how immature DGCs contribute to the function of the DG in learning and memory.
Memory formation requires not just synaptic activity but also new transcription and translation (Alberini & Kandel, 2015; Costa-Mattioli et al., 2009; Yap & Greenberg, 2018). An initial wave of activity-induced transcription is detectable within minutes of a stimulus and results in the expression of immediate early genes (IEGs) such as Fos and Arc (Yap & Greenberg, 2018). Additional transcriptional changes occur over the next several hours, and transcriptional and translational inhibitors can continue to impair memory over 1–2 days (Alberini & Kandel, 2015; Costa-Mattioli et al., 2009; Yap & Greenberg, 2018). To explore the transcriptional mechanisms downstream of neuronal activity in the DG in an unbiased manner, we previously used single-nuclei RNA sequencing (snRNA-Seq). We exposed mice to a novel environment (NE), isolated individual DGCs, and classified them as active or inactive based on the expression of IEG proteins (Jaeger et al., 2018; Lacar et al., 2016). We observed that DGCs, whose firing and IEG expression are quite sparse in comparison to other hippocampal cell types, have a more dramatic transcriptional response to activation than CA1 pyramidal cells (Jaeger et al., 2018). Enhanced activity-dependent gene expression in DGCs may support the development of highly selective responses to specific stimuli. However, our previous work did not address the contribution of immature cells.
In the current study, we sought to compare activity-related changes in immature and mature DGCs by addressing two questions. First, are immature DGCs more or less likely than mature DGCs to express IEG markers of activation in response to a NE? Second, among immature and mature DGCs that express IEG proteins, what transcriptional changes are observed in active cells? We hypothesized that hyperexcitable immature DGCs would show enhanced IEG expression and activity-dependent transcriptional change. Counterintuitively, we observed that immature DGCs are both less likely to show IEG markers of activity and less transcriptionally modified after activation.
2 |. MATERIALS AND METHODS
2.1 |. Animals and drug treatment
All animal procedures were approved by the Institutional Animal Care and Use Committee of The Salk Institute for Biological Studies. Experiments were conducted in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals and the US Public Health Service’s Policy on Humane Care and Use of Laboratory Animals. To generate Ascl1CreERT2 × LSL-Sun1-sfGFP mice, hemizygous Ascl1tm1.1(Cre/ERT2)Jejo/J mice (Jackson Laboratory stock #012882, RRID: IMSR_JAX:012882) (Kim et al., 2011) were bred with homozygous B6;129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/MmbeJ (Jackson laboratory stock #030952, RRID:IMSR_JAX:030952, a generous gift from Dr. Margarita Behrens) (Mo et al., 2015). All mice were housed under a standard 12-hr light/dark cycle with ad libitum access to rodent chow and water. Male and female mice were 6–10 weeks of age at the beginning of drug treatment. To induce Cre-recombination and label immature cells, TAM (Sigma-Aldrich, T5648) was delivered at 200 mg/kg in 90% corn oil/10% ethanol by oral gavage daily for 7 days.
2.2 |. NE exposure
To stimulate behaviorally relevant neuronal activity, mice were exposed to a NE for 1 hr. NE cages contained huts, tunnels, running wheels, and sawdust bedding or a textured plastic floor. Cage dimensions were 54 in. × 34 in. base, 12 in. height or 42 in. × 42 in. base, and 12 in. height. At the conclusion of NE exposure, mice were returned to the home cage until sacrifice.
2.3 |. Seizure treatment
To induce widespread IEG activity in the DG, PTZ (Sigma-Aldrich, P6500) was administered i.p. at 50 mg/kg in PBS. PBS-injected animals served as controls for staining conditions. Mice were observed for 90 min before perfusion.
2.4 |. Histology
For histological analysis, mice were deeply anesthetized with ketamine/xylazine/acepromazine i.p. and transcardially perfused with 0.9% NaCl followed by 4% paraformaldehyde (PFA). Brains were extracted, post-fixed in PFA overnight, and then sunk in 30% sucrose. Coronal sections (20–40 μm) were obtained on a sliding microtome and stored at −20°C until staining. For staining, a 1 in 12 series of sections was washed in TBS, blocked with 0.25% Triton X-100 in TBS with 3% horse serum and incubated with primary antibody diluted in blocking buffer for 3 nights at 4°C. Sections were washed and incubated with fluorophore-conjugated secondary antibodies for 2 hr at room temperature. DAPI was applied at 1:1000 in TBS wash following secondary incubation. At the conclusion of TBS washes, sections were mounted with Immu-Mount mounting media.
2.5 |. Antibodies
Antibodies used in this study were:
Mouse anti-PROX1 (EMD Millipore #MAB5654, RRID:AB_2170714).
Rat anti-CTIP2 (Abcam #ab18465, RRID:AB_2064130).
Chicken anti-GFP (Aves Labs #GFP-1020, RRID:AB_10000240).
Guinea pig anti-ARC (Synaptic Systems #156005, RRID:AB_2151848 and #156004, RRID:AB_2619853).
Rabbit anti-FOS (Synaptic Systems #226003, RRID:AB_2231974).
Donkey anti-mouse AF647 (Jackson ImmunoResearch #715-605-151, RRID:AB_2340863).
Donkey anti-rat Dyl405 (Jackson ImmunoResearch #712-475-153, RRID:AB_2340681).
Donkey anti-chicken AF488 (Jackson ImmunoResearch #703-545-155, RRID:AB_2340375).
Donkey anti-guinea pig Cy3 (Jackson ImmunoResearch #706-165-148, RRID:AB_2340460).
Donkey anti-rabbit Cy3 (Jackson ImmunoResearch #711-165-152, RRID:AB_2307443).
Donkey anti-rabbit AF647 (Jackson ImmunoResearch #711-605-152, RRID:AB_2492288).
2.6 |. Imaging and quantification
Images of the DG were obtained on a Zeiss laser scanning confocal microscope (710 or 880) using a 20× objective. Z-stacks were obtained through the full section thickness, and tiles spanning the DG were stitched using ZEN software. Active cells (ARC+ or FOS+) and colocalization with markers of maturity (GFP, CTIP2) were quantified manually by a blinded observer. For NE experiments, 3–4 sections were analyzed per mouse. For PTZ experiments, two sections were analyzed per mouse. All GFP+ immature cells in each section were quantified. Among GFP− cells, 100 mature CTIP2 +GFP− cells were randomly selected blind to FOS status, avoiding the inner edge of the GCL to avoid inadvertently selecting immature cells. Because the goal of the PTZ experiment was to assess IEG rates in immature cells under widespread activation conditions, only samples that showed a high IEG response (>20% of mature DGCs activated) were included in the quantification.
2.7 |. Nuclei dissociation
Nuclei dissociation was performed as previously described (Jaeger et al., 2018; Lacar et al., 2016). Briefly, the hippocampus was dissected bilaterally after deep anesthesia with ketamine/xylazine/acepromazine and cervical dislocation. Hippocampal tissue was immediately placed into a nuclei isolation medium (sucrose 0.25 M, KCl 25 mM, MgCl2 5 mM, Tris-HCl 10 mM, dithiothreitol 100 μM, 0.1% Triton, and protease inhibitors). Tissue was dounce homogenized and then samples were washed, resuspended in nuclei storage buffer (0.167 M sucrose, MgCl2 5 mM, and Tris-HCl 10 mM, dithiothreitol 100 μM, and protease inhibitors) and filtered. Antibody staining proceeded in the nuclei storage buffer. Solutions and samples were kept cold throughout the protocol. For RNA-seq experiments, tools and solutions were made RNAse-free and RNAse inhibitors were used (Ambion #AM2684 at 1:1000 in both isolation and storage buffers).
2.8 |. Flow cytometry
For analysis of activation rates in immature and mature cells, dissociated nuclei were stained for dentate markers PROX1 and CTIP2, cell age indicator GFP, and the IEGs ARC or FOS. Data were collected on a BD LSRII cytometer (BD Biosciences) and analyzed with FlowJo software (Tree Star). Nuclei were gated based on forward and side scatter, and DGCs were identified based on co-labeling with PROX1 and CTIP2. Nuclei from mice lacking the Ascl1-CreERT2 transgene and not exposed to the NE were used as negative controls to validate gating strategy for immature (GFP+) and active (ARC or FOS+) cells. Quantifications were based on capturing an average of 42,694 ± 11,936 mature DGCs, which yielded 1004 ± 385 immature GFP+ DGCs per mouse. For nuclei isolation before RNA-Seq, samples were run on a BD Influx cell sorter and collected directly into 384-well plates pre-filled with 1 μL lysis buffer (0.1% Triton-X, 1 U/μL RNAse inhibitor, 2.5 mM dNTP mix, 2.5 μM anchored oligo-dT primer) and frozen at −80°C until further processing.
2.9 |. Single-nuclei RNA-seq
Libraries were prepared using the Smart-seq2 protocol as previously described with minor modifications (Jaeger et al., 2018; Picelli et al., 2013, 2014). First-strand synthesis with ProtoScript II reverse transcriptase (New England Biolabs), PCR preamplification with KAPA Hifi HotStart ReadyMix (Kapa), and bead cleanup (Agencourt AMPure XP beads, Beckman Coulter) were performed with the aid of a Mosquito HV liquid handler (SPT Labtech). The quality of cDNA was assessed by Bioanalyzer chip (Agilent) and concentration quantified by Qubit assay (Invitrogen). Single-nuclei libraries were prepared with a Nextera XT DNA library prep kit (Illumina) and pooled together for sequencing. The quality of pooled libraries was assessed by TapeStation assay (Agilent). Libraries were sequenced on an Illumina HiSeq using single-end 50 bp reads by the Salk Institute Next-Generation Sequencing Core. Reads were trimmed using Solexa-QA++ dynamic trim and aligned to the mm10 (GRCm38) reference genome with Ensembl gene annotation using RSEM (bowtie). Transcripts per million values calculated by RSEM were log2 + 1 transformed. A total of 401 nuclei pooled from 14 mice (9 males and 5 females) were included in the analysis.
2.10 |. Single-nuclei RNA-seq data preprocessing
Seurat was used for cell clustering analysis. The top 2000 highly variable genes were determined by the variance stabilizing transformation (vst) method. Significant principal components (PCs) were estimated independently for each dataset. Clustering was performed based on these significant PCs. For UMAP plots, the default parameters were used, except the number of PCs was determined using the JackStraw function in Seurat. DEGs between cell types were determined using a Wilcoxon test with false discovery rate (FDR) < 0.05. GO pathway analysis was done using DAVID (https://david.ncifcrf.gov/) and significant GO terms were determined with FDR < 0.05.
2.11 |. Statistics
Cell activation was analyzed in GraphPad Prism. Specific t test or ANOVA results are described in Section 3. Distributions were assumed to be normal, with no explicit normality tests performed. Significance thresholds were set at p < .05.
3 |. RESULTS
We previously studied how neuronal activation arising from NE exposure (Jaeger et al., 2018; Lacar et al., 2016) triggered activity-related transcription in DGCs, but we did not discriminate between immature and mature DGCs. To identify immature and mature DGCs, cells were birthdated in adult mice expressing a nuclear membrane-targeted GFP under the control of an Ascl1-driven Cre recombinase (Ascl1CreERT2 × LSL-Sun1-sfGFP mice, Figure 1a). In response to tamoxifen (TAM) treatment, GFP is expressed in Ascl1+ neural progenitors within the DG, and the GFP signal persists in cells as they differentiate and mature, remaining clearly visible within the subgranular zone and granule cell layer up to 13 weeks later (Figure 1b).
FIGURE 1.
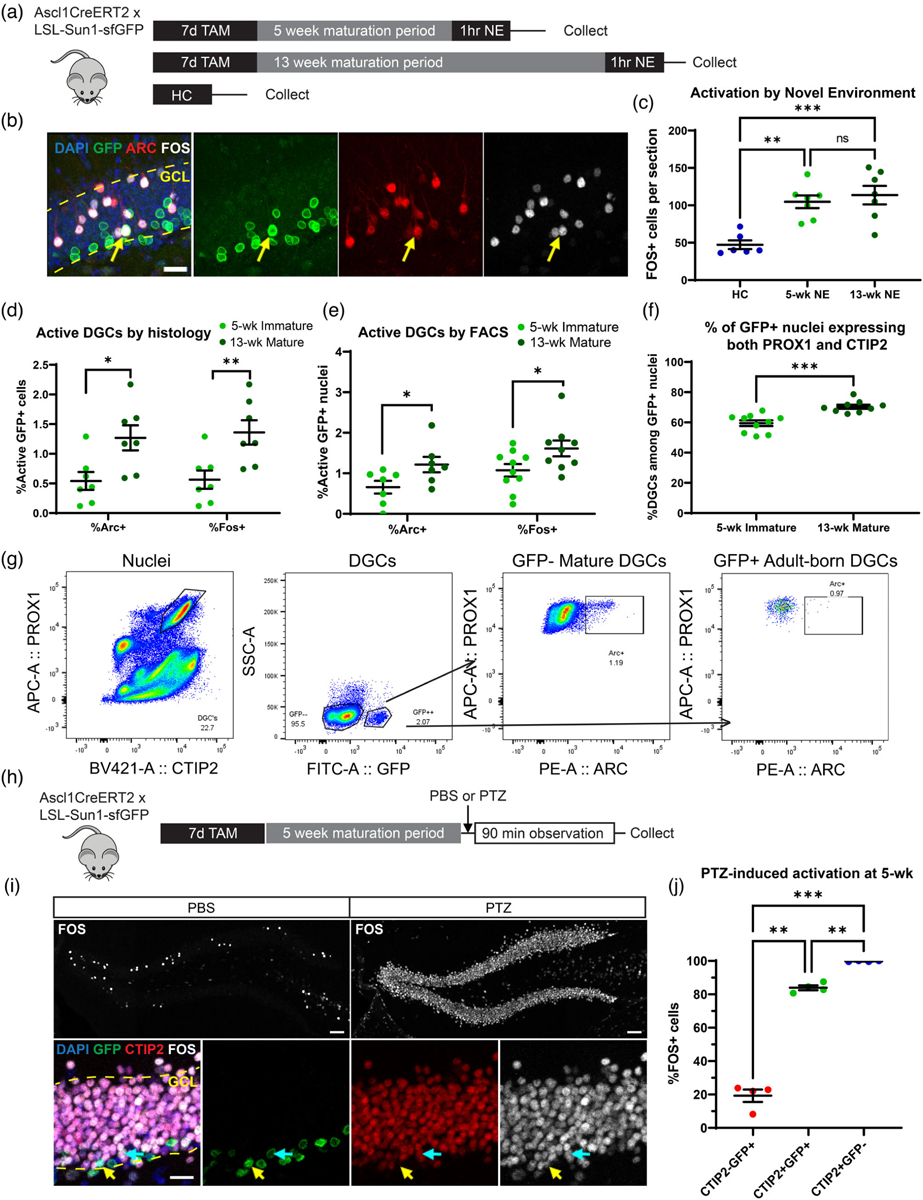
Reduced immediate early gene (IEG) protein expression in immature DGCs. (a) Experimental timeline. Mice received tamoxifen (TAM) gavage for 7 days to label DGCs and were exposed to a novel environment (NE) to stimulate activation 5 or 13 weeks (wk) later. Untreated littermates collected directly from the home cage (HC) were used as a control. (b) Sample of durable GFP labeling 13 weeks after TAM in the DG. Scale bar: 20 μm. Yellow arrows indicate a cell triple labeled for GFP, ARC, and FOS. Dashed yellow line indicates the boundaries of the granule cell layer (GCL). (c) NE exposure induces significantly more FOS and ARC expression than the home cage control condition (n’s: HC = 6, 5-wk NE = 7, 13-wk NE = 7). (d) Quantification of active 5-week and 13-week DGCs in coronal tissue sections shows greater IEG expression in more mature cells (n’s: 5-wk = 7, 13-wk = 7). (e) Quantification of active 5-week and 13-week DGCs via flow cytometry (FACS) also shows greater IEG expression in more mature cells (n’s: Arc 5-wk = 7, Arc 13-wk = 7, Fos 5-wk = 10, Fos 13-wk = 9). (f) Co-expression of DG marker proteins PROX1 and CTIP2 occurs in the majority of GFP+ nuclei assessed by FACS, but co-expression continues to increase between 5 and 13 weeks (n’s: 5-wk = 10, 13-wk = 9). (g) Sample FACS gating strategy for identifying active immature DGCs. DGCs were identified by co-labeling with marker proteins PROX1 and CTIP2. Cell age was identified by GFP. Cell activation was assessed with ARC or FOS. (h) Experimental timeline for seizure treatment. Mice received TAM gavage for 7 days to label DGCs and were given pentylenetetrazole (PTZ) 5 or 13 weeks later. PBS-injected littermates served as staining controls. Mice were perfused following a 90-min observation period after the injection. (i) Top: Sample of FOS labeling following either PBS (left) or PTZ (right). Scale bar: 50 μm. Bottom: Sample of cell classification in PTZ-treated mice. Turquoise arrows indicate a cell triple labeled for GFP, CTIP2, and FOS. Yellow arrows indicate a cell double labeled for GFP and FOS, negative for CTIP2. Dashed yellow line indicates GCL boundaries. Scale bar: 20 μm. (j) Quantification reveals that CTIP2−GFP+ immature cells activate at significantly lower rates than CTIP2+GFP+ immature cells. CTIP2+GFP+ immature cells still have reduced FOS relative to mature CTIP2+GFP− cells at 5 weeks after TAM even under seizure conditions (n = 4, within-subject comparison). Points and n’s in dot plots represent mice. Error bars are mean ± S.E.M. *p < .05, **p < .01, ***p < .001, ns = not significant.
Because immature DGCs are electrophysiologically hyperexcitable, the ability of this immature population to express IEGs in response to a behaviorally relevant stimulation was measured and compared to mature cells. Ascl1CreERT2 × LSL-Sun1-sfGFP mice were treated with TAM to label newborn cells and exposed to a NE 5 or 13 weeks later, when labeled cells were immature or mature, respectively (Figure 1a). The NE was a large arena filled with objects previously unknown to the mice, including tunnels, huts, and running wheels. The NE provided opportunities for spatial exploration and interaction with these novel objects, both processes that engage the hippocampus. We previously observed habituation of exploration upon reexposure to the same environment 4 hr later, suggesting that mice encode a memory of the NE that impacts their behavior at this later time point (Jaeger et al., 2018). We extended the time of NE exposure out to 1 hr, compared to 15 min for previous studies, because pilot experiments suggested slightly higher rates of IEG expression with the longer exposure. Mice were perfused immediately after NE exposure, and expression of IEG proteins was quantified in coronal sections of the DG. Untreated wild-type littermates collected directly from their home cages were used as a control. As we previously observed, 1 hr of NE exposure led to a significant increase in FOS+ cells compared to home cage controls (HC) (Figure 1c; one-way ANOVA F(2,17) = 13.24, p = .0003, Tukey’s multiple comparisons test p = .0019 for HC vs. 5-week, p = .0005 for HC vs. 13-week, p = ns for 5-week vs. 13-week). Surprisingly, both of these IEG proteins were expressed at a lower rate in immature (5-week) compared to mature (13-week) GFP+ cells (Figure 1d; ARC: Welch’s t test t = 2.797, p = .0176; FOS: Welch’s t test t = 3.103, p = .0099). Activation rates of immature and mature GFP+ cells were also compared using flow cytometry (Figure 1e–g). Mature GFP- nuclei were used as a control to validate the gating strategy for adult-born GFP+ nuclei (Figure 1g). At 5 weeks of age, immature GFP+ nuclei expressed ARC and FOS at a reduced rate compared to mature 13-week-old nuclei (Figure 1e; ARC: Welch’s t test t = 2.261, p = .0438; FOS: Welch’s t test t = 2.166, p = .0461). We also observed that the majority of GFP+ nuclei expressed both DGC markers PROX1 and CTIP2 by 5 weeks of age, but the fraction of the GFP+ population expressing both markers continued to increase between 5 and 13 weeks, suggesting ongoing maturation of the adult-born cell population beyond 5 weeks (Figure 1f; Welch’s t test t = 4.754, p = .0002). These results indicate that immature DGCs, despite their electrophysiological hyperexcitability, are actually less likely to express protein markers of activation than mature DGCs, further suggesting that the vast majority of ARC+ and FOS+ cells observed in the DG during memory encoding are likely to be mature DGCs.
To assess whether variability in maturation status within the immature cell population impacted rates of activation, we used the GABA-A receptor antagonist pentylenetetrazole (PTZ). PTZ is a seizure-inducing drug that we have previously observed to elicit a widespread IEG response throughout the DG (Lacar et al., 2016). Ascl1CreERT2 × LSL-Sun1-sfGFP mice received TAM treatment and were then injected with saline (PBS) or PTZ (50 mg/kg, i.p.) 5 weeks later (Figure 1h). All PTZ-injected mice had visible signs of seizure, including altered postures, twitching, and motor spasms. Mice were perfused for histological analysis 90 min after drug injection. PBS-injected animals were used as a control to validate staining conditions (Figure 1i). We classified 5-week-old GFP+ cells based on the presence or absence of CTIP2. Based on our flow cytometry results that the fraction of PROX1+CTIP2+ nuclei continued to increase after 5 weeks, we considered cells lacking CTIP2 expression to be more immature than their CTIP2+ neighbors. In response to PTZ treatment, immature GFP+ cells without CTIP2 had a dramatically lower activation rate than either CTIP2+GFP+ immature cells or mature CTIP2+-GFP− cells (Figure 1j; one-way repeated measures ANOVA F(1.026, 3.078) = 270.5, p = .0004; Tukey’s multiple comparisons test p = .0021 for CTIP2−GFP+ vs. CTIP2+GFP+, p = .0004 for CTIP2−GFP+ vs. CTIP2+GFP−). The immature cells expressing CTIP2 responded robustly to PTZ but still expressed FOS at a lower rate than mature GFP− cells (Tukey’s multiple comparisons test p = .0040 for CTIP2+GFP+ vs. CTIP2+GFP−). These results indicate that, despite variability in the maturation of cells tagged by the GFP reporter, immature DGCs appear less likely to express IEG proteins than mature DGCs. Before CTIP2 expression, immature DGCs are particularly resistant to activation.
We next asked whether different activity-related transcriptional programs were triggered in mature DGCs and the subset of immature DGCs that did successfully generate ARC protein in response to a NE. As above, newborn cells were labeled in Ascl1CreERT2 × LSL-Sun1-sfGFP mice and allowed to mature to 5 weeks of age. Mice were exposed to a NE for 1 hr and then sacrificed for hippocampus dissection either immediately (1-hr group) or after 3 additional hr in the home cage (4-hr group) (Figure 2a). These time points were selected to capture a first wave of transcription corresponding to IEG expression and a second wave corresponding to late response genes. The dissected hippocampus was dounce homogenized and stained for DGC marker proteins PROX1 and CTIP2, cell age marker GFP, and activation marker ARC. Nuclei from four populations were isolated for sequencing (Figure 2b) at each of these time points: (1) Mature Inactive cells; (2) Immature Inactive cells; (3) Mature Active cells; and (4) Immature Active cells. A total of 401 nuclei pooled from 14 mice (9 males and 5 females) were included in our final analysis. As suggested by the above histology and flow cytometry quantifications in 5- versus 13-week cells, the Immature Active population was extremely small (Figure S1). After nuclei isolation and RNA-Seq library preparation by SmartSeq2, these four populations were compared within each time point. Prior work in our lab has demonstrated that the activity-related transcriptional response differs at 1 and 4 hr after the activation event, as some IEGs return to baseline levels, others exhibit sustained changes at both time points, and some late response genes are activated only at the 4-hr time point.
FIGURE 2.
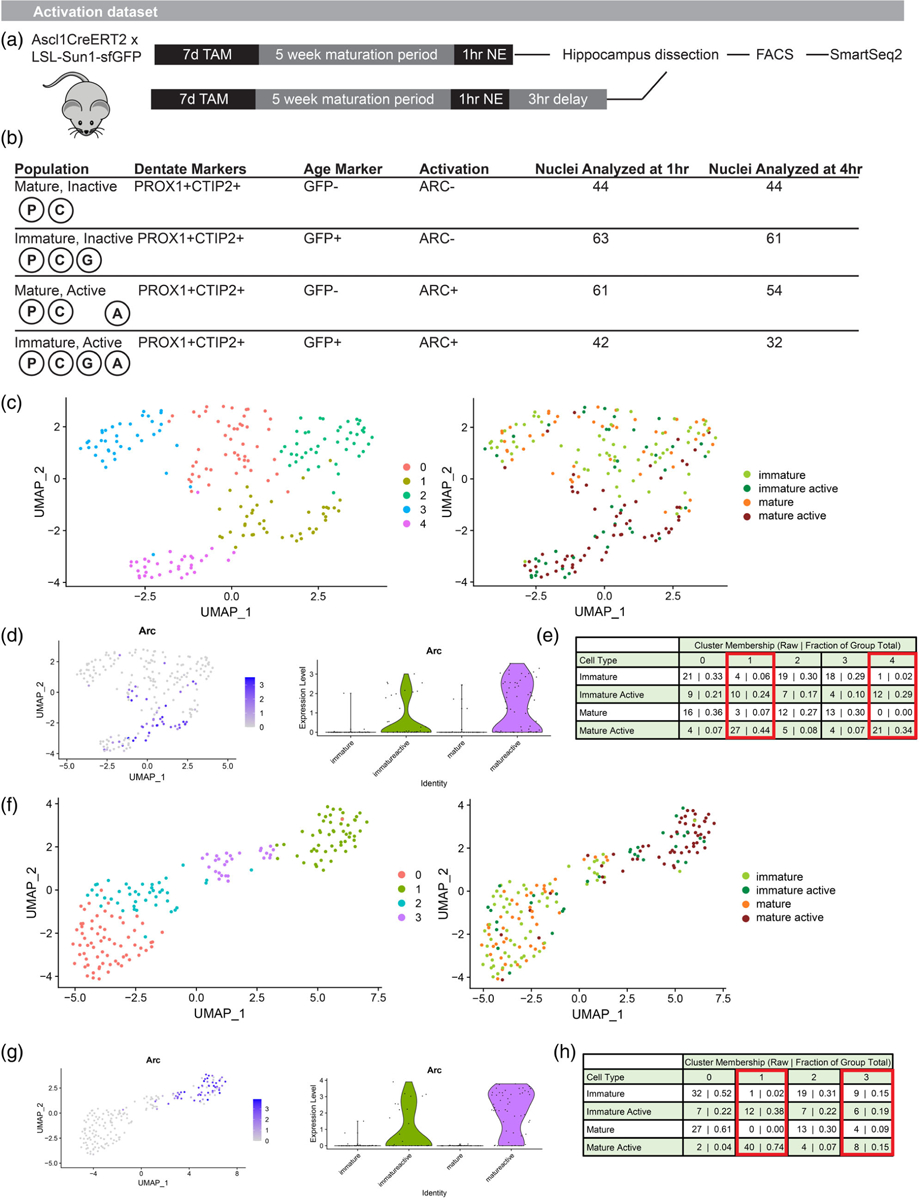
Reduced transcriptional response to activation in immature DG neurons. (a) Experimental timeline. Mice received TAM to label DGCs, were exposed to a NE 5 weeks later, and then were sacrificed for hippocampus dissection and nuclei preparation immediately or after an additional 3-hr delay. (b) Diagram of nuclear cell markers used to isolate specific populations via FACS and total cells per population included in the analysis. (c) Unbiased clustering of nuclei isolated at 1 hr after the start of NE. (d) Arc expression in nuclei isolated at 1 hr identifies active clusters. (e) Mature Active cells preferentially localize to clusters 1 and 4. Immature Active cells are more broadly distributed across clusters despite isolation based on ARC protein expression. (f) Unbiased clustering of nuclei isolated at 4 hr after the start of NE. (g) Arc expression in nuclei isolated at 4 hr identifies active clusters. (h) Mature Active cells preferentially localize to clusters 1 and 3. Immature Active cells are more broadly distributed across clusters despite isolation based on ARC protein expression. DG, dentate granule; DGC, dentate granule cell; NE, novel environment.
At 1 hr after activation, unbiased clustering segregated most Mature Active cells, featuring ARC protein expression, into 2 clusters that also exhibited high relative levels of Arc transcripts (Figure 2c,d). As expected, few Mature Inactive or Immature Inactive cells were found within these active clusters. In contrast, Immature Active cells did not preferentially localize with Mature Active cells. Instead, Immature Active cells were broadly distributed across clusters (Figure 2e; Fisher’s exact test for Mature Active versus Immature Active in active clusters 1 and 4 versus inactive clusters 0, 2, and 3, p = .0093). Similar results were observed at 4 hr after activation. Again, the vast majority of Mature Active cells identified by ARC protein expression occupied active clusters with higher levels of Arc transcripts (Figure 2f,g). Immature Active cells were again broadly distributed across clusters instead of preferentially belonging to active clusters (Figure 2h; Fisher’s exact test for Mature Active versus Immature Active in active clusters 1 and 3 versus inactive clusters 0 and 2, p = .0011). At both time points, we also observed that Immature Inactive cells occupied clusters populated by Mature Inactive cells. Immature and mature DGCs thus appeared to have broadly similar transcriptional profiles at baseline, and Immature Inactive cells did not have transcriptional signs of activation at baseline in the absence of ARC protein. These results suggest that, even among the 5-week-old DGCs that activated in response to a NE, the overall transcriptional response to activation was blunted relative to the response observed in mature cells.
Further analysis of active versus inactive cells revealed that the number of differentially expressed genes (DEGs) detected was far greater in mature than immature cells at both the 1- and 4-hr time points (Figure 3a). Among the DEGs, three were common to both immature and mature cells at 1 hr and five were common to both immature and mature cells at 4 hr (Figure 3b). The remaining DEGs were unique to mature cells. No DEGs were unique to the Immature Active versus Immature Inactive comparison. Activation in mature cells significantly upregulated the expression of IEGs previously identified in active DGCs such as Homer1 and Bdnf by 1 hr, but these genes fell short of significance in immature cells (Figure 3c). Sustained upregulation of some genes, including Bdnf, Spry2, and Pim1, was observed only in mature cells at 4 hr, along with late response genes we previously identified in active DGCs such as Sorcs3 and Blnk. GO-term analysis of the activity-response genes unique to mature cells showed significant enrichment of terms related to postsynaptic membrane, cell junction, and postsynaptic density (Figure 3d,e). Overall, these results demonstrate that activity-related transcriptional change in the DG occurs mainly in mature cells. Among immature DGCs sufficiently active to express ARC protein, their transcriptional state is activated to a lesser degree than mature cells, failing to show the same level of activity-induced gene expression.
FIGURE 3.
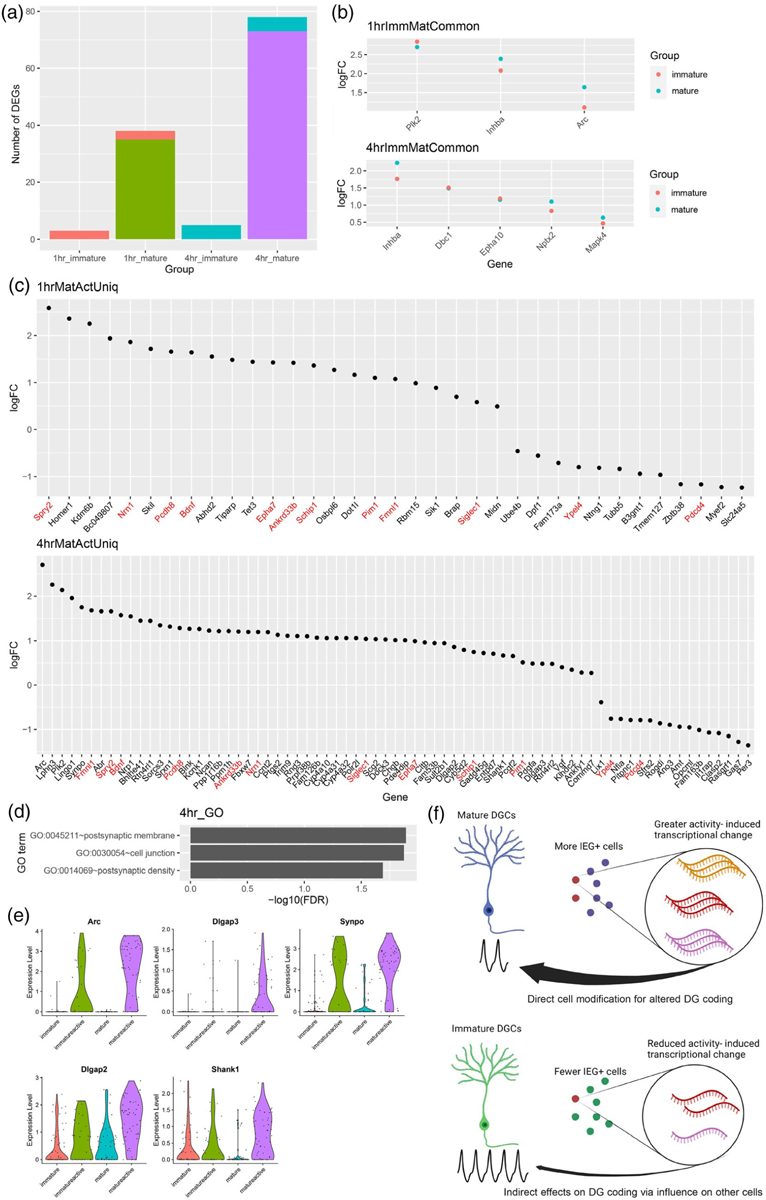
Immature DG neurons show activity-related changes in transcription of only a subset of genes observed in mature. (a) Number of differentially expressed genes (DEGs) between Active and Inactive nuclei based on maturation status and time point, for example, 1-hr immature indicates the number of DEGs between Immature Active and Immature Inactive cells isolated at 1 hr after the start of NE. Coral bar indicates genes shared between 1-hr immature and 1-hr mature comparisons. Blue bar indicates genes shared between 4-hr immature and 4-hr mature comparisons. (b) Log2 fold change (logFC) between active and inactive nuclei of DEGs common to both immature and mature nuclei from A (ImmMatCommon genes). (c) FC of DEGs uniquely identified in mature, but not immature, active, and inactive nuclei comparisons (Uniq genes). DEGs detected in mature cells at both 1 and 4 hr after the start of NE are labeled in red. No DEGs were uniquely identified in immature cells. (d) GO terms significantly enriched among DEGs uniquely identified in mature cells 4 hr after activation. (e) Violin plots of DEGs presented in both “postsynaptic membrane” and “postsynaptic density” terms from (d). (f) Model for contribution of immature DGCs to activity-dependent changes (created with BioRender.com). Mature cells respond by engaging cell intrinsic mechanisms of transcriptional change, whereas physiologically hyperactive immature cells may contribute more indirectly by influencing other cells. DG, dentate granule; DGC, dentate granule cell; NE, novel environment.
4 |. DISCUSSION
We observed reduced in vivo recruitment of immature 5-week-old DGCs compared to more mature cells when they were stimulated by NE exploration. Even among those 5-week-old DGCs that expressed activation marker ARC, activity-induced transcriptional change in immature DGCs was less pronounced than in their mature counterparts. These results were surprising, given the consistent finding from electrophysiological studies that immature DGCs under about 6–7 weeks of age are hyperexcitable to perforant path stimulation or direct current injection in ex vivo slices. Functionally, our results demonstrate that the coupling of behaviorally induced cortical input to the DG, expression of IEG proteins, and downstream consequences of IEG expression differs between mature and immature cells. These differences further suggest that the role of immature DGCs in memory encoding may be an indirect one. Mature DGCs may serve as the main substrate for cellular modification in the first few hours after a memorable experience. Immature DGCs may contribute to hippocampal function by modifying the response of mature cells rather than undergoing dramatic activity-dependent changes to directly encode a memory engram during this timeframe (Figure 3f).
We decided to focus on 5-week-old DGCs to target an age when immature neurons are synaptically connected both via inputs from the entorhinal cortex and outputs to CA3, but still show unique physiological properties (Dieni et al., 2016; Ge et al., 2007; Toni et al., 2008; Yang et al., 2015; Zhao et al., 2006). We thus could have missed an effect that occurs earlier in their development. However, given previous studies showing minimal IEG expression before 3–4 weeks of age (Jessberger & Kempermann, 2003; Jungenitz et al., 2014; Snyder et al., 2009), this effect would have to be quite transient. The low responsiveness of the subset of 5-week-old cells lacking the DG marker CTIP2, which we believe are more immature than their CTIP2+ counterparts, also argues against an earlier high-IEG phase. A few papers have reported increased in vivo recruitment of immature DGCs at age ranges similar to those studied here (Kee et al., 2007), but other groups have published conflicting results (Snyder et al., 2009; Stone et al., 2011). Our results add to the literature suggesting that immature DGCs are gradually incorporated into the circuitry of the DG, with synaptic connectivity and IEG expression converging on mature levels over a period of 1–2 months, rather than showing a hyperactive phase of IEG induction.
This study is, to the best of our knowledge, the first to use snRNA-Seq to compare activity-dependent transcription in adult-born DGCs. We elected to study nuclei to avoid inconsistent recovery of dendritic compartments and to prevent spurious IEG expression from whole-cell dissociation methods (Lacar et al., 2016; Wu et al., 2017). However, our approach will miss differences in dendritic RNA. It is possible that immature and mature DGCs differ in RNA localization either at rest or following activation. Additionally, the low fraction of immature DGCs within the dentate compared to mature DGCs, combined with their lower rate of ARC and FOS expression, means that IEG-positive immature DGCs are extraordinarily rare. Estimating that approximately 1%–3% of DGCs are tagged as immature with the GFP reporter and approximately 0.5%–1.0% of those are ARC+, the active immature population comprises somewhere between 0.005% and 0.03% of all DGCs. The rarity of this population makes it challenging to obtain large sample sizes, limiting our statistical power and precluding use of droplet-based sequencing methods to tease out any subpopulations of immature DGCs with different levels of responsiveness. Spatial transcriptomics may be fruitful in future studies to correlate activity-dependent transcription in immature DGCs with other key developmental features such as their migration into the granule cell layer or position along the dorsal-ventral axis. Other stimulation methods, such as seizure-inducing drugs, optogenetics, or chemogenetics, may be valuable in future studies to obtain larger populations for analysis. However, these methods all have the tradeoff of being less reflective of endogenous activation processes, and we elected not to pursue them as part of our snRNA-Seq experiments. Note that while PTZ-induced seizures did activate a larger fraction of immature DGCs than the NE exposure, immature DGCs still became FOS+ at a lower rate than mature DGCs even under these severe conditions.
We instead allotted 1 hr of exploration in the NE in an attempt to maximize IEG expression from a naturalistic experience, but this timeline does introduce some variability in attempting to timestamp the activation of specific cells. Some DGCs may have preferentially activated towards the start or end of this exploration window, potentially leaving the most recently active cells without time to produce ARC protein. However, Arc RNA can be generated within minutes, and we did not observe widespread Arc expression among nuclei that lacked ARC protein, suggesting our inactive cells were correctly classified.
Some of the reduced activity-induced transcriptional response in immature DGCs may reflect an inherent difference between adult-born and developmentally born DGCs. Although adult-born DGCs resemble their developmentally born counterparts at maturity, reports have emerged of some persistent differences, including increased spine density and mossy fiber bouton size among adult-born cells (Cole et al., 2020), changes in dendritic arbor structure (Kerloch et al., 2019), and increased ability of spatial learning tasks to cause structural remodeling of dendrites (Lemaire et al., 2012). Future studies might address these questions by labeling specific cohorts of developmentally born cells as a comparison.
The functional role of immature DGCs remains a subject of active debate. Although our results show that activity-dependent transcription in immature DGCs is reduced, prior studies demonstrate that immature DGCs still make an important functional contribution. Immature DGCs influence the ability to discriminate between similar experiences, known as behavioral pattern separation, as well as mood-related behaviors (Anacker & Hen, 2017; Toda et al., 2019). Exactly how this influence occurs is unclear. It has been hypothesized that the role of hyperexcitable immature DGCs is not to carry a specific signal but rather to modulate the activity of mature cells in the DG (Anacker & Hen, 2017; Drew et al., 2016). Several groups have reported that a loss of immature DGCs increases activation of the DG, including greater spread of current injection via the perforant path or greater IEG activation (Anacker et al., 2018; Burghardt et al., 2012; Ikrar et al., 2013). If this modulatory effect occurs at levels of activity that are insufficient to trigger IEG expression, it may explain how immature DGCs can be physiologically hyperactive without showing the same downstream modifications observed in mature DGCs. Even DGCs within the first 2–3 weeks of life can be stimulated to produce spikes, before any reliable induction of IEG proteins (Li et al., 2017; Schmidt-Hieber et al., 2004). Action potentials may thus still be generated in immature DGCs, allowing them to recruit inhibitory feedback via hilar interneurons, but these action potentials may not be converted into IEG expression as efficiently as in mature cells.
Immature DGCs may also provide a response that is important due to its quality rather than its quantity. Immature DGCs, whose developing dendritic arbors are smaller than those of mature cells, may provide less correlated output (Dieni et al., 2016; Li et al., 2017). Less correlated output would in turn provide unique computational power to the DG circuit even if the transcriptional response is reduced. Immature DGCs may also have a longer-term impact on DG circuitry as they compete for synaptic connections with existing cells (Akers et al., 2014; McAvoy et al., 2016). Environmental conditions as immature DGCs are developing are known to influence their survival (Tashiro et al., 2007), and different cohorts of immature DGCs maturing over different intervals may add temporal information not detectable over the short time course examined here (Rangel et al., 2014). Further studies of activity-dependent transcription under conditions where neurogenesis is ablated or enhanced could provide evidence of how this small fraction of cells can influence the DG and related behaviors.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Mary Lynn Gage for editorial assistance, Caz O’Connor and Lara Boggeman for flow cytometry guidance, and Amanda Argoncillo for rodent technical help. This work was supported by NIH Grant #MH114030 (F.H.G.), the JPB Foundation, American Heart Association/Paul G. Allen Frontiers Group Brain Health & Cognitive Impairment Initiative (19PABHI34610000), Annette C. Merle-Smith, the Dolby Family, and the Brinson Foundation. The authors also thank the Salk Institute Core facilities, in particular: the Waitt Advanced Biophotonics Core Facility with funding from NIH-NCI CCSG: P30 014195 and the Waitt Foundation; the Flow Cytometry Core Facility with funding from NIH-NCI CCSG: P30 014195; and the NGS Core Facility with funding from NIH-NCI CCSG: P30 014195, the Mary K. Chapman Foundation and the Helmsley Charitable Trust.
Funding information
Annette C. Merle-Smith; Brinson Foundation; JPB Foundation; National Institute of Mental Health, Grant/Award Number: MH114030; Paul G. Allen Frontiers Group, Grant/Award Number: 19PABHI34610000; The Dolby Family; NIH-NCI CCSG, Grant/Award Number: P30 014195; Waitt Foundation; Mary K. Chapman Foundation; Helmsley Charitable Trust
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
RNA-Seq data have been submitted to GEO (accession number GSE214309). Other data supporting the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Akers KG, Martinez-Canabal A, Restivo L, Yiu AP, De Cristofaro A, Hsiang H-LL, Wheeler AL, et al. (2014). Hippocampal neurogenesis regulates forgetting during adulthood and infancy. Science, 344(6184), 598–602. 10.1126/science.1248903 [DOI] [PubMed] [Google Scholar]
- Alberini CM, & Kandel ER (2015). The regulation of transcription in memory consolidation. Cold Spring Harbor Perspectives in Biology, 7(1), a021741. 10.1101/cshperspect.a021741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anacker C, & Hen R (2017). Adult hippocampal neurogenesis and cognitive flexibility—Linking memory and mood. Nature Reviews Neuroscience, 18(6), 335–346. 10.1038/nrn.2017.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anacker C, Luna VM, Stevens GS, Millette A, Shores R, Jimenez JC, Chen B, & Hen R (2018). Hippocampal neurogenesis confers stress resilience by inhibiting the ventral dentate gyrus. Nature, 559(7712), 98–102. 10.1038/s41586-018-0262-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghardt NS, Park EH, Hen R, & Fenton AA (2012). Adult-born hippocampal neurons promote cognitive flexibility in mice. Hippocampus, 22(9), 1795–1808. 10.1002/hipo.22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JD, Espinueva DF, Seib DR, Ash AM, Cooke MB, Cahill SP, O’Leary TP, Kwan SS, & Snyder JS (2020). Adult-born hippocampal neurons undergo extended development and are morphologically distinct from neonatally-born neurons. Journal of Neuroscience, 40(30), 5740–5756. 10.1523/JNEUROSCI.1665-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, & Sonenberg N (2009). Translational control of long-lasting synaptic plasticity and memory. Neuron, 61(1), 10–26. 10.1016/j.neuron.2008.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson NB, Kaifosh P, Zaremba JD, Lovett-Barron M, Tsai J, Denny CA, Balough EM, Goldberg AR, Drew LJ, Hen R, Losonczy A, & Kheirbek MA (2016). Distinct contribution of adult-born hippocampal granule cells to context encoding. Neuron, 90(1), 101–112. 10.1016/j.neuron.2016.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieni CV, Nietz AK, Panichi R, Wadiche JI, & Overstreet-Wadiche L (2013). Distinct determinants of sparse activation during granule cell maturation. Journal of Neuroscience, 33(49), 19131–19142. 10.1523/JNEUROSCI.2289-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieni CV, Panichi R, Aimone JB, Kuo CT, Wadiche JI, & Overstreet-Wadiche L (2016). Low excitatory innervation balances high intrinsic excitability of immature dentate neurons. Nature Communications, 7(1), 11313. 10.1038/ncomms11313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew LJ, Kheirbek MA, Luna VM, Denny CA, Cloidt MA, Wu MV, Jain S, Scharfman HE, & Hen R (2016). Activation of local inhibitory circuits in the dentate gyrus by adult-born neurons. Hippocampus, 26(6), 763–778. 10.1002/hipo.22557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Yang C. h., Hsu K. s., Ming G. l., & Song H (2007). A critical period for enhanced synaptic plasticity in newly generated neurons of the adult brain. Neuron, 54(4), 559–566. 10.1016/j.neuron.2007.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikrar T, Guo N, He K, Besnard A, Levinson S, Hill A, Lee H-K, Hen R, Xiangmin X, & Sahay A (2013). Adult neurogenesis modifies excitability of the dentate gyrus. Frontiers in Neural Circuits, 7, 204. 10.3389/fncir.2013.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger BN, Linker SB, Parylak SL, Barron JJ, Gallina IS, Saavedra CD, Fitzpatrick C, Lim CK, Schafer ST, Lacar B, Jessberger S, & Gage FH (2018). A novel environment-evoked transcriptional signature predicts reactivity in single dentate granule neurons. Nature Communications, 9(1), 3084. 10.1038/s41467-018-05418-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger S, & Kempermann G (2003). Adult-born hippocampal neurons mature into activity-dependent responsiveness. European Journal of Neuroscience, 18(10), 2707–2712. 10.1111/j.1460-9568.2003.02986.x [DOI] [PubMed] [Google Scholar]
- Jungenitz T, Radic T, Jedlicka P, & Schwarzacher SW (2014). High-frequency stimulation induces gradual immediate early gene expression in maturing adult-generated hippocampal granule cells. Cerebral Cortex, 24(7), 1845–1857. 10.1093/cercor/bht035 [DOI] [PubMed] [Google Scholar]
- Kee N, Teixeira CM, Wang AH, & Frankland PW (2007). Preferential incorporation of adult-generated granule cells into spatial memory networks in the dentate gyrus. Nature Neuroscience, 10(3), 355–362. 10.1038/nn1847 [DOI] [PubMed] [Google Scholar]
- Kempermann G, Song H, & Gage FH (2015). Neurogenesis in the adult hippocampus. Cold Spring Harbor Perspectives in Biology, 7(9), a018812. 10.1101/cshperspect.a018812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerloch T, Clavreul S, Goron A, Abrous DN, & Pacary E (2019). Dentate granule neurons generated during perinatal life display distinct morphological features compared with later-born neurons in the mouse hippocampus. Cerebral Cortex, 29(8), 3527–3539. 10.1093/cercor/bhy224 [DOI] [PubMed] [Google Scholar]
- Kim EJ, Ables JL, Dickel LK, Eisch AJ, & Johnson JE (2011). Ascl1 (Mash1) defines cells with long-term neurogenic potential in subgranular and subventricular zones in adult mouse brain. PLoS One, 6(3), e18472. 10.1371/journal.pone.0018472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacar B, Linker SB, Jaeger BN, Krishnaswami SR, Barron JJ, Kelder MJE, Parylak SL, Paquola ACM, Venepally P, Novotny M, O’Connor C, Fitzpatrick C, Erwin JA, Hsu JY, Husband D, McConnell MJ, Lasken R, & Gage FH (2016). Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nature Communications, 7(1), 11022. 10.1038/ncomms11022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplagne DA, Soledad Espósito M, Piatti VC, Morgenstern NA, Zhao C, van Praag H, Gage FH, & Schinder AF (2006). Functional convergence of neurons generated in the developing and adult hippocampus. PLoS Biology, 4(12), e409. 10.1371/journal.pbio.0040409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire V, Tronel S, Montaron M-F, Fabre A, Dugast E, & Abrous DN (2012). Long-lasting plasticity of hippocampal adult-born neurons. Journal of Neuroscience, 32(9), 3101–3108. 10.1523/JNEUROSCI.4731-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Sultan S, Heigele S, Schmidt-Salzmann C, Toni N, & Bischofberger J (2017). Silent synapses generate sparse and orthogonal action potential firing in adult-born hippocampal granule cells. ELife, 6, e23612. 10.7554/eLife.23612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Burgin A, Mongiat LA, Belén Pardi M, & Schinder AF (2012). Unique processing during a period of high excitation/inhibition balance in adult-born neurons. Science, 335(6073), 1238–1242. 10.1126/science.1214956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAvoy KM, Scobie KN, Berger S, Russo C, Guo N, Decharatanachart P, Vega-Ramirez H, Miake-Lye S, Whalen M, Nelson M, Bergami M, Bartsch D, Hen R, Berninger B, & Sahay A (2016). Modulating neuronal competition dynamics in the dentate gyrus to rejuvenate aging memory circuits. Neuron, 91(6), 1356–1373. 10.1016/j.neuron.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, Eddy SR, Ecker JR, & Nathans J (2015). Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron, 86(6), 1369–1384. 10.1016/j.neuron.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongiat LA, Soledad Espósito M, Lombardi G, & Schinder AF (2009). Reliable activation of immature neurons in the adult hippocampus. PLoS One, 4(4), e5320. 10.1371/journal.pone.0005320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, & Sandberg R (2013). Smart-Seq2 for sensitive full-length transcriptome profiling in single cells. Nature Methods, 10(11), 1096–1098. 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, & Sandberg R (2014). Full-length RNA-seq from single cells using Smart-Seq2. Nature Protocols, 9(1), 171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Rangel LM, Alexander AS, Aimone JB, Wiles J, Gage FH, Chiba AA, & Quinn LK (2014). Temporally selective contextual encoding in the dentate gyrus of the hippocampus. Nature Communications, 5, 3181. 10.1038/ncomms4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Hieber C, Jonas P, & Bischofberger J (2004). Enhanced synaptic plasticity in newly generated granule cells of the adult hippocampus. Nature, 429(6988), 184–187. 10.1038/nature02553 [DOI] [PubMed] [Google Scholar]
- Snyder JS, Choe JS, Clifford MA, Jeurling SI, Hurley P, Ashly Brown J, Kamhi F, & Cameron HA (2009). Adult-born hippocampal neurons are more numerous, faster maturing, and more involved in behavior in rats than in mice. Journal of Neuroscience, 29(46), 14484–14495. 10.1523/JNEUROSCI.1768-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SSD, Teixeira CM, Zaslavsky K, Wheeler AL, Martinez-Canabal A, Wang AH, Sakaguchi M, Lozano AM, & Frankland PW (2011). Functional convergence of developmentally and adult-generated granule cells in dentate gyrus circuits supporting hippocampus-dependent memory. Hippocampus, 21(12), 1348–1362. 10.1002/hipo.20845 [DOI] [PubMed] [Google Scholar]
- Tashiro A, Makino H, & Gage FH (2007). Experience-specific functional modification of the dentate gyrus through adult neurogenesis: A critical period during an immature stage. Journal of Neuroscience, 27(12), 3252–3259. 10.1523/JNEUROSCI.4941-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Parylak SL, Linker SB, & Gage FH (2019). The role of adult hippocampal neurogenesis in brain health and disease. Molecular Psychiatry, 24(1), 67–87. 10.1038/s41380-018-0036-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toni N, Laplagne DA, Zhao C, Lombardi G, Ribak CE, Gage FH, & Schinder AF (2008). Neurons born in the adult dentate gyrus form functional synapses with target cells. Nature Neuroscience, 11(8), 901–907. 10.1038/nn.2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praag H, Schinder AF, Christie BR, Toni N, Palmer TD, & Gage FH (2002). Functional neurogenesis in the adult hippocampus. Nature, 415(6875), 1030–1034. 10.1038/4151030a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YE, Pan L, Zuo Y, Li X, & Hong W (2017). Detecting activated cell populations using single-cell RNA-seq. Neuron, 96(2), 313–329.e6. 10.1016/j.neuron.2017.09.026 [DOI] [PubMed] [Google Scholar]
- Yang SM, Alvarez DD, & Schinder AF (2015). Reliable genetic labeling of adult-born dentate granule cells using Ascl1CreERT2 and Glast-CreERT2 murine lines. Journal of Neuroscience, 35(46), 15379–15390. 10.1523/JNEUROSCI.2345-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap E-L, & Greenberg ME (2018). Activity-regulated transcription: Bridging the gap between neural activity and behavior. Neuron, 100(2), 330–348. 10.1016/j.neuron.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Matthew Teng E, Summers RG, Ming G. l., & Gage FH (2006). Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. Journal of Neuroscience, 26(1), 3–11. 10.1523/JNEUROSCI.3648-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data have been submitted to GEO (accession number GSE214309). Other data supporting the findings of this study are available from the corresponding author upon reasonable request.


