Abstract
Walleye discrete epidermal hyperplasia (WEH) is a hyperproliferative skin disease that is prevalent on adult walleye fish throughout North America. We have identified two retroviruses associated with WEH, designated here as walleye epidermal hyperplasia virus type 1 and type 2 (WEHV1 and WEHV2), that are closely related to one another (77% identity) and to walleye dermal sarcoma virus (64% identity) within the polymerase region. WEHV1 and/or WEHV2 viral DNA was readily detected by PCR in hyperplastic tissue samples, but only low levels of viral DNA were detected in uninvolved skin. Southern blot analysis showed one to three copies of integrated WEHV2 viral DNA in lesions but did not detect WEHV2 viral DNA in uninvolved skin from the same fish. Northern blots detected abundant levels of WEHV1 and/or WEHV2 virion RNA transcripts of approximately 13 kb in hyperplastic tissue, but virion RNA was not observed in uninvolved skin and muscle. These results suggest that WEHV1 and WEHV2 are the causative agents of discrete epidermal hyperplasia.
Four virus-associated skin lesions have been observed on walleyes (Stizostedion vitreum) inhabiting lakes throughout North America: lymphocystis, dermal sarcoma, diffuse epidermal hyperplasia, and discrete epidermal hyperplasia (reviewed in reference 30). These diseases can be found singly or in any combination. Lymphocystis is caused by an iridovirus, and diffuse epidermal hyperplasia is associated with a percid herpesvirus (12, 25). Walleye dermal sarcoma (WDS) is associated with a retrovirus, walleye dermal sarcoma virus (WDSV) (13, 24). Walleye discrete epidermal hyperplasia (WEH), like WDS, is thought to have a retroviral etiology based on the observation by electron microscopy of retroviruslike particles in lesions (24, 29, 30).
Walleye discrete epidermal hyperplasia is a hyperproliferative skin disease characterized by plaques of thickened epidermis that can be found on any part of the body (24). The disease has been observed on approximately 10% of adult breeding walleyes in Oneida Lake, N.Y., and on up to 20% of the walleyes in some lakes in Canada in a given year (3, 24, 30). Like many neoplastic skin diseases of fish, including WDS, WEH is observed on sexually mature fish and the lesions appear and regress on a seasonal basis (1, 4, 17); they are present from fall through spring and are absent in the summer months (3). This seasonality may be caused by environmental factors such as water temperature that affect host endocrine activity and immunocompetency (1).
Although 13 neoplastic diseases of fish are believed to have a retroviral etiology (17), WDSV and the snakehead fish retrovirus (SnRV) are the only piscine retroviruses that have been molecularly cloned and sequenced (10, 11, 13). Furthermore, WDSV is the only piscine retrovirus that is etiologically associated with a neoplasia; it is a large complex retrovirus (12.7 kb) that contains three open reading frames in addition to gag, pol, and env, and it appears to have a complex life cycle, i.e., dramatically differing levels of gene expression are observed at different stages of disease (5, 11, 18). The putative proteins encoded by the additional open reading frames may be involved in the regulation of viral gene expression and the induction of WDS. Although dermal sarcomas often appear to be malignant histologically, the lesions regress, and neither local invasion nor metastasis has been observed in feral fish (4, 15). However, inoculation of 9-week-old walleye fingerlings with cell-free filtrates from dermal sarcomas resulted in locally invasive tumors (8).
Since retroviruslike particles have been observed in hyperplastic lesions by electron microscopy (29), we were interested in cloning and characterizing this virus and comparing it with WDSV. As a step toward this goal, we cloned two retroviruses from walleye hyperplasias, designated walleye epidermal hyperplasia type 1 and type 2 (WEHV1 and WEHV2). These viruses are closely related to each other and to WDSV. We report here the amino acid sequences of the pro-pol open reading frames of these viruses as well as evidence to suggest that they are the causative agents of WEH.
Cloning and characterization of retroviral pol sequences.
Preliminary experiments showed that virion preparations made from hyperplastic lesions had associated reverse transcriptase (RT) activity (data not shown). To amplify a segment of the pol genes from the retroviruses present in hyperplastic lesions, PCR amplification was accomplished with degenerate pol primers as described by Donehower et al. (7). These primers encode the amino acid sequences VLPQG (5′-ctcggatccGTNYTNCCNCARGG-3′) and YMDD (5′-ctcgtcgacRTCRTCCATRTA-3′) and generate an amplicon of approximately 135 bp (lowercase letters represent added restriction sites) from retroviral pol templates. Virion RNA from sucrose gradient purifications or total RNA from hyperplastic tissue was isolated with RNAzol (Tel-test Laboratories). The RNA was treated with 1 U of RNase-free DNase (Boehringer) in 1× first-strand synthesis buffer (Life Technologies) for 1 h at 37°C followed by phenol-chloroform extraction. cDNA was prepared using 0.5 μg of the YMDD primer or 1 μg of random hexamers (Boehringer) and 400 U of Superscript II (Life Technologies). The products were digested with BamHI and SalI and cloned into pBluescript II SK− (Stratagene). Clones were manually sequenced with Sequenase version 2.0 (U.S. Biochemicals) with T3 and T7 primers on double-stranded DNA templates. Two 114-bp retroviral pol segments were cloned from both RT PCR experiments (data not shown). The sequences of the two cDNA clones were 78% identical, suggesting that they were derived from two different but closely related retroviruses, designated walleye epidermal hyperplasia virus type 1 and type 2 (WEHV1 and WEHV2, respectively).
To obtain larger segments of the viral DNA flanking the VLPQG-YMDD interval in the pol gene, 5′ and 3′ rapid amplifications of cDNA ends (RACE) were employed with WEHV1- or WEHV2-specific primers. 5′ RACE was carried out according to the manufacturer’s directions (Life Technologies). Briefly, cDNA synthesis was performed with the 3′ YMDD primer and 1 μg of total RNA isolated from hyperplastic tissue. After RNase H digestion, the cDNA was purified by centrifugation and filtered through Millipore Ultrafree-MC 30,000 NMWL filters. A poly(dC) tail was added onto the 3′ end of the purified cDNA with terminal deoxynucleotidyltransferase. PCR amplification was performed with a 5′ GI anchor primer and the 3′ YMDD primer. A second round of PCR was done with an adapter primer (5′-GGCCACGCGTCGACTAGTAC-3′) and 3′ specific pol primers for WEHV1 (5′-cattgaattcggatcCAAATTTCCGAAGTCAGAGA-3′) or WEHV2 (5′-ca ttgaattcggatccACATACTTCCGAAGTTAAGCT-3′). Template DNA was denatured for 5 min at 96°C, followed by the addition of 2.5 U of Taq polymerase (Life Technologies). The amplification program consisted of 30 s at 94°C, 30 s at 50°C, and 2 min at 72°C for 35 cycles. The WEHV1 and WEHV2 5′ RACE products were digested with BamHI/SalI and EcoRI/SalI, respectively, and cloned into Bluescript II SK− (Stratagene). 5′ RACE yielded products of 269 and 533 bp for WEHV1 and WEHV2, respectively (data not shown).
First-strand cDNA for 3′ RACE was synthesized with an oligo(dT) primer (5′-ggccacgcgtcgactagtacT[12]-3′) as the anchor primer and 1 μg of total RNA. 3′ RACE was used in combination with the Elongase amplification system (Life Technologies) with the goal of obtaining the entire 3′ end of the viral genomes. Amplification of the cDNA was done with the adapter primer (above) and 5′ specific pol primers for WEHV1 (5′-cattgaattcggatccAATCTATCGCCAGATATGAC-3′) or WEHV2 (5′cattgaattcggatccAGGCAGTATACTTGGACAGTGT-3′). The PCR program consisted of 30 s at 94°C, 30 s at 55°C, and 6 min at 68°C for 35 cycles, yielding WEHV1 and WEHV2 products that were smaller than anticipated (1.9 and 1.6 kb, respectively). The 3′ RACE products were cloned into pBluescript II SK− after digestion with BamHI and SpeI. Subsequent DNA sequencing showed that small 3′ RACE products were the result of the oligo(dT) primer, used for cDNA synthesis, annealing to an A-rich sequence within the WEHV1 and WEHV2 pol genes.
The 3′ RACE fragments derived from WEHV1 and WEHV2 were labeled with 32P by random priming (Boehringer) and used as probes to isolate genomic clones from a lambda library (Stratagene’s lambda DASH II/BamHI vector kit and Gigapak II XL packaging extract). The WEHV1 and WEHV2 pol sequences were determined from overlapping 5′ and 3′ Bluescript SK II− subclones. The WEHV1 pol overlap consisted of 2,922 bp and the WEHV2 pol overlap consisted of 1,517 bp.
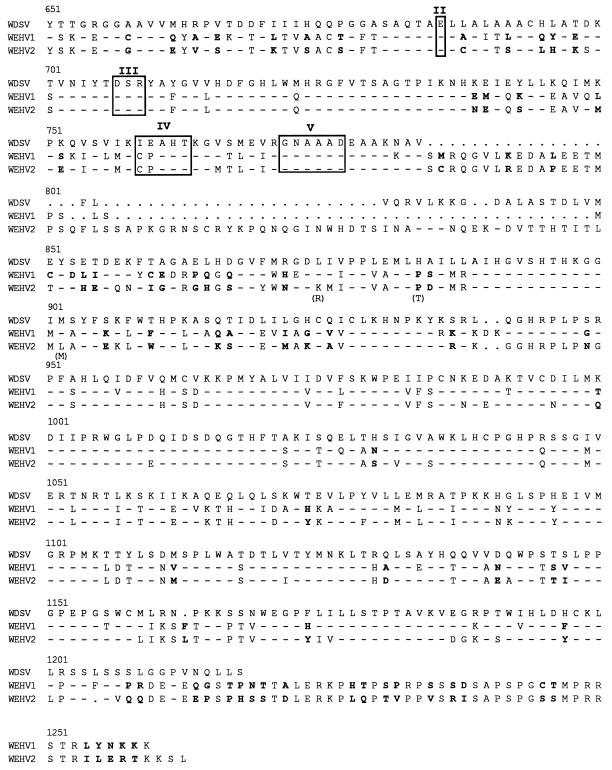
Figure 1 shows an alignment of the predicted amino acid sequences of the pro-pol open reading frames of WEHV1 and WEHV2 compared with that of WDSV. The pro-pol products consist of 1,200, 1,247, and 1,169 amino acids, respectively. Sequence analysis of the WEHV1 pol overlap region (amino acid positions 227 to 1200) showed that the 5′ and 3′ subclones were identical. However, the WEHV2 5′ and 3′ subclones differed in the overlap region (636 to 1140) by 18 nucleotides (1.2%), resulting in three amino acid substitutions: K to R (position 862), P to T (871), and L to M (889). The similarity of the overlapping regions in pol for WEHV1 and WEHV2 suggests that their respective 5′ and 3′ subclones are derived from the same strain of virus, although a minor heterogeneity exists within the population of WEHV2 proviruses.
FIG. 1.
Alignment of the predicted amino acid sequences of the pro-pol open reading frame in WDSV, WEHV1, and WEHV2. The seven conserved domains in RT are shown with brackets. LPQG and YMDD are indicated by four asterisks. The five conserved regions of RNase H are outlined in blocks labeled I through V. Identity with respect to WDSV (-), gaps (.), and differences between WEHV1 and WEHV2 are in bold-faced type. Amino acid changes between WEHV2 subclones are indicated in parentheses.
Like WDSV, the pro and pol genes of the hyperplasia viruses are in the same open reading frame (Fig. 1). Throughout the entire pro-pol gene, WEHV1 and WEHV2 have 77% amino acid identity, and each shares 64% identity with WDSV. The seven conserved domains in RT described by Xiong and Eickbush (28) are depicted in Fig. 1. Within this conserved region of RT, WEHV1 and WEHV2 have 90% amino acid identity and both have 70% identity with WDSV. The signature sequences for protease (LVDTG) and RT (LPQG and YMDD) are shown in Fig. 1 (7, 19). The five conserved regions of RNase H, as described by McClure (16), are shown as blocks I through V. Region I (FS/TDGS) is located at amino acid position 646. The three pol genes are the most divergent in the region located downstream of block V (GNAAAD) in RNase H between amino acids 784 and 950. This region contains a stretch of amino acid residues in WEHV2 pol that is not found in WEHV1 or WDSV.
Phylogeny.
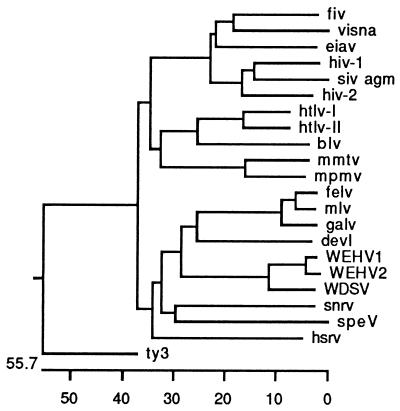
A phylogenetic analysis of WEHV1 and WEHV2 was performed with the two previously described complex piscine retroviruses, WDSV and SnRV (10), and with representatives of the seven retroviral genera. The pol sequences used in the analysis extended 132 amino acids, from domain 1 through the center of domain 5 (YXDD), as previously described (22), i.e., the conserved region of RT (28). This analysis was included so that the RT sequences of retroviral fragments isolated from two lower vertebrates, the lizard-like reptile tuatara (Sphenodon) and the poison dart frog (Dendrobates ventrimaculatus), could be placed in the alignment (22, 23). The yeast retrotransposon Ty3 was chosen as an outgroup. The tree was generated with the Megalign application in the DNAstar (Madison, Wis.) series of programs, in which a modification of the neighbor-joining method is used (20).
The analysis showed that WEHV1, WEHV2, and WDSV have a common ancestor (Fig. 2). WEHV1 and WEHV2 have 95% amino acid identity in this small region of RT, suggesting that they are different strains of the same virus or distinct species. We favor the latter possibility because (i) the amino acid sequences are only 77% identical throughout the entire pro-pol open reading frame, (ii) preliminary sequence analysis of WEHV1 and WEHV2 suggests that they are significantly different in other regions of the genome, and (iii) the percentage of identity between WEHV1 and WEHV2 in the conserved region is similar to that in RT between feline leukemia virus and murine leukemia virus (MLV) (92% identity) (Fig. 2), two viruses that are considered to be distinct and cause similar diseases in different species. WEHV1 and WEHV2 have 83 and 81% amino acid identity with WDSV, respectively, over the same region of pol (Fig. 2). The relationship between the WEHVs and WDSV may be analogous to that of human T-cell leukemia virus types 1 and 2 (82% identity), two viruses considered to be distinct that cause different diseases in humans (9). The three walleye viruses as a group are distantly related to SnRV (41% identity) and to members of the MLV group of retroviruses (i.e., MLV, 45% identity). Based on the limited current database, it appears that the retroviruses of fish, reptiles, and amphibians may ultimately form their own distinct genera within the retrovirus family.
FIG. 2.
Phylogenetic tree. The conserved RT domains spanning domain 1 through the middle of domain 5 (YMDD)—132 residues total—were analyzed and compared among the viruses shown with software from DNAstar. fiv, feline immunodeficiency virus; visna, visna virus; eiav, equine infectious anemia virus; hiv-1, human immunodeficiency virus type 1; siv agm, simian immunodeficiency virus of African green monkeys; htlv-I, human T-cell leukemia type 1; blv, bovine leukemia virus; mmtv, mouse mammary tumor virus; mpnv, Mason-Pfizer monkey virus; felv, feline leukemia virus; galv, gibbon ape leukemia virus; devl, Dendrobates ventrimaculatus; speV, Sphenodon virus; hsrv, human spumaretrovirus.
WEHV1 and/or WEHV2 DNA can be detected in walleye hyperplasias by PCR and Southern blotting.
A preliminary screen for the presence of WEHV1 and WEHV2 in hyperplastic lesions collected from 34 fish was done by PCR. To obtain sufficient material for analysis, multiple lesions from individual fish were pooled to form individual samples. Genomic DNA was isolated from approximately 100 mg of hyperplastic tissue with DNA isolator reagent (Genosys). PCR was done with 500 ng of DNA and the WEHV1- and WEHV2-specific pol primer pairs described above (without restriction sites). PCR was carried out as described for 5′ RACE, except that a 60-s extension at 72°C was used. The WEHV1 and WEHV2 primer sets amplified products of 226 and 116 bp, respectively, and did not cross-amplify (data not shown).
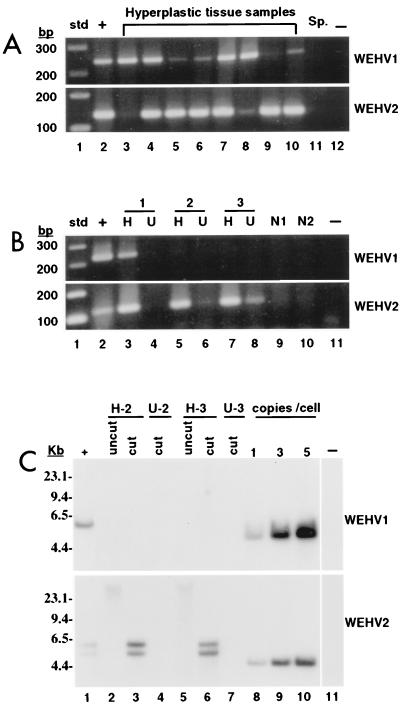
Representative PCR data are shown in Fig. 3A. Importantly, WEHV1 and WEHV2 were not detected in sperm DNA (lane 11), indicating that these viruses are exogenous to walleye. Both WEHV1 and WEHV2 viral DNA were found in 30 hyperplasia samples represented in lanes 2 to 10, whereas four hyperplasia lesions contained only WEHV2 viral DNA (data not shown). WEHV1 viral DNA was not found to be independent of WEHV2 viral DNA by PCR. Some samples showed an abundance of both viruses (lanes 4 and 7), and others showed an abundance of WEHV1 (lanes 3 and 8) or WEHV2 (lanes 5, 6, 9, and 10). The weak PCR signal observed for WEHV1 and WEHV2 in these samples may represent amplification of low levels of viral DNA due to background infection of the skin. To test this possibility, hyperplasias and uninvolved skin from three affected animals and skin from two clinically normal fish were tested for both viruses by PCR. As shown in Fig. 3B, PCR signals for WEHV1 (lane 3) or WEHV2 viral DNA (lanes 3, 5, and 7) were strong in the diseased tissue relative to the signals for uninvolved skin samples (lanes 4, 6, and 8). Importantly, WEHV1 and WEHV2 viral DNAs were not detected in skin from normal fish (lanes 9 and 10). Although only semiquantitative, these PCR data suggest that low levels of viral DNA are present in the uninvolved skin of diseased animals, whereas high levels of viral DNA are present in hyperplastic tissue. The data also demonstrate that the two viruses are commonly found together in diseased fish.
FIG. 3.
Detection of WEHV1 and WEHV2 in hyperplastic lesions. (A) PCR of viral DNA from lesions (10 μl loaded). Positive control, 5′ RACE clones (+); lesions (lanes 3 to 10); negative genomic DNA control, walleye sperm DNA (Sp.); negative buffer control (−). std, standard. (B) PCR of viral DNA from lesions (H) and associated uninvolved skin (U) from three diseased fish. Positive control (+); negative buffer control (−); H (lanes 3, 5, and 7); and U (lanes 4, 6, and 8). Skin from two clinically normal fish (N1 and N2) (lanes 9 and 10). std, standard. (C) Southern blot analysis. Positive control, pool of lesions from four fish that were positive for both viruses by PCR (+); undigested DNA from lesions (lanes 2 and 5); BamHI-EcoRI (WEHV1)- or PstI (WEHV2)-digested DNA from lesions (lanes 3 and 6); digested DNA from uninvolved skin (lanes 4 and 7); copy-number controls of 1, 3, and 5 copies/cell (lanes 8, 9, and 10); negative control (−), 5 copies of WEHV1 or WEHV2 3′ RACE clones, bottom and top, respectively.
The finding that viral DNA could be amplified from uninvolved skin, albeit at a low level, suggests that qualitative PCR alone is not sufficient to differentiate proviral DNA associated with diseased tissue and proviral DNA present in uninvolved skin. Therefore, Southern blot analysis was used to quantify viral DNA in these tissues. Our assumptions were that there would be at least one proviral DNA copy per cell in lesions if the viruses are etiologically associated with disease and that there would be a significantly lower level in uninvolved skin. Hyperplasias and uninvolved skin from two of the three fish that were positive for WEHV2 by PCR (Fig. 3B, lanes 5 to 8) were used for this analysis; the tissues from the fish harboring both viruses (lanes 1 and 2) were insufficient to isolate a sufficient amount of genomic DNA for this assay. Genomic DNA was isolated from hyperplasias and uninvolved skin by standard procedures (21). DNA (15 μg) was electrophoresed on a 0.9% gel, blotted onto nitrocellulose (21), and hybridized with the WEHV1- and WEHV2-specific pol probes described above. The probes, which have only 67% nucleic acid identity, did not cross-hybridize (Fig. 3C, lane 11 top and lane 11 bottom). WEHV1 and WEHV2 were detected in a positive control sample made from a pool of several lesions previously found to be positive for both viruses by PCR (lane 1). WEHV2 DNA was detected at 1 to 3 copies per cell (lanes 3 and 6) and comigrated with uncut genomic DNA, indicating that it is integrated into the chromosome (lanes 2 and 5). In contrast to the PCR results, WEHV2 DNA was not detected in uninvolved skin by Southern analysis (lanes 4 and 7). These data imply that the weakly positive results obtained by PCR from uninvolved skin (Fig. 3B, lanes 6 and 8) represent viral DNA that is present at less than one copy per cell. We infer that the same is true for the weakly positive results for the hyperplasias shown above (Fig. 3A, lanes 5, 6, 9, and 10 for WEHV1 and lanes 3 and 8 for WEHV2) and that only the more abundant species is etiologically associated with the lesions. From the PCR and Southern data, we conclude that WEHV1 and WEHV2 proviruses are abundant in diseased tissue, relatively rare in uninvolved skin, and undetectable in clinically normal fish.
The PstI digest of WEHV2 genomic DNA resulted in two bands of approximately 5.3 and 6.0 kb that hybridized with the WEHV2 pol probe (Fig. 3C, lanes 1, 3, and 6). The 5.3-kb band is consistent with that predicted from the sequence of the WEHV2 genomic clone. The larger band may result from restriction fragment length polymorphism, but this possibility needs to be investigated. Since pools of lesions from individual fish were used in the analysis, it is possible that variants of WEHV2 with PstI polymorphisms are present in the same sample, as suggested by the nucleic acid mutations observed in the WEHV2 pol overlap sequence (above). Restriction fragment length polymorphisms are common among retroviral isolates.
Full-length WEHV1 and/or WEHV2 RNAs are abundant in hyperplastic lesions.
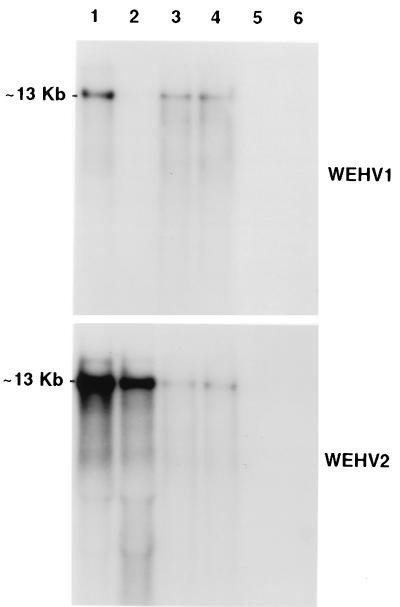
To determine if WEHV1 and WEHV2 genes are expressed in hyperplastic lesions, 26 of the 34 samples analyzed by PCR were analyzed by Northern blotting. Total RNA was isolated from hyperplastic lesions with RNAzol, and 10 μg was electrophoresed in formaldehyde gels and blotted onto nitrocellulose (21). The blots were hybridized with WEHV1 and WEHV2 pol-specific probes to detect full-length genomic viral RNA. As shown in Fig. 4, WEHV1 and WEHV2 genomic RNAs are abundant and largely undegraded in spring lesions (lanes 1 to 4). The apparent size of WEHV1 and WEHV2 full-length RNAs is approximately 13 kb, similar to that of WDSV (12.7 kb). Both WEHV1 and WEHV2 viral RNAs were detected in 13 samples, whereas WEHV1 RNA was detected alone in five samples and WEHV2 RNA was detected alone in six samples. A sample in which only WEHV2 RNA was detected is shown in lane 2. Two samples did not contain detectable levels of full-length viral RNA, perhaps due to sample degradation. Importantly, no viral RNA was detected in uninvolved skin or muscle of the infected fish (lanes 5 and 6, respectively), whose lesions were analyzed in lane 1.
FIG. 4.
Representative sample of Northern blot analysis of total RNA isolated from hyperplastic tissue. WEHV1 (top)- and WEHV2 (bottom)-specific pol probes were used for hybridization. Hyperplastic tissue samples (lanes 1 to 4). Each sample contains two or three lesions collected from an individual fish. RNA isolated from uninvolved skin and muscle (lanes 5 and 6) from the fish lesion is shown in lane 1.
We have identified two related exogenous retroviruses, WEHV1 and WEHV2, that are associated with walleye discrete epidermal hyperplasia. The sequences of the putative pro-pol open reading frames of WEHV1 and WEHV2 show that they are closely related but distinct from each other and that they are related to WDSV (11). By analogy with WDSV, WEHV1 and WEHV2 are presumed to be complex retroviruses.
PCR was used as a preliminary screen to test for the presence of WEHV1 and/or WEHV2 in hyperplastic lesions. However, our data suggest that PCR was not ideal for these studies because it did not unambiguously discriminate between viral DNA associated with diseased tissue and proviruses present in uninvolved skin at low copy numbers. Southern blot analysis clearly showed that the copy number and integrated status of WEHV2 is consistent with a retroviral etiology, i.e., at least one proviral copy was present per cell in lesions, whereas uninvolved skin did not contain detectable levels of viral DNA. Consistent with these results, Northern blot analysis showed that abundant levels of WEHV1 and/or WEHV2 genomic RNA were present in lesions but not in uninvolved tissues. Together, these data provide evidence that WEHV1 and WEHV2 are expressed specifically in diseased tissue and are involved in tumorogenesis.
The abundance of WEHV1 and WEHV2 viral RNA observed in spring hyperplasias is similar to that of WDSV viral RNA observed in spring dermal sarcomas (5, 17). In contrast, one to three copies per cell of proviral DNA were present in spring hyperplasias, whereas up to 50 copies per cell of unintegrated viral DNA (UVD) were found in spring dermal sarcomas (13). This difference in viral copy number may reflect the conditions of the lesions at the time of sampling. Typically, spring dermal sarcomas collected during the spawning run in April are in the late stages of regression, characterized by tumor shedding and necrosis (15). The hyperplasias used for this analysis were also collected in April but were not visibly regressing. The regression of hyperplastic lesions has not been documented, but it is assumed to occur because hyperplasias are not seen in the summer (3). WEHV1 and WEHV2 UVD may accumulate in cells of hyperplastic lesions as they begin to regress later in the season.
The high levels of UVD seen in spring dermal sarcomas may contribute to tumor regression by inducing cell death, analogous to the suggestion that UVD contributes to the cytopathic effects of avian leukosis virus subgroups B, D, and F (26, 27). However, it has been shown recently that the envelope proteins of avian leukosis virus subgroups B and D interact with a cellular receptor, inducing apoptosis, possibly explaining the cytopathology associated with these viruses (6). Therefore, the effect of UVD on cell viability in ALV and also on that in walleye retrovirus systems remains unclear.
In summary, our data strongly suggest that WEHV1 and WEHV2 are the causative agents of discrete epidermal hyperplasia. This hypothesis is supported by a recent experimental transmission of epidermal hyperplasia to walleye fingerlings inoculated with cell-free filtrates of hyperplastic lesions. As with the WDSV transmission model (14), the injected fingerlings developed epidermal hyperplasia in 20 to 24 weeks, and the WEHV1 and WEHV2 viral DNAs were found in samples of the transmitted hyperplasias which were assayed by PCR (2).
The discovery of two retroviruses associated with walleye epidermal hyperplasia was unexpected. Although most of the fish analyzed by PCR harbored both viruses, the observation that some hyperplasias contained only WEHV2 viral DNA suggests that this virus is capable of causing disease independently of WEHV1. Because we pooled multiple lesions from individual fish, it is not clear if both WEHV1 and WEHV2 are naturally found together in the same lesion or if they exist independently in different lesions on the same fish. To address this issue, individual hyperplastic lesions will be characterized to determine their virus profiles. Additionally, experimental transmission of epidermal hyperplasia to walleye fingerlings with WEHV1 or WEHV2 preparations will be necessary to demonstrate their ability to independently induce disease. Further molecular characterization of WEHV1 and WEHV2 will provide insight about the role they play in the pathogenesis of discrete epidermal hyperplasia.
Nucleotide sequence accession numbers.
The accession numbers for the WEHV1 and WEHV2 pro-pol sequences are AF014792 and AF014793, respectively.
Acknowledgments
We thank R. Colesante and M. Babenzein of the New York State Department of Environmental Conservation, Oneida Hatchery, for providing fish and V. Vogt and A. Eaglesham for critical reading of the manuscript.
The work was supported in part by a USDA grant (93-37204-9214) to P.R.B. and by funds from the USDA Animal Health and Disease Program administered by the College of Veterinary Medicine, Cornell University, to D.L.H. L.A.L. was supported by an NIH training grant (CA09682).
REFERENCES
- 1.Anders K, Yoshimizu M. Role of viruses in the induction of skin tumours and tumour-like proliferations of fish. Dis Aquat Org. 1994;19:215–232. [Google Scholar]
- 2.Bowser, P. R., K. Earnest-Koons, G. A. Wooster, L. A. LaPierre, D. L. Holzschu, and J. W. Casey. Experimental transmission of discrete epidermal hyperplasia in walleyes. J. Aquat. Anim. Health, in press.
- 3.Bowser P R, Wolfe M J, Forney J L, Wooster G A. Seasonal prevalence of skin tumors from walleye (Stizostedion vitreum) from Oneida Lake, New York. J Wildl Dis. 1988;24:292–298. doi: 10.7589/0090-3558-24.2.292. [DOI] [PubMed] [Google Scholar]
- 4.Bowser P R, Wooster G A. Regression of dermal sarcoma in adult walleyes (Stizostedion vitreum) J Aquat Anim Health. 1991;3:147–150. [Google Scholar]
- 5.Bowser R N, Wooster G A, Quackenbush S L, Casey R N, Casey J W. Comparison of fall and spring tumors as inocula for experimental transmission of walleye dermal sarcoma. J Aquat Anim Health. 1996;8:78–81. [Google Scholar]
- 6.Brojatsch J, Naughton J, Rolls M M, Zingler K, Young J A T. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 7.Donehower L A, Bohannon R C, Ford R J, Gibbs R A. The use of primers from highly conserved pol regions to identify uncharacterized retroviruses by the polymerase chain reaction. J Virol Methods. 1990;28:33–46. doi: 10.1016/0166-0934(90)90085-t. [DOI] [PubMed] [Google Scholar]
- 8.Earnest-Koons K, Wooster G A, Bowser P A. Invasive walleye dermal sarcoma in laboratory-maintained walleyes Stizostedion vitreum. Dis Aquat Org. 1996;24:227–232. [Google Scholar]
- 9.Green P L, Chen I S Y. Molecular features of the human T-cell leukemia virus. In: Levy J A, editor. The retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 277–311. [Google Scholar]
- 10.Hart D, Frerichs G N, Rambaut A, Onions D E. Complete nucleotide sequence and transcriptional analysis of the snakehead fish retrovirus. J Virol. 1996;70:3606–3616. doi: 10.1128/jvi.70.6.3606-3616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holzschu D L, Martineau D, Fodor S K, Vogt V M, Bowser P R, Casey J W. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J Virol. 1995;69:5320–5331. doi: 10.1128/jvi.69.9.5320-5331.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly R K, Nielsen O, Mitchell S C, Yamamoto T. Characterization of Herpesvirus vitreum isolated from hyperplastic epidermal tissue of walleye, Stizostedion vitreum vitreum (Mitchill) J Fish Dis. 1983;6:249. [Google Scholar]
- 13.Martineau D, Bowser P R, Renshaw R R, Casey J W. Molecular characterization of a unique retrovirus associated with a fish tumor. J Virol. 1992;66:596–599. doi: 10.1128/jvi.66.1.596-599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martineau D, Bowser P R, Wooster G A, Armstrong G A. Experimental transmission of a dermal sarcoma in fingerling walleyes (Stizostedion vitreum vitreum) Vet Pathol. 1990;27:230–234. doi: 10.1177/030098589002700403. [DOI] [PubMed] [Google Scholar]
- 15.Martineau D, Bowser P R, Wooster G A, Forney J L. Histologic and ultrastructural studies of dermal sarcoma of walleye (Pisces: Stizostedion vitreum) Vet Pathol. 1990;27:340–346. doi: 10.1177/030098589002700506. [DOI] [PubMed] [Google Scholar]
- 16.McClure M A. Evolution of retroposons by acquisition or deletion of retrovirus-like genes. Mol Biol Evol. 1991;8:835–856. doi: 10.1093/oxfordjournals.molbev.a040686. [DOI] [PubMed] [Google Scholar]
- 17.Poulet F M, Bowser P R, Casey J W. Retroviruses of fish, reptiles, and molluscs. In: Levy J A, editor. The retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 1–38. [Google Scholar]
- 18.Quackenbush, S. L., D. L. Holzschu, P. R. Bowser, and J. W. Casey. Transcriptional analysis of walleye dermal sarcoma virus (WDSV). Virology 237:107–112. [DOI] [PubMed]
- 19.Rao J K M, Erikson J W, Wlodawer A. Structural and evolutionary relationships between retroviral and eucaryotic aspartic proteinases. Biochemistry. 1991;30:4663–4671. doi: 10.1021/bi00233a005. [DOI] [PubMed] [Google Scholar]
- 20.Saitou N, Nei M. The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–525. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 22.Tristem M, Herniou E, Summers K, Cook J. Three retroviral sequences in amphibians are distinct from those in mammals and birds. J Virol. 1996;70:4864–4870. doi: 10.1128/jvi.70.7.4864-4870.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tristem M, Myles T, Hill F. A highly divergent retroviral sequence in the tuatara (Sphenodon) Virology. 1995;210:206–211. doi: 10.1006/viro.1995.1333. [DOI] [PubMed] [Google Scholar]
- 24.Walker R. Virus associated with epidermal hyperplasia in fish. Natl Cancer Inst Monogr. 1969;31:195–207. [PubMed] [Google Scholar]
- 25.Weissenberg R. Fifty years of research on lymphocystis virus disease of fish. Ann N Y Acad Sci. 1965;126:362–374. doi: 10.1111/j.1749-6632.1965.tb14286.x. [DOI] [PubMed] [Google Scholar]
- 26.Weller S K, Joy A E, Temin H M. Correlation between cell killing and massive second-round superinfection by members of some subgroups of avian leukosis virus. J Virol. 1980;33:494–506. doi: 10.1128/jvi.33.1.494-506.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weller S K, Temin H M. Cell killing by avian leukosis viruses. J Virol. 1981;39:713–721. doi: 10.1128/jvi.39.3.713-721.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong Y, Eickbush T H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto T, Kelly R K, Nielsen O. Epidermal hyperplasia of walleye, Stizostedion vitreum vitreum (Mitchill), associated with retrovirus-like type-C particles: prevalence, histologic and electron microscopic observations. J Fish Dis. 1985;8:425–436. [Google Scholar]
- 30.Yamamoto T, Kelly R K, Nielsen O. Morphological differentiation of virus-associated skin tumors of walleye (Stizostedion vitreum vitreum) Fish Pathol. 1985;20:361–372. [Google Scholar]