ABSTRACT
Schistosomiasis japonica is one of the neglected tropical diseases characterized by chronic hepatic, intestinal granulomatous inflammation and fibrosis, as well as dysbiosis of intestinal microbiome. Previously, the probiotic Bacillus amyloliquefaciens has been shown to alleviate the pathological injuries in mice infected with Schistosoma japonicum by improving the disturbance of the intestinal microbiota. However, the underlying mechanisms involved in this process remain unclear. In this study, metagenomics sequencing and functional analysis were employed to investigate the differential changes in taxonomic composition and functional genes of the intestinal microbiome in S. japonicum-infected mice treated with B. amyloliquefaciens. The results revealed that intervention with B. amyloliquefaciens altered the taxonomic composition of the intestinal microbiota at the species level in infected mice and significantly increased the abundance of beneficial bacteria. Moreover, the abundance of predicted genes in the intestinal microbiome was also significantly changed, and the abundance of xfp/xpk and genes translated to urease was significantly restored. Further analysis showed that Limosilactobacillus reuteri was positively correlated with several KEGG Orthology (KO) genes and metabolic reactions, which might play important roles in alleviating the pathological symptoms caused by S. japonicum infection, indicating that it has the potential to function as another effective therapeutic agent for schistosomiasis. These data suggested that treatment of murine schistosomiasis japonica by B. amyloliquefaciens might be induced by alterations in the taxonomic composition and functional gene of the intestinal microbiome in mice. We hope this study will provide adjuvant strategies and methods for the early prevention and treatment of schistosomiasis japonica.
IMPORTANCE
Targeted interventions of probiotics on gut microbiome were used to explore the mechanism of alleviating schistosomiasis japonica. Through metagenomic analysis, there were significant changes in the composition of gut microbiota in mice infected with Schistosoma japonicum and significant increase in the abundance of beneficial bacteria after the intervention of Bacillus amyloliquefaciens. At the same time, the abundance of functional genes was found to change significantly. The abundance of genes related to urease metabolism and xfp/xpk related to D-erythrose 4-phosphate production was significantly restored, highlighting the importance of Limosilactobacillus reuteri in the recovery and abundance of predicted genes of the gut microbiome. These results indicated potential regulatory mechanism between the gene function of gut microbiome and host immune response. Our research lays the foundation for elucidating the regulatory mechanism of probiotic intervention in alleviating schistosomiasis japonica, and provides potential adjuvant treatment strategies for early prevention and treatment of schistosomiasis japonica.
KEYWORDS: Bacillus amyloliquefaciens, Schistosoma japonicum, intestinal microbiome, functional genes
INTRODUCTION
Schistosomiasis, a zoonotic parasitic disease that seriously endangered the health of humans and animals, was mainly prevalent in 78 countries and regions of Asia, Africa, and South America. More than 700 million people worldwide were threatened by the risk of schistosomiasis infection, and almost 240 million people were affected by schistosomiasis, of which 120,000 had severe clinical symptoms. Therefore, schistosomiasis severely damaged human health and greatly hindered social and economic development (1). The main pathogens of human schistosomiasis include Schistosoma haematobium, Schistosoma japonicum, and Schistosoma mansoni (2). Schistosomiasis japonica, which is caused by S. japonicum, is currently endemic in China (particularly in the Yangtze River Valley), Indonesia, and the Philippines (3). During the progression of infection with S. japonicum, various organs of the host were damaged to various degrees. Allergic dermatitis was caused by the invasion of cercariae from the host skin into the host body. Afterwards, cercariae reached the hepatic portal venous system along with the host’s systemic circulatory system, where they developed into adults and caused fibrosis and enlargement of the liver (4). After developing into adults, S. japonicum would move in pairs to the mesenteric veins, combine to lay eggs, and thus disrupt the intestinal mucosa to cause intestinal granulation (3). Praziquantel (PZQ) has been the first-choice drug for the clinical treatment of schistosomiasis japonica and has been used worldwide for more than 30 years. Due to its wide range and high frequency of use, schistosomiasis has become less susceptible and has developed resistance to PZQ, which resulted in lower treatment efficacy of PZQ and impeded schistosomiasis control and prevention (5, 6). Based on the current situation of schistosomiasis control, the development and application of novel therapeutic means are crucial.
Probiotic therapy had a long history, and cases of using human feces to treat infections or food poisoning were presented in ancient China (7). Probiotic therapy, which could significantly improve the course of disease exacerbations through the intervention of probiotics, had no adverse effects and no problem of drug resistance (8). After entering the host, probiotics were able to compete for niches through colonization to modulate the microbiota and inhibit adhesion of pathogenic bacteria through the production of bacteriocins, short-chain fatty acids (SCFAs), and biosurfactants (9). They were also able to interact with the host immune system to improve immunity by suppressing proinflammatory cytokines, stimulating secretory IgA production and enhancing communication between the gut and the brain (10). The use of probiotics was also highly recommended in clinics to promote the alleviation of diseases and the repair of organs. For example, the intervention of Akkermansia muciniphila in patients with obesity was able to promote the outer membrane protein AMUC_1100 to interact directly with TLR-2, thereby strengthening the intestinal barrier, reducing inflammation, and ultimately improving health (11). Currently, probiotic therapies are also gradually being considered for application in the treatment of schistosomiasis japonica. Based on the recent studies, the intervention of Bacillus subtilis significantly alleviated the pathological damage caused by schistosomiasis japonica through modulating the host intestinal microbiome, and the transcriptome analysis revealed significant changes in the expression of genes associated with differentiation of Th1, Th2, and Th17 cells (12). It is indicated that the intestinal microenvironment in the host challenged with S. japonicum could be reshaped after the probiotic intervention, but the specific mechanism is still not clear. According to related studies, the intervention of probiotics changed the composition of the microbiota in different parts of the host, thus changing the metabolites of the microbiome, and ultimately effectively alleviating the disease condition by intervening in the host’s immune regulatory system (13). Pentanoate, a type of SCFAs metabolically produced by the gut microbiota, could promote glycolysis and activate mTOR activity by inducing the AMPK pathway, which may reduce Th1- and Th2-type immune responses by stimulating secretion of IL-10, ultimately leading to granuloma suppression, but the specific regulatory mechanisms required further studies (13, 14). In conclusion, microbial metabolites were able to affect the disease course of schistosomiasis japonica by modulating the host’s immune response, and probiotic therapy could also be an effective and safe strategy for the prevention and treatment of schistosomiasis.
Bacillus amyloliquefaciens, a nonpathogenic and nontoxic probiotic, had highly stable physicochemical properties and could survive in extreme environments and tolerate acid and high temperatures (15–17). In recent years, B. amyloliquefaciens has gradually been applied in clinical disease treatment to improve human health. According to relevant studies, it has been found that B. amyloliquefaciens could regulate the homeostasis of the intestinal microbiota, downregulate metabolic pathways related to bile acid synthesis, lower levels of inflammatory factors, and restore the repair of the intestinal barrier, ultimately alleviating the symptoms of intestinal inflammatory diseases (18–20). In a previous study, we found that B. amyloliquefaciens could reduce the degree of liver fibrosis and intestinal granuloma in S. japonicum-infected mice, maintain the homeostasis of the intestinal microenvironment, remodel the intestinal microbiota of S. japonicum-infected mice, and modulate the relative abundance of potentially pathogenic bacteria (e.g., Escherichia-Shigella) and beneficial bacteria (e.g., Muribaculaceae) (21). It was demonstrated that the intervention of B. amyloliquefaciens was able to alleviate the pathological condition of schistosomiasis japonica by restoring intestinal homeostasis as well as modulating the relative abundance of beneficial and pathogenic bacteria. However, the mechanisms by which B. amyloliquefaciens alleviated the symptoms of S. japonicum infection by altering the composition of the microbiota as well as regulating the host’s immune response remain unknown and require further investigation.
In this study, shotgun metagenomic sequencing was conducted to analyze the fecal samples collected from the mice acutely infected with S. japonicum before and after treatment with B. amyloliquefaciens. The changes in composition and gene function of the intestinal microbiome in S. japonicum-infected mice after B. amyloliquefaciens intervention were analyzed. Based on these results, the potential mechanism by which the intervention of B. amyloliquefaciens alleviates the pathological condition of S. japonicum-infected mice was proposed. We expect that our findings could provide a safe and effective strategy for the treatment of Schistosomiasis japonica.
MATERIALS AND METHODS
Preparation for suspensions of B. amyloliquefaciens, mice infection, and sample collection
The detailed protocols about the preparation for suspensions of B. amyloliquefaciens, mouse infection, and intragastric administration of B. amyloliquefaciens followed the methods reported in our previous research (21). Briefly, B. amyloliquefaciens was centrifuged at 8,000 rpm for 10 min at 25°C (Glanlab, Changsha, China). Mice were given 0.3-mL suspension of B. amyloliquefaciens every 3 days at a fixed time by intragastric administration according to groups. Specific groups were described as follows: (i) PBS: healthy mice intragastric administrated by phosphate-buffered saline (PBS); (ii) BA: healthy mice intragastric administrated by suspension of B. amyloliquefaciens; (iii) SJ: S. japonicum-infected mice intragastric administrated by PBS; (iv) SJBA: S. japonicum-infected mice intragastric administrated by suspension of B. amyloliquefaciens. All the mice were sacrificed by cervical dislocation at day 45 post-infection. The collection of fecal samples and the measurement of weight were conducted 1 day before sacrifice.
Metagenome DNA extraction and shotgun sequencing
Three samples from each group at day 45 post-infection were selected for shotgun sequencing. Total microbial genomic DNA of fecal samples was extracted following the instructions of OMEGA Soil DNA Kit (D5625-01) (Omega Bio-Tek, Norcross, GA, USA) and was stored at −20°C prior to further assessment. The quantity and quality of extracted DNAs were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively. The extracted microbial DNA was processed to construct metagenome shotgun sequencing libraries with insert sizes of 400 bp by using Illumina TruSeq Nano DNA LT Library Preparation Kit. Each library was sequenced by Illumina HiSeq X-ten platform (Illumina, USA) with PE150 strategy at Personal Biotechnology Co., Ltd. (Shanghai, China).
Metagenomics analysis
Raw sequencing reads were processed to obtain quality-filtered reads for further analysis. Cutadapt was used for removing sequencing adapters from raw sequencing reads (22). Fastp was used for trimming low-quality reads by using a sliding window algorithm (23). Bowtie2 was used for filtering out the sequence from mice by using the reference genome of mice (mm39) (24). Kraken2 was used for taxonomic classification of the remaining high-quality-filtered reads by using the custom Kraken2 microbiological database, including bacteria and fungi with default settings (25). Bracken was used to estimate the relative abundance of microorganisms in different samples (26). Then, the taxonomic table with relative abundance was used for further analysis. After quality control, the remaining high-quality reads were used for the analysis of the function of genes. Megahit was used for assembling filtered-out reads to contigs (27). Prodigal was used for predicting the coding sequences (CDSs) from the generated contigs (28). CD-HIT was used to remove redundancy from all predicted genes (29). Salmon was used to estimate the read coverage of each gene in different samples, and the abundance of genes normalized by Salmon was used for further analysis (30). eggNOG-mapper was used for function annotation for genes (31). Then, the abundance of the same orthologs (OGs) annotated by eggNOG-mapper was added up, and the OGs table with abundance was used for further analysis. The annotation tables with the abundance about Clusters of Orthologous Genes (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were generated from the result of the annotation from eggNOG-mapper. To determine the host of the predicted genes, Kraken2 was used to align the predicted CDSs to the custom Kraken2 microbiological database, including bacteria. A relatively conservative discrimination method was used to determine the host of genes (genes were considered to be one clade only when genes were determined in the same clades).
Analysis of species with significant difference in relative abundance
The relative abundance of species was calculated by dividing the reads count of each species by the sum of the reads counts of all species in one sample. The relative abundance was used to perform differential analysis further. The radar plot that was used to show the relative abundance of composition of the intestinal microbiota was performed by the “fmsb” package (32). Species with significant changes in relative abundance were discovered by linear discriminant analysis (LDA) effect size (LEfSe) (http://huttenhower.sph.harvard.edu/galaxy/) (33). The thresholds of P value and LDA scores were set at 0.05 and 2.0. The density of distribution in LDA scores was analyzed by the “ggExtra” package (34). Heatmap was used to present the relative abundance of species with significant changes in relative abundance using the “pheatmap” package (35). The relative abundance of species shown in heatmap was normalized by log10 calculation. All statistical analyses were performed using the R environment (version 4.2.1) (36). The statistical results were visualized by the “ggplot2” package (37).
Discrimination of orthologs with significant changes in abundance
A Wilcoxon test was performed to calculate the P value for detecting OGs with significant changes in abundance. The Benjamin and Hochberg false discovery rate (FDR) was used to correct the P value. The “log2 FoldChange” was explained by log2 FoldChange = log2[(A + 1)/(B + 1)], where A and B represented the abundance of OGs in different groups, respectively. The thresholds of PFDR, P value, and absolute value of log2 FoldChange were set at 0.385, 0.05, and 1.0 (38). Significant changes in the abundance of OGs in the four groups were explained by the fact that the trend of significant changes in the abundance of OGs in the SJ group compared to the PBS and BA groups is opposite to that in the SJBA group compared to the SJ group. A Venn diagram using “ggVennDiagram” package was performed to discover OGs with significant changes in abundance, and the definition of significant changes was based on the descriptions above (39). The Sankey plot showed the annotation information about COG of OGs with significant changes in abundance in the SJBA group by using the “networkD3” package (40). All statistical analyses were performed using the R environment. The statistical results were visualized by the “ggplot2” package.
Detection of KEGG pathway with significant changes in abundance
A Wilcoxon test was performed to calculate the P value for detecting the KEGG pathway with significant changes in abundance. The Benjamin and Hochberg FDR was used to correct the P value. The “log2 FoldChange” was explained by log2 FoldChange = log2[(A + 1)/(B + 1)], where A and B represented the abundance of the KEGG pathway in different groups, respectively. The thresholds of PFDR, P value, and |log2 FoldChange| were set at 0.385, 0.05, and 1.0. The definition of significant changes was the same as that in the previous section. The “UpSetR” package was used to determine the KEGG pathway with significant changes in abundance (41). The “ggalluvial” package was used to show the association between phyla, families, KEGG Orthology (KO) genes, and metabolic reactions (42). Heatmap was used to present the relative abundance of KO genes and KEGG reactions, which was normalized by z-score. The correlation of KO genes, metabolic reaction, and species was shown by a network plot, which was calculated by packages of “Hmisc” and “igraph” and visualized by Gephi software (version 0.10.0) (43–45). The threshold of the P value corrected by FDR and the absolute value of the Spearman correlation were 0.05 and 0.85.
RESULTS
Treatment of B. amyloliquefaciens significantly changes the composition of intestinal microbiota in mice infected with S. japonicum
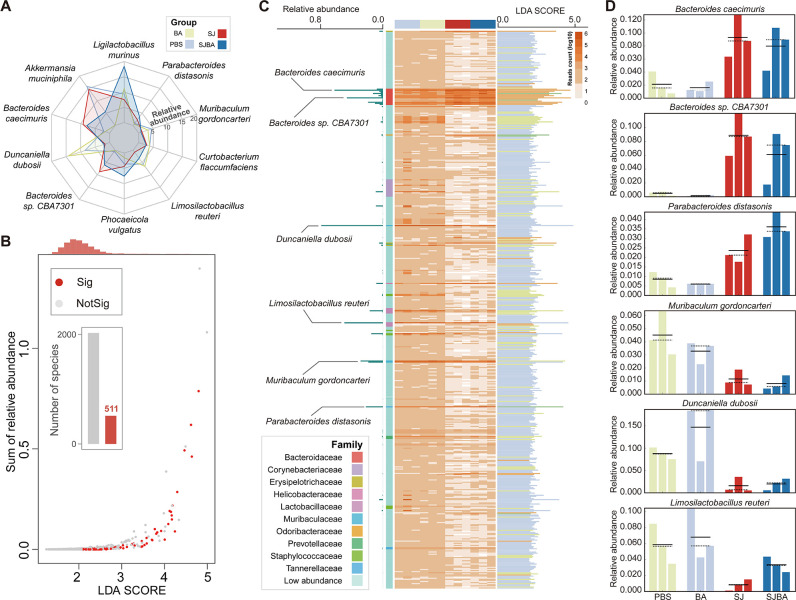
To explore the impact of B. amyloliquefaciens intervention on the intestinal microbiota of mice infected with S. japonicum, a radar plot was performed to present the composition of the intestinal microbiota within the four groups. Species that were in the top 10 in relative abundance were used to display. According to the results, Akkermansia muciniphila, Bacteroides caecimuris, and Bacteroides sp. CBA7301 obviously increased in the SJ group; whereas, Ligilactobacillus murinus, Phocaeicola vulgatus, and Limosilactobacillus reuteri obviously increased in the SJBA group compared to the SJ group (Fig. 1A). Then, LEfSe analyses were performed to discover the species with significant changes in relative abundance. LDA scores and P values were calculated by LEfSe. We detected a total of 511 species with significant differences in relative abundance, whose P value was smaller than 0.05 and whose LDA scores were greater than 2.0. The LDA scores were observed to be centrally distributed at 2.0 (Fig. 1B). Subsequently, the relative abundance of species with significant changes in relative abundance was shown by the heatmap. We found that six species showed significant changes in relative abundance among the top 10 species in terms of relative abundance, and the LDA scores of these six species were very high, indicating extremely significant differences among the four groups (Fig. 1C). Finally, the relative abundance of the six species in each group is shown in Fig 1D. These data demonstrated that B. amyloliquefaciens treatment significantly changes the composition of the intestinal microbiota in S. japonicum-infected mice.
Fig 1.
Alteration in the composition of the intestinal microbiome in the S. japonicum-infected mice after treatment with B. amyloliquefaciens. (A) Changes in the relative abundance of the top 10 species by relative abundance in each of the four groups. Different colors distinguish the four groups. (B) The scatterplot shows the relationships between relative abundance and LDA scores of the species. The thresholds of the P value and LDA scores were set at 0.05 and 2.0. Red points represent species with a significant difference in relative abundance in which the P value was less than 0.05 and the LDA scores were greater than 2.0; whereas, gray points mean species did not differ in relative abundance. The red columns in the histogram represent the number of species with a significant difference in relative abundance. The orange column at the top of the plot represents the density of distribution in the LDA scores. “Sig” means significance, and “NotSig” means not significance. (C) The relative abundance and LDA scores of species with a significant difference in relative abundance. The heatmap shows the relative abundance of species with a significant difference in relative abundance. Color depth and relative abundance are proportional. The left of the plot illustrates the relative abundances of species with significant differences in relative abundances among the top 10 species by relative abundance. The right of the plot presents the LDA scores of species with a significant difference in relative abundance. Different colors of columns represent different groups. (D) The right of the plot shows the relative abundance of species with significant differences in relative abundance among the top 10 species by relative abundance in the four groups. Different colors of columns represent different groups.
Functional gene changes of intestinal microbiome in S. japonicum-infected mice after treating with B. amyloliquefaciens
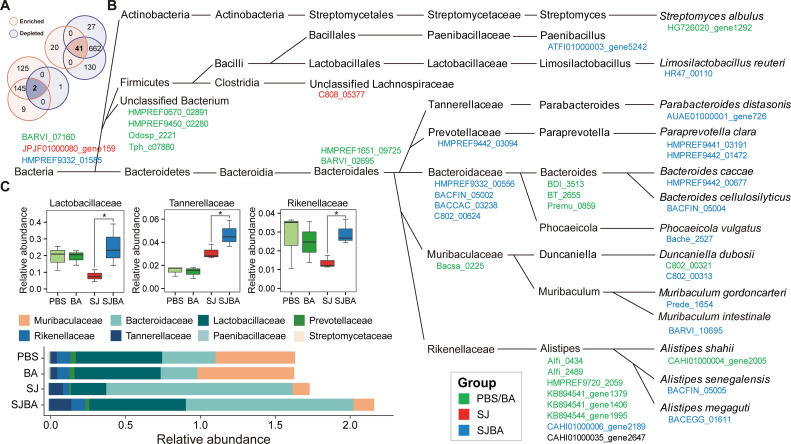
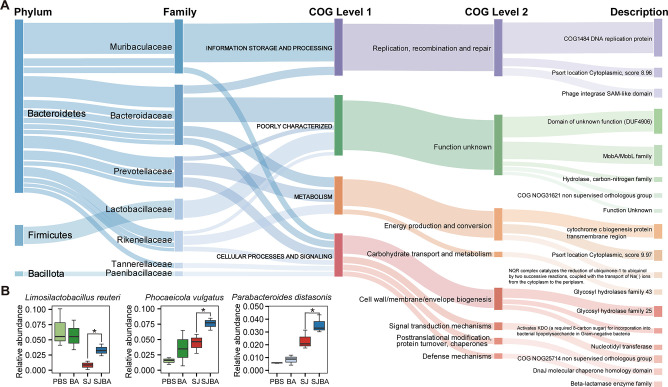
The Wilcoxon test and log2 FoldChange were used to determine whether OGs were significantly changed in the SJBA group. After screening, we found a total of 2,055 OGs with significant changes in abundance, of which |log2 FoldChange| was greater than 1.0 (Fig. S1A). Then, a Venn diagram was created to discover the OGs that meet the definition of significant changes (refer to the Materials and Methods). We also found 43 OGs significantly increased in the SJ groups but decreased in the SJBA group, or decreased in the SJ groups but increased in the SJBA group (Fig. 2A). Based on the heatmap about the abundance of 43 OGs, 20 OGs were observed to have a high abundance in the SJBA group (Fig. S1B). To figure out the host of OGs, the highest taxonomic classifications of OGs were shown. As suggested by the results, most OGs were generated by Bacteroidales, and most OGs that have a high abundance in the PBS and BA groups were observed to be expressed by Alistipes (Fig. 2B). We also found eight families expressed most OGs with significant changes in abundance, and Lactobacillaceae, Tannerellaceae, and Rikenellaceae were found to be significantly increased in the SJBA group (Fig. 2C). It meant that they might play important roles in maintaining intestinal homeostasis. Finally, OGs with significant changes in abundance, which had a higher abundance in the SJBA group, were annotated by the COG. The taxonomic classification of OGs was also related to their functions. According to the results, the OG that was annotated as “COG1484 DNA replication protein,” “domain of unknown function (DUF 4906),” and “MobA/MobL family” in the COG database had a high abundance in the SJBA group (Fig. 3A). We also defined the host of OGs with significant changes in abundance that had a higher abundance in the SJBA group. We found that L. reuteri, P. vulgatus, and Alistipes senegalensis were significantly increased in the SJBA group compared to the SJ group (Fig. 3B).
Fig 2.
The taxonomic classification levels of genes with a significant difference in abundance. (A) The Venn diagram shows the number of shared and unique OGs with the opposite trend of significant change in abundance among different groups. Orange represents that OGs were significantly enriched, and purple represents that OGs were significantly depleted. The number of OGs in this plot came from Figure S1. (B) The highest taxonomic classification levels of OGs with a significant difference in abundance. Different colors of OGs represent that OGs were enriched in different groups. (C) The relative abundance of family in the four groups. Data were analyzed by Wilcoxon test. *P < 0.05.
Fig 3.
COG annotations of OGs enriched in the SJBA group. (A) Detailed information of COG annotations of OGs that were enriched in the SJBA group. (B) The relative abundance of species of OGs that had a significant difference in abundance in the SJBA group. Data were analyzed by Wilcoxon test. *P < 0.05.
Significant recovery of abundance for the KEGG pathway after treating with B. amyloliquefaciens on mice infected with S. japonicum
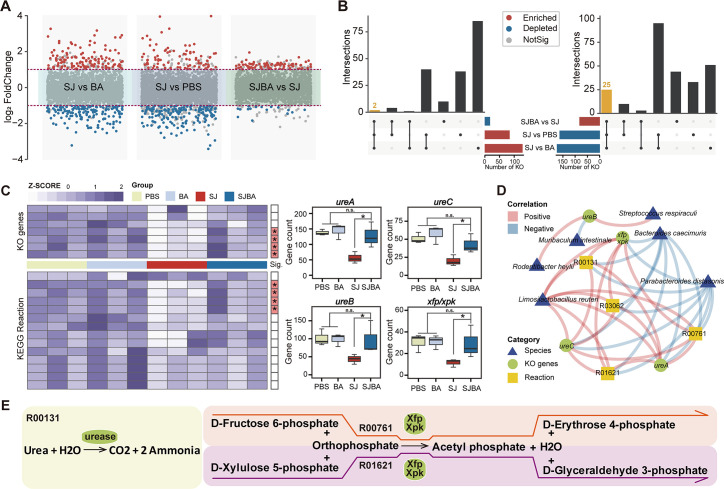
To make it clear which metabolic pathway has significantly changed after treatment with B. amyloliquefaciens, differential analysis on the KEGG pathway was performed. Wilcoxon test was performed to detect the metabolic pathways with significant changes among the groups of SJ and PBS, SJ and BA, and SJBA and SJ (Fig. 4A). Then, the significant changes with opposite trends in the comparison of different groups were intersected to obtain the KEGG pathway with a significant difference in abundance that meets the definition of significant change (refer to the Materials and Methods). Based on the results, 27 KEGG pathways with significant changes in abundance were screened (Fig. 4B). Suggested by the information in the KEGG database, the number of KO genes had descriptions that contained the information about enzymes, while enzymes could catalyze metabolic reactions. All the information about enzymes and metabolic reactions came from the KEGG database. At the same time, nucleotide sequences, which could be annotated to KO genes, could also be used for taxonomic annotation. Based on the above associations, KO genes (which came from 27 KEGG pathways with significant changes in abundance and which are involved in the catalysis of metabolic reactions), metabolic reactions, and their information about taxonomic classification that can be annotated were associated. Finally, 8 KO genes, 16 metabolic reactions, and 8 species, which could be correlated based on the association analysis method described above, were obtained (Fig. S2). To determine which KO genes and metabolic reactions changed significantly after treatment with B. amyloliquefaciens, Wilcoxon tests were performed. Four KO genes and four metabolic reactions selected by the association analysis were found to be significantly different in the SJBA group compared to the SJ group (Fig. 4C). Six species selected by the association analysis were found to have significant changes in relative abundance based on the results of LEfSe (Fig. 1C). To verify the results of the association analysis, the correlation between four KO genes, four metabolic reactions, and six species was analyzed by Spearman correlation analysis. Based on the results, L. reuteri was found to have significant positive correlation with all the KO genes and metabolic reactions, and all the KO genes also had significant positive correlation with all the metabolic reactions. Meanwhile, B. caecimuris and Parabacteroides distasonis had significant negative correlation with all the KO genes and metabolic reactions (Fig. 4D). Eventually, the relationships among four KO genes and four metabolic reactions were analyzed based on the information from the KEGG database and on the results from Spearman correlation analysis. We found that ureases translated by ureA, ureB, and ureC could catalyze the decomposition of urea into ammonia, and xylulose-5-phosphate/fructose-6-phosphate phosphoketolase (Xfp/Xpk) translated by xfp/xpk could catalyze the metabolic reaction to produce D-erythrose 4-phosphate and D-glyceraldehyde 3-phosphate (Fig. 4E).
Fig 4.
The relationship between the KEGG pathway and species in the SJBA group. (A) Difference analysis of the KEGG pathway in abundance among different groups. Red points represent significant enrichment of the KEGG pathway in abundance in different groups. Blue points represent significant depletion of the KEGG pathway in abundance. Gray points represent the KEGG pathway in abundance but had no significance. Data were analyzed by the Wilcoxon test. The thresholds of P value and log2 FoldChange were set at 0.05 and 1.0. (B) The UpSet diagram shows the KEGG pathway with significant difference in abundances. Different colors of columns represent different trends of significant change. (C) Screening for KO genes and metabolic reactions with significant differences in abundance. KO genes and metabolic reactions were picked up from Figure S2. The heatmap shows the relative abundance of KO genes and metabolic reactions. The red color on the right of the plot means KO genes or metabolic reactions were significantly different in abundance in the SJBA group. Data were analyzed by the Wilcoxon test. “n.s.” means no significance. *P < 0.05. (D) The correlation network between KO genes, metabolic reactions, and species. The positive and negative correlations are separated by the colors red and blue. Triangles in blue indicate species, circles in green indicate KO genes, and squares in yellow indicate metabolic reactions. (E) Metabolic reactions involving KO genes which differed significantly in abundance. The ellipse in green represents KO genes. Different modules in different colors represent different KEGG metabolic reactions.
DISCUSSION
Schistosomiasis japonica, a parasitic disease mainly endemic in China, seriously threatens human health. After 70-year effort, high-profile achievements have been made in the prevention and treatment of schistosomiasis japonica in China. However, several problems and challenges still remain, such as the increasingly prominent side effects and drug resistance problems of PZQ (46–48). Earlier studies proved that altering the intestinal microbiota of S. japonicum-infected mice was able to alleviate the pathological symptoms of schistosomiasis japonica, indicating that it could be a potential therapeutic strategy for schistosomiasis (49). Previously, we found that the pathological injuries in the liver and intestine caused by infection with S. japonicum were significantly reduced after B. amyloliquefaciens intervention, which was closely associated with the significant recovery of the composition and homeostasis of the intestinal microbiota (21). However, there were no studies elucidating the related mechanisms between the alteration of the intestinal microbiome and the alleviation of symptoms of schistosomiasis japonica in the host. Therefore, in this study, shotgun metagenomic sequencing and functional analysis were performed to evaluate the changes in the taxonomic composition at the species level and gene functions of the intestinal microbiome in S. japonicum-infected mice after B. amyloliquefaciens treatment. Additionally, we expected to lay the foundation for the elucidation of the regulatory mechanisms between the intervention of B. amyloliquefaciens and the immune regulation of the host in response to S. japonicum infection. Our results showed that the intestinal microbiota at the species level of S. japonicum-infected mice changed significantly after B. amyloliquefaciens intervention, and that the relative abundance of beneficial bacteria increased significantly. Meanwhile, the gene expression of the intestinal microbiome was significantly higher in the SJBA group.
After S. japonicum infection, symptoms of intestinal granulomatous disease were induced in the host, which resembled those of inflammatory bowel disease (IBD), typically characterized by hematochezia and unformed stools. At the same time, intestinal granulomas destroyed the intestinal microenvironment and significantly affected the colonizing microbiome of the intestine (2, 50). After B. amyloliquefaciens intervention, we found that the relative abundance of bacteria at the species level with the highest abundance was significantly changed in the intestinal microbiome of S. japonicum-infected mice. The increasing relative abundance of beneficial bacteria L. murinus in the SJBA group was the most obvious, but not significantly different when compared to the SJ group (51). Next, LEfSe analysis identified a significant elevation in the relative abundance of L. reuteri and P. distasonis in the SJBA group. L. reuteri is a well-studied beneficial bacterium that has been applied in various foods and food supplements and has been used as a potential cell factory (52–54). Meanwhile, L. reuteri had been clinically proven to have a positive palliative effect on a number of diseases, such as ameliorating diarrhea in children, promoting intestinal epithelial regeneration to repair damaged intestinal mucosa, and promoting the development and function of the regulatory immune system (55–59). In conclusion, L. reuteri was important for the promotion of human health, which made it of increasing interest (60). The role of P. distasonis in intestinal diseases is mixed. A study of colorectal cancer (CRC) found that IL-10, TGF-β, and tight-junction proteins were significantly higher in the colons of mice that were given P. distasonis on a regular basis. This suggests that P. distasonis played a protective role in stopping the growth of colon tumors (61). However, P. distasonis has been observed to promote colitis in studies of Crohn’s disease and ulcerative colitis (UC) (62). Meanwhile, we observed that the abundance of P. vulgatus was significantly increased in the SJBA group compared to the SJ group in the analysis of differential genes. P. vulgatus, which was capable of disrupting colonic epithelial integrity through the expression of proteases, had been reported to have a potential pathogenic role and could aggravate UC (63–65). However, the prebiotic properties of P. vulgatus have also been reported in related studies. P. vulgatus had an enzymatic system that degrades complex polysaccharides and was able to form SCFAs (66–68). Currently, there are no relevant studies of P. distasonis and P. vulgatus in schistosomiasis, so more in-depth studies are needed to explain our findings. There were a lot more B. caecimuris and Bacteroides sp. CBA7301 in the SJ group, but not many studies have looked at these two bacteria in intestinal diseases.
Therefore, based on the compositional changes of intestinal microbiota at the species level, we found that the abundance of beneficial bacteria in the intestine was significantly upregulated after B. amyloliquefaciens intervention, suggesting that restoring beneficial bacteria in abundance due to colonization of B. amyloliquefaciens played important roles in alleviating the symptoms of intestinal granulomas caused by S. japonicum infection. However, the properties (pathogenic or beneficial) of these bacteria differed significantly in abundance remains to be explored, and their role in schistosomiasis japonica also needs more research to reveal.
Metagenomic shotgun sequencing was not only able to show a more detailed and accurate taxonomic information about the microbiota, but also provided a more relevant information about functional pathways and the abundance of genes at the taxonomic level of all species in the whole microbial community (69–71). Tracing the taxonomic classification of differential genes expressed by intestinal microbiome, we found that L. reuteri, P. vulgatus, and P. distasonis were significantly elevated in the SJBA group, indicating their active and important roles in the intestinal microbiome. Subsequently, we annotated the identified genes with the KEGG database, and finally we found that the expression of the genes annotated as urease related to urea lysis and xfp/xpk was recovered in the SJBA group, and was significantly elevated compared to the SJ group through association analysis and differential analysis. At the same time, under strict validation of Spearman correlation analysis, we found a significantly positive correlation between L. reuteri and all genes expressing urease and xfp/xpk. Urease can hydrolyze host urea to release ammonia (72). The results of relevant studies demonstrated the pro-inflammatory mechanism of urease, which decomposed urea to produce ammonia, leading to ecological imbalance of the intestinal microbiota and exacerbating the deterioration of colitis (72, 73). These results indicated that the expression of urease genes was not conducive to the recovery of intestinal diseases. However, in this study, we conducted differential analysis on the expression level of urease genes. We found that there were already some genes related to urease expression that exercise normal physiological functions in the host in the control group (PBS and BA groups). At the same time, the expression of genes related to urease in the SJBA group did not have significant differences compared to the control group, but had significantly increased compared to the SJ group. The results illustrated that the intervention of B. amyloliquefaciens significantly restored the gene expression related to urease of the intestinal microbiome in S. japonicum-infected mice. At the same time, we also conducted taxonomic annotation on the host expressing the gene about urease and found that L. reuteri was one of the species that expresses genes related to urease and was the only species with significant differences in abundance. The relevant studies also showed that the supernatant of L. reuteri culture medium stopped the growth of pathogenic bacteria in vitro, and ammonia was found in the supernatant (74).
Meanwhile, L. reuteri has also been shown to be able to produce ammonia (55, 74, 75). Therefore, based on our experimental results, we speculated that the intervention of B. amyloliquefaciens promoted significant upregulation of the abundance of L. reuteri while restoring the production of ammonia by enhancing the expression of urease genes in L. reuteri. The production of ammonia changed the pH value of the intestinal microenvironment, inhibited the colonization of pathogenic bacteria, and ultimately hindered the deterioration of schistosomiasis japonica (76). In parallel, we also observed a significant recovery of abundance of genes, which could be translated to Xfp/Xpk. Moreover, Xfp/Xpk was able to catalyze the generation of D-erythrose 4-phosphate according to the KEGG database. Xfp/Xpk had been reported to be associated with the utilization of prebiotic fiber ingredients, and restoration of their abundance was beneficial for the host to use prebiotic fiber ingredients to restore intestinal health (77). D-erythrose 4-phosphate, one of the products that could be generated by Xfp/Xpk catalysis, was observed to be significantly increased in the mouse model of colitis recovery, suggesting potential relationship between increased levels of D-erythrose 4-phosphate and reduced symptoms of intestinal inflammatory disease (78). Similarly, we traced the microbial host of xfp/xpk, and L. reuteri was found to be one of the species capable of expressing the xfp/xpk and the only one species that differed significantly in abundance. Consulting the KEGG database, we found that D-erythrose 4-phosphate was one precursor molecule for the biosynthesis of aromatic amino acids (e.g., tryptophan, phenylalanine, tyrosine). Also, L. reuteri was found to metabolize tryptophan into indole derivatives. Some of these could activate the aryl hydrocarbon receptor (AHR), which could help innate lymphoid cells (ILC) make more IL-22 (79–81). IL-22 could inhibit the colonization of fungi in the intestinal mucosa and enhance the repair of the intestinal mucosa (81). Therefore, the findings suggested potential interaction between the restored abundance of xfp/xpk in L. reuteri and the host’s immune regulatory system, which ultimately alleviates the pathological damage caused by S. japonicum infection by improving the strength of the intestinal barrier and strengthening host self-healing of the intestine after injury. However, more detailed and credible regulatory mechanisms involved in this hypothesis needed further studies to be elucidated.
Our results showed significant changes of L. reuteri in the composition and abundance of predicted genes of the intestinal microbiome in S. japonicum-infected mice, highlighting its importance in maintaining homeostasis of intestinal microbiome. Our results, along with those from other studies, showed that the genes that could be translated to urease and Xfp/Xpk are important in metabolic pathways and in the host immune response to S. japonicum infection. However, the exact regulatory mechanisms needed to be elucidated and supported by more experiments. In addition, metabolomic sequencing and immunohistochemical techniques will be used in our future studies, to analyze the differential alterations of physiological metabolites and the expression of inflammatory factors in the intestine and liver of S. japonicum-infected mice after B. amyloliquefaciens intervention, thereby further elucidating the interactive relationship between the intestinal microbiome and the host immune response, as well as validating our metagenomic results. We are also aware that one limitation of this study is the relatively small sample size in the experimental design. We will expand the number of model mice to increase the credibility and accuracy of the results in the subsequent in-depth analysis. On the other hand, the importance of L. reuteri in the gut microbiome of S. japonicum-infected mice is highlighted in our results. As one of the colonizing bacteria in the intestine, its ability to secrete reuterin and histamine played important roles in the host’s self-protection, repair and reinforcement of the intestinal barrier, and activation of the host’s immune response (82, 83). What’s more, the abundance of L. reuteri was recovered after the intervention of B. amyloliquefaciens. If we directly infected the S. japonicum-infected mice with L. reuteri, would it have better recovery and inhibitory effects on the pathological symptoms caused by the S. japonicum infection? Meanwhile, the biosynthesis and metabolism of aromatic amino acids were also one of the mechanisms underlying the regulation of the host immune response, based on our results. At the same time, it has been reported that intervention through the administration of L. reuteri to mice accompanied by a tryptophan-rich diet was able to induce the expression of CD4CD8αα T cells++ in the intestinal epithelium, which then mediate Th1- and Th2-type immune responses (84). Therefore, intervention with L. reuteri treatment and a diet rich in aromatic amino acids might become more effective treatment of schistosomiasis japonica, which would also be further investigated in our subsequent experiments.
In summary, metagenomic analysis showed that the composition of the intestinal microbiota at the species level in S. japonicum-infected mice significantly changed after B. amyloliquefaciens intervention, and the abundance of intestinal beneficial bacteria significantly increased. At the same time, the abundance of predicted functional genes in the gut microbiome also significantly changed after the intervention. The abundance of genes involved in urease metabolism and Xfp/Xpk related to D-erythrose 4-phosphate production were significantly restored in the SJBA group, highlighting the importance of L. reuteri in the recovery and gene expression of the intestinal microbiome. These results also suggested a potential regulatory mechanism between the gene function of the intestinal microbiome and the host immune response. Our study lays the foundation for the elucidation of the regulatory mechanisms of probiotic intervention to alleviate schistosomiasis japonica and provides potential adjuvant therapeutic strategies for the early prevention and treatment of schistosomiasis japonica.
Supplementary Material
ACKNOWLEDGMENTS
We would like to express our gratitude to Ying Xiao and Qingqun Wang for their valuable assistance during the process of sacrificing mice. Their contributions were instrumental in ensuring the smooth execution of this essential procedure. We also give our thanks to Umar for his assistance in polishing the language of this manuscript.
This work was funded by the National Natural Science Foundation of China (32170071, 32300051, and 82102428), and the Natural Science Foundation of Hunan Province (2022JJ40663).
J.H. and Z.Y. conceived the study; J.W. isolated and characterized B. amyloliquefaciens; H.C., S.H., and S.Y. performed the experiments; H.C. and Y.Z. analyzed the data; H.C. and R.S. visualized the statistical results; H.C. and S.H. wrote the draft; H.C. and S.H. wrote and edited the final manuscript. All authors contributed to the article and approved the submitted version.
Contributor Information
Jing Huang, Email: jing_huang@csu.edu.cn.
Zheng Yu, Email: yuzheng@csu.edu.cn.
Patricia Albuquerque, Universidade de Brasilia, Brazil.
DATA AVAILABILITY
The datasets analyzed during the current study are available in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), under accession number PRJNA973216.
ETHICS APPROVAL
Experiments about the isolation and identification of B. amyloliquefaciens were reviewed and approved by the IRB of the School of Basic Medical Science at Central South University (No. 2021-KT75). All experiments on mice in this study were conducted in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal experiments were reviewed and approved by the IRB of the School of Basic Medical Science at Central South University (No. 2021-KT25).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.03735-23.
Fig. S1 and S2.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Ke XD, Shen S, Song LJ, Yu CX, Kikuchi M, Hirayama K, Gao H, Wang J, Yin X, Yao Y, Liu Q, Zhou W. 2017. Characterization of Schistosoma japonicum CP1412 protein as a novel member of the ribonuclease T2 molecule family with immune regulatory function. Parasit Vectors 10:89. doi: 10.1186/s13071-016-1962-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McManus DP, Dunne DW, Sacko M, Utzinger J, Vennervald BJ, Zhou XN. 2018. Schistosomiasis. Nat Rev Dis Primers 4:13. doi: 10.1038/s41572-018-0013-8 [DOI] [PubMed] [Google Scholar]
- 3. Colley DG, Bustinduy AL, Secor WE, King CH. 2014. Human schistosomiasis. Lancet 383:2253–2264. doi: 10.1016/S0140-6736(13)61949-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li XH, DeMarco R, Neves LX, James SR, Newling K, Ashton PD, Cao JP, Wilson RA, Castro-Borges W. 2018. Microexon gene transcriptional profiles and evolution provide insights into blood processing by the Schistosoma japonicum esophagus. PLoS Negl Trop Dis 12:e0006235. doi: 10.1371/journal.pntd.0006235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doenhoff MJ, Cioli D, Utzinger J. 2008. Praziquantel: mechanisms of action, resistance and new derivatives for schistosomiasis. Curr Opin Infect Dis 21:659–667. doi: 10.1097/QCO.0b013e328318978f [DOI] [PubMed] [Google Scholar]
- 6. Melman SD, Steinauer ML, Cunningham C, Kubatko LS, Mwangi IN, Wynn NB, Mutuku MW, Karanja DMS, Colley DG, Black CL, Secor WE, Mkoji GM, Loker ES. 2009. Reduced susceptibility to praziquantel among naturally occurring Kenyan isolates of Schistosoma mansoni. PLoS Negl Trop Dis 3:e504. doi: 10.1371/journal.pntd.0000504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wieërs G, Belkhir L, Enaud R, Leclercq S, Philippart de Foy J-M, Dequenne I, de Timary P, Cani PD. 2019. How probiotics affect the microbiota. Front Cell Infect Microbiol 9:454. doi: 10.3389/fcimb.2019.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RB-Z, et al. 2018. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174:1388–1405. doi: 10.1016/j.cell.2018.08.041 [DOI] [PubMed] [Google Scholar]
- 9. Simon E, Călinoiu LF, Mitrea L, Vodnar DC. 2021. Probiotics, prebiotics, and synbiotics: implications and beneficial effects against irritable bowel syndrome. Nutrients 13:2112. doi: 10.3390/nu13062112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vieira AT, Teixeira MM, Martins FS. 2013. The role of probiotics and prebiotics in inducing gut immunity. Front Immunol 4:445. doi: 10.3389/fimmu.2013.00445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KCH, Aalvink S, Martinez LO, Dumas M-E, Maiter D, Loumaye A, Hermans MP, Thissen J-P, Belzer C, de Vos WM, Cani PD. 2017. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 23:107–113. doi: 10.1038/nm.4236 [DOI] [PubMed] [Google Scholar]
- 12. Lin D, Song Q, Zhang Y, Liu J, Chen F, Du S, Xiang S, Wang L, Wu X, Sun X. 2021. Bacillus subtilis attenuates hepatic and intestinal injuries and modulates gut microbiota and gene expression profiles in mice infected with Schistosoma japonicum. Front Cell Dev Biol 9:766205. doi: 10.3389/fcell.2021.766205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Michaudel C, Sokol H. 2020. The gut microbiota at the service of immunometabolism. Cell Metab 32:514–523. doi: 10.1016/j.cmet.2020.09.004 [DOI] [PubMed] [Google Scholar]
- 14. Gasaly N, de Vos P, Hermoso MA. 2021. Impact of bacterial metabolites on gut barrier function and host immunity: a focus on bacterial metabolism and its relevance for intestinal inflammation. Front Immunol 12:658354. doi: 10.3389/fimmu.2021.658354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen YC, Huang SD, Tu JH, Yu JS, Nurlatifah AO, Chiu WC, Su YH, Chang HL, Putri DA, Cheng HL. 2020. Exopolysaccharides of Bacillus amyloliquefaciens modulate glycemic level in mice and promote glucose uptake of cells through the activation of Akt. Int J Biol Macromol 146:202–211. doi: 10.1016/j.ijbiomac.2019.12.217 [DOI] [PubMed] [Google Scholar]
- 16. Lee NK, Kim WS, Paik HD. 2019. Bacillus strains as human probiotics: characterization, safety, microbiome, and probiotic carrier. Food Sci Biotechnol 28:1297–1305. doi: 10.1007/s10068-019-00691-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. WoldemariamYohannes K, Wan Z, Yu Q, Li H, Wei X, Liu Y, Wang J, Sun B. 2020. Prebiotic, probiotic, antimicrobial, and functional food applications of Bacillus amyloliquefaciens. J Agric Food Chem 68:14709–14727. doi: 10.1021/acs.jafc.0c06396 [DOI] [PubMed] [Google Scholar]
- 18. Chen J, Lv Z, Cheng Z, Wang T, Li P, Wu A, Nepovimova E, Long M, Wu W, Kuca K. 2021. Bacillus amyloliquefaciens B10 inhibits aflatoxin B1-induced cecal inflammation in mice by regulating their intestinal flora. Food Chem Toxicol 156:112438. doi: 10.1016/j.fct.2021.112438 [DOI] [PubMed] [Google Scholar]
- 19. Li X, Lv Z, Chen J, Nepovimova E, Long M, Wu W, Kuca K. 2021. Bacillus amyloliquefaciens B10 can alleviate liver apoptosis and oxidative stress induced by aflatoxin B1. Food Chem Toxicol 151:112124. doi: 10.1016/j.fct.2021.112124 [DOI] [PubMed] [Google Scholar]
- 20. Zeng Z, Zhou Y, Xu Y, Wang S, Wang B, Zeng Z, Wang Q, Ye X, Jin L, Yue M, Tang L, Zou P, Zhao P, Li W. 2022. Bacillus amyloliquefaciens SC06 alleviates the obesity of ob/ob mice and improves their intestinal microbiota and bile acid metabolism. Food Funct 13:5381–5395. doi: 10.1039/d1fo03170h [DOI] [PubMed] [Google Scholar]
- 21. Chen H, Sun R, Wang J, Yao S, Batool SS, Yu Z, Huang S, Huang J. 2023. Bacillus amyloliquefaciens alleviates the pathological injuries in mice infected with Schistosoma japonicum by modulating intestinal microbiome. Front Cell Infect Microbiol 13:1172298. doi: 10.3389/fcimb.2023.1172298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j 17:10. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 23. Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langdon WB. 2015. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min 8:1. doi: 10.1186/s13040-014-0034-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu J, Breitwieser FP, Thielen P, Salzberg SL. 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 3:e104. doi: 10.7717/peerj-cs.104 [DOI] [Google Scholar]
- 27. Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033 [DOI] [PubMed] [Google Scholar]
- 28. Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. doi: 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122. doi: 10.1093/molbev/msx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakazawa M. 2023. fmsb: functions for medical statistics book with some demographic data. R package version 0.7.5. Available from: https://CRAN.R-project.org/package=fmsb
- 33. Galaxy C. 2022. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res 50:W345–W351. doi: 10.1093/nar/gkac247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Attali D, Baker C. 2022. ggExtra: add marginal histograms to “ggplot2”, and nore “ggplot2” enhancements. R package version 0.10.0. Available from: https://CRAN.R-project.org/package=ggExtra
- 35. Kolde R. 2019. pheatmap: pretty heatmaps. R package version 1.0.12. Available from: https://CRAN.R-project.org/package=pheatmap
- 36. R Core Team . 2022. R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. Available from: https://www.R-project.org
- 37. Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag New York. Available from: http://link.springer.com/10.1007/978-3-319-24277-4 [Google Scholar]
- 38. Zhao Y, Chen L, Chen L, Huang J, Chen S, Yu Z. 2022. Exploration of the potential relationship between gut microbiota remodeling under the influence of high-protein diet and Crohn’s disease. Front Microbiol 13:831176. doi: 10.3389/fmicb.2022.831176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao C. 2022. ggVennDiagram: a “ggplot2” implement of Venn diagram. R Package Version 1.2.2. Available from: https://CRAN.R-project.org/package=ggVennDiagram
- 40. Allaire J, G C, Russell K, Yetman C. 2017. networkd3: D3 Javascript network graphs from R. R package version 0.4. Available from: https://CRAN.R-project.org/package=networkD3
- 41. Gehlenborg N. 2019. UpSetR: a more scalable alternative to Venn and Euler diagrams for visualizing intersecting sets. R package version 1.4.0. Available from: https://CRAN.R-project.org/package=UpSetR
- 42. Brunson JC, Read QD. 2023. “ggalluvial: alluvial plots in 'ggplot2'” R package version 0.12.5. Available from: http://corybrunson.github.io/ggalluvial/
- 43. Harrell JF. 2023. Hmisc: Harrell miscellaneous. R package version 5.0-1. Available from: https://CRAN.R-project.org/package=Hmisc
- 44. Csardi G, Nepusz T. 2006. The igraph software package for complex network research. Interjournal, *complex systems*, 1695. https://igraph.org.
- 45. Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks. ICWSM
- 46. Dai SM, Guan Z, Zhang LJ, Lv S, Cao CL, Li SZ, Xu J. 2020. Imported schistosomiasis, China, 2010-2018. Emerg Infect Dis 26:179–180. doi: 10.3201/eid2601.191250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu J, Li SZ, Zhang LJ, Bergquist R, Dang H, Wang Q, Lv S, Wang TP, Lin DD, Liu JB, Ren GH, Yang K, Liu Y, Dong Y, Zhang SQ, Zhou XN. 2020. Surveillance-based evidence: elimination of schistosomiasis as a public health problem in the peoples’ Republic of China. Infect Dis Poverty 9:63. doi: 10.1186/s40249-020-00676-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang GJ, Liu L, Zhu HR, Griffiths SM, Tanner M, Bergquist R, Utzinger J, Zhou XN. 2014. China’s sustained drive to eliminate neglected tropical diseases. Lancet Infect Dis 14:881–892. doi: 10.1016/S1473-3099(14)70727-3 [DOI] [PubMed] [Google Scholar]
- 49. Zhang B, Wu X, Song Q, Ning A, Liang J, Song L, Liu J, Zhang Y, Yuan D, Sun X, Wu Z. 2020. Gut microbiota modulates intestinal pathological injury in Schistosoma japonicum-infected mice. Front Med (Lausanne) 7:588928. doi: 10.3389/fmed.2020.588928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barnett R. 2018. Schistosomiasis. Lancet 392:2431. doi: 10.1016/S0140-6736(18)33008-3 [DOI] [PubMed] [Google Scholar]
- 51. Petitfils C, Maurel S, Payros G, Hueber A, Agaiz B, Gazzo G, Marrocco R, Auvray F, Langevin G, Motta J-P, et al. 2023. Identification of bacterial lipopeptides as key players in IBS. Gut 72:939–950. doi: 10.1136/gutjnl-2022-328084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Giraffa G, Chanishvili N, Widyastuti Y. 2010. Importance of lactobacilli in food and feed biotechnology. Res Microbiol 161:480–487. doi: 10.1016/j.resmic.2010.03.001 [DOI] [PubMed] [Google Scholar]
- 53. Kristjansdottir T, Bosma EF, Branco Dos Santos F, Özdemir E, Herrgård MJ, França L, Ferreira B, Nielsen AT, Gudmundsson S. 2019. A metabolic reconstruction of Lactobacillus reuteri JCM 1112 and analysis of its potential as a cell factory. Microb Cell Fact 18:186. doi: 10.1186/s12934-019-1229-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Luo H, Li P, Wang H, Roos S, Ji B, Nielsen J. 2021. Genome-scale insights into the metabolic versatility of Limosilactobacillus reuteri. BMC Biotechnol 21:46. doi: 10.1186/s12896-021-00702-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mu Q, Tavella VJ, Luo XM. 2018. Role of Lactobacillus reuteri in human health and diseases. Front Microbiol 9:757. doi: 10.3389/fmicb.2018.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Savino F, Cordisco L, Tarasco V, Palumeri E, Calabrese R, Oggero R, Roos S, Matteuzzi D. 2010. Lactobacillus reuteri DSM 17938 in infantile colic: a randomized, double-blind, placebo-controlled trial. Pediatrics 126:e526–33. doi: 10.1542/peds.2010-0433 [DOI] [PubMed] [Google Scholar]
- 57. Schepper JD, Collins FL, Rios-Arce ND, Raehtz S, Schaefer L, Gardinier JD, Britton RA, Parameswaran N, McCabe LR. 2019. Probiotic Lactobacillus reuteri prevents postantibiotic bone loss by reducing intestinal dysbiosis and preventing barrier disruption. J Bone Miner Res 34:681–698. doi: 10.1002/jbmr.3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Urbańska M, Gieruszczak-Białek D, Szajewska H. 2016. Systematic review with meta-analysis: Lactobacillus reuteri DSM 17938 for diarrhoeal diseases in children. Aliment Pharmacol Ther 43:1025–1034. doi: 10.1111/apt.13590 [DOI] [PubMed] [Google Scholar]
- 59. Wu H, Xie S, Miao J, Li Y, Wang Z, Wang M, Yu Q. 2020. Lactobacillus reuteri maintains intestinal epithelial regeneration and repairs damaged intestinal mucosa. Gut Microbes 11:997–1014. doi: 10.1080/19490976.2020.1734423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Alayande KA, Aiyegoro OA, Nengwekhulu TM, Katata-Seru L, Ateba CN. 2020. Integrated genome-based probiotic relevance and safety evaluation of Lactobacillus reuteri PNW1. PLoS One 15:e0235873. doi: 10.1371/journal.pone.0235873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Koh GY, Kane AV, Wu X, Crott JW. 2020. Parabacteroides distasonis attenuates tumorigenesis, modulates inflammatory markers and promotes intestinal barrier integrity in azoxymethane-treated A/J mice. Carcinogenesis 41:909–917. doi: 10.1093/carcin/bgaa018 [DOI] [PubMed] [Google Scholar]
- 62. Dziarski R, Park SY, Kashyap DR, Dowd SE, Gupta D. 2016. Pglyrp-regulated gut microflora Prevotella falsenii, parabacteroides distasonis and Bacteroides eggerthii enhance and Alistipes finegoldii attenuates colitis in mice. PLoS One 11:e0146162. doi: 10.1371/journal.pone.0146162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Abath FGC, Morais CNL, Montenegro CEL, Wynn TA, Montenegro SML. 2006. Immunopathogenic mechanisms in schistosomiasis: what can be learnt from human studies? Trends Parasitol 22:85–91. doi: 10.1016/j.pt.2005.12.004 [DOI] [PubMed] [Google Scholar]
- 64. Mills RH, Dulai PS, Vázquez-Baeza Y, Sauceda C, Daniel N, Gerner RR, Batachari LE, Malfavon M, Zhu Q, Weldon K, Humphrey G, Carrillo-Terrazas M, Goldasich LD, Bryant M, Raffatellu M, Quinn RA, Gewirtz AT, Chassaing B, Chu H, Sandborn WJ, Dorrestein PC, Knight R, Gonzalez DJ. 2022. Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides vulgatus proteases with disease severity. Nat Microbiol 7:262–276. doi: 10.1038/s41564-021-01050-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Onderdonk AB, Franklin ML, Cisneros RL. 1981. Production of experimental ulcerative colitis in gnotobiotic guinea pigs with simplified microflora. Infect Immun 32:225–231. doi: 10.1128/iai.32.1.225-231.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fischbach MA, Sonnenburg JL. 2011. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10:336–347. doi: 10.1016/j.chom.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. doi: 10.4161/gmic.19897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lück R, Deppenmeier U. 2022. Genetic tools for the redirection of the central carbon flow towards the production of lactate in the human gut bacterium Phocaeicola (Bacteroides) vulgatus. Appl Microbiol Biotechnol 106:1211–1225. doi: 10.1007/s00253-022-11777-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang J, Jia H. 2016. Metagenome-wide association studies: fine-mining the microbiome. Nat Rev Microbiol 14:508–522. doi: 10.1038/nrmicro.2016.83 [DOI] [PubMed] [Google Scholar]
- 71. Liao Z, Yang S, Li Q, Lu H. 2022. The role of metagenomic next-generation sequencing as a promising technology for diagnosing HIV-TB coinfection. Zoonoses 2. doi: 10.15212/ZOONOSES-2022-0032 [DOI] [Google Scholar]
- 72. Ni J, Shen T-CD, Chen EZ, Bittinger K, Bailey A, Roggiani M, Sirota-Madi A, Friedman ES, Chau L, Lin A, et al. 2017. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci Transl Med 9:416. doi: 10.1126/scitranslmed.aah6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pan L, Han P, Ma S, Peng R, Wang C, Kong W, Cong L, Fu J, Zhang Z, Yu H, Wang Y, Jiang J. 2020. Abnormal metabolism of gut microbiota reveals the possible molecular mechanism of nephropathy induced by hyperuricemia. Acta Pharm Sin B 10:249–261. doi: 10.1016/j.apsb.2019.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yu H, Ganas P, Schwendicke F. 2020. Environment-specific probiotic supernatants modify the metabolic activity and survival of Streptococcus mutans in vitro. Front Microbiol 11:1447. doi: 10.3389/fmicb.2020.01447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wilson CM, Loach D, Lawley B, Bell T, Sims IM, O’Toole PW, Zomer A, Tannock GW. 2014. Lactobacillus reuteri 100-23 modulates urea hydrolysis in the murine stomach. Appl Environ Microbiol 80:6104–6113. doi: 10.1128/AEM.01876-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Stewart GS, Smith CP. 2005. Urea nitrogen salvage mechanisms and their relevance to ruminants, non-ruminants and man. Nutr Res Rev 18:49–62. doi: 10.1079/NRR200498 [DOI] [PubMed] [Google Scholar]
- 77. Kang JW, Tang X, Walton CJ, Brown MJ, Brewer RA, Maddela RL, Zheng JJ, Agus JK, Zivkovic AM. 2022. Multi-omic analyses reveal bifidogenic effect and metabolomic shifts in healthy human cohort supplemented with a prebiotic dietary fiber blend. Front Nutr 9:908534. doi: 10.3389/fnut.2022.908534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fu YP, Li CY, Peng X, Wangensteen H, Inngjerdingen KT, Zou YF. 2022. Pectic polysaccharides from Aconitum carmichaelii leaves protects against DSS-induced ulcerative colitis in mice through modulations of metabolism and microbiota composition. Biomed Pharmacother 155:113767. doi: 10.1016/j.biopha.2022.113767 [DOI] [PubMed] [Google Scholar]
- 79. Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, Colonna M. 2011. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of notch. Nat Immunol 13:144–151. doi: 10.1038/ni.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Schiering C, Wincent E, Metidji A, Iseppon A, Li Y, Potocnik AJ, Omenetti S, Henderson CJ, Wolf CR, Nebert DW, Stockinger B. 2017. Feedback control of AHR signalling regulates intestinal immunity. Nature 542:242–245. doi: 10.1038/nature21080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L. 2013. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39:372–385. doi: 10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 82. Thomas CM, Hong T, van Pijkeren JP, Hemarajata P, Trinh DV, Hu W, Britton RA, Kalkum M, Versalovic J. 2012. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS One 7:e31951. doi: 10.1371/journal.pone.0031951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Walter J, Britton RA, Roos S. 2011. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci USA 108 Suppl 1:4645–4652. doi: 10.1073/pnas.1000099107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, Cella M, Gordon JI, Hsieh CS, Colonna M. 2017. Lactobacillus Reuteri induces gut intraepithelial Cd4+Cd8Αα+ T cells. Science 357:806–810. doi: 10.1126/science.aah5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2.
An accounting of the reviewer comments and feedback.
Data Availability Statement
The datasets analyzed during the current study are available in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), under accession number PRJNA973216.