ABSTRACT
Fruit bodies (sporocarps) of wild mushrooms growing in natural environments play a substantial role in the preservation of microbial communities, for example, clinical and food-poisoning bacteria. However, the role of wild mushrooms as natural reservoirs of plant pathogenic bacteria remains almost entirely unknown. Furthermore, bacterial transmission from a mushroom species to agricultural plants has rarely been recorded in the literature. In September 2021, a creamy-white Gram-negative bacterial strain was isolated from the sporocarp of Suillus luteus (slippery jack) growing in Bermuda grass (Cynodon dactylon) lawn in Southern Iran. A similar strain was isolated from the same fungus in the same area in September 2022. Both strains were identified as Burkholderia gladioli based on phenotypic features as well as phylogeny of 16S rRNA and three housekeeping genes. The strains were not only pathogenic on white button mushrooms (Agaricus bisporus) but also induced hypersensitive reaction (HR) on tobacco and common bean leaves and caused soft rot on a set of diverse plant species, that is, chili pepper, common bean pod, cucumber, eggplant, garlic, gladiolus, narcissus, onion, potato, spring onion, okra, kohlrabi, mango, and watermelon. Isolation of plant pathogenic B. gladioli strains from sporocarp of S. luteus in two consecutive years in the same area could be indicative of the role of this fungus in the preservation of the bacterium in the natural environment. B. gladioli associated with naturally growing S. luteus could potentially invade neighboring agricultural crops, for example, vegetables and ornamentals. The potential role of wild mushrooms as natural reservoirs of phytopathogenic bacteria is further discussed.
IMPORTANCE
The bacterial genus Burkholderia contains biologically heterogeneous strains that can be isolated from diverse habitats, that is, soil, water, diseased plant material, and clinical specimens. In this study, two Gram-negative pectinolytic bacterial strains were isolated from the sporocarps of Suillus luteus in September 2021 and 2022. Molecular phylogenetic analyses revealed that both strains belonged to the complex species Burkholderia gladioli, while the pathovar status of the strains remained undetermined. Biological investigations accomplished with pathogenicity and host range assays showed that B. gladioli strains isolated from S. luteus in two consecutive years were pathogenic on a set of diverse plant species ranging from ornamentals to both monocotyledonous and dicotyledonous vegetables. Thus, B. gladioli could be considered an infectious pathogen capable of being transmitted from wild mushrooms to annual crops. Our results raise a hypothesis that wild mushrooms could be considered as potential reservoirs for phytopathogenic B. gladioli.
KEYWORDS: clinical bacteria, cross-kingdom pathogen, edible mushroom, soft rot bacteria, Suillus luteus
INTRODUCTION
Deciphering the survival modes of plant pathogenic bacteria in the absence of their main host plant plays a pivotal role in understanding the corresponding disease cycle (1). Most ecological studies on the survival of bacterial phytopathogens are focused on the role of seeds, plant debris, soil, non-host plants, weeds, and machinery in this phenomenon (2–4). Depending on the fundamental biological features of each bacterial pathogen, one or some of these habitats act as a natural reservoir of primary inoculum for the establishment of the disease. Within the past few decades, there has been an increasing interest in elucidating the role of non-agronomic biological niches in the survival of plant pathogenic bacteria (5). For instance, Morris et al. (6) showed that the life cycle of phytopathogenic Pseudomonas syringae includes a wide range of natural reservoirs, for example, rain, snow, alpine streams, and lakes as well as wild plants and epilithic biofilms. Recently, microbiome studies based on the metagenomics of non-agronomic environments revealed that a number of plant pathogenic bacteria—or their very close relatives—were present in lichens (symbiotic associations of fungi, algae, and bacteria) (7, 8). While plant pathogenicity has not been tested for most of these microbial communities associated with environmental vegetation, Vilhelmsson et al. (5) noted that these new habitats could be considered potential reservoirs for established or emerging plant diseases. All this evidence raised the question of whether the fungal constituents of agronomic environments, for example, sporocarps of wild mushrooms, could act as reservoirs of plant pathogenic bacteria. On the other hand, the potential transmission of an infectious bacterium from a sporocarpous fungus to an agricultural plant remains unclear.
From 2018 to 2022, we have conducted a series of comprehensive surveys to monitor the microbial communities associated with edible and wild mushrooms in Iran. These mushrooms were either produced under a protected environment or grown under natural conditions (9–11). Samplings from the natural environments were conducted year-round and repeated for two years to provide an inclusive insight into the microbial dynamics of sporocarps of wild mushrooms in each area. In this framework, apart from dozens of environmental strains, that is, enterobacteria, pseudomonads, and bacilli (11, 12), a Gram-negative bacterial strain (named Ir1503) was isolated from the sporocarp of the wild mushroom Suillus luteus (slippery jack) naturally grown in Bermuda grass (Cynodon dactylon) lawn in Shiraz county in September 2021. In the subsequent year (September 2022), another strain (Ir1504) similar to that isolated in 2021 was isolated from the sporocarps of the same mushroom (S. luteus) in the same location. Preliminary investigations, that is, colony color, morphology, and pigmentation suggested that both strains belonged to Burkholderia sp. (13).
Plant pathogenic members of Burkholderia include Burkholderia cepacia, Burkholderia cenocepacia, two pathovars of Burkholderia gladioli (i.e., B. gladioli pv. gladioli and B. gladioli pv. alliicola), and Burkholderia glumae. Furthermore, B. gladioli pv. agaricicola causes soft rot of edible mushrooms (14). B. glumae is pathogenic on rice [causing panicle blight (15)], while B. cepacia, B. cenocepacia, and B. gladioli induce soft rot diseases on a set of taxonomically diverse vegetables, ornamentals, and mushrooms (16–19). Since the beginning of the current century, classification of Burkholderia spp. has undergone various changes both in the species and infra-species levels. Pathogenicity and host range of the species have also been studied on various hosts (20–22). For instance, Jones et al. (23) analyzed genome sequences of 206 B. gladioli strains from diverse origins, showing that B. gladioli pv. alliicola and food-poisoning B. gladioli pv. cocovenenans strains were distinct, while B. gladioli pv. gladioli and B. gladioli pv. agaricicola were indistinguishable based on genomic and phylogenomic analyses. Moreover, it has been shown that soft-rot activity is a universal feature across B. gladioli lineages where pathovars of the species have overlapping host ranges and the pathotype strains could not be distinguished based on experimental host range assay (23).
Preliminary in vitro screenings showed that the two bacterial strains isolated from S. luteus in this study were capable of inducing soft rot on fleshy plant tissues. These observations raised a hypothesis that the wild mushroom S. luteus could be a potential reservoir of plant pathogenic bacteria, for example, Burkholderia sp. under natural conditions. Thus, the purpose of this study was to investigate the biological characteristics, pathogenicity, and host range, as well as taxonomic status of the Burkholderia strains isolated from S. luteus. Furthermore, we performed a series of biological assays to see if mushroom-associated Burkholderia strains could act as a potential plant pathogen under experimental conditions.
RESULTS
Bacterial strains
The Gram-negative bacterial strains Ir1503 and Ir1504 were isolated from brown-colored sporocarps of S. luteus in September 2021 and September 2022, respectively. The sporocarps of S. luteus had brown pits while no tissue maceration and malformation were observed. The two bacterial strains had circular creamy-white colonies with yellowish-green non-fluorescent pigmentation on the agar medium, with smooth (Ir1503) and wrinkled (Ir1504) margins, and were 1 to 2 mm in diameter. The two bacterial strains were oxidase-negative, catalase-positive, and obligate aerobic. The strains were positive in urease production, Tween 80 hydrolysis, and utilization of D-sorbitol, raffinose, inositol, and D-mannitol. They also had pectinase and protease activity, hydrolyzed casein, and induced hypersensitive reaction (HR) on tobacco (Nicotiana tabacum cv. Turkish) and common bean leaves (Fig. S1A through C). However, they were negative in amylolytic activity and levan production. Based on these phenotypic characteristics, the two strains were preliminarily identified as members of Burkholderia spp.
Pathogenicity on edible mushroom
Both strains Ir1503 and Ir1504 induced brown pitting and tissue rot on button mushroom caps 24 hours post inoculation (hpi) while initial symptoms on the specimens inoculated with both strains started as soon as 6 hpi (Fig. 1A and B). Severe soft rot and brown cavities were observed on mushroom caps inoculated with both Ir1503 and Ir1504 36–48 hpi (Fig. 1A and B). Surprisingly, the pathotype strain of B. gladioli pv. alliicola (CFBP 2422PT; Fig. 1C) also induced soft rot symptoms similar to those caused by the strains Ir1503 and Ir1504. Initial symptoms of CFBP 2422PT started on 12 hpi and extended to the fleshy tissues making pitted slippery cavities on the cap. Pathotype strain of B. gladioli pv. gladioli (CFBP 2427PT), however, induced brown discoloration and blotch symptoms on mushroom caps with no tissue maceration and pitting even 48 hpi (Fig. 1D). As for type strain of B. cepacia CFBP 2227T (Fig. 1E), inoculation of mushroom caps by CFBP 2227T resulted in discoloration of the inoculated areas even 6 hpi and continued to become deep brown and slightly pitted. Considering the colony color and pigmentation of the strain CFBP 2227T which is yellowish green with non-fluorescent pigmentation on the agar medium, the discoloration noted at 6 hpi for CFBP 2227T could be the inoculum itself. Minimal disease symptoms were induced on the mushroom caps inoculated with B. cenocepacia strain CFBP 8617 where only faint creamy discoloration was observed on the specimens with no pitting, malformation, and soft rot (Fig. 1F). While brown blotch symptoms were observed on the mushroom caps inoculated with Pseudomonas tolaasii CFBP 8707 (positive control), the caps inoculated with non-pathogenic strain of Escherichia coli dh5α and sterile distilled water (SDW) remained symptomless until 72 hpi (Fig. S2).
Fig 1.
Pathogenicity of B. gladioli strains isolated from the sporocarp of S. luteus on Agaricus bisporus cap under controlled conditions. The strains Ir1503 (A) and Ir1504 (B) as well as the pathotype strain of B. gladioli pv. alliicola CFBP 2422PT (C) induced severe pitting and soft rot on the sporocarp of button mushrooms. However, the pathotype strain of B. gladioli pv. gladioli (CFBP 2427PT, D) and the type strain of B. cepacia (CFBP 2227T, E) induced only brown blotch on the mushroom caps, while B. cenocepacia CFBP 8617 (F) had only a slight effect on the cap. hpi, Hours post inoculation.
Plant pathogenicity and host range
The strain Ir1503 isolated from S. luteus in 2021 was pathogenic on onion (both white and red onions) as well as taxonomically related species, that is, garlic, narcissus, and spring onion (Fig. 2A). In all plant species inoculated with the latter strain, water-soaked areas and tissue maceration were observed in the site of inoculation. In onions and spring onions, infected tissues turned to gray or light brown, while an unpleasant smell was associated with severe infections. In garlic and narcissus, infected tissues turned brown and dark brown, respectively, as shown in Fig. 2A. Brown rot discoloration and maceration were observed on the ovary of gladiolus flowers. The strain Ir1504 which was isolated in the subsequent year (September 2022) from the same fungus in the same location showed a similar virulence scheme on onions, garlic, narcissus, spring onion, and gladiolus as shown in Fig. 2B. However, the severity of the symptoms on narcissus, spring onion, and gladiolus was less than that observed on the plant’s inoculation with the strain Ir1503. Interestingly, pathotype strains of B. gladioli pv. alliicola (CFBP 2422PT; Fig. 2C) and B. gladioli pv. gladioli (CFBP 2427PT; Fig. 2D) induced soft rot and maceration on all tested plant species including the main host of each other, confirming that the pathovar designation within B. gladioli is not supported by experimental host range assays. None of the strains could induce soft rot on Persian shallot bulbs.
Fig 2.
Plant pathogenicity of B. gladioli strains Ir1503 (A) and Ir1504 (B) isolated from the sporocarp of S. luteus compared to the pathotype strains of B. gladioli pv. alliicola (CFBP 2422PT, C) and B. gladioli pv. gladioli (CFBP 2427PT, D). All strains induced soft rot and tissue maceration on onion (the main host of B. gladioli pv. alliicola), garlic, spring onion, and gladiolus (main host of B. gladioli pv. gladioli). Except for the strain Ir1503 which induced black discoloration on the narcissus bulb, the other strains had a slight effect (faint discoloration) on this plant.
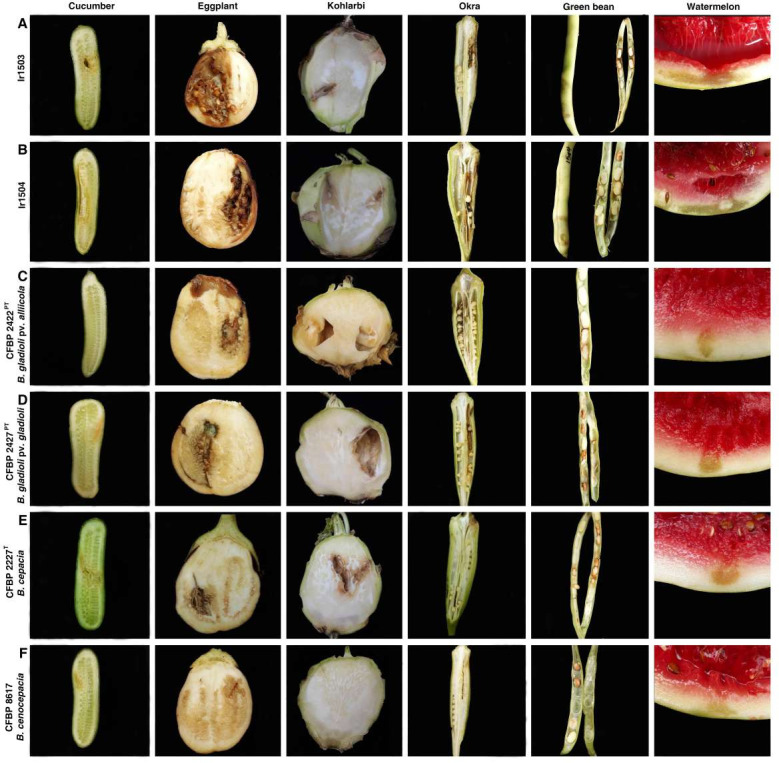
In order to further investigate the host range and plant pathogenicity of the mushroom-associated strains beyond monocotyledonous plants, they were inoculated on a set of dicotyledonous vegetables and fruits. Both strains Ir1503 and Ir1504 induced soft rot and tissue maceration on chili pepper, cucumber, eggplant, green bean pods, kohlrabi, okra, and watermelon fruit as shown in Fig. 3A and B; Fig. S3. Brown discoloration of the inoculated tissues and cavity was observed in cucumber, eggplant, and kohlrabi. On okra and green bean pods, soft rot and discoloration were observed on green immature seeds besides the fleshy tissues of the pod. In watermelon, soft rot was observed on the rind (inoculation site), which then extended into the fleshy pulp 48 hpi, leading to the destruction of the entire fruit. Similar symptoms were observed when B. gladioli pv. alliicola CFBP 2422PT (Fig. 3C), B. gladioli pv. gladioli CFBP 2427PT (Fig. 3D), and B. cepacia CFBP 2227T (Fig. 3E) were inoculated on eggplant, kohlrabi, and green bean. Pathotype strain of B. gladioli pv. alliicola did not induce soft rot on the cucumber while B. gladioli pv. gladioli and B. cenocepacia caused faint discoloration on the site of inoculation. The type strain of B. cepacia induced soft rot and cavity on cucumber similar to that observed in the specimens inoculated with Ir1503 and Ir1504. On the other hand, pathotype strains of B. gladioli pv. alliicola and B. gladioli pv. gladioli induced discoloration and soft rot on okra pods while the type strain of B. cepacia had no effect on this plant species. On watermelon, symptoms caused by the strains Ir1503 and Ir1504 were more severe compared to those induced by the reference strains of B. gladioli, B. cepacia, and B. cenocepacia as shown in Fig. 3F. Extended rind rot and discoloration followed by destruction of pulp tissues were observed on the watermelon fruits inoculated by the former strains, while symptoms caused by the latter strains were limited to water-soaked area and faint discoloration in the site of inoculation. Overall, among the strains investigated in this study, B. cenocepacia strain CFBP 8617 had the narrowest host range where none of the inoculated plant species showed severe soft rot and maceration. On the other hand, the strains Ir1503 and Ir1504 isolated from the wild mushroom S. luteus were the most aggressive strains where they induced soft rot on all plant species (except for Persian shallot) tested in this study.
Fig 3.
In vitro host range assay on cucumber, eggplant, kohlrabi, okra, green bean, and watermelon inoculated with Ir1503 (A) and Ir1504 (B) as well as the pathotype strain of B. gladioli pv. alliicola CFBP 2422PT (C), pathotype strain of B. gladioli pv. gladioli CFBP 2427PT (D), type strain of B. cepacia CFBP 2227T (E), and B. cenocepacia CFBP 8617 (F). All strains induced soft rot and tissue maceration on cucumber, eggplant, green bean pods, kohlrabi, okra, and watermelon fruit except for B. cenocepacia strain CFBP 8617 which had the most restricted host range where none of the inoculated plant species showed severe soft rot and maceration.
Phylogenetic analyses
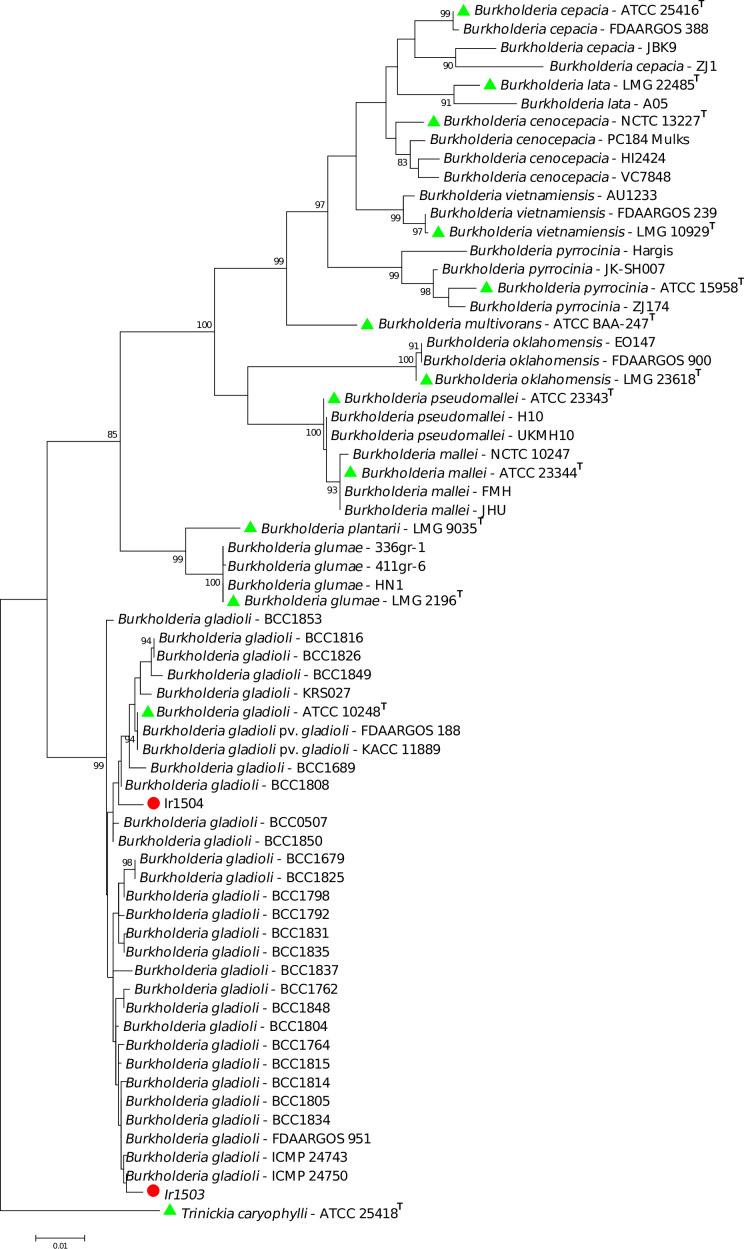
BLAST search using the sequences of individual genes (i.e., 16S rRNA, atpD, gyrB, and lepA) suggested that the bacterial strains isolated in this study belong to the complex species B. gladioli (Fig. 4). Phylogenetic analyses using the concatenated sequences of three housekeeping genes showed that the strain Ir1503 was clustered in a sub-clade next to the environmental and garlic pathogenic strains of the species. However, the strain Ir1504 was placed in another sub-clade along with the type strain of the species (ATCC 10248T) as well as gladiolus-pathogenic strains of B. gladioli pv. gladioli FDAARGOS188 and KACC 11889 (Fig. 4).
Fig 4.
Phylogeny of B. gladioli strains isolated from the sporocarp of S. luteus among the type/standard strains of Burkholderia spp. based on the concatenated sequences of atpD, gyrB, and lepA genes. Phylogenetic analyses were performed using MEGA7 software via the Maximum Likelihood method. The strains Ir1503 and Ir1504 were identified as B. gladioli while Ir1504 was placed in the clade where the type strain of the species was clustered. The strain Ir1503 was clustered in another sub-clade next to the environmental and garlic pathogenic strains of the species. Trinickia caryophylli ATCC 25,418T (formerly known as Burkholderia caryophylli) was used as an out-group in the phylogenetic tree. Green triangles indicate the type strains of each species, while red circles show the strains isolated in this study.
DISCUSSION
In this study, a Gram-negative pectinolytic bacterial strain was isolated from the sporocarps of S. luteus in September 2021. In the subsequent year, surveys were conducted in the same location on the same fungus which resulted in the isolation of another Gram-negative pectinolytic strain in September 2022 (strains Ir1503 and Ir1504, respectively). Molecular phylogenetic analyses revealed that both strains belonged to the complex species B. gladioli while pathovar status of the strains remained undetermined. Biological investigations accomplished with pathogenicity and host range assays showed that B. gladioli strains isolated from S. luteus in two consecutive years were pathogenic on a set of diverse plant species ranging from monocotyledonous vegetables and ornamentals, that is, onions and gladiolus to dicotyledonous annual crops, that is, chili pepper, cucumber, eggplant, green bean, kohlrabi, okra, mango, and watermelon. These results raise a hypothesis that wild mushrooms could serve as potential reservoirs for phytopathogenic B. gladioli. However, more directed efforts are needed to justify this conclusion.
In 1991, Pseudomonas gladioli pv. agaricicola causing bacterial soft rot of Agaricus bitorquis was described by Lincoln et al. (16), which was later reclassified as B. gladioli pv. agaricicola (24). The latter pathogen has rarely been investigated for its host range and biological characteristics compared with the other two pathovars in the species (23). B. gladioli strains isolated from S. luteus were aggressively pathogenic on button mushrooms, while the symptoms were indistinguishable from those induced by the pathotype strain of B. gladioli pv. alliicola CFBP 2422PT. Inclusion of the pathotype strains of B. gladioli pv. alliicola and B. gladioli pv. gladioli in the pathogenicity assays deciphered a mixed host range of phytopathogenic and mycopathogenic B. gladioli strains. Recently, we have isolated B. gladioli strains from garlic bulbs possessing soft rot symptoms. The garlic strains showed a wide host range including bulbous monocotyledons, dicotyledonous vegetables, and cacti, as well as wild and edible mushrooms (25). On the other hand, B. gladioli strains from distinct environmental and clinical sources have been reported to be pathogenic on mushrooms (23). These indications suggest that the assignment of plant-pathogenic and mushroom-pathogenic B. gladioli strains into different pathovars will be misleading—if not impossible due to their mixed host range—in terms of pathogen identification and disease management.
Within the past few decades, several plant pathologists have focused their studies on the role of non-agronomic biological niches in the survival of plant pathogens. For instance, Vilhelmsson et al. (5) noted that phytopathogenic bacteria including P. syringae, Burkholderia glathei [now known as Paraburkholderia glathei (26)], and Xanthomonas spp. were present in significant numbers in or on lichen thalli. A number of bacterial taxa were retrieved frequently in different lichen species sampled in the same or different sites. Paenibacillus sp. and Burkholderia sp. seem to be common in lichens (27). Furthermore, Bartoli et al. (28) noted that environmental strains of P. syringae were capable of causing symptoms on kiwifruit and showed a wide host range revealing their potential as future pathogens of a variety of hosts. Some strains of B. cepacia are adapted to the ecological niche of the rhizosphere capable of fixing nitrogen or producing indole-3-acetic acid hormone (29). Burkholderia spp. could survive in non-agronomic niches even in the presence of hazardous materials (30). In the present study, host range assays showed that the B. gladioli strains isolated from S. luteus are invasive pathogens of fleshy fruits, vegetables, and edible mushrooms. While the dissemination and spread of B. gladioli from the sporocarps of S. luteus to agricultural crops still remains unknown, precautions need to be taken to protect the infestation of agricultural areas with this pathogen.
In another perspective, mushroom microflora plays a pivotal role in both crop production and public health. The probability of wild mushrooms to become contaminated with plant pathogens and clinical bacterial pathogens is relatively greater than that of cultivated varieties. Contamination of wild mushrooms with bacterial pathogens can occur directly or indirectly via animals or insects. Venturini et al. (31) showed that mushrooms carry clinical bacterial species Listeria monocytogenes and Yersinia enterocolitica in Spain. In the UK, mushrooms purchased from supermarkets, originally grown in five different countries, were all positive for the presence of clinical bacterial species Pseudomonas aeruginosa. The number of Colony-forming unit (CFU) per gram in the outer layer of the caps was much higher than those in the whole mushrooms. In Zaragoza (Spain), 22 species of cultivated and wild fresh mushrooms sold in retail markets and supermarkets were studied by Venturini et al. (31) to quantify their microbial load. The most prevalent microbial groups included Enterobacteriaceae, pseudomonads, and lactic acid bacteria (31). In a study on golden chanterelle mushroom (Cantharellus cibarius), fluorescent pseudomonads represented 78% of the microbial load (31, 32).
The genus Burkholderia comprises versatile bacterial pathogens that cause severe diseases in humans (33), animals (34), and plants (35). Genetic distinctions between plant- and human-pathogenic Burkholderia strains are not clear (36). B. cepacia causes fatal pulmonary infections in cystic fibrosis patients (37). Pneumonia infections caused by B. gladioli and B. glumae in patients with chronic granulomatous disease were reported (38, 39). Septicemia caused by B. cenocepacia in cystic fibrosis patients was also reported (36). A number of Burkholderia species, that is, B. cepacia, B. cenocepacia, and B. gladioli are common pathogens of both animals and plants. Yet, little is known about the molecular basis of the infection, their spatial distribution, and the biological role of toxic agents involved in their virulence (40). We used reference strains of the latter three species in our biological investigations alongside our strains isolated in this study and noted that they were not only pathogenic on a diverse set of plant species but also could invasively infect edible mushrooms. All these facts highlight on the one hand the necessity of substantial investigations in the ecology of cross-kingdom Burkholderia species. On the other hand, reliable, efficient, and low-cost microbiological methods need to be developed for the detection, diagnosis, and differentiation of human-, plant-, and mushroom-pathogenic Burkholderia strains on a commercial scale.
MATERIALS AND METHODS
Sampling and bacterial isolation
Samplings from naturally grown wild mushrooms were conducted in September 2021 and at the same time in the subsequent year in Shiraz, Iran. Fully-grown sporocarps of wild mushrooms either grown in soil or in association with trees and shrubs were sampled and immediately transferred to the laboratory for further analyses. Microbial isolation was conducted on yeast-extract peptone glucose agar (YPGA) medium via multiple streaking as recommended by Schaad et al. (13). In brief, pieces of sporocarps were aseptically cut and macerated in a few drops of SDW using a sterile mortar and a pestle. A loopful of the resulting suspensions was streaked on YPGA medium and the plates were incubated at 27°C for 3–4 days. The resulting purified bacterial strains were re-suspended in SDW and stored at 4°C for further use or maintained in 15% glycerol at −70°C for long-term storage.
In order to determine whether the mushroom-associated bacterial strains possess phytopathogenic features, all bacterial strains were initially evaluated for pectinolytic activity and induction of HR. The HR test was conducted on tobacco (N. tabacum cv. Turkish) and common bean (Phaseolus vulgaris cv. Derakhshan) leaves using the bacterial suspension from a 48-h old culture on YPGA medium at a concentration of 108 CFU/mL (41). The pectinolytic activity was confirmed by the potato disks test, and the intensity of pectinolytic activity of the strains was assessed based on the intensity of rotting on the disks (42). All the tests were repeated twice. Based on the latter assays, a Gram-negative pectinolytic bacterial strain was isolated from the sporocarp of S. luteus in September 2021, while surveys in the subsequent year (September 2022) yielded another pectinolytic strain similar to that isolated in the previous year. Phenotypic features and biochemical characteristics of the strains were investigated using the procedure described by Schaad et al. (13). Details of the experimental procedure were described previously (43–45). In brief, Gram reaction, oxidase and catalase activities, aerobic/anaerobic growth, and colony characteristics on yeast extract-dextrose-calcium carbonate agar medium were determined. Enzymatic activity of the strains, hydrolyses of different substrates, and utilization of organic compounds were also investigated using the standard procedure (13). Based on the phenotypic characteristics of the strains, they were preliminarily identified as members of Burkholderia sp., while positive pectinase and HR activity suggested that they could possess phytopathogenic features. Thus, type/pathotype strains of B. cepacia CFBP 2227T, B. gladioli pv. gladioli CFBP 2427PT, and B. gladioli pv. alliicola CFBP 2422PT and standard strain B. cenocepacia CFBP 8617 were used as controls in all the biochemical and phenotypic tests. All the tests were repeated twice.
Pathogenicity test on edible mushroom
The strains Ir1503 and Ir1504 isolated from sporocarps of S. luteus under natural conditions could have had pathogenicity features on sporocarpous fungi including edible mushrooms. Thus, they were evaluated for their virulence on the sporocarps of white button mushrooms along with the reference strains of Burkholderia spp. and the mushroom pathogen P. tolaasii (46). Pathogenicity of the strains was evaluated (repeated twice) on the caps (sporocarps) of fresh white button mushrooms using the cut-cap method (9). In brief, the bacterial suspension was inoculated using a micropipette onto the cut surface of the caps (1 × 107 CFU/mL in SDW; four spots/cap; 20 µL/spot). The inoculated specimens were incubated in a dark moist chamber at 24°C–27°C up to 72 hpi and periodically monitored for symptom development. Reference strains of B. cepacia CFBP 2227T, B. gladioli pv. alliicola CFBP 2422PT, B. gladioli pv. gladioli CFBP 2427PT, and B. cenocepacia CFBP 8617 were also used in the pathogenicity tests in the same manner. The brown blotch pathogen of mushroom P. tolaasii CFBP 8707 was used as a positive control, while the non-pathogenic strain of E. coli (dh5α) and SDW was used as negative controls. Koch’s postulates were accomplished by re-isolation and identification of bacterial strains from the symptomatic caps using colony morphology and Gram staining.
Plant pathogenicity and host range assays
In order to decipher the pathogenicity and host range of the two mushroom-associated Burkholderia strains, they were inoculated on onion (the main host of B. cepacia and B. gladioli pv. alliicola) and gladiolus (the host of B. gladioli pv. gladioli) as well as taxonomically related species, that is, garlic (Allium sativum), narcissus (Narcissus jonquilla), spring onion (Allium fistulosum), and Persian shallot (Allium stipitatum). This assay would shed light not only on the pathogenicity features of the strains but also on their host range. Plant inoculation and virulence assessment were conducted according to the procedure described previously (44, 47). In brief, the vegetables and fruits were superficially disinfected with 1% sodium hypochlorite. Then, 10 µL of the bacterial suspension (107 CFU/mL) was inoculated using a micropipette into the fleshy parts of these plants. The inoculated specimens were incubated in a moist chamber with a humidity of 80%–90% at 28°C for 72 hours.
Furthermore, the pathogenicity of the two strains was evaluated on the fleshy tissues of several vegetables and annual crops outside the established host range of phytopathogenic Burkholderia spp., that is, chili pepper (Capsicum annuum; Solanaceae), cucumber (Cucumis sativus; Cucurbitaceae), eggplant (Solanum melongena; Solanaceae), green bean (P. vulgaris; Fabaceae), kohlrabi (Brassica oleracea; Brassicaceae), mango (Mangifera indica; Anacardiaceae), okra (Abelmoschus esculentus; Malvaceae), potato (Solanum tuberosum; Solanaceae), and watermelon (Citrullus lanatus; Cucurbitaceae). The preparation of plant materials and inoculation procedure were the same as described above. Three replicates were used for each strain. Furthermore, B. cepacia CFBP 2227T, B. cenocepacia CFBP 8617, B. gladioli pv. alliicola CFBP 2422PT, and B. gladioli pv. gladioli CFBP 2427PT were used as controls. The experiments were repeated twice.
Phylogenetic analyses
Phenotypic features and colony characteristics suggested that the two strains had high similarity to Burkholderia spp. To decipher the exact taxonomic position of the strains, a phylogenetic analysis was conducted using the sequences of 16S rRNA along with atpD, gyrB, and lepA housekeeping genes (22). Sequences of the three housekeeping genes atpD, gyrB, and lepA have been shown to robustly delineate the phylogenetic position of different Burkholderia lineages (19, 22, 48). Bacterial DNAs were extracted using the Expin Combo GP (GeneAll) DNA extraction kit via a procedure recommended by the manufacturer. For PCR reactions, the Universal PCR Kit, AmpliqonTaq DNA Polymerase Master Mix Red (Ampliqon A/S, Odense, Denmark), was applied according to the manufacturer’s recommendations. For each strain, a 20 µL PCR including 50 ng of total DNA and 1 µL of each primer (10 pmol/µL) were used. The annealing temperatures and the corresponding primer sequences were described in the previous work (25). The PCR products were subjected to nucleotide sequencing via Sanger technology, and the resulting sequences were assembled, edited, and aligned using the combination of BioEdit, Clustal W, and MEGA7 software (49). The sequences of the three genes were concatenated in the alphabetic order of the genes, and phylogenetic trees were constructed using Maximum Likelihood method with 1,000 bootstrap replicates (50).
ACKNOWLEDGMENTS
We thank CIRM-CFBP culture collection (https://cirm-cfbp.fr) for strain deposition and preservation as well as for providing reference strains.
This study was supported by Shiraz University, the University of Tehran, and CIRM-CFBP. The work of E.O. was supported by the University of Tehran.
E.O. conceived and designed the study. M.H. carried out the experiments with assistance from S.M.T., A.S., H.A., and M.B. P.P. contributed to the strain preservation and provided the reference strains. E.O. analyzed and interpreted the data and prepared the manuscript with assistance from M.H. All the authors revised the final version of the manuscript while E.O. acted as the corresponding author.
Contributor Information
Ebrahim Osdaghi, Email: eosdaghi@ut.ac.ir.
Kevin Loren Hockett, Pennsylvania State University, University Park, Pennsylvania, USA.
DATA AVAILABILITY
The strains Ir1503 and Ir1504 isolated in this study are available in the French Collection for Plant-associated Bacteria (CFBP) and the International Collection of Microorganisms from Plants (ICMP) under the accession numbers lr1503 = CFBP 9103 = ICMP 24728 and lr1504 = ICMP 24720. Nucleotide sequences obtained in this study are deposited in the NCBI GenBank database under the following accession numbers OR263462-OR263463 for 16S rRNA, OR396931-OR396932 for atpD, OR424415-OR424416 for gyrB, and OR424417-OR424418 for lepA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.03395-23.
Figure S1 (Pectinolytic activity of the strain Ir1503 on potato disk [A], and hypersensitive reaction of the same strain on tobacco [B] and common bean [C] leaves), Figure S2 (Inoculation of mushroom caps with non-pathogenic strain of E. coli dh5α [left] and brown blotch pathogen of mushroom P. tolaasii CFBP 8707 [right]; while brown blotch symptoms were observed on the mushroom caps inoculated with P. tolaasii CFBP 8707 [positive control], the caps inoculated with E. coli remained symptomless until 72 hours post inoculation), and Figure S3 (Pathogenicity of Burkholderia gladioli strains Ir1503 and Ir1504 on chili pepper).
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Cooke BM, Jones DG, Kaye B, eds. 2006. The epidemiology of plant diseases. vol 2. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 2. Nelson EB. 2004. Microbial dynamics and interactions in the spermosphere. Annu Rev Phytopathol 42:271–309. doi: 10.1146/annurev.phyto.42.121603.131041 [DOI] [PubMed] [Google Scholar]
- 3. Gitaitis R, Walcott R. 2007. The epidemiology and management of seedborne bacterial diseases. Annu Rev Phytopathol 45:371–397. doi: 10.1146/annurev.phyto.45.062806.094321 [DOI] [PubMed] [Google Scholar]
- 4. Osdaghi E, van der Wolf JM, Abachi H, Li X, De Boer SH, Ishimaru CA. 2022. Bacterial ring rot of potato caused by Clavibacter sepedonicus: a successful example of defeating the enemy under international regulations. Mol Plant Pathol 23:911–932. doi: 10.1111/mpp.13191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vilhelmsson O, Sigurbjörnsdóttir A, Grube M, Höfte M. 2016. Are lichens potential natural reservoirs for plant pathogens? Mol Plant Pathol 17:143–145. doi: 10.1111/mpp.12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffière A, Yan S, Dominguez H, Thompson BM. 2008. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J 2:321–334. doi: 10.1038/ismej.2007.113 [DOI] [PubMed] [Google Scholar]
- 7. Cardinale M, Vieira de Castro J Jr, Müller H, Berg G, Grube M. 2008. In situ analysis of the bacterial community associated with the reindeer lichen Cladonia arbuscula reveals predominance of Alphaproteobacteria. FEMS Microbiol Ecol. 66:63–71. doi: 10.1111/j.1574-6941.2008.00546.x [DOI] [PubMed] [Google Scholar]
- 8. Sigurbjörnsdóttir MA, Heiðmarsson S, Jónsdóttir AR, Vilhelmsson O. 2014. Novel bacteria associated with Arctic seashore lichens have potential roles in nutrient scavenging. Can J Microbiol 60:307–317. doi: 10.1139/cjm-2013-0888 [DOI] [PubMed] [Google Scholar]
- 9. Hamidizade M., Taghavi SM, Martins SJ, Herschlag RA, Hockett KL, Bull CT, Osdaghi E. 2020. Bacterial brown pit, a new disease of edible mushrooms caused by Mycetocola sp. Plant Dis. 104:1445–1454. doi: 10.1094/PDIS-10-19-2176-RE [DOI] [PubMed] [Google Scholar]
- 10. Hamidizade M, Taghavi SM, Moallem M, Aeini M, Fazliarab A, Abachi H, Herschlag RA, Hockett KL, Bull CT, Osdaghi E. 2023. Ewingella americana: an emerging multifaceted pathogen of edible mushrooms. Phytopathology 113:150–159. doi: 10.1094/PHYTO-08-22-0299-R [DOI] [PubMed] [Google Scholar]
- 11. Moallem M. 2022. Investigation of some important bacterial diseases of edible button mushroom in some areas of Iran. MSc thesis. Shahid Chamran University, Iran. [Google Scholar]
- 12. Hamidizade M. 2019. Feasibility of biocontrol of bacterial brown blotch of button mushroom (Agaricus bisporus) using antagonistic bacteria. MSc Thesis, Shiraz University, Iran [Google Scholar]
- 13. Schaad NW, Jones JB, Chun W. 2001. Laboratory guide for the identification of plant pathogenic bacteria. 3rd ed. American Phytopathological society (APS press). [Google Scholar]
- 14. Bull CT, De Boer SH, Denny TP, Firrao G, Saux MFL, Saddler GS, Takikawa Y. 2010. Comprehensive list of names of plant pathogenic bacteria, 1980-2007. J Plant Pathol 92:551–592. doi: 10.4454/JPP.V92I3.302 [DOI] [Google Scholar]
- 15. Ortega L, Rojas CM. 2021. Bacterial panicle blight and Burkholderia glumae: from pathogen biology to disease control. Phytopathology 111:772–778. doi: 10.1094/PHYTO-09-20-0401-RVW [DOI] [PubMed] [Google Scholar]
- 16. Lincoln SP, Fermor TR, Stead DE, Sellwood JE. 1991. Bacterial soft rot of Agaricus bitorquis. Plant Pathol 40:136–144. doi: 10.1111/j.1365-3059.1991.tb02302.x [DOI] [Google Scholar]
- 17. Parke JL. 2000. Burkholderia cepacia: friend and foe? Plant Health Instr 66:124–125. doi: 10.1094/PHI-I-2000-0926-01 [DOI] [Google Scholar]
- 18. Coenye T, Vandamme P. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol 5:719–729. doi: 10.1046/j.1462-2920.2003.00471.x [DOI] [PubMed] [Google Scholar]
- 19. Ansari M, Taghavi SM, Zarei S, Miri K, Portier P, Osdaghi E. 2019. Pathogenicity and molecular phylogenetic analysis reveal a distinct position of the banana fingertip rot pathogen among the Burkholderia cenocepacia genomovars. Plant Pathology 68:804–815. doi: 10.1111/ppa.12976 [DOI] [Google Scholar]
- 20. Parke JL, Gurian-Sherman D. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu Rev Phytopathol 39:225–258. doi: 10.1146/annurev.phyto.39.1.225 [DOI] [PubMed] [Google Scholar]
- 21. Vandamme P, Holmes B, Coenye T, Goris J, Mahenthiralingam E, LiPuma JJ, Govan JRW. 2003. Burkholderia cenocepacia sp. nov. a new twist to an old story. Res Microbiol 154:91–96. doi: 10.1016/S0923-2508(03)00026-3 [DOI] [PubMed] [Google Scholar]
- 22. Baldwin A, Mahenthiralingam E, Thickett KM, Honeybourne D, Maiden MCJ, Govan JR, Speert DP, Lipuma JJ, Vandamme P, Dowson CG. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J Clin Microbiol 43:4665–4673. doi: 10.1128/JCM.43.9.4665-4673.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones C, Webster G, Mullins AJ, Jenner M, Bull MJ, Dashti Y, Spilker T, Parkhill J, Connor TR, LiPuma JJ, Challis GL, Mahenthiralingam E. 2021. Kill and cure: genomic phylogeny and bioactivity of Burkholderia gladioli bacteria capable of pathogenic and beneficial lifestyles. Microb Genom 7:mgen000515. doi: 10.1099/mgen.0.000515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Young JM, Saddler GS, Takikawa Y, De Boer SH, Vauterin L, Gardan L, Gvozdyak RI, Stead DE, De . 1996. Names of plant pathogenic bacteria 1864-1995. Rev Plant Pathol 75:721–763. [Google Scholar]
- 25. Abachi H, Moallem M, Taghavi SM, Hamidizade M, Soleimani A, Fazliarab A, Portier P, Osdaghi E. 2023. Garlic bulb decay and soft rot caused by the cross-kingdom pathogen Burkholderia gladioli. Plant Dis. doi: 10.1094/PDIS-08-23-1603-RE [DOI] [PubMed] [Google Scholar]
- 26. Sawana A, Adeolu M, Gupta RS. 2014. Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet 5:429. doi: 10.3389/fgene.2014.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cardinale M, Puglia AM, Grube M. 2006. Molecular analysis of lichen-associated bacterial communities. FEMS Microbiol Ecol 57:484–495. doi: 10.1111/j.1574-6941.2006.00133.x [DOI] [PubMed] [Google Scholar]
- 28. Bartoli C, Lamichhane JR, Berge O, Guilbaud C, Varvaro L, Balestra GM, Vinatzer BA, Morris CE. 2015. A framework to gauge the epidemic potential of plant pathogens in environmental reservoirs: the example of kiwifruit canker. Mol Plant Pathol 16:137–149. doi: 10.1111/mpp.12167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bevivino A, Tabacchioni S, Chiarini L, Carusi MV, Del Gallo M, Visca P. 1994. Phenotypic comparison between rhizosphere and clinical isolates of Burkholderia cepacia. Microbiology (Reading) 140 (Pt 5):1069–1077. doi: 10.1099/13500872-140-5-1069 [DOI] [PubMed] [Google Scholar]
- 30. Peñaloza-Vazquez A, Mena GL, Herrera-Estrella L, Bailey AM. 1995. Cloning and sequencing of the genes involved in glyphosate utilization by Pseudomonas pseudomallei. Appl Environ Microbiol 61:538–543. doi: 10.1128/aem.61.2.538-543.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Venturini ME, Reyes JE, Rivera CS, Oria R, Blanco D. 2011. Microbiological quality and safety of fresh cultivated and wild mushrooms commercialized in Spain. Food Microbiol 28:1492–1498. doi: 10.1016/j.fm.2011.08.007 [DOI] [PubMed] [Google Scholar]
- 32. Danell E, Alström S, Ternström A. 1993. Pseudomonas fluorescens in association with fruit bodies of the ectomycorrhizal mushroom Cantharellus cibarius. Mycol Res 97:1148–1152. doi: 10.1016/S0953-7562(09)80519-4 [DOI] [Google Scholar]
- 33. Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N Engl J Med 367:1035–1044. doi: 10.1056/NEJMra1204699 [DOI] [PubMed] [Google Scholar]
- 34. Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3:144–156. doi: 10.1038/nrmicro1085 [DOI] [PubMed] [Google Scholar]
- 35. Angus AA, Agapakis CM, Fong S, Yerrapragada S, Estrada-de los Santos P, Yang P, Song N, Kano S, Caballero-Mellado J, de Faria SM, Dakora FD, Weinstock G, Hirsch AM. 2014. Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS One 9:e83779. doi: 10.1371/journal.pone.0083779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Springman AC, Jacobs JL, Somvanshi VS, Sundin GW, Mulks MH, Whittam TS, Viswanathan P, Gray RL, Lipuma JJ, Ciche TA. 2009. Genetic diversity and multihost pathogenicity of clinical and environmental strains of Burkholderia cenocepacia. Appl Environ Microbiol 75:5250–5260. doi: 10.1128/AEM.00877-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Govan JRW, Hughes JE, Vandamme P. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol 45:395–407. doi: 10.1099/00222615-45-6-395 [DOI] [PubMed] [Google Scholar]
- 38. Ross JP, Holland SM, Gill VJ, DeCarlo ES, Gallin JI. 1995. Severe Burkholderia (Pseudomonas) gladioli infection in chronic granulomatous disease: report of two successfully treated cases. Clin Infect Dis 21:1291–1293. doi: 10.1093/clinids/21.5.1291 [DOI] [PubMed] [Google Scholar]
- 39. Weinberg JB, Alexander BD, Majure JM, Williams LW, Kim JY, Vandamme P, LiPuma JJ. 2007. Burkholderia glumae infection in an infant with chronic granulomatous disease. J Clin Microbiol 45:662–665. doi: 10.1128/JCM.02058-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dose B, Thongkongkaew T, Zopf D, Kim HJ, Bratovanov EV, García-Altares M, Scherlach K, Kumpfmüller J, Ross C, Hermenau R, Niehs S, Silge A, Hniopek J, Schmitt M, Popp J, Hertweck C. 2021. Multimodal molecular imaging and identification of bacterial toxins causing mushroom soft rot and cavity disease. ChemBioChem 22:2901–2907. doi: 10.1002/cbic.202100330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mafakheri H, Taghavi SM, Zarei S, Kuzmanović N, Osdaghi E. 2022. Occurrence of crown gall disease on Japanese spindle (Euonymus japonicas var. Green Rocket) caused by Agrobacterium rosae in Iran. Plant Dis 106:313. doi: 10.1094/PDIS-03-21-0580-PDN [DOI] [PubMed] [Google Scholar]
- 42. Osdaghi E, Taghavi SM, Hamzehzarghani H, Fazliarab A, Lamichhane JR. 2017. Monitoring the occurrence of tomato bacterial spot and range of the causal agent Xanthomonas perforans in Iran. Plant Pathol 66:990–1002. doi: 10.1111/ppa.12642 [DOI] [Google Scholar]
- 43. Mafakheri Hamzeh, Taghavi SM, Zarei S, Rahimi T, Hasannezhad MS, Portier P, Fischer-Le Saux M, Dimkić I, Koebnik R, Kuzmanović N, Osdaghi E. 2022. Phenotypic and molecular-phylogenetic analyses reveal distinct features of crown gall-associated Xanthomonas strains. Microbiol Spectr 10:e0057721. doi: 10.1128/spectrum.00577-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parvin SMR, Taghavi SM, Osdaghi E. 2023. Field surveys indicate taxonomically diverse Pectobacterium species inducing soft rot of vegetables and annual crops in Iran . Plant Pathol 72:1260–1271. doi: 10.1111/ppa.13735 [DOI] [Google Scholar]
- 45. Sedighian N, Taghavi SM, Hamzehzarghani H, van der Wolf JM, Wicker E, Osdaghi E. 2020. Potato-infecting Ralstonia solanacearum strains in Iran expand knowledge on the global diversity of brown rot ecotype of the pathogen. Phytopathology 110:1647–1656. doi: 10.1094/PHYTO-03-20-0072-R [DOI] [PubMed] [Google Scholar]
- 46. Osdaghi E, Martins SJ, Ramos-Sepulveda L, Vieira FR, Pecchia JA, Beyer DM, Bell TH, Yang Y, Hockett KL, Bull CT. 2019. 100 years since Tolaas: bacterial blotch of mushrooms in the 21st century. Plant Dis 103:2714–2732. doi: 10.1094/PDIS-03-19-0589-FE [DOI] [PubMed] [Google Scholar]
- 47. Yaripour Z, Mohsen Taghavi S, Osdaghi E, Lamichhane JR. 2018. Host range and phylogenetic analysis of Xanthomonas alfalfae causing bacterial leaf spot of alfalfa in Iran. Eur J Plant Pathol 150:267–274. doi: 10.1007/s10658-017-1271-0 [DOI] [Google Scholar]
- 48. Spilker T, Baldwin A, Bumford A, Dowson CG, Mahenthiralingam E, LiPuma JJ. 2009. Expanded multilocus sequence typing for Burkholderia species. J Clin Microbiol 47:2607–2610. doi: 10.1128/JCM.00770-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version. 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hall BG. 2011. Phylogenetic trees made easy: a how-to manual. 4th ed. Sinauer Associates, Sunderland (MA). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 (Pectinolytic activity of the strain Ir1503 on potato disk [A], and hypersensitive reaction of the same strain on tobacco [B] and common bean [C] leaves), Figure S2 (Inoculation of mushroom caps with non-pathogenic strain of E. coli dh5α [left] and brown blotch pathogen of mushroom P. tolaasii CFBP 8707 [right]; while brown blotch symptoms were observed on the mushroom caps inoculated with P. tolaasii CFBP 8707 [positive control], the caps inoculated with E. coli remained symptomless until 72 hours post inoculation), and Figure S3 (Pathogenicity of Burkholderia gladioli strains Ir1503 and Ir1504 on chili pepper).
Data Availability Statement
The strains Ir1503 and Ir1504 isolated in this study are available in the French Collection for Plant-associated Bacteria (CFBP) and the International Collection of Microorganisms from Plants (ICMP) under the accession numbers lr1503 = CFBP 9103 = ICMP 24728 and lr1504 = ICMP 24720. Nucleotide sequences obtained in this study are deposited in the NCBI GenBank database under the following accession numbers OR263462-OR263463 for 16S rRNA, OR396931-OR396932 for atpD, OR424415-OR424416 for gyrB, and OR424417-OR424418 for lepA.