ABSTRACT
The fattening of calves is often associated with high antimicrobial use and the selection of antimicrobial resistance (AMR). The objective of this observational longitudinal study was to describe the AMR and strain dynamics, using whole-genome sequencing (WGS), of fecal Escherichia coli in a cohort of 22 calves. All calves received antimicrobial group treatments on Day (D) 1 (oxytetracycline, intramuscularly) and on D4 through D12 (doxycycline, in-feed). Additionally, eight calves received individual parenteral treatments between D7 and D59, including florfenicol, amoxicillin, marbofloxacin, and gamithromycin. Rectal swabs were collected from all calves on D1 (prior to treatment), D2, D9, and D82. The swabs were spread onto Enterobacterales-selective agar, and three E. coli colonies per plate were subjected to WGS. Out of 264 isolates across all calves and sampling times, 80 unique strains were identified, a majority of which harbored genes conferring resistance to tetracyclines, streptomycin, and sulfonamides. The diversity of strains decreased during the in-feed antimicrobial group treatment of the calves. On D82, 90% of isolates were strains that were not isolated at previous sampling times, and the median number per strain of AMR determinants to tetracyclines, florfenicol, β-lactams, quinolones, or macrolides decreased compared to D9. Additionally, clonal dissemination of some strains represented the main transmission route of AMR determinants. In this study, WGS revealed important variations in strain diversity and genotypic AMR of fecal E. coli over time in calves subjected to group antimicrobial treatments.
IMPORTANCE
The continued emergence and spread of antimicrobial resistance (AMR) determinants are serious global concerns. The dynamics of AMR spread and persistence in bacterial and animal host populations are complex and not solely driven by antimicrobial selection pressure. In calf fattening, both antimicrobial use and carriage prevalence of antimicrobial-resistant bacteria are generally recognized as high. This study provides new insights into the short-term, within-farm dynamics and transmission of AMR determinants in Escherichia coli from the dominant fecal flora of calves subjected to antimicrobial group treatments during the rearing period. The diversity of E. coli strains decreased over time, although, in contrast to previous observations in extended-spectrum β-lactamase-producing Enterobacterales, the predominance of a few clones was not observed. The spread of AMR determinants occurred through the dissemination of clonal strains among calves. The median number per strain of AMR determinants conferring resistance to selected antimicrobials decreased toward the end of the rearing period.
KEYWORDS: Escherichia coli, fecal carriage, antimicrobial resistance, molecular epidemiology, cattle
INTRODUCTION
In most countries, the fattening of veal calves involves the transport and commingling of calves from multiple dairy farms of origin to the fattening facility. There may be variations between countries with regard to feeding, housing conditions, and duration of the fattening period. In Switzerland, calves are purchased around the age of 30 days, housed in groups, fed mainly milk or milk substitute, with constant free access to water and roughage, and slaughtered around the age of 160 days (1). The commingling of large numbers of calves from multiple origins and of various health and immune status is a risk factor for increased morbidity and mortality (1, 2). Thus, group treatment with antimicrobials administered via feed or parenteral injections at the beginning of the fattening period is common practice and results in high antimicrobial use (AMU) (3–5). In Switzerland, antimicrobial prescription data for 2020 revealed that fattening cattle was the animal category that was attributed the highest rank in terms of the amount of active substance prescribed, and the second highest rank in terms of the number of treatments (6). In turn, AMU in general, as well as group or oral treatment specifically, has been associated with increased antimicrobial resistance (AMR) in commensal and pathogenic bacteria in veal calves (3, 7, 8).
The dynamics of AMR in bacteria isolated from veal calves are complex. In investigation reports on the dynamics of phenotypic AMR over the course of the fattening period, changes in AMR varied according to the AMR indices used, bacterial species, and study farms. For instance, based on the antimicrobial drug concentration inhibiting the growth of 50% of tested isolates (MIC50), AMR increased among Escherichia coli (for azithromycin) and Mannheimia haemolytica isolates (for clindamycin and tiamulin) (9). In another study from arrival to departure from the farm, in which 98% of treatments were group treatments, the proportion of E. coli isolates resistant to amoxicillin, tetracyclines, streptomycin, and sulfonamides increased, whereas the proportion of isolates resistant to β-lactams other than amoxicillin and to quinolones decreased (10). A reduction in other AMR indices, such as the antimicrobial resistance index (the proportion of tested drugs to which an isolate shows resistance) or the proportion of multidrug-resistant isolates (fecal E. coli, nasal Pasteurella multocida, and M. haemolytica), through the fattening period was also reported (3, 9). In other studies with a focus on extended-spectrum β-lactamase (ESBL)-producing Enterobacterales, the AMR dynamics in veal calf populations were investigated using strain typing of isolates [based on either pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), or a combination of ESBL gene sequence, MLST, and plasmid replicon typing]. Upon arrival at the fattening farms, a diversity of isolates was observed, which was attributed to the multiple farms of origin of the calves (10, 11). Some clonal variants were detected at subsequent sampling time points that were not detected upon arrival, which could be explained by their clonal expansion following antimicrobial group treatment or by their source being the fattening farm environment (11, 12). Over the fattening period, the diversity decreased, and the dominance of a limited number of clones was observed. These results, along with those of studies on dairy farms, suggest that the dynamics of ESBL gene dissemination are the result of a combination of the clonal spread of resistant strains, horizontal transfer of plasmids between different E. coli strains, and transfer of genes between plasmids (10, 11, 13–15). Mathematical modeling of ESBL carriage (determined phenotypically) further supported that both between-animal transmission and sporadic introduction affect AMR dynamics, whereas AMU affects the clearance of the ESBL phenotype rather than its acquisition (16). Nevertheless, it was previously pointed out that the phenotypic AMR dynamics observed among the subpopulation of ESBL-producing isolates do not necessarily reflect those of the dominant E. coli flora (isolated on non-selective medium) (10, 17), and this might also be the case for genotypic AMR dynamics.
As aforementioned, previous reports on the AMR dynamics of the dominant E. coli population in calves over the course of the fattening period were mainly based on phenotypic AMR, which provides limited information on the strain dynamics underlying the observed phenotypes. Therefore, the objective of this study was to describe the dynamics of whole-genome sequencing (WGS)-based AMR in commensal fecal E. coli isolated from a cohort of calves subjected to antimicrobial group treatments.
RESULTS
Study population
Twenty-two calves were studied longitudinally from arrival at a research facility (D0), where they were housed with 22 other calves, until the end of the rearing period (week 13). The calves were weaned at week 7. The median (range) age and weight of the study calves upon arrival were 27 (19–72) days and 75 (55–89) kg. All animals were crossbreds of a Brown Swiss dam and a Simmental (n = 11), Limousin (n = 7), or Angus (n = 4) sire. On D1 (before any treatment), D2, D9, and D82, rectal swab samples were collected from each calf. All calves received antimicrobial group treatments consisting of long-acting oxytetracycline intramuscularly on D1, followed by in-feed doxycycline from D4 to D12. Additionally, over the study period, eight calves received individual systemic treatments after a clinical diagnosis of infectious bronchopneumonia, including two calves treated twice each with different drugs (Table 1). The antimicrobial drugs used included florfenicol (n = 5 calves), amoxicillin (n = 2), marbofloxacin (n = 2), and gamithromycin (n = 1). One individual treatment occurred between the second (D2) and third (D9) samplings, whereas the remaining individual treatments occurred after the third sampling (D9) and a minimum of 23 days prior to the fourth sampling (D82).
TABLE 1.
Individual calf antimicrobial treatments
| Day(s) of treatment | Calf identification number | Antimicrobial drug | Administration routea | Recommended dosing intervalb |
|---|---|---|---|---|
| D7–10 | 604 | Marbofloxacin | IM | Once daily for 3 days |
| D13 + D15 | 621 | Amoxicillin | IM | Twice at 48-h interval |
| D16 + D18 | 610 and 616 | Florfenicol | IM | Twice at 48-h interval |
| D17 + D19 | 618c | Florfenicol | IM | Twice at 48-h interval |
| D17 + D19 | 607c | Amoxicillin | IM | Twice at 48-h interval |
| D21 | 614 | Gamithromycin | SC | Once |
| D22 | 607c | Florfenicol | IM | Twice at 48-h interval |
| D28–30 | 618c | Marbofloxacin | IM | Once daily for 3 days |
| D55 + D57+ D59 | 605 | Florfenicol | IM | Twice at 48-h interval |
IM, intramuscular and SC, subcutaneous.
Swiss Compendium of Veterinary Products (Institut für Veterinärpharmakologie und –toxikologie. Tierarzneimittelkompendium der Schweiz, Zurich, Switzerland. https://www.vetpharm.uzh.ch/tak/. Accessed 27 July 2023).
Two calves each treated twice.
Strains
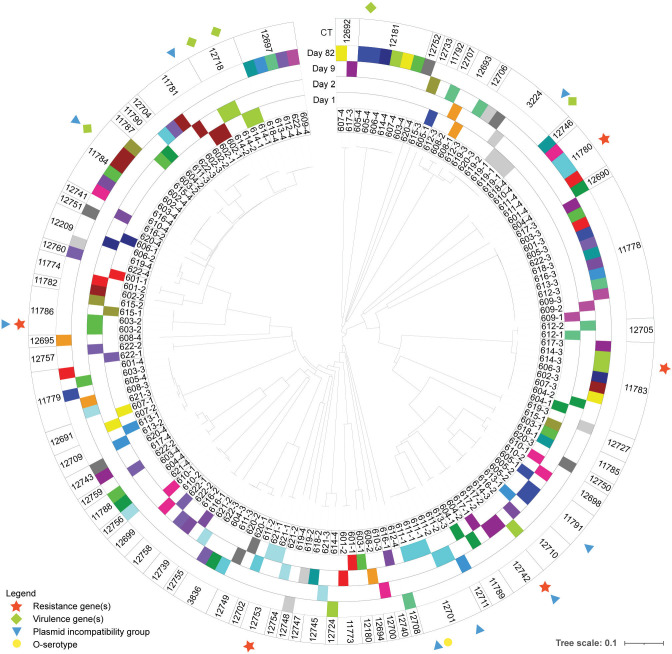
From 88 rectal swab samples, a total of 264 E. coli isolates (three per sample) and their respective genome sequences were obtained (Table S1). In 35 samples, all three isolates were identical [i.e., three isolates with identical core genome MLST complex type (CT), in silico serotype, AMR and virulence genes, and plasmid incompatibility groups within the same calf and time]. Those were distributed over time as 8, 8, 11, and 8 samples on D1, D2, D9, and D82, respectively. In 39 samples (12, 9, 7, and 11 samples on D1, D2, D9, and D82, respectively), two different strains were isolated, whereas 14 samples (2, 5, 4, and 3 samples on D1, D2, D9, and D82, respectively) yielded three different strains. After the removal of within-sample replicates, 155 isolates remained (38, 41, 37, and 39 isolates from D1, D2, D9, and 82, respectively), represented in a phylogenetic tree in Fig. 1. Across all calves and sampling times, 80 unique strains (unique set of CT, AMR, and virulence genes) were obtained. Of those, 39 were isolated each from one single sample, whereas the remaining had clones isolated from 2 up to 11 samples. The 80 unique strains belonged to 39 sequence types (ST), the most common being ST58 and ST10. The detailed distribution of strains is shown in Table 2. The sequence types ST13057, ST13058, and ST13059 represented newly assigned STs.
Fig 1.
Phylogenetic tree representing the genetic relationship between the 155 Escherichia coli isolated from calves, based on core genome MLST complex type (CT; outer circle). Isolate numbers (inner circle) are composed of the calf ID and sampling time, respectively. Each color represents a calf, and the position of each colored band represents the sampling time at which the strain was isolated. Within a CT, when non-identical within-sample isolates (same calf and time) are shown, their difference is indicated as the presence or absence of resistance genes (star), virulence genes (diamond), plasmid incompatibility group (triangle), or O type (circle).
TABLE 2.
Distribution of sequence type and core genome multi-locus sequence typing based CT among the 80 unique E. coli strains isolated from 22 calvesa
| Sequence type | Number of strains | CT |
|---|---|---|
| 58 | 15 | 11781, 11784, 11787, 11790, 12209, 12697, 12704, 12718, 12751 |
| 10 | 10 | 11785, 11789, 12698, 12701, 12711, 12742, 12750 |
| 69 | 5 | 3224, 12693, 12706, 12707 |
| 2522 | 4 | 11779, 12691, 12709 |
| 162 | 3 | 3836, 12702, 12749 |
| 301 | 3 | 11783, 12705 |
| 446 | 3 | 11788, 12743, 12759 |
| 101 | 2 | 12699, 12758 |
| 329 | 2 | 11780 |
| 8580 | 2 | 12181 |
| 10850 | 2 | 11791, 12710 |
| 13057 | 2 | 12753 |
The remaining ST were represented by a single strain: ST21 (CT 12745), ST56 (CT 11786), ST57 (CT 12692), ST117 (CT 11792), ST120 (CT 12740), ST124 (CT 12755), ST155 (CT 11774), ST306 (CT 12700), ST339 (CT 12746), ST361 (CT 11778), ST410 (CT 12180), ST448 (CT 12748), ST675 (CT 12739), ST718 (CT 12756), ST723 (CT 12724), ST730 (CT 12690), ST783 (CT 11773), ST949 (CT 11782), ST1302 (CT 12727), ST1308 (CT 12754), ST2521 (CT 12757), ST3057 (CT 12752), ST3249 (CT 12694), ST3695 (CT 12695), ST6126 (CT 12708), ST13058 (CT 12733), and ST13059 (CT 12747).
AMR determinants
A majority of the 80 strains harbored genes conferring resistance to tetracyclines (n = 57), streptomycin (n = 49), and sulfonamides (n = 42). The 80 strains also harbored genes conferring resistance to β-lactams (n = 38), kanamycin (n = 22), trimethoprim (n = 19), chloramphenicol (n = 19), florfenicol (n = 15), quinolones (n = 11), gentamicin (n = 10), erythromycin/telithromycin/tylosin (n = 6), tobramycin (n = 4), hygromycin (n = 3), fosfomycin (n = 3), and lincosamides (n = 1). The most frequent plasmid incompatibility groups were IncFIB (AP001918) (n = 38 strains), IncQ1 (n = 28), and IncI1-Iα (n = 24). Up to six incompatibility groups per strain were detected. In 16 unique strains, no known plasmid incompatibility group was detected, including strains such as those belonging to CT 11778 (ST361) and 12692 (ST57) harboring numerous AMR genes.
Dynamics of AMR determinants over time
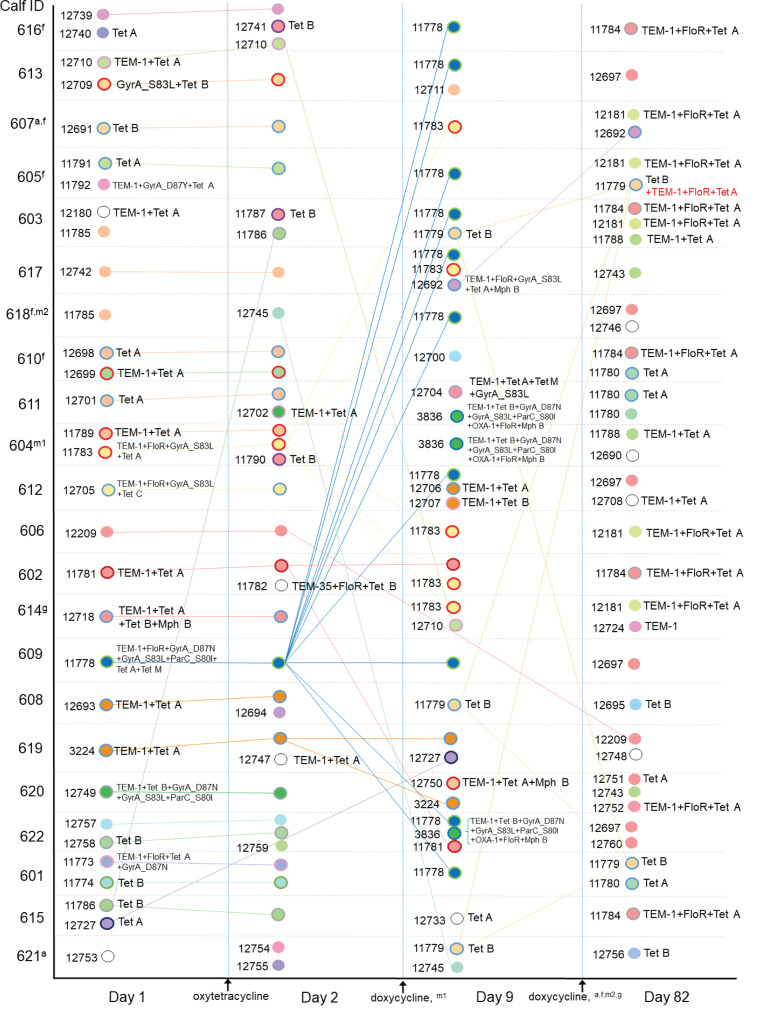
The distribution of strains per calf and sampling time was determined based on cgMLST phylogenetic relationship analysis and their AMR determinants to antimicrobial drugs or classes used for group or individual treatments during the study (tetracyclines, florfenicol, β-lactams, quinolones, and macrolides; Fig. 2). Among calves, the most prevalent AMR determinants were those conferring resistance to β-lactams, tetracyclines, florfenicol, and quinolones, including TEM-1, Tet(A), FloR, GyrA_S83L, and Tet(B), detected at least once in 21, 21, 19, 17, and 16 calves, respectively. The distribution of AMR determinant combinations among the 80 unique strains is shown in Table 3. Among the strains isolated at each sampling time, the distribution of the number of antimicrobial drugs or classes (tetracyclines, florfenicol, β-lactams, quinolones, and macrolides) against which a strain carried at least one resistance determinant [e.g., a combination of Tet(A) and Tet(B) counted as 1] is shown in Fig. 3. The median number of drugs or classes was 1.5, 1, 4, and 1 on D1, D2, D9, and D82, respectively. The distribution of selected AMR determinants among calves over time is illustrated in Fig. 4. The number of calves carrying E. coli strains with the combination of GyrA_D87N, GyrA_S83L, and ParC_S80I increased from two calves on D1 and D2 to 12 calves on D9 (including one calf treated with marbofloxacin between D2 and D9), then decreased to 0 on D82. Similarly, the number of calves carrying E. coli containing FloR increased from 4 and 5 calves on D1 and D2, respectively, to 16 and 10 calves on D9 and D82, respectively. Of note, the carriage of E. coli containing FloR increased before the incidence of any individual calf treatment with florfenicol (i.e., between D9 and D82). On D82, four of the five florfenicol-treated calves had E. coli carrying FloR, three of which already had strains carrying FloR on D9 (but of different STs). However, FloR carriage was also common among calves not directly exposed to florfenicol (6 out of 17 calves on D82). The carriage prevalence of the remaining AMR determinants of interest was relatively stable over time.
Fig 2.
Distribution among calves and dynamics over time of Escherichia coli isolates, with respective core genome MLST CT, sequence type (inner circle color), and determinants of resistance to antimicrobial drugs used during the study (outer circle color). For instance, on D1, calves 606, 602, and 614 carried isolates of different CT belonging to ST58 (same inner circle color) and harbored different resistance determinants’ combinations (different outer circle colors) or lack thereof. In cases of within-sample isolates with identical CT and shown resistance determinants (clonal or not), a single one of the strains is shown for improved clarity. Resistance determinants written in red represent an addition to the temporally previously observed combination of CT and resistance determinants. Connecting lines between isolates at different time points indicate clonal isolates based on CT. Superscripts indicate which calves and at which time interval between samplings received individual antimicrobial treatments: amoxicillin (a), florfenicol (f), gamithromycin (g), and marbofloxacin (m; first and second treatment incidence).
TABLE 3.
Distribution of determinants of antimicrobial resistance to tetracyclines, florfenicol, β-lactams, quinolones, and macrolides, alone or in combination, with corresponding sequence types, among 80 unique E. coli strains isolated from 22 calves
| AMR determinanta combinations | n | Sequence types |
|---|---|---|
| Tet(A) | 10 | 10 (n = 4), 58, 120, 329, 1302, 10850, and 13058 |
| Tet(B) | 10 | 56, 58 (n = 3), 101, 718, 2522 (n = 2), 3695, and 155 |
| Tet(B) + GyrA_S83L | 1 | 2522 |
| TEM-1 | 1 | 723 |
| TEM-1 + Tet(A) | 13 | 10, 58, 69 (n = 4), 101, 162, 410, 446, 6126, 10850, and 13059 |
| TEM-1 + Tet(B) | 1 | 69 |
| TEM-1 + Tet(A) + Mph(B) | 1 | 10 |
| TEM-1 + Tet(A) + Tet(B) + Mph(B) | 3 | 58 (n = 3) |
| TEM-1 + Tet(A) + FloR | 6 | 3057, 8580 (n = 2), and 58 (n = 3) |
| TEM-1 + Tet(A) + Tet(B) + FloR | 1 | 2522 |
| TEM-35 + Tet(B) + FloR | 1 | 949 |
| TEM-1 + Tet(A) + FloR + GyrA_S83L | 2 | 301 (n = 2) |
| TEM-1 + Tet(A) + FloR + GyrA_S83L + Mph(B) | 1 | 57 |
| TEM-1 + Tet(C) + FloR + GyrA_S83L | 1 | 301 |
| TEM-1 + Tet(A) + FloR + GyrA_D87Y | 1 | 783 |
| TEM-1 + Tet(A) + GyrA_D87Y | 1 | 117 |
| TEM-1 + Tet(A) + Tet(M) + GyrA_S83L | 1 | 58 |
| TEM-1 + Tet(B) + GyrA_D87N + GyrA_S83L + ParC_S80I | 1 | 162 |
| TEM-1 + Tet(A) + Tet(M) +FloR + GyrA_D87N + GyrA_S83L + ParC_S80I | 1 | 361 |
| OXA-1 +TEM-1 +Tet(B) +FloR + Mph(B) +GyrA_D87N + GyrA_S83L + ParC_S80I | 1 | 162 |
Antibiotic resistance determinants and their function: GyrA_D87N and GyrA_D87Y, DNA gyrase subunit A with substitution of aspartic acid (D) to asparagine (N) or to tyrosine (Y) at codon 87 for quinolone resistance; GyrA_S83L, DNA gyrase subunit A with substitution of serine (S) to leucine (L) at codon 83 for quinolone resistance; ParC_S80I, DNA topoisomerase 4 subunit A with substitution of serine (S) to isoleucine (I) at codon 80 for quinolone resistance; Tet(A), Tet(B), and Tet(C), tetracycline efflux; Tet(M), ribosomal protection for tetracycline resistance; OXA-1, TEM-1, and TEM-35, β-lactamase; FloR, phenicol efflux; and Mph(B), macrolide phosphotransferase.
Fig 3.
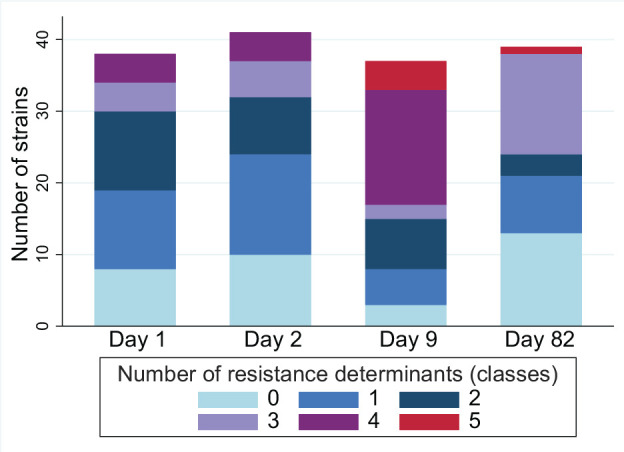
Distribution of the number of E. coli strains according to the number of resistance determinants they harbored against different antimicrobial drugs or classes (tetracyclines, florfenicol, β-lactams, quinolones, and macrolides) at four sampling times. Within a strain, multiple determinants against a single class were counted as one. After the removal of within-sample replicates, the total number of isolates on D1, D2, D9, and D82 was 38, 41, 37, and 39, respectively.
Fig 4.
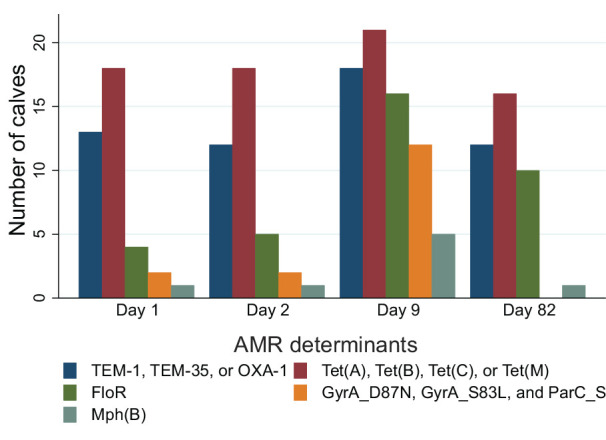
Distribution of the number of calves carrying at least one E. coli harboring selected AMR determinants or combination thereof, at four sampling times. Antibiotic resistance determinants and their function: GyrA_D87N, DNA gyrase subunit A with substitution of aspartic acid (D) to asparagine (N) at codon 87 for quinolone resistance; GyrA_S83L, DNA gyrase subunit A with substitution of serine (S) to leucine (L) at codon 83 for quinolone resistance; ParC_S80I, DNA topoisomerase 4 subunit A with substitution of serine (S) to isoleucine (I) at codon 80 for quinolone resistance; Tet(A), Tet(B), and Tet(C), tetracycline efflux; Tet(M), ribosomal protection for tetracycline resistance; OXA-1, TEM-1, and TEM-35, β-lactamase; FloR, phenicol efflux; and Mph(B), macrolide phosphotransferase.
Diversity and dissemination of E. coli strains
Overall, there was a high level of similarity between E. coli strains from D1 and D2 samplings, where isolates from D2 were clones of isolates from D1 in 19 calves (as indicated by connecting lines in Fig. 2). Conversely, only 4 out of 39 strains from D82 were clones of isolates identified at previous sampling times. The diversity of strains varied over time. On D1 and D2, 34 and 37 unique strains were identified, belonging to 20 and 21 STs, respectively. At each of these samplings, a single one of these strains [CT 11785 (ST10) on D1 and CT 11786 (ST56) on D2] had a clone also isolated from a different calf. At samplings on D9 and D82, 18 unique strains (13 STs) and 24 unique strains (14 STs), respectively, were identified, and 19 and 15 clones of these strains (all strains combined), respectively, were simultaneously found in other calves. Dispersion of some of the clones among calves is illustrated in Fig. 1 and 2. For instance, two strains [CT 11778 (ST361) and CT 11783 (ST301)] carrying resistance determinants to tetracyclines, β-lactams, florfenicol, and quinolones were initially isolated from a single calf (ID 609 and 604, respectively) at D1 and D2, and from 10 and 6 calves, respectively, on D9. Together, these two strains were the main drivers (16/21 strains) for the increased prevalence of florfenicol and quinolone resistance determinant carriage on D9. However, neither of these clones were detected in any calf on D82. Also noticeable in Fig. 2 is the fact that a combination of AMR determinants, TEM-1, FloR, and Tet(A), appeared on D82 in multiple calves harboring clones that were not observed at previous samplings [CT 11784 (ST58) and CT 12181 (ST8580)]. On the other hand, the clonal spread of strains without AMR determinants was also observed, such as strains of CT 12697 (ST58) detected in five calves on D82.
Of 36 groups of clonal isolates (based on identical CT) that were either repeatedly isolated in a calf over time or shared between calves or both, the isolates were identical within group for 26 CT, whereas the isolates belonging to 10 CT differed within CT with regards to AMR genes (n = 5), virulence genes (n = 4), or O-serotype (n = 1). One isolate [CT 11779 (ST2522)] at D82 carried the AMR determinants TEM-1, FloR, and Tet(A), which were absent from the other strains of CT 11779 (which all were identical clones) isolated on D9 and D82. Inspection of plasmid incompatibility groups by PlasmidFinder revealed that this isolate carried a plasmid IncI1-Iα, whereas no plasmid incompatibility group was detected in other CT 11779 clones. The strain was further investigated by comparing its pangenome with that of another CT 11779 isolate from D9. Acquired singleton gene clusters, representing the acquired material that differed between the two strains, were extracted as concatenated DNA and compared with the GenBank database using the nucleotide-BLAST algorithm (www.ncbi.nlm.nih.gov). There was a 100% similarity with the sequence of an E. coli plasmid p1514kTc1_N_I1_ST113 from the database (GenBank accession no. NZ_MW800641), indicating the acquisition of a plasmid. To investigate whether the AMR genes acquired by this strain were located on a plasmid, the contigs from the assembly were aligned with the plasmid p1514kTc1_N_I1_ST113 using Mauve (option MCM). As a result, determinants of AMR to β-lactams (TEM-1), aminoglycosides [APH(3″)-Ib and APH(6)-Id], and sulfonamides (Sul2) were identified, flanked by the insertion sequence IS26 (associated with the mobilization of AMR genes), along with the plasmid replication initiation protein RepZ (associated with IncI plasmids). Thus, the acquisition of these AMR determinants was confirmed to be associated with the acquisition of a plasmid.
DISCUSSION
The objectives of this study were to describe the AMR dynamics in 264 commensal fecal E. coli isolates collected longitudinally from calves in the rearing period subjected to antimicrobial group treatments and to characterize them using WGS for the identification of AMR genes and strain typing. Already upon arrival, a majority of calves (18/22) harbored E. coli carrying AMR genes. Genotypic AMR to tetracyclines, β-lactams, streptomycin, and sulfonamides, which are antimicrobial classes commonly used in cattle, was predominant among the study calves and among the collection of unique strains. This population of resistant strains is likely well established in calves, possibly belonging to the normal flora, and maintained through frequent AMU. This finding is similar to previous reports of phenotypic AMR in commensal E. coli in veal calves in Switzerland, France, and Belgium (3, 9, 10). In these previous studies, variations in the proportions of isolates showing phenotypic AMR between two different time points were observed. In the present study, variations in the number of calves carrying isolates with specific AMR determinants were also observed. The larger number of sampling times than in aforementioned studies (four vs two) provided additional detail, for instance, the observed initial increase in the number of calves carrying isolates with quinolone resistance determinants, followed by a decrease at the last sampling time, namely 82 and 81 days after the calves’ arrival to the research facility and the first antimicrobial treatment, respectively. However, it cannot be excluded that short-term changes in AMR carriage between sampling times could have been missed. Moreover, the selection of three colonies per sample, rather than a single one as in the aforementioned studies, allowed to capture a wider diversity of E. coli strains, within samples and overall.
The overall diversity of E. coli strains varied over time (range of 18–37 unique strains per sampling time). A large diversity of strains was observed upon arrival, consistent with previous reports and likely attributable to the diverse origins of the calves (10, 11). There was limited variation between D1 and D2, which would suggest that the level and duration of exposure of the fecal flora to oxytetracycline administered via parenteral injection to the calves between these two samplings were insufficient to elicit a major shift in the dominant flora. In contrast, on D9 (i.e., 5 days after the start of in-feed doxycycline treatment), the number of unique strains decreased, and the number of clonal strains shared among calves increased. In studies on ampicillin-resistant E. coli in beef calf cohorts (not subjected to antimicrobial group treatment), the predominant genotypes (assessed by PFGE fingerprinting) showed temporal variations, but the number of different genotypes at each sampling remained relatively stable (18, 19). Based on the dynamics of phenotypic AMR profiles, Catry et al. (3) suggested that oral antimicrobial group treatment in veal calves results in the selection of a large number of resistant strains rather than a small number of resistant clones. This was partially supported by our findings based on WGS. In contrast, in studies on ESBL-producing E. coli subpopulations in veal calves, strain typing revealed an important reduction in strain diversity and the dominance of a few clones at the end of the study period (10 weeks and 5 months, respectively) (10, 11). This contrast suggests that AMR dynamics may differ, not only based on the diversity of animal sources and antimicrobial treatment but also depending on whether the bacterial populations of interest are E. coli from the dominant flora or the ESBL-producing subdominant flora.
An additional observation in the present study was that only 10% of isolates on D82 were clones of previously identified strains. Similar observations have been reported for ESBL-producing strains in veal calves (11). Some strains may have initially been part of the calves’ subdominant flora but gone undetected due to the isolate selection strategy since only three colonies per plate were selected. Other strains may have been acquired from the barn environment. These previously undetected strains carried on average fewer AMR determinants than the strains isolated on D9, which they had displaced or replaced (1.56 vs 2.95 AMR determinants).
In previous studies with a focus on ESBL-producing Enterobacterales, the spread of AMR genes was reported to occur through clonal dissemination of strains, although horizontal transfer of plasmids between strains and of genes between plasmids was also suspected (10–13). In the present study, strains were not isolated using a medium selecting specific resistant strains such as ESBL-producing Enterobacterales, but AMR determinants in the dominant E. coli fecal flora also appeared to spread among calves predominantly by clonal strains. The concurrent acquisition of a plasmid and AMR genes was suspected in one isolate of CT 11779, although not all of the acquired AMR genes could be confirmed to be plasmid associated. However, it cannot be excluded that the transfer of plasmids and associated AMR genes also occurred between clones of different CT. Further studies using a more systematic, in-depth characterization of isolates with long-reads WGS may provide supporting evidence for this AMR determinant transmission pathway. Such genomic characterization has allowed the identification of different patterns of AMR gene dissemination dynamics between dairy farms in a previous study (15). The generalizability of the present study’s findings regarding AMR determinant transmission dynamics is limited by the inclusion of a single-rearing facility and cohort. Indeed, based on a high genetic diversity of ESBL-producing E. coli strains from slaughtered animals, other authors postulated that the spread of ESBL genes among E. coli in animal populations occurs either by plasmids or by a diversity of pathways (17, 20). This means that different modes of transmission of AMR determinants may concomitantly be at play, and the predominant mode may vary according to the studied bacterial population and host population (e.g., at herd or national level).
Antimicrobial use is recognized as a major driver for AMR, and the association of antimicrobial group treatments with decreased phenotypic antimicrobial susceptibility in commensal E. coli in veal and dairy calves was reported in previous studies (8, 21). In the present study, all calves were treated parenterally with oxytetracycline and orally with doxycycline, and a third (8/22) of calves were additionally treated with a variety of other antimicrobial drugs. Tetracycline resistance determinants were present in numerous strains among different calves already prior to treatment, and the selection pressure of treatment may have contributed to their maintenance in the E. coli population until D82. Additionally, the proportion of calves carrying isolates harboring florfenicol or quinolone resistance determinants increased over time, including prior to florfenicol treatments and in calves that were not treated with these antimicrobial classes. Similarly, in a previous study in stocker calves receiving macrolide metaphylaxis, the proportion of quinolone-resistant M. haemolytica increased 7 days after treatment, compared to pre-treatment (22). In another study in preweaned calves, those receiving macrolide metaphylaxis had an increased proportion of quinolone-resistant E. coli compared to untreated calves from day 2 to day 7 after treatment (23). A possible explanation for these observations was co-selection, where the use of an antimicrobial drug results in the selection of resistance to drugs of different classes. In the present study, all isolates carrying florfenicol or quinolone resistance determinants also carried tetracycline resistance determinants and showed an increase in prevalence, which could be the result of co-selection following oxytetracycline and doxycycline treatments. Regarding the impact of individual calf treatments on AMR carriage or maintenance, a clear trend was neither identified among calves that received additional individual treatments nor in other calves in this study.
A strength of this longitudinal study is that multiple sampling points over an extended time period were included. However, due to the large time gap between the last day of group treatment (D12), or individual treatments where applicable, and the next sample collection (D82), short-term changes in AMR carriage during that period might have been missed. Additionally, three colonies per fecal sample culture (morphologically different whenever possible) were selected for characterization, which increased the total number of different strains to 155 (compared to 88 if a single isolate per sample had been selected). This selection method provided a measure of within-sample strain diversity as it revealed that ≥2 strains could be retrieved from a majority (53/88) of samples. The optimal number of isolates needed to capture the full spectrum of E. coli strains in the calves’ flora is unknown. In a previous study where 20 colonies per swab were characterized based on a combination of phenotypic AMR pattern, biotype (based on fermentation pattern), and O-serogroup (where possible), an overall mean of 5.17 strains per swab was reported (24). Although it is conceivable that the selection of five or six isolates per sample in the present study could have resulted in a larger diversity of strains, the proportion of replicate strains would likely also have been higher. Nevertheless, the selection of three isolates allowed for the detection of variations in strain diversity and in their clonal spread during and after a course of antimicrobial treatment.
In this observational study, fecal samples were longitudinally collected from a group of calves reared in conditions reflecting those of Swiss veal-fattening facilities (e.g., multiple origins, group housing, and antimicrobial group treatment). In the calf fattening industry, antimicrobial group treatments represent a major portion of AMU and are therefore a key target for reducing AMU (2–5). Strain-typing data from the present study suggest that the clonal spread of commensal fecal E. coli easily occurs among commingled calves. In addition to the spread of AMR determinants by clonal strains, the location of AMR determinants on mobile genetic elements such as plasmids facilitates their transfer between bacteria, including potential transfer from commensal to pathogenic and zoonotic bacteria. With regards to calf health, whether AMR selection and their maintenance in commensal bacteria have a negative impact on response to therapeutic antimicrobial treatment of diseases caused by pathogenic bacteria remains to be determined (22).
In conclusion, in this cohort of calves subjected to antimicrobial group treatments, analysis of the genotypic AMR dynamics among commensal fecal E. coli isolates revealed a reduction in strain diversity during treatment. Furthermore, clonal dissemination of strains among calves was the primary mode for the spread of AMR determinants, although the acquisition of plasmid-mediated AMR genes by a strain was also suspected. Despite the rapid initial clonal expansion of some strains, their replacement by other strains by the time of the last sampling led to a reduction in carriage prevalence for some AMR determinants, while others were maintained, underlining the complexity of AMR dynamics in the dominant fecal E. coli flora of calves during the rearing period.
MATERIALS AND METHODS
Animals and sample collection
This was an observational longitudinal study. Twenty-two calves were gathered for rearing in a research facility in Switzerland. A total of 8 calves first transited through a livestock market, and the remaining 14 calves were collected from 12 different farms of origin and transported together. Upon arrival, the calves were examined for clinical signs of disease, weighed, and assigned unique identification numbers. Four calves had diarrhea (undetermined cause). The calves were then housed together in one pen, which had been previously cleaned and disinfected, and remained in this pen for the entire study period. In the same barn were housed 22 other calves that were delivered the following day (but not included in the present study), 5 of which had fever upon clinical examination at arrival. The calves were fed milk replacer through an automatic distribution system. The daily amount of milk replacer was gradually increased from 4 L upon arrival to 6 L at 3 weeks after arrival, then gradually decreased until weaning, at 7 weeks after arrival. Concentrates, hay, and corn silage were offered from week 1, week 2, and week 5, respectively. The calves were reared over 13 weeks until they reached 160 kg on average. Upon arrival, all calves received iron (563 mg/calf, orally) and selenium (1.6 mg/calf, subcutaneously) supplementation. On the day after arrival (D1), all study calves received metaphylactic antimicrobial treatment with long-acting oxytetracycline, 20 mg/kg of body weight, intramuscularly. From D4 to D12, the calves received doxycycline in milk replacer at a daily rate of 17.5 mg/kg of body weight. Any additional antimicrobial treatment of individual calves during the rearing period was administered at the discretion of the treating veterinarian and was recorded. Rectal swab samples were collected on D1 (before the first treatment), D2 (after intramuscular antibiotic treatment), D9 (during oral antibiotic treatment), and D82 (end of the rearing period). The swabs (TRANSWAB Gel Amies Plain, mwe, UK) were transported at room temperature in Amies medium to the laboratory for processing within 4 hours. The sampling period spanned from February to April 2020.
Bacterial culture and species identification
The swabs were spread directly onto Enterobacterales-selective BROLAC agar (Thermo Fisher Scientific, Waltham, USA), and the plates were incubated at 37°C for 24 hours. Three lactose-fermenting colonies exhibiting a dissimilar morphology were selected per plate to take the within-sample strain diversity into account. If no difference in morphology was observed, the colonies were chosen randomly. Species identification was assigned using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (Microflex LT, Bruker Daltonics GmbH, Bremen, Germany). If one of the selected colonies was not identified as E. coli (in less than 10 instances), an additional colony was selected in order to obtain a total of three E. coli per sample. Isolates identified as E. coli were regrown in pure culture on trypticase soy agar containing 5% sheep blood (TSA-SB; Becton, Dickinson and Company, NJ, USA) and stored in 30% glycerol at −80°C.
Whole-genome sequencing data
For library preparation, the isolates were grown on TSA-SB, and bacterial DNA was extracted using proteinase K and mechanical disruption using glass beads (PowerBead, Qiagen, Hilden, Germany). The extracted DNA was purified using the AMPure XP paramagnetic bead-based chemistry (Beckman Coulter, Brea, CA, USA). Library preparation was performed using Nextera DNA Flex Library Prep Kit according to the manufacturer’s instructions (Illumina Inc., San Diego, CA, USA). All isolates were subjected to WGS on an Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) at the Next Generation Sequencing Platform, Institute of Genetics, University of Bern. The software SeqSphere+ (version 7, Ridom GmbH, Münster, Germany) was used for the determination of ST, CT, in silico serotype, as well as the identification of resistance and virulence genes. The sequences were also screened for acquired resistance genes using the databases ResFinder (https://bitbucket.org/genomicepidemiology/resfinder_db.git, accessed 14 October 2021) and for resistance-associated chromosomal mutations using PointFinder (https://bitbucket.org/genomicepidemiology/pointfinder_db.git, accessed 22 April 2022). Determination of plasmid incompatibility groups was conducted by in silico replicon typing using the PlasmidFinder database (https://bitbucket.org/genomicepidemiology/plasmidfinder_db.git, accessed 20 March 2022).
The sequence data set was first inspected for within-sample (same calf and time) replicate isolates based on identical CT, in silico serotype, AMR genes, virulence genes, and plasmid incompatibility groups. Replicates were removed from the data set in order to prevent the overall strain diversity from being affected by the within-sample dominance of a strain. A phylogenetic tree of the remaining 155 isolates was generated using Ridom SeqSphere+ (version 8.5.1, Ridom GmbH, Münster, Germany) and visualized and annotated using iTOL (version 6.7.5, https://itol.embl.de). The data set was then inspected for clonal isolates within calves over time, as well as between calves. Isolates were defined as clones based on identical CT, whereas identical clones were additionally defined based on AMR genes and virulence genes. Replicates of identical clones were removed to create a data set of unique strains to prevent a disproportionate weight of clonal strains on the reported distribution of AMR determinants. When clonal isolates carried combinations of AMR determinants that differed from one another within a group (sharing the same CT), differences in plasmid incompatibility groups were inspected.
ACKNOWLEDGMENTS
The authors thank the experimental farm staff of Agroscope, Posieux, Switzerland for their excellent support and collaboration.
The study was financed by internal funds of the Institute of Veterinary Bacteriology, University of Bern, Switzerland (REF 660-50) to V.P. and internal funds of the Clinic for Ruminants, University of Bern, Switzerland (REF 33-014) to M.M. and V.B.G.
Contributor Information
Véronique Bernier Gosselin, Email: veronique.bernier@unibe.ch.
Aude A. Ferran, Innovations Therapeutiques et Resistances, Toulouse, France
DATA AVAILABILITY
The Illumina raw reads were deposited into the Sequence Read Archive (SRA) database of the National Center for Biotechnology Information (NCBI) under BioProject accession number PRJNA1005565 with BioSample accession numbers SAMN36988384 to SAMN36988647.
ETHICS APPROVAL
The procedures were approved by the competent animal care and use authority (authorization for animal experimentation FR-31589).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.03214-23.
Genome sequences of 264 Escherichia coli isolated from 22 calves.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Lava M, Schüpbach-Regula G, Steiner A, Meylan M. 2016. Antimicrobial drug use and risk factors associated with treatment incidence and mortality in Swiss veal calves reared under improved welfare conditions. Prev Vet Med 126:121–130. doi: 10.1016/j.prevetmed.2016.02.002 [DOI] [PubMed] [Google Scholar]
- 2. Lava M, Pardon B, Schüpbach-Regula G, Keckeis K, Deprez P, Steiner A, Meylan M. 2016. Effect of calf purchase and other herd-level risk factors on mortality, unwanted early slaughter, and use of antimicrobial group treatments in Swiss veal calf operations. Prev Vet Med 126:81–88. doi: 10.1016/j.prevetmed.2016.01.020 [DOI] [PubMed] [Google Scholar]
- 3. Catry B, Dewulf J, Maes D, Pardon B, Callens B, Vanrobaeys M, Opsomer G, de Kruif A, Haesebrouck F. 2016. Effect of antimicrobial consumption and production type on antibacterial resistance in the bovine respiratory and digestive tract. PLoS One 11:e0146488. doi: 10.1371/journal.pone.0146488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pardon B, Catry B, Dewulf J, Persoons D, Hostens M, De Bleecker K, Deprez P. 2012. Prospective study on quantitative and qualitative antimicrobial and anti-inflammatory drug use in white veal calves. J Antimicrob Chemother 67:1027–1038. doi: 10.1093/jac/dkr570 [DOI] [PubMed] [Google Scholar]
- 5. Schnyder P, Schönecker L, Schüpbach-Regula G, Meylan M. 2019. Effects of management practices, animal transport and barn climate on animal health and antimicrobial use in Swiss veal calf operations. Prev Vet Med 167:146–157. doi: 10.1016/j.prevetmed.2019.03.007 [DOI] [PubMed] [Google Scholar]
- 6. BLV (Bundesamt für Lebensmittelsicherheit und Veterinärwesen) . 2022. IS ABV Erste Übersicht der Verschreibungen von Antibiotika bei Nutztieren in der Schweiz 2020. Available from: https://www.blv.admin.ch/dam/blv/de/dokumente/tiere/tierkrankheiten-und-arzneimittel/tierarzneimittel/jahresbericht-isabv-2020.pdf.download.pdf/JAHRESBERICHT_ISABV_Daten_2020_final_Hauptteil_D.pdf. Retrieved 9 Apr 2022.
- 7. Di Labio E, Regula G, Steiner A, Miserez R, Thomann A, Ledergerber U. 2007. Antimicrobial resistance in bacteria from Swiss veal calves at slaughter. Zoonoses Public Health 54:344–352. doi: 10.1111/j.1863-2378.2007.01071.x [DOI] [PubMed] [Google Scholar]
- 8. Schönecker L, Schnyder P, Overesch G, Schüpbach-Regula G, Meylan M. 2019. Associations between antimicrobial treatment modalities and antimicrobial susceptibility in Pasteurellaceae and E. coli isolated from veal calves under field conditions. Vet Microbiol 236:108363. doi: 10.1016/j.vetmic.2019.07.015 [DOI] [PubMed] [Google Scholar]
- 9. Becker J, Perreten V, Steiner A, Stucki D, Schüpbach-Regula G, Collaud A, Rossano A, Wüthrich D, Muff-Hausherr A, Meylan M. 2022. Antimicrobial susceptibility in E. coli and Pasteurellaceae at the beginning and at the end of the fattening process in veal calves: comparing ‘outdoor veal calf’ and conventional operations. Vet Microbiol 269:109419. doi: 10.1016/j.vetmic.2022.109419 [DOI] [PubMed] [Google Scholar]
- 10. Gay E, Bour M, Cazeau G, Jarrige N, Martineau C, Madec J-Y, Haenni M. 2019. Antimicrobial usages and antimicrobial resistance in commensal Escherichia coli from veal calves in France: evolution during the fattening process. Front Microbiol 10:792. doi: 10.3389/fmicb.2019.00792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hordijk J, Mevius DJ, Kant A, Bos MEH, Graveland H, Bosman AB, Hartskeerl CM, Heederik DJJ, Wagenaar JA. 2013. Within-farm dynamics of ESBL/AmpC-producing Escherichia coli in veal calves: a longitudinal approach. J Antimicrob Chemother 68:2468–2476. doi: 10.1093/jac/dkt219 [DOI] [PubMed] [Google Scholar]
- 12. Bello Gonzalez TDJ, Kant A, Dijkstra Q, Marcato F, van Reenen K, Veldman KT, Brouwer MSM. 2022. Changes in fecal carriage of extended-spectrum beta-lactamase producing Enterobacterales in Dutch veal calves by clonal spread of Klebsiella pneumoniae. Front Microbiol 13:866674. doi: 10.3389/fmicb.2022.866674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liebana E, Batchelor M, Hopkins KL, Clifton-Hadley FA, Teale CJ, Foster A, Barker L, Threlfall EJ, Davies RH. 2006. Longitudinal farm study of extended-spectrum beta-lactamase-mediated resistance. J Clin Microbiol 44:1630–1634. doi: 10.1128/JCM.44.5.1630-1634.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hordijk J, Fischer EAJ, van Werven T, Sietsma S, Van Gompel L, Timmerman AJ, Spaninks MP, Heederik DJJ, Nielen M, Wagenaar JA, Stegeman A. 2019. Dynamics of faecal shedding of ESBL- or AmpC-producing Escherichia coli on dairy farms. J Antimicrob Chemother 74:1531–1538. doi: 10.1093/jac/dkz035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tello M, Ocejo M, Oporto B, Lavín JL, Hurtado A. 2022. Within-farm dynamics of ESBL-producing Escherichia coli in dairy cattle: resistance profiles and molecular characterization by long-read whole-genome sequencing. Front Microbiol 13:936843. doi: 10.3389/fmicb.2022.936843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bastard J, Haenni M, Gay E, Glaser P, Madec JY, Temime L, Opatowski L. 2021. Drivers of ESBL-producing Escherichia coli dynamics in calf fattening farms: a modelling study. One Health 12:100238. doi: 10.1016/j.onehlt.2021.100238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haenni M, Châtre P, Métayer V, Bour M, Signol E, Madec J-Y, Gay E. 2014. Comparative prevalence and characterization of ESBL-producing Enterobacteriaceae in dominant versus subdominant enteric flora in veal calves at slaughterhouse, France. Vet Microbiol 171:321–327. doi: 10.1016/j.vetmic.2014.02.023 [DOI] [PubMed] [Google Scholar]
- 18. Hoyle DV, Davison HC, Knight HI, Yates CM, Dobay O, Gunn GJ, Amyes SGB, Woolhouse MEJ. 2006. Molecular characterisation of bovine faecal Escherichia coli shows persistence of defined ampicillin resistant strains and the presence of class 1 integrons on an organic beef farm. Vet Microbiol 115:250–257. doi: 10.1016/j.vetmic.2006.01.006 [DOI] [PubMed] [Google Scholar]
- 19. Hoyle DV, Yates CM, Chase-Topping ME, Turner EJ, Davies SE, Low JC, Gunn GJ, Woolhouse MEJ, Amyes SGB. 2005. Molecular epidemiology of antimicrobial-resistant commensal Escherichia coli strains in a cohort of newborn calves. Appl Environ Microbiol 71:6680–6688. doi: 10.1128/AEM.71.11.6680-6688.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geser N, Stephan R, Kuhnert P, Zbinden R, Kaeppeli U, Cernela N, Haechler H. 2011. Fecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae in swine and cattle at slaughter in Switzerland. J Food Prot 74:446–449. doi: 10.4315/0362-028X.JFP-10-372 [DOI] [PubMed] [Google Scholar]
- 21. Berge ACB, Moore DA, Sischo WM. 2006. Field trial evaluating the influence of prophylactic and therapeutic antimicrobial administration on antimicrobial resistance of fecal Escherichia coli in dairy calves. Appl Environ Microbiol 72:3872–3878. doi: 10.1128/AEM.02239-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Woolums AR, Karisch BB, Frye JG, Epperson W, Smith DR, Blanton J, Austin F, Kaplan R, Hiott L, Woodley T, Gupta SK, Jackson CR, McClelland M. 2018. Multidrug resistant Mannheimia haemolytica isolated from high-risk beef stocker cattle after antimicrobial metaphylaxis and treatment for bovine respiratory disease. Vet Microbiol 221:143–152. doi: 10.1016/j.vetmic.2018.06.005 [DOI] [PubMed] [Google Scholar]
- 23. Pereira RV, Altier C, Siler JD, Mann S, Jordan D, Warnick LD. 2020. Longitudinal effects of enrofloxacin or tulathromycin use in preweaned calves at high risk of bovine respiratory disease on the shedding of antimicrobial-resistant fecal Escherichia coli. J Dairy Sci 103:10547–10559. doi: 10.3168/jds.2019-17989 [DOI] [PubMed] [Google Scholar]
- 24. Hinton M, Rixson PD, Allen V, Linton AH. 1984. The persistence of drug resistant Escherichia coli strains in the majority faecal flora of calves. J Hyg (Lond) 93:547–557. doi: 10.1017/s0022172400065128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genome sequences of 264 Escherichia coli isolated from 22 calves.
Data Availability Statement
The Illumina raw reads were deposited into the Sequence Read Archive (SRA) database of the National Center for Biotechnology Information (NCBI) under BioProject accession number PRJNA1005565 with BioSample accession numbers SAMN36988384 to SAMN36988647.