Abstract
The TAATGARAT motif in the herpes simplex virus (HSV) immediate-early (IE) gene promoters plays a key role in their activation by the Oct-1–Vmw65 complex, but its role in mediating inhibitory effects of cellular octamer-binding proteins is less clear. We have used indicator viruses containing reporter constructs with different IE promoters driving a reporter β-galactosidase gene within the viral genome to investigate this. We showed that deletion of the upstream IE promoter region containing the TAATGARAT motifs abolishes the inhibitory effect of the cellular octamer-binding proteins Oct-2.4 and Oct-2.5 on the viral IE promoter. This inhibitory effect can be restored by addition of a single TAATGARAT motif to the minimal promoter within the viral genome. Hence, the TAATGARAT motif can indeed mediate both positive and negative effects of cellular transcription factors when it is located within the viral genome.
The herpes simplex virus (HSV) immediate-early (IE) gene promoters contain multiple copies of the sequence TAATGARAT (R = purine) (1), which is related to the octamer motif (ATGCAAAT) found in a number of cellular gene promoters (5) (Fig. 1). In addition, the IE1 (ICP0) promoter contains multiple composite motifs consisting of overlapping octamer-TAATGARAT sequences (17) (Fig. 1). Such motifs play a critical role in the viral lytic cycle. Thus, they act as a target for transactivation by a complex consisting of the cellular octamer-binding protein Oct-1 and the virion transactivator Vmw65 (VP16, α-TIF) (7, 16, 18). This complex binds to the TAATGARAT motifs in the IE genes and greatly stimulates their transcription, causing the high-level IE gene expression that occurs during the normal lytic cycle.
FIG. 1.
Comparison of the consensus octamer sequences found in cellular gene promoters (5) with the consensus TAATGARAT sequence found in HSV IE gene promoters (1). The oligonucleotides used in this study are also indicated. They contain an overlapping octamer-TAATGARAT motif from the IE1 promoter (OT) or a simple TAATGARAT motif from the IE3 promoter (T).
It is also possible, however, that the binding of cellular transcription factors to the TAATGARAT motif is involved in silencing of the IE promoters in neuronal cells, resulting in the absence of IE gene expression that is observed when such cells are latently infected with HSV (4, 20) (for reviews, see references 9 and 19). Although the precise nature of the octamer-binding proteins that might mediate this inhibitory effect remains unclear, it has been demonstrated that cellular octamer-binding proteins can inhibit viral growth and the activity of the IE promoters.
Thus, BHK fibroblast cell lines (14) artificially engineered (by transfection of appropriate cDNA clones) to express either the Oct-2.4 or Oct-2.5 isoform of the cellular octamer-binding protein Oct-2 (which are normally expressed in neuronal cells [13]) show dramatically reduced permissiveness for the HSV lytic cycle compared to control BHK cells (12). Similarly, both we (11, 13) and others (15) have shown that specific isoforms of Oct-2 can repress the basal activity of the IE promoters and their transactivation by Vmw65 in cotransfection assays, and a similar effect on transactivation has also been demonstrated for the IE promoter of a related virus, varicella-zoster virus (15).
Although such studies indicate that specific isoforms of Oct-2 can repress the HSV IE promoters and inhibit the viral lytic cycle, they do not indicate that these effects are mediated via the TAATGARAT motifs in the IE promoters. Thus, it has not yet been demonstrated that deletion of the TAATGA RAT-containing region from the IE promoters abolishes the inhibitory effect. Similarly, it has not been shown that linkage of a TAATGARAT motif to a minimal IE promoter will confer repression by Oct-2 or that such effects can be demonstrated within the context of the viral genome.
We have therefore prepared recombinant indicator viruses in which various IE promoter constructs driving the expression of a β-galactosidase reporter gene have been integrated into the nonessential viral gene UL43 in HSV type 1 (HSV-1) strain 17. These viruses have been used to infect BHK cell lines expressing different isoforms of Oct-2 in order to determine the role of the TAATGARAT motif in mediating responses to Oct-2 within the context of the viral genome.
Initially, four constructs were prepared, each containing an IE promoter linked to a lacZ reporter gene. The basic construct (IE1LacZ) contains the IE1 promoter sequences from nucleotide −585 to +150 relative to the transcriptional start site. It thus contains three octamer-TAATGARAT motifs (6). In contrast, the IEminLacZ construct has been truncated to nucleotide −185 relative to the transcriptional start site so that it lacks all octamer-TAATGARAT motifs. The IEmin+OTLacZ and IEmin+TLacZ constructs are identical to the IEminLacZ construct except that they contain (respectively) a single overlapping octamer-TAATGARAT motif (IEmin+OT) or a TAATGARAT motif (IEmin+T) cloned immediately upstream of the minimal IE (IEmin) promoter.
All four constructs were cloned into flanking regions of the UL43 gene and introduced into the UL43 gene of wild-type virus (HSV-1 strain 17) by standard recombination techniques (2, 3). Recombinant viruses were purified and grown as previously described (8). All viruses were shown to be free of contamination with nonrecombinant viruses by staining of plaques with the chromogenic β-galactosidase substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and the structure of each recombinant was confirmed by Southern blotting of viral DNA (data not shown). Since the UL43 gene is not essential for virus growth, these indicator viruses retain the full growth potential of the wild-type virus but can be used to study the processes regulating viral promoter activity within the context of the viral genome.
These viruses were then used to infect a series of cell lines, derived from BHK-21 cells (8), which had been obtained by stable transfection with expression vectors containing cDNA clones encoding different isoforms of Oct-2 (12). These isoforms were generated by alternative splicing of the transcript derived from the single gene encoding Oct-2 (23); such alternative splicing has been shown to take place in a cell type-specific manner and to produce isoforms with different effects on the activity of octamer-TAATGARAT-containing promoters (13). Thus, the predominant form of Oct-2 produced in B lymphocytes, Oct-2.1, has a strong C-terminal activation domain and activates the IE promoters in cotransfection assays (10, 13). In contrast, the predominant forms expressed in neuronal cells, Oct-2.4 and Oct-2.5, lack this domain and repress the IE promoters (10, 13). In agreement with this, we previously showed that these BHK cell lines expressing Oct-2.4 or Oct-2.5 showed dramatically reduced permissiveness for the growth of HSV compared to parental BHK cells or cells expressing Oct-2.1 (12).
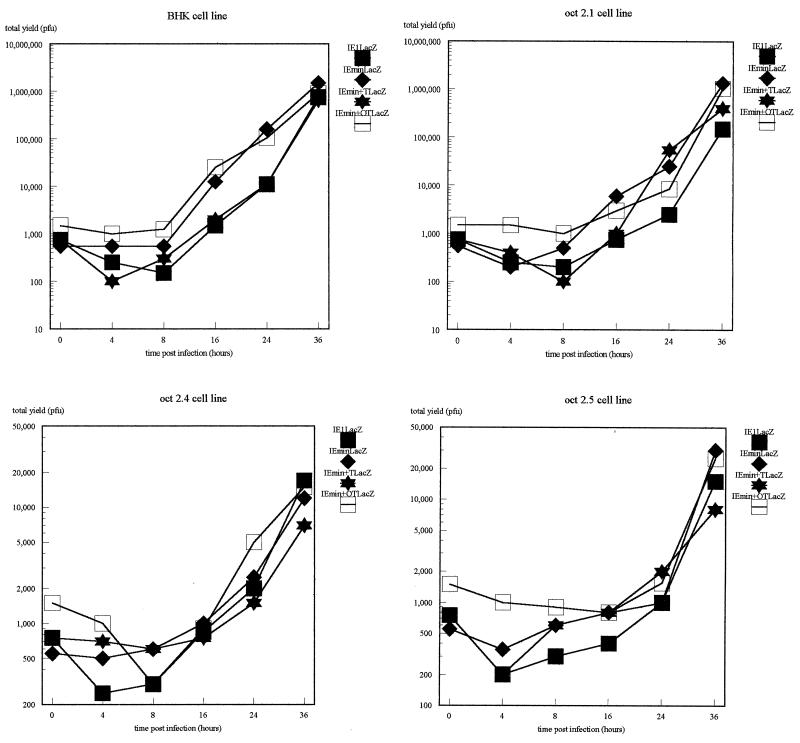
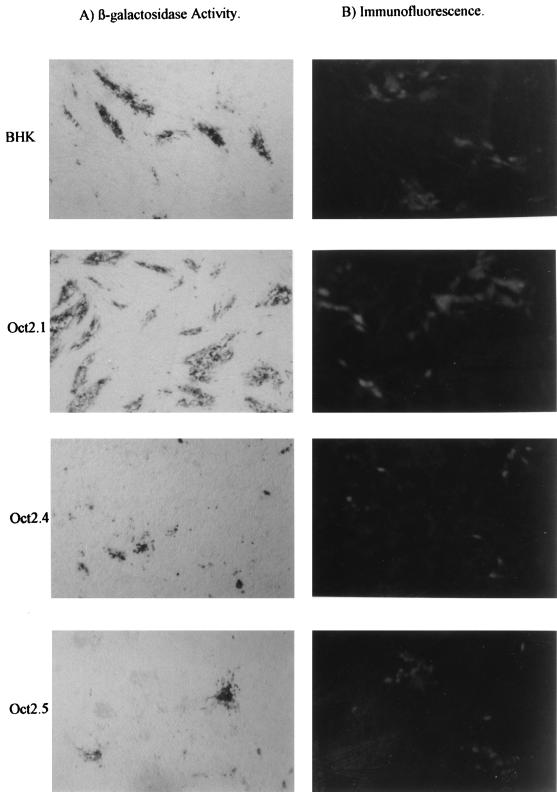
When these cell lines were infected with the indicator viruses, we observed, as expected, differences in permissiveness between the different cell lines; these differences, as expected, affected all of the viruses equally since the only difference between them lies in the indicator gene inserted into the UL43 locus. This is clearly evident in the growth curves for the viruses on each cell line, which show both the similar growth of all four viruses on the same cell line and the drastically reduced growth of all viruses on the Oct-2.4 and Oct-2.5 cell lines (Fig. 2). The differences in plaque size (as determined both by staining with an anti-HSV-1 polyclonal antibody and by the size of the area staining positively for β-galactosidase) for one of the four viruses on all four cell lines are illustrated in Fig. 3.
FIG. 2.
Growth curves of the IE1LacZ (solid boxes), IEmin (diamonds), IEmin+T (stars), and IEmin+OT (open boxes) viruses for each of the four cell lines.
FIG. 3.
Representative plaques stained with anti-HSV antibody or for β-galactosidase activity after the IEmin+OT virus was plated on parental BHK cells or cells engineered to express the different isoforms of Oct-2.
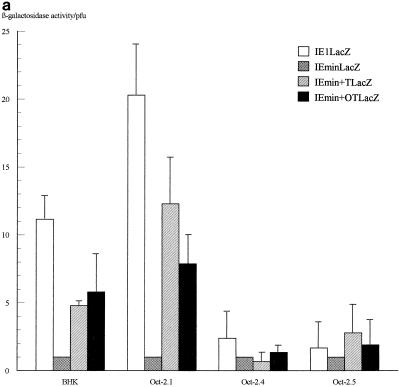
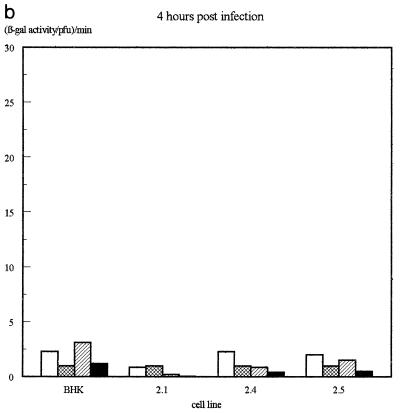
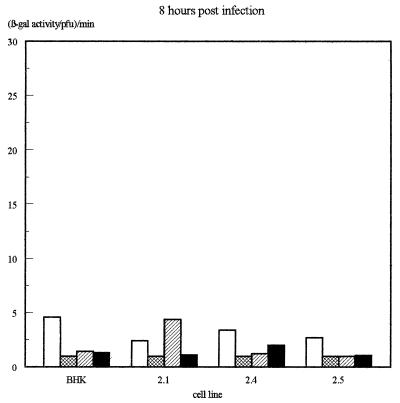
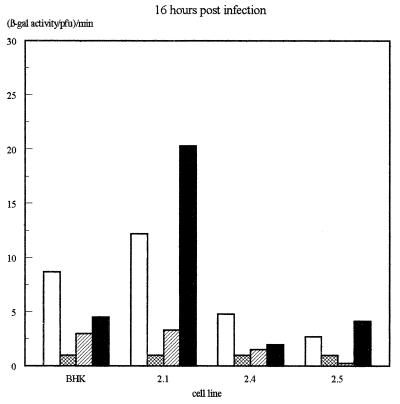
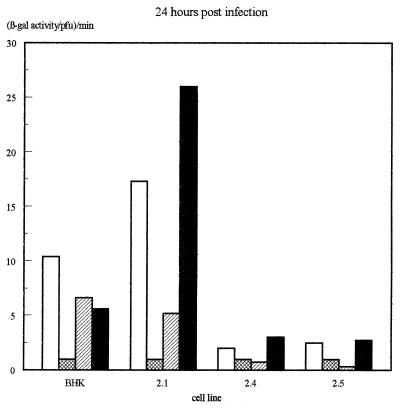
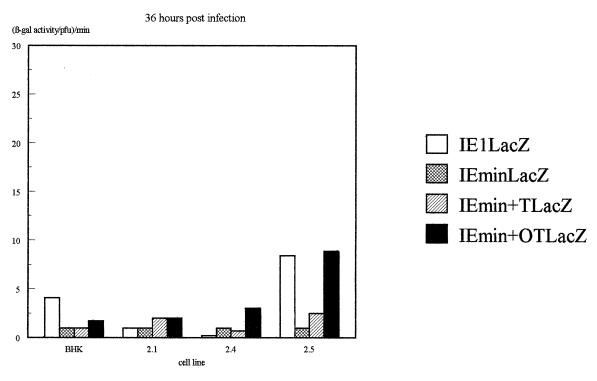
The β-galactosidase activity produced by the different IE promoters in each virus on each of the four cell lines was then assessed quantitatively by measuring total β-galactosidase activity in infected-cell extracts and normalizing for virus growth as determined by titrating the virus produced in each infection. The results of several infections, determined 16 h after infection, are illustrated in Fig. 4a, while the β-galactosidase levels obtained at various time points in viral growth experiments (Fig. 2) are illustrated in Fig. 4b.
FIG. 4.
Assay of β-galactosidase activity produced by each virus on each of the cell lines. (a) β-Galactosidase activities assayed at 16 h postinfection. Shown are averages of three independent experiments. Error bars indicate standard deviations. (b) Results of a time course experiment. In each case, values have been expressed as β-galactosidase activity per PFU of virus as assayed by titrating the virus harvested at each time point on permissive Vero cells.
In these experiments, the IELacZ virus resulted in strong β-galactosidase activity on the parental BHK cells and the Oct-2.1-expressing cells but much-reduced activity on the Oct-2.4- or Oct-2.5-expressing cells even when the latter was normalized for the reduced growth of virus on these cells. This parallels our previous finding (12) that the transcription of the natural viral IE genes themselves within the viral genome is reduced during infection of Oct-2.4- or Oct-2.5-expressing cells. Interestingly, however, our present experiments show that this effect is due to the upstream elements of the IE promoter. Thus, their deletion in the IEminLacZ construct resulted in the same low β-galactosidase activity in all the cell lines. This was still approximately 10-fold greater than the minimum detectable activity, however, indicating that any inhibition by Oct-2.4 or Oct-2.5 would have been detectable. Hence, the removal of the upstream IE promoter sequences not only reduces promoter activity by deleting positively acting sequences but also abolishes the inhibitory effect on promoter activity observed in the Oct-2.4- and Oct-2.5-expressing cell lines.
These positive and negative effects evidently could be mediated by any of the sequences present in the large region of the IE1 promoter which was removed in producing the IEmin promoter. However, the experiments with the IEmin+OT and IEmin+T viruses directly prove that such effects are dependent on the TAATGARAT motifs. Thus, addition of a single overlapping octamer-TAATGARAT motif or a simple TAATGARAT motif to the IEmin promoter greatly enhanced its activity in the BHK cells and the Oct-2.1-expressing cells, although the level of promoter activity was not as high as with the full IE1 promoter. Hence, most, but not all, of the positive effect of the upstream IE promoter sequences within the context of the viral genome can be produced by a single octamer-TAATGARAT or TAATGARAT motif.
Most interestingly, the addition of a single octamer-TAATGARAT or TAATGARAT motif to the IEmin promoter also restored its ability to be repressed by Oct-2.4 or Oct-2.5. Thus, when the IEmin+OT or IEmin+T virus was used to infect the Oct-2.4 and Oct-2.5 cell lines, β-galactosidase activity was not significantly different from that observed with IEmin or with the IELacZ virus containing the full IE1 promoter. Moreover, the β-galactosidase levels produced by these viruses on the Oct-2.5 or Oct-2.4 cell lines were much lower than those produced by the same viruses on the Oct-2.1 cell line or parental BHK cells. Hence, a single octamer-TAATGARAT or TAATGARAT motif can indeed mediate the inhibitory effect of Oct-2.4 and Oct-2.5 on an IE promoter within the context of a viral genome infecting a cell. Similar results were obtained at 16 and 24 h postinfection, although it appeared that repression was partially overcome in the Oct-2.5 cell line by 36 h (Fig. 4b).
The experiments presented here thus clearly indicate that the octamer-TAATGARAT and TAATGARAT motifs in the IE promoters can act as a target for inhibitory octamer-binding proteins as well for the stimulating effect of the Oct-1–Vmw65 complex. Thus, in addition to playing a critical role in the high-level expression of the IE genes during the lytic cycle, these motifs might also play a critical role in the silencing of the IE genes which occurs during latency, acting as a target for inhibitory octamer-binding proteins such as Oct-2.4 and Oct-2.5 or other related members of the POU family of transcription factors which are expressed in neuronal cells (for reviews, see references 21 and 22). Further studies involving the establishment of latent infections in vivo with our indicator viruses will be necessary to explore this possibility as well as to determine why the IE1 promoter contains overlapping octamer-TAATGARAT motifs whereas the other IE promoters contain only simple TAATGARAT motifs.
Acknowledgments
This work was supported by Glaxo/Wellcome.
REFERENCES
- 1.Batterson W, Roizman B. Characterization of the herpes simplex virion-associated factor responsible for the induction of α genes. J Virol. 1983;46:371–377. doi: 10.1128/jvi.46.2.371-377.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffin R S, Maclean A R, Latchman D S, Brown S M. Safe delivery of a transgene to the mouse central or peripheral nervous system using HSV1 ICP34.5 deletion mutant vectors. Gene Ther. 1996;3:886–891. [PubMed] [Google Scholar]
- 3.Coffin R S, Howard M K, Cumming D V E, Dollery C M, McEwan J, Yellon D M, Marber M S, Maclean A R, Brown S M, Latchman D S. Gene delivery to cardiac cells in vitro and in vivo using herpes simplex virus vectors. Gene Ther. 1996;3:560–566. [PubMed] [Google Scholar]
- 4.Croen K D, Ostrove J M, Dragovic L J, Smialek J E, Straus S E. Latent herpes simplex virus in human trigeminal ganglia. Detection of an immediate early gene anti-sense transcript by in situ hybridization. N Engl J Med. 1987;317:1427–1432. doi: 10.1056/NEJM198712033172302. [DOI] [PubMed] [Google Scholar]
- 5.Falkner F G, Moickat R, Zachau H G. Sequences closely related to an immunoglobulin promoter/enhancer element occur also upstream of other eukaryotic and prokaryotic genes. Nucleic Acids Res. 1986;13:7847–7863. doi: 10.1093/nar/14.22.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelman I H, Silverstein S. Herpes simplex virus immediate-early promoters are responsive to virus and cell trans-acting factors. J Virol. 1987;61:2286–2296. doi: 10.1128/jvi.61.7.2286-2296.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goding C R, O’Hare P. Herpes simplex virus Vmw65-octamer binding protein interaction: a paradigm for combinatorial control of transcription. Virology. 1989;173:363–367. doi: 10.1016/0042-6822(89)90548-5. [DOI] [PubMed] [Google Scholar]
- 8.Latchman D S, Kemp L M. Growth of herpes simplex virus and purification of viral DNA. Methods Mol Biol. 1991;8:191–200. doi: 10.1385/0-89603-191-8:191. [DOI] [PubMed] [Google Scholar]
- 9.Latchman D S. Molecular biology of herpes simplex virus latency. Int J Exp Pathol. 1990;71:133–141. [PMC free article] [PubMed] [Google Scholar]
- 10.Lillycrop K A, Dawson S J, Estridge J K, Gerster T, Matthias P, Latchman D S. Repression of a herpes simplex virus immediate-early promoter by the Oct-2 transcription factor is dependent on an inhibitory region at the N terminus of the protein. Mol Cell Biol. 1994;14:7633–7642. doi: 10.1128/mcb.14.11.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lillycrop K A, Estridge J K, Latchman D S. The octamer binding protein oct-2 inhibits transactivation of the herpes simplex virus immediate early genes by the virion protein Vmw65. Virology. 1993;196:888–891. doi: 10.1006/viro.1993.1552. [DOI] [PubMed] [Google Scholar]
- 12.Lillycrop K A, Howard M K, Estridge J K, Latchman D S. Inhibition of herpes simplex virus infection by ectopic expression of neuronal splice variants of the Oct-2 transcription factor. Nucleic Acids Res. 1994;22:815–820. doi: 10.1093/nar/22.5.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lillycrop K A, Latchman D S. Alternative splicing of the Oct-2 transcription factor is differentially regulated in B cells and neuronal cells and results in protein isoforms with opposite effects on the activity of octamer/TAATGARAT-containing promoters. J Biol Chem. 1992;267:24960–24966. [PubMed] [Google Scholar]
- 14.Macpherson, I., and M. Stoker. Polyoma transformation of hamster cell clones—an investigation of the genetic factors affecting cell competence. Virology 16:147–151. [DOI] [PubMed]
- 15.Moriuchi H, Moriuchi M, Cohen J I. Proteins and cis-acting elements associated with transactivation of the varicella-zoster virus (VZV) immediate-early gene 62 promoter by VZV open reading frame 10 protein. J Virol. 1995;69:4693–4701. doi: 10.1128/jvi.69.8.4693-4701.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Hare P, Goding C R. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell. 1988;52:435–445. doi: 10.1016/s0092-8674(88)80036-9. [DOI] [PubMed] [Google Scholar]
- 17.Perry L J, Rixon F J, Everett R D, Frame M C, McGeogh D J. Characterization of the IE110 gene of herpes simplex virus type 1. J Gen Virol. 1986;67:2365–2380. doi: 10.1099/0022-1317-67-11-2365. [DOI] [PubMed] [Google Scholar]
- 18.Preston C M, Frame M C, Campbell M E M. A complex formed between cell components and a herpes simplex virus structural polypeptide binds to a viral immediate early gene regulatory DNA sequence. Cell. 1988;52:425–434. doi: 10.1016/s0092-8674(88)80035-7. [DOI] [PubMed] [Google Scholar]
- 19.Roizman B, Sears A E. An inquiry into the mechanisms of herpes simplex virus latency. Annu Rev Microbiol. 1987;41:543–571. doi: 10.1146/annurev.mi.41.100187.002551. [DOI] [PubMed] [Google Scholar]
- 20.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L, Feldman L T. RNA complementary to a herpes virus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 21.Verrijzer C P, van der Vliet P C. POU domain transcription factors. Biochim Biophys Acta. 1993;1173:1–21. doi: 10.1016/0167-4781(93)90237-8. [DOI] [PubMed] [Google Scholar]
- 22.Wegner M, Drolet D W, Rosenfeld M G. POU-domain proteins: structure and function of developmental regulators. Curr Opin Cell Biol. 1993;5:488–498. doi: 10.1016/0955-0674(93)90015-i. [DOI] [PubMed] [Google Scholar]
- 23.Wirth T, Priess A, Annweiler A, Zwilling S, Oeler B. Multiple Oct-2 isoforms are generated by alternative splicing. Nucleic Acids Res. 1991;19:43–51. doi: 10.1093/nar/19.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]