Horesh et al. use forward and reverse genetics to identify four novel gain-of-function JAK1 mutations in 59 patients. These individuals present with autoimmunity, atopy, colitis, and/or dermatitis, suggestive of a novel syndrome termed JAK1-associated atopy colitis and/or dermatitis (JAACD).
Abstract
Inborn errors of immunity lead to autoimmunity, inflammation, allergy, infection, and/or malignancy. Disease-causing JAK1 gain-of-function (GoF) mutations are considered exceedingly rare and have been identified in only four families. Here, we use forward and reverse genetics to identify 59 individuals harboring one of four heterozygous JAK1 variants. In vitro and ex vivo analysis of these variants revealed hyperactive baseline and cytokine-induced STAT phosphorylation and interferon-stimulated gene (ISG) levels compared with wild-type JAK1. A systematic review of electronic health records from the BioME Biobank revealed increased likelihood of clinical presentation with autoimmunity, atopy, colitis, and/or dermatitis in JAK1 variant-positive individuals. Finally, treatment of one affected patient with severe atopic dermatitis using the JAK1/JAK2-selective inhibitor, baricitinib, resulted in clinically significant improvement. These findings suggest that individually rare JAK1 GoF variants may underlie an emerging syndrome with more common presentations of autoimmune and inflammatory disease (JAACD syndrome). More broadly, individuals who present with such conditions may benefit from genetic testing for the presence of JAK1 GoF variants.
Introduction
Inborn errors of immunity (IEI) are a group of heterogeneous disorders that affect the establishment, homeostasis, and/or regulation of the immune system. They are caused by germline mutations in immune or immune-regulatory genes and can result in early-onset severe infections, autoimmunity, autoinflammation, allergy, and/or malignancy (Tangye et al., 2022). The unique causative genetic variants are thought to range from exceedingly rare (private) to ultrarare (minor allele frequency [MAF] < 0.1%); however, current studies estimate the aggregate frequency of these disorders to be between ultrarare and rare (0.1% > MAF < 1%), affecting nearly 1 in 1,000 individuals and accounting for a growing healthcare burden (Picard et al., 2015; Tangye et al., 2020, 2022).
The JAK–STAT pathway encompasses an evolutionarily conserved signaling cascade by which cells respond to exogenous ligands and transmit their signals into transcriptional regulation, thereby orchestrating various biological functions including cell growth, differentiation, innate immune sensing, and adaptive immune activation. In brief, the pathway starts with the engagement of a transmembrane receptor by an extracellular ligand, which triggers dimerization or oligomerization of partnering receptors. This induces the preassociated Janus kinases (JAK1-4, TYK2) to cis- and trans-phosphorylate tyrosine residues of their own and the partnering receptor–JAK complexes. This phosphorylation signature induces the recruitment of signal transducer and activator of transcription (STAT1-4, STAT5a–b, STAT6) protein monomers, which are also phosphorylated, enabling the formation of homo- or heterodimers. These active STAT–STAT complexes then translocate to the nucleus where they initiate specific gene expression programs that carry out the desired cellular response.
Within the JAK–STAT pathway, JAK1 stands out as a master regulator of the cascade in that it mediates signals for at least 28 different cytokines, accounting for almost half of all ligands that signal through the JAK–STAT pathway (Hammarén et al., 2019). The JAK1 gene (chr1:64,833,223-65,067,754; GRCh38/hg38) encodes a broadly expressed mRNA transcript that is translated into an evolutionarily conserved tyrosine kinase consisting of four core domains—FERM, SH2, pseudokinase, and kinase (Yamaoka et al., 2004; Fagerberg et al., 2014). Once translated, JAK1 traffics to the cell membrane where it binds its cognate intracellular receptor tails, forming the readied complex necessary for cytokine signaling. We and others have previously used a forward genetics approach to identify and characterize four families with exceedingly rare IEIs (Del Bel et al., 2017; Gruber et al., 2020; Takeichi et al., 2022; Fayand et al., 2023) that are caused by GoF variants in the JAK1 gene. In these cases, the affected individuals initially presented in infancy and experienced lifelong, treatment-refractory inflammatory disease with heterogeneous features ranging from atopic dermatitis and asthma to gastrointestinal (GI) inflammation, autoimmunity, and growth defects. The four families were found to each harbor a non-synonymous single-nucleotide variant (SNV) in the JAK1 gene, resulting in the amino acid changes A634D (CADD = 32) (Del Bel et al., 2017), S703I (CADD = 26.1) (Gruber et al., 2020), H596D (CADD = 24.1) (Takeichi et al., 2022), and C787F (CADD = 28) (Fayand et al., 2023), all within the pseudokinase domain of the JAK1 protein. Of the four variants, A634D and S703I were tested biochemically in the human in vitro system, exhibiting remarkably high levels of basal and cytokine-induced JAK and STAT phosphorylation, as well as hyperactive induction of interferon (IFN)-stimulated genes (ISGs) compared with wild-type (WT) JAK1. This in vitro GoF phenotype was mirrored by detailed molecular characterization of patient samples ex vivo, in which hyperactive baseline and cytokine-induced JAK–STAT pathway activation was determined to be the likely cause of the severe disease phenotypes.
Given the four previous cases of exceedingly rare JAK1 GoF variants leading to high levels of biochemical activity and ensuing severe inflammatory disease, we hypothesized that less rare JAK1 variants with mild-to-moderate GoF activity could be associated with more common presentations of inflammatory, allergic, and/or autoimmune diseases. Here, we use both forward and reverse genetic approaches to describe four novel JAK1 GoF variants with a spectrum of in vitro and ex vivo activity, all leading to a syndromic disease pattern encompassing autoimmunity, atopy, colitis, and/or dermatitis.
Results
Six families with a spectrum of inflammatory disease were found to harbor heterozygous variants in JAK1
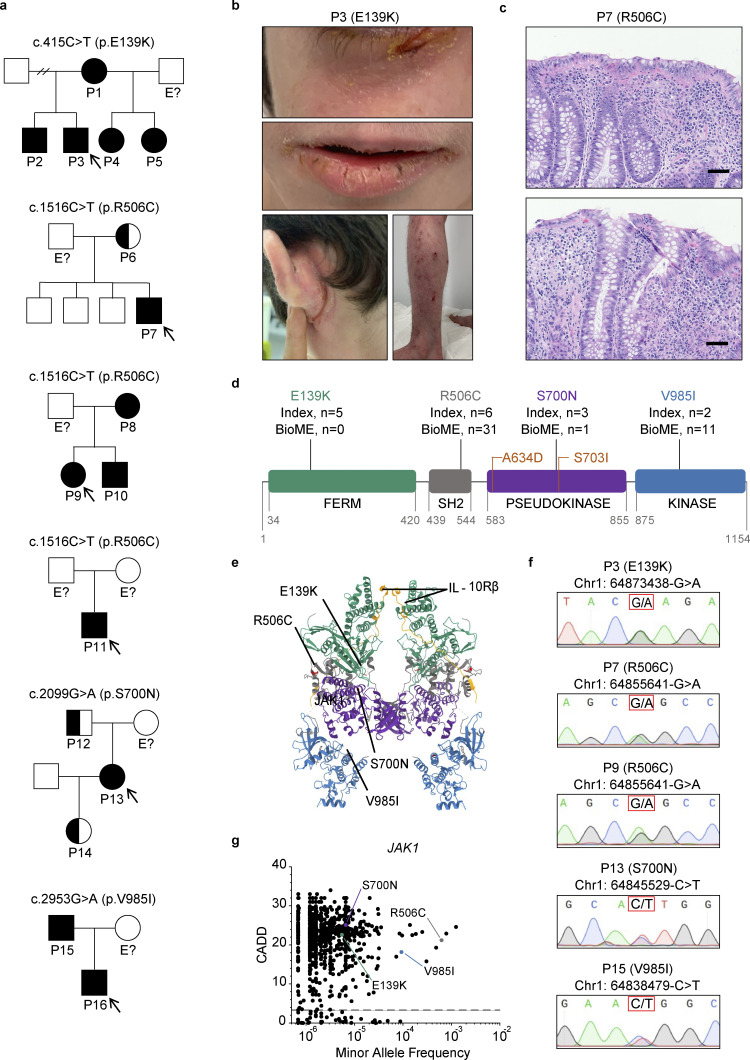
Initially, six unrelated probands and seven affected family members (Fig. 1 a) presented to specialty clinicians in dermatology, gastroenterology, or clinical immunology with the spectrum of autoimmunity, atopy, colitis, and dermatitis (Table 1 and Table S1). P3, who is of self-reported French-European descent, presented in infancy with severe eczema (Fig. 1 b), moderate persistent asthma, persistent diarrhea and constipation, and multiple food allergies, akin to the a priori presentation in his brother, P2, and mother, P1. P7, who is of self-reported African-American descent, presented in early childhood with very-early-onset inflammatory bowel disease (VEO-IBD) with perianal fistula (Fig. 1 c), atopic dermatitis, growth deceleration, and food allergy. P9, who is of self-reported Ashkenazi Jewish descent, presented in middle childhood with autoimmune periocular muscle weakness (myasthenia gravis–like) and Crohn’s disease. P11, who is of self-reported Ashkenazi Jewish descent, presented in early childhood with severe atopic dermatitis, recurrent cutaneous infections, moderate persistent asthma, recurrent diarrhea, growth deceleration, and multiple food allergies. P13, who is of self-reported Mestizo South American descent, presented in adulthood with nonspecific colitis, recurrent respiratory infections, autoimmune thrombocytopenia, and autoimmune hepatitis, suggesting a diagnosis of combined variable immune deficiency (CVID). Finally, P16, who is of self-reported Ashkenazi Jewish descent, presented in infancy with eczema, recurrent otitis media, VEO-IBD, and suspected eosinophilic gastrointestinal disease, while her father, P15, presented with adult-onset type I diabetes. Given the complex and often treatment-refractory state of disease in these individuals, genetic workup was sought by the clinical teams caring for each. In P1, P2, and P3, exome sequencing revealed a heterozygous SNV in the FERM domain of JAK1 (chr1: 64873438-G>A; c.415C>T; p.E139K). In P6, P7, P8, P9, P10, and P11, genetic testing with a commercial primary immunodeficiency (PID) panel revealed a heterozygous SNV in the JAK1 SH2 domain (chr1: 64855641-G>A; c.1516C>T; p.R506C). In P13, exome sequencing identified a heterozygous SNV in the JAK1 pseudokinase domain (chr1: 64845529-C>T; c.2099G>A; p.S700N). Finally, in P15 and P16, PID panel revealed a heterozygous SNV in the JAK1 kinase domain (chr1: 64838479-C>T; c.2953G>A; p.V985I). In total, these SNVs result in single-amino acid changes spanning all four domains of the JAK1 protein (Fig. 1, d and e) (Glassman et al., 2022). Genomic DNA for each patient and their kin who provided a sample was confirmed to be heterozygous at the indicated loci (Fig. 1 f) by Sanger sequencing.
Figure 1.
Six families with inflammatory and autoimmune diseases are found to harbor heterozygous JAK1 variants affecting all four JAK1 protein domains. (a) Pedigrees for each kindred with the identified JAK1 variants. Affected individuals (shaded) or unaffected carriers (half-shaded) are numbered (P#). Unshaded individuals labeled with “E?” have an unknown JAK1 genotype status. All other unshaded individuals are JAK1 WT. Initially presenting probands are identified by arrows. Male sex is indicated by squares and female sex by circles. (b) Severe atopic dermatitis (SCORAD = 60) affecting P3. Upper panel represents affected areas underneath the eyes. Middle panel represents affected areas of the lower face. Lower left panel represents affected areas of the lower extremities and feet. Lower right panel represents affected areas of the upper extremities and hands. (c) H&E-stained tissue sections of small intestine from P7 (JAK1 R506C) depicting focal neutrophilic inflammation (upper and lower), granuloma (upper), and cryptitis (lower). 100× magnification. Representative micrographs shown from six individual images. Black scale bars are equivalent to 100 μm. (d) Location of four amino acid substitutions arising from the identified JAK1 SNVs along the four domains of the JAK1 protein. Counts of subjects from index families (Index) and subjects present in the Mount Sinai Biobank. Previously described GoF variants are indicated in orange. Digits below domains represent amino acid numbers bordering each domain. (e) Full crystal structure of murine JAK1 dimer, colored by domain, complexed with intracellular tails of IFN-λR1/IL-10Rb in yellow (PDB ID 7T6F) (Glassman et al., 2022). Domains colored as follows: FERM = green, SH2 = gray, pseudokinase = purple, kinase = blue. Corresponding murine-to-human amino acid substitutions depicted in red and labeled by protein consequence. (f) Chromatograms from Sanger sequencing of genomic DNA sampled from affected individuals. Corresponding genomic locations and nucleotide substitutions are indicated above each chromatogram. P11 and P16 did not provide a biological sample and are therefore not included in the data set. (g) Minor allele frequencies for missense JAK1 variants present in gnomAD (v4.0) mapped against corresponding CADD scores (99% confidence interval, HGMD). Prototype JAK1 variants are labeled for reference. The dotted line represents JAK1 mutation significance cutoff (MSC) of 3.313.
Table 1.
Syndromic features of JAK1-mediated disease in six affected probandsa
| Proband | P3 | P7 | P9 | P11 | P13 | P16 |
|---|---|---|---|---|---|---|
| Age in years (at time of study) | 14 | 4 | 21 | 7 | 39 | 7 |
| Race/ethnicity (self-reported) | European/French | African American | Ashkenazi Jewish | Ashkenazi Jewish | Mestizo S. American | Ashkenazi Jewish |
| JAK1 variant | c.415C>T; p.E139K | c.1516C>T; p.R506C | c.1516C>T; p.R506C | c.1516C>T; p.R506C | c.2099G>A; p.S700N | c.2953G>A; p.V985I |
| Neonatal/developmental | ||||||
| Recurrent infections | ✓ | − | − | ✓ | ✓ | ✓ |
| Growth deceleration | ✓ | ✓ | − | ✓ | − | − |
| Adverse reactions to vaccines | − | − | − | ✓ | ✓ | − |
| Autoimmunity | ||||||
| Myasthenia gravis | − | − | ✓ | − | − | − |
| Autoimmune hepatitis | − | − | − | − | ✓ | − |
| Autoimmune thrombocytopenia | − | − | − | − | ✓ | − |
| Other autoimmune | ✓ | − | − | − | ✓ | − |
| Atopy (excluding dermatitis) | ||||||
| Food allergy | ✓ | ✓ | − | ✓ | − | ✓ |
| Asthma | ✓ | − | − | ✓ | ✓ | − |
| Allergic rhinitis | − | − | − | − | ✓ | − |
| Colitis (GI inflammation) | ||||||
| IBD | − | ✓ | ✓ | − | − | ✓ |
| Diarrhea/constipation (non-IBD) | ✓ | − | − | ✓ | ✓ | ✓ |
| Eosinophilic esophagitis | − | − | − | − | − | ✓ |
| Dermatitis | ||||||
| Atopic dermatitis | ✓ | ✓ | − | ✓ | − | ✓ |
| Hematologic | ||||||
| Peripheral leukocyte anomaly | − | − | − | − | ✓ | − |
| Peripheral eosinophilia | ✓ | ✓ | − | ✓ | − | ✓ |
| Ig abnormalities | ✓ | ✓ | − | ✓ | ✓ | ✓ |
| Family history (of any category) | ✓ | − | ✓ | ✓ | − | ✓ |
| Neonatal/developmental sum | 2 | 1 | 0 | 3 | 2 | 1 |
| Autoimmunity sum | 1 | 0 | 1 | 0 | 3 | 0 |
| Atopy sum | 2 | 1 | 0 | 2 | 2 | 1 |
| Colitis sum | 1 | 1 | 1 | 1 | 1 | 2 |
| Dermatitis sum | 1 | 1 | 0 | 1 | 0 | 1 |
| Hematologic sum | 2 | 2 | 0 | 2 | 2 | 2 |
| Family history | 1 | 0 | 1 | 1 | 0 | 0 |
| Grand total | 10 | 6 | 3 | 10 | 10 | 7 |
Check marks (✓) indicate pertinent positives in medical history. Dashes (−) indicate pertinent negatives in medical history. Results reported in binary terms (yes/no) (i.e., whether or not a sign/symptom was present, not how many times a given sign/symptom was present).
Allele counts and variant frequencies from the genome aggregation database (gnomAD, v4.0), which holds genetic data from 807,162 individuals (Karczewski et al., 2020; Koenig et al., 2023, Preprint), are as follows: for JAK1 E139K (P1, P2, P3, P4, P5; kindred I), allele count n = 8, frequency = 5.47 × 10−6. For JAK1 R506C (P6, P7, P8, P9, P10, P11; kindreds II, III, and IV), allele count n = 1,008, frequency = 6.25 × 10−4. For JAK1 V985I (P15, P16; kindred VI) (Fig. 1 g), allele count n = 150, frequency = 7.73 × 10−5. For JAK1 S700N (P12, P13, P14; kindred V), allele count n = 1, frequency = 6.57 × 10−6. In silico damage prediction by Combined Annotation Dependent Depletion (CADD) (Kircher et al., 2014; Rentzsch et al., 2019), which is based on sequence context, gene annotations, epigenetic markers, functional predictions, and accounts for evolutionary constraint, indicated a probable functional consequence from these variants (CADD: E139K = 22.6; R506C = 21.2; S700N = 25.1; V985I = 18.2) as are all above the JAK1 mutation significance cutoff (MSC) of 3.313 (99% CI, HGMD) (Fig. 1 g). Each variant was further tested specifically for predicted GoF activity using a novel in silico approach that employs natural language processing of published abstracts and iterative machine-learning to predict whether a given SNV will be neutral or cause gain or loss of function (Sevim Bayrak et al., 2021). This additional in silico approach identified the variants that encode JAK1 E139K, R506C, and S700N as likely GoF (Table 2). The results of both in silico damage predictions combined with a lack of other plausible candidate variants in the genetic analysis (Table S2) implicated these genetic variants as the most likely causes of the syndromic clinical phenotypes.
Table 2.
In silico prediction of GoF activity in JAK1 variants
| Variant class | JAK1 variant | Predicted deleterious (Y/N) | Frequency, BioME (v1.2) |
||
|---|---|---|---|---|---|
| Transcript | Protein | (CADD) (Rentzsch et al., 2019, Kircher et al., 2014) | (Sevim Bayrak et al., 2021) | n = 30,814 | |
| Exceedingly rare Severe GoF |
c.1901C>A c.2108G>T |
p.Ala634Asp p.Ser703Ile |
Y (32) Y (26.1) |
Y (GoF) Y (GoF) |
Private, n = 0 Private, n = 0 |
| Ultra-rare or rare Mild-to-moderate GoF |
c.415C>T c.1516C>T c.2099G>A c.2953G>A |
p.Glu139Lys p.Arg506Cys p.Ser700Asn p.Val985Ile |
Y (22.6) Y (21.2) Y (25.1) Y (18.2) |
Y (GoF) Y (GoF) Y (GoF) N (neutral) |
Private, n = 0 5.63 × 10−04, n = 31 1.66 × 10−05, n = 1 1.82 × 10−04, n = 11 |
| Total = 43 (0.14%) | |||||
| Ultra-rare or rare Predicted GoF |
c.287C>T c.296A>G c.379T>A c.779G>C c.1078C>T c.1439C>T c.1889A>G c.1951G>A c.2005T>G c.2111A>G c.2414T>A c.2498A>G c.2617C>T c.2675G>A c.3230A>G |
p.Thr96Ile p.Asp99Gly p.Trp127Arg p.Ser260Thr p.Arg360Trp p.Thr480Ile p.Asp630Gly p.Val651Met p.Phe669Val p.Tyr704Cys p.Phe805Tyr p.Asn833Ser p.Arg873Cys p.Cys892Tyr p.Tyr1077Cys |
Y (21.7) Y (23.3) Y (25.4) Y (20.4) Y (25.1) Y (22.5) Y (33.0) Y (27.6) Y (28.2) Y (27.2) Y (29.7) Y (15.8) Y (32.0) Y (24.8) Y (31.0) |
Y (GoF) Y (GoF) Y (GoF) Y (GoF) Y (GoF) Y (GoF) Y (GoF) Y (GoF) Y (GoF) Y (GoF) Y (GoF) Y (GoF) Y (GoF) Y (GoF) Y (GoF) |
1.66 × 10−05, n = 1 1.66 × 10−05, n = 1 1.66 × 10−05, n = 1 1.66 × 10−05, n = 1 4.97 × 10−05, n = 3 1.66 × 10−05, n = 1 1.66 × 10−05, n = 1 9.77 × 10−04, n = 59 1.66 × 10−05, n = 1 1.66 × 10−05, n = 1 1.66 × 10−05, n = 1 1.36 × 10−03, n = 80 1.66 × 10−05, n = 1 1.66 × 10−05, n = 1 4.97 × 10−05, n = 3 |
| Total = 156 (0.51%) | |||||
Systematic review of electronic health records (EHRs) reveals enrichment of syndromic autoimmune and inflammatory phenotypes in JAK1 variant-positive individuals
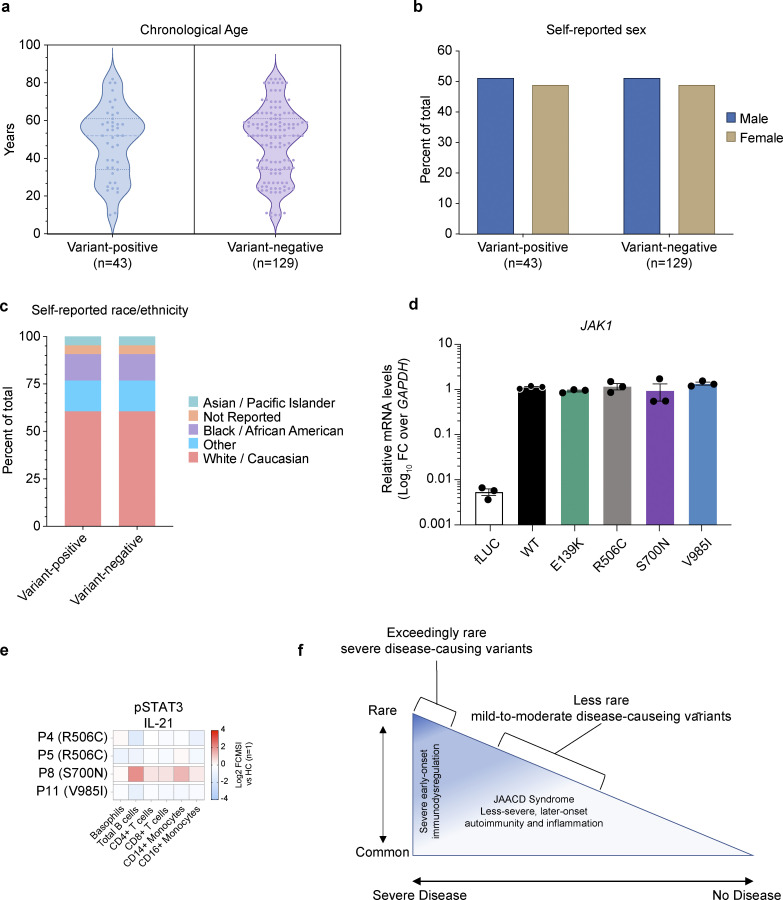
Despite the ultrarare or rare nature of these SNVs, we hypothesized that these candidate JAK1 variants might be contributing, at least in part, to more common presentations of autoimmunity and autoinflammation. To test this, we first mined the EHR-linked Mount Sinai BioME Biobank, which includes over 30,000 individuals with exome sequencing data and identified 43 additional individuals harboring either of the prototype JAK1 variants in the heterozygous state: E139K (n = 0), R506C (n = 31), S700N (n = 1), and V985I (n = 11). To determine whether these JAK1 variants were associated with autoimmune or inflammatory diseases, we devised a blinded systematic review that employed a comprehensive search of EHRs for key terms representing signs and symptoms of autoimmune and inflammatory disease (Table S3)—comparing EHR findings from JAK1 variant-positive individuals (n = 43) to variant-negative individuals (n = 129) who were age-, sex-, and self-reported race/ethnicity matched (Fig. S1, a–c). The results of this blinded systematic review unveiled over ninefold enrichment (OR = 9.6, P = 0.04) of Hashimoto thyroiditis and an eightfold enrichment of rheumatic disease (OR = 8.3, P = 0.01) in EHRs from JAK1 variant-positive individuals compared with variant-negative individuals. Moreover, EHRs from JAK1 variant-positive individuals were more likely to contain evidence of syndromic phenotypes (2+ diagnoses) encompassing autoimmunity, atopy, colitis, and/or dermatitis (Table 3 and Table S3). Collectively, the use of forward and reverse genetics, along with clinical characteristics, suggest that these JAK1 variants might be the drivers of this complex syndromic immune disease.
Figure S1.
Demographics of individuals included in a systematic review of EHRs, in vitro, and ex vivo assessment of JAK1 variants. (a) Chronological age in years (calculated from the date of birth) of each JAK1 variant-positive and variant-negative individual at the time of systematic review (2022). Each dot represents a single subject; the width of the shaded area is proportionate to the number of points at that age. Dotted lines delineate quartiles; middle dotted line represents median age. (b) Sex (self-reported) for variant-positive and variant-negative individuals at the time of systematic review (2022). Proportions represented as percent of the total for each genotype. (c) Self-reported race/ethnicity for variant-positive and variant-negative individuals at the time of systematic review (2023). Proportions represented as percent of total for each genotype status. (d) JAK1 mRNA levels in stably transduced U4C cells expressing doxycycline-inducible negative control (fLUC) or JAK1 constructs. JAK1 levels are represented as log10-transformed RNA levels, normalized to the endogenous control transcript, GAPDH. Each individual point represents the average of technical triplicates from a single well (n = 3 wells per variant). Lines and whiskers represent the mean ± SEM from three wells. Representative data shown from three independent experiments for each variant. (e) Canonical pSTAT3 signaling in patient whole blood stimulated with IL-21 (100 ng/ml). Results expressed as log2-transformed fold-change versus stimulated HCs (n = 1 per run). (f) Rare-to-common JAK1 GoF variants lie on a correlated frequency and phenotype spectrum with JAACD syndrome accounting for more common presentations that are often less severe and later onset.
Table 3.
Overrepresented syndromic features identified in JAK1 variant-positive individuals by blinded systematic review of EHRsa
| Odds ratio (95% CI) | P value (Fisher’s exact) | Variant positive (n = 43) | Variant negative (n = 129) | |
|---|---|---|---|---|
| Sex (female) | 1.098 (0.5504–2.204) | 0.8609 | 22 | 63 |
| Autoimmunity | 2.203 (1.066–4.551) | 0.0412* | 21 | 39 |
| Hashimoto thyroiditis | 9.600 (1.377–125.4) | 0.0487* | 3 | 1 |
| Rheumatic disease | 8.355 (1.659–42.67) | 0.0112* | 5 | 2 |
| Autoimmunity and ≥ any one category | 2.335 (1.106–4.880) | 0.0235* | 20 | 35 |
| Autoimmunity and ≥ any two categories | 2.47 (1.167–5.044) | 0.0252* | 17 | 27 |
| Atopy | 1.499 (0.7531–3.050) | 0.2908 | 23 | 56 |
| Atopy and ≥ any one category | 1.665 (0.8196–3.372) | 0.1553 | 21 | 47 |
| Atopy and ≥ any two categories | 2.183 (1.074–4.626) | 0.0513 | 18 | 32 |
| Colitis (GI inflammation) | 1.499 (0.7554–3.083) | 0.2922 | 24 | 59 |
| Colitis and ≥ any one category | 1.649 (0.8265–3.365) | 0.1619 | 23 | 53 |
| Colitis and ≥ any two categories | 2.43 (1.147–5.101) | 0.0219* | 20 | 34 |
| Dermatitis | 1.323 (0.6304–2.701) | 0.5798 | 30 | 82 |
| Dermatitis and ≥ any one category | 1.675 (0.8299–3.466) | 0.2143 | 28 | 68 |
| Dermatitis and ≥ any two categories | 2.559 (1.256–5.011) | 0.0105* | 23 | 40 |
| Any 3+ categories (autoimmunity, atopy, colitis, or dermatitis) | 2.559 (1.256–5.011) | 0.0105* | 23 | 40 |
Odds ratios with 95% confidence interval (CI) generated using Baptista Pike method. P values generated using Fisher’s exact test (two-tailed) test. P values <0.05 are indicated by an asterisk (*). Variant-positive and variant-negative columns indicate the number of EHRs with at least one finding present for each row.
In vitro testing reveals baseline and cytokine-induced GoF phenotype in cells expressing JAK1 variants
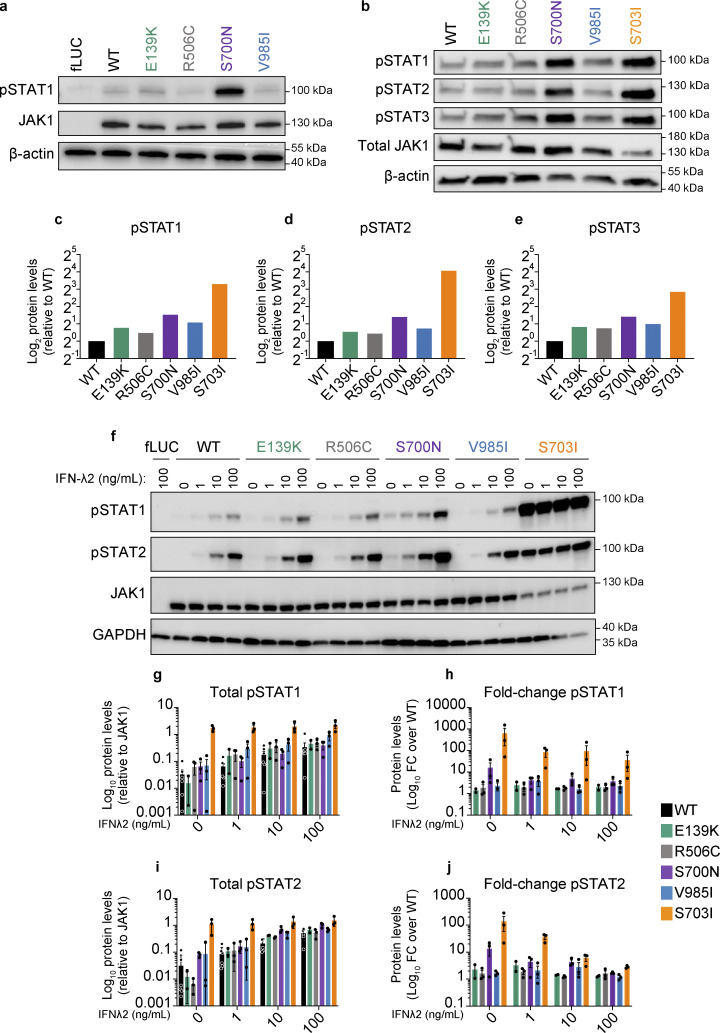
As clinical phenotypes in affected individuals were akin to those of previously described JAK1 GOF cases, we sought to screen each prototype variant for JAK1 hyperactivity. We stably transduced U4C (JAK1-null) cells (Pellegrini et al., 1989) with doxycycline-inducible WT or variant JAK1 constructs and measured the levels of mRNA expression and protein production. All cells carrying a prototype variant expressed JAK1 mRNA (Fig. S1 d) and produced JAK1 protein (Fig. 2 a) at comparable levels with that of WT. Next, we assessed the baseline and cytokine-induced biochemical activity of JAK1 by measuring the amounts of hallmark downstream signals, pSTAT1, pSTAT2, and pSTAT3 (Philips et al., 2022), and calculating their stoichiometric levels relative to the amount of JAK1 present. In the absence of any cytokine, cells expressing JAK1 E139K, R506C, S700N, and V985I were capable of inducing increased pSTAT1 (Fig. 2, a–c and f–h), pSTAT2 (Fig. 2, b, d, f, i, and j), and pSTAT3 (Fig. 2, b and e) signals as compared with WT. To assess the cytokine-induced levels of JAK1 activity, cells expressing JAK1 E139K, R506C, S700N, and V985I were stimulated with IFN-λ2, a type III IFN, and one of the 25+ cytokines that uses JAK1 for signal transduction (Philips et al., 2022). In this setup, IFN-λ2–stimulated cells expressing each variant had dose-dependent augmented levels of JAK1-normalized pSTAT1 (Fig. 2, f–h) and JAK1-normalized pSTAT2 (Fig. 2, f, i, and j) compared with stimulated cells expressing the WT allele.
Figure 2.
In vitro assessment of prototype JAK1 variants reveals baseline and cytokine-induced GoF in STAT phosphorylation and ISG production. (a) Western blot analysis of pSTAT1, JAK1, and β-actin (endogenous loading control) protein levels in stably transduced U4C cells expressing doxycycline-inducible fLUC or JAK1 constructs. Representative images are shown from three independent experiments for each variant. (b) Western blot analysis of pSTATs1-3, JAK1, and β-actin (endogenous loading control) protein levels in transfected U4C cells (JAK1 null) expressing either WT or variant JAK1 constructs. Representative images are shown from two independent experiments for each variant. (c–e) Band densitometry quantifying the levels of JAK1-relative (c) pSTAT1, (d) pSTAT2, and (e) pSTAT3 in lanes from panel b. Protein levels represented as log2-transformed β-actin–normalized protein levels relative to β-actin–normalized JAK1 levels from the same lane. (f) Western blot analysis of pSTAT1, pSTAT2, JAK1, and GAPDH (endogenous loading control) protein levels in stably transduced U4C cells expressing doxycycline-inducible fLUC or JAK1 constructs. Each lane corresponds to independent wells, which were doxycycline-induced then cytokine stimulated with the indicated doses of IFN-λ2 (0, 1, 10, 100 ng/ml). Representative images are shown from three independent experiments for each variant. (g–j) Band densitometry quantifying pSTAT1 (g and h) and pSTAT2 (i and j) protein levels in cells expressing JAK1 variants. Protein levels represented as GAPDH-normalized pSTAT levels relative to GAPDH-normalized JAK1 levels from the same lane (g and i), and as fold change in GAPDH-normalized relative pSTAT levels versus the WT condition (h and j). Each point represents an individual replicate for each variant from n = 7 total experiments. Lines and whiskers represent the mean ± SEM. Source data are available for this figure: SourceData F2.
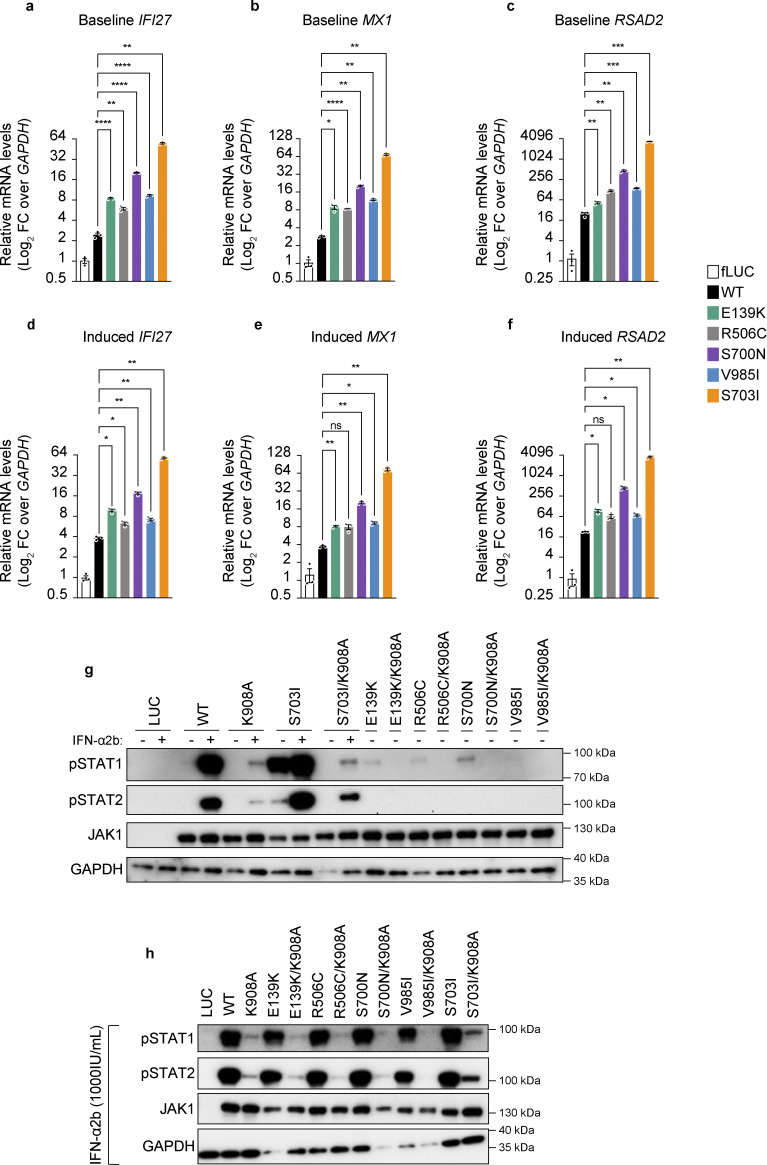
To determine whether this proximal pSTAT hyperactivity translates into hyperactive downstream JAK1-mediated signals, we assessed the levels of ISGs (Levy et al., 1986; Schoggins et al., 2011), which are normally induced in response to IFN-α2b, a type I IFN. In line with the baseline GoF in pSTAT signaling, we detected increased baseline (no cytokine) levels of the canonical ISGs, IFI27, MX1, and RSAD2 (Fig. 3, a–c) in cells expressing either of the JAK1 variants compared with cells expressing WT JAK1. This was true even in cells expressing JAK1 R506C, which was the mildest variant in terms of baseline pSTAT levels (Fig. 2 a). Furthermore, upon stimulation with IFN-α2b, cells expressing any of the prototype SNVs exhibited increased IFI27 levels (Fig. 3 d) compared with cells expressing WT JAK1, while cells expressing JAK1 E139K, S700N, or V985I had increased MX1 and RSAD2 levels (Fig. 3, e and f) compared with cells expressing WT JAK1.
Figure 3.
In vitro assessment of baseline pSTAT activity on transcription and trans-regulation by JAK1 variants. (a–f) ISG (IFI27, MX1, and RSAD2) mRNA levels assessed by RT-qPCR of RNA from stably transduced and induced U4C cells that were stimulated with (a–c) mock or (d–f) IFN-α2b (1,000 IU/ml) for 30 min, then rested for 12 h. ISG levels are represented as log2-transformed relative mRNA levels over the endogenous control transcript, GAPDH. Each individual point represents the average of technical triplicates from a single well (n = 3 wells per condition). Lines and whiskers represent the mean ± SEM from three wells. P values indicated above bars were obtained by statistical testing by one-way ANOVA with Dunnett’s T3 comparison test, with statistical assessment as follows: *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0005; ns = not significant. Representative data shown from experiments conducted three independent times for each condition. (g and h) Western blot analysis of pSTAT1-2, JAK1, and GAPDH (endogenous loading control) protein levels in stably transduced U4C cells expressing doxycycline-inducible variant JAK1 constructs (single mutants) or catalytically dead variant JAK1 constructs (double mutants). In panel g, minus signs (−) indicate mock stimulated cells, and plus signs (+) indicate IFN-α2b (1,000 IU/ml) stimulated cells. In panel h, all lanes represent cells stimulated with IFN-α2b (1,000 IU/ml). Source data are available for this figure: SourceData F3.
We next sought to determine whether this increased pSTAT and ISG levels were mediated through trans-regulation of the partnering JAK as is seen with the previously described JAK1 S703I variant (Gruber et al., 2020). To test this, we stably transduced cells with inducible constructs carrying either JAK1 variants alone, or in tandem (double mutants; same DNA strand) with the catalytically inactivating amino acid change, K908A (Haan et al., 2011). In this system, cells expressing JAK1 K908A are catalytically dead JAK1, therefore any STAT phosphorylation that arises from a given membrane–receptor complex must originate from the partnering JAK’s kinase activity. The presence of hyperactive STAT phosphorylation in cells expressing double mutants (JAK1 variant + K908A) compared with K908A alone would therefore be due to trans-activation of the partnering JAK’s kinase activity by the variant carrying JAK1 protein. Upon testing this with cells expressing JAK1 E139K, R506C, S700N, and V985I double mutants (variant + K908A), there are no higher levels of STAT phosphorylation at baseline (Fig. 3 g) or in the presence of IFN-α2b (Fig. 3 h), thereby implicating cis-activation as the mechanism by which these JAK1 variants hyper-phosphorylate their STAT targets.
In summary, cells expressing each prototype JAK1 variant in vitro induce baseline hyperactive STAT1–3 phosphorylation along with dose-dependent IFN-λ2–induced STAT1 and STAT2 phosphorylation compared with cells expressing JAK1 WT. This GoF in pSTAT signaling translates into a clear buildup of baseline and IFN-α2b induced ISGs, which are again detected at higher levels in cells expressing these JAK1 variants than those expressing JAK1 WT. Overall, each JAK1 variant elicits biochemically assessed levels of pSTATs and ISGs that are higher than WT but lower than that of the early-onset severe disease-causing variant, JAK1 S703I. Therefore, these findings define, at least in vitro, JAK1 E139K, R506C, S700N, and V985I as clear GoF variants that are mild alleles compared with that of JAK1 S703I (Figs. 2 and 3).
Ex vivo characterization of patient whole blood reveals GoF in proximal JAK–STAT signaling and immune cell dysregulation
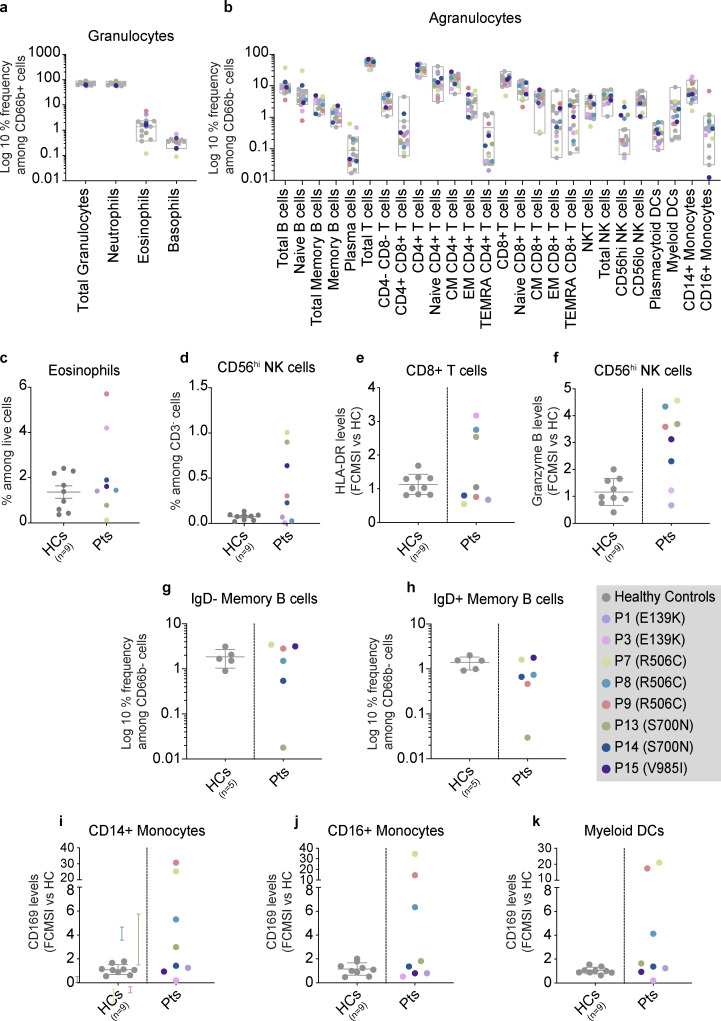
We then sought to investigate basal and induced JAK1 signaling pathways ex vivo. First, whole blood from individuals harboring JAK1 E139K, R506C, S700N, or V985I was evaluated for the abundance of 30 distinct peripheral blood cell types (Table S4) and their respective levels of basal pSTAT1, pSTAT3, pSTAT5, and pSTAT6. The percent frequencies of all cell types were within age-matched healthy control ranges (Fig. 4, a and b), except for eosinophils, which had elevated proportions in two out of eight patients (Fig. 4 c), and CD56hi natural killer (NK) cells, which had elevated proportions in five out of eight patients (Fig. 4 d), consistent with previous reports of JAK1 GoF (Del Bel et al., 2017; Gruber et al., 2020). However, assessment of specific lymphocyte populations in these patients revealed dysregulation of some immune cells. First, CD8+ T cells from E139K, R506C, and S700N genetic backgrounds had increased levels of surface HLA-DR as compared with healthy controls (Fig. 4 e). We also documented increased baseline levels of granzyme B in CD56hi NK cells in patients with JAK1 R506C, S700N, and V985I (Fig. 4 f). IgD-positive and IgD-negative B cell proportions in all patients except P13, who have late-onset hypogammaglobulinemia, were found to be comparable with the healthy control ranges indicating normal class switching (Fig. 4, g and h). Finally, CD169, which is encoded by the canonical ISG, SIGLEC1, levels were elevated in CD14+ monocytes, CD16+ monocytes, and myeloid DCs of R506C-carrying individuals (Fig. 4, i–k).
Figure 4.
Ex vivo assessment of individuals with JAK1 variants by whole blood CyTOF. (a and b) Granulocytes (a) and agranulocytes (b) in whole blood from individuals with JAK1 variants and HCs. Results are depicted as log10-transformed percent frequencies of CD66b+ cells and CD66b− cells, respectively. Gray box plots represent HC ranges (min-max) with the middle bar representing the average for HCs. For panels a and b, number of HCs for each patient are as follows: P3 n = 3; P7 n = 4; P9 n = 2; P13 n = 4; P15 n = 4. For panel b, CM = central memory; EM = effector memory; TEMRA = T effector memory CD45RA+; NKT = NK T cells; NK = natural killer. Populations labeled “granulocytes” comprise all CD66b+ cells (including neutrophils, eosinophils, and basophils). (c and d) Percent frequencies of manually gated (c) eosinophils and (d) NK cells in whole blood from individuals with JAK1 variants and healthy controls (HCs). HC group (n = 9) labeled as “HCs.” Patient group (n = 8) labeled as “Pts.” Results are expressed as a percent of live cells for eosinophils and as a percent of CD3− cells for CD56hi NK cells. Each point represents a single subject. Bars and whiskers represent the mean ± SEM for that group. (e and f) Baseline levels of (e) HLA-DR and (f) granzyme B in manually gated CD8+ T cells and CD56hi NK cells respectively. Levels measured in whole blood from individuals with JAK1 variants compared to HCs. For panels a and b, results are expressed as fold-change in mean signal intensity (FCMSI) over the average of nine HCs. Number of HCs for each patient are as follows: P3 n = 3; P7 n = 4; P9 n = 2; P13 n = 4; P15 n = 4. Results are from four independent runs with 1 (P1 and P3) or 2 (P7, P8, P9, P13, P14, P15) technical replicates. Gray bars and whiskers represent the mean ± SEM for HCs. (g and h) Percent frequencies of (g) IgD negative and (h) IgD positive memory B cells in whole blood from individuals with JAK1 variants. Results expressed as the log10-transformed percent frequencies of CD66b− cells (agranulocytes). The number of HCs from three independent runs are as follows: P7 and P15, n = 2; P8 and P9, n = 2; P13 and P14, n = 1. There was no biological sample available for this experiment from Kindred I (P1 and P3), therefore IgD status was not assessed. (i–k) Baseline CD169 levels in (i) CD14+ monocytes, (j) CD16+ monocytes, and (k) myeloid DCs in whole blood from individuals with JAK1 variants compared to HCs. Results expressed as FCMSI over the average of nine HCs. Number of HCs for each patient are as follows: P3 n = 3; P7 n = 4; P9 n = 2; P13 n = 4; P15 n = 4. Results are from four independent runs with 1 (P1 and P3) or 2 (P7, P8, P9, P13, P14, P15) technical replicates. Gray bars and whiskers represent the mean ± SEM for HCs.
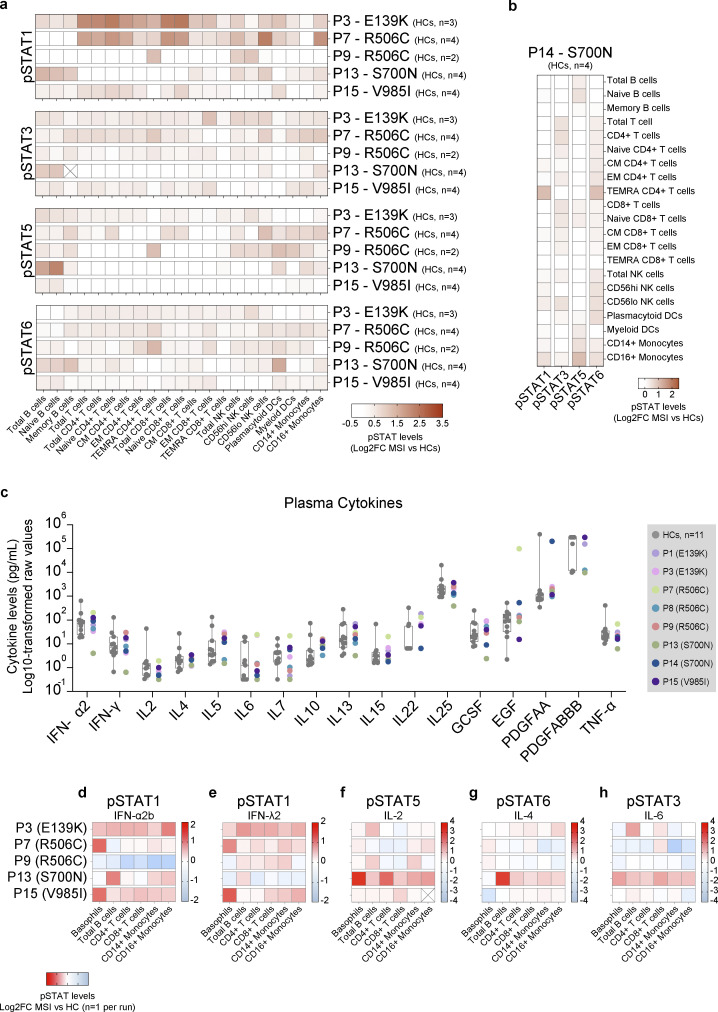
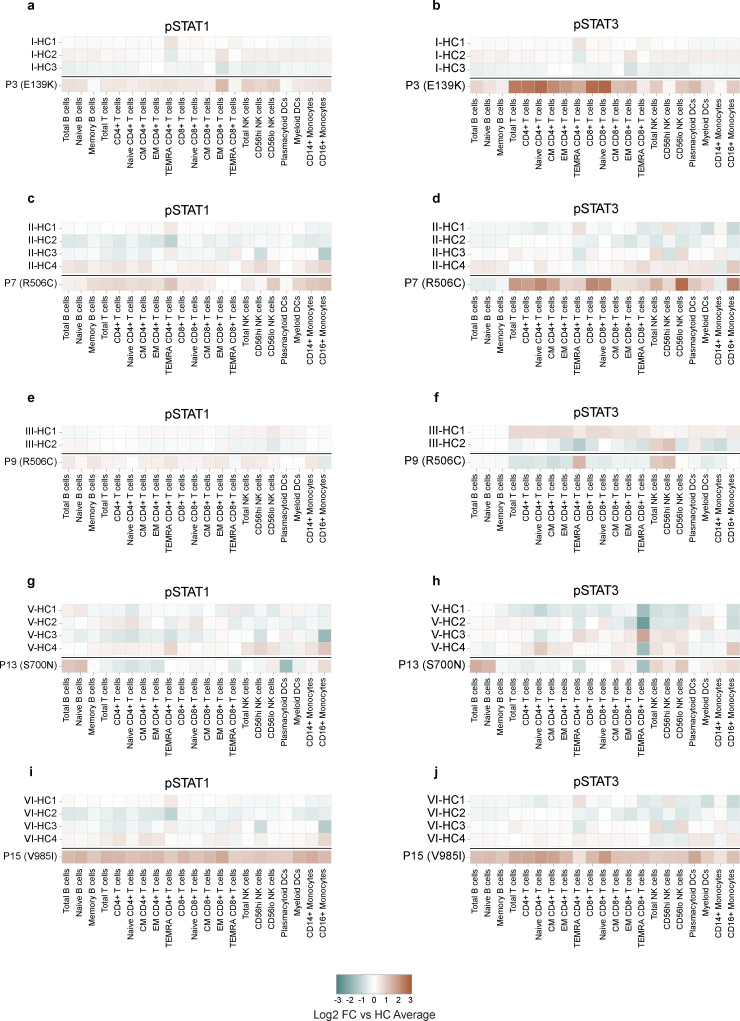
In contrast to the largely normal immune cell proportions, the basal levels of pSTAT1 were elevated across most cell types in all genotypes except S700N, in which basal pSTAT1 levels were elevated only in B cells, monocytes, and NK cells (Fig. 5 a and Fig. S2). Basal pSTAT3 levels (Fig. 5 a and Fig. S2) were the most prominently detected of any pSTAT tested, particularly in patients with JAK1 E139K and R506C variants. Basal pSTAT5 and pSTAT6 were also detected in all genotypes (Fig. 5 a), albeit with differing cell type specificity. Interestingly, baseline pSTAT levels were also elevated in B cells (pSTAT5), T cells (pSTAT3, 5), NK cells (pSTAT1, 3, 6), DCs (pSTAT1, 3, 5, 6), and monocytes (pSTAT1, 5, 6) in whole blood from P14 (Fig. 5 b), who harbors the JAK1 S700N variant but has not yet developed clinical signs of disease. These baseline pSTAT signals in JAK1 variant-positive individuals were present despite the absence of JAK1-stimulating cytokines in their plasma beyond healthy control levels (Fig. 5 c), pointing to a JAK1 GoF-intrinsic process.
Figure 5.
Ex vivo assessment of whole blood from individuals with JAK1 variants reveals cell type–specific GoF in STAT phosphorylation. (a) Baseline pSTAT1, 3, 5, and 6 levels in manually gated cells from unstimulated whole blood from individuals with JAK1 variants. Each row represents an individual run with the number of HCs for each run labeled. Manually gated cell types for each run are labeled by column. Cells marked with an X indicate no cells were captured for that run. Results are depicted as log2-transformed fold-change in mean signal intensity (MSI) versus the mean signal intensity of HCs from the same run. Number of HCs for each patient are as follows: P3 n = 3; P7 n = 4; P9 n = 2; P13 n = 4; P15 n = 4. (b) Baseline pSTAT1, 3, 5, and 6 levels in manually gated cells from P14. Results depicted as log2-transformed fold change in mean signal intensity (MSI) compared with the average of (HCs from the same run). Each row represents an individual run with HC (n = 4). (c) Multiplex Luminex assay on plasma from individuals with JAK1 variants (n = 8) and HCs (n = 11). Results are expressed as the log10-transformed raw values (pg/ml). All results depicted are from a single run. Gray boxes represent mean ± SD, whiskers represent minimum to maximum range of HCs. For panels a and b: CM = central memory; EM = effector memory; TEMRA = T effector memory CD45RA+; NKT = natural killer T cell; NK = natural killer, DC = dendritic cells. (d–h) Canonical pSTAT levels in stimulated whole blood from individuals with JAK1 variants. Cytokine concentrations as follows: IFN-α2b (P3 = 100 IU/ml; all other patients = 1,000 IU/ml), IFN-λ2, IL-2, IL4, or IL-6 (P3 = 10 ng/ml; all other patients = 100 ng/ml). Each row represents an individual run. Manually gated cell types for each run are labeled by column. Cells marked with an X indicate no cells were captured for that run. Results expressed as log2-transformed fold-change in mean signal intensity versus mean signal intensity of stimulated HCs from the same run (n = 1 HC per run).
Figure S2.
Ex vivo assessment baseline pSTAT levels compared to individual healthy controls from the same run. (a–j) pSTAT1 (left) and pSTAT3 (right) levels in manually gated cells from unstimulated whole blood from individuals with JAK1 variants encoding JAK1 E139K (a and b), R506C (c and d), R506C (e and f), S700N (g and h), or V985I (i and j). Each panel represents a single run with the corresponding HCs labeled accordingly. Results expressed as log2-transformed fold-change in mean signal intensity (MSI) compared with the average of HCs for that run. HC counts are as follows: P3 n = 3; P7 n = 4; P9 n = 2; P13 n = 4; P14 n = 4; P15 n = 4.
To assess the plasticity of induced JAK-STAT signaling in individuals with JAK1 variants, we stimulated the patients’ whole blood with IFN-α2b, IFN-λ2, interleukin (IL)-2, IL-4, IL-6, and IL-21, and then measured levels of induced pSTATs. Following IFN-α2b or IFN-λ2 stimulation, patients’ cells, compared with the stimulated healthy controls, reached increased levels of pSTAT1 in most cell types (Fig. 5, d and e), with the strongest phenotype evident in basophils, B cells, T cells, and monocytes. In the same experimental setting, stimulation with IL-2 mostly resulted in WT-like pSTAT5 levels (Fig. 5 f), except in B cells in which pSTAT5 levels were elevated in most genotypes, and in JAK1 S700N in which most cell types exhibited GoF signaling. The same was also true for stimulation with IL-4 in that JAK1 S700N exhibited elevated levels of pSTAT6 (Fig. 5 g). Stimulation with IL-6 (Fig. 5 h) or IL-21 (Fig. S1 e) also yielded elevated levels of pSTAT3 in JAK1 S700N as compared with stimulated healthy control.
In summary, there are increased basal levels of pSTAT1 and pSTAT3 across genotypes which, upon stimulation with IFN-α2b or IFN-λ2, or with IL-6 or IL-21 respectively, are further augmented compared with healthy controls. Likewise, increased basal levels of pSTAT5 and pSTAT6 are enhanced further with IL-2 or IL-4, respectively, although a bit less pronounced. Individuals harboring these hyperactive JAK1 variants have elevated levels of cell-type-specific activation markers in T cells and NK cells, and higher levels of JAK-STAT activation markers in monocytes compared with healthy controls. Together, these data suggest that individuals harboring these GoF JAK1 variants experience steady-state inflammation that can worsen further upon exposure to cytokine triggers, leading to hyperactive cellular JAK–STAT signaling and the capacity for broader immune dysregulation—although in a genotype- and cell type–specific manner.
JAK1 GoF–associated atopic dermatitis resolves in one patient treated with a JAK1/JAK2-selective inhibitor
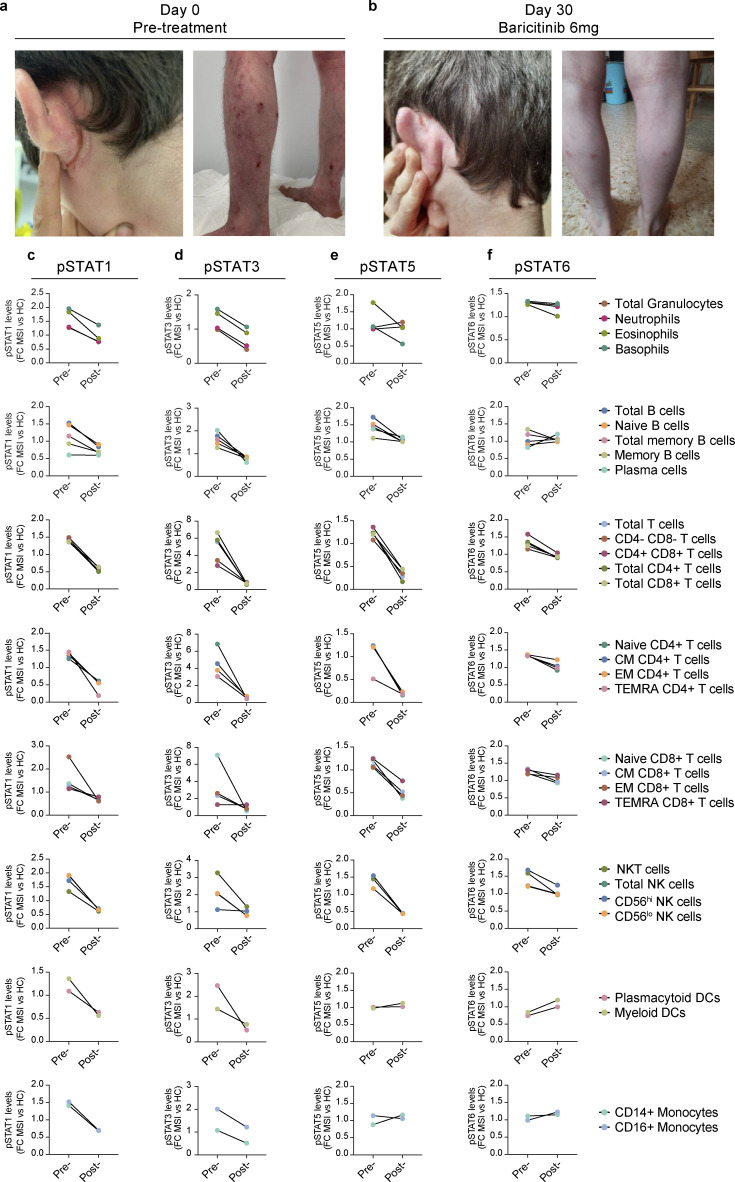
Considering the in vitro and ex vivo findings highlighting JAK1 hyperactivity as a likely potentiator of the clinical phenotypes observed, P3 (kindred I, JAK1 E139K), who experienced severe reductions in quality of life due to his severe atopic dermatitis (Fig. 1 b and Fig. 6 a), was considered for precision therapy with a JAK-inhibitor. The severity of atopic dermatitis in P3, which was present since infancy and was resistant or refractory to multiple therapies (topical steroids, systemic methotrexate, and systemic cyclosporine), was weighed against the risks of serious adverse events with JAK inhibitor use (Singh, 2022; Taylor et al., 2022), with the potential benefit of treatment ultimately outweighing the potential risks. The FDA-approved JAK1/JAK2-selective inhibitor, baricitinib (Mayence and Vanden Eynde, 2019), was prescribed in oral form at a daily dose of 6 mg (4 mg in the morning, then 2 mg at night) together with continued topical betamethasone use. The treatment regimen was well-tolerated and induced clinically significant resolution of the patient’s severe atopic dermatitis (Fig. 6 b), with the patient’s Scoring Atopic Dermatitis (SCORAD) index (European Task Force on Atopic Dermatitis, 1993) reducing from SCORAD = 60 at treatment initiation to SCORAD = 16 after 30 days. To assess the in vivo effects of baricitinib on JAK1-mediated signaling events, we again measured basal pSTAT1 (Fig. 6 c), pSTAT3 (Fig. 6 d), pSTAT5 (Fig. 6 e), and pSTAT6 (Fig. 6 f) signals in whole blood from the treated patient harboring the JAK1 E139K-causing allele. Concurrent with the resolution of his severe atopic dermatitis, baseline pSTAT1, pSTAT3, pSTAT5, and pSTAT6, signals in posttreatment patient cells normalized to levels comparable with the untreated healthy control. Together, these clinical and ex vivo findings implicate JAK1 E139K as a GoF mutant associated with the clinical features of this syndrome.
Figure 6.
JAK-inhibitor therapy resolves atopic dermatitis and abrogates hyperactive STAT phosphorylation in whole blood. (a–f) Severe atopic dermatitis in P3 (a) before (SCORAD = 60) and (b) after (SCORAD = 16) 30 days of in vivo treatment with baricitinib. Levels of pSTAT1 (c), pSTAT3 (d), pSTAT5 (e), and pSTAT6 (f) from P3 (E139K), who were treated with baricitinib in vivo. Results depicted as pretreatment (pre-) and posttreatment (post-) levels of pSTATs in granulocyte subsets, B cell subsets, major T cells subsets, CD4+ T cells subsets, CD8+ T cell subsets, NK cells subsets, dendritic cell subsets, and monocyte subsets. Results represented as fold-change in mean signal intensity (FCMSI) compared with non-treated HC (n = 1 for pre-treatment run and n = 1 for post-treatment run). For all panels, CM = central memory; EM = effector memory; TEMRA = T effector memory CD45RA+; NK = natural killer, DC = dendritic cell. Populations labeled “granulocytes” comprise all CD66b+ cells (including neutrophils, eosinophils, and basophils).
Discussion
To date, four disease-causing inborn GoF variants have been identified in JAK1: A634D, S703I, H596D, and C787F (Del Bel et al., 2017; Gruber et al., 2020; Takeichi et al., 2022; Fayand et al., 2023), two of which, JAK1 A634D and S703I, have undergone detailed biochemical and molecular assessment. Given that these variants are not present in public genetic databases such as gnomAD, they are deemed private variants and therefore considered, by historical standards (Tangye et al., 2022), to be exceedingly rare in the human population. However, the signs and symptoms of the 59 individuals we studied, all harboring one of four novel JAK1 variants, comprise more clinically common presentations of autoimmune and inflammatory diseases, which may explain the clinical features of 0.14% of all individuals in our healthcare system (Table 2). These clinical phenotypes include both autoimmune and inflammatory features and can molecularly exhibit cellular immune JAK–STAT hyperactivity, although this hyperactivity may even predate the development of the disease. The spectrum of disease includes variable genotype-specific immune dysregulation ranging from intermittent peripheral eosinophilia, for which there remains no clear temporal or molecular link with JAK1 GOF, to myeloid and lymphoid hyperactivation, even in the context of immunosuppressive treatments. Despite these molecular phenotypes being mild, the identification and study of the patients herein demonstrates that even rare or ultrarare variants could be contributing to more common presentations of autoimmunity and inflammation than previously thought (Fig. S1 f). Indeed, this work expands the number of bona fide JAK1 GOF-driven disease cases from four individuals (JAK1 A634D and S703I) to include 59 additional individuals. This demonstrates the importance and power of integrating long-standing forward genetics approaches used in IEI discovery with emerging technologies that enable reverse genetics-guided diagnosis and characterization of these diseases. Inspired by the marriage of these two approaches, we posit that these individually rare (<1:1,000) or ultrarare (<1:10,000) genotypes represent risk alleles of varying penetrance which, collectively, act as harbingers of a JAK1 GoF-associated autoimmunity, atopy, colitis, and dermatitis (JAACD) syndrome, as indicated by the clearly enriched syndromic phenotypes in our systematic EHR review.
Akin to the previously described disease-causing JAK1 variants, the four genotypes identified here are predicted to be deleterious by CADD assessment, with prediction scores for every variant (H596D = 24.1; A634D = 32; S703I = 26.1; C787F = 28; E139K = 22.6; R506C = 21.2; S700N = 25.1; V985I = 18.2) rising well above the JAK1 MSC of 3.313. However, the directionality of protein damage, whether GoF or loss of function (LoF), is impossible to predict using traditional methods such as CADD. As such, we used a novel in silico framework, which utilized natural language processing to identify features of published disease-causing alleles, which could, in turn, be used to iteratively train a JAK1-specific algorithm that could predict whether a given JAK1 allele is a neutral (no effect), LoF, or GoF (Kircher et al., 2014; Rentzsch et al., 2019; Sevim Bayrak et al., 2021). This tool accurately identified JAK1 A634D and S703I as likely GoF, which was expected given that they are published variants. More importantly, this tool accurately labeled JAK1 E139K, R506C, and S700N as likely GoF, and V985I as neutral, thus suggesting these four genotypes are likely to represent only a sampling of potential JAK1 GoF variants. Expanding this further, the identification of 15 additional JAK1 variants that are predicted to be GoF by the same in silico approach suggests that many more individuals may harbor variants with disease-causing GoF activity, potentially explaining up to 0.5% of all individuals in our healthcare system (Table 2). This hypothesis must, of course, be rigorously documented with masked case-control studies tested biochemically and evaluated immunologically. However, our identification of overrepresented syndromic features suggests that in patients presenting with these clinical phenotypes, especially when multiple family members are affected, JAK1 genotyping could lead to a substantial increase in precision therapy for a variety of inflammatory disorders. This is especially true for clinical presentations that might not have prompted genetic testing using current guidelines.
The clinical and immunological success in treating severe JAK1 GoF-mediated disease with JAK inhibitors in this study and previous ones (Del Bel et al., 2017; Gruber et al., 2020; Takeichi et al., 2022; Fayand et al., 2023) makes a strong case for a clinical trial where inclusion criteria could incorporate the presence of a JAK1 GoF variant. This could be timely given the growing use of next-generation genomics and EHR-linked genetic databases in broader and earlier genetic diagnosis of disease-causing JAK1 GOF cases.
Materials and methods
Human experimental guidelines approval statement
This research was conducted in accordance with the Medical Research Guidelines (https://www.ukri.org/councils/mrc/) and with the principles expressed in the Helsinki Declaration. Informed consent was obtained from all subjects under protocols approved by the Mount Sinai Institutional Review Board (IRB) (study #16-01286), the Baylor College of Medicine Institutional Review Board (study #H-29697), or the Imagine Institute Institutional Review Board (study #DC-2020-3994).
Patient identification and genotyping
Each proband initially presented to either a dermatologist (P3, E139K), gastroenterologist (P7, R506C), or clinical immunologist (P9, R506C; P9, R506C; P11, R506C; P13, S700N; P16, V985I) with complex clinical phenotypes, prompting genetic testing in the probands and their families. All tested patients and relatives underwent informed consent under IRB-approved protocols, and samples were collected in accordance therein. For exome sequencing of P1, P2, and P3 (Kindred I), genomic DNA was isolated under IRB-approved protocols (Imagine Institute IRB, study #DC-2020-3994) from patient whole blood (I Prep, Thermo Fisher Scientific) and then sequenced (Sure Select AllExon v5+UTR; Agilent), after which sequences were aligned (BWA aligner) with the human reference genome GRCh37/UCSC hg19. For exome sequencing of P12 and P13 (kindred V), samples were collected under IRB-approved protocols (Baylor College of Medicine IRB, study #H-29697) as previously described (Lam et al., 2019). Briefly, genomic DNA was isolated from the patient sample, from which exomes were captured (Sure Select AllExon v5+UTR; Agilent), then sequenced (HiSeq, Illumina), and aligned to the human genome GRCh37/UCSC hg19. For targeted genotyping of P6, P7, P8, P9, P10, P11, P15, and P16, the PID Panel (InVitae) was used clinically to probe for causative variants from a set of 474 genes. Following the initial identification of JAK1 VUS’, patient whole blood was obtained from P1, P3, P6, P7, P8, P9, P13, P14, and P15 under IRB-approved protocols (study #16-01286; Mount Sinai IRB), and genomic DNA was isolated for validation of each JAK1 variant by Sanger sequencing. In short, DNA was isolated (cat. no. 51304; Qiagen) from Ficoll-isolated granulocytes from the whole blood of each subject and then amplified by PCR using targeted probes. PCR products were then purified (cat. no. K0831; Thermo Fisher Scientific) and Sanger sequenced using targeted primers and analyzed using Snapgene (v4.2.11). Outside of the patients presented in this study, there were no others referred to us with JAACD features, nor did any of the patients in this study have additional JAK1 GoF variants on top of those described herein.
Variant analysis
Allele counts and variant frequencies were obtained from gnomAD (v4.0) (https://gnomad.broadinstitute.org/). In silico damage prediction was carried out using either combined annotation-dependent depletion (CADD) or a natural language processing and machine learning–based GoF prediction tool (Sevim Bayrak et al., 2021). The CADD framework functions by integrating sequence-derived context, gene annotations, epigenetic markers, and functional predictions into a single metric by contrasting evolutionarily stable variants with simulated variants (Kircher et al., 2014; Rentzsch et al., 2019). The resulting score for any given SNV is compared with the gene-specific mutation significance cutoff (MSC), which is considered the upper threshold of non-deleteriousness for that gene (Kircher et al., 2014; Rentzsch et al., 2019). The natural language processing and machine learning–based tool for GoF prediction functions by using natural language processing to identify discriminative gene- and protein-level features of published disease-causing SNVs to train a machine-learning algorithm that can predict whether a given SNV will be neutral (no effect), LoF or GoF in terms of protein activity (Sevim Bayrak et al., 2021). To do this for JAK1 specifically, over 150,000 published abstracts were text-mined using natural language processing to extract the discriminative features of pathogenic JAK1 variants. The extracted features were then annotated at the variant, gene, and protein levels, and the annotations were integrated into an iteratively trained in silico model. The final trained tool was then used to test whether candidate SNVs would confer neutral, LoF, or GoF phenotypes on JAK1 activity.
Visualization of JAK1 crystal structure
The full crystal structure of murine JAK1 dimer complexed with intracellular tails of IFN-λR1/IL-10Rβ (PDB ID 7T6F) (Glassman et al., 2022) was visualized using UCSF ChimeraX (v1.6.1), developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases (Pettersen et al., 2021).
Systematic EHR review
The Mount Sinai BioME Biobank is an EHR-linked biobank in New York City with exome sequence data available for over 30,000 subjects in the New York City metropolitan area (The BioME Biobank Program of the Charles Bronfman Institute for Personalized Medicine. GCO #07-0529—MSSM PPHS approved until 6/09/2023). This biobank was mined in accordance with IRB-approved methods (study #18-00893; Mount Sinai IRB) to identify subjects harboring any of the four prototype variants: E139K (n = 0), R506C (n = 31), S700N (n = 1), and V985I (n = 11). Variant-negative control subjects were identified in the same biobank from the remaining pool of JAK1 SNV-negative participants. Control subjects were age-, sex-, self-reported race/ethnicity–, and EHR density–matched to the variant-positive subjects at a magnitude of 3× (n = 129).
The systematic review search terms were defined using level 1 and 2 morbidity classification codes from the US Centers for Medicare and Medicaid Services (CMS) International Classification of Diseases (ICD)-10 schema as a guide for comprehensive phenotype inclusion (https://www.cms.gov). The major categories for search terms included signs and symptoms of any inflammatory or autoimmune disease, as well as signs and symptoms present in the index families from this study and from previously described JAK1 GOF cases. As an internal control measure, we included several control search terms which we would not expect to see overrepresented based on JAK1 genotype such as iatrogenic diseases, or potential phenocopies of JAK1 GOF such as irritable bowel syndrome (IBS)—none of which were enriched in JAK1 variant-negative or variant-positive groups.
To prevent detection bias in the systematic review, EHR identification numbers for all individuals were pooled, scrambled, and blinded with respect to genotype status. The systematic review was then conducted by a single investigator who documented the presence of any of these terms in binary (yes/no) for each EHR. Control search terms for each category were included and assessed using the same methods. Following EHR review completion, total counts for major categories and individual diagnoses were computed and, upon unblinding, compared between JAK1 variant-positive and variant-negative EHRs. Statistical significance was determined by Fisher’s exact test (P value, two-sided) and Baptista-Pike method (OR).
Cell lines and in vitro JAK1 expression
U4C (JAK1 null) fibrosarcoma cells were either transiently transfected or stably transduced to express JAK1 as follows: for transient expression, HEK293T (ATCC) cells were transfected with plasmids (pTRIP-X-IRES-RFP) encoding either WT or variant JAK1 for 30 h with the Transit-X2 dynamic delivery system (cat. no. 6005; Mirus Bio). For stable expression, lentiviral vectors carrying tetracycline-inducible plasmids (FUW-tetO-BFP) with either Firefly Luciferase (fLUC) (negative control) or JAK1 were first produced in HEK293T cells (ATCC). U4C cells were then stably transduced in the presence of polybrene for 48 h. Cells were then negatively selected with hygromycin B (cat. no. HG-80; Omega Scientific) until mock transduced control cells were completely negatively selected (∼2 wk). JAK1 expression was induced with 48-h exposure of transduced U4Cs to 2 µg/ml of doxycycline-HCL (cat. no. D3072; Sigma-Aldrich).
In vitro cytokine stimulations
Cells were stimulated with PBS (cat. no. 10010023; Thermo Fisher Scientific) for mock conditions, or with IFN-α2b (NDC no. 0085-4350-01; Merck) or IFN-λ2 (cat. no. 754202; BioLegend) at the indicated concentrations.
RT-qPCR
For JAK1 levels, doxycycline-induced cells were lysed and RNA was harvested (cat. no. 74106; Qiagen) and then reverse transcribed into cDNA (cat. no. 43-688-13; Thermo Fisher Scientific). qPCR was conducted for JAK1 (cat. no. 4331182 Assay ID Hs01026996_m1; Thermo Fisher Scientific) using GAPDH (Assay ID Hs01922876_u1) as a control gene on the QuantStudio 6 Pro (Thermo Fisher Scientific). Results were analyzed on Graphpad Prism 9 (v9.4) software for Windows. For ISG levels, doxycycline-induced cells were stimulated with PBS (mock) or IFN-α2b, 1,000 IU/ml (NDC no. 0085-4350-01; Merck) for 30 min, then allowed to rest for 12 h. RNA was harvested from cells (cat. no. 74106; Qiagen), and reverse transcribed into cDNA (cat. no. 43-688-13; Thermo Fisher Scientific). qPCR was conducted for JAK1 (cat. no. 4331182 Assay ID Hs01026996_m1; Thermo Fisher Scientific), IFI27 (cat. no. 4331182 Assay ID Hs01086373_g1; Thermo Fisher Scientific), MX1 (Assay ID Hs00895608_m1), and RSAD2 (cat. no. 4351370 Assay ID Hs00369813_m1; Thermo Fisher Scientific) using GAPDH (Assay ID Hs01922876_u1) as a control gene on the QuantStudio 6 Pro device (Thermo Fisher Scientific). Results were analyzed on GraphPad Prism 9 (v9.4) software for Windows by one-way analysis of variance (ANOVA) with Dunnett’s T3 comparison test.
Western blot
For western blot, doxycycline-induced cells were stimulated with PBS (mock) or IFN-λ2 (cat. no. 754202; BioLegend) for 15 min, then lysed (cat. no. 89901; Thermo Fisher Scientific), and clarified by centrifugation. The resulting clarified lysates were resuspended (cat. no. NP0007; Invitrogen; Thermo Fisher Scientific) and denatured/size-separated by boiling and SDS-PAGE (cat. no. 5671085; Bio-Rad). Samples were transferred to a membrane (cat. no. 10600021; Cytiva) under semidry conditions and then probed for protein targets. Antibodies and dilutions used for western blot are as follows: pSTAT1—1:1,000 (clone no. 58D6, cat. no. 9167S; Cell Signaling), pSTAT2—1:1,000 (clone no. D3P2P, cat. no. 88410; Cell Signaling), pSTAT3—1:2,000 (clone no. D3Aq, cat. no. 9145; Cell Signaling), JAK1—1:1,000 (clone no. D1T6W, cat. no. 50996; Cell Signaling), β-actin—1:20,000 (clone no. 8H10D10, cat. no. 3700; Cell Signaling), and GAPDH—1:20,000 (cat. no. MAB374; Millipore). Band quantification was conducted on ImageJ (v1.53t) software and analyzed on GraphPad Prism 9 (v9.4) software for Windows (Schneider et al., 2012). Uncropped scans and source data are provided in source data files.
Patient samples and whole blood stimulations
Whole blood was obtained under IRB-approved protocols (study #16-01286; Mount Sinai IRB) in sodium heparin tubes for both JAK1 variant-positive subjects (P1, P3, P7, P8, P9, P13, P14, and P15) and healthy controls (n = 9). The only probands from whom a whole blood specimen was not available were P11 (R506C) and P16 (V985I), although specimens from affected individuals with the same genotypes were represented in the study (P7, P8, and P9 for R506C; P15 for V985I). Whole blood was either mock-treated with PBS (cat. no. 10010023; Thermo Fisher Scientific)) or stimulated with IFN-α2b (NDC no. 0085-4350-01; Merck), IFN-λ2 (cat. no. 754202; BioLegend), IL-2 (cat. no. 589102; BioLegend), IL-4 (cat. no. 574002; BioLegend), IL-6 (cat. no. 570802; BioLegend), or IL-21 (cat. no. 571202; BioLegend) at the indicated concentrations for 30 min at 37°C, 5%CO2. Samples were then stabilized (cat. no. 501351689; Smart Tube) and rapidly cryopreserved at −80°C.
Whole blood assessment by mass cytometry
Samples were thawed, lysed, stained, and processed for mass cytometry (CyTOF) at the Human Immune Monitoring Center of the Icahn School of Medicine at Mount Sinai as previously described (Geanon et al., 2021). Briefly, samples were thawed and red blood cells were lysed (cat. no. 501351689; Smart Tube) and removed by centrifugation. Individual samples were then tagged with unique identifying barcodes (cat. no. 201060; MaxPar), pooled, and stained for surface markers for 30 min on ice. Cells were then fixed and permeabilized with methanol, followed by washing, resuspension, and staining for intracellular markers. All antibodies in the panel were preconjugated or conjugated at the Human Immune Monitoring Core with the X8 MaxPar conjugation kit (cat. no. 201300; Fluidigm). Antibody panels for all runs were validated, titrated, and premixed by the Human Immune Monitoring Center of the Icahn School of Medicine at Mount Sinai with each antibody at a final dilution of 1 µl/3 × 106 cells. The antibodies used are as follows: 89Y-CD45 (clone no. HI30, cat. no. 3089003B; Fluidigm), 115In-CD11c (clone no. Bu15, cat. no. MA182142; Invitrogen), 142Nd-CD19 (clone no. HIB19, cat. no. 302202; BioLegend), 143Nd-CD45RA (clone no. HI100, cat. no. 304102; BioLegend), 144Nd-CD141 (clone no. M80, cat. no. 344102; BioLegend), 145Nd-CD4 (clone no. RPA-T4, cat. no. 300502; BioLegend), 146Nd-CD8 (clone no. RPA-T8, cat. no. 301002; BioLegend), 147Sm-pSTAT5 (clone no. 47, cat. no. 3147012A; Fluidigm), 148Nd-CD16 (clone no. 3G8, cat. no. 302014; BioLegend), 149Sm-pSTAT6 (clone no. 18, cat. no. 3149004A; Fluidigm), 150Nd-CD1c (clone no. L161, cat. no. 331502; BioLegend), 151Eu-CD123 (clone no. 6H6, cat. no. 306002; BioLegend), 152Sm-CD66b (clone no. G10F5, cat. no. 305102; BioLegend), 153Eu-pSTAT1 (clone no. 4a, cat. no. 3153005A; Fluidigm), 154Sm-CD25 (clone no. M-A251, cat. no. 356102; BioLegend), 155Gd-CD27 (clone no. O323, cat. no. 302802; BioLegend), 156Gd-p38 (clone no. D3F9, cat. no. 3156002A; Fluidigm), 158Gd-pSTAT3 (clone no. 4, cat. no. 3158005A; Fluidigm), 159Tb-pMAPKAP2 (clone no. 27B7, cat. no. 3159010A; Fluidigm), 160Gd-CD14 (clone no. M5E2, cat. no. 301810; BioLegend), 161Dy-CD56 (clone no. B159, cat. no. 555513; BD Biosciences), 162Dy-TCRgd (clone no. B1, cat. no. 331202; BioLegend), 164Dy-CD161 (clone no. HP-3G10, cat. no. 339902; BioLegend), 166Er-CD169 (clone no. 7-239, cat. no. 346002; BioLegend), 168Er-CD3 (clone no. UCHT1, cat. no. 300402; BioLegend), 169Tm-STAT1 (clone no. 10C4B40, cat. no. 661002; BioLegend), 170Er-CD38 (clone no. HB-7, cat. no. 356602; BioLegend), 171Yb-EPX (clone no. AHE-1, cat. no. AB190715; Abcam), 173Yb-Perforin-FITC (clone no. dG9, cat. no. 308105; BioLegend), 174Yb-HLA-DR (clone no. L243, cat. no. 307602; BioLegend), 175Lu-pS6 (clone no. N7-548, cat. no. 3175009A; Fluidigm), and d176Yb-GranzymeB (clone no. GB11, cat. no. 561142; BD Biosciences). Stained samples were acquired on a CyTOF2 Mass Cytometer (Fluidigm). Data files were normalized using a standard bead-based algorithm (CyTOF software; Fluidigm). Cells were manually gated, and the resulting populations were visualized in lower dimensions using viSNE in Cytobank (v8.1) (https://www.cytobank.org/).
Targeted treatment of P3 with JAKinib
The patient was treated with a JAK1/JAK2-selective inhibitor, baricitinib (Olumiant, Eli Lilly) (Mayence and Vanden Eynde, 2019)—formulated as 2 mg film-coated tablets—taken orally at a dose of 6 mg/day (two tablets in the morning, one tablet at night) with or without food.
Online supplemental material
Fig. S1 includes information about the demographics of individuals included in the systematic review of EHRs, the expression of the JAK1 variants at the mRNA level, a schematic figure of the spectrum of autoimmune and inflammatory diseases caused by rare to mild JAK1 GOF variants, and the levels of pSTAT3 in whole blood after IL-21 stimulation. Fig. S2 shows the ex vivo assessment of baseline pSTAT levels compared with individual healthy controls from the same CyTOF run. Table S1 shows variants of uncertain significance identified in probands. Table S2 shows variants of uncertain significance identified in probands. Table S3 lists search terms used in a blinded systematic review of EHRs. Table S4 shows the gating strategy for ex vivo immune profiling of whole blood.
Supplementary Material
shows variants of uncertain significance identified in probands.
shows variants of uncertain significance identified in probands.
lists of search terms used in blinded systematic review of EHRs.
shows gating strategy for ex vivo immune profiling of whole blood.
is the source file for Fig. 2.
is the source file for Fig. 3.
Acknowledgments
We thank all participants and their families for taking part in this study. We thank Geoffrey Kelly and Seunghee Kim-Schulze from the Human Immune Monitoring Center (HIMC) at the Icahn School of Medicine for their technical expertise in mass cytometry. We also thank Amanda Merkelson for her guidance on access and navigation of the Mount Sinai BioME Biobank.
M.E. Horesh was supported by the National Institute of General Medical Sciences grant 5T32GM007280, National Institute of Allergy and Infectious Diseases (NIAID) grant 5T32AI078892, and NIAID grant 5R01AI148963. D. Bogunovic was supported by NIAID grant 5R01AI148963.
Author contributions: M.E. Horesh designed and performed most of the experiments, analyzed the data, and together with D. Bogunovic, wrote the manuscript. M. Martin-Fernandez, S. Buta, T. Le Voyer, A. Richardson, S. Hodeib, M. Youssef, J. Rosain, and O. Stewart helped to prepare and stimulate whole blood and process samples for mass cytometry. M. Martin-Fernandez, C. Gruber, R. Patel, V. Sancho-Shimizu, J.-L. Casanova, N.S. Abul-Husn, Y. Itan, J.D. Milner, and J. Bustamante helped design the experiments and analyze and interpret the data. Y. Wu, D. Stein, and Y. Itan helped design and carry out machine-learning guided in silico damage prediction. N.S. Abul-Husn and Y. Itan guided the design and analysis of the systematic EHR review. Y. Wu generated and retrieved EHR and demographic data for variant-positive individuals and retrieved demographic and EHR data for variant-negative matched controls. A. Richardson, A.E. Lee, R. Rubinstein, L. Ewald, and N. Maheshwari assisted with variant cloning, cell line generation, and in vitro testing of JAK1 activity. E. Puzenat, H. Lesmana, J.A. Kurowski, E. Feuille, L.A. Pedroza, R.L. Fuleihan, A Haseley, A Hovnanian, P. Quartier, G. Duartier, D. Mullan, V. Rahming, I.K. Chinn, J.R. Lupski, J.S. Orange, J.-L. Casanova, J.D. Milner, and J. Bustamante identified patients, genotyped patients, documented medical history, and/or prepared clinical reports. D. Bogunovic helped to design the experiments, analyze, and interpret the data, and write the manuscript. All authors commented on and edited the manuscript and figures.
Data availability
The exome sequencing data from P1, P3, and P13 are not available for deposition in the public domain due to IRB restrictions Imagine Institute IRB Protocol no. DC-2020-3994. The exome sequencing data for P13 have been deposited in dbGAP under accession code phs000711.v7.p2 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000711.v7.p2). The exome sequencing data for variant-positive subjects in the BioME Biobank have been deposited in dbGAP under accession code phs003453.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003453.v1.p1). Genotyping data from the remaining subjects were generated using clinical genetic panels and are considered protected health information, therefore are not available for deposition in the public domain. The Sanger sequencing data confirming these genotypes has been deposited in Mendeley Data under doi: https://doi.org/10.17632/2vfgyfyjfh.1.
References
- Del Bel, K.L., Ragotte R.J., Saferali A., Lee S., Vercauteren S.M., Mostafavi S.A., Schreiber R.A., Prendiville J.S., Phang M.S., Halparin J., et al. 2017. JAK1 gain-of-function causes an autosomal dominant immune dysregulatory and hypereosinophilic syndrome. J. Allergy Clin. Immunol. 139:2016–2020.e5. 10.1016/j.jaci.2016.12.957 [DOI] [PubMed] [Google Scholar]
- European Task Force on Atopic Dermatitis . 1993. Severity scoring of atopic dermatitis: The SCORAD index. Consensus report of the European Task Force on atopic dermatitis. Dermatology. 186:23–31. 10.1159/000247298 [DOI] [PubMed] [Google Scholar]
- Fagerberg, L., Hallström B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 13:397–406. 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayand, A., Hentgen V., Posseme C., Lacout C., Picard C., Moguelet P., Cescato M., Sbeih N., Moreau T.R.J., Zhu Y.Y.J., et al. 2023. Successful treatment of JAK1-associated inflammatory disease. J. Allergy Clin. Immunol. 152:972–983. 10.1016/j.jaci.2023.06.004 [DOI] [PubMed] [Google Scholar]
- Geanon, D., Lee B., Gonzalez-Kozlova E., Kelly G., Handler D., Upadhyaya B., Leech J., De Real R.M., Herbinet M., Magen A., et al. 2021. A streamlined whole blood CyTOF workflow defines a circulating immune cell signature of COVID-19. Cytometry. 99:446–461. 10.1002/cyto.a.24317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassman, C.R., Tsutsumi N., Saxton R.A., Lupardus P.J., Jude K.M., and Garcia K.C.. 2022. Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation. Science. 376:163–169. 10.1126/science.abn8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, C.N., Calis J.J.A., Buta S., Evrony G., Martin J.C., Uhl S.A., Caron R., Jarchin L., Dunkin D., Phelps R., et al. 2020. Complex autoinflammatory syndrome unveils fundamental principles of JAK1 kinase transcriptional and biochemical function. Immunity. 53:672–684.e11. 10.1016/j.immuni.2020.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan, C., Rolvering C., Raulf F., Kapp M., Drückes P., Thoma G., Behrmann I., and Zerwes H.G.. 2011. Jak1 has a dominant role over Jak3 in signal transduction through γc-containing cytokine receptors. Chem. Biol. 18:314–323. 10.1016/j.chembiol.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Hammarén, H.M., Virtanen A.T., Raivola J., and Silvennoinen O.. 2019. The regulation of JAKs in cytokine signaling and its breakdown in disease. Cytokine. 118:48–63. 10.1016/j.cyto.2018.03.041 [DOI] [PubMed] [Google Scholar]
- Karczewski, K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 581:434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig, Z., Yohannes M.T., Nkambule L.L., Zhao X., Goodrich J.K., Kim H.A., Wilson M.W., Tiao G., Hao S.P., Sahakian N., et al. 2023. A harmonized public resource of deeply sequenced diverse human genomes. bioRxiv. 10.1101/2023.01.23.525248 (Preprint posted February 28). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, M.T., Coppola S., Krumbach O.H.F., Prencipe G., Insalaco A., Cifaldi C., Brigida I., Zara E., Scala S., Di Cesare S., et al. 2019. A novel disorder involving dyshematopoiesis, inflammation, and HLH due to aberrant CDC42 function. J. Exp. Med. 216:2778–2799. 10.1084/jem.20190147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, D., Larner A., Chaudhuri A., Babiss L.E., and Darnell J.E. Jr.. 1986. Interferon-stimulated transcription: Isolation of an inducible gene and identification of its regulatory region. Proc. Natl. Acad. Sci. USA. 83:8929–8933. 10.1073/pnas.83.23.8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayence, A., and Vanden Eynde J.J.. 2019. “Baricitinib: A 2018 novel FDA-approved small molecule inhibiting Janus kinases,” pharmaceuticals (basel, Switzerland). Pharmaceuticals. 12:37. 10.3390/ph12010037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, S., John J., Shearer M., Kerr I.M., and Stark G.R.. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 9:4605–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., and Ferrin T.E.. 2021. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30:70–82. 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips, R.L., Wang Y., Cheon H., Kanno Y., Gadina M., Sartorelli V., Horvath C.M., Darnell J.E. Jr., Stark G.R., and O’Shea J.J.. 2022. The JAK-STAT pathway at 30: Much learned, much more to do. Cell. 185:3857–3876. 10.1016/j.cell.2022.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard, C., Al-Herz W., Bousfiha A., Casanova J.L., Chatila T., Conley M.E., Cunningham-Rundles C., Etzioni A., Holland S.M., Klein C., et al. 2015. Primary immunodeficiency diseases: An update on the classification from the International Union of Immunological Societies expert committee for primary immunodeficiency. J. Clin. Immunol. 35:696–726. 10.1007/s10875-015-0201-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch, P., Witten D., Cooper G.M., Shendure J., and Kircher M.. 2019. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47:D886–D894. 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins, J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., and Rice C.M.. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 472:481–485. 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevim Bayrak, C., Stein D., Jain A., Chaudhary K., Nadkarni G.N., Van Vleck T.T., Puel A., Boisson-Dupuis S., Okada S., Stenson P.D., et al. 2021. Identification of discriminative gene-level and protein-level features associated with pathogenic gain-of-function and loss-of-function variants. Am. J. Hum. Genet. 108:2301–2318. 10.1016/j.ajhg.2021.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, J.A. 2022. Risks and benefits of Janus kinase inhibitors in rheumatoid arthritis - past, present, and future. N. Engl. J. Med. 386:387–389. 10.1056/NEJMe2117663 [DOI] [PubMed] [Google Scholar]
- Takeichi, T., Lee J.Y.W., Okuno Y., Miyasaka Y., Murase Y., Yoshikawa T., Tanahashi K., Nishida E., Okamoto T., Ito K., et al. 2022. Autoinflammatory keratinization disease with hepatitis and autism reveals roles for JAK1 kinase hyperactivity in autoinflammation. Front. Immunol. 12:737747. 10.3389/fimmu.2021.737747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye, S.G., Al-Herz W., Bousfiha A., Chatila T., Cunningham-Rundles C., Etzioni A., Franco J.L., Holland S.M., Klein C., Morio T., et al. 2020. Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J. Clin. Immunol. 40:24–64. 10.1007/s10875-019-00737-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye, S.G., Al-Herz W., Bousfiha A., Cunningham-Rundles C., Franco J.L., Holland S.M., Klein C., Morio T., Oksenhendler E., Picard C., et al. 2022. Human inborn errors of immunity: 2022 update on the classification from the international union of immunological societies expert committee. J. Clin. Immunol. 42:1473–1507. 10.1007/s10875-022-01289-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, P.C., Takeuchi T., Burmester G.R., Durez P., Smolen J.S., Deberdt W., Issa M., Terres J.R., Bello N., and Winthrop K.L.. 2022. Safety of baricitinib for the treatment of rheumatoid arthritis over a median of 4.6 and up to 9.3 years of treatment: Final results from long-term extension study and integrated database. Ann. Rheum. Dis. 81:335–343. 10.1136/annrheumdis-2021-221276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka, K., Saharinen P., Pesu M., Holt V.E., Silvennoinen O., O’Shea J.J.. 2004. The Janus kinases (Jaks). Genome Biol. 5:253. 10.1186/gb-2004-5-12-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows variants of uncertain significance identified in probands.
shows variants of uncertain significance identified in probands.
lists of search terms used in blinded systematic review of EHRs.
shows gating strategy for ex vivo immune profiling of whole blood.
is the source file for Fig. 2.
is the source file for Fig. 3.
Data Availability Statement
The exome sequencing data from P1, P3, and P13 are not available for deposition in the public domain due to IRB restrictions Imagine Institute IRB Protocol no. DC-2020-3994. The exome sequencing data for P13 have been deposited in dbGAP under accession code phs000711.v7.p2 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000711.v7.p2). The exome sequencing data for variant-positive subjects in the BioME Biobank have been deposited in dbGAP under accession code phs003453.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003453.v1.p1). Genotyping data from the remaining subjects were generated using clinical genetic panels and are considered protected health information, therefore are not available for deposition in the public domain. The Sanger sequencing data confirming these genotypes has been deposited in Mendeley Data under doi: https://doi.org/10.17632/2vfgyfyjfh.1.