Abstract
Among the functions of the replicase of equine arteritis virus (EAV; family Arteriviridae, order Nidovirales) are important viral enzyme activities such as proteases and the putative RNA polymerase and RNA helicase functions. The replicase is expressed in the form of two polyproteins (open reading frame 1a [ORF1a] and ORF1ab), which are processed into 12 nonstructural proteins by three viral proteases. In immunofluorescence assays, the majority of these cleavage products localized to the perinuclear region of the cell. A dense granular and vesicular staining was observed, which strongly suggested membrane association. By using confocal microscopy and double-label immunofluorescence, the distribution of the EAV replicase was shown to overlap with that of PDI, a resident protein of the endoplasmic reticulum and intermediate compartment. An in situ labeling of nascent viral RNA with bromo-UTP demonstrated that the membrane-bound complex in which the replicase subunits accumulate is indeed the site of viral RNA synthesis. A number of ORF1a-encoded hydrophobic domains were postulated to be involved in the membrane association of the arterivirus replication complex. By using various biochemical methods (Triton X-114 extraction, membrane purification, and sodium carbonate treatment), replicase subunits containing these domains were shown to behave as integral membrane proteins and to be membrane associated in infected cells. Thus, contribution to the formation of a membrane-bound scaffold for the viral replication-transcription complex appears to be an important novel function for the arterivirus ORF1a replicase polyprotein.
Equine arteritis virus (EAV) (13) is the prototype of the Arteriviridae, a recently established family of positive-strand RNA viruses (for reviews, see references 27 and 35) which also includes lactate dehydrogenase-elevating virus, porcine reproductive and respiratory syndrome virus, and simian hemorrhagic fever virus. The gene encoding the arterivirus replicase (or nonstructural proteins [nsps]) is related to that of coronaviruses (9, 36), and the two virus families have recently been united in a new order, the Nidovirales (6). Despite a considerable size difference, the common ancestry of the arterivirus and coronavirus replicases is evident from the presence of an array of conserved domains and from the use of identical genome replication and expression strategies, including the discontinuous transcription of a nested set of subgenomic mRNAs to express the genes in the 3′ end of the genome (6, 12, 36).
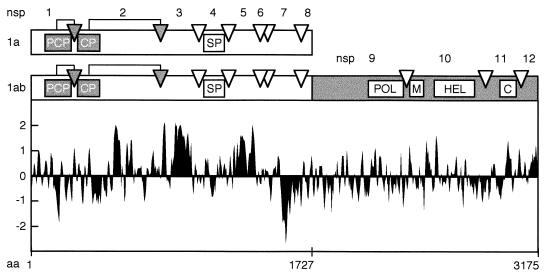
The EAV replicase gene covers the 5′-terminal three-quarters of the 12.7-kb genome (9) and is composed of two open reading frames (ORFs), ORF1a and ORF1b, which are both expressed from the genomic RNA. The replicase ORFs encode two multidomain precursor proteins: the 1,727-amino-acid ORF1a protein (187 kDa) and the 3,175-amino-acid ORF1ab protein (345 kDa). The latter is produced by means of a ribosomal frameshift in the ORF1a/ORF1b overlap region, which occurs with an estimated efficiency of between 15 and 20% during genome RNA translation (9). The EAV ORF1a and ORF1ab polypeptides are processed extensively by three ORF1a-encoded proteases (38, 40, 41). Our current understanding of EAV replicase processing is summarized in Fig. 1. The ORF1a protein can be cleaved at seven sites (39, 41, 52), yielding eight processing end products (named nsp1 to nsp8) and a number of processing intermediates. The N-terminal nsp1 contains a papainlike cysteine protease, which cleaves the nsp1/2 site (38). Likewise, nsp2 is generated by an autoproteolytic event that is carried out by a second cysteine protease, which is located in the N-terminal domain of nsp2 (40). The remaining nsp3–8 precursor (96 kDa) can be processed at five sites by the nsp4 chymotrypsinlike serine protease (SP) (41, 52). Cleavage of the nsp4/5 junction in nsp3–8 requires association of this processing intermediate with cleaved nsp2, which acts as a cofactor (52). As a result, two different pathways are used to process the C-terminal half of the ORF1a protein: a major pathway leading to cleavage of the nsp4/5 site (upon association of nsp2 and nsp3–8), and a minor pathway in which the nsp4/5 site remains uncleaved and the nsp5/6 and nsp6/7 sites are processed instead.
FIG. 1.
Proteolytic processing scheme, hydrophobicity plot, and subunit nomenclature of the EAV ORF1a and ORF1ab replicase polyproteins (39, 49, 52). The three EAV protease domains (papainlike cysteine protease [PCP], cysteine protease [CP], and serine protease [SP]) and their cleavage sites (arrows and arrowheads) are shown. In the ORF1b-encoded polypeptide, the four major domains conserved in nidoviruses are depicted: POL, putative RNA-dependent RNA polymerase; M, putative metal-binding domain; HEL, putative RNA helicase; C, conserved C-terminal domain specific for nidoviruses. The hydrophobicity plot was generated by the method of Kyte and Doolittle (24). Above the axis is hydrophobic. aa, amino acid.
The ORF1b-encoded part of the EAV replicase is assumed to contain functions essential for viral RNA replication and mRNA transcription (9, 47). Its processing by the nsp4 SP yields a set of cleavage intermediates and four end products (nsp9 to nsp12), including those that carry the putative viral RNA polymerase (nsp9) and helicase (nsp10) activities (48, 49). Immunofluorescence studies revealed that nsp9 and nsp10 localize to the perinuclear region of EAV-infected cells, where they are probably associated with intracellular membranes (49). However, an analysis of the ORF1b-encoded part of the replicase does not reveal domains of significant hydrophobicity (Fig. 1). Remarkably, such domains are present in a number of ORF1a-derived cleavage products, in particular nsp2, nsp3, and nsp5 (39, 52). This suggests that these replicase subunits may mediate the membrane association of a replication complex which also includes ORF1b-encoded proteins. To study the putative role of the hydrophobic ORF1a-encoded subunits in the membrane association of the EAV replication complex, we have analyzed their subcellular localization by using immunofluorescence assays and biochemical analyses. Our results show that a number of ORF1a-encoded replicase subunits behave as integral membrane proteins and that they are part of a membrane-bound complex that is involved in viral RNA synthesis.
MATERIALS AND METHODS
Cells, virus, and antisera.
Baby hamster kidney (BHK-21), rabbit kidney (RK-13), and African green monkey kidney (Vero) cells were used for infection experiments with the EAV Bucyrus strain (13) by the method described by de Vries et al. (11). The EAV replicase-specific rabbit antisera used in this study have been described previously (39, 49). Due to the recent revision of the EAV nsp nomenclature (52), the names of some antisera have been adapted to correspond to the number of the cleavage products which they recognize. Thus, the anti-nsp5 (39), anti-B1, and anti-B2 sera (49) are now referred to as anti-nsp7–8, anti-nsp9, and anti-nsp10, respectively. As before, the anti-nsp4 serum is a 1:1 mixture of the anti-4C and anti-4M peptide antisera (39). Mouse monoclonal antibodies (MAbs) were used to visualize the localization of the EAV ORF5-encoded glycoprotein GL (MAb 93B [18]) and the cellular enzyme protein disulfide isomerase (PDI; MAb 1D3 [50]). A rat MAb (BU1/75 [ICR1]; Harlan Sera-Lab Ltd., Loughborough, England) raised against bromodeoxyuridine (BrdU) was used to detect viral RNA that had been metabolically labeled with bromo-UTP (BrUTP) (see below).
BrUTP labeling.
The newly synthesized viral RNA in EAV-infected RK-13 or BHK cells (infected with a multiplicity of infection of 10 to 20) was labeled from 6.5 to 7.5 h postinfection (p.i.) with 10 mM BrUTP (Sigma). At 30 min before labeling, 10 μg of dactinomycin (Sigma) per ml was added to the medium to shut down host cell mRNA transcription. BrUTP was introduced into the cells by using cationic liposomes (Lipofectin; Life Technologies) and the manufacturer’s method for transfection of plasmid DNA. Briefly, Lipofectin and BrUTP (a freshly prepared 400 mM stock solution) were mixed in a small volume of serum-free medium and incubated for 15 min at room temperature. Subsequently, the mixture was diluted fivefold in medium containing 1% fetal calf serum and 10 μg of dactinomycin per ml and added to infected cells.
Immunofluorescence assays.
Cells were grown on coverslips, infected with EAV, and incubated at 39.5°C. The cells were fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS; pH 7.4) and washed with PBS containing 10 mM glycine. Following permeabilization with 0.1% Triton X-100 in PBS, indirect immunofluorescence assays were carried out with the anti-replicase rabbit antisera at dilutions of between 1:150 and 1:300 in PBS containing 5% fetal calf serum. Tissue culture supernatants of the hybridoma cell lines producing the anti-GL (93B) and anti-PDI (1D3) MAbs were each used at a dilution of 1:40. The anti-BrdU MAb tissue culture supernatant was used at a 1:10 dilution. As secondary antibodies, a Cy3-conjugated donkey anti-rabbit immunoglobulin G (IgG) antibody (Jackson ImmunoResearch Laboratories; 1:800 dilution), a fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG antibody (Dianova-Immunotech GmbH, Hamburg, Germany) (1:300 dilution), and an FITC-conjugated rabbit anti-rat IgG antibody (DAKO, Glostrup, Denmark) (1:100 dilution) were used.
Fluorescence microscopy.
For conventional fluorescence microscopy, samples were examined with a Zeiss Axiophot or Olympus microscope. Confocal fluorescence microscopy was performed with the confocal scanning laser beam microscope developed at the European Molecular Biology Laboratory, Heidelberg, Germany (42). The 476-nm (FITC) and 529-nm (Cy3) laser lines of an argon-ion laser (Spectra-Physics), an Axiophot microscope (Zeiss), and a 100× Plan-Neofluar objective (Zeiss) were used for imaging. The laser power, photomultiplier sensitivity, and number of averages (usually 32) were adjusted to generate images with sufficient contrast.
Radioactive labeling and immunoprecipitation of EAV nonstructural proteins.
RK-13 cells were infected with EAV (multiplicity of infection, 10 to 20) or mock infected, incubated at 39.5°C, and starved in medium without methionine or cysteine for 15 min before being labeled. Proteins were labeled from 5 to 8 h p.i. with 200 μCi of [35S]methionine and 80 μCi of [35S]cysteine per ml of medium (Express label; NEN). After cell lysis and cell fractionation (see below), EAV nsps were immunoprecipitated (39, 49) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography as described previously (39).
Cell fractionation and sodium carbonate extraction.
Intracellular membranes were isolated from EAV-infected and mock-infected RK-13 cells essentially as described by Fujiki et al. (16). After metabolic 35S labeling, the cells were washed with ice-cold PBS, scraped from the dish in PBS, and gently pelleted by centrifugation for 5 min at 500 × g. The cells were resuspended at 2 × 106 cells per ml in hypotonic buffer (1 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, 15 mM NaCl) containing the protease inhibitors leupeptin (2 μg/ml) and phenylmethylsulfonyl fluoride (0.4 mM) and broken by 15 to 20 strokes of a Dounce cell homogenizer. The nuclei were removed by centrifugation at 4°C for 5 min at 1,500 × g. The membranes were pelleted from the postnuclear supernatant (PNS) by ultracentrifugation through a 6% (wt/vol) sucrose cushion in a Beckman SW55 rotor (150,000 × g for 30 min at 4°C). Alternatively, the PNS was used for a Na2CO3 extraction. The pH was adjusted to 11 by addition of an equal volume of 200 mM Na2CO3, the sample was incubated on ice for 30 min, and membranes were pelleted as described above. Membrane pellets were dissolved in immunoprecipitation (IP) buffer (0.5% [vol/vol] Nonidet P-40, 0.1% [wt/vol] sodium desoxycholate, 0.5% [wt/vol] SDS, 150 mM NaCl, 5 mM EDTA, 20 mM Tris [pH 7.6]). The same concentrations of detergents were added to the supernatant fractions. In addition, the pH of the supernatant of the Na2CO3 extraction was brought to 7 by the addition of HCl. Finally, samples were diluted twice in immunoprecipitation buffer and used for immunoprecipitation as described previously (39).
TX-114 extraction.
Triton X-114 (TX-114) extractions were carried out essentially as described by Bordier (4). After metabolic 35S labeling, EAV-infected and mock-infected RK-13 cells were put on ice, washed twice with ice-cold PBS, and lysed with an ice-cold solution of 1% TX-114 in PBS. The nuclear fraction was removed by centrifugation in a microcentrifuge. The cytosolic fraction was incubated on ice for 15 min and was subsequently loaded on a cushion of 6% (wt/vol) sucrose in PBS containing 1% TX-114. The phases were separated by incubation at 37°C for 5 min and centrifugation at room temperature for 3 min at 2,700 × g. The supernatant was subjected to one or two additional rounds of TX-114 extraction. The detergent pellets were pooled and dissolved in IP buffer. The immunoprecipitation conditions for both fractions were equalized by adding concentrated detergents to the supernatant fraction. Samples were diluted fourfold in IP buffer and used for immunoprecipitations as described previously (39).
RESULTS
ORF1a- and ORF1b-encoded replicase subunits colocalize in the perinuclear region of EAV-infected cells.
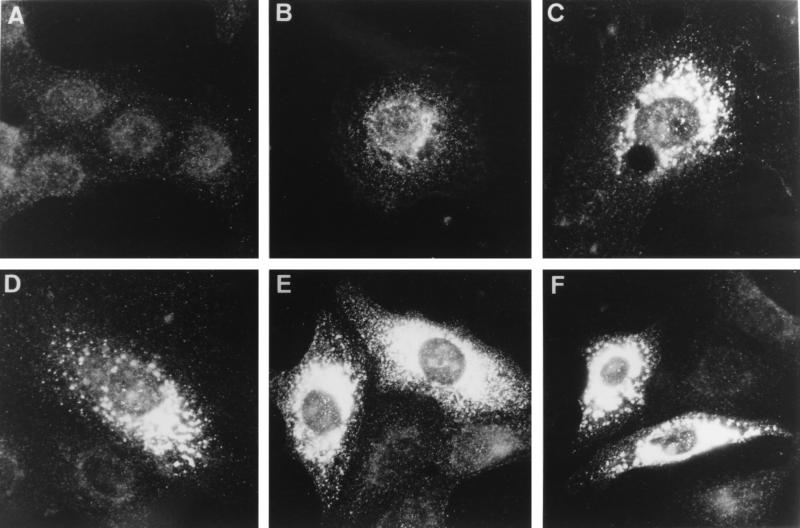
We have previously reported that at late time points in infection the ORF1b-encoded cleavage products nsp9 and nsp10, which contain the putative viral RNA polymerase and helicase activities, respectively, localize to the perinuclear region of EAV-infected BHK-21, RK-13, and Vero cells (49). The nsp9 and nsp10 staining in immunofluorescence assays strongly suggested the association of these proteins with intracellular membranes. By using antisera recognizing nsp1, nsp2, nsp4, nsp7–8, and nsp8 (39), this analysis was now extended to the ORF1a-encoded replicase subunits. With the exception of the anti-nsp1 serum, the staining of infected cells obtained with the ORF1a protein-specific antisera was identical to the pattern previously observed with the anti-nsp9 and anti-nsp10 sera (Fig. 2 and data not shown). These results are consistent with colocalization of most ORF1a- and ORF1b-encoded replicase subunits, suggesting that they assemble into a large complex which is likely to be involved in viral RNA synthesis. nsp1 colocalized only partially with the other replicase subunits and was also seen in the rest of the cytoplasm and—more interestingly—in the nucleus. The properties of this N-terminal replicase cleavage product will be discussed elsewhere (37).
FIG. 2.
Immunofluorescence analysis showing the time course of the intracellular distribution of the EAV replicase in Vero cells. Cells were mock infected (A) or EAV infected and fixed at 4 h (B), 6 h (C), 8 h (D), 10 h (E), and 12 h (F) p.i. Subsequently, the cells were processed for indirect immunofluorescence analysis with an anti-nsp2 antiserum (39). Photographs were generated with the same exposure times for recording and printing. Essentially identical results were obtained with antisera recognizing nsp4, nsp7–8, nsp8, nsp9, nsp10, and nsp12 (reference 49 and data not shown).
Some minor differences were observed among the three cell lines used for these studies (Fig. 3B, E, and H), most noticeably a concentration of the signal on one side of the nucleus in RK-13 cells. To analyze the development of the replicase pattern throughout the infection cycle, time course experiments were carried out, as illustrated in Fig. 2 for Vero cells labeled with the anti-nsp2 serum. The first signal, a punctate labeling of the perinuclear region, was detected at 3 h p.i. in BHK-21 and RK-13 cells (data not shown) and at 4 h p.i. in Vero cells (Fig. 2B), in which EAV replication is somewhat delayed. Subsequently, the signal rapidly increased (Fig. 2C and D) to give a dense staining in the area around the nucleus (Fig. 2E to F). A less dense, punctate pattern was observed toward the edges of the cells, suggesting labeling of vesicular structures. At later time points, cells exhibited pronounced cytopathic effects in the form of large cytoplasmic vacuoles, which did not stain with any of the EAV replicase antisera (data not shown).
FIG. 3.
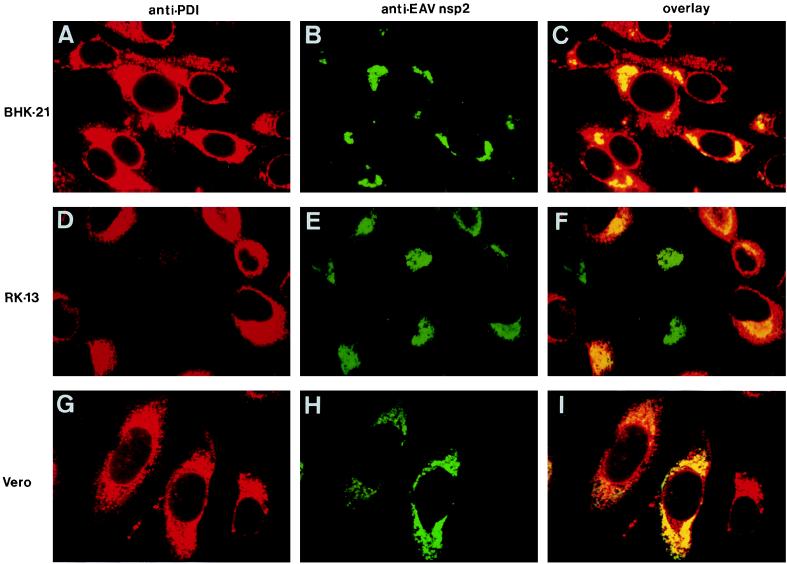
Colocalization of the EAV replicase and the cellular protein PDI, a marker for ER and IC. Three different cell lines were used: BHK-21 (A to C), RK-13 (D to F), and Vero (G to I). These were EAV infected, fixed at 8 h p.i., and processed for double-label immunofluorescence analysis with a mouse MAb directed against PDI (50), an anti-nsp2 rabbit antiserum (39), and appropriate secondary antibodies conjugated to fluorescent tags. PDI is shown in red (A, D, and G), and EAV nsp2 is shown in green (B, E, and H). For each sample, the differentially fluorescing images were recorded from the same optical section by using a confocal microscope. A computer-generated overlay of the PDI and EAV nsp2 images is shown (C, F, and I).
The EAV replicase is associated with membranes of the ER and/or IC.
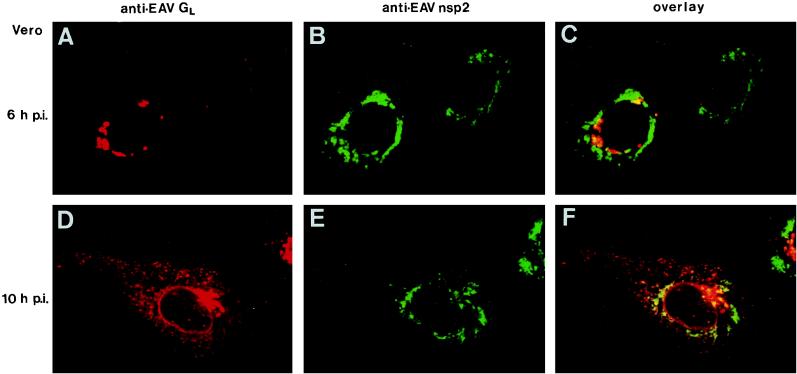
Confocal immunofluorescence microscopy (42) was used to analyze the subcellular localization of EAV replicase proteins in more detail. Double-labeling experiments were carried out with various rabbit antisera directed against the EAV replicase and mouse MAbs recognizing the cellular protein PDI (50) or the EAV ORF5 glycoprotein GL (18). PDI is a resident luminal protein of the endoplasmic reticulum (ER) and the intermediate compartment (IC) (Fig. 3A, D, and G) (23, 50). The anti-GL MAb can be used to stain the Golgi complex of EAV-infected cells. Double-label experiments with this antibody and an antiserum directed against the medial Golgi marker mannosidase II have shown that at the onset of EAV structural protein synthesis (6 to 8 h p.i. [Fig. 4A]), GL localizes exclusively to the Golgi complex (46). At later time points, probably due to intracellular virus assembly and transport, an additional vesicular staining throughout the cell and occasionally labeling of the nuclear envelope could be observed (Fig. 4D).
FIG. 4.
Intracellular distribution of the EAV replicase (nsp2) and the major EAV glycoprotein GL, which can, at the early stages of infection, be used as a marker for the Golgi complex (see the text). Vero cells were EAV infected, fixed at 6 h (A to C) and 10 h (D to F) p.i., and processed for double-label immunofluorescence analysis with a mouse MAb directed against GL (18), the anti-nsp2 rabbit antiserum (39), and appropriate fluorescent conjugates. GL is shown in red (A and D), and EAV nsp2 is shown in green (B and E). For each sample, the differentially fluorescing images were recorded from the same optical section by using a confocal microscope. Computer-generated overlays of the GL and nsp2 images are also shown (C and F).
Time course experiments with three cell lines revealed that throughout the infection cycle, the staining that is typical for the EAV replicase overlapped with a substantial part of the region of the cell that labeled for PDI (Fig. 3C, F, and I). During the course of infection, slight changes in the distribution of PDI were observed in a subpopulation of the cells, in particular a concentration of the signal in the perinuclear region. In some infected RK-13 cells, the labeling for PDI disappeared completely (Fig. 3D and F). As illustrated in Fig. 4, the areas which labeled for the EAV replicase were almost completely separated from the Golgi complex, which was stained with the anti-GL MAb. Thus, our data suggested that the EAV replicase complex interacts with the PDI-positive membrane compartments of the ER and/or IC.
Colocalization of replicase proteins and viral RNA synthesis.
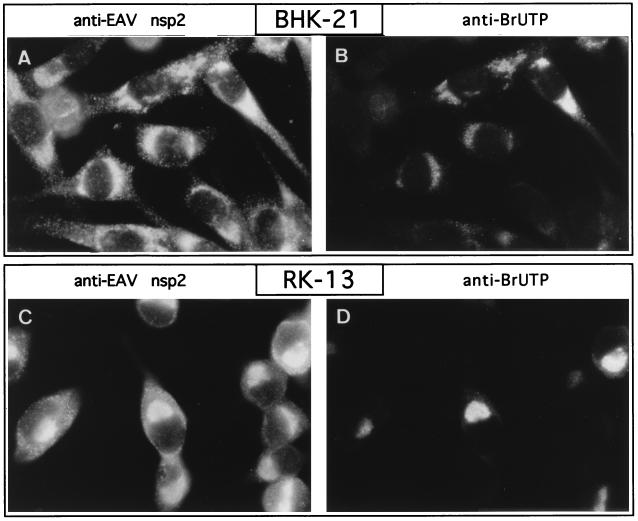
The fact that most replicase subunits, including nsp9, which contains the putative RNA polymerase function, localize to the same region of infected cells suggests the formation of a membrane-associated, multicomponent complex involved in viral RNA synthesis. To investigate whether the localization of the replicase complex indeed corresponds to the site of viral RNA synthesis in infected cells, a metabolic in vivo labeling of de novo-synthesized EAV RNA was performed by using BrUTP. It has been shown that this UTP analog can be incorporated into nascent RNA transcripts by both cellular (22, 51) and viral (29) DNA-dependent RNA polymerases and also by viral RNA-dependent RNA polymerases (30). Antibodies originally raised to detect BrdU-labeled DNA also recognize BrUTP-labeled RNA transcripts (45). This makes it possible to visualize de novo-synthesized RNA by using indirect immunofluorescence techniques. A disadvantage is that BrUTP cannot be taken up directly by intact cells (22, 51). However, Haukenes et al. (20) recently reported that it is possible to introduce BrUTP into cells by using the cationic liposomes which are commonly used for the transfection of plasmid DNA. By using a similar approach, EAV-infected BHK-21 and RK-13 cells were given a BrUTP-liposome mixture at 6.5 h p.i., just before the peak of viral RNA synthesis (10). Before and during the labeling, the cels were given dactinomycin to shut off host cell mRNA synthesis. After a 1-h incubation, the cells were fixed and processed for immunofluorescence with a rat MAb directed against BrdU and BrUTP, which convincingly labeled a subset (20 to 40%) of the cells. No labeling was observed when mock-infected cell cultures were used (data not shown). Following these initial single-label experiments, double-label experiments were carried out with the anti-BrUTP MAb and the EAV anti-nsp2 rabbit antiserum (Fig. 5). In addition to a population of double-positive cells, these experiments revealed the presence of EAV-infected (nsp2-positive) cells that had not been labeled with BrUTP. This nicely confirmed that the BrUTP staining shown in Fig. 5A and C is indeed specific and not due to, e.g., an unwanted cross-reaction between the anti-BrUTP MAb or the anti-rat IgG conjugate with any of the other antibodies used. Most importantly, Fig. 5 shows that the position of EAV nsp2 in the infected cell (and thus the position of most replicase subunits) corresponds very closely to the site of viral RNA synthesis. Thus, these in situ RNA labelings confirmed that the membrane-associated complex in which most arterivirus replicase proteins accumulate is the site of viral RNA synthesis.
FIG. 5.
Colocalization of the EAV replicase (nsp2) and viral RNA synthesis. BHK-21 (A and B) and RK-13 (C and D) cells were EAV infected, and de novo-synthesized viral RNA was labeled in situ by using BrUTP. Dactinomycin was used to shut off host cell RNA synthesis, and BrUTP was subsequently introduced into the cells by using liposomes. The cells were fixed at 7.5 h p.i. and processed for double-label immunofluorescence analysis with the anti-nsp2 rabbit antiserum (39), a rat MAb recognizing BrUTP-labeled RNA, and appropriate fluorescent conjugates. The labeling of the same cells for nsp2 (A and C) and BrUTP-labeled RNA (B and D) is shown. The presence of nsp2-positive but BrUTP-negative cells is explained by the fact that BrUTP could be introduced into only 20 to 40% of the cells (see the text).
Characterization of EAV replicase subunits by Triton X-114 extraction.
As outlined in the introduction and Fig. 1, the hydrophobicity profile of a number of ORF1a-encoded subunits strongly suggests that these subunits may associate with membranes and thereby play an important role in the formation of a membrane-associated scaffold for the arterivirus replication complex. In addition to immunofluorescence assays (Fig. 2 to 5), we used biochemical methods to characterize the properties of ORF1a- and ORF1b-encoded replicase subunits in more detail.
EAV replicase proteins from infected cells were subjected to TX-114 extraction. This detergent has been widely used for an extraction procedure which separates proteins mainly on the basis of their hydrophobic properties (4). Proteins that contain substantial hydrophobic domains, like most membrane proteins, usually partition into the detergent phase during TX-114 extraction, whereas hydrophilic proteins remain in the aqueous phase. Following 35S protein labeling, EAV-infected RK-13 cells were lysed in a buffer containing 1% TX-114. Cell lysates were kept on ice to avoid phase separation, and the nuclear fraction was removed by centrifugation. An immunoprecipitation analysis revealed that the nuclear pellet contained only small amounts of EAV replicase proteins (data not shown). Subsequently, the cell lysate was subjected to two or three rounds of TX-114 extraction (see Materials and Methods). The detergent phases from these extractions were combined and dissolved in immunoprecipitation buffer. The aqueous and detergent phases were equalized with respect to volume and to detergent and salt concentrations and were used for immunoprecipitations with antisera recognizing nsp1, nsp2, nsp4, nsp7–8, nsp9, and nsp10. The immunoprecipitation results of a typical TX-114 extraction analysis are shown in Fig. 6. The TX-114 lysis and extraction procedure did not interfere with the immunoprecipitation of the previously described EAV replicase subunits and processing intermediates (39, 49, 52). Nevertheless, the efficiency with which some of the antisera immunoprecipitated certain cleavage products appeared to be influenced (compared to our standard lysis and immunoprecipitation protocol [see below]). The strong interaction between cleaved nsp2 and nsp3 (or nsp3-containing processing intermediates), which leads to extensive mutual coimmunoprecipitation (39), was not affected.
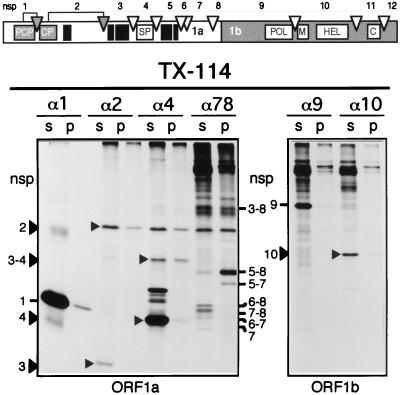
FIG. 6.
TX-114 extraction analysis of ORF1a- and ORF1b-encoded EAV replicase subunits. The replicase processing scheme and nsp nomenclature are shown at the top of the figure, with solid boxes representing hydrophobic domains (Fig. 1). RK-13 cells were EAV infected, and viral proteins were 35S labeled from 5 to 8 h p.i. Cells were lysed with 1% TX-114, and the lysate was subjected to three rounds of TX-114 extraction (see Materials and Methods) (4). Detergent pellets (p) and supernatants (s) were pooled and diluted in immunoprecipitation buffer. EAV replicase subunits were immunoprecipitated with a set of previously described rabbit antisera (39, 49) recognizing nsp1 (α1), nsp2 (α2), nsp4 (α4), nsp7–8 (α78), nsp9 (α9), and nsp10 (α10). The samples were analyzed by SDS-PAGE and autoradiography.
As expected, the proteins which lack hydrophobic domains of substantial size (the ORF1a-encoded nsp1 and nsp4 subunits, and the ORF1b-encoded nsp9 and nsp10 products [Fig. 1]) were recovered almost exclusively from the aqueous phase. Despite the presence of a central hydrophobic region (Fig. 1), nsp2 was recovered mainly from the aqueous phase when immunoprecipitated directly with the anti-nsp2 serum. The extremely hydrophobic nsp3 could not be studied directly due to the lack of an nsp3-specific antiserum. It was surprising, however, that the fraction of nsp3 which coimmunoprecipitated with nsp2, and thus can be assumed to be complexed with this upstream cleavage product, was found exclusively in the aqueous phase. It was also noteworthy that the nsp2 fraction which coprecipitated with the anti-nsp7–8 serum, probably in the form of an nsp2/nsp3–8 complex, showed an increased affinity for the detergent phase.
Although the hydrophilic nsp4 was recovered from the aqueous phase, a substantial part of the nsp3–4 processing intermediate, in which nsp4 is N-terminally extended with the hydrophobic nsp3, partitioned with the detergent phase. The anti-nsp4 serum also immunoprecipitated a set of bands in the 35- to 40-kDa region, which were specific for virus-infected lysates (data not shown). These bands may represent C-terminally extended nsp4-containing cleavage products (e.g. nsp4–5 and nsp4–6), although these were previously seen as minor bands (52). Possibly, the presence of TX-114 affects the efficiency with which these proteins were immunoprecipitated, since the fully cleaved nsp4 also appeared to be precipitated more efficiently from TX-114 lysates than from lysates produced by our standard lysis method (39).
The analysis of the cleavage products derived from the complex, alternative processing of the nsp5 to nsp8 region (52) provided convincing evidence that the hydrophobic region in nsp5 strongly influences the biochemical properties of specific processing intermediates which contain nsp5 at their N terminus. nsp5–8 and nsp5–7 were recovered almost exclusively from the detergent phase, but, strikingly, N-terminally truncated cleavage products that lacked nsp5 (nsp6–8, nsp7–8, nsp6–7, and nsp7) remained predominantly in the aqueous phase. The nsp3–8 processing intermediate was more or less equally divided between the two phases, but it should be noticed that the precipitated amount of this normally quite abundant intermediate (see Fig. 7) was remarkably small, suggesting that a substantial part of the nsp3–8 molecules either may not have been immunoprecipitated or may have aggregated and remained at the top of the gel.
FIG. 7.
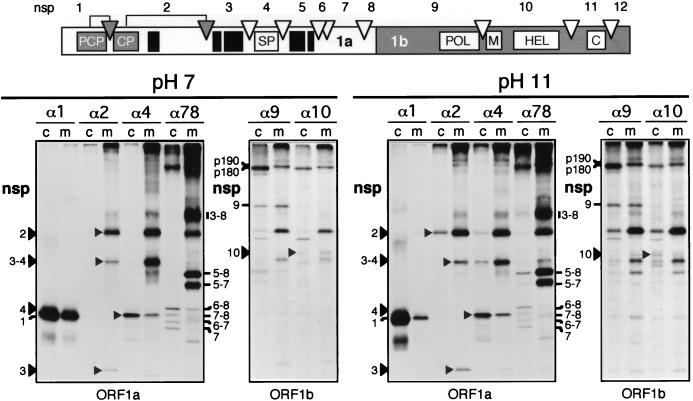
Membrane association of ORF1a- and ORF1b-encoded EAV replicase subunits. The replicase-processing scheme and nsp nomenclature are shown at the top of the figure, with solid boxes representing hydrophobic domains (Fig. 1). RK-13 cells were EAV infected, and viral proteins were 35S labeled from 5 to 8 h p.i. Cells were broken with a Dounce cell homogenizer, and a PNS was prepared. Half of the PNS was kept at pH 7, whereas the other half was treated with 100 mM sodium carbonate at pH 11 to remove integral and peripheral proteins from the membranes (16). Subsequently, membrane (m) and cytoplasmic (c) fractions were prepared by ultracentrifugation, diluted in immunoprecipitation buffer, and used for an immunoprecipitation analysis with the same set of antisera described in the legend to Fig. 6. The immunoprecipitation samples were analyzed by SDS-PAGE and autoradiography.
Identification of EAV replicase subunits in membrane fractions from infected cells.
To characterize the membrane association of EAV replicase subunits in more detail, intracellular membranes were isolated from infected cells and treated with sodium carbonate. This treatment can be used to differentiate between proteins that are peripherally associated with membranes and proteins that are integrated into the lipid bilayer (16). By using 100 mM Na2CO3 (at pH 11), microsomes can be stripped of their luminal and peripheral proteins, and subsequently the remaining membrane sheets, along with their integral proteins, can be pelleted and analyzed.
Following 35S protein labeling, EAV-infected RK-13 cells were broken by using hypotonic conditions and a Dounce cell homogenizer and a PNS was prepared by low-speed centrifugation. An immunoprecipitation analysis (data not shown) of PNS and nuclear pellet revealed that, in contrast to methods involving lysis by detergents, a substantial amount of EAV replicase proteins was present in the (crude) nuclear fraction. As a result, approximately 50% of the amount of nsp2, nsp3, nsp3–4, nsp3–8, nsp5–8, and nsp5–7 was lost. For the less hydrophobic proteins (nsp1, nsp4, nsp6–8 and its derivatives, nsp9, and nsp10) the loss was less dramatic but was nevertheless substantial. Subsequently, the PNS was divided into two equal parts, one of which was kept at pH 7 and the other was treated with Na2CO3 at pH 11 for 30 min and subsequently neutralized with HCl. Membranes were isolated from both samples by ultracentrifugation through a sucrose cushion. An immunoprecipitation analysis of membrane and cytoplasmic fractions was carried out with the same antisera used in the TX-114 extraction analysis (see above).
As shown in Fig. 7 (pH 7 panel), most of the EAV replicase-processing products were recovered from the membrane fraction of infected cells. The subunits which displayed affinity for the detergent fraction in the TX-114 extraction analysis (nsp2, nsp3–8, nsp3–4, nsp5–8, and nsp5–7) (Fig. 6) were almost exclusively membrane associated. However, substantial amounts of the hydrophilic nsp1 and nsp4 subunits were also recovered from the membrane fraction. Of the ORF1a-encoded proteins, only the C-terminal products produced by the minor processing pathway (nsp6–8 and its derivatives) were largely cytoplasmic. The ORF1b-derived nsp9 and nsp10 subunits, which are assumed to be at the heart of the viral RNA transcription machinery, were recovered largely from the membrane fraction, despite their presence in the aqueous phase upon TX-114 extraction (Fig. 6). Finally, there was a remarkable difference between the behavior of two closely related precursor proteins, which were previously named p190 and p180 on the basis of their mobility in SDS-PAGE (48). These abundant processing intermediates both reacted with the anti-nsp7–8, anti-nsp9, anti-nsp10, anti-nsp11, and anti-nsp12 antisera but not with the anti-nsp4 antiserum (Fig. 7) (49). In view of these data, we previously postulated that the p190 precursor consisted of nsp5–8 extended with the complete ORF1b-encoded protein (nsp9–12) and that p180 was an N-terminally truncated version of p190 (49). It was remarkable that p190, which contains the hydrophobic nsp5 domain, was recovered exclusively from the membrane fraction whereas the slightly smaller p180 molecule was present only in the cytoplasmic fraction. This result seems to confirm the hypothesis that p180 is an N-terminally truncated version of p190, which probably results from processing of the closely spaced nsp5/6 and/or nsp6/7 junctions. These data again underline the strong influence of nsp5 on the localization of precursor proteins of which it is part.
The immunoprecipitation analysis of the other half of the cell lysate, which had been treated at pH 11 before separation of the cytoplasmic and membrane fractions, revealed only minor changes compared to the results obtained with the pH 7 sample (Fig. 7, pH 11 panel). Small amounts of most cleavage products were released from the membranes, with the nsp1 and nsp2 immunoprecipitations showing the largest differences from the results of the analysis at pH 7. Nevertheless, the bulk of the nsp2, nsp3–8, nsp3–4, nsp5–8, nsp5–7, and p190 processing products remained associated with the membrane fraction. In combination with the data from the TX-114 extraction analysis, these results suggest that a number of these proteins contain one or multiple membrane-spanning domains.
DISCUSSION
The genome replication and mRNA transcription of eukaryotic positive-strand RNA viruses depends on a unique process of RNA-dependent RNA synthesis which occurs in the cytoplasm of the infected cell. A common property among these viruses appears to be the intimate association of their RNA-synthesizing machinery with intracellular membranes. Early in infection, a macromolecular complex is generated, which consists of viral RNA, replicative proteins, host cell-derived membranes, and probably host cell proteins. The reasons for the membrane association of viral RNA synthesis are poorly understood. It is generally assumed that the membranes play a structural and/or organizational role in the complex, possibly by offering a suitable microenvironment for viral RNA synthesis and/or by facilitating the use of membrane-bound host enzymes. Different virus groups use different intracellular membrane sites or compartments, e.g., endosomes and lysosomes (15), the endoplasmic reticulum (30, 32), or chloroplasts (7). Some viruses modify host cell membranes extensively and induce specific vesicular membrane structures which carry the viral replication complex (2, 14, 33, 53).
Little is known about the subcellular localization of the ribonucleoprotein complexes involved in nidovirus RNA replication and mRNA transcription. By using biochemical methods, the majority of coronavirus minus-strand RNAs was recently shown to be membrane associated (34), probably in the form of double-stranded replicative intermediates. Furthermore, ORF1b-derived replicase cleavage products of both the arterivirus EAV (nsp9, nsp10, and nsp12 [49]) and the human coronavirus 229E (21) have been reported to localize to intracellular membranes, a conclusion based on their subcellular distribution as observed in immunofluorescence studies. In this investigation, we have extended our previous studies (49) on the intracellular distribution of EAV replicase subunits by showing that most ORF1a-encoded cleavage products localize to the same region of infected cells as nsp9 and nsp10, the presumed viral polymerase and helicase proteins. Using a metabolic in situ RNA labeling (Fig. 5), we have provided convincing evidence that the complex in which the replicase subunits accumulate is indeed the site of viral RNA synthesis. At the resolution of the confocal microscope, the localization of the EAV replication complex partially overlaps with that of the cellular protein PDI (Fig. 3), suggesting the association with a subdomain of the ER or IC or the formation of ER- and IC-derived vesicular structures with which the replication complex associates. The latter idea is supported by the vesicular replicase staining which was observed in the perinuclear region of infected cells at early stages of infection (Fig. 2B) and toward the edges of the cell at later stages (e.g. Fig. 2D). Interestingly, the formation of paired membranes and large numbers of double-membrane vesicles at 3 to 6 h p.i. was previously described as a typical feature of arterivirus infection (5, 28, 43, 54). So far, the origin of these membrane structures has remained unclear, but they are unlikely to play a role in virus assembly. By using immunoelectron microscopy, we are currently investigating whether they are part of the macromolecular complex that is involved in arterivirus RNA replication and mRNA transcription.
The biochemical studies presented in Fig. 6 and 7 clearly implicate the hydrophobic domains in nsp2, nsp3, and especially nsp5 in membrane association. Although our results suggest that these subunits contain transmembrane domains, it should be noticed that the interpretation of our data is complicated by the presence of apparently strong protein-protein (and possibly protein-RNA) interactions within the replication complex. These interactions were not disrupted by, e.g., pH 11 treatment or immunoprecipitation in the presence of 0.5% SDS. Thus, the presence of certain replicase subunits in the TX-114 phase or in purified membrane fractions could result from their association with other viral (or even host) proteins which contain membrane-spanning domains. In particular, the interaction between nsp2 and nsp3 or nsp3-containing processing intermediates leads to extensive, mutual coimmunoprecipitation (39), which could not be disrupted by a treatment with 2% SDS, 1.5 M KCl, or 3 M urea or at pH 13 (data not shown). Furthermore, small amounts of nsp3–4, nsp4, nsp5–8, and nsp5–7 routinely coimmunoprecipitated with nsp9 and nsp10 during the membrane purification analyses (Fig. 7). We also observed that a number of hydrophilic proteins (nsp1, nsp4, nsp9, and nsp10) remained in the aqueous phase (as expected) upon TX-114 extraction (Fig. 6) but partially separated with the membrane fraction upon membrane purification (Fig. 7). Also, these results suggest the presence of multiple protein-protein interactions between replicase subunits in the replication complex.
The most conclusive results were obtained for the hydrophobic domain in nsp5. The presence of this subunit in the N terminus of a processing intermediate seems to result in almost complete membrane association. This was true not only for the relatively small products derived from the C-terminal part of the ORF1a protein (nsp5–8 and nsp5–7) but also for very large processing intermediates like p190, which is assumed to represent nsp5–12 (49). Otherwise comparable proteins that lack the nsp5 domain, like p180, nsp6–8, and nsp6–7, were recovered almost exclusively from the cytoplasmic fraction (Fig. 7). We recently described the existence of two proteolytic pathways for the processing of the nsp5-containing region of the EAV replicase polyprotein (52). The “major” pathway, which requires the association of nsp3–8 with nsp2, leads to processing of the nsp4/5 junction and generates processing products with a hydrophobic N-terminal nsp5 domain, which have now been shown to become membrane associated. The alternative, “minor” processing pathway results in cleavage of the nsp5/6 or nsp6/7 sites and can now be predicted to generate a set of equivalent but probably cytoplasmic proteins. Since we have recently developed an infectious cDNA clone for EAV (47), we may be able to investigate the significance of the minor processing pathway and its products in the near future.
At present it is unclear at which stage the various EAV replicase subunits associate with membranes. The translation, proteolytic processing, and membrane association of the replicase are likely to occur in a highly coordinated fashion. However, the absence of substantial hydrophobic stretches from the first 500 residues of the arterivirus replicase polyprotein (Fig. 1) rules out the use of the conventional, signal sequence-dependent mechanism for cotranslational translocation. Thus, the insertion of the transmembrane regions probably occurs posttranslationally, as has been proposed for certain other replicative proteins of positive-strand RNA viruses (32, 44). This mechanism may (partially) depend on prior proteolytic cleavages within the EAV replicase polyproteins. A computer analysis of the nsp5 sequence with PHDtm software (31) predicts up to five potential transmembrane helices in the region between residues 1280 and 1410 of the ORF1a protein. For the hydrophobic regions in nsp2 and nsp3, the same method predicts two and four transmembrane segments, respectively. In both nsp3 and nsp5, hydrophobic domains are located in the N terminus of the protein, just downstream of the nsp2/3 and nsp4/5 cleavage sites, which supports the concept of a relationship between proteolytic processing and membrane insertion. For example, the cleavage at the nsp4/5 site may be a crucial regulatory step, which could liberate the hydrophobic domain in nsp5 and allow its membrane insertion. Previously, we observed that expression products carrying (parts of) nsp5 in their N terminus could be translocated relatively efficiently (41). Although this property is an artifact of the expression system, it again underlines the affinity of the nsp5 region for membranes. Recent transient-expression experiments with nsp2 suggest that this protein is able to associate with membranes in the absence of other viral proteins. However, the expression of nsp2 to nsp7 was required to obtain the dense perinuclear staining (Fig. 2) that is typical for EAV-infected cells (data not shown). Future protein expression studies may shed more light on the membrane topology and protein-protein interactions of the various ORF1a-encoded replicase subunits.
Protein sequence comparisons (9, 12, 19, 36) and experimental data (47) have implicated the replicase ORF1b-encoded proteins of nidoviruses in viral RNA replication and subgenomic mRNA transcription. However, information on the functions of the more abundantly expressed ORF1a-encoded subunits is limited thus far. Only the role of this part of the replicase in proteolytic processing has been firmly established by the identification of multiple protease domains in all nidovirus ORF1a proteins (1, 3, 8, 17, 25, 26, 38, 40, 41, 55). The results presented in this paper indicate that contributing to the formation of a membrane-associated scaffold for the viral replication complex is an important additional function of the ORF1a polyprotein. Interestingly, arterivirus and coronavirus ORF1a polyproteins contain hydrophobic regions in comparable positions, which strongly suggests that they are important for the basic set of replicase functions which have been conserved during nidovirus evolution.
ACKNOWLEDGMENTS
We gratefully acknowledge Leonie van Dinten, Fred Wassenaar, Sasha Gorbalenya, and Maria Restrepo-Hartwig (University of Wisconsin, Madison) for technical assistance, helpful discussions, and/or critical review of the manuscript. Y.M. and E.J.S. thank Gareth Griffiths and the members of the 1995 Griffiths laboratory at the European Molecular Biology Laboratory (Heidelberg, Germany) for their hospitality. We are indebted to Janis Burkhardt for assistance with antibody selection and immunofluorescence microscopy, to Stephen Fuller (EMBL) for the anti-PDI monoclonal antibody, and to Amy Glaser (Cornell University) for the anti-EAV GL monoclonal antibody.
E.J.S. was supported by an EMBL travel grant from the Netherlands Organization for Scientific Research (NWO).
REFERENCES
- 1.Baker S C, Yokomori K, Dong S, Carlisle R, Gorbalenya A E, Koonin E V, Lai M M C. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J Virol. 1993;67:6056–6063. doi: 10.1128/jvi.67.10.6056-6063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bienz K, Egger D, Pfister T, Troxler M. Structural and functional characterization of the poliovirus replication complex. J Virol. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonilla P J, Hughes S A, Pinon J D, Weiss S R. Characterization of the leader papain-like proteinase of MHV A-59: identification of a new in vitro cleavage site. Virology. 1995;209:489–497. doi: 10.1006/viro.1995.1281. [DOI] [PubMed] [Google Scholar]
- 4.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 5.Breese S S, Jr, McCollum W H. Electron microscopic characterization of equine arteritis virus. In: Bryans J T, Gerber H, editors. Proceedings of the 2nd International Conference on Equine Infectious Diseases. S. Basel, Switzerland: Karger; 1970. pp. 133–139. [Google Scholar]
- 6.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 7.de Graaff M, Coscoy L, Jaspars E M. Localization and biochemical characterization of alfalfa mosaic virus replication complexes. Virology. 1993;194:878–881. doi: 10.1006/viro.1993.1335. [DOI] [PubMed] [Google Scholar]
- 8.den Boon J A, Faaberg K S, Meulenberg J J M, Wassenaar A L M, Plagemann P G W, Gorbalenya A E, Snijder E J. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Boon J A, Snijder E J, Chirnside E D, de Vries A A F, Horzinek M C, Spaan W J M. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Boon J A, Spaan W J M, Snijder E J. Equine arteritis virus subgenomic RNA transcription: UV inactivation and translation inhibition studies. Virology. 1995;213:364–372. doi: 10.1006/viro.1995.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries A A F, Chirnside E D, Horzinek M C, Rottier P J M. Structural proteins of equine arteritis virus. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doll E R, Bryans J T, McCollum W H M, Wallace M E. Isolation of a filterable agent causing arteritis of horses and abortion of mares. Its differentiation from the equine (abortion) influenza virus. Cornell Vet. 1957;47:3–41. [PubMed] [Google Scholar]
- 14.Egger D, Pasamontes L, Bolten R, Boyko V, Bienz K. Reversible dissociation of the poliovirus replication complex: functions and interactions of its components in viral RNA synthesis. J Virol. 1996;70:8675–8683. doi: 10.1128/jvi.70.12.8675-8683.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiki Y, Hubbard A L, Fowler S, Lazarow P B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao H Q, Schiller J J, Baker S C. Identification of the polymerase polyprotein products p72 and p65 of the murine coronavirus MHV JHM. Virus Res. 1996;45:101–109. doi: 10.1016/S0168-1702(96)01368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaser A L, de Vries A A F, Dubovi E J. Comparison of equine arteritis virus isolates using neutralization monoclonal antibodies and identification of sequence changes in GL associated with neutralization resistance. J Gen Virol. 1995;76:2223–2233. doi: 10.1099/0022-1317-76-9-2223. [DOI] [PubMed] [Google Scholar]
- 19.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haukenes G, Szilvay A M, Brokstad K A, Kanestrom A, Kalland K H. Labeling of RNA transcripts of eukaryotic cells in culture with BrUTP using a liposome transfection reagent (DOTAP) BioTechniques. 1997;22:308–312. doi: 10.2144/97222st03. [DOI] [PubMed] [Google Scholar]
- 21.Heusipp G, Grotzinger C, Herold J, Siddell S G, Ziebuhr J. Identification and subcellular localization of a 41 kda, polyprotein 1ab processing product in human coronavirus 229E-infected cells. J Gen Virol. 1997;78:2789–2794. doi: 10.1099/0022-1317-78-11-2789. [DOI] [PubMed] [Google Scholar]
- 22.Jackson D A, Hassan A B, Errington R J, Cook P R. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krijnse Locker J, Ericsson M, Rottier P J M, Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from RER to the Golgi complex requires only one vesicular transport step. J Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 25.Liu D X, Brown T D K. Characterisation and mutational analysis of an ORF 1a-encoding proteinase domain responsible for proteolytic processing of the infectious bronchitis virus 1a/1b polyprotein. Virology. 1995;209:420–427. doi: 10.1006/viro.1995.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y, Lu X, Denison M R. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J Virol. 1995;69:3554–3559. doi: 10.1128/jvi.69.6.3554-3559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plagemann P G W. Lactate dehydrogenase-elevating virus and related viruses. In: Fields B N, Knipe P M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1105–1120. [Google Scholar]
- 28.Pol J M, Wagenaar F, Reus J E G. Comparative morphogenesis of three PRRS virus strains. Vet Microbiol. 1997;55:203–208. doi: 10.1016/s0378-1135(96)01329-6. [DOI] [PubMed] [Google Scholar]
- 29.Pombo A, Ferreira J, Bridge E, Carmo-Fonseca M. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 1994;13:5075–5085. doi: 10.1002/j.1460-2075.1994.tb06837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Restrepo-Hartwig M A, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rost B, Casadio R, Fariselli P, Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaad M C, Jensen P E, Carrington J C. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 1997;16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlegel A, Giddings T H, Ladinsky M S, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sethna P B, Brian D A. Coronavirus genomic and subgenomic minus-strand RNAs copartition in membrane-protected replication complexes. J Virol. 1997;71:7744–7749. doi: 10.1128/jvi.71.10.7744-7749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snijder E J, Meulenberg J J M. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 36.Snijder E J, Spaan W J M. The coronaviruslike superfamily. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 239–255. [Google Scholar]
- 37.Snijder, E. J., Y. van der Meer, G. J. van Tol, and A. L. M. Wassenaar. 1998. Unpublished data.
- 38.Snijder E J, Wassenaar A L M, Spaan W J M. The 5′ end of the equine arteritis virus replicase gene encodes a papainlike cysteine protease. J Virol. 1992;66:7040–7048. doi: 10.1128/jvi.66.12.7040-7048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snijder E J, Wassenaar A L M, Spaan W J M. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snijder E J, Wassenaar A L M, Spaan W J M, Gorbalenya A E. The arterivirus nsp2 protease: an unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J Biol Chem. 1995;270:16671–16676. doi: 10.1074/jbc.270.28.16671. [DOI] [PubMed] [Google Scholar]
- 41.Snijder E J, Wassenaar A L M, van Dinten L C, Spaan W J M, Gorbalenya A E. The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases. J Biol Chem. 1996;271:4864–4871. doi: 10.1074/jbc.271.9.4864. [DOI] [PubMed] [Google Scholar]
- 42.Stelzer E H, Wacker I, DeMey J R. Confocal fluorescence microscopy in modern cell biology. Semin Cell Biol. 1991;2:145–152. [PubMed] [Google Scholar]
- 43.Stueckemann J A, Holth M, Swart W J, Kowalchyk K, Smith M S, Wolstenholme A J, Cafruny W A, Plagemann P G W. Replication of lactate dehydrogenase-elevating virus in macrophages. 2. Mechanism of persistent infection in mice and cell culture. J Gen Virol. 1982;59:263–272. doi: 10.1099/0022-1317-59-2-263. [DOI] [PubMed] [Google Scholar]
- 44.Towner J S, Ho T V, Semler B L. Determinants of membrane association for poliovirus protein 3AB. J Biol Chem. 1996;271:26810–26818. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- 45.Vanderlaan M, Thomas C B. Characterization of monoclonal antibodies to bromodeoxyuridine. Cytometry. 1985;66:501–505. doi: 10.1002/cyto.990060603. [DOI] [PubMed] [Google Scholar]
- 46.van der Meer, Y., and E. J. Snijder. 1998. Unpublished data.
- 47.van Dinten L C, den Boon J A, Wassenaar A L M, Spaan W J M, Snijder E J. An infectious arterivirus cDNA clone: identification of a replicase point mutation which abolishes discontinuous mRNA transcription. Proc Natl Acad Sci USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Dinten, L. C., S. Rensen, W. J. M. Spaan, A. E. Gorbalenya, and E. J. Snijder. Unpublished data.
- 49.van Dinten L C, Wassenaar A L M, Gorbalenya A E, Spaan W J M, Snijder E J. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J Virol. 1996;70:6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaux D, Tooze J, Fuller S. Identification by anti-idiotypic antibodies of an intracellular membrane protein that recognizes a mammalian endoplasmic reticulum retention signal. Nature. 1990;345:495–502. doi: 10.1038/345495a0. [DOI] [PubMed] [Google Scholar]
- 51.Wansink D G, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wassenaar A L M, Spaan W J M, Gorbalenya A E, Snijder E J. Alternative proteolytic processing of the arterivirus ORF1a polyprotein: evidence that nsp2 acts as a cofactor for the nsp4 serine protease. J Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westaway E G, Mackenzie J M, Kenney M T, Jones M K, Khromykh A A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2b with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood O, Tauraso N M, Liebhaber H. Electron microscopic study of tissue cultures infected with simian haemorrhagic fever virus. J Gen Virol. 1970;7:129–136. doi: 10.1099/0022-1317-7-2-129. [DOI] [PubMed] [Google Scholar]
- 55.Ziebuhr J, Herold J, Siddell S G. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J Virol. 1995;69:4331–4338. doi: 10.1128/jvi.69.7.4331-4338.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]