The poor prognosis of M3 uveal melanomas generates a local adaptive immune response, which does not recirculate, while the better prognosis of SF3B1-mutated uveal melanomas induces a systemic immune response in the absence of a detectable local response.
Abstract
Uveal melanoma (UM) is the most common cancer of the eye. The loss of chromosome 3 (M3) is associated with a high risk of metastases. M3 tumors are more infiltrated by T-lymphocytes than low-risk disomic-3 (D3) tumors, contrasting with other tumor types in which T cell infiltration correlates with better prognosis. Whether these T cells represent an antitumor response and how these T cells would be primed in the eye are both unknown. Herein, we characterized the T cells infiltrating primary UMs. CD8+ and Treg cells were more abundant in M3 than in D3 tumors. CD39+PD-1+CD8+ T cells were enriched in M3 tumors, suggesting specific responses to tumor antigen (Ag) as confirmed using HLA-A2:Melan-A tetramers. scRNAseq-VDJ analysis of T cells evidenced high numbers of proliferating CD39+PD1+CD8+ clonal expansions, suggesting in situ antitumor Ag responses. TCRseq and tumor-Ag tetramer staining characterized the recirculation pattern of the antitumor responses in M3 and D3 tumors. Thus, tumor-Ag responses occur in localized UMs, raising the question of the priming mechanisms in the absence of known lymphatic drainage.
Introduction
Uveal melanoma (UM) is a rare and aggressive form of melanoma that arises from melanocytes in the uveal tract of the eye (Rodrigues et al., 2019). Although UM accounts for only 5% of all melanoma cases, it represents the most common primary intraocular malignancy in adults (Bronkhorst and Jager, 2013). Contrasting with the progress made in the understanding and management of cutaneous melanoma, the prognosis for patients with UM remains poor (Demkowicz et al., 2023). The primary tumor is usually treated efficiently, but one-third of the patients eventually develop metastases, involving the liver in 90% of the cases (Rodrigues et al., 2019). Contrary to skin melanoma, a low mutation rate is found in UMs, which also do not harbor the recurrent mutations observed in cutaneous melanoma (Robertson et al., 2017). The initial oncogenic event in UM is an activating mutation of the GNAQ or GNA11 proteins (Van Raamsdonk et al., 2009). A second genetic event characterizes the different subtypes of UM: (i) deleterious mutations in BRCA1-associated protein 1 (BAP1) on chromosome 3, accompanied by loss of one copy of chromosome 3 (monosomy 3 [M3] or isodisomy 3 [iso3]); (ii) mutations in splicing factor 3b subunit 1 (SF3B1); or (iii) mutations occurring in the 5′ region of the eukaryotic translation initiation factor 1A X-linked (EIF1AX).
Thus, the genetic status of chromosome 3 defines two distinct subsets of UM. M3 tumors are associated with BAP1 mutations, which lead to the complete inactivation of BAP1 (BAP1ina). M3 UM is associated with the poorest prognosis, as over 80% of cases exhibit early metastases (Harbour et al., 2010). An additional poor prognosis factor is the gain of q-arm of chromosome 8 (Cassoux et al., 2014; Robertson et al., 2017). On the other hand, disomy 3 (D3) tumors carry SF3B1 or EIF1AX mutations and are associated with better clinical outcomes. SF3B1-mutated D3 tumors show a propensity for late metastases, while EIF1AX-mutated D3 tumors have a good prognosis (Yavuzyigitoglu et al., 2016). The molecular and immune mechanisms underlying the distinct clinical outcomes between M3 and D3 tumors remain unclear.
Consistent with the low mutation burden of UMs (Furney et al., 2013), immune checkpoint inhibitors have a low response rate in metastatic patients. Unlike cutaneous melanoma, UMs do not express most oncotestis antigens (Ags) (Mulcahy et al., 1996), indicating that their epigenetic landscape is different from that of skin melanoma. Despite the scarcity of classical sources of tumor Ags, we have observed an increase in the frequency of effector CD4+ and CD8+ T cells in the peripheral blood of metastatic UM patients as compared with healthy donors (Péguillet et al., 2014). Moreover, the frequencies of effector CD4+ and CD8+ T cells were correlated in patients, suggesting a coordinated immune response toward unknown tumor Ags. As in cutaneous melanoma, an immune response against HLA-A2–restricted epitopes derived from differentiation melanocyte Ags such as Melan-A or gp100 or against the oncotestis Ag NY-ESO-1 is detected in metastatic UM (Karlsson et al., 2020). Until recently, no tumor-specific neo-Ags have been characterized.
This has just changed as we showed that SF3B1 mutations generate public neo-Ags specifically expressed by tumor cells, leading to spontaneous CD8+ T cell responses in the blood of patients bearing metastatic SF3B1mut D3 tumors (Bigot et al., 2021). These CD8+ T cells were identified using HLA-A2 tetramers loaded with the SF3B1mut-related neo-epitopes. Notably, the tumor cells were recognized and killed by neo-epitope–specific CD8+ T cells. However, the intratumoral immune response was barely characterized. Besides therapeutic opportunities, these public neo-Ags can be used to monitor the spontaneous antitumor Ag-specific immune responses in patients at different stages of their disease. In contrast, the T cell response against tumor-specific neo-Ags in patients with M3 tumors has not been investigated in the absence of known neo-Ags.
Yet, in addition to its impact on prognosis, the inactivation of BAP1 in M3 tumors has been associated with increased immune infiltration within the primary tumor (Maat et al., 2008). In particular, regulatory T cells (Tregs) and myeloid-derived suppressor cells are more abundant in M3 than in D3 tumors, which might contribute to the establishment of an immunosuppressive microenvironment (Bronkhorst et al., 2012). Not much is known about the Ag specificity of the CD8+ T cells found in the primary tumors, which are often not excised nor biopsied. We found circulating memory Melan-A–specific CD8+ T cells in metastatic patients (Bigot et al., 2021). The presence in the blood of cells responding to Melan-A, gp100, and tyrosinase has been suggested by indirect means (Triozzi et al., 2015) in some patients with primary or metastatic disease. The exact nature and the Ag specificity of the CD8+ T cells found in the eye tumor or the liver metastasis of UM remain unclear. In other tumor types, simultaneous expression of both PD-1 and CD39 by the tumor-infiltrating CD8+ T cells indicates an immune response toward tumor neo-Ags (Simoni et al., 2018; Duhen et al., 2018). Whether CD8+ T cells with such characteristics are found in the different types of primary UM is unknown.
Herein, we studied the phenotype of the T cells found in primary UM at the time of diagnosis using multicolor flow cytometry, single-cell transcriptomic (single-cell RNA sequencing [scRNAseq]) associated with single-cell TCR repertoire analysis with multiplex tetramer staining, and bulk TCR sequencing (TCRseq) for alpha and beta chains. We evidenced differences in the number and phenotype of the T cells according to the M3 versus D3 status of the tumor. We also characterized the systemic T cell phenotype and looked for tumor Ag–specific immune response both in the tumor and the blood. Intriguingly, we observed an early tumor Ag–specific systemic response at the time of primary treatment in some patients.
Results
Characterization of the main T cell subsets present in primary UMs
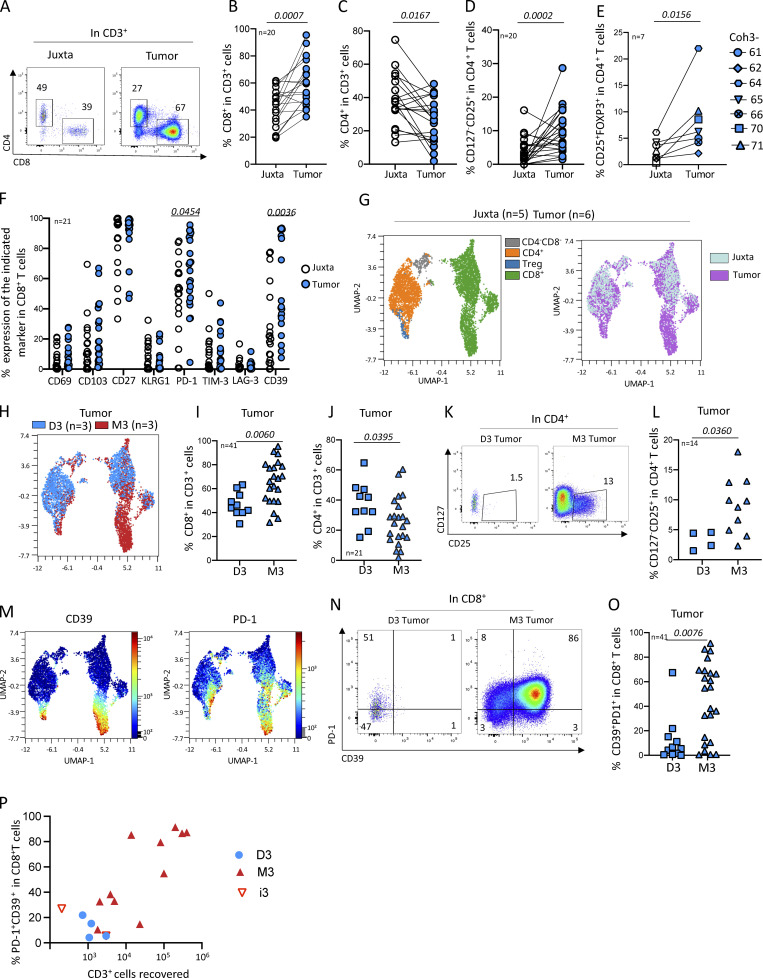
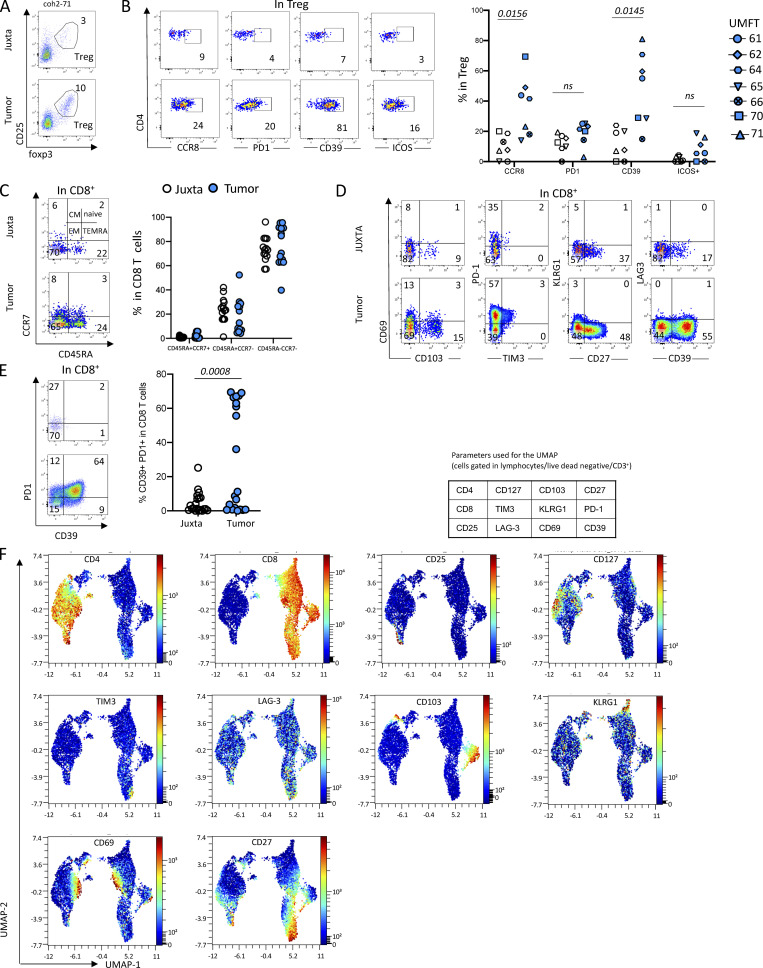
To investigate the adaptive immune response in localized UM, we first quantified and determined the phenotype of the main T cell subsets (CD8+, CD4+, and regulatory CD4+ [Tregs]) isolated from primary tumors as compared to juxta-tumoral tissue in 20 enucleated patients (Table S1). The proportion of CD8+ T cells was higher in the tumor than in the juxta-tumor site (Fig. 1, A and B), whereas it was the opposite for CD4+ T cells (Fig. 1 C). To assess the proportion of Tregs, we first measured the frequency of CD25+CD127− (Seddiki et al., 2006) in CD4+ T cells Fig. 1 D), which was higher (P = 0.0002) in the tumor than in the juxta-tumor site (Fig. 1 D). As the CD25+CD127− phenotype of Tregs has not been as well validated in tissues as in blood, we confirmed this result using a FOXP3 staining of CD4+ T cells (Fig. S1 A) in seven primary UM tumors. The proportion of Tregs (CD25+FOXP3+) in CD4+ T cells was significantly (P = 0.0156) higher in tumors than in the juxta-tumor site (Fig. 1 E). To assess the activation status of Tregs in the tumor, we measured the expression of CCR8, PD-1, CD39, and ICOS (Fig. S1 B). The proportion of Tregs expressing CCR8 or CD39 was higher in the tumor as compared with the juxta-tumoral site (P = 0.0156 and P = 0.0145, respectively), but the expression of PD-1 and ICOS was similar (Fig. S1 B). Notably, the patient (coh3_64) with the highest (22%) proportion of Tregs rapidly became metastatic 92 days after enucleation (Table S1). Whether the high proportion of Tregs in the tumor of this patient reflects a rapidly growing tumor or is instrumental in tumor progression is unknown.
Figure 1.
Characterization of the T cells infiltrating enucleated primary UMs. (A) Example of CD4 versus CD8 expression in T (CD3+) cells in juxta and tumor sites. (B–E) Frequency of the indicated cell populations in juxta-tumor and tumor tissues. (D and E) Frequency of Tregs is defined as CD127−CD25+ (D) or CD25+FOXP3+ (E) in CD4+ T cells. (F) Frequency of CD8+ T cells expressing CD69, CD103, CD27, KLRG1, PD-1, TIM3, LAG3, and CD39 in juxta-tumor and tumor tissues (representative staining in Fig. S1 D). (G, H, and M) UMAP of flow cytometry data in five juxta-tumor and six primary tumors, gated in T cells. (G) UMAPs of color-coded surface expression of CD4, CD127−CD25+CD4+ (Treg), and CD8+ in CD3+ (left) and distribution of CD3+ cells from either juxta-tumor (light blue) or tumor sites (violet) (right). (H) UMAP of CD3+ cells color-coded for D3 (blue) or M3 (red) tumors. (I and J) Frequency of CD8+ T cells (I) and CD4+ T cells (J) in D3 and M3 tumors. (K) Example of CD127 versus CD25 staining of CD4+ T cells in D3 or M3 tumors. (L) Frequency of Tregs in D3 or M3 tumors (only samples with >10 CD4+ cells were considered). (M) UMAP of color-coded CD39 (left) and PD-1 (right) surface expression in CD3+ cells. (N) Example of PD-1 versus CD39 expression in CD8+ T cells from D3 and M3 tumors. (O) Frequency of CD8+ T cells expressing both CD39 and PD-1 in M3 and D3 tumors. (P) Distribution of PD-1+CD39+ in CD8+ T cells according to tumor T cell infiltration. Coh1 (n = 20) was used in A–C. Coh2 (n = 7) was used in E. Coh3 (n = 33) was used in D, F–N, and P. Coh1 and coh3 were used in O. Non-parametric paired Wilcoxon test was used in B–E. Non-parametric Mann–Whitney test was used in F, J, L, and O. Only P < 0.05 is shown.
Figure S1.
Characterization of the Tregs and CD8+ cells in primary UMs. (A) Representative dot plots of Treg (CD25+FoxP3+) in CD4+ T cells in juxta-tumor and tumor tissues (frequencies in Fig. 1 B). (B) Representative dot plots of CCR8, PD-1, CD39, and ICOS expression in Treg defined as CD4+CD25+FoxP3+ (left) and frequency of Treg expressing these markers in juxta-tumor and tumor tissues (right). (C) Representative dot plots of CD45RA and CCR7 expression defining: naïve: CD45RA+CCR7+; TEMRA: CD45RA+CCR7−; effector memory (EM): CD45RA−CCR7−; central memory (CM): CD45RA−CCR7+) (left) and frequency of these subsets in CD8+ T cells in juxta-tumor and tumor tissues (right). (D) Representative dot plots showing CD8+ T cells expressing CD69, CD103, CD27, KLRG1, PD-1, TIM3, LAG3, CD39 in juxta-tumor and tumor (frequencies in Fig. 1 F). (E) Representative dot plots of PD-1 and CD39 expression in CD8+ T cells (left) and frequency of CD39+PD-1+ cells in CD8+ T cells from juxta-tumor and tumor tissues (right). (F) Unsupervised analysis of CD3+ cells in five juxta-tumor and six tumor tissues. Expression of the indicated markers in the UMAP representation. Coh1 (n = 20) was used in C. Coh2 (n = 7) was used in B. Coh3 (n = 33) was used in D. Coh1 and coh3 were used in E. Coh3 was used in F. Non-parametric Mann–Whitney test was used in B−E. Only P < 0.05 are shown.
Using another cohort of primary UMs (Table S1), we characterized the phenotype of intratumoral CD8+ T cells. In both the tumor and the juxta-tumoral sites, only a small fraction of the CD8+ T cells expressed CD69 or CD103, characteristics of tissue residency (Fig. 1 F and Fig. S1 D). The high proportion of CD8+ T cells negative for CCR7 in both the tumor and juxta-tumor sites is consistent with a memory phenotype, as expected for tissue-located cells (Fig. S1 C). To better understand the CD8 T cell functions, we further studied exhaustion markers, such as KLRG1, LAG3, and TIM3. KLRG1, whose lack of expression has been linked to T cell exhaustion and antitumor activity in blood CD8+ T cells (Luoma et al., 2022; Wherry et al., 2007), was infrequently expressed in the tumor and juxta-tumor sites. Very few CD8+ T cells expressed LAG3 (Fig. 1 F) whose increased expression at the RNA level has been linked to high-risk UMs (Durante et al., 2020). Few CD8+ T cells expressed TIM3 (Fig. 1 F). The expression of these two last markers was not different between the tumor and the juxta tumor sites. In contrast, PD-1 and CD39 were more expressed by CD8+ T cells infiltrating tumors than in the juxta tumoral site (P = 0.0454 and 0.0036, respectively) (Fig. 1 F). Notably, a larger proportion of CD8+ T cells simultaneously expressed PD-1 and CD39 in the tumor as compared with the juxta tumor site (Fig. S1 E), suggesting a tumor-Ag–specific immune response (Simoni et al., 2018; Duhen et al., 2018). Furthermore, the distribution of the CD39+PD-1+ double expression by T cells in tumors was bimodal: 45% (9/20) (in coh1) of the primary tumors and not in the corresponding juxta-tumoral tissue (Fig. S1 E), suggesting a specific tumor-Ag response in a subgroup of patients.
Since many of the phenotypic variables were correlated in the different samples, we turned to a dimensionality reduction analysis. CD4, CD8, and Tregs were projected onto different regions of the uniform manifold approximation and projection (UMAP) (Fig. 1 G and Fig. S1 F). The CD8 T cells and Tregs were more abundant in the tumor (violet, Fig. 1 G, right panel). Strikingly, a tumor-specific cluster of CD8 T cells was apparent in the bottom right region of the UMAP (Fig. 1 G, right panel).
Comparison of T cells in M3 versus D3 tumors
As it was reported that tumors harboring a chromosome 3 monosomy (M3) contained more myeloid and lymphoid cells than D3 tumors (Bronkhorst et al., 2012), we compared the T cell distribution in the UMAP according to M3 versus D3 status. Strikingly, the tumor-specific CD8+ cluster seen above was mostly found in M3 tumors (Fig. 1 H, red dots). Accordingly, the proportion of CD8+ T cells was more abundant than CD4+ T cells in M3 than in D3 tumors (P = 0.006) while it was the opposite for CD4+ T cells, which were more abundant in D3 than in M3 tumors (P = 0.039) (Fig. 1, I and J). An increased proportion of Treg (CD127−CD25+) was also observed in M3 tumors as compared with D3 tumors (P = 0.0360) (Fig. 1, K and L). We then focused on the analysis of the PD-1 and CD39 on D3 versus M3 tumors. Notably, the distribution of the proportion of CD8+ T cells expressing both PD-1 and CD39 was bimodal and much higher in M3 (>25% in 16 out of 23) than in D3 (>25% in 1 out of 10) tumors (P = 0.0076) (Fig. 1, M–O). Finally, a strong correlation between the proportion of PD-1+CD39+ in CD8+ T cells and the number of T cells recovered from the tumor was observed in M3 tumors (Fig. 1 P).
Thus, primary M3 UM tumors harbor increased T cell cellularity and proportion of both Tregs and CD39+PD-1+CD8+ T cells, suggesting the occurrence of an antitumor Ag-specific in situ immune response.
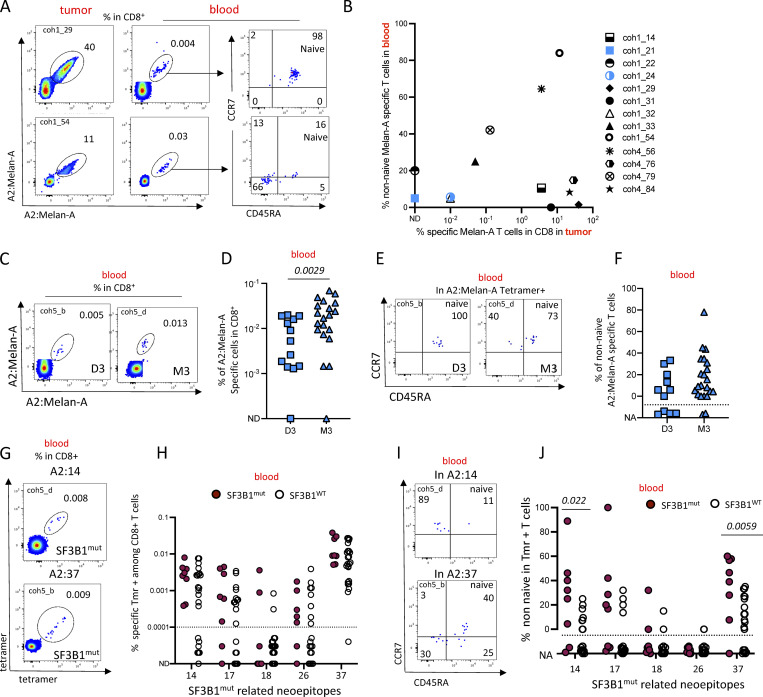
Abundant Melan-A–specific CD8+ T cells are present in primary UMs
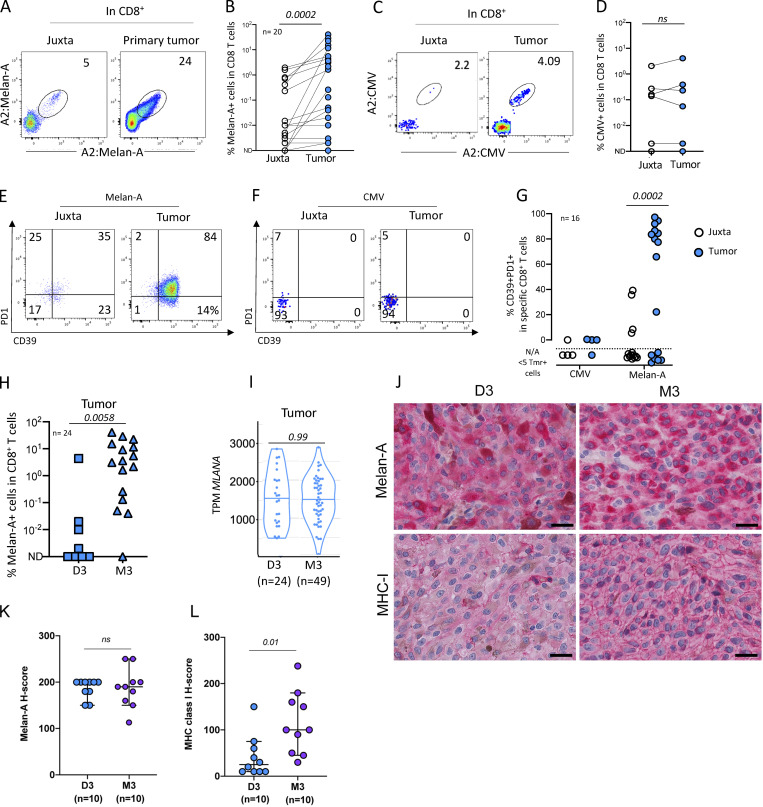
To identify the Ag specificity of the PD-1+CD39+ CD8+ T cells seen above, we quantified the frequency of CD8+ T cells specific for the tumor differentiation Ag Melan-A. As a control, we also characterized the response against a viral Ag (CMV). HLA-A2:Melan-A tetramer-positive CD8 T cells were abundant, reaching up to 24% of the CD8+ T cells in the tumor (Fig. 2, A and B). Melan-A–specific T cells were more frequent in the tumor than in the juxta tumoral site (P = 0.0002) (Fig. 2 B). HLA-A2:CMV–specific CD8+ T cells were found in 4/7 juxta-tumors and tumors at a similar frequency, consistent with the prevalence of CMV infection in our patient population (Fig. 2, C and D). As expected, no difference in HLA-A2:CMV–specific CD8+ T cell frequencies was observed between juxta and tumor tissues (Fig. 2 D). In the tumor, the Melan-A–specific CD8+ T cells expressed CD39 and PD-1 while it was not the case in the juxta tumoral site (Fig. 2 E) nor for the CMV-specific CD8+ T cells in the tumor or the juxta tumoral site (Fig. 2 F). Thus, Melan-A–specific T cells were positive for both PD-1 and CD39 in 43% of UMs (10/23) (Fig. 2 G) while the CMV-specific T cells were negative for these markers (Fig. 2 G). Altogether, these results suggest that the Melan-A–specific CD8+ T cells found in the tumor correspond to an ongoing antitumor response.
Figure 2.
Detection of tumor-specific T cells infiltrating primary UMs. (A) Example of dual color staining of Melan-A–specific CD8+ T cells in juxta-tumor and tumor tissues. (B) Frequency of HLA-A2:Melan-A-tetramer–positive cells in CD8+ T cells in juxta-tumor and tumor tissues. (C) Representative dot plot of HLA-A2:pp65 CMV–specific CD8+ T cells. (D) Frequency of HLA-A2:CMV-tetramer–positive CD8+ T cells in juxta-tumor and tumor samples. (E and F) Example of PD-1 and CD39 expression in tetramer+CD8+ T cells in juxta-tumor and tumor tissues. (E) HLA-A2:Melan-A-tetramer–positive CD8+ T cells. (F) HLA-A2:CMV-tetramer–positive CD8+ T cells. (G) Frequency of tetramer-positive CD8+ T cells expressing both CD39 and PD-1 in tetramer-positive cells in the juxta-tumor and tumor tissues. (H) Frequency of HLA-A2:Melan-A-tetramer–positive cells in CD8+ T cells infiltrating D3 or M3 tumors. (I) MLANA gene expression (RNAseq) in 24 D3 and 49 M3 enucleated primary UMs. (J) Examples of immunohistochemistry for Melan-A and MHC class I in a representative case of D3 and M3 UM, respectively. Scale bars, 25 μm. (K and L) Results of semiquantitative evaluation of Melan-A (K) and MHC class I (L) immunostaining in tumor cells in D3 and M3 UM expressed as H-score (median, 95% CI). Coh1 (n = 20) and coh4 (n = 4) were used in A–H. Independent UM cohorts for J–L. ND: not detected. N/A: not applicable; the phenotype was not calculated when less than five tetramer+ cells were detected. Non-parametric paired Wilcoxon or unpaired Mann–Whitney tests were used as appropriate.
Interestingly, the frequency of Melan-A–specific T cells was much higher in M3 than in D3 tumors (Fig. 2 H) despite similar MLANA mRNA expression in both tumor types (Fig. 2 I), suggesting that the lower frequency of Melan-A–specific T cells in D3 tumors is not due to lack of Ag. This was confirmed by analyzing Melan-A protein by immune chemistry in 10 D3 and 10 M3 primary tumors (Fig. 2, J and L). The level of Melan-A protein was similar in both tumor types. Notably, HLA membrane expression, as measured by immunochemistry, was more important in M3 than in D3 tumors (Fig. 2, K and L), confirming that M3 tumors are more inflamed than D3 tumors (Bronkhorst et al., 2012) as correlated with a stronger IFN-γ signature (Robertson et al., 2017). Altogether, our results indicate that the increased number of CD8+ T cells observed in M3 tumors is probably driven by specific antitumor Ag responses.
Single cell transcriptomic and TCR analysis in primary UM tumors
To further analyze the T cell response in the tumor and characterize the tumor Ag–specific immune response, we used scRNAseq coupled with VDJ-TCRseq technology on three primary HLA-A2+ UMs. We focused on M3 tumors as they are infiltrated by a high number of T cells, contrary to the D3 ones. To maximize the information, we sorted the following: (1) Melan-A–specific CD8 T cells using barcoded HLA-A2:Melan-A tetramers; (2) HLA-A2:Melan-A tetramer–negative CD39+PD-1+ CD8+ T cells; (3) CD8+ T cells that displayed neither of the previous characteristics; (4) CD4+ T cells (see Materials and methods). These cells were mixed in a 10%, 30%, 30%, and 30% proportions, respectively, before input into a 10X apparatus for 5′ transcriptomic and VDJ sequencing. After the exclusion of doublets, debris, and dying cells, we retrieved 6,668, 9,545, and 5,338 cells for coh4_84, coh4_56, and coh4_76 samples, respectively. The median number of detected genes was 2,179, 2,408, and 1,952, respectively. The number of cells labeled with Melan-A tetramer was 712 (10.7% for coh4_84), 696 (7.3% for coh4_56), and 1,368 (25.6% for coh4_76).
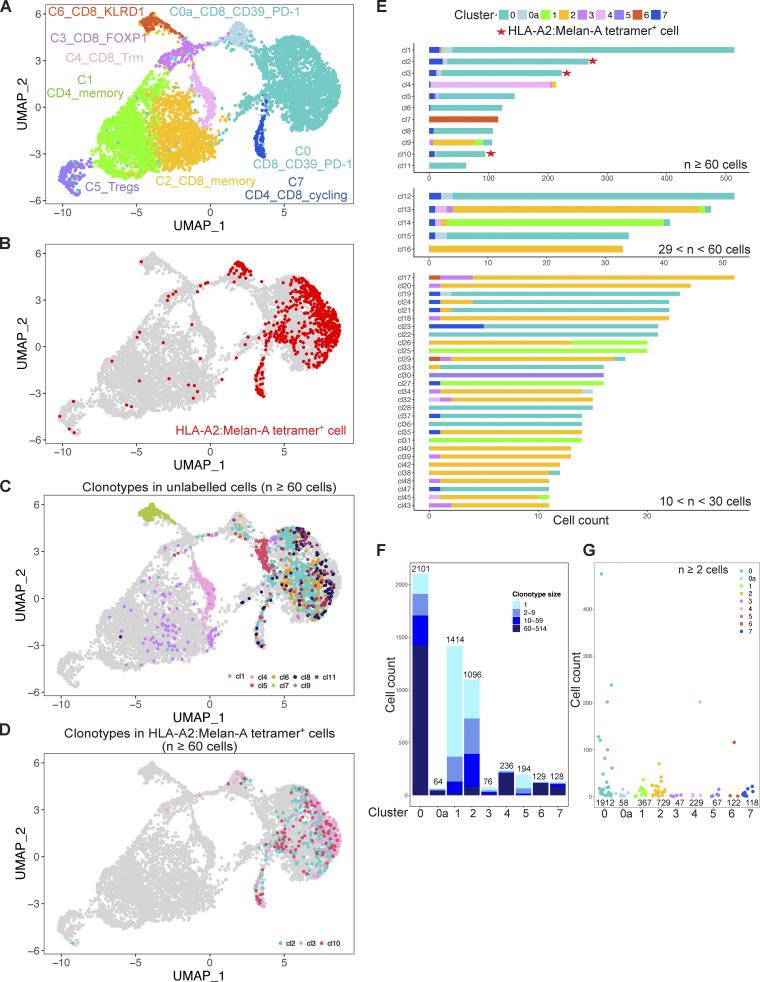
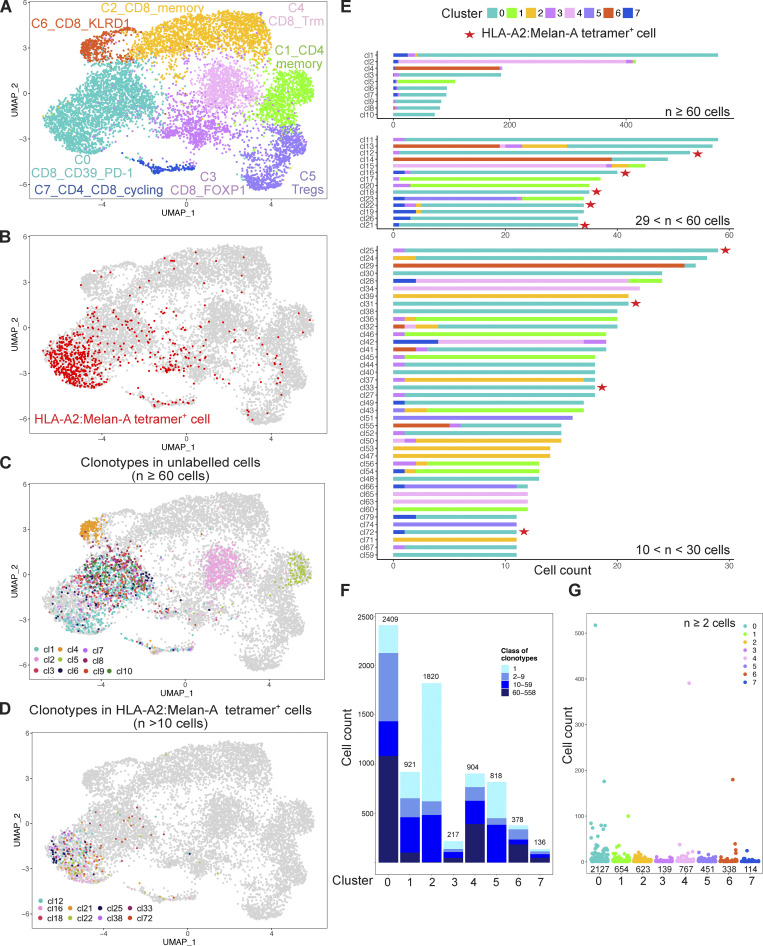
We first focused on the coh4_84 sample to illustrate our analysis pipeline (Fig. 3). We performed a clustering with the Louvain algorithm (resolution 0.2), followed by UMAP. These unsupervised graph-based clustering of transcriptome data partitioned the cells into nine subsets (Fig. 3 A and Fig. S2). These subsets were named according to the differential expression of key genes (Fig. 4 and Table S2). Cluster C1 corresponded to CD4+ memory T cells with the expression of CD4, IL7R, and CD40LG while cluster C5 corresponded to Tregs characterized by the expression of CD4, FOXP3, CCR8, and CTLA4. Cluster C7 corresponded to cycling CD4+ and CD8+ T cells expressing MKI67 and TOP2A, indicating that some T cells proliferate in the tumor bed.
Figure 3.
VDJ scRNAseq of T cells found in a primary M3 UM. CD3+ T cells isolated from an enucleated primary M3 UM tumor (coh4_84) were FACSorted into HLA-A2:Melan-A tetramer+ CD8+ T cells, HLA-A2:Melan-A tetramerneg CD39+PD-1+CD8+ T cells, HLA-A2:Melan-A tetramer−CD39− or PD-1− CD8+ T cells, CD4+ T cells that were mixed in a 10, 30, 30, and 30% proportion, respectively, before analysis using the 10X VDJ 5′ scRNAseq technology. (A) UMAP of the scRNAseq dataset colored by inferred cluster identity. (B) Projection on the transcriptomic UMAP of the cells labelled by the HLA-A2:Melan-A tetramer+ cells (red). (C and D) Projection on the UMAP of the most expanded clonotypes (n ≥ 60 cells) corresponding to unlabeled (C, others) and labeled (D, HLA-A2:Melan-A) cells by the Melan-A tetramer. (E) Cluster of origin for cells expressing the most expanded clonotypes (n > 10 cells). Red stars indicate HLA-2A:Melan-A tetramer–specific clonotypes. (F) Distribution of the clonotype size in the different clusters. One clonotype can be represented in multiple clusters. (G) Clonal size of the expanded (n ≥ 2 cells) clonotypes in the different clusters.
Figure S2.
VDJ scRNAseq of T cells found in primary UM from patient coh4_56. See the Fig. 3 legend for descriptions of individual panels.
Figure 4.
Differential expression of key genes in the three scRNAseq UM data sets. (A) Dot plot shows scaled expression of selected genes (see Materials and methods) for each cluster colored by average expression in each cluster. Dot size represents the percentage of cells in each cluster with more than one read of the corresponding gene. (B) Hierarchical clustering of the different clusters from patients coh4_84, coh4_56, and coh4_79.
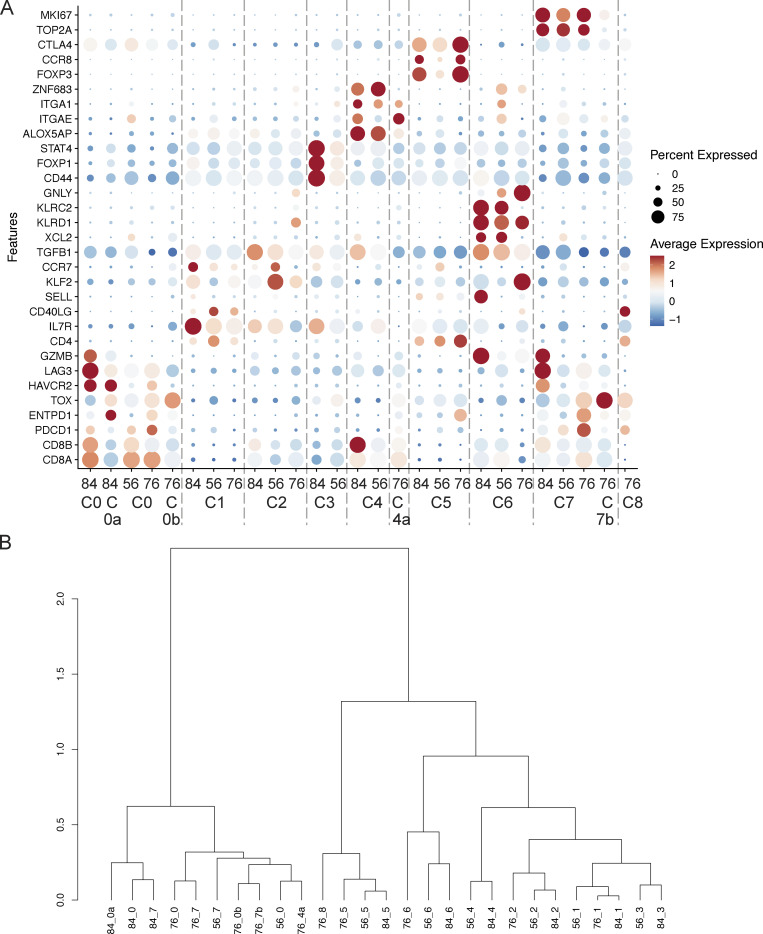
Six other clusters encompassed CD8+ T cells. The most abundant cluster C0 corresponded to the expression of PDCD1 (PD-1), ENTPD1 (CD39), HAVCR2 (TIM3), LAG3, GZMB, and TOX (Figs. 4 and S3), suggesting chronic activation by tumor Ags. Confirming this hypothesis, barcoded Melan-A tetramer-positive cells projected mostly onto this C0 cluster (Fig. 3 B). The other large CD8+ cluster C2, corresponded to expression of IL7R, KLF2, CCR7, and TGFB1 suggesting a less differentiated state and a memory phenotype. Four smaller CD8+ T cell clusters were observed: C0a, C3, C6, and C4. Cluster C0a was close to cluster C0 with similar expression of ENTPD1, HAVCR2, and PDCD1 but decreased expression of the ribosomal protein genes, indicating lower protein biosynthesis. Cluster C3 was characterized by increased expression of the transcription factor FOXP1 which prevents spontaneous T cell activation and preserves memory potential (Kaminskiy et al., 2022). A high level of CD44 expression was also found suggesting particular interactions with the extracellular matrix. Like in the previous cluster (C0a), expression of ribosomal protein genes was decreased, indicating low protein biosynthesis. Cluster C6 was characterized by the expression of XCL2 and XCL1 (Table S2), two chemokines implicated in mediating interactions between dendritic cells and T cells and induction of CD8+ effector T cell responses (Fox et al., 2015). The inhibitory KIR receptors KLRC2 (NKG2-C) and KLRD1 (CD94) and the corresponding natural killer (NK) adapter TYROBP (encoding DAP12) were also expressed. SELL expression was high indicating the possibility of entering the lymph node while GNLY, an antimicrobial protein present in cytotoxic granules, suggested cytotoxic functions. Notably, the integrins ITGA1 (CD49a) and ITGAE (CD103) as well as the transcription factor ZNF683 (Hobit) were also expressed, suggesting some level of tissue residency. Cluster C4 highly expressed the integrins ITGA1 and ITGAE (CD103), as well as ALOX5AP (Kortekaas et al., 2020), indicating tissue residency in accordance with the expression of ZNF683.
Figure S3.
VDJ scRNAseq of T cells found in primary UM from patient coh4_76. See the Fig. 3 legend for descriptions of individual panels.
As the proportion of the different populations of the UMAP matched what was put into the 10X apparatus, the equilibrated numbers of cells in the different clusters allowed us to investigate the relationship between phenotype, TCR clonal expansions, and putative antitumor Ag reactivity in primary UM tumors. Notably, almost all the cells labeled with the HLA-A2:Melan-A tetramer projected onto cluster C0, and a few belonged to the related cluster C0a and to the cycling cluster C7 (Fig. 3 B).
Large clonal expansions are found in specific transcriptomic clusters
Clonal expansion may reflect an antitumor response or non-specific recruitment to the tumor of effector T cells bearing other specificities such as antiviral responses. For TCR analysis, we assigned cells from our dataset into clonotypes (set of cells sharing identical nucleotide sequences of TCRα and/or TCRβ chains) (Table S3). For the TCR repertoires, we took into account the different possibilities: 112 cells with only a TCRα chain; 423 cells with two TCRα and one TCRβ chains; 863 cells with one TCRβ chain; 4,040 cells with one TCRα and one TCRβ chains.
TCR repertoire analysis detected 2,136 clonotypes in 5,438 cells in which a TCR could be assigned. The projection onto the UMAP of the largest (≥60 cells) clonotypes specific or not for HLA-A2:MelanA is displayed in Fig. 3, C and D. Strikingly, most of the clonotypes projected on the UMAP predominantly to a given cluster except for a few exceptions. Several of the biggest clonotypes (≥60 cells) were found in cluster C0 while C2, C4, and C6 clusters encompassed each one a predominant clonotype (cl9 in C2, cl4 in C4, and cl7 in C6) (Fig. 3 E). Notably, some cells from clonotypes found in C0 projected also onto the related cluster C0a (CD8+CD39+PD-1+) and onto the cycling cluster C7, indicating the relatedness of these three clusters. Three (cl2, cl3, and cl10) of the most expanded clonotypes were specific for Melan-A and expressed the TRAV12-2 gene as expected (Cole et al., 2009). 12 other singleton clonotypes were HLA-A2:Melan-A tetramer+ cells while the rest of the cells were distributed among smaller clonotypes which were partially (<50%) labeled, suggesting non-specific staining or lower avidity. The presence of Melan-A–specific cells in cluster C0 confirmed our working hypothesis that the tumor-Ag–specific CD8+ T cells belonged to this cluster. Notably, the largest clonotype (cl1) belonged to cluster C0 and is not specific for Melan-A.
The smaller clonotypes (11–60 cells) were distributed throughout different clusters (Fig. 3 E), including a Treg clonotype (cl30 in C5). The CD8+ memory C2 and CD4+ memory C1 encompassed mostly small (2–60 cells) clonotypes, especially for the latter.
Interestingly, most if not all clonotypes encompassing cycling cells (C7) also belonged to C0 with five exceptions (cl4, 13, 14, 27, and 35) in which, however, only one cycling cell was observed. This could be doublet cells remaining after filtering. Thus, C0 encompassed large clonotypes, many of which were proliferating in the tumor bed despite the absence of known tertiary lymphoid structure in primary UM (Mariani et al., 2023). These data suggest that the 309 clonotypes found in C0 represent an antitumor CD8+ T cell response. The specificity for Melan-A is known for three clonotypes while the Ag specificities for the 306 other TCRs remain to be determined. In particular, the largest clonotype (cl1, 514 cells) probably recognizes a strongly immunogenic tumor-related peptide–MHC complex.
Clonal size ranged from 1 to 514 cells and the clonal size distribution varied according to the clusters (Fig. 3, F and G). Singletons were above 15% only in C1 (CD4+ memory), C2 (CD8+ memory), C3 (FOXP1), and C5 (Tregs), indicating higher TCR diversity in these clusters while the proportion of expanded clonotypes was very high in the other clusters. The average size of the expanded (≥2 cells) clonotypes was higher in C0, C4, and C6 (Fig. 3 G), consistent with an in situ immune response for C0 as many clonotypes are shared with the proliferative C7.
scRNAseq VDJ analysis of two other primary tumors
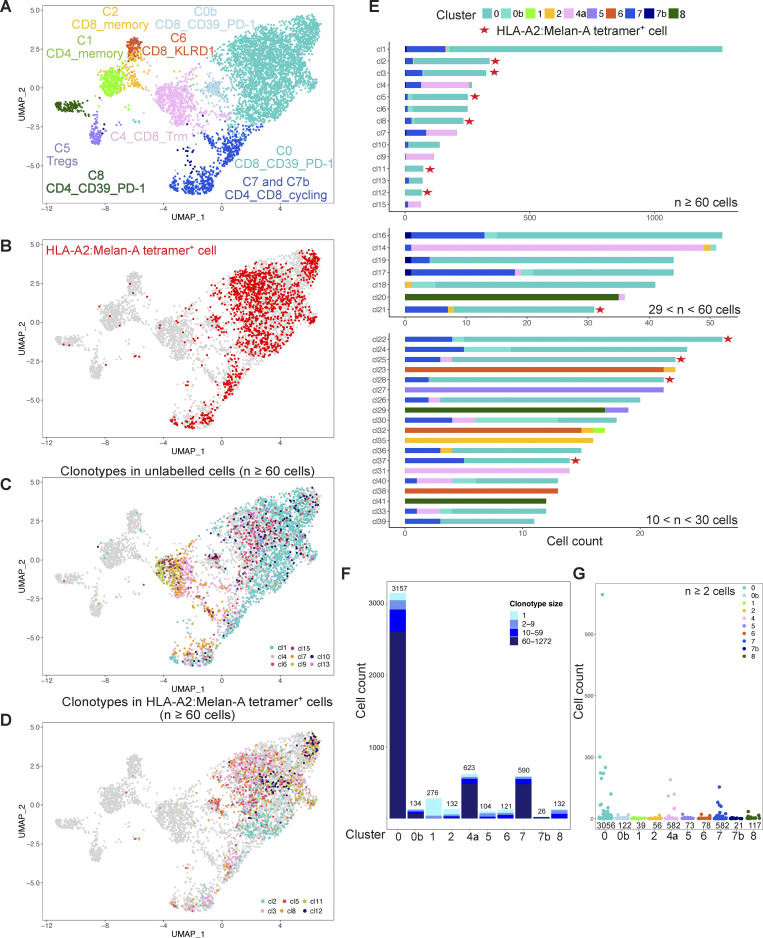
We next studied two other primary tumor samples, coh4_56 and coh4_76. The data were analyzed separately without integration to avoid artificial homogenization (Figs. S2 and S3). To allow a common nomenclature of the observed clusters, the expression of key genes was also analyzed in the three datasets (Fig. 4 A and Table S2). The distribution of the clusters in coh4_56 was very similar to coh4_84. For coh4_76, despite the experimental design, the proportion of memory CD4+ T cells (C1) was low, but an additional cluster (C8) corresponding to CD4+ T cells expressing PDCD1 and CD40LG was identified. The frequency of Melan-A–specific CD8+ T cells in coh4_76 was also higher, 26%, well above the targeted 10% value. The proportion of cycling cells (C7) was higher in coh4_76 than in the two other samples. Notably, the particularly exhausted CD39+PD-1+CD8+ C0a was not found in coh4_56 and coh4_76 while C3 was not observed in coh4_76. In addition, an additional cluster C4a was identified in coh4_76 as the differential expression of the genes of our candidate list indicated similarity to the tissue resident memory (Trm) C4 (ITGAE, ITGA1) cluster. However, the expression of ZNF683 was low whereas TOX and PDCD1 were also expressed. These mixed Trm and exhausted phenotypes lead to clustering to the C0 branch of the hierarchical dendrogram (Fig. 4 B). Additional small clusters were identified in coh4_76: C0b and C7b. Still, the three tumors harbored similar CD8+ and CD4+ T cell subsets displaying common (but not identical) features (Fig. 4). Most of the Melan-A–specific CD8+ T cells belonged to C0 and the related C0a and C0b in the three datasets (Figs. S2 and S3), confirming that these clusters probably correspond to antitumor Ag-specific T cells.
TCR repertoire analysis detected 3,082 and 734 clonotypes in 7,603 and 5,295 cells in which a TCR could be assigned for the coh4_56 and coh4_76 samples, respectively. As above, in these two samples, the biggest clonotypes (≥ 60 cells) were mostly found in C0. Notably, in the coh4_76 sample, 4 of the 14 biggest clonotypes (≥ 60 cells) belonged to the CD8+ Trm/exhausted C4a and also included some proliferative cells (C7), suggesting in situ proliferation. Similarly, in the coh4_56 sample, several large clones also belonged to C4, and some of them also displayed proliferation (C7). While the Melan-A–specific CD8+ T cells corresponded to only three large clones in coh4_84, several clones of intermediate size (10–60 cells) or smaller (2–9 cells) were observed in the two other tumors (Fig. S3 E, Fig. S4 E, and Table S2). In coh4_76, six (cl2, cl3, cl5, cl8, cl11, and cl12) of the most expanded (≥ 60 cells) clonotypes represented >80% of the cells specific for Melan-A and mostly expressed the TRAV12-2 gene as expected (Fig. S3 E and Table S2).
Figure S4.
Pattern of recirculation between the blood and the tumor according to cluster and clonotype size in the tumor. (A and B) Representation of the clonotype identified in the blood by their TCRβ (A) or TCRα (B) chains according to their number of read (Nread) detected in the bulk TCRseq (x axis) and plotted according to their clonal size in the tumor identified in single cells (y axis). Each clonotype is colored by its cluster of origin in the single-cell tumor scRNAseq. In each panel, the number of reads in the blood corresponding to the tumor singletons is split according to their cluster of origin in the tumor. The percentages indicated above the triangles on the left (tumor clonotypes not found in the blood) indicate the percentage of non-circulating clonotypes.
The proportion of singletons in the three tumors, indicating smaller clonal expansions in a given cluster, was higher in Tregs (C5), in CD4+ and CD8+ T memory cells (C1 and C2), and also in FOXP1+CD8+ T cells (C3) as compared with other clusters (Fig. 3 F, Fig. S2 F, and Fig. S3 F). C0 encompassed several large and medium size clonotypes. In contrast, C4 and C6 were composed of one or two large clonotypes in the three tumors (Fig. 3 G, Fig. S2 G, and Fig. S3 G). Altogether, these results suggest that CD8+ T cells with specific differentiation status either accumulate or proliferate in the tumor and confirm that the cells belonging to the C0-related clusters (C0a, C0b, C7, C7b) probably recognize tumor Ags.
Pattern of recirculation of the TCRs found in the three primary tumors according to the cluster of origin
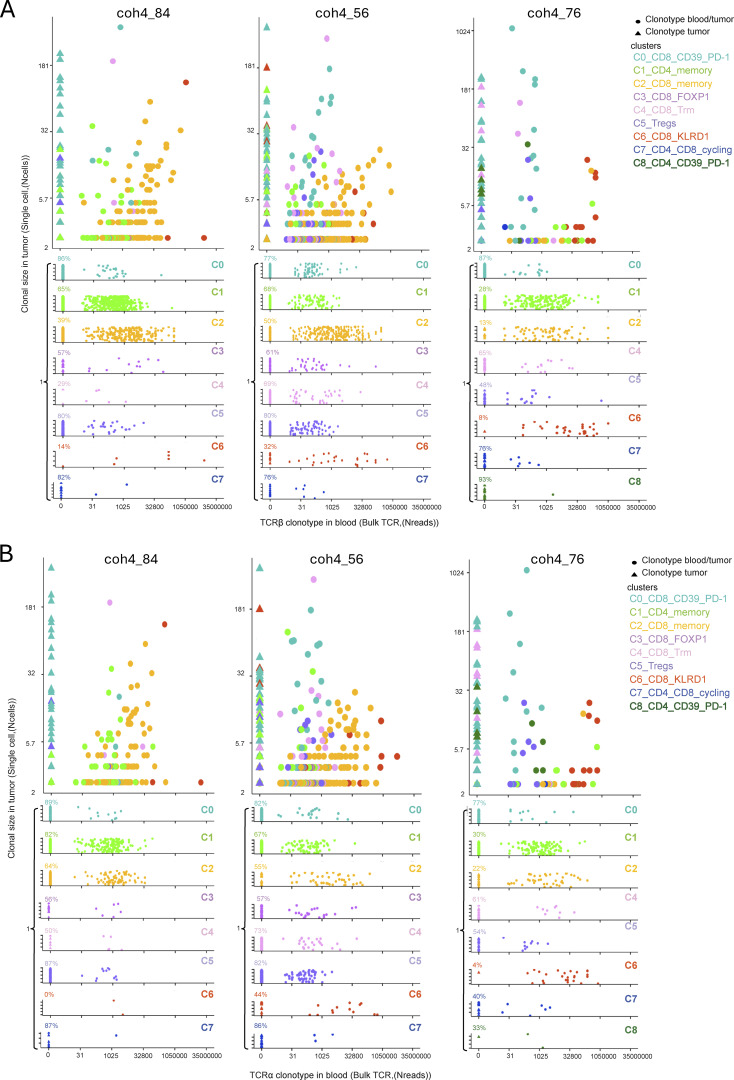
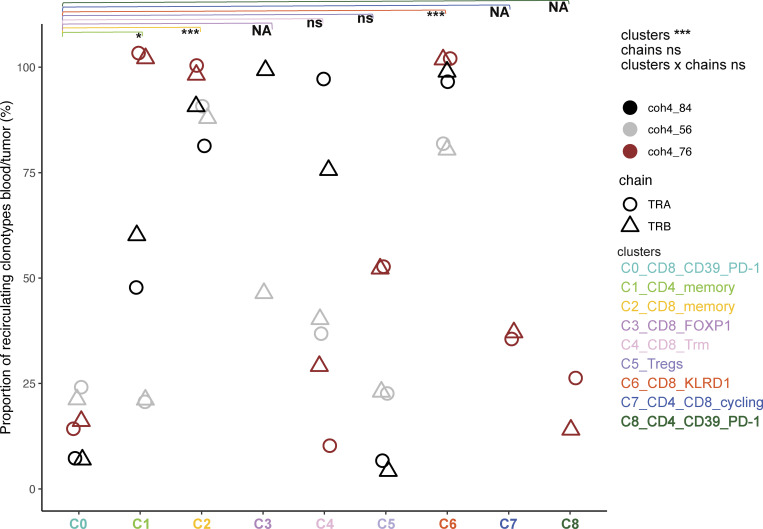
One important parameter characterizing memory T cell subsets is the ability to recirculate systemically. We, therefore, sequenced the bulk of TCRs in peripheral blood mononuclear cell (PBMC) from the three patients to quantify the number of clonotypes identified in the tumor scRNAseq that would also be found in the blood according to cluster of origin (Figs. 5 and S4; Tables S3 and S4). As the clones and the clusters differ widely in size, to avoid complex normalizations or sub-sampling approaches, we only scored scRNA clonotypes present in the blood in each cluster (Fig. 5) and ranked them according to the number of reads in the blood and their clonal size in the tumor (Fig. S4). A given clonotype was assigned to the cluster to which the majority of the cells belonged. Except for a few cases, the TCRα and TCRβ analysis gave very consistent results, validating the approach (Figs. 5 and S4).
Figure 5.
Frequency of tumor clonotypes found in the blood according to assigned cluster and clonotype size in the tumor. The frequency was computed as the number of tumor clonotypes found in the blood (Boolean variable) using bulk RNAseq divided by the total number of clonotypes assigned to a given cluster for each patient for each TCR chain (TCRα [circle] and TCRβ [triangle]). A given clonotype was assigned to the cluster in which the majority of the cells was found. Only clonotypes ≥2 cells were analyzed. The effects of the TCR chain nature (TCRα or TCRβ) and clusters were evaluated using an ANOVA. Tukey’s post-hoc test to identify differences between clusters. The statistical differences are indicated as: *, P < 0.05; ***, P < 0.001 or ns for non-significant, and NA for non-applicable when data points were missing leading to underpower.
Consistent with their differentiation state, despite their large clonal size, the proportion of TCRs found in the blood was low for the CD39+PD-1+CD8+ T cells (C0), and the related cycling (C7) clusters, indicating that the immune response was mostly localized. Most of the TCRs corresponding to Tregs (C5) were not found in the blood (except for a few clones in coh4_76, Figs. 5 and S4), suggesting also local proliferation. Few TCRs expressed by memory CD4+ T cells (C1) were found in the blood apart from patient coh4_76 whose CD4+ T cells cluster were split into two clusters, C1 and C8. The C1 cells did recirculate while C8 did not and may correspond to an intratumor response as it expresses ENTPD1 and PDCD1 like C0. In contrast, TCRs corresponding to the memory CD8+ T cells (C2) were found in the blood. This difference between memory CD4+ and CD8+ T cells’ apparent recirculation pattern may be related to the higher clonal size of CD8+ T cells (Seder and Ahmed, 2003). The TCRs belonging to the FOXP1+CD8+ T cells (C3) were found in the blood despite their relatively low clonal size in the tumor, suggesting specific trafficking pattern. Notably, many TCRs corresponding to C4 were also found in the blood despite a Trm program. The TCRs expressed by KLRD1+ effector CD8+ T cells (C6) were found in large numbers in the blood indicating a particular circulatory pattern for these cells as TCRs corresponding to singleton in the tumors were frequently found in the blood. Consistent with their C0 assignment, the Melan-A–specific T cells did not recirculate much in the blood except for patient coh4_56 in which ≈20% Melan-A–specific T cells displayed a particular differentiation status (C3) and were found as memory cells in the blood according to cytometry analysis (Fig. 6 B, below).
Figure 6.
Antitumor systemic responses detected in the blood of patients with localized UM tumors. (A and B) Analysis of matched blood and tumor samples. (A) Dual color HLA-A2:Melan-A staining of CD8+ T cells in the tumor and blood in two patients. (B) Proportion of non-naïve HLA-A2:Melan-A-tetramer+ T cells in the blood according to the number of Melan-A–specific T cells in the primary tumor. (C–F) Frequency and phenotype of Melan-A–specific CD8+ T cells in the blood of patients with D3 or M3 tumors. (C) Dot plots of Melan-A–specific CD8+ T cells in the blood of two patients. (D) Frequency of HLA-A2:Melan-A-tetramer+ in blood CD8+ T cells. (E) Examples of CCR7-CD45RA staining of blood A2:Melan-A CD8+ T cells. (F) Frequency of non-naïve HLA-A2:Melan-A T cells in the blood of patients with D3 or M3 tumors. (G–J) Detection of CD8+ T cells specific for SF3B1mut related neo-epitopes in the blood of patients with UM tumors mutated (SF3B1mut) or not (SF3B1WT) for SF3B1. (G) Example of HLA-A2:14 (top) and HLA-A2:37 (bottom) tetramer staining. (H) Frequency of tetramer-positive cells in blood CD8+ T cells of patients with SF3B1mut or SF3B1WT tumors. The dashed line indicated the quantitation limit. (I) Examples of naïve/memory phenotype of tetramer-positive T cells. (J) Frequency of non-naïve tetramer-positive T cells in the blood of patients with SF3B1mut or SF3B1WT tumors. (K) Frequency of patients with memory responses against SF3B1mut-related neo-peptides according to mutational status of SF3B1. Coh1 (n = 20) was used in A. Coh1 and coh4 were used in B. Coh5 was used in C–K. Non-parametric Mann–Whitney test was used in D, F, H, and J. Only P < 0.05 are shown. ND: non detected; NA: not applicable; the phenotype was not calculated when less than five tetramer-positive cells were detected.
Thus, many T cells found in the tumor corresponded to clones with specific differentiation (C2 or C6 for instance) and large clonal size in the blood, which may indicate antiviral specificity. Altogether, these results indicate that in M3 UM primary tumors, the T cells most likely responding to tumor-Ags do not recirculate much in the blood.
Systemic tumor Ag–specific immune responses in untreated UM patients
The presence of tumor Ag–specific T cell response at the time of primary treatment indicates some form of priming somewhere in the body. In a prototypic immune response, naïve T cells do not have access to tissue and circulate between the blood and secondary lymphoid organs where they are primed by incoming Ag presented by Ag-presenting cells. After priming, effector T cells circulate in the blood to reach tissues to mediate their effector functions while central memory T cells circulate between the lymphoid organs, the lymph, and the blood. Since we observed a local antitumor immune response in UM patients (Fig. 1), we expected to find tumor Ag–specific T cells circulating in the blood. We therefore characterized the lymphoid compartment in the blood of 86 patients before surgical treatment of their eye tumor (Table S1).
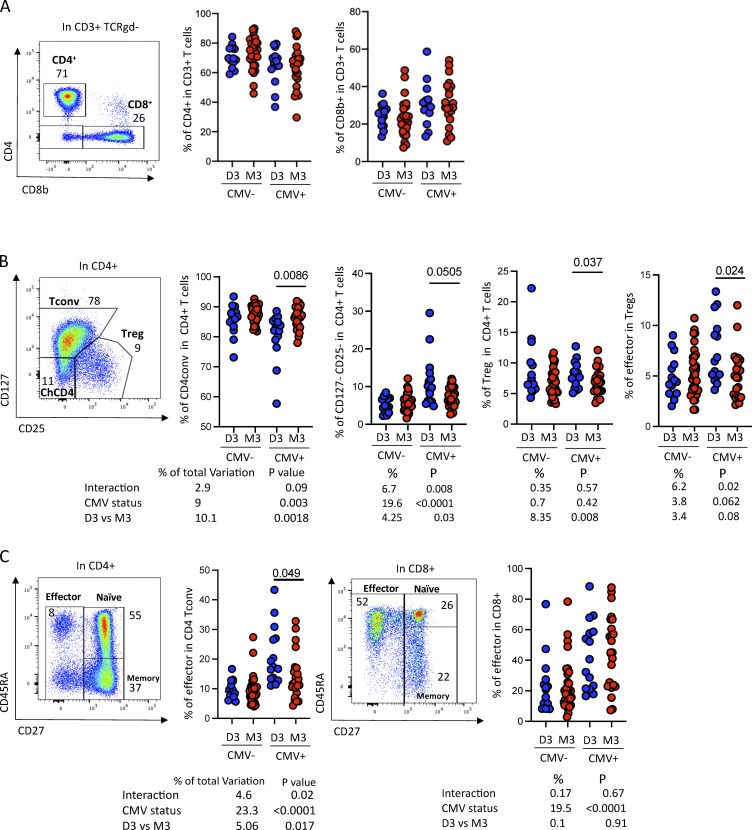
We firstly measured the proportions of the main T cell subsets (CD4, CD8, Tregs (CD127−CD25+), naïve, effector, memory, chronically activated (CD127−CD25−CD4+ (Péguillet et al., 2014) subsets as well as the main innate-like T cells (MAIT, NKT, and γδ) and also the expression of a couple of activation markers (PD-1, ICOS) using a 15-parameter flow cytometry panel according to the gating strategy displayed in Fig. S5). As CMV latent infection strongly impacts the distribution of many peripheral blood lymphocyte subsets with an increased proportion of effector cells in both CD4+ and CD8+ T cells (Patin et al., 2018) and the CMV-driven accumulation of terminally differentiated T cells can negatively impact response to ICI (Naigeon et al., 2023), we stratified our analysis according to CMV serological status. As we had observed a strong impact of the D3/M3 status in the primary tumor, we also analyzed the data according to the D3 or M3 status. We found no difference neither in the proportion of CD4+ or CD8+ T cells (Fig. S5 A), nor in CD4 subsets (Treg, conventional T cells, or chCD4) (Fig. S5 B) in CMV-negative patients. Notably, CMV+ patients with D3 tumors harbored increased frequency of effector in both Tregs and convCD4+ T cells as compared to patients with M3 tumors (Fig. S5, B and C), suggesting increased stimulation, possibly reflecting an antitumor response.
Figure S5.
Characterization of the T cells circulating in patients with primary UMs. (A) Example of CD4 versus CD8 expression in T (CD3+) cells in the peripheral blood (left) and frequency of the indicated cell populations in patients with D3 or M3 UM tumors stratified according to CMV status (right). (B) Example of CD127 versus CD25 staining in CD4+ T cells in the blood (left) and frequency of the indicated populations in D3 or M3 patients (right). (C) Example of CD45RA versus CD27 staining. Frequency of effector (CD27−), naïve (CD45RA+CD27+), and memory (CD45RA−CD27+) among CD4+ (left) or CD8+ (right) T cells. A two-way ANOVA was performed and is displayed below the relevant panel.
The level of granularity imparted by generic subset phenotyping in the blood is certainly not sufficient to assess antitumor responses. To study the relationship between local and circulating tumor Ag–associated specific T cells, we compared the frequency and phenotype of Melan-A–specific T cells in patients for whom paired tumor-blood samples were available (n = 13). In two patients (coh1_54 and coh4_56), Melan-A–specific T cells were found in the tumor and the Melan-A–specific T cells in the blood displayed a memory phenotype (Fig. 6, A and B). For two other patients (coh4_79 and coh1_33), the same pattern was observed to a lower extent. In five patients (coh1_14, coh1_29, coh1_31, coh4_76 and coh4_84), Melan-A–specific T cells were found in the tumor but were naive in the blood while in the other four patients, Melan-A–specific T cells were not present in the tumor nor were memory in the blood (Fig. 6, A and B). Thus, 5 out of 13 of the patients displayed an abundant in situ immune response against a tumor-associated Ag (TAA) without displaying a systemic response.
To further examine the occurrence of an antitumor-associated Ag immune response at primary treatment, we studied PBMCs of a large cohort (n = 33) of HLA-A*02:01 patients undergoing primary tumor resection. We stratified the results according to D3 and M3 status. Notably, the frequency of Melan-A–specific T cells was higher in the blood of patients bearing M3 tumors as compared with D3 tumors (P = 0.0029) (Fig. 6, C and D). Since the acquisition of an effector or memory (CD45RA− or CCR7−CD45RA+) phenotype by CD8+ T cells indicates previous contact with the Ag, we quantified the proportion of non-naive cells in the Melan-A–specific T cells. The proportion of Melan-A–specific CD8+ T displaying an effector/memory phenotype was similar in patients bearing D3 or M3 tumors (Fig. 6, E and F). The absence of statistical significance between the two groups may be related to a lack of statistical power as the low number of Melan-A–specific T cells available for the naïve/memory analysis was insufficient in 4 (out of 11) and 2 (out of 21) patients in the D3 and M3 groups, respectively.
To study the immune response against tumor-specific Ag, we turned to the neo-Ags related to mutations in the splicing factor SF3B1 which is mutated in ∼20% of UMs. The multiple (>500) neo-Ags are shared between patients and are tumor-specific as they are not expressed in normal tissues (Alsafadi et al., 2016). We previously characterized the T cell response against these neo-Ags in a small cohort of metastatic patients (Bigot et al., 2021). In the current study, the tetramer staining on tumor-infiltrating lymphocytes (TILs) was performed on 20 fresh enucleation samples, which unfortunately were found afterward to be WT for SF3B1. Thus, we could not analyze the in situ response of T cells toward SF3B1mut-related neo-Ags. We therefore studied the immune response against SF3B1mut-related neo-epitopes in the peripheral blood of these patients. Blood CD8+ T cells from HLA-A*02:01 patients with SF3B1mut (n = 8) (Table S1) or SF3B1WT (n = 26) localized UM were stained with HLA-A2 tetramers loaded with SF3B1mut related A2:14, A2:17, A2:18, A2:26, and A2:37 neo-peptides (Fig. 6 G). The frequency of SF3B1mut-related NeoAg-specific T cells was similar in patients bearing SF3B1mut or SF3B1WT tumors. We then quantified the proportion of non-naive cells in the neo-epitope-specific T cells (Fig. 6 H). In patients bearing SF3B1WT tumors, SF3B1mut-related neoepitope specific T cells were mostly naïve, while in several patients bearing SF3B1mut tumors, the proportion of non-naïve was above 40%. The presence of a few memory cells in the blood of patients bearing SF3B1WT tumors may be explained by the crossreactivity of T cells toward environmental Ags. Still, the proportion of non-naive cells for epitopes A2:14 and A2:37 was higher (P = 0.02 and 0.006, respectively) in patients bearing SF3B1mut tumors than in SF3B1WT tumors. For the other epitopes, A2:17, A2:18, and A2:26, the frequency of specific T cells was lower and had an effector/memory phenotype in only one or two patients (Fig. 6, I and J). Thus, tumor Ag–specific T cells were detected in the blood of patients with UM at primary treatment. Since metastasis occurs later in SF3B1mut tumors than in M3 tumors, the systemic antitumor response observed in D3 tumors could be related to the detection of disseminated tumor cells and may be involved in metastasis containment.
Discussion
In this study, we characterized the in situ immune response toward TAA in primary UM tumors. In the blood, we observed responses against this same class of Ags (Melan-A) as well as against tumor-specific Ags (SF3B1mut related). The immune infiltrate including tumor Ag–specific CD8 T cells was more important in M3 tumors than in D3 tumors despite M3 tumors having the worst prognosis. Notably, no systemic response was observed in ≈50% of the patients with M3 tumors despite the presence of abundant Melan-A–specific T cells in the tumor. Surprisingly, a tumor Ag–specific immune response was observed systemically in some patients with D3 tumors in the absence of a local immune response.
Thus, an immune response was present in many patients in the eye, which is usually considered an immune-privileged site. In the absence of known lymphatic drainage of the posterior chamber of the eye (Dickinson and Gausas, 2006; Reyes et al., 2017) and the absence of lymph node invasion in this disease, the way tumor Ags would reach a draining lymph node to prime naïve T cells is mysterious. Yet, more recent studies point to a functional lymphatic drainage of the retina and vitreum (Grüntzig and Hollmann, 2019). Still, the macrophage and dendritic cells associated with the remodeling of the tumor stroma may capture tumor Ags and migrate to the draining lymph node or directly to the blood to reach the spleen and prime naïve T cells in both sites. The conditioning of the Ag-presenting cells (APCs) by the eye microenvironment may lead to inadequate priming as we showed in an autochthonous lung tumor model in mice (Alonso et al., 2018). The resulting primed T cells would be able to circulate back to the eye tumor without being able to mediate efficient effector functions. Inflammation may be more prominent in M3 tumors leading to both accumulation of effector CD8+ T cells and an increase in Tregs (Bronkhorst et al., 2012). Whatever the site of priming, CD8+ T cells were recruited inside the tumor while their circulating effector counterparts were not detected in the blood in most cases. High expression of HLA class I and II has been observed in UM of patients with decreased survival (Ericsson et al., 2001). This is compatible with the proliferation of tumor-specific CD8+ T cells we observed in high-risk M3 tumors indicating that tumor Ags are presented in the tumor bed.
In contrast, the number and frequency of CD8+ T cells in D3 tumors was lower but a systemic immune response toward Melan-A and SF3B1mut-related neo-Ags was often detected. One hypothesis to reconcile this set of data is that M3 tumors are permissive for T cell infiltration but inhibit the effector capacity of CD8+ T cells in situ leading to exhaustion and deletion while D3 tumors are not permissive for infiltration, and the primed antitumor CD8+ T cells continue to circulate in the blood. Another hypothesis stems from one important limitation of this work related to the exclusive study of enucleated patients corresponding to more advanced and maybe more aggressive tumors displaying a higher risk of metastasis than common UM. In fact, the disease may already be disseminated in some patients with localized UM at the time of study. Dormant micrometastases infiltrated by immune cells have been found in patients without macrometastases and deceased from UM-unrelated causes (Gill et al., 2023). This disseminated disease may induce strong priming, leading to the important tumor infiltration we observed in M3 tumors. In line with a reduced metastases occurrence, the disseminated tumor mass may be lower in D3 tumors. This would induce poor priming of tumor Ag–specific T cells that would not be activated strongly enough to translocate to the D3 tumors, as efficient priming is required to reach tissues (Joncker et al., 2006).
Primary UM has been often described as poorly infiltrated (Mariani et al., 2023) which is in apparent contradiction with our findings. This may be due to different experimental approaches, as in most studies, immunohistochemistry (IHC) was used while we applied flow cytometry and scRNAseq. However, our study lacks spatial information on the distribution of the T cells in tumoral versus peritumoral areas. On the other hand, sample preparation for flow cytometry uses bigger tissue volume than thin tissue sections, leading to the recovery of larger numbers of infiltrating cells. Although our approach cannot distinguish peri versus intratumoral cells, we observed striking differences between tumor and juxta-tumoral tissues.
The nature of the tumor Ags recognized by the CD8 T cells found in the eye tumor is only partially known. The low mutation burden of UM suggests that the immune response we observed should be directed to TAA such as Melan-A, PRAME, PMEL, and TYR or possibly to other Ags related to dysregulation of mRNA expression or processing (Xie et al., 2023). In the TILs of cutaneous melanoma, the TCRs specific for highly expressed melanoma-associated Ag are less avid for their cognate peptides than the TCRs that are specific for tumor-specific neo-Ags which are expressed at low levels (Oliveira et al., 2021). Which of these cells are instrumental to control tumor growth is not known, but highly mutated tumors are associated with better survival during ICI treatment (Rizvi et al., 2015). As UM is poor in point mutations, it is to be expected that the main response should be directed against tumor-specific Ags. An exception is the D3 SF3B1mut tumors that elicit immune responses directed to peptides generated by aberrant splicing (Bigot et al., 2021). We evidenced a response against the differentiation melanoma Ag Melan-A in M3 tumors, but we did not have access to HLA-A2+ SF3B1mut tumors due to the rarity of enucleation in this clinical setting. The number of Melan-A–specific clones in the tumor bed varied a lot among the different patients: in all cases, large clones were present but smaller clones were also observed in some patients. Still, a more diversified antitumor response toward other tumor Ags is very likely as most expanded clonotypes belonging to cluster C0 were not labeled by the Melan-A tetramer.
The scRNAseq study provided interesting clues on the intratumoral responses: in the great majority of the cases, the only proliferating cells belonged to cluster C0 CD39+PD-1+CD8+ T cells, suggesting that the other expanded clonotypes belonging to other clusters were not tumor specific. Besides the chronically activated cluster C0 CD39+PD-1+CD8+ T cells, smaller clonal expansions were observed in both CD8+ and CD4+ T cells. As expected (Seder and Ahmed, 2003), the size of the clonal expansions was higher in CD8+ than in CD4+ T cells. In many cases, expanded clonotypes projected on very discrete regions (some cluster corresponded to one clone) of the UMAP indicating particular and discrete differentiation programs as previously found in lung cancers (Gueguen et al., 2021). This striking result indicates a strong link between effector differentiation and TCR specificity. This could be related to the context of initial priming or to the characteristics of TCR MHC:epitope interaction as proposed by Achar et al. (2022).. In the three patients studied, we observed at least eight different differentiation states. Additional differentiation states may exist as we studied only three samples.
As expected, the pattern of recirculation of the T cells varied according to their differentiation state in the tumor. A large proportion of the cells belonging to C0 were not found in the circulation despite their high clonal size. This may preclude the isolation of tumor-Ag–specific T cells in the blood. It remains to be determined whether the C0 T cells that recirculate differ from those that do not, regarding Ag specificity or TCR avidity. Notably, we did not find consistent differences in the three patients regarding the transcriptome of circulating versus non-circulating clonotypes in the C0 cluster. Surprisingly, the large clones from the Trm CD8+ C4 cluster were found in the blood suggesting they represent very large clones in the body leading to some level of “leakiness” in their tissue residency program. Notably, even very small clones from the KLRD1+CD8+ C6 were found in the blood, suggesting also high clonal size in the body or a strong recirculation pattern. Altogether, the differential recirculation pattern of the T cells found in the tumor according to their differentiation state indicates that studying the blood provides only a biased view of the antitumor response. This result justifies making efforts to characterize the T cell response inside tumors even though only large primary UMs are surgically treated. Indeed, identifying the Ag specificity of the expanded TCRs expressed by C0 cells and shared between patients may lead to new therapeutic targets accessible to bispecific T cell engagers (Nathan et al., 2021), TCR, T cell therapies, or therapeutic vaccines.
Materials and methods
Human samples
Blood and tumor samples were collected from patients with localized and untreated UM followed at Institut Curie. Some clinical characteristics of the patient and the genomic status of the samples used in the different figures are listed in Table S1. Written informed consent had been obtained from the patients. The samples were obtained either through the institutional review board (IRB)–approved (CPP: 19.12.19.62211), protocol Salome (IC 2019-13) or during diagnosis or therapeutic procedures. All patients were informed that tissue and blood specimens harvested during diagnosis and therapeutic procedures might be used for research purposes and had signed an informed consent form.
HLA-A2 typing by flow cytometry
Fresh blood was incubated with HLA-A2 FITC (clone BB7.2) and the cells were analyzed in a BD Fortessa cytometer.
Tumor mutations and chromosome 3/8 analysis
DNA was extracted from frozen tissues by phenol-chloroform. The SureSelect XT HS enrichment kit (AGILENT, DRAGON custom panel) was used followed by sequencing on an ILLUMINA sequencer (NextSeq or NovaSeq). Bioinformatics analysis encompassed the following steps: (1) Demultiplexing of raw data (BCL) to Fastq with bcl2fastq; (2) alignment to the human reference genome (GRCh37/hg19) with BWA mem; (3) establishment of the genomic profile to highlight copy number alterations using an adaptation of the Facets script; (4) detection and annotation of point variants using VarScan2 and intermediate size indels with transIndel; and (5) in silico analysis of identified nucleotide variants (Alamut, Cosmic).
Tissue processing
The tumors and the juxta-tumor tissue were cut into small fragments and dissociated in CO2-independent media containing 5% fetal bovine serum (FBS), 2 mg/ml collagenase, 25 µg/ml DNAse, and 2 mg/ml hyaluronidase. The tissues were then agitated in a shaker at 37°C, 180–200 rpm for 35 min. Then the cells were washed twice in PBS+ (D-PBS, 2 mM EDTA, 1% human serum) and filtered through a 40-µm mesh. Then the cells were stained for flow cytometry analysis or frozen in 90% FBS 10% DMSO for further analysis.
HLA-A2 tetramers
Biotinylated recombinant HLA-A*02:01 molecules were purchased from immuneAware as easYmers (catalog #1002-1) or produced and folded with a photo-cleavable peptide KILGFVFJV by the recombinant protein core facility P2R as previously described (Rodenko et al., 2006). Peptides were synthetized at >95% purity (GeneCust): UM-A2-14 LLIRWQHFL, UM-A2-17 AALPILFQV, UM-A2-18 ALLLQLFTL, UM-A2-26 ALLPGLPAA, UM-A2-37 RLPGVLPRA, A2-MelanA ELAGIGILTV (mutated version), A2-FLU MP 58-66 GILGFVFTL, A2-CMV pp65 495-503 NLVPMVATV (Bigot et al., 2021). Monomers were loaded with peptides by incubation at 18°C for 48 h (easYmers) or by exposing them to 366 nm UV light at 4°C for 1 h. For each tetramer, MHC/peptide complex (100 μmol/liter) was combined for 1 h at room temperature with fluorescent streptavidin (BioLegend) or oligo-tagged streptavidin (BioLegend) for a single-cell experiment. Tetramers were stored at 4°C for a maximum of 3 mo.
HLA-A2 tetramers and antibody staining
PBMC were isolated using standard Ficoll-gradient procedures and either studied fresh for phenotyping or frozen as above. PBMC were thawed in RPMI medium (GIBCO) containing 10% FBS. Cell suspensions were incubated for 30 min in a culture medium containing 50 nmol/liter dasatinib (Lissina et al., 2009) to improve tetramer staining. CD8+ T cells from PBMC were enriched using a human CD8+ T cell enrichment kit (catalog #19053; Stemcell) following the manufacturer’s instructions. Cells from tumors were stained without enrichment. Dead cells were stained with live/dead AQUA (Invitrogen). For tetramer staining, tetramers for each specificity were labeled separately with two different fluorochromes to allow the use of six different tetramer/peptide complexes in the same experiment and to decrease the noise related to nonspecific binding (Andersen et al., 2012). Briefly, cells were incubated for 20 min with tetramer complexes in brilliant stain buffer (BD) and then cells were stained for 20 min with surface antibodies (Table S5). Staining for transcription factors was performed on cells using the Foxp3/Transcription Factor Staining Buffer Set (catalog #00-5523; Thermo Fisher Scientific). Cells were then washed and analyzed in a BD Fortessa cytometer. Data were analyzed using FlowJo V10.2 software (Tree star).
Unsupervised analysis of flow cytometry data
The FCS files were first cleaned in FlowJo by removing debris, dead cells positive for live/dead AQUA, and CD3− cells. The processed FCS files were imported into the OMIQ program for Arcsinh scaling, subsampling (maximum 1,000 cells per sample), gating for CD4+ and CD8+, and UMAP construction using the following features CD69, LAG-3, KLRG1, CD25, CD103, CD4, CD39, CD8, PD-1, CD127, CD27, and Tim3 using default UMAP settings.
IHC
IHC was performed on 3 μm–thick sections of formalin-fixed paraffin-embedded patient tumor tissue from primary UMs (enucleation specimens), with D3 or M3 status confirmed by molecular studies. It was performed on a Leica BOND RX autostainer (Leica) using the Bond Polymer Refine RED Detection Kit (Leica) and the following antibodies and conditions: HLA Class 1 ABC (clone MHC-I/8147R, pH 9 retrieval, 1/10,000 dilution, 15-min incubation; Thermo Fischer Scientific) and Melan-A (clone A103, pH9, 1/200, 45 min; Dako). Staining was evaluated in tumor cells in a semiquantitative manner by a pathologist (J. Cyrta) blinded to the molecular data and the biological hypothesis. Only cytoplasmic staining was scored for the anti-Melan-A IHC, and only membranous staining was scored for the anti-MHC-I IHC. The result for each case was expressed as an H-score (defined as the sum of each of the staining intensities [range, 0–3] multiplied by the percentage of stained tumor cells at that intensity level). Results between the two molecular groups were compared using the Student’s t test.
Single-cell transcriptomic and VDJ analysis
Thawed tumor-infiltrating lymphocytes from three patients (coh4_84, coh4_56, and coh4_76) were stained with tetramers loaded with peptide Melan-A (ELAGIGILTV) associated with fluorochromes phycoerythrin (PE) and allophycocyanin (APC). PE tetramers were performed using TotalSeq-C0951 PE streptavidin (BioLegend) and APC tetramers using conventional APC streptavidin. Cells were then stained with CD4 FITC, CD3 A700, CD8 PECy7, PD-1 APC-Cy7, CD39 PercP eFluor710, and finally with DAPI. The CD4+, CD8+ MelanA+, CD8+CD39+PD-1+ cells, and the rest of the CD8+ T cells were sorted using a FACS ARIA (BD). The four sorted populations were counted and mixed at 30%, 10%, 30%, and 30%, respectively. A total of 15,000 cells per donor were loaded onto a Chromium controller using a Chromium Next GEM Single Cell 5′ GEX and VDJ reagent kit with feature barcoding technology according to the manufacturer’s instructions.
scRNAseq analysis
For each sample, raw reads were processed using Cell Ranger (version 7.0.1). The reference genome was GRCh38-2020-A for the scRNAseq and vdj_GRCh38_alts_ensembl-3.1.0-3.1.0 for sc VDJ-seq. The median number of unique molecular identifiers (UMIs) for TRα and TRβ contig per cell was 3, 3, and 4 and 9, 13, and 12 for coh4_84, coh4_56, and coh4_76, respectively. All analyses were performed using R version 4.2.3 and the following packages: Seurat_4.1.3, clustree_0.4.4. Based on the distribution of the numbers of genes and molecules detected per cell, the following filters were applied to remove outliers: For coh4_84, the filters were set at nFeature_RNA > 1,100 and nCount_RNA > 2,400 while for coh4_56 and coh4_76 filters were set at 1,500 >nFeature_RNA < 26,000 and 1,500 > nCount_RNA > 6,000, respectively. Cells containing >8% for coh4_84 and >10% for the two other samples of mitochondrial genes were considered dying cells and filtered out.
For scRNAseq analysis, all TCR-related genes were excluded to avoid interfering in the clusterization (TRAV, TRBV, TRAJ, TRAC, TRBD, TRBJ, TRBC). 2,000 highly variable features were considered. Graph-based clustering (Louvain method) was performed using the default parameters and a UMAP (dims = 15, 18, 20 for the 3 samples, respectively) was constructed with a resolution of 0.2, 0.3, and 0.2 respectively based on the stability observed with the clustree package. The differentially expressed genes (DEGs) were determined using the FindAllMarkers() function (using a logistic regression base 10, min.pct = 0.1 and Fold-change >0.25, Table S2). To characterize the different clusters from the three patients, we used a candidate gene supervised approach (including CD8A, CD8B, PDCD1, ENTPD1, TOX, HAVCR2, LAG3, GZMB, CD4, IL7R, CD40LG, SELL, KLF2, CCR7, TGFB1, XCL2, KLRD1, KLRC2, GNLY, CD44, FOXP1, STAT4, ALOX5AP, ITGAE, ITGA1, ZNF683, FOXP3, CCR8, CTLA4, TOP2A, and MKI67) coupled with hierarchical clustering. For the hierarchical clustering, the average expression of each candidate gene was calculated by cluster and by the patient after respective scaling. Then, a Pearson correlation followed by a ward.D2 method was performed (h.clust, stats V4.2.3).
A subclustering was necessary for coh4_84 and coh4_76 samples to split a cluster that exhibited heterogeneity for several genes of interest such as CD4, FOXP1, ENTPD-1, and PDCD1 for the coh4_84 and CD4, XCL2, and ENTPD-1 for the coh4_76 samples. A resolution of 0.1 and 0.2 was selected for both samples, respectively, and the corresponding subclusters were reinjected into the final UMAP.
Hash-tag oligos (HTO) were used to multiplex cells labeled by the HLA-A2:Melan-A tetramers. Demultiplexing of HTO data was performed using CITE-seq-Count (version 1.4.3). HTO data was transformed using centered log-ratio (CLR) normalized and cell barcodes were assigned to mice of origin using Seurat function HTODemux() with default threshold for classification (positive quantile = 0.99). In total, 6,668; 9,545; and 5,338 cells for coh4_84, coh4_56, and coh4_76 respectively, with a barcode detected in both RNA and HTO arrays were considered for downstream analysis.
A clonotype was defined as a set of cells having identical CDR3-nt sequences of (i) both TCRα and TCRβ chains, (ii) either a TCRα/TCRβ with no sequences available for another TCRβ or TCRα chain respectively, and (iii) 2 TCRα and 1 TCRβ. Clonotypes with 2 TCRβ chains associated or not to a TCRα chain were discarded from the TCR analysis.
The distribution of the clonotype sizes in the different clusters (in panel F of Figs. 3, S3, and S5) was calculated according to the following procedure. Each clonotype was assigned to a specific category according to its total number of cells represented in the entire dataset: 1 cell (singleton), 2–9 cells, 10–59 cells, and over 60 cells. Then the cells belonging to a specific clonotype were ranked according to their previously defined categories among each cluster. Thus, the cells of an expanded clonotype “dark blue” can be considered in several clusters.
The average clonal size in each cluster is shown in panel G of Figs. 3, S3, and S4, and was computed on the expanded (n ≥ 2 cells) clonotypes as the number of cells divided by the number of clonotypes inside each cluster.
Bulk TCRseq
Frozen PBMC drawn at the time of the enucleation were studied. After thawing, 1.7 × 106, 2.16 × 106, and 2.32 × 106 peripheral blood lymphocytes from coh4_56, coh4_76, and coh4_84 patients, respectively, were retrieved for RNA extraction using Trizol chloroform technique. Human TCR RNA Multiplex-Thrm-001 Kit (MiLaboratories) on 500 ng RNA was used for amplification of TCRα and β cDNA to make Illumina libraries using the combination of highly sensitive multiplex PCR and UMIs to generate 38 × 106 to 67 × 106 reads per sample. TCRseq was performed on a Novaseq sequencer with pair-end, 2 × 150 base pair reads. The number of UMI obtained was TRA = 279,354 and TRB = 345,102 for coh4_56, TRA = 275,183 and TRB = 453,231 for coh4_76, and TRA = 95,874 and TRB = 235,315 for coh4_84. Mixcr (version 4.4.2) was used to perform the alignment and assemble the reads with the following parameters: milab-human-rna-tcr-umi-multiplex. Then the data were analyzed using dplyr (version 1.0.1.0). For this section, the cdr3-nt sequences of the bulk PBMC dataset and the VDJ-scRNAseq for each patient were joined to identify common clonotypes for both TCRα and TCRβ chains (Table S4).
We estimated the recirculation rate in the PBMC of the clonotype by attributing a clonotype defined by its CDR3-nt chains α and β to its most expanded cluster identified previously in the VDJ-scRNAseq analysis. To be homogenous among the three samples and according to their transcriptional profile proximity, the following clusters were pooled for this analysis in Fig. 5: cluster C0 and C0a of the coh4_84, cluster C0 and C0b, and cluster C7 and 7b of the coh4_76, respectively.
Quantification and statistical analysis
For each experiment, the statistical tests used are indicated in the figure legends. The following statistical tests were used and calculated by GraphPad Prism v8 (GraphPad): Mann–Whitney and Wilcoxon.
Online supplemental material
Five figures and five tables are provided. Fig. S1 shows the characterization of Tregs and CD8+ cells in primary UMs. Figs. S2 and S3 display the VDJ scRNAseq of T cells in primary UM from patient coh4_56 and patient coh4_76, respectively. Fig. S4 shows the pattern of recirculation between the blood and the tumor according to cluster and clonotype size in the tumor. Fig. S5 characterizes the phenotype of the T cells found in the blood of patients with primary UMs. Table S1 displays the main characteristics of the patients and samples included in the study and lists the samples used for each figure. Table S2 lists the DEGs for each patient. Table S3 lists the TCRs characterized in the different scRNAseq samples. Table S4 shows the results of the bulk TCRseq from the PBMC of a patient and by TCRα and TCRβ chains. Table S5 lists the antibodies used in this study.
Supplementary Material
shows the main characteristics of the patients and samples included in the study and lists the samples used for each figure.
shows the DEGs for each patient.
lists the TCRs characterized in the different scRNAseq samples.
lists the bulk TCRseq from the PBMC by patient and by TCRα and TCRβ chains.
lists the antibodies used in this study.
Acknowledgments
We thank the patients who have agreed to participate in this study. We thank the flow cytometry core and the ICGex NGS platform of the Institut Curie for technical help with scRNAseq experiments. We thank Maude Delost for the extensive reviewing of the paper and Sergio Roman-Roman for editing and support. We thank Josh Waterfall for discussion and Leanne De Koning for the support. We thank Vadim Karnaukhov for suggestions in data processing. We thank the Clinic Immunology laboratory members for their help in blood sample purification and phenotyping. We thank the pathology unit for fresh tissue samples. ICGex is supported by the grants ANR10EQPX03 (Equipex) and ANR10INBS0908 (France Génomique Consortium) from the Agence Nationale de la Recherche (“Investissements d’Avenir” program), by the ITMO-Cancer Aviesann (Plan Cancer III) and the SiRIC-Curie program (INCa-DGOS-465 and INCa-DGOS-Inserm_12554).
Funding was provided by the Institut National de la Santé et de la Recherche Médicale (O. Lantz), Institut Curie (O. Lantz), Ligue contre le Cancer (doctoral fellowship to F. Lucibello), and the Institut Curie MD PhD program (F. Lucibello). This study was supported by ANR-10-IDEX-0001-02 PSL and LabEx DCBIOL; the program France 2030 launched by the French Government; SIRIC2 INCa-DGOS-Inserm_12554; Canceropole Ile-de-France (Emergence program); and the European Union’s Horizon 2020 Research and Innovation program (UM Cure 2020 project, grant agreement no. 667787).
Author contributions: Conceptualization: O. Lantz and A.I. Lalanne; Methodology: F. Lucibello and A.I. Lalanne; Investigation: F. Lucibello, A.I. Lalanne, A.-L. Le Gac, A. Soumare, S. Aflaki, J. Cyrta, C. Jamet, C. Ekwegbara, L.B, M. Mestdagh, A. Houy, A. Le Ven, and G. Pierron; Formal analysis: O. Lantz, F. Lucibello, A.-L. Le Gac, and A.I. Lalanne; Funding acquisition: O. Lantz; Critical material provision: K. Bernardeau, N. Cassoux, M. Rodrigues, A. Matet, D. Malaise, S. Piperno-Neumann, and S. Gardrat; Supervision: O. Lantz, A.I. Lalanne, and M. Salou; Writing—original draft: F. Lucibello, A.-L. Le Gac, A.I. Lalanne, and O. Lantz; Writing—review & editing: O. Lantz, F. Lucibello, A.-L. Le Gac, A.I. Lalanne, M.-H. Stern, and M. Rodrigues.
Data availability
Sequencing data have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE248838. Flow cytometry raw data will be made accessible to investigators for reanalysis upon reasonable request.
References
- Achar, S.R., Bourassa F.X.P., Rademaker T.J., Lee A., Kondo T., Salazar-Cavazos E., Davies J.S., Taylor N., François P., and Altan-Bonnet G.. 2022. Universal antigen encoding of T cell activation from high-dimensional cytokine dynamics. Science. 376:880–884. 10.1126/science.abl5311 [DOI] [PubMed] [Google Scholar]
- Alonso, R., Flament H., Lemoine S., Sedlik C., Bottasso E., Péguillet I., Prémel V., Denizeau J., Salou M., Darbois A., et al. 2018. Induction of anergic or regulatory tumor-specific CD4+ T cells in the tumor-draining lymph node. Nat. Commun. 9:2113. 10.1038/s41467-018-04524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsafadi, S., Houy A., Battistella A., Popova T., Wassef M., Henry E., Tirode F., Constantinou A., Piperno-Neumann S., Roman-Roman S., et al. 2016. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 7:10615. 10.1038/ncomms10615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, R.S., Kvistborg P., Frøsig T.M., Pedersen N.W., Lyngaa R., Bakker A.H., Shu C.J., Straten P., Schumacher T.N., and Hadrup S.R.. 2012. Parallel detection of antigen-specific T cell responses by combinatorial encoding of MHC multimers. Nat. Protoc. 7:891–902. 10.1038/nprot.2012.037 [DOI] [PubMed] [Google Scholar]
- Bigot, J., Lalanne A.I., Lucibello F., Gueguen P., Houy A., Dayot S., Ganier O., Gilet J., Tosello J., Nemati F., et al. 2021. Splicing patterns in SF3B1-mutated uveal melanoma generate shared immunogenic tumor-specific neoepitopes. Cancer Discov. 11:1938–1951. 10.1158/2159-8290.CD-20-0555 [DOI] [PubMed] [Google Scholar]
- Bronkhorst, I.H.G., and Jager M.J.. 2013. Inflammation in uveal melanoma. Eye. 27:217–223. 10.1038/eye.2012.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronkhorst, I.H.G., Vu T.H.K., Jordanova E.S., Luyten G.P.M., Burg S.H.V.D., and Jager M.J.. 2012. Different subsets of tumor-infiltrating lymphocytes correlate with macrophage influx and monosomy 3 in uveal melanoma. Invest. Ophthalmol. Vis. Sci. 53:5370–5378. 10.1167/iovs.11-9280 [DOI] [PubMed] [Google Scholar]
- Cassoux, N., Rodrigues M.J., Plancher C., Asselain B., Levy-Gabriel C., Lumbroso-Le Rouic L., Piperno-Neumann S., Dendale R., Sastre X., Desjardins L., and Couturier J.. 2014. Genome-wide profiling is a clinically relevant and affordable prognostic test in posterior uveal melanoma. Br. J. Ophthalmol. 98:769–774. 10.1136/bjophthalmol-2013-303867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, D.K., Yuan F., Rizkallah P.J., Miles J.J., Gostick E., Price D.A., Gao G.F., Jakobsen B.K., and Sewell A.K.. 2009. Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. J. Biol. Chem. 284:27281–27289. 10.1074/jbc.M109.022509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demkowicz, P., Pointdujour-Lim R., Miguez S., Lee Y., Jones B.S.C.L., Barker C.A., Bosenberg M., Abramson D.H., Shoushtari A.N., Kluger H., et al. 2023. Determinants of overall survival in patients with metastatic uveal melanoma. Cancer. 129:3275–3286. 10.1002/cncr.34927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, A.J., and Gausas R.E.. 2006. Orbital lymphatics: Do they exist? Eye. 20:1145–1148. 10.1038/sj.eye.6702378 [DOI] [PubMed] [Google Scholar]
- Duhen, T., Duhen R., Montler R., Moses J., Moudgil T., de Miranda N.F., Goodall C.P., Blair T.C., Fox B.A., McDermott J.E., et al. 2018. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 9:2724. 10.1038/s41467-018-05072-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante, M.A., Rodriguez D.A., Kurtenbach S., Kuznetsov J.N., Sanchez M.I., Decatur C.L., Snyder H., Feun L.G., Livingstone A.S., and Harbour J.W.. 2020. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat. Commun. 11:496. 10.1038/s41467-019-14256-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson, C., Seregard S., Bartolazzi A., Levitskaya E., Ferrone S., Kiessling R., and Larsson O.. 2001. Association of HLA class I and class II antigen expression and mortality in uveal melanoma. Invest. Ophthalmol. Vis. Sci. 42:2153–2156. [PubMed] [Google Scholar]
- Fox, J.C., Nakayama T., Tyler R.C., Sander T.L., Yoshie O., and Volkman B.F.. 2015. Structural and agonist properties of XCL2, the other member of the C-chemokine subfamily. Cytokine. 71:302–311. 10.1016/j.cyto.2014.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney, S.J., Pedersen M., Gentien D., Dumont A.G., Rapinat A., Desjardins L., Turajlic S., Piperno-Neumann S., de la Grange P., Roman-Roman S., et al. 2013. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 3:1122–1129. 10.1158/2159-8290.CD-13-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, V.T., Norrman E., Sabazade S., Karim A., Lardner E., and Stålhammar G.. 2023. Multiorgan involvement of dormant uveal melanoma micrometastases in postmortem tissue from patients without coexisting macrometastases. Am. J. Clin. Pathol. 160:164–174. 10.1093/ajcp/aqad029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüntzig, J., and Hollmann F.. 2019. Lymphatic vessels of the eye: Old questions: New insights. Ann. Anat. 221:1–16. 10.1016/j.aanat.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Gueguen, P., Metoikidou C., Dupic T., Lawand M., Goudot C., Baulande S., Lameiras S., Lantz O., Girard N., Seguin-Givelet A., et al. 2021. Contribution of resident and circulating precursors to tumor-infiltrating CD8+ T cell populations in lung cancer. Sci. Immunol. 6:eabd5778. 10.1126/sciimmunol.abd5778 [DOI] [PubMed] [Google Scholar]
- Harbour, J.W., Onken M.D., Roberson E.D.O., Duan S., Cao L., Worley L.A., Council M.L., Matatall K.A., Helms C., and Bowcock A.M.. 2010. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 330:1410–1413. 10.1126/science.1194472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joncker, N.T., Helft J., Jacquet A., Premel V., and Lantz O.. 2006. Intratumor CD4 T-cell accumulation requires stronger priming than for expansion and lymphokine secretion. Cancer Res. 66:5443–5451. 10.1158/0008-5472.CAN-05-3526 [DOI] [PubMed] [Google Scholar]
- Kaminskiy, Y., Kuznetsova V., Kudriaeva A., Zmievskaya E., and Bulatov E.. 2022. Neglected, yet significant role of FOXP1 in T-cell quiescence, differentiation and exhaustion. Front. Immunol. 13:971045. 10.3389/fimmu.2022.971045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson, J., Nilsson L.M., Mitra S., Alsén S., Shelke G.V., Sah V.R., Forsberg E.M.V., Stierner U., All-Eriksson C., Einarsdottir B., et al. 2020. Molecular profiling of driver events in metastatic uveal melanoma. Nat. Commun. 11:1894. 10.1038/s41467-020-15606-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortekaas, K.E., Santegoets S.J., Sturm G., Ehsan I., van Egmond S.L., Finotello F., Trajanoski Z., Welters M.J.P., van Poelgeest M.I.E., and van der Burg S.H.. 2020. CD39 identifies the CD4+ tumor-specific T-cell population in human cancer. Cancer Immunol. Res. 8:1311–1321. 10.1158/2326-6066.CIR-20-0270 [DOI] [PubMed] [Google Scholar]
- Lissina, A., Ladell K., Skowera A., Clement M., Edwards E., Seggewiss R., van den Berg H.A., Gostick E., Gallagher K., Jones E., et al. 2009. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J. Immunol. Methods. 340:11–24. 10.1016/j.jim.2008.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma, A.M., Suo S., Wang Y., Gunasti L., Porter C.B.M., Nabilsi N., Tadros J., Ferretti A.P., Liao S., Gurer C., et al. 2022. Tissue-resident memory and circulating T cells are early responders to pre-surgical cancer immunotherapy. Cell. 185:2918–2935.e29. 10.1016/j.cell.2022.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maat, W., Ly L.V., Jordanova E.S., de Wolff-Rouendaal D., Schalij-Delfos N.E., and Jager M.J.. 2008. Monosomy of chromosome 3 and an inflammatory phenotype occur together in uveal melanoma. Invest. Ophthalmol. Vis. Sci. 49:505–510. 10.1167/iovs.07-0786 [DOI] [PubMed] [Google Scholar]
- Mariani, P., Torossian N., van Laere S., Vermeulen P., de Koning L., Roman-Roman S., Lantz O., Rodrigues M., Stern M.-H., Gardrat S., et al. 2023. Immunohistochemical characterisation of the immune landscape in primary uveal melanoma and liver metastases. Br. J. Cancer. 129:772–781. 10.1038/s41416-023-02331-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy, K.A., Rimoldi D., Brasseur F., Rodgers S., Liénard D., Marchand M., Rennie I.G., Murray A.K., McIntyre C.A., Platts K.E., et al. 1996. Infrequent expression of the MAGE gene family in uveal melanomas. Int. J. Cancer. 66:738–742. [DOI] [PubMed] [Google Scholar]
- Naigeon, M., Roulleaux Dugage M., Danlos F.-X., Boselli L., Jouniaux J.-M., de Oliveira C., Ferrara R., Duchemann B., Berthot C., Girard L., et al. 2023. Human virome profiling identified CMV as the major viral driver of a high accumulation of senescent CD8+ T cells in patients with advanced NSCLC. Sci. Adv. 9:eadh0708. 10.1126/sciadv.adh0708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan, P., Hassel J.C., Rutkowski P., Baurain J.-F., Butler M.O., Schlaak M., Sullivan R.J., Ochsenreither S., Dummer R., Kirkwood J.M., et al. 2021. Overall survival benefit with tebentafusp in metastatic uveal melanoma. N. Engl. J. Med. 385:1196–1206. 10.1056/NEJMoa2103485 [DOI] [PubMed] [Google Scholar]
- Oliveira, G., Stromhaug K., Klaeger S., Kula T., Frederick D.T., Le P.M., Forman J., Huang T., Li S., Zhang W., et al. 2021. Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature. 596:119–125. 10.1038/s41586-021-03704-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patin, E., Hasan M., Bergstedt J., Rouilly V., Libri V., Urrutia A., Alanio C., Scepanovic P., Hammer C., Jönsson F., et al. 2018. Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat. Immunol. 19:302–314. 10.1038/s41590-018-0049-7 [DOI] [PubMed] [Google Scholar]
- Péguillet, I., Milder M., Louis D., Vincent-Salomon A., Dorval T., Piperno-Neumann S., Scholl S.M., and Lantz O.. 2014. High numbers of differentiated effector CD4 T cells are found in patients with cancer and correlate with clinical response after neoadjuvant therapy of breast cancer. Cancer Res. 74:2204–2216. 10.1158/0008-5472.CAN-13-2269 [DOI] [PubMed] [Google Scholar]
- Reyes, N.J., O’Koren E.G., and Saban D.R.. 2017. New insights into mononuclear phagocyte biology from the visual system. Nat. Rev. Immunol. 17:322–332. 10.1038/nri.2017.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi, N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S., et al. 2015. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 348:124–128. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, A.G., Shih J., Yau C., Gibb E.A., Oba J., Mungall K.L., Hess J.M., Uzunangelov V., Walter V., Danilova L., et al. 2017. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 32:204–220.e15. 10.1016/j.ccell.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenko, B., Toebes M., Hadrup S.R., van Esch W.J.E., Molenaar A.M., Schumacher T.N.M., and Ovaa H.. 2006. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 1:1120–1132. 10.1038/nprot.2006.121 [DOI] [PubMed] [Google Scholar]
- Rodrigues, M., Koning L., Coupland S.E., Jochemsen A.G., Marais R., Stern M.-H., Valente A., Barnhill R., Cassoux N., Evans A., et al. 2019. So close, yet so far: Discrepancies between uveal and other melanomas. A position paper from UM Cure 2020. Cancers. 11:1032. 10.3390/cancers11071032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddiki, N., Santner-Nanan B., Martinson J., Zaunders J., Sasson S., Landay A., Solomon M., Selby W., Alexander S.I., Nanan R., et al. 2006. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J. Exp. Med. 203:1693–1700. 10.1084/jem.20060468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder, R.A., and Ahmed R.. 2003. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Immunol. 4:835–842. 10.1038/ni969 [DOI] [PubMed] [Google Scholar]
- Simoni, Y., Becht E., Fehlings M., Loh C.Y., Koo S.-L., Teng K.W.W., Yeong J.P.S., Nahar R., Zhang T., Kared H., et al. 2018. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 557:575–579. 10.1038/s41586-018-0130-2 [DOI] [PubMed] [Google Scholar]
- Triozzi, P.L., Aldrich W., Crabb J.W., and Singh A.D.. 2015. Spontaneous cellular and humoral tumor antigen responses in patients with uveal melanoma. Melanoma Res. 25:510–518. 10.1097/CMR.0000000000000207 [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk, C.D., Bezrookove V., Green G., Bauer J., Gaugler L., O’Brien J.M., Simpson E.M., Barsh G.S., and Bastian B.C.. 2009. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 457:599–602. 10.1038/nature07586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry, E.J., Ha S.-J., Kaech S.M., Haining W.N., Sarkar S., Kalia V., Subramaniam S., Blattman J.N., Barber D.L., and Ahmed R.. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 27:670–684. 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Xie, N., Shen G., Gao W., Huang Z., Huang C., and Fu L.. 2023. Neoantigens: Promising targets for cancer therapy. Signal Transduct. Target. Ther. 8:9. 10.1038/s41392-022-01270-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavuzyigitoglu, S., Koopmans A.E., Verdijk R.M., Vaarwater J., Eussen B., van Bodegom A., Paridaens D., Kiliç E., de Klein A., and Rotterdam Ocular Melanoma Study Group . 2016. Uveal melanomas with SF3B1 mutations: A distinct subclass associated with late-onset metastases. Ophthalmology. 123:1118–1128. 10.1016/j.ophtha.2016.01.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the main characteristics of the patients and samples included in the study and lists the samples used for each figure.
shows the DEGs for each patient.
lists the TCRs characterized in the different scRNAseq samples.
lists the bulk TCRseq from the PBMC by patient and by TCRα and TCRβ chains.
lists the antibodies used in this study.
Data Availability Statement
Sequencing data have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE248838. Flow cytometry raw data will be made accessible to investigators for reanalysis upon reasonable request.