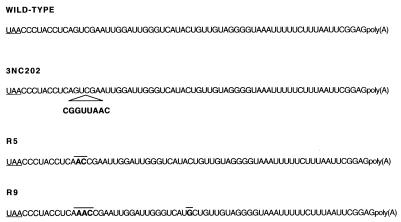Abstract
The poliovirus 3′ noncoding region (3′NCR) is involved in the efficient synthesis of viral negative-stranded RNA molecules. A strong interaction between a 105-kDa host protein and the wild-type 3′NCR, but not with a replication-defective mutant 3′NCR, was detected. This 105-kDa protein was identified as nucleolin which predominantly resides in the nucleolus and has been proposed to function in the folding of rRNA precursor molecules. A functional role for nucleolin in viral genome amplification was examined in a cell-free extract which has been shown to support the assembly of infectious virus from virion RNA. At early times of viral gene expression, extracts depleted of nucleolin produced less infectious virus than extracts depleted of fibrillarin, another resident of the nucleolus, indicating a functional role of nucleolin in the early stages of the viral life cycle in this in vitro system. Immunofluorescence analysis of uninfected and infected cells showed a nucleocytoplasmic relocalization of nucleolin, but not of fibrillarin, in poliovirus-infected cells. Relocalization of nucleolin was not simply a consequence of virally induced inhibition of translation or transcription, because inhibitors of translation or transcription did not induce nucleolar-cytoplasmic relocalization of nucleolin. These findings suggest a novel virus-induced mechanism by which certain nucleolar proteins are selectively redistributed in infected cells.
Poliovirus is a positive-stranded RNA virus whose genome, approximately 7,440 nucleotides in length, is translated into a 220-kDa polyprotein (30, 41). The polyprotein is proteolyzed by virus-encoded proteases to yield the structural capsid proteins and the nonstructural proteins required for the amplification of the viral RNA genome in infected cells (44). The mechanisms by which positive- and negative-strand RNAs are synthesized in infected cells are currently under scrutiny. Much work has concentrated on the identification and characterization of RNA-protein complexes located at the 5′ terminus of the positive-strand and at the 3′ end of the negative-strand viral RNA. Both viral 3CD and host cell poly(rC) binding proteins are associated with a cloverleaf RNA structure located at the extreme 5′ end of the viral genome (1, 2, 21, 26, 38). This ribonucleoprotein complex is thought to play a role in the synthesis of viral positive-strand RNAs. An interaction of viral polypeptide 2C (5) and cellular p36 and p38 proteins with the 3′ end of viral negative-strand RNAs, complementary to the 5′ end of the positive strand, has been implicated in the synthesis of viral RNAs (42, 43). Curiously, only p36 and p38 proteins isolated from infected cells bind to viral RNA. Recently, it has been shown that viral protease 3C can promote the formation of p38-RNA complexes, likely by proteolytic processing of an as-yet-unidentified p38 precursor protein with different RNA binding affinity or specificity (42).
The 3′ noncoding region (3′NCR) of poliovirus is 65 nucleotides (nt) in length, is highly conserved among enteroviruses (53), and is likely to contain sequences involved in the synthesis of negative-strand RNA molecules (for recent reviews, see references 29 and 56). Structural and genetic analysis of the 3′NCR has revealed the presence of a possibly multidomain RNA structure whose integrity is needed for efficient viral RNA synthesis (28, 39, 40). For example, analysis of a poliovirus mutant, 3NC202, which is temperature sensitive for RNA replication (46), has pointed to a role of RNA structures, located in the wild-type 3′NCR, in RNA replication (28). However, the 3′NCR is not absolutely required for RNA replication. Mutant poliovirus genomes that completely lack these sequences can replicate at lower efficiency and are likely to bear additional uncharacterized mutations elsewhere in the genome (52).
Using wild-type, mutant 3NC202, and revertant RNA genomes as tools, we have searched for specific RNA-protein complexes in the viral 3′NCR. This study describes the identification of a specific complex between the wild-type 3′NCR and nucleolin. Immunodepletion of nucleolin from cell-free extracts that support the poliovirus replicative cycle resulted in a diminished yield of infectious virus. Nucleolin resides predominantly in the nucleolus but shuttles between the nucleolus and the cytoplasm. Unexpectedly, upon infection with poliovirus, nucleolin dramatically relocalizes to the cytoplasm. The implications of these findings in poliovirus-host interactions and the uses of poliovirus as a tool to study nucleocytoplasmic trafficking are discussed.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were grown as monolayers in Dulbecco’s modified Eagle’s medium supplemented with 10% calf serum (Gibco BRL). Suspension cultures of HeLa S3 cells (American Type Culture Collection) were maintained in minimum essential medium Eagle (Sigma) supplemented with 5% calf serum and 2% fetal calf serum (GIBCO-BRL). Stocks of wild-type Mahoney type 1 virus and of poliovirus mutant 3NC202 were prepared at 37 and at 32.5°C, respectively (45). Revertants R5 and R9 were selected after infection with 3NC202 at the nonpermissive temperature of 39.5°C. The nucleotide changes in the 3′NCR of the revertant R5 genome were inserted into a wild-type full-length cDNA, and the recombinant virus yielded the expected revertant phenotype after isolation of individual plaques following transfection under agar (28). Revertants R5 and R9 were propagated at 39.5°C in HeLa cells.
Plasmids.
Plasmids that harbor a T7 promoter which directs the synthesis of RNA molecules containing various wild-type and mutant 3′NCRs were constructed by PCR, using Taq polymerase (Promega Biotec) as described previously (4). The following template DNAs utilized for PCR were described previously (28): T7-WT(B/S) pA, T7-R5(B/S) pA, T7-R9(B/S) pA, and T7pGem3NC202. Primers used for PCR were 5′-AAGCTTCAGGAGTGTGCC-3′ (the nucleotides in bold indicate a HindIII site followed by poliovirus nt 7294 to 7305) and 5′-TGAATTCT20CTCCGAATTAAAG-3′ [the sequence in bold indicates an EcoRI site followed by a poly(T) tract and sequences complementary to poliovirus sequences from nt 7440 to 7428]. Following PCR, the double-strand DNA product was purified (4), ligated into the pGemT plasmid (Promega Biotec), and transformed into TG1 cells. The sequences of the inserted 3′NCRs were verified by dideoxy nucleotide sequencing (Sequenase; U.S. Biochemicals). The 3′NCR-containing cDNAs were then excised after digestion with HindIII and EcoRI, isolated, and ligated into the vector pGem4 (Promega Biotec) which was previously digested with HindIII and EcoRI. The resulting plasmids contained a T7 promoter upstream of wild-type or mutant poliovirus nt 7294 to 7440, followed by a motif of 20 thymidine residues and an EcoRI site.
Measurement of viral RNA synthesis.
HeLa cells (2 × 106) were grown to 80 to 90% confluence as monolayer cultures, infected with wild-type Mahoney type 1 poliovirus at a multiplicity of infection of 20 in the presence of 5 μg of actinomycin D (Gibco-BRL)/ml and 10 μCi of [3H]uridine (Amersham)/ml, and incubated at 37°C. At different times after infection, the cells were harvested by scraping into 1 ml of ice-cold phosphate-buffered saline (PBS). Next, 5 ml of ice-cold 10% trichloroacetic acid (TCA; Mallinckrodt) was added, and the reaction mixtures were placed at 4°C for 15 h (4). The TCA-precipitated material was adsorbed to glass microfiber filters (Whatman) by filtration, using a vacuum manifold (Millipore, Inc.). The filters were air dried, and 5 ml of Ecolume scintillation fluid (ICN) was added. The samples were counted in a Beckman LS6000 IC liquid scintillation counter. This experiment was performed several times, producing data which were similar to those shown in Fig. 1.
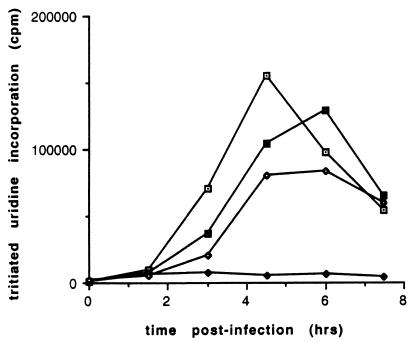
FIG. 1.
Viral RNA synthesis in cells infected with polioviruses containing various 3′NCR mutations. [3H]uridine incorporation in cells infected with the wild type (⊡), mutant 3NC202 (⧫), revertant R5 (▪), and revertant R9 (◊) is shown at the indicated times postinfection.
In vitro RNA synthesis.
RNA molecules were synthesized in the presence of [32P]CTP and 4-thio-UTP by T7 RNA polymerase as described previously (51), with minor modifications. Briefly, EcoRI-linearized plasmids (150 μg/ml) were incubated in transcription buffer (40 mM Tris-HCl [pH 7.5], 12 mM MgCl2, 10 mM dithiothreitol [DTT], and 4 mM spermidine) in the presence of 26 μCi of [α-32P]CTP (800 Ci/mmol), 250 μg of bovine serum albumin (BSA)/ml, 100 μM CTP, 500 μM GTP, 500 μM UTP, 250 μM 4-thio-UTP, and 40 U of RNasin (Promega Biotec). The 4-thio-UTP was synthesized by the method of Stade et al. (51). Transcription was initiated by the addition of 4 μg of T7 RNA polymerase/ml (kindly provided by Bruce Burnett and Charles McHenry, University of Colorado Health Sciences Center), followed by incubation for 2 h at 37°C. One unit of RQ1 DNase (Promega Biotec) per μg of DNA template was added, and incubation was continued for an additional 30 min at 37°C. The samples were extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and nucleic acids were precipitated in ethanol, a step which removed unincorporated nucleotides. The RNAs were resuspended in water and quantitated by scintillation counting. The integrity of the transcripts was analyzed by denaturing polyacrylamide gel electrophoresis (PAGE).
Unlabeled competitor RNAs were synthesized according to standard protocols (Promega Biotec). After treatment with RNase-free DNase RQ1 (Promega Biotec), the RNAs were extracted with phenol-chloroform and precipitated by the addition of 2.5 volumes of ethanol. The RNAs were resuspended in water and quantitated by measuring the A260 (40 μg/ml/A260 unit).
RNA ligands used for the RNA affinity matrix were synthesized according to the manufacturer’s protocol (Boehringer Mannheim), using 50 μg of EcoRI-linearized T7pGem4-3′NCRpA20 plasmid/ml, 40 U T7 RNA polymerase, and 350 μM biotin-16-UTP (Boehringer Mannheim) in a 20-μl reaction volume. After incubation for 2 h at 37°C, the biotinylated RNAs were treated with RQ1 DNase (see above) and purified according to the manufacturer’s instructions. The RNAs were resuspended in water and quantified spectrophotometrically.
Preparation of HeLa cell extracts.
HeLa extracts were prepared from approximately 106 cells by a Nonidet P-40 (NP-40) lysis method as described previously (45), with minor modifications. Briefly, cells were washed twice with ice-cold PBS and scraped with a rubber policeman into 0.5 ml of PBS. Next, the cells were sedimented by centrifugation at 500 × g for 1 min at 25°C. After removal of the supernatant, 0.5 ml of NP-40 lysis buffer (50 mM Tris-HCl [pH 7.9], 5 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1% NP-40) was added to the pellet, and incubation proceeded for 5 min on ice with occasional gentle mixing. The extract was centrifuged at 500 × g for 1 min at 25°C, and the supernatant was used as a source of soluble proteins. The amount of protein was quantitated by using a standard protein assay according to the manufacturer’s instructions (BioRad), using immunoglobulin G (IgG) protein (Sigma) as a standard.
UV cross-linking assay.
UV cross-linking assays were carried out as described previously (33), with some modifications. Briefly, 11.0 μg of NP-40 lysate or 20 ng of a monomethylsulfonate (Mono S) fraction containing nucleolin (see next section) was incubated in cross-linking buffer (8.5 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4], 3.0 mM MgCl2, 1.3 mM ATP, 5.0 mM creatine phosphate, 2.7 mM KCl, 1.0 mM DTT, and 4% glycerol) in the presence or absence of unlabeled competitor RNA in a 40-μl reaction volume for 10 min at 30°C. After the addition of 4-thio-UTP-containing radiolabeled RNA (4 × 105 cpm; 1 nM final concentration) the reaction mixtures were further incubated for 15 min at 30°C. Next, the samples were covered with glass and irradiated with UV light at 312 nm, using a Stratalinker model 1800 (Stratagene) at 3,000 μW/cm2 for 30 min at 4°C. After irradiation, the samples were digested with 0.14 mg of pancreatic RNase A (United State Biochemicals)/ml for 30 min at 37°C. The cross-linked RNA-protein complexes were resolved on sodium dodecyl sulfate (SDS)-containing 7.5 or 10% polyacrylamide gels and visualized by autoradiography. Competition experiments were quantitated by using the PhosphorImager screen and Molecular Dynamics Imaging software (Molecular Dynamics).
Purification of the 105-kDa RNA binding protein.
Ten liters of suspension HeLa cells (generous gift of Jerry Schaack, University of Colorado Health Sciences Center) was grown to a density of 5 × 105 cells/ml. Cells were washed twice with ice-cold PBS and collected by low-speed centrifugation at 3,500 rpm for 5 min at 4°C. Next, the cell pellet was resuspended in five packed-cell volumes of NP-40 lysis buffer and incubated for 10 min on ice with occasional gentle mixing. The extracts were clarified twice by centrifugation at 10,000 rpm in a Sorvall RC5C, for 10 min at 4°C, and the supernatant was used as a source of soluble proteins. This supernatant was fractionated by precipitation with 55 to 70% (wt/vol) ammonium sulfate as described previously (17). The precipitate was resuspended in NP-40 lysis buffer and dialyzed in a microdialysis apparatus (Pierce) against NP-40 lysis buffer overnight at 4°C. Glycerol (Sigma) was added to 15% (vol/vol), and aliquots were stored at −80°C. An RNA affinity column was utilized to purify the 105-kDa binding protein further. Biotin-16-UTP-containing wild-type 3′NCR RNA (200 μg; see above) was incubated for 15 min at 30°C with the dialyzed ammonium sulfate fraction (6 mg of protein total) in the presence of cross-linking buffer, adjusted to 0.23 M KCl, and supplemented with 200 U of RNasin (Promega Biotec), 1 mM PMSF (Gibco BRL), 1 μM pepstatin A (Sigma), and 1 μM leupeptin (Sigma). Next, 100 μl (packed bead volume) of streptavidin agarose (Gibco BRL) was added, and the samples were rotated in 1.5-ml Eppendorf tubes overnight at 4°C. The streptavidin agarose beads were washed twice with 1.0 ml of ice-cold SAA buffer (8.5 mM HEPES [pH 7.4], 3 mM MgCl2, 4% glycerol), and bound proteins were eluted sequentially with SAA buffer containing 0.3, 0.5, and 1.0 M KCl. Six micrograms of protein from each fraction was analyzed in SDS-containing 10% polyacrylamide gels followed by silver staining according to Blum et al. (9). Four micrograms of protein from each fraction was used in each cross-linking reaction. The KCl concentrations in cross-linking reactions were adjusted to 0.23 M, except for the reaction containing the 1.0 M eluate, which was adjusted to 0.5 M KCl.
A fraction of purified nucleolin was obtained from Nancy Maizels (Yale University). This sample of nucleolin was purified from human B-cell nuclei as a component of the B-cell-specific transcription factor LR1 after Mono Q and Mono S chromatography as described previously (25).
Amino acid sequence determination of the 105-kDa protein.
Forty-five micrograms of protein from the 0.5 M KCl fraction was separated on SDS-containing 7.5% polyacrylamide gels and transferred to a polyvinylidene difluoride (PVDF) membrane (BioRad) in transfer buffer (25 mM Tris-OH, 192 mM glycine, and 10% methanol) at 25 V for 17 h at 4°C as described previously (4). The PVDF membrane was stained with Ponceu S (Sigma) according to the manufacturer’s instructions, and the 105-kDa band was excised and transferred to an Eppendorf tube. After being rinsed five times with deionized distilled water at 25°C, the sample was stored at −80°C. Peptide sequence determination was performed at the Harvard microchemistry facility. The sequences of two peptides, generated by treatment of the sample with the endoproteinase LysC (Harvard microchemistry facility), are displayed in Table 1.
TABLE 1.
Partial amino acid sequences of two LysC-generated peptides derived from the 105-kDa RNA-binding protein from HeLa cells
| Peptide | Amino acid sequence | Corresponding amino acid position in human nucleolina |
|---|---|---|
| 1 | VEGTEPTTAFNLFVGNLNFNK | 298–318 |
| 2 | TLVLSNLSYSATEETLQEVFEK | 487–508 |
From reference 50.
Immunoprecipitation of cross-linked complexes.
Immunoprecipitation experiments were performed as described previously (47), with minor modifications. Cross-linking was performed as described above, using 2 × 105 cpm of 32P-labeled wild-type poliovirus 3′NCR that contained 4 thiouridine residues and 4 μg of protein from the 0.5 M KCl fraction obtained from the RNA affinity column. After RNase A digestion, monoclonal antibodies directed against human nucleolin (kindly provided by Ning-Hsing Yeh, National Yang-Ming University, Taiwan) or polyclonal antibodies recognizing poliovirus polypeptide 2A were added to the cross-linking reaction mixes and incubated on ice for 30 min with occasional mixing. A 50% slurry of protein A-Sepharose (PAS; Sigma) in IP buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% NP-40, and 1 mM EDTA [pH 8.0]) was added, and incubation proceeded for 15 min on ice with occasional mixing. The PAS was pelleted, and the supernatants were saved for analysis by SDS-PAGE. The PAS was washed two times with ice-cold IP buffer and resuspended in SDS loading buffer. Samples were then analyzed in SDS-containing 7.5% polyacrylamide gels.
Preparation of HeLa S10 extracts and de novo synthesis of poliovirus.
Extracts from HeLa S3 cells were prepared by the method of Barton et al. (7), with the following modifications. S10 extracts were prepared from 2.5 liters of HeLa S3 cells, grown to a density of 4 × 105 cells/ml. The first wash was carried out with 2.5 liters of isotonic buffer (35 mM HEPES-KOH [pH 7.4], 146 mM NaCl, 11 mM glucose). Nuclease treatment was performed with 30 U of micrococcal nuclease (Boehringer Mannheim)/ml of extract for 15 min at 20°C.
De novo synthesis was initiated with virion RNA as described by Barton et al. (6). Briefly, 15 μl of immunodepleted (see below) HeLa S10 extract, resuspended in 1× PB (20 mM HEPES-KOH [pH 7.4], 120 mM potassium acetate, 4 mM magnesium acetate, 5 mM DTT), was incubated with 1.0 μg of purified virion RNA (see below) in a final volume of 30 μl, with final concentrations of 17.75 mM HEPES-KOH (pH 7.4), 90 mM potassium acetate, 2.0 mM magnesium acetate, 2.6 mM DTT, 1.0 mM ATP, 0.25 mM GTP, 0.25 mM CTP, 0.25 mM UTP, 30 mM creatine phosphate, and 0.4 mg of creatine kinase/ml for 6 h at 34°C. It should be noted that the HeLa S10 extract constitutes 50% of the total reaction volume, and the endogenous concentration of salts, nucleoside triphosphates, and other components in the extract is unknown as pointed out by Barton et al. (6). After incubation, samples were treated with 0.8 μg of RNase A (Sigma) and 76 U of RNase T1 (Gibco BRL) for 20 min at 25°C. Three volumes of PBS was added, and the samples were assayed for PFU on HeLa cell monolayers (28).
Immunodepletion of HeLa S10 extracts.
Approximately 2 × 107 Dynabeads M-450 coated with sheep anti-mouse IgG (Dynal) were washed with 1.0 ml of PB buffer containing 5 mg of BSA (Sigma)/ml at 25°C and pelleted in a magnet (Dynal MPC). The supernatant was discarded, and 520 μg of a tissue culture supernatant containing monoclonal anti-nucleolin or monoclonal anti-fibrillarin antibodies (generous gift from K. Michael Pollard, Scripps Research Institute) in 1.0 ml of PB buffer was added and incubated for 20 h at 4°C. Next, the beads were pelleted and washed twice with 1.0 ml of PB buffer at 25°C. Thirty microliters of S10 extract was added to the pelleted beads and incubated for 30 min on ice with occasional mixing. The beads were pelleted, and the supernatant was incubated three more times with antibody-containing beads. These four-time-depleted extracts were the sources of the immunodepleted S10 extracts used in the cell-free de novo synthesis system.
Western blotting.
Western blotting was performed using the ECL detection kit (Amersham Life Science) according to the manufacturer’s recommendations, with some modifications. Briefly, 10 to 20 μg of extract was separated on a 7.5% SDS-containing polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane (Micron Separations Inc.) in 192 mM glycine, 25 mM Tris base, and 20% methanol for 1 h at 25°C. The membrane was blocked with 5% milk-PBS plus 0.1% Tween 20 for 20 h at 4°C. Primary antibody incubation was performed with 5 μg of polyclonal nucleolin antibody/ml in PBS for 1.5 h at 25°C. Secondary antibody incubation was done with a 1:5,000 dilution of anti-rabbit antibody conjugated to horseradish peroxidase (Cappel) in PBS for 2 h at 25°C. Next, the membrane was washed once in PBS–0.1% Tween 20 buffer for 15 min followed by four washes of 5 min each. The membrane was then treated with ECL reagents and subjected to autoradiography.
Immunofluorescence assays.
Immunofluorescence experiments were performed using HeLa cells as described previously (34), with the following modifications. Briefly, the cells were grown on glass coverslips to 50% confluence and infected with wild-type poliovirus at a multiplicity of infection of 50. At different times postinfection, the coverslips were washed twice with PBS at 37°C. For staining with anti-nucleolin antibody, anti-TATA-binding protein (TBP) antibody (provided by Judith Jaehning, University of Colorado Health Sciences Center) or anti-Sam68 antibody (Santa Cruz Technologies), cells were fixed by incubation in methanol-glacial acetic acid (3:1, Fisher Scientific) for 10 min at 25°C. For staining with anti-fibrillarin antibodies, cells were fixed by incubation in ice-cold acetone-methanol (3:1, Mallinckrodt) for 3 min at −20°C, followed by removal of the fixation solution and air drying for 5 min at 25°C. Fixed cells were washed three times with PBS for 5 min each at 25°C and incubated with primary antibodies either undiluted (anti-fibrillarin) or diluted in PBS–0.1% BSA (Sigma) at the following ratios: anti-nucleolin (1:5), anti-TBP (1:100), anti-Sam68 (1:1,000). Following incubation with primary antibodies at 4°C overnight, the cells were washed three times with PBS for 5 min each at 25°C. The secondary antibodies were diluted 1:100 in PBS–0.1% BSA; cells which had been stained with anti-nucleolin or anti-fibrillarin were incubated with fluorescein isothiocyanate-conjugated goat anti-mouse IgG (Caltag); cells which had been stained with anti-TBP or anti-Sam68 were incubated with Texas red-conjugated goat anti-rabbit IgG (Cappel). The secondary antibody was incubated for 1 h at 25°C. Next, the cells were washed three times with PBS for 5 min each at 25°C and mounted onto glass slides using Vectashield (Vector Laboratories). Cells were viewed and photographed using an Olympus BX60 system microscope.
RESULTS
Isolation and characterization of poliovirus genomes bearing mutations in the 3′NCR.
We reported previously the isolation and phenotypes of polioviruses carrying mutations in the 3′NCR (46). One of those, mutant 3NC202, contains an 8-nt insertion at nucleotide position 7387 and displays a temperature-sensitive phenotype for RNA synthesis (46). Subsequently, several phenotypic revertants of 3NC202 were isolated based on their abilities to grow at the nonpermissive temperature. Figure 1 shows that, in contrast to mutant 3NC202, the two revertants, R5 and R9, accumulated viral RNA at the nonpermissive temperature, although not as much as wild-type. However, both revertant viruses still displayed a delay in viral RNA accumulation compared to that of wild-type virus (Fig. 1).
The genotypes of the wild-type, mutant, and revertant genomes are listed in Fig. 2. By deleting 8 nt adjacent to the 3NC202 insertion, the R5 genome differs from the wild-type by only 2 nt (i.e., G7386→A7386 and U7387→C7387; Fig. 2). In contrast, R9 differs from the wild-type by 3 nt and the 3′NCR of R9 is one nucleotide longer than the 3′NCR of the wild-type (Fig. 2). Because of their different phenotypes in viral RNA synthesis, we decided to use wild-type, mutant, and revertant 3′NCRs as tools to search for specific RNA-protein complexes that may be involved in viral RNA synthesis.
FIG. 2.
Nucleotide sequences of wild-type and mutant 3′NCRs. Shown are the sequences from nt 7373 to the 3′ terminal polyadenosine sequences of wild-type, mutant (3NC202), and revertant (R5 and R9) viral RNA genomes. The translation termination codons are underlined. The mutant 3NC202 has an 8-nt insertion, and the revertants R5 and R9 have nucleotide changes relative to the wild-type poliovirus indicated in bold and overlined.
Interaction of host cell proteins with the poliovirus 3′NCR.
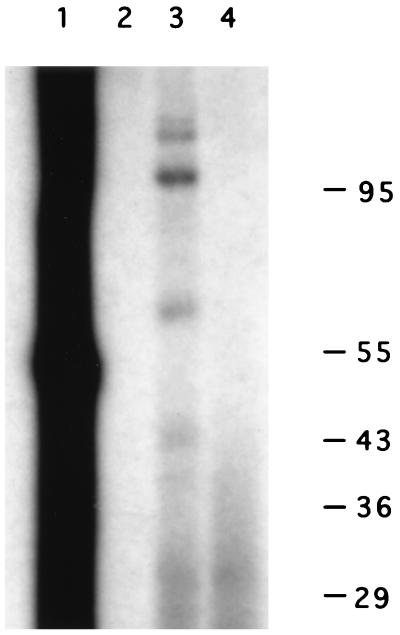
Radiolabeled, thiouridine-containing RNAs representing the 3′-terminal 108 nt of the wild-type viral genome were mixed with cytoplasmic extracts prepared from human HeLa cells. After irradiation at 312 nm and digestion with RNases, cross-linked samples were separated in SDS-polyacrylamide gels and visualized by autoradiography to identify any transfer of labeled nucleotides to protein. Figure 3 (lane 3) shows that several cellular factors which displayed apparent molecular weights of 120,000, 105,000, 68,000, and 45,000 could be cross-linked to the viral 3′NCR. Digestion of the reaction mixture with proteinase K abolished the appearance of the species (lane 4), confirming that they are proteins. A similar cross-linking pattern was observed with the extracts from infected HeLa cells (55).
FIG. 3.
Interactions of proteins with the poliovirus 3′NCR RNA. UV cross-linking of cellular proteins to the wild-type 3′NCR was performed as described in Materials and Methods. Lanes 1 and 2 contain samples of the radiolabeled RNA before and after treatment with RNase A, respectively. Lanes 3 and 4 contain nuclease-treated, cross-linked reaction mixtures without and with treatment with proteinase K, respectively. Numbers on the right indicate the migration of marker proteins of known molecular weights (in thousands). The autoradiograph was scanned using a Microtek ScanMaker E6, and the resulting image was labeled using Adobe Photoshop version 3.0.
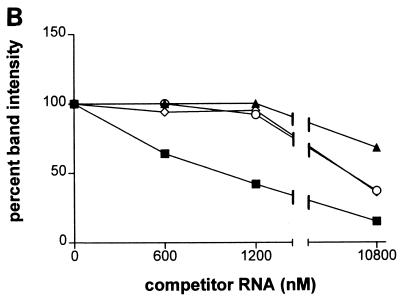
To examine the specificity of binding of these proteins to wild-type 3′NCR-containing RNAs, various amounts of wild-type, mutant, or revertant competitor 3′NCR RNAs were added to the incubations before cross-linking with UV light. The formation of 105-kDa-wild-type 3′NCR complexes was inhibited by approximately 40% in the presence of a 100-fold molar excess of unlabeled wild-type 3′NCR RNA in the binding reaction (Fig. 4A and B). In contrast, similar concentrations of mutant 3NC202 or revertant 3′NCRs did not significantly inhibit the formation of 105-kDa protein-RNA complexes under the same conditions. Formation of 105-kDa protein-3′NCR complexes could be inhibited by higher concentrations (875-fold molar excess) of either mutant or revertant 3′NCRs. At this concentration, the revertant 3′NCRs competed more efficiently than the mutant 3′NCR for binding of the 105-kDa protein to the wild-type 3′NCR (Fig. 4B). Also, the binding of the 68-kDa protein was more specific to wild-type than to mutant 3NC202 and revertant RNAs. Curiously, formation of 68-kDa protein-viral RNA complexes was dependent on the presence of 3′ terminal polyadenosine residues on the RNAs (55). Binding of the 120-kDa protein to wild-type RNA was not significantly affected in the presence of excess wild-type, mutant, or revertant RNAs; in contrast, binding of the 45-kDa protein to wild-type RNA was diminished in the presence of similar amounts of both mutant and revertant RNAs. Thus, the 120- and 45-kDa protein-wild-type 3′NCR complexes are likely to represent unspecific RNA-protein complexes.
FIG. 4.
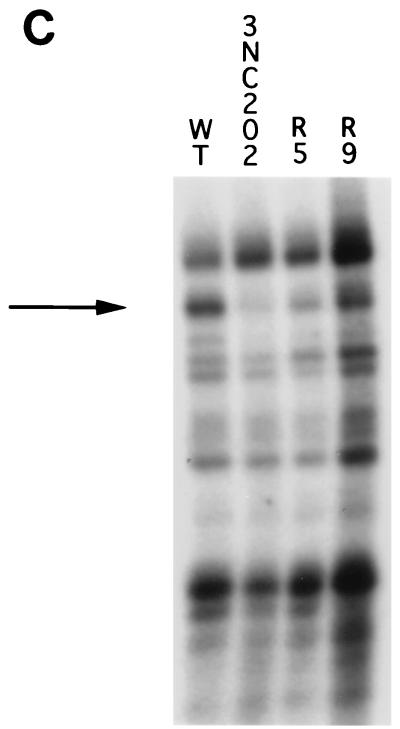
(A) Formation of wild-type-105-kDa complexes in the presence of wild-type, mutant, or revertant 3′NCR-containing competitor RNAs. UV cross-linking was performed as described in Materials and Methods, except that unlabeled wild-type (WT), mutant (3NC202), or revertant (R5, R9) RNAs were preincubated with the HeLa extract prior to the addition of radiolabeled wild-type RNA (12 nM). The triangles at the top represent the molar excess (50-, 100-, and 875-fold) of each unlabeled RNA over the radiolabeled RNA. The arrow identifies the 105-kDa RNA-binding protein. (B) Quantitation of the formation of 105-kDa protein-RNA complexes in the presence of competitor RNAs. The amount of UV-cross-linked complexes formed was set to 100% in the absence of competitor RNA. The x axis indicates the concentrations of wild-type (▪), mutant (▴), or revertant (R5, ○; R9, ◊) RNAs relative to that of radiolabeled wild-type RNA (12 nM). (C) Formation of wild-type-, mutant-, and revertant-105-kDa protein complexes. UV cross-linking was performed as described above with labeled wild-type (WT), mutant (3NC202), or revertant (R5, R9) RNAs. The arrow denotes the migration of the 105-kDa RNA binding activity. The autoradiographs in panels A and C were scanned using a Microtek ScanMaker E6, and the resulting images were labeled using Adobe Photoshop version 3.0.
Next, we determined the direct binding of host cell proteins to mutant and revertant RNA molecules. UV cross-linking of cytoplasmic extracts to radiolabeled mutant 3NC202 RNAs displayed a diminished binding particular to the 105-kDa protein compared to wild-type RNA (Fig. 4C). In contrast, the 105-kDa protein could be cross-linked with higher efficiencies to both R5 and R9 revertant RNAs than to 3NC202 mutant RNA; although not to the same level seen with wild-type RNA (Fig. 4C). Although UV cross-linking experiments are in general only semiquantitative, the binding of the 105- and 68-kDa proteins to the viral 3′NCRs correlated somewhat with the phenotypes of the revertant and mutant genomes in RNA synthesis (Fig. 1). Clearly, the in vivo phenotype in RNA synthesis of the 3NC202 virus is much more striking than the in vitro phenotype in nucleolin binding of the 3NC202 3′NCR. Nevertheless, because the 105-kDa protein could be cross-linked most efficiently, we first decided to determine the identity of the 105-kDa protein and to study its potential role in the viral infectious cycle.
Purification of the 105-kDa protein and its identification as nucleolin.
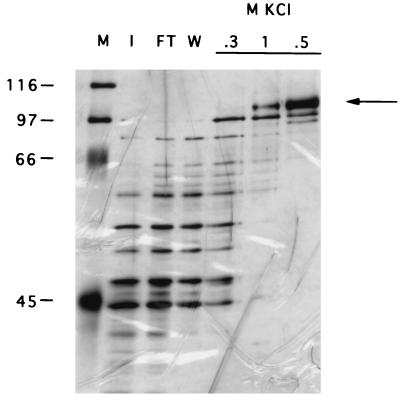
The 105-kDa protein was partially purified by ammonium sulfate precipitation and RNA affinity chromatography (see Materials and Methods). Figure 5 shows that a major 105-kDa protein could be eluted from the affinity matrix after treatment with 0.5 M KCl. In hopes that this protein was identical to the 105-kDa protein that could be cross-linked to wild-type 3′NCRs (Fig. 4), the 105-kDa band from the 0.5 M KCl fraction was excised from the gel and prepared for microsequencing (see Materials and Methods). Table 1 shows that the partial amino acid sequence of the two peptides matched, with 100% identity, the contiguous amino acid sequences present in human nucleolin (13, 50). Thus, the 105-kDa protein excised from the gel is closely related or identical to nucleolin.
FIG. 5.
Purification of the 105-kDa protein using affinity chromatography with viral 3′NCR RNA. A silver-stained SDS-polyacrylamide gel which displays various fractions of 105-kDa protein-containing extracts after RNA affinity chromatography is shown. Lanes: M, molecular weight markers; I, input fraction; FT, flowthrough fraction; W, wash fraction. Fractions after elution with 0.3, 0.5, and 1.0 M KCl are shown. The arrow identifies the 105-kDa protein that was prepared for microsequencing. The migration of marker proteins of known molecular weights (in thousands) is indicated at the left. Note that the 1 M eluate was erroneously loaded prior to the 0.5 M eluate. The autoradiograph was scanned using a Microtek ScanMaker E6, and the resulting image was labeled using Adobe Photoshop version 3.0.
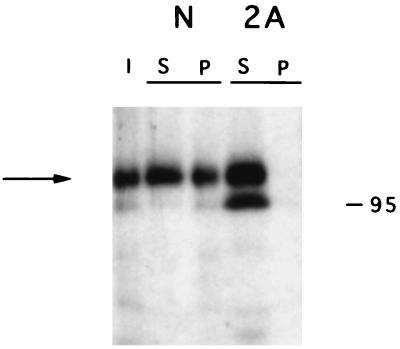
Two experiments were performed to test whether nucleolin was the 105-kDa poliovirus 3′NCR RNA-binding protein. First, the 0.5 M KCl-RNA affinity chromatography fraction was cross-linked to radiolabeled, thiouridine-containing viral 3′NCR sequences; the reaction was then immunoprecipitated with antibodies directed against nucleolin or poliovirus protein 2A. Figure 6 shows that antibodies directed against nucleolin could immunoprecipitate cross-linked RNA-protein complexes (lane N, fraction P), whereas antibodies directed against 2A did not (lane 2A, fraction P). In a second experiment, a sample containing purified nucleolin (see Materials and Methods) was mixed with radiolabeled, thiouridine-containing 3′NCRs in the absence or presence of unlabeled competitor 3′NCRs and irradiated at 312 nm. Figure 7 shows that the purified nucleolin could be cross-linked to the viral RNA (Fig. 7, lane 1). Curiously, the purified nucleolin sample migrated somewhat faster than the nucleolin present in the 0.5 M RNA affinity eluate (Fig. 7, lane 4). Nucleolin is known to be subjected to autoproteolysis (15, 20). Thus, the faster-migrating, purified nucleolin (i.e., 95-kDa nucleolin) is likely to be a product of this reaction. Formation of the 95-kDa nucleolin-RNA complexes, like the formation of the 105-kDa protein-RNA complexes from the 0.5 M KCl fraction (see above), could be inhibited by the addition of increasing amounts of unlabeled viral 3′NCR sequence elements (lanes 2 and 3). These data strongly suggest that the 105-kDa poliovirus 3′NCR-binding protein is authentic, unproteolyzed nucleolin.
FIG. 6.
Immunoprecipitation of cross-linked RNA-protein complexes with antinucleolin and anti-2A antibodies. The 0.5 M KCl fraction obtained after 3′NCR-RNA affinity chromatography was cross-linked to the wild-type poliovirus 3′NCR, and immunoprecipitation experiments were performed. Lane I, cross-linked RNA-protein complex before immunoprecipitation. The supernatant (S) and pellet (P) fractions following immunoprecipitation with antinucleolin (N) or anti-2A (2A) antibodies are displayed. The arrow indicates the migration of the 105-kDa protein-RNA. The migration of a marker protein of known molecular weight (in thousands) is indicated at the right. The autoradiograph was scanned using a Microtek ScanMaker E6, and the resulting image was labeled using Adobe Photoshop version 3.0.
FIG. 7.
Binding of purified nucleolin to the viral 3′NCR. Cross-linking was performed as described in Materials and Methods, except that various amounts of unlabeled wild-type 3′NCR was preincubated with purified nucleolin prior to the addition of radiolabeled wild-type RNA. Lanes 1, 2, and 3 contain 0, 10-, and 100-fold molar excess, respectively, of the unlabeled RNA over the radiolabeled RNA (12 nM). Lane 4 displays the pattern when the 0.5 M KCl fraction, obtained after 3′NCR RNA affinity chromatography, was cross-linked to the wild-type 3′NCR and displayed in the same gel. The arrow indicates the 105-kDa RNA binding activity. The autoradiograph was scanned using a Microtek ScanMaker E6, and the resulting image was labeled using Adobe Photoshop version 3.0.
Cell extracts, immunodepleted of nucleolin, display decreased virus production at early times of viral gene expression.
To examine whether the binding of nucleolin to the viral 3′NCR has a functional role in the viral infectious cycle, virus production was measured in a cell-free system which has been shown to de novo synthesize infectious poliovirions when programmed with genomic RNA (7, 36). First, it was confirmed by UV cross-linking that nucleolin bound to the 3′NCR in this cell-free system (data not shown). Next, the extracts were incubated in four successive rounds with monoclonal antibodies directed against either fibrillarin or nucleolin (see Materials and Methods). The depletion of nucleolin was then monitored by Western blotting. Figure 8 shows that nucleolin was greatly depleted from the extract after four rounds of incubation with the monoclonal antibody directed against nucleolin (lane 3). In contrast, incubation of the extract with the monoclonal antibodies directed against fibrillarin did not significantly remove nucleolin (lane 2). Next, the efficiencies of these depleted extracts for virion production were assayed. It was observed that both fibrillarin- and nucleolin-depleted extracts synthesized and assembled the same amount of infectious virus after programming the depleted extracts with virion RNA and incubating for 20 h (not shown). To test whether nucleolin might have an effect on virus production at an earlier time of incubation in this cell-free assay, virus yields from fibrillarin- and nucleolin-depleted extracts were measured after only 6 h of incubation. Table 2 shows that virus yield was repeatedly at least 10-fold reduced in nucleolin-depleted extracts compared to that of fibrillarin-depleted extracts. However, virus yield in the nucleolin-depleted extract was only reduced sixfold compared to that of the fibrillarin-depleted extract after a 10-h incubation (not shown). Thus, the absence of a significant amount of nucleolin diminished viral yield at early times in the cell-free assay, suggesting that nucleolin increases the efficiency of viral gene expression at times when viral RNA and viral proteins are limiting. However, the absence of nucleolin was overcome at later times in this cell-free assay. Whether nucleolin enhances the rate of viral mRNA translation or RNA replication or if it affects RNA stability is unclear at present.
FIG. 8.
Immunodepletion of HeLa S10 cell extracts. Extracts (lane 1) were subjected to four successive rounds of immunodepletion using monoclonal antibodies directed against fibrillarin (lane 2) or monoclonal antibodies directed against nucleolin (lane 3). The amount of nucleolin was visualized by Western blotting using an antibody directed against nucleolin, as described in Materials and Methods. The autoradiograph was scanned using a Microtek ScanMaker E6, and the resulting image was labeled using Adobe Photoshop version 3.0.
TABLE 2.
Virion production in immunodepleted HeLa S10 extracts after 6 h of incubation
| Expt | Virus yield (PFU/ml)
|
Fold difference | |
|---|---|---|---|
| Fibrillarin-depleted extracta | Nucleolin-depleted extracta | ||
| 1 | 3700 | 350 | 11 |
| 2 | 5900 | 300 | 20 |
| 3 | 650 | 50 | 13 |
Note that the activities of the depleted extracts in virion formation are reduced by 10- to 20-fold compared to those of nondepleted extracts.
So far, the addition of various nucleolin-containing fractions to nucleolin-depleted extracts failed to restore virion production (not shown). These results seem to indicate that either nucleolin in these purified fractions was inactive or that essential nucleolin-associated factors were removed from the extracts during immunodepletion. Of course, any of such nucleolin-associated factors could have been responsible for the observed reduction in virus yield in the nucleolin-depleted extracts.
Nucleocytoplasmic relocalization of nucleolin in poliovirus-infected cells.
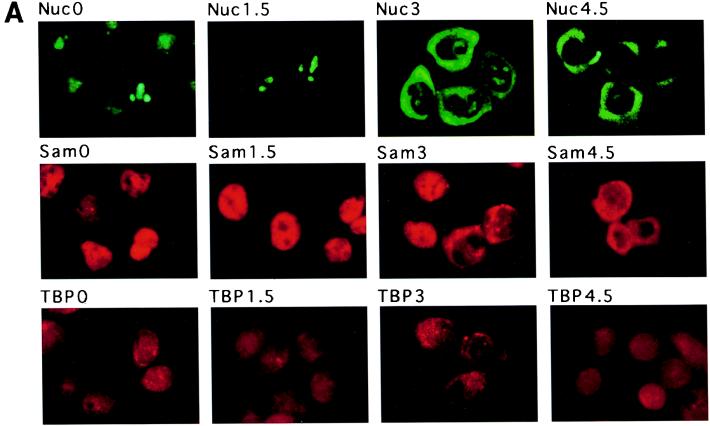
Nucleolin is a nucleolar protein that shuttles between the nucleolus and the cytoplasm. Its nucleolar functions include roles in the regulation of rDNA transcription and rRNA processing (19, 35). Because the poliovirus infectious cycle takes places in the cytoplasm, one would expect an accumulation of nucleolin in the cytoplasm of infected cells if the binding of nucleolin to the 3′NCR of the viral RNA has a significant role in the viral infectious cycle. We examined the intracellular distribution of nucleolin in uninfected and poliovirus-infected cells by immunofluorescence, using a monoclonal antibody directed against nucleolin. Cells showed punctate, predominantly nucleolar staining of nucleolin at the beginning of infection (Fig. 9A, panel Nuc0). However, a dramatic relocalization of nucleolin into the cytoplasm commenced at 3 h after infection (panel Nuc3). As a positive control, the relocalization of Sam68 (Src-associated in mitosis, 68-kDa) protein from the nucleus in mock-infected cells (panel Sam0) to the cytoplasm in infected cells (panels Sam3 and Sam4.5) was monitored (34). In contrast, the TATA-binding protein TBP localized to the nucleus in both mock-infected (TBP0) and infected cells (TBP4.5), as noted by McBride et al. (34).
FIG. 9.
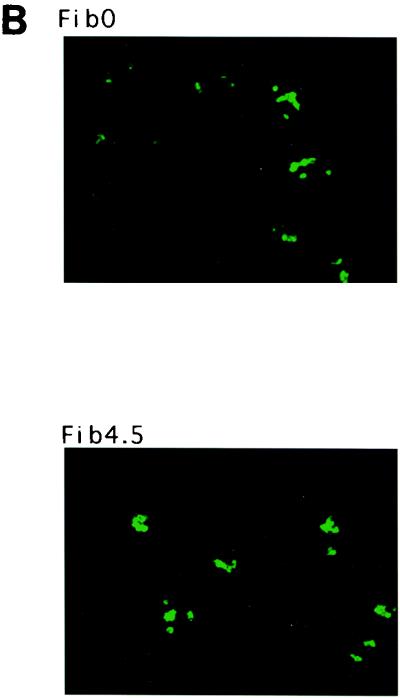
(A) Immunolocalization of nucleolin (Nuc), Sam68 (Sam), and TATA-binding protein (TBP) during poliovirus infection. Cells were stained with anti-nucleolin (Nuc), anti-Sam68 (Sam), or anti-TBP (TBP) at 0, 1.5, 3, and 4.5 h after infection with wild-type poliovirus. (B) Immunolocalization of fibrillarin during wild-type poliovirus infection. Cells were stained with anti-fibrillarin antibodies at 0 (Fib0) and 4.5 (Fib4.5) h after infection with wild-type poliovirus. Color slides were scanned using a Nikon 35-mm film scanner LS-1000, and the resulting image was labeled using Adobe Photoshop version 3.0.
To determine whether other nucleolar proteins relocated from the nucleolus to the cytoplasm during poliovirus infection, the intracellular distribution of the nucleolus-resident fibrillarin (3, 37) was monitored. Figure 9B shows that fibrillarin remained associated with the nucleolus during the infectious cycle (panel Fib4.5). Therefore, the relocalization of nucleolin was not due to an overall nucleolar-cytoplasmic redistribution of proteins.
Subcellular localization of nucleolin in cells during inhibition of protein synthesis.
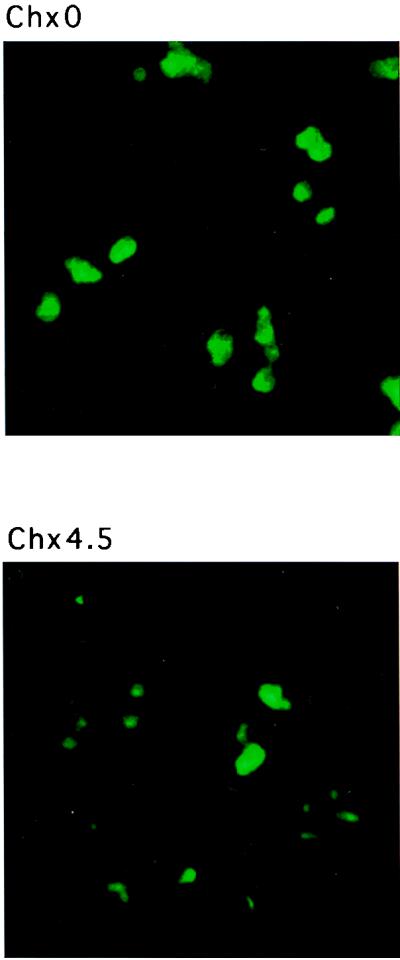
Infection of cells with poliovirus results in the selective inhibition of cellular cap-dependent protein synthesis (reviewed in references 18 and 24). Thus, it is possible that the inhibition of protein synthesis was primarily responsible for the nucleolar-cytoplasmic relocalization of nucleolin. To test whether this is the case, cells were treated with cycloheximide, which inhibits the elongation of polypeptide chains by inhibiting the peptidyltransferase activity of 60S ribosomal subunits. The relocalization of nucleolin in treated cells was then monitored by immunofluorescence analysis. Figure 10 shows that the localization of nucleolin remained exclusively nucleolar in both untreated (panel Chx0) and cycloheximide-treated (panel Chx4.5) cells. Thus, the nucleolar-cytoplasmic relocalization of nucleolin requires ongoing viral gene expression and is not simply a consequence of the inhibition of host cell translation.
FIG. 10.
Immunolocalization of nucleolin in cycloheximide-treated cells. Cells were stained with monoclonal antibodies directed against nucleolin after 0 (Chx0) or 4.5 (Chx4.5) h of incubation with cycloheximide (20 μg/ml). Color slides were scanned using a Nikon 35-mm film scanner LS-1000, and the resulting image was labeled using Adobe Photoshop version 3.0.
Subcellular localization of nucleolin in cells during inhibition of host cell transcription.
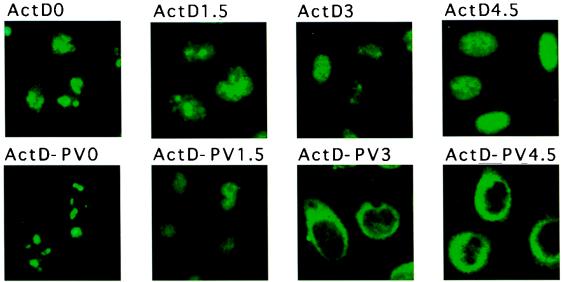
It is known that infection by poliovirus induces the inhibition of cellular RNA synthesis (reviewed in reference 48). Because the activities of all three RNA polymerases are inhibited in infected cells, the overall cessation of RNA synthesis could lead to release of nucleolin into the cytoplasm. Thus, the outcome of inhibition of DNA-dependent RNA synthesis on the subcellular localization of nucleolin was examined in cells treated with actinomycin D. Figure 11 shows that a nucleolar-nuclear redistribution of nucleolin could be observed at 1.5 h after inhibition of transcription (compare panel ActD0 with panel ActD1.5). Nucleolin resided almost entirely in the nucleus at 4.5 h after transcriptional inhibition (panel ActD4.5) with concomitant loss of the nucleoli, likely due to the inhibition of RNA polymerase I activity. However, nucleolin did not relocalize to the cytoplasm under these conditions. Because the viral RNA-dependent RNA polymerase is insensitive to actinomycin D, we could test whether infection of actinomycin D-treated cells with poliovirus induces a nucleolar-nuclear redistribution of nucleolin. As was observed in uninfected actinomycin D-treated cells (panel ActD1.5), infection of actinomycin D-treated cells resulted in a nucleolar-nuclear relocalization of nucleolin (panel ActD-PV1.5). By 3 h after infection, however, nucleolin further relocalized from the nucleus into the cytoplasm in infected cells (panel ActD-PV3). These findings indicate that inhibition of transcription results in a redistribution of nucleolin from the nucleolus to the nucleus. However, the nuclear-cytoplasmic redistribution of nucleolin requires infection of the cells by poliovirus. Because a nuclear accumulation of nucleolin was not observed in cells infected with poliovirus in the absence of actinomycin D (Fig. 9A), it is unlikely that the virus-induced inhibition of cellular transcription plays a role in the nucleolar-cytoplasmic relocalization of nucleolin in infected cells.
FIG. 11.
Immunolocalization of nucleolin in uninfected (ActD; top panels) and poliovirus-infected (ActD-PV; bottom panels) cells which were treated with actinomycin D (5 μg/ml). Cells were stained with monoclonal antibodies directed against nucleolin after 0, 1.5, 3, or 4.5 h, as indicated. Color slides were scanned using a Nikon 35-mm film scanner LS-1000, and the resulting image was labeled using Adobe Photoshop version 3.0.
DISCUSSION
We have identified an interaction of human nucleolin with RNA sequences derived from the 3′NCR of the wild-type poliovirus RNA genome. Although the exact cellular functions of nucleolin are not known, its peculiar domain structure has suggested that nucleolin is likely to be a multifunctional protein (31). The amino-terminal third of nucleolin contains repeated TPXKK motifs, predicted to be sites of phosphorylation by p34cdc2 (8) and CK2 protein kinases (32). The central domain of the protein contains four RNA recognition motifs (RRM) (12), each 80 to 100 amino acids in length, often found in proteins that interact with RNA (14). The carboxy-terminal domain of nucleolin and several other RRM-containing proteins is unusually rich in glycine, arginine, and phenylalanine residues, the so-called GAR (or RGG) domain. Curiously, a 10-kDa GAR-containing fragment of nucleolin interacts nonspecifically with RNA and can destabilize RNA helices (22), supposedly by an associated RNA helicase activity (54). These biochemical properties have implicated a role for nucleolin in ribosomal DNA transcription and in the formation and processing of pre-rRNA molecules in the nucleolus. Thus, the interaction of nucleolin with a cytoplasmically located viral RNA was surprising.
What determines the specificity of interaction of nucleolin with the viral 3′NCR?
Nucleolin binds pre-rRNAs at multiple sites which contain short stem-loop structures called nucleolin recognition elements or NREs (49). Nucleolin interacts with an NRE through a highly conserved UCCCGA motif, located in the loop of the hairpin (23). Mutations that destroy any of the cytosine residues in the binding motif greatly reduce the binding of nucleolin to the NRE (23). Inspection of the poliovirus 3′NCR sequences which contain the nucleolin-binding sites fails to reveal a UCCCGA motif. Thus, nucleolin must contact sequences in the viral 3′NCR which differ from the canonical NRE. Interestingly, Zaidi and Malter described the specific interaction of nucleolin cleavage products with a conserved 29-nt sequence present in the 3′NCR of amyloid precursor protein (APP) mRNA (59, 60); it was postulated that the interaction of nucleolin with the 3′NCR of APP mRNA resulted in an increase in mRNA stability (58). The nucleolin-binding site in APP mRNA contains the sequence CAUUUUGGU (58). A very similar sequence (CAUUUUAGU; nt 7365 to 7373) can be found at the very 3′ end of the coding region of the poliovirus genome. This sequence encompasses the terminal codons of the viral polymerase and the stop codon. Curiously, this sequence is located in a loop of a predicted pseudoknot structure whose stability was proposed to be affected in mutant 3NC202 and revertant genomes (28). While the exact binding site of nucleolin in the viral 3′NCR is not yet known, it is interesting to note that two different structural models of the viral 3′NCR predict that the viral nt 7365 to 7371 sequence element either resides in a loop of a predicted pseudoknot, as mentioned above, or is base paired with the terminal polyadenosine sequences (40). It is of course possible that these two alternative structures both exist in an equilibrium that could be affected by sequence- and structure-specific RNA-binding proteins. Binding of nucleolin to the 3′-terminal sequences of the viral genome bearing defined mutations should identify the structural features that govern the formation of nucleolin-3′NCR complexes.
Functional role of nucleolin or nucleolin-containing complexes in viral gene expression.
Immunodepletion of nucleolin from cell extracts capable of de novo synthesis of poliovirions (6, 36) diminished the efficiency with which virions were synthesized when assayed at early times of incubations. Interestingly, the presence of nucleolin was not required when virion production was assayed at late times of incubation in the cell extract. That the effect of nucleolin on viral gene expression was seen only at early times suggests that nucleolin may aid in the assembly of complexes that are needed for RNA replication or packaging when viral RNA or protein is limiting. For example, nucleolin could act as an RNA chaperone that could increase the steady-state level of replication-competent RNA molecules. Later in infection, when substantial amounts of RNAs have been synthesized, the level of replication-competent RNAs may not be rate limiting. Preliminary experiments, in which the levels of radiolabeled, newly synthesized viral RNA were monitored, have indicated that less RNA was synthesized in nucleolin-depleted extracts than in fibrillarin-depleted extracts (55).
Although an in vivo function for nucleolin in the viral life cycle remains to be elucidated, there are two putative roles that nucleolin could fulfill at early stages in the viral life cycle. It is known that infectious viral cycles can be initiated by viral positive-stranded RNAs in enucleated cells (16). However, enucleated cells produce approximately 10-fold less virions than do nucleated cells when infection is initiated with positive-stranded RNA molecules (16). This finding implies that nuclear functions may be needed to enhance virion production. This notion is further supported by the observation that the double-stranded replicative form of the viral RNA is unable to initiate an infectious cycle in enucleated cells in contrast to single-stranded virion RNA (16). If the RNA helicase activity of nucleolin is needed to unwind double-stranded RNA molecules, a decrease in the steady-state level of nucleolin in the cytoplasm of enucleated cells may explain the poor infectivity of both single- and double-stranded viral RNA. Second, nucleolin could function as an RNA chaperone that is needed to speed up the attainment of structures that make the viral RNA replication competent. That RNA chaperones can lead to accelerated attainment of thermodynamically favored RNA structures has been shown in hammerhead ribozyme catalysis (27).
Nucleolar-cytoplasmic relocalization of nucleolin in infected cells.
Nucleolin is known to shuttle between the nucleolus and the cytoplasm (11). Several scenarios could explain why nucleolin is relocated to the cytoplasm in poliovirus-infected cells. One could envisage that the inhibition of several host macromolecular pathways in infected cells results in the relocalization of nucleolin but curiously not of fibrillarin. However, this scenario is less likely because the addition of actinomycin D and cycloheximide, inhibitors of host cell transcription and translation, respectively, did not induce nucleolar-cytoplasmic relocalization of nucleolin. Another possibility is that the nuclear import pathway is inhibited by poliovirus infection. As a consequence, nucleolin would be unable to reenter the nucleus in infected cells. There are at least three distinct nuclear import pathways (10, 57). Nucleolin is thought to share the karyopherin β-dependent pathway used by mRNA-binding protein A1 (57). Whether karyopherin β-dependent pathways are operational in poliovirus-infected cells remains to be tested. It is also possible that nucleolin becomes trapped in the cytoplasm by virus-induced ligands which could be viral proteins, modified cellular proteins, or, most likely, the viral RNA itself. Finally, the interaction of nucleolin with pre-rRNA in the nucleolus could be inhibited in infected cells. Nucleolin would then relocalize into the cytoplasm due to loss of affinity for its nucleolar ligand. Using poliovirus to distinguish between these possibilities will provide a novel experimental means of studying the mechanism of intracellular distribution of host cell proteins.
ACKNOWLEDGMENTS
We thank Gregg Johannes and Marshall Kosovsky for insightful discussions during the course of this work. We thank Karla Kirkegaard for valuable discussions and critical reading of the manuscript. We are grateful to Laurie Dempsey and Nancy Maizels (Yale University) for a sample of purified nucleolin, Ning-Hsing Yeh (National Yang-Ming University, Taiwan), Judith Jaehning (University of Colorado Health Sciences Center) and K. Michael Pollard (Scripps Research Institute) for antibodies, and Anne McBride (Stanford University) for help with immunofluorescence.
This work was supported by grants AI25105 and T32 NS07321 from the National Institutes of Health. P.S. acknowledges the receipt of a Faculty Research Award from the American Cancer Society.
REFERENCES
- 1.Andino R, Rieckhof G E, Achacosco P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 3.Aris J P, Blobel G. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc Natl Acad Sci USA. 1991;88:931–935. doi: 10.1073/pnas.88.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates, Inc., John Wiley and Sons, Inc.; 1989. [Google Scholar]
- 5.Banerjee R, Echeverri A, Dasgupta A. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J Virol. 1997;71:9570–9578. doi: 10.1128/jvi.71.12.9570-9578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton D J, Black E P, Flanegan J B. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J Virol. 1995;69:5516–5527. doi: 10.1128/jvi.69.9.5516-5527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton D J, Morasco B J, Flanegan J B. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 1996;275:35–57. doi: 10.1016/s0076-6879(96)75005-x. [DOI] [PubMed] [Google Scholar]
- 8.Belenguer P, Caizergues-Ferrer M, Labbe J C, Doree M, Amalric F. Mitosis-specific phosphorylation of nucleolin by p34cdc2 protein kinase. Mol Cell Biol. 1990;10:3607–3618. doi: 10.1128/mcb.10.7.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 10.Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borer R A, Lehner C F, Eppenberger H M, Nigg E A. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 12.Bugler B, Bourbon H, Lapeyre B, Wallace M O, Chang J H, Amalric F, Olson M O. RNA binding fragments from nucleolin contain the ribonucleoprotein consensus sequence. J Biol Chem. 1987;262:10922–10925. [PubMed] [Google Scholar]
- 13.Bugler B, Caizergues-Ferrer M, Bouche G, Bourbon H, Amalric F. Detection and localization of a class of proteins immunologically related to a 100-kDa nucleolar protein. Eur J Biochem. 1982;128:475–480. doi: 10.1111/j.1432-1033.1982.tb06989.x. [DOI] [PubMed] [Google Scholar]
- 14.Burd C, Dreyfuss G. Conserved structures and diversity of function of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 15.Chen C M, Chiang S Y, Yeh N H. Increased stability of nucleolin in proliferating cells by inhibition of self-cleaving activity. J Biol Chem. 1991;266:7754–7758. [PubMed] [Google Scholar]
- 16.Detjen B M, Lucas J, Wimmer E. Poliovirus single-stranded RNA and double-stranded RNA: differential infectivity in enucleate cells. J Virol. 1978;27:582–586. doi: 10.1128/jvi.27.3.582-586.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deutscher M P. Maintaining protein stability. Methods Enzymol. 1990;182:83–89. doi: 10.1016/0076-6879(90)82010-y. [DOI] [PubMed] [Google Scholar]
- 18.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–573. [Google Scholar]
- 19.Escande-Geraud M L, Azum M C, Tichadou J L, Gas N. Correlation between rDNA transcription and distribution of a 100 kD nucleolar protein in CHO cells. Exp Cell Res. 1985;161:353–363. doi: 10.1016/0014-4827(85)90092-8. [DOI] [PubMed] [Google Scholar]
- 20.Fang S H, Yeh N H. The self-cleaving activity of nucleolin determines its molecular dynamics in relation to cell proliferation. Exp Cell Res. 1993;208:48–53. doi: 10.1006/excr.1993.1221. [DOI] [PubMed] [Google Scholar]
- 21.Gamarnik A V, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 22.Ghisolfi L, Joseph G, Amalric F, Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J Biol Chem. 1992;267:2955–2959. [PubMed] [Google Scholar]
- 23.Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J Mol Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- 24.Haller A A, Semler B L. Translation and host cell shutoff. In: Rotbart H A, editor. Human enterovirus infections. Washington, D.C: American Society for Microbiology; 1995. pp. 113–133. [Google Scholar]
- 25.Hanakahi L A, Dempsey L A, Li M J, Maizels N. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc Natl Acad Sci USA. 1997;94:3605–3610. doi: 10.1073/pnas.94.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 27.Herschlag D, Khosla M, Tsuchihashi Z, Karpel R L. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. . (Erratum, 13:3926.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobson S J, Konings D A M, Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3′ terminus of the poliovirus RNA genome and its role in viral RNA amplification. J Virol. 1993;67:2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson K L, Sarnow P. Viral RNA synthesis. In: Rotbart H A, editor. Human enterovirus infections. Washington, D.C: American Society for Microbiology; 1995. pp. 95–112. [Google Scholar]
- 30.Kitamura N, Semler B L, Rothberg P G, Larsen G R, Adler C J, Dorner A J, Emini E A, Hanecak R, Lee J J, van der Werf S, Anderson C W, Wimmer E. Primary structure, gene organization, and polypeptide expression of poliovirus RNA. Nature. 1981;291:547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- 31.Lapeyre B, Bourbon H, Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Acad Sci USA. 1987;84:1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li D, Dobrowolska G, Krebs E G. The physical association of casein kinase 2 with nucleolin. J Biol Chem. 1996;271:15662–15668. doi: 10.1074/jbc.271.26.15662. [DOI] [PubMed] [Google Scholar]
- 33.McBratney S, Sarnow P. Evidence for involvement of trans-acting factors in selection of the AUG start codon during eukaryotic translational initiation. Mol Cell Biol. 1996;16:3523–3543. doi: 10.1128/mcb.16.7.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McBride A E, Schlegel A, Kirkegaard K. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc Natl Acad Sci USA. 1996;93:2296–2301. doi: 10.1073/pnas.93.6.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medina F J, Cerdido A, Fernandez-Gomez M E. Components of the nucleolar processing complex (pre-rRNA, fibrillarin, and nucleolin) colocalize during mitosis and are incorporated to daughter cell nucleoli. Exp Cell Res. 1995;221:111–125. doi: 10.1006/excr.1995.1358. [DOI] [PubMed] [Google Scholar]
- 36.Molla A, Paul A V, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 37.Ochs R L, Lischwe M A, Sphon W H, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54:123–134. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 38.Parsley T B, Towner J S, Blyn L B, Ehrenfeld E, Semler B L. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 39.Pilipenko E V, Maslova S V, Sinyakov A N, Agol V I. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 1992;20:1739–1745. doi: 10.1093/nar/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilipenko E V, Poperechny K V, Maslova S V, Melchers W J G, Slot H J B, Agol V I. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (“kissing”) interactions. EMBO J. 1997;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- 41.Racaniello V R, Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci USA. 1981;78:4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roehl H H, Parsley T B, Ho T V, Semler B L. Processing of a cellular polypeptide by 3CD proteinase is required for poliovirus ribonucleoprotein complex formation. J Virol. 1997;71:578–585. doi: 10.1128/jvi.71.1.578-585.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roehl H H, Semler B L. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand RNA. J Virol. 1995;69:2954–2961. doi: 10.1128/jvi.69.5.2954-2961.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rueckert R R. Picornaviridae and their replication. In: Fields B N, et al., editors. Virology. 2nded. Vol. 1. New York, N.Y: Raven Press; 1990. pp. 507–548. [Google Scholar]
- 45.Sarnow P. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J Virol. 1989;63:467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarnow P, Bernstein H D, Baltimore D. A poliovirus temperature-sensitive RNA synthesis mutant located in a noncoding region of the genome. Proc Natl Acad Sci USA. 1986;83:571–575. doi: 10.1073/pnas.83.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 48.Schlegel A, Kirkegaard K. Cell biology of enterovirus infection. In: Rotbart H A, editor. Human enterovirus infections. Washington, D.C: American Society for Microbiology; 1995. pp. 135–153. [Google Scholar]
- 49.Serin G, Joseph G, Faucher C, Ghisolfi L, Bouche G, Amalric F, Bouvet P. Localization of nucleolin binding sites on human and mouse pre-ribosomal RNA. Biochimie. 1996;78:530–538. doi: 10.1016/0300-9084(96)84759-6. [DOI] [PubMed] [Google Scholar]
- 50.Srivastava M, Fleming P J, Pollard H B, Burns A L. Cloning and sequencing of the human nucleolin cDNA. FEBS Lett. 1989;250:99–105. doi: 10.1016/0014-5793(89)80692-1. [DOI] [PubMed] [Google Scholar]
- 51.Stade K, Rinke-Appel J, Brimacombe R. Site-directed cross-linking of mRNA analogues to the Escherichia coli ribosome; identification of 30S ribosomal components that can be cross-linked to the mRNA at various points 5′ with respect to the decoding site. Nucleic Acids Res. 1989;23:9889–9908. doi: 10.1093/nar/17.23.9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Todd S, Towner J S, Brown D M, Semler B L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toyoda H, Kohara M, Kataoka Y, Suganuma T, Omata T, Imura N, Nomoto A. Complete nucleotide sequences of all three poliovirus serotype genomes: implication for genetic relationship, gene function and antigenic determinants. J Mol Biol. 1984;174:561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- 54.Tuteja N, Huang N W, Skopac D, Tuteja R, Hrvatic S, Zhang J, Pongor S, Joseph G, Faucher C, Amalric F, Falaschi A. Human DNA helicase IV is nucleolin, an RNA helicase modulated by phosphorylation. Gene. 1995;160:143–148. doi: 10.1016/0378-1119(95)00207-m. [DOI] [PubMed] [Google Scholar]
- 55.Waggoner, S., and P. Sarnow. Unpublished observation.
- 56.Wimmer E, Hellen C U T, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 57.Yaseen N R, Blobel G. Cloning and characterization of human karyopherin beta 3. Proc Natl Acad Sci USA. 1997;94:4451–4456. doi: 10.1073/pnas.94.9.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaidi S H, Denman R, Malter J S. Multiple proteins interact at a unique cis-element in the 3′-untranslated region of amyloid precursor protein mRNA. J Biol Chem. 1994;269:24000–24006. [PubMed] [Google Scholar]
- 59.Zaidi S H, Malter J S. Amyloid precursor protein mRNA stability is controlled by a 29-base element in the 3′-untranslated region. J Biol Chem. 1994;269:24007–24013. [PubMed] [Google Scholar]
- 60.Zaidi S H, Malter J S. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyloid protein precursor mRNA. J Biol Chem. 1995;270:17292–17298. doi: 10.1074/jbc.270.29.17292. [DOI] [PubMed] [Google Scholar]