Abstract
Appendiceal Adenocarcinoma (AA) is unique from other gastrointestinal malignancies in that it almost exclusively metastasizes to the peritoneal cavity. However, few studies have investigated the molecular interaction of the peritoneal microenvironment and AA. Here, we use a multi-omics approach with orthotopic and flank-implanted patient-derived xenografts (PDXs) to study the effect of the peritoneal microenvironment on AA.
AA tumors implanted in the peritoneal microenvironment tended to grow faster and displayed greater nuclear expression of KI-67 relative to the same tumors implanted in the flank. Comparing the tumor-specific transcriptome (excluding stromal transcription), the peritoneal microenvironment relatively upregulated genes related to proliferation, including MKI67 and EXO1. Peritoneal tumors were also enriched for proliferative genesets, including E2F and Myc Targets. Proteomic studies found a 2.5-fold increased ratio of active-to-inactive YAP in peritoneal tumors, indicating downregulation of Hippo signaling.
Introduction
Appendiceal adenocarcinoma (AA) is a rare cancer, with an age-adjusted incidence rate of 1.3 (per 100,000 persons) (1). AA is usually discovered in one of two ways: incidentally during pathological review of appendectomy samples or only after the tumor has spread to the peritoneal space, leading to peritoneal carcinomatosis (PC). AA is initially managed using cytoreductive surgery and hyperthermic intraperitoneal chemotherapy (HIPEC) (2). Upon progression, AA responds poorly to systemic chemotherapy (3). Diffuse peritoneal carcinomatosis—through bowel obstruction and cancer cachexia—is the primary cause of mortality (4).
Despite clear clinical and molecular differences, AA is often treated similarly to colorectal cancer (CRC) (5). AA generally has a more indolent natural history compared to CRC. Additionally, AA is usually not diagnosed via colonoscopies since the appendix is not easily visualized and AA itself rarely invades into the cecum. Relative to CRC, AA has more GNAS mutations and fewer APC and TP53 mutations (6).
Importantly, unlike other gastrointestinal malignancies such as CRC, non-peritoneal metastases of AA are rare. So, it is difficult to comparatively study the effect of the peritoneal microenvironment using patient samples of AA. Anatomically, cancer cells from appendiceal tumors can directly enter the peritoneal space once they have invaded through the full thickness of the bowel wall (T4). However, the degree to which interaction between tumor cells and the Tumor Microenvironment (TME) in the peritoneal cavity promotes the proliferation of AA is limited. Here we bypass the constraints imposed by the natural history of AA by taking the same human tumor and implanting it in both peritoneum and flank of mice to generate PDXs with similar tumor components but different TME. It is hoped that a better understanding of how AA interacts with the TME will identify potential therapeutic targets in this orphan disease.
In this study, we implanted three different AA tumors into both the peritoneal cavity and the flank of immunocompromised NSG mice and used growth rate measurements, bulk RNA-Sequencing, Immunohistochemistry (IHC), and protein expression quantification using Reverse Phase Protein Arrays (RPPAs) to comparatively study the effect of the peritoneal microenvironment on AA.
Methods
Establishment of PDX models, PDX Tumor Growth Measurement, and PDX Tumor Collection
AA tumors from different sources (Jackson Laboratory for TM00351, Dr. Kjersti Flatmark [University of Oslo] for PMCA-3 (7,8), and the MD Anderson Cancer Center [MDACC] for AAP16) were implanted into female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice in the peritoneal cavity or flank. Details regarding the three tumors are located in Table S1. The tumors were visualized throughout the entire region of the peritoneal cavity in each mouse using a 4.7T small animal MRI system (Bruker Biospin MRI, Billerica, MA, USA) using 40 sections of 0.75 mm thickness with a 0.25 mm gap between sections. TM00351 and AAP16 growth were evaluated by tumor size (mm) while PMCA-3 growth was evaluated using mass due to its mucinous nature (g). Tumors were collected for histology, immunohistochemistry, RNA-Seq, and reverse phase protein arrays (RPPA). See Supplemental Methods.
Immunohistochemistry
Immunohistochemistry was conducted using Ku80, Vimentin, CDX2, Ki-67, and MUC1; individual stained slices of Ku80, Vimentin, and CDX2 were computationally merged. Antibody information is included in Table S2. See Supplemental Methods.
RNA-Seq and Transcriptome Cluster Analysis
Count matrices were normalized using DESeq2’s variance-stabilizing transform before cluster analysis. All heatmaps were made using the R Package pheatmap. All heatmap clustering was conducted using the ward.D method with Euclidean distance measurements. Principal component analysis was conducted using prcomp or DESeq2’s application of prcomp (9).
Estimation of Stromal and Immune Cells in Malignant Tumor Tissues Using Expression Data (ESTIMATE) Stromal Score
Counts were first normalized using DESeq2’s normalization algorithm. The normalized count matrix was then input into the R package hacksig (10) to implement the ESTIMATE algorithm to calculate the stromal score (11).
Differential Gene Expression Analysis (DGEA)
All DGEAs were conducted using the R Package DESeq2, which normalizes counts and then calculates the log2foldchange (LFC) for each gene using a generalized linear model (9). DESeq2 filters outliers and low-read genes. In all combined DGEAs run, the specific tumor and number of samples for each tumor was controlled for. LFC values were shrunken using the apeglm algorithm to prevent the outsize effect of small count numbers on the LFC (12).
Gene Ontology Overrepresentation Analysis (ORA)
All ORAs were conducted using ClueGO (v2.5.9) (13), a Cytoscape (v3.9.1) (14) plug-in that clusters significantly enriched gene terms based on their similarity, as measured by their kappa score. Two separate networks were generated from genes identified in the DGEA to be significantly upregulated in peritoneal PDXs and genes significantly upregulated in flank PDXs. See Supplemental Methods.
Gene Set Enrichment Analysis (GSEA)
All genes output from DGEA were ranked by their pi-value: (15). The ranked list was then used for pre-ranked GSEA analysis (16) using the Hallmark Genesets (h.all.v2023.1.Hs.symbols) from the MSigDB Collections database (17). An additional gene set was created from 351 genes elevated in intestinal goblet cells and added to the GSEA (data available from v23.0.proteinatlas.org) (18). Broad Institute’s standalone GSEA (version 4.2.3) software was used to perform enrichment analysis (http://www.gsea-msigdb.org/gsea/index.jsp) with 1000 permutations, a weighted enrichment statistic, excluding gene sets with less than 15 or greater than 500 genes, and collapsing gene symbols. Leading edge analysis was conducted using the same GSEA standalone software. Only leading-edge genes that passed a significance cutoff (|pi-value|≥0.5) are presented.
RPPA and Differential Protein Expression Analysis (DPEA)
DPEA was conducted on the linearized EC50 values from RPPA using the limma-voom pipeline from the R Package edgeR(19–21). The specific tumor and batch effects were controlled for.
Statistical Analyses
All statistical analyses were performed in RStudio 2022.07.01 using R v4.2.2. Results were considered significant when the multiple hypothesis corrected p-value ≤ 0.05 for all analyses except GSEA, where results were considered significant when FDR.q.val ≤ 0.25, as recommended by mSigDB (https://www.gsea-msigdb.org/gsea/doc/GSEAUserGuideFrame.html).
Data and Code Availability
All data and associated R code are available in the linked CodeOcean capsule (Capsule ID: 7745616).
Results
PDX models of Appendiceal Adenocarcinoma show a separation of human tumor and mouse TME
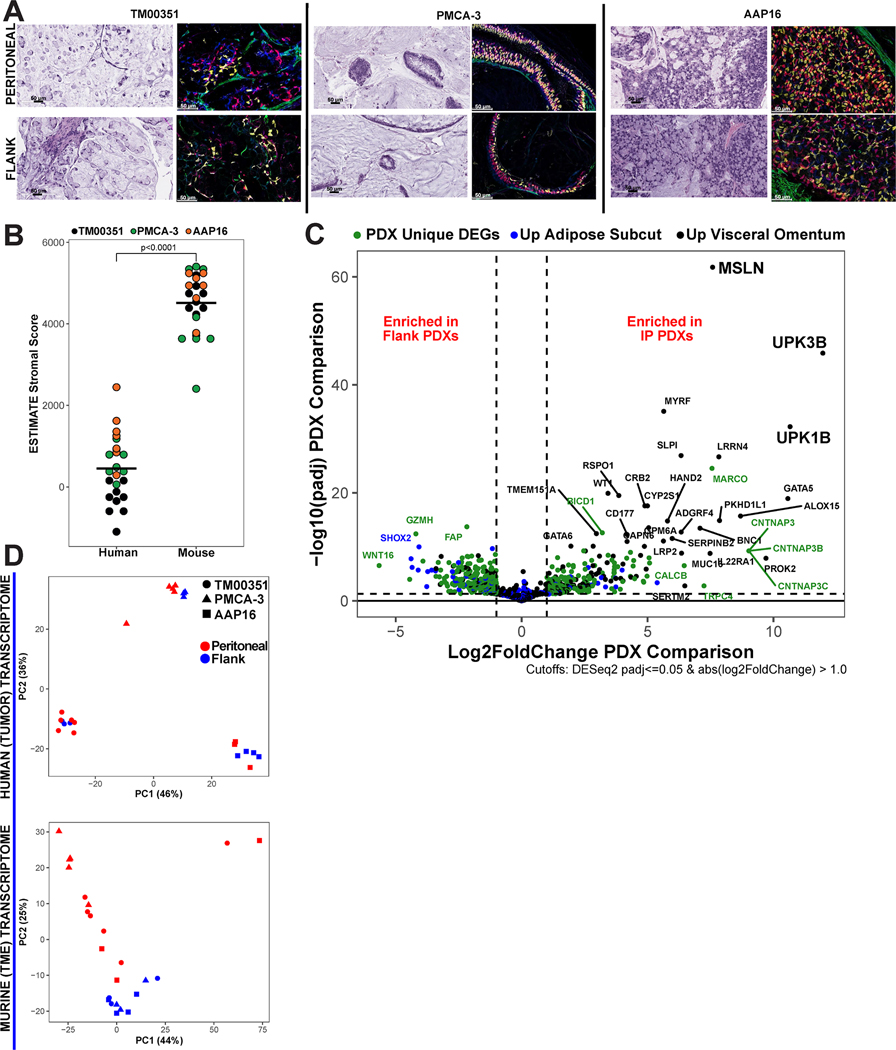
When human tumors are implanted into mice and passaged to form PDXs, the human TME cells are progressively replaced by host cells(22,23). To evaluate the relative contribution of human and mouse cells in our PDX models, we performed differential staining of the tumor samples using Ku80, a universal marker for human cells; CDX2, a marker for gastrointestinal (GI) epithelial tissue; and Vimentin, a marker for stromal tissue (Figures 1a, S1, Table S2). The Ku80 and CDX2-stained cells were highly overlapping and there was minimal overlap of Vimentin and Ku80 consistent with a clear separation of human tumor cells and murine stromal cells. Evaluating the architecture of the three PDX models, PMCA-3 showed less cellularity, more mucin, and greater differentiation (lower grade) with tumor cells still organized into thin layers, relative to TM00351 and AAP16, likely attributed to its GNASR201C mutation status (24).
Figure 1. AA PDX models are comprised of human tumor and mouse stroma.
(A) Computationally merged 40x magnification immunohistochemistry (IHC) staining of peritoneal and flank-implanted tumors using human marker KU80 (red), GI epithelial marker CDX2 (gold), and stromal marker Vimentin (green). Overlap of KU80 and CDX2 is orange. Representative H&E images are also shown. (B) ESTIMATE Stromal Score for each sample’s transcriptome split by assignment to either human reads or mouse reads, black bars represent mean (Wilcoxon p<0.0001). (C) Volcano plot displaying differentially expressed genes (DEGs) from peritoneal tumor microenvironment (TME) vs. flank TME. Points were colored based on overlap with differentially expressed genes (DEGs) from normal human visceral omentum vs. human subcutaneous adipose tissues. PDX unique DEGs are genes that were DEGs in the PDX TME comparison but were not differentially expressed in the human tissue comparison (D) PCA plots made using the top 500 genes by variance displaying clustering of human tumor reads (top) and mouse TME reads (bottom).
Computational deconvolution of transcriptome identifies that flank and peritoneal PDX models recapitulate differences in human tissue
The goal of computational deconvolution of human and mouse reads was to isolate the contributions of the tumor and TME to the total transcriptome so that each could be analyzed independently. As a control, we quantified the stromal component of the computationally deconvoluted transcriptomes using the ESTIMATE algorithm stromal score(11), which confirmed that the stromal transcriptome was murine for both flank and peritoneal PDXs (449.86 vs. 4514.65; p<0.0001) (Figure 1b). Model AAP16, which was established in the peritoneum and had only 3 passages, retained some human stromal cells, while TM00351 which was established in mouse flank and passaged 7 times had essentially no stromal cells (Table S1). Additionally, there was no significant relationship between implant site and tumor purity (TM00351 p=0.55, PMCA-3 p = 0.071, AAP16 p > 0.999) (Figure S2). To test the validity of our assumption that the mouse tissues are similar to the human tissues, the list of differentially expressed genes (DEGs) from two differential gene expression analyses (DGEA) were compared. The first compared the mouse peritoneal TME to the mouse flank TME. The second compared human visceral omentum to human subcutaneous tissue (25). The list of genes, especially genes that are known to be specific to the peritoneal cavity, should be found in both lists of DEGs (Figure 1c). The DEGs between these two comparisons were highly overlapping, including known peritoneal genes MSLN (padj<0.001, Log2FoldChange=7.58), LRNN4 (padj<0.001, Log2FoldChange=7.83), and UPK3B (padj<0.001, Log2FoldChange=11.96). When the same analysis was run on each tumor individually, key peritoneal genes were once again identified (Figures S3–S5). When subset to only DEGs with a log-fold change≥1 and an adjusted p-value≤0.05, 35.8% of DEGs enriched in the peritoneal PDX TME were also enriched in the visceral omentum (Figure S6). GSEA was run on the tumor microenvironment, which showed that the flank TME was relatively enriched in the EMT gene set (Figure S7).
Tumor Transcriptomes Cluster by Model and TME Transcriptomes Cluster by Implant Site
We next performed a cluster analysis of both tumor and TME transcriptomes from the two implant sites (Figure 1d). Principal Component Analysis (PCA) of the tumor transcriptomes revealed that the majority of differences are accounted for by inter-model variances, as expected given the substantial transcriptomic differences anticipated between tumors from distinct patients with different somatic mutation profiles. We employed ward.D, an alternative unsupervised hierarchical clustering method, which validated our results (Figure S8). Applying the same PCA and ward.D clustering techniques to the TME transcriptome revealed that samples primarily distinguish by implant site as opposed to inter-model differences (Figure 1d).
Peritoneal Microenvironment influences tumor-intrinsic transcription and promotes a faster growth rate
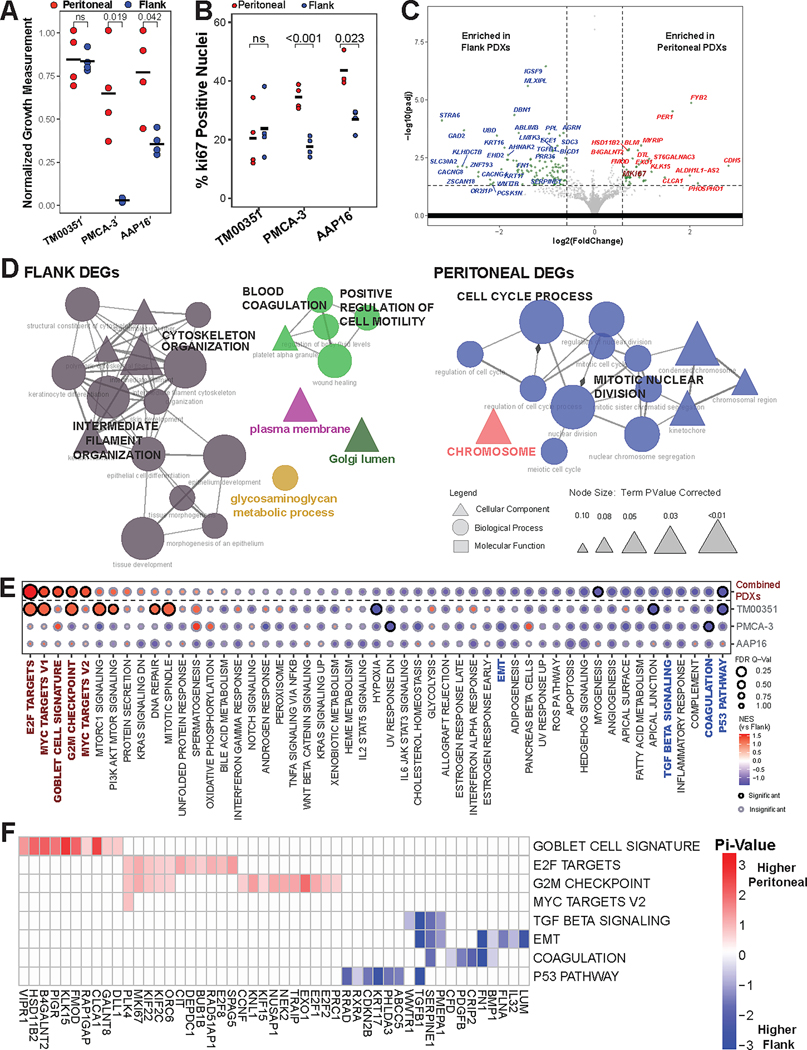
The growth rate of peritoneal tumors was significantly faster than flank for the PMCA-3 (2.68g tumor size vs. 0.04g, p = 0.019) and AAP16 (2.54mm tumor size vs 1.17mm, p=0.042), but not for TM00351 (19.4mm vs 19.0mm, p = 0.856) (Figure 2a). This is potentially because TM00351 was initially established as a flank PDX by the Jackson Lab (and thus had a greater number of passages as a flank PDX relative to other models) prior to this flank vs. peritoneal implantation experiment (Table S1). The samples were subsequently stained for Ki-67, a proliferative marker in colorectal and other gastrointestinal cancers (26) (Figure 2b, Fig S10). Both PMCA-3 (34.4% vs. 17.7%, p<0.001) and AAP16 (43.5% vs. 26.9%, p = 0.023) displayed a higher percentage of positive nuclei when grown in the peritoneal cavity compared to when they were grown in the flank; TM00351 did not (20.5% vs 23.8%, p = 0.668).
Figure 2. Peritoneal microenvironment influences tumor transcriptome to promote tumor growth.
(A) Normalized growth measurements separated by implant site for the three PDX models (pval by Student’s t-test, black bar = mean). Growth measurements were normalized to the highest measured value for each tumor and then plotted. (B) Quantification and comparison of percent of nuclei positive for Ki-67 (pval by Student’s t-Test, black bars = mean). (C) Volcano plot displaying differentially expressed genes (DEGs) from comparing intraperitoneal (IP) PDX tumor transcriptomes compared to flank PDX tumor transcriptomes. MKI67 is emphasized by size and color. (D) Individually generated ClueGO networks made from gene ontologies using either the flank enriched DEGs or the peritoneally enriched DEGs. Edge width denotes strength of overlap measured by the kappa score while node size denotes adjusted p-value. (E) Gene set enrichment analysis (GSEA) results for either all three models combined (top row) or each model separately (bottom three rows). Emphasized gene sets (color, bold) are further analyzed using leading edge analysis (LEA). (F) LEA results for emphasized gene sets from Fig 2E, only including genes with |pi-value|≥0.5.
To identify a potential transcriptomic basis for increased growth, we conducted DGEA comparing the tumor transcriptome from peritoneal and flank PDXs. 223 differentially expressed genes (DEGs) were identified (padj≤0.05). Notably the gene for KI67, MKI67 (padj=0.025, Log2FoldChange=0.66), and EXO1 (padj=0.0081, Log2FoldChange=0.88) were both relatively upregulated in peritoneal PDXs (Figures 2c, S11–S15, Table S3). Additionally, CLCA1 (padj=0.0002, Log2FoldChange=1.47), KLK15 (padj<0.0001, Log2FoldChange=1.11), and HSD11B2(padj<0.0001, Log2FoldChange=0.69), three genes with increased expression in intestinal goblet cells, were also upregulated in peritoneal PDXs (27). When expression of the 223 DEGs was used to perform cluster analysis on the human tumor transcriptomes, we found that instead of clustering solely by the tumor, samples additionally clustered by implant site (Figure S9). This indicates that the DGEA is identifying differences in the tumor-intrinsic transcriptome resulting from the different implant sites.
Peritoneal PDXs overexpress cell cycle ontologies
We performed GO ORA on the 223 tumor DEGs separated into those genes that were relatively upregulated in the peritoneal PDXs and those that were relatively upregulated in the flank PDXs. Two groupings of significantly overrepresented (padj≤0.05) GOs were formed from the peritoneal PDX DEGs: cell cycle process/mitotic nuclear division (13 ontologies) and chromosome (1 ontology). In comparison, the flank PDX DEGs were enriched in GO groups labeled as intermediate filament organization/cytoskeleton organization (15 ontologies), blood coagulation/positive regulation of cell motility (5 ontologies), glycosaminoglycan metabolic process (1 ontology), Golgi lumen (1 ontology), and plasma membrane (1 ontology) (Figure 2d).
GSEA of Human Transcriptome Shows that the Peritoneal Microenvironment Enriches Cell Division Pathways and Goblet Cell (GC) Associated Genes
The results from GSEA further show that cell division pathways were relatively enriched in the peritoneal PDX tumors. These included E2F TARGETS (NES=1.60, FDR q-val=0.004), MYC TARGETS V1 (NES=1.42, FDR q-val=0.21), MYC TARGETS V2 (NES=1.35, FDR q-val=0.21), and G2M CHECKPOINT (NES=1.39, FDR q-val=0.17) (Figure 2e). Additionally, the peritoneal microenvironment had a relative enrichment of the GOBLET CELL SIGNATURE gene set created using genes elevated in intestinal goblet cells (NES=1.40, FDR q-val =0.20) (18). GSEA on individual samples shows that although proliferative gene sets like E2F and MYC were only significantly enriched in TM00351, both PMCA-3 and AAP16 displayed the same direction and a similar effect size for these proliferative gene sets (Figures S16–S18). This indicates that the PMCA-3 and AAP16 comparisons were likely underpowered. Meanwhile, gene sets like P53 PATHWAY (NES=−1.38, FDR q-val=0.19) and MYOGENESIS (NES=−1.30, FDR q-val=0.25) were enriched in the flank PDXs (Figure 2e). EPITHELIAL-MESENCHYMAL TRANSITION (EMT) and COAGULATION were both relatively enriched in the flank tumors (although not significantly).
The results from GSEA were further analyzed using Leading Edge Analysis, which determines which genes are driving the enrichment of specified gene sets (Figure 2f). The genes driving the GOBLET CELL SIGNATURE gene set were CLCA1 and KLK15, along with genes like FMOD, B4GALNT2, and HSD11B2. These were among the most differentially expressed genes in the peritoneum (Figure 2b). There was a lack of overlap between the cell division-related gene sets (E2F, MYC V1, MYC V2, and G2M) and the GOBLET CELL SIGNATURE leading edge genes, indicating that the expression of goblet cell genes is not also driving the cell division enrichment. Although tumor-level differences in expression do exist, differential expression of many of these genes is not tumor-specific (Figure S7, S8).
Proteomic analysis implicates decreased Hippo pathway signaling in the peritoneal microenvironment
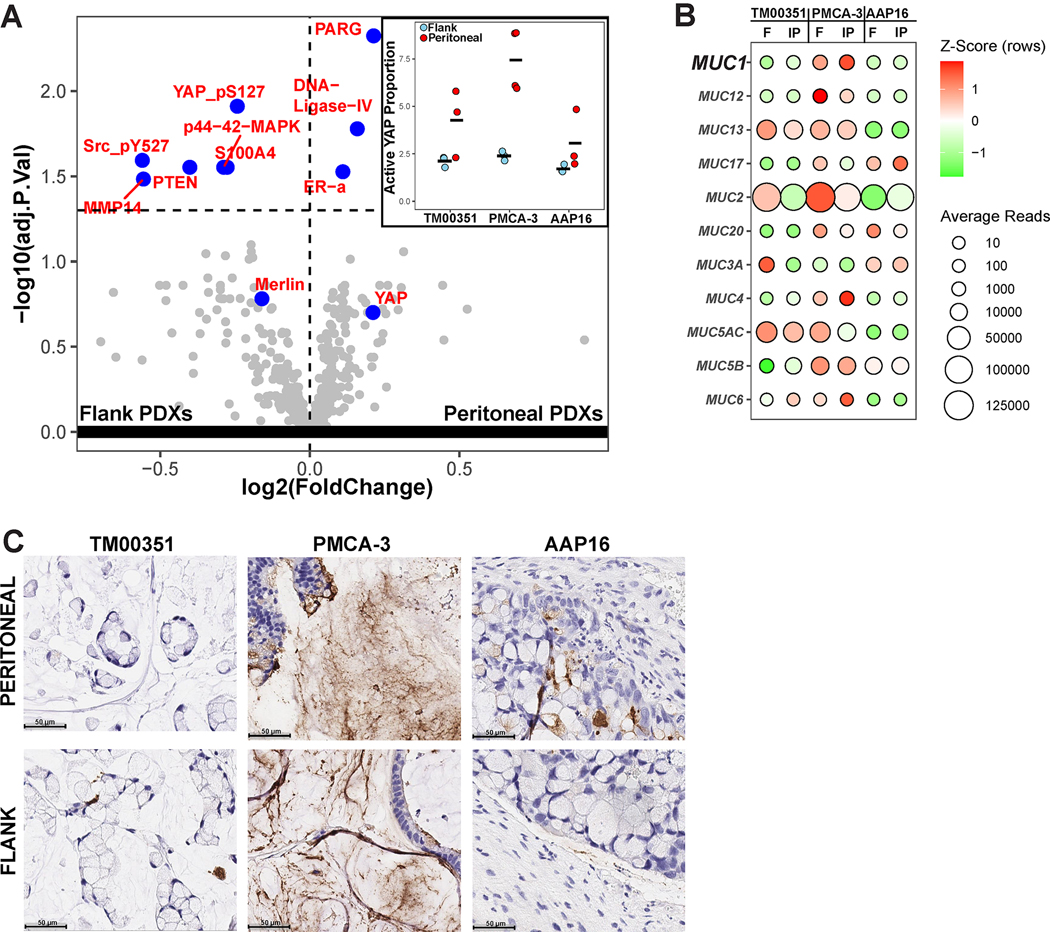
After controlling for batch and model type, we identified 9 differentially expressed proteins (DEPs) (Fig 3a, Figure S19). DNA-Ligase-IV (padj<0.0001, Log2FoldChange=0.21) and Poly(ADP-ribose) glycohydrolase (PARG) (padj=0.0001, Log2FoldChange=0.15) were both enriched in the peritoneal PDXs. DNA-Ligase-IV performs double-strand break repair, which EXO1, one of the genes relatively upregulated in the peritoneal PDXs, is also involved in (28). High levels of PARG are also associated with proliferation (29). Proteins and phosphoforms that are related to upregulated Hippo pathway signaling or are upregulated by deficient YAP signaling (YAP_pS127 (30), Src_pY527(30), Merlin(30), PTEN(31), MMP14 (32)) have higher proteomic expression in the flank PDXs. Moreover, the ratio of active non-phosphorylated YAP to inactive phosphorylated YAP is significantly higher in the peritoneal PDXs compared to the flank PDXs (5.18 vs. 2.07, p = 0.0033) (Figure 3a inset). Additionally, both MMP14 and S100A4 are associated with increased invadopodia formation, which enhances the EMT and invasion processes (33). This shows concordance with the transcriptomic analysis, where flank PDX tumors had higher expression of the EMT pathway.
Figure 3. Peritoneal microenvironment promotes Hippo pathway inhibition but does not affect tumor-intrinsic expression of Mucin Genes.
(A) Volcano Plot from differential protein expression analysis comparing implant site using reverse phase protein array (RPPA) data. Inset shows the ratio of active, unphosphorylated YAP to inactive, phosphorylated YAP for each of the samples, as measured by normalized and linearized EC50 values. (B) Average normalized counts from RNA-Seq for all mucin genes expressed (total normalized counts ≥ 100), split by tumor implanted and implant site. The color shows the z-transformed average reads for each gene while the size shows the average normalized counts without z-transformation. (C) Chromogenic Immunohistochemistry for MUC1 (brown) split by implanted model (columns) and implant site (rows).
Mucin gene expression is not influenced by microenvironment
Finally, given the potential importance of mucin production for peritoneal malignancies, we used the differential implantation experiment to examine the effect of the peritoneal microenvironment on mucin expression(34). Using data from RNA-Seq, we show that tumor-level differences outweigh the effect of the microenvironment. None of the mucins were differentially expressed when comparing implant site (Table S3). However, inter-model differences still existed. As expected by its histopathological features (Table S1), PMCA-3 had high levels of mucin expression relative to the other tumors, regardless of implant site. The next highest was TM00351, which was also mucinous. The sole non-mucinous tumor, AAP16, had the lowest relative expression of mucin (Figure 3b).
To better show that mucin expression is relatively independent of microenvironment, we used IHC to explore MUC1 production. From the RNA-Seq, it is observed that PMCA-3 expresses much more MUC1 relative to the other models. IHC confirms this is the trend: PMCA-3 is the only tumor that has high levels of staining for MUC1 (Figure 3c). The level of MUC1 staining is not dependent on the implant site. Together with the RNA-Seq data, this shows that something intrinsic to the tumor is likely driving mucin expression.
Discussion
Previous studies have shown that AA rarely metastasizes out of the peritoneal cavity. However, it remained unknown if the peritoneal microenvironment directly benefits AA relative to other microenvironments, or if AA selectively metastasizes to the peritoneal cavity solely due to spatial proximity.
By performing a controlled experiment by implanting the same appendiceal tumor into different anatomic sites, we identify that the peritoneal microenvironment can influence the tumor intrinsic expression of AA, and in doing so, promotes the proliferation of AA tumors. In addition to finding downstream gene programs associated with cell division and proliferation, we also identify some of the potential drivers of tumorigenesis, including upregulation of Myc Targets and downregulation of pathways related to apoptosis, such as P53 signaling.
Proteomic and phosphoproteomic analysis provided additional insight and a potential explanation for these transcriptomic differences. The peritoneally implanted AA tumors had a higher ratio of active unphosphorylated YAP to inactive phosphorylated YAP compared to flank-implanted AA tumors. YAP is a known oncogene, with higher YAP activity known to upregulate proliferation and Myc signaling (35,36). YAP activation has also been shown to inhibit apoptosis, anoikis, and potentially plays a role in the survival of metastases in their new site (37).
Notably, YAP activity is connected to the extracellular environment via the Hippo tumor suppressor pathway (30). The Hippo pathway inactivates YAP’s transcriptional activity by phosphorylation and subsequent cytosolic localization. The Hippo pathway responds to interactions with the cell’s extracellular environment, including interactions with other cells, the extracellular matrix, and other mechanical cues (30). A possible explanation for our findings is that peritoneal TME-specific proteins like MSLN, which aids in cell-cell binding, inhibit the Hippo pathway in AA, leading to a higher ratio of active to inactive YAP (38). This leads to higher growth rates and proliferative pathway expression in the peritoneal AA tumors along with inhibition of anoikis/apoptotic pathways. These data suggest that YAP inhibition may be considered as a therapeutic strategy in appendiceal cancer.
We acknowledge several limitations of this study. NSG mice were used to improve the take rate of PDX models; they are however heavily immunosuppressed. Moreover, due to a limited number of AA PDX models available, PDXs with different passage numbers and initial implantation sites were used. Because of this, the PDX tumors with higher passage numbers like PMCA-3 or initial establishment in the flank like TM00351 likely adapted to their environment more than AAP16. Finally, although appendiceal cancer is a heterogeneous disease with important differences between histologic subtype, we were limited to selecting only AA tumors that could grow in both the peritoneal microenvironment and the flank microenvironment.
In summary, the omics data in conjunction with the differential establishment rate/growth rate suggests the importance of the peritoneal microenvironment “soil” for AA to effectively grow, rather than the peritoneal microenvironment being chosen solely due to spatial proximity.
Supplementary Material
Implications:
The peritoneal microenvironment promotes growth of appendiceal tumors and expression of proliferative pathways in PDXs.
Acknowledgments
This research was supported by the National Cancer Institute (K22 CA234406 to J.P.S., Cancer Center Support Grant P30 CA016672, SPORE program P50 CA221707), the Cancer Prevention & Research Institute of Texas (RR180035 to J.P.S.; J.P.S. is a CPRIT Scholar in Cancer Research), the Linda Walmsley Fund and the Col Daniel Connelly Memorial Fund. This study was also supported by the UT MD Anderson Cancer Center Colorectal Cancer Moonshot Program.
We thank the following core facilities at MD Anderson Cancer Center for their services used in this study: Reverse Phase Protein Array Core (Supported by NCI grant # CA16672 and #R50CA221675), Advanced Technology Genomics Core (Supported by NCI Grant CA016672(ATGC)), Biospecimen Extraction Facility for sample processing and DNA/RNA/protein extraction, and Small Animal Imaging Facility (Supported by the Cancer Center Support Grant CA16672) for in vivo live imaging. Finally, we thank Dr. Kjersti Flatmark for generously providing us with and characterizing the PMCA-3 PDX model.
Footnotes
COI Statements/Disclosures: JPS – Scientific Advisory Board for Engine Biosciences, NaDeNo Nanoscience; Research funding from BostonGene, Celsius Therapeutics. All other authors declare no COI.
References
- 1.Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–64. [DOI] [PubMed] [Google Scholar]
- 2.Sugarbaker PH, Zhu BW, Sese GB, Shmookler B. Peritoneal carcinomatosis from appendiceal cancer: Results in 69 patients treated by cytoreductive surgery and intraperitoneal chemotherapy. Dis Colon Rectum. 1993. Apr;36(4):323. [DOI] [PubMed] [Google Scholar]
- 3.Shen JP, Yousef AM, Zeineddine FA, Zeineddine MA, Tidwell RS, Beaty KA, et al. Efficacy of Systemic Chemotherapy in Patients With Low-grade Mucinous Appendiceal Adenocarcinoma: A Randomized Crossover Trial. JAMA Netw Open. 2023. Jun 1;6(6):e2316161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugarbaker PH. Are There Curative Options to Peritoneal Carcinomatosis? Ann Surg. 2005. Nov;242(5):748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raghav K, Shen JP, Jácome AA, Guerra JL, Scally CP, Taggart MW, et al. Integrated clinico-molecular profiling of appendiceal adenocarcinoma reveals a unique grade-driven entity distinct from colorectal cancer. Br J Cancer. 2020. Oct;123(8):1262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ang CSP, Shen JP, Hardy-Abeloos CJ, Huang JK, Ross JS, Miller VA, et al. Genomic Landscape of Appendiceal Neoplasms. JCO Precis Oncol. 2018;2:PO.17.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flatmark K, Davidson B, Kristian A, Stavnes HT, Førsund M, Reed W. Exploring the peritoneal surface malignancy phenotype—a pilot immunohistochemical study of human pseudomyxoma peritonei and derived animal models. Hum Pathol. 2010. Aug 1;41(8):1109–19. [DOI] [PubMed] [Google Scholar]
- 8.Fleten KG, Lund-Andersen C, Waagene S, Abrahamsen TW, Mørch Y, Boye K, et al. Experimental Treatment of Mucinous Peritoneal Metastases Using Patient-Derived Xenograft Models. Transl Oncol. 2020. Aug 1;13(8):100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014. Dec 5;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carenzo A, Pistore F, Serafini MS, Lenoci D, Licata AG, De Cecco L. hacksig: a unified and tidy R framework to easily compute gene expression signature scores. Bioinformatics. 2022. May 13;38(10):2940–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013. Oct 11;4(1):2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019. Jun 15;35(12):2084–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009. Apr 15;25(8):1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003. Nov;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao Y, Hsiao TH, Suresh U, Chen HIH, Wu X, Wolf SE, et al. A novel significance score for gene selection and ranking. Bioinformatics. 2014. Mar 15;30(6):801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005. Oct 25;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015. Dec 23;1(6):417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson M, Zhang C, Méar L, Zhong W, Digre A, Katona B, et al. A single–cell type transcriptomics map of human tissues. Sci Adv. 2021. Jul 28;7(31):eabh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma Oxf Engl. 2010. Jan 1;26(1):139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015. Apr 20;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014. Feb 3;15(2):R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blomme A, Van Simaeys G, Doumont G, Costanza B, Bellier J, Otaka Y, et al. Murine stroma adopts a human-like metabolic phenotype in the PDX model of colorectal cancer and liver metastases. Oncogene. 2018. Mar;37(9):1237–50. [DOI] [PubMed] [Google Scholar]
- 23.Fujii E, Kato A, Suzuki M. Patient-derived xenograft (PDX) models: characteristics and points to consider for the process of establishment. J Toxicol Pathol. 2020. Jul;33(3):153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ideno N, Yamaguchi H, Ghosh B, Gupta S, Okumura T, Steffen DJ, et al. GNASR201C Induces Pancreatic Cystic Neoplasms in Mice That Express Activated KRAS by Inhibiting YAP1 Signaling. Gastroenterology. 2018. Nov;155(5):1593–1607.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013. Jun;45(6):580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullwinkel J, Baron-Lühr B, Lüdemann A, Wohlenberg C, Gerdes J, Scholzen T. Ki-67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J Cell Physiol. 2006. Mar;206(3):624–35. [DOI] [PubMed] [Google Scholar]
- 27.Liu CL, Shi GP. Calcium-activated chloride channel regulator 1 (CLCA1): More than a regulator of chloride transport and mucus production. World Allergy Organ J. 2019. Nov 29;12(11):100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieber MR. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End-Joining Pathway. Annu Rev Biochem. 2010;79(1):181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrision D, Gravells P, Thompson R, Bryant HE. Poly(ADP-Ribose) Glycohydrolase (PARG) vs. Poly(ADP-Ribose) Polymerase (PARP) – Function in Genome Maintenance and Relevance of Inhibitors for Anti-cancer Therapy. Front Mol Biosci [Internet]. 2020. [cited 2023 Jun 30];7. Available from: https://www.frontiersin.org/articles/10.3389/fmolb.2020.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015. Nov 5;163(4):811–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, et al. YAP mediates crosstalk between the Hippo and PI(3)K–TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol. 2012. Dec;14(12):1322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venghateri JB, Dassa B, Morgenstern D, Shreberk-Shaked M, Oren M, Geiger B. Deciphering the involvement of the Hippo pathway co-regulators, YAP/TAZ in invadopodia formation and matrix degradation. Cell Death Dis. 2023. Apr 25;14(4):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy DA, Courtneidge SA. The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol. 2011. Jul;12(7):413–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hugen N, Velde CJH van de, Wilt JHW de, Nagtegaal ID. Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann Oncol. 2014. Mar 1;25(3):651–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziosi M, Baena-López LA, Grifoni D, Froldi F, Pession A, Garoia F, et al. dMyc Functions Downstream of Yorkie to Promote the Supercompetitive Behavior of Hippo Pathway Mutant Cells. PLOS Genet. 2010. Sep 23;6(9):e1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiemer SE, Zhang L, Kartha VK, Packer TS, Almershed M, Noonan V, et al. A YAP/TAZ-Regulated Molecular Signature Is Associated with Oral Squamous Cell Carcinoma. Mol Cancer Res MCR. 2015. Jun;13(6):957–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012. Jan 1;26(1):54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gubbels JA, Belisle J, Onda M, Rancourt C, Migneault M, Ho M, et al. Mesothelin-MUC16 binding is a high affinity, N-glycan dependent interaction that facilitates peritoneal metastasis of ovarian tumors. Mol Cancer. 2006. Oct 26;5:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and associated R code are available in the linked CodeOcean capsule (Capsule ID: 7745616).