Abstract
Influenza A virus (IAV) pandemics result from interspecies transmission events within the avian reservoir and further into mammals including humans. Receptor incompatibility due to differently expressed glycan structures between species has been suggested to limit zoonotic IAV transmission from the wild bird reservoir as well as between different bird species. Using glycoproteomics, we have studied the repertoires of expressed glycan structures with focus on putative sialic acid-containing glycan receptors for IAV in mallard, chicken and tufted duck; three bird species with different roles in the zoonotic ecology of IAV. The methodology used pinpoints specific glycan structures to specific glycosylation sites of identified glycoproteins and was also used to successfully discriminate α2-3- from α2-6-linked terminal sialic acids by careful analysis of oxonium ions released from glycopeptides in tandem MS/MS (MS2), and MS/MS/MS (MS3). Our analysis clearly demonstrated that all three bird species can produce complex N-glycans including α2-3-linked sialyl Lewis structures, as well as both N- and O- glycans terminated with both α2-3- and α2-6-linked Neu5Ac. We also found the recently identified putative IAV receptor structures, Man-6P N-glycopeptides, in all tissues of the three bird species. Furthermore, we found many similarities in the repertoires of expressed receptors both between the bird species investigated and to previously published data from pigs and humans. Our findings of sialylated glycan structures, previously anticipated to be mammalian specific, in all three bird species may have major implications for our understanding of the role of receptor incompatibility in interspecies transmission of IAV.
Keywords: birds, glycoproteomics, influenza virus, interspecies transmission, sialic acid receptor
Introduction
Zoonotic viruses have the capacity to cause severe pandemics (Kreuder Johnson et al. 2015). Viruses of the three latest influenza A virus (IAV) pandemics (H2N2 1957, H3N2 1968, and H1N1 2009) (Horimoto and Kawaoka 2001; Katz et al. 2009; Yen and Webster 2009) have all contained genetic material originating from avian influenza viruses (AIVs). These events have most likely involved spread from wild to domestic bird species and further into pigs and humans. Key barriers for interspecies transmission are molecular interactions between host and virus, including recognition of host receptors by virus surface proteins, adaptation to changes in pH and temperature, as well as avoiding the host’s innate and adaptive immune response to infection (Short et al. 2015; Subbarao 2019). Hence, one of the biological traits regarded to be important for interspecies transmission of IAV is a change in receptor binding specificity. Hemagglutinin (HA), the receptor binding protein of AIVs bind to α2-3- and α2-6-linked sialic acid (Sia) containing glycans, whereas human IAVs bind more exclusively to α2-6-linked Sia-containing glycans (Gottschalk 1952; Gottschalk 1954; Blix et al. 1957; Rogers and Paulson 1983; Rogers and D'Souza 1989; Eriksson et al. 2018; Verhagen et al. 2021). Sialic acid, or more precisely the 5-N-acetylneuraminic acid (Neu5Ac) is a nine-carbon monosaccharide found at terminal positions of many cell surface glycan structures and is utilized by IAVs for cell recognition and attachment. The Sia residue is commonly linked to the penultimate galactose (Gal) via either an α2-3- or an α2-6- linkage, whereas two Sia in a series bind to each other via an α2-8- linkage. However, the isomeric structures of the Sia-Gal linkage are not the exclusive determinant for IAV receptor binding (Gambaryan et al. 2005; Stevens et al. 2006; Gambaryan et al. 2018). Rather, the glycan structure underneath the terminal Sia together with the Sia–Gal linkage, are regarded as responsible for creating the binding epitope recognized by the virus, and that will determine the affinity of the HA-receptor interaction.
Originally it was believed that α2-3-linked Sia containing glycans were exclusive to bird tissues, whereas α2-6-linked Sia were exclusive to humans (Gambaryan et al. 2005; Stevens et al. 2006; Gambaryan et al. 2018). However, more recent studies have revealed that α2-3-linked Sia can indeed be found in the human airway epithelium (Shinya et al. 2006; Walther et al. 2013), and similarly screenings with lectins have indicated the presence of α2-6-linked Sia in avian tissues (Ellström et al. 2009; Liu et al. 2009; Yu et al. 2011; Costa et al. 2012; Franca et al. 2013; Naguib et al. 2019). Earlier studies have reported 3′STF (Neu5Acα2-3Galβ1-3GalNAc) and 3′-sialylneolacto 3′SLN (Neu5Acα2-3Galβ1-4GlcNAc), as preferred receptor structures for AIVs isolated from ducks, whereas AIVs isolated from chickens reportedly have higher affinity for sulfated and/or fucosylated structure analogs (Gambaryan et al. 2005; Eriksson et al. 2018; Gambaryan et al. 2018). Species-specific differences in the sialylated glycan structures serving as receptors for IAVs have been proposed as barriers for transmission between bird species (Gambaryan et al. 2005; Gambaryan et al. 2008; Gambaryan et al. 2018).
For most bird species, little is known in detail about the preferred site of replication for low pathogenic avian influenza virus (LPAIV). In mallards (Anas platyrhynchos), the main animal reservoir for IAVs in nature, the replication of LPAIV mainly takes place in the surface epithelium of the intestinal tract and transmission occurs via the fecal-oral route, although replication in the respiratory tract has also been reported, both for LPAIV and highly pathogenic avian influenza virus (HPAIV) (Webster et al. 1978; Ellström et al. 2008; Wille et al. 2018). In chickens (Gallus gallus), the respiratory tract is reportedly more important for replication of both LPAIV and HPAIV, although replication in the intestinal tract has also been reported for this species (Slemons and Swayne 1990; Mundt et al. 2009; Alqazlan et al. 2021). Indirect evidence suggest that this is the case also in an array of wild bird species, including tufted ducks (Aythya fuligula) (Bröjer et al. 2009; Jourdain et al. 2011; Bergervoet et al. 2019; Naguib et al. 2019), particularly in the case of HPAIV infection.
Since previous Sia characterizations of bird tissue only made use of lectin detection, knowledge is lacking regarding the presence of glycoprotein-specific Sia structures. Therefore, we have in this study characterized and compared the IAV glycan receptor repertoire displayed on cell surfaces of putative target organs of mallards, chickens and tufted ducks using LC-MS/MS based glycoproteomics. Our selection of bird species was based on the facts that i) mallard is the most well described wild avian host for IAV, ii) the tufted duck is a representative of diving ducks that are known to have lower prevalence of IAV compared to mallards and iii) chicken represents a key link to zoonotic transmission. Furthermore, the whole genome sequences are available for all three species, which is a prerequisite for the glycoproteomic analysis pinpointing specific glycan structures to specific glycosylation sites of glycoproteins identified in complex biological samples. An earlier limitation of glycoproteomics was that determination of isomers, such as α2-3- or α2-6-linked Sia, has not been possible to address. However, by careful analysis of oxonium ions released from glycopeptides during tandem MS/MS (MS2), and further into MS/MS/MS (MS3) it has become possible to discriminate α2-3 from α2-6-linked Sia (Yu et al. 2016; Pett et al. 2018), and this methodology is now used here in the glycoproteomic analysis of bird tissues.
Material and methods
Ethical statement
All animal experiments were carried out in strict accordance with a protocol legally approved by the regional board of the animal ethics committee, Sweden (permission number 5.8.18-07998/2017). All animal experiments were conducted in BSL2 animal facilities at the Swedish National Veterinary Institute (SVA).
Epithelial cell isolation for glycoproteomic analysis
Epithelial cells were isolated from tissue samples collected from trachea, lung, ileum, and colon of captive healthy mallards, chickens and tufted ducks (5 individuals per species) (Arike et al. 2020). All mallards and chickens were ascertained seronegative for AIV. However, for glycoproteomic analysis on tufted ducks we had to use individuals that were seropositive for AIV upon arrival to the animal facility. Importantly, after arrival all flocks were PCR negative for AIV as determined by regular testing, and the tufted ducks used for structural analysis were observed at the animal facility for four weeks before autopsy and collection of epithelial cells for analyses. Hence, the glycoproteomic structures identified in all three species should be considered as unaffected by any ongoing AIV infection. The biopsies were rinsed in chilled (~8 °C) PBS (Sigma-Aldrich) upon collection to remove any debris. Adipose tissue, blood vessels, and other macroscopic tissue remains were carefully manually removed from trachea and intestine before the tissue sample was cut open. The lung samples were rinsed, cut open, and rinsed again to wash away blood remains. Intestinal specimens were cut open and intestinal content carefully removed. The loosely attached outer mucus layer was gently removed from the intestinal specimens, while retaining the inner firmly attached mucus layer, enabling us to reveal receptor structures of importance for virus attachment and uptake into the epithelial cells. Rinsed tissue samples were incubated at 37 °C for 1 h in 10 mL DPBS with 1 mM DTT and 3 mM EDTA (Sigma-Aldrich). After incubation, the liquid was carefully aspirated and replaced with PBS (Sigma-Aldrich). The samples were vortexed for 5 × 20 s and the tissue was then removed. The remaining cell slurry was spun at 1,000 g for 5 min at 4 °C and the supernatant was discarded. The pellets from each organ and bird were individually snap-frozen in liquid nitrogen and stored at −80 °C.
Protein digestion, enrichment and fractionation
The samples were homogenized using the lysis matrix D on a Fastprep-24 instrument (MP Biomedicals) in lysis buffer (50 mM triethylammonium bicarbonate (TEAB), 2% sodium dodecyl sulfate (SDS)) and 5 cycles 40 s each. The samples were centrifuged at maximum speed for 15 min, and supernatants harvested. The protein concentration was determined using Pierce BCA Protein Assay (Thermo Fisher Scientific) and the Benchmark Plus microplate reader (BIO-RAD) with BSA solutions as standards.
Sample aliquots (500–1,000 μg) were digested with trypsin using the filter-aided sample preparation (FASP) method (Wisniewski et al. 2009), with small modifications. Briefly, samples were reduced with 100 mM dithiothreitol at 60 °C for 30 min, spin-filtered (30 kDa MWCO Pall Nanosep centrifugation filters, Sigma-Aldrich), washed repeatedly with 8 M urea and followed by digestion buffer (1% sodium deoxycholate (SDC) in 50 mM TEAB) prior to alkylation with 10 mM methyl methanethiosulfonate in digestion buffer for 20 min. Protein digestion was performed in digestion buffer by addition of 5 μg Pierce MS grade Trypsin (Thermo Fisher Scientific) at 37 °C overnight, followed by an additional 2 h incubation with new trypsin added the consecutive day. Peptides were isolated by centrifugation and SDC was removed by acidification with 10% trifluoroacetic acid. Glycopeptides were enriched with hydrophilic interaction liquid chromatography (HILIC) according to (Parker et al. 2013), with slight modifications. In short, peptides were loaded onto an in-house zwitterionic Zic-HILIC SPE cartridge containing 20 mg of Zic-HILIC particles (10 μm, 200 Å; Sequant/Merck). The flow-through was collected and re-circulated through the column an additional three times. The column was washed with totally 1.2 mL of 80% (v/v) acetonitrile and 1% (v/v) trifluoroacetic acid. Enriched glycopeptides were eluted with 4 times 50 μL 0.1% (v/v) trifluoroacetic acid followed by 50 μL of 25 mM NH4HCO3 and finally 50 μL of 50% (v/v) acetonitrile and dried by vacuum centrifugation. Each sample was fractionated into six fractions (5.0%–17.5% acetonitrile in 0.1% trimethylamine), using Pierce high pH reversed-phase peptide fractionation kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. After an initial analysis of the peptide amounts in each of the six fractions for a few test samples, fractions 1, 5 and 6 were pooled; fractions 2 and 3 were pooled; and fraction 4 was left intact, to constitute the three samples with similar protein amounts. For the MS3 approach, the samples were used unfractionated. Samples from each tissue and each individual bird were dried in a vacuum centrifuge and reconstituted in 15 μL of 3% acetonitrile, 0.1% formic acid for LC-MS/MS analysis.
NanoLC-MS/MS analysis
Peptide samples were individually analyzed on an Orbitrap Fusion Tribrid mass spectrometer interfaced with Easy-nLC1200 liquid chromatography system (Thermo Fisher Scientific). Peptides were trapped on an Acclaim Pepmap 100 C18 trap column (100 μm × 2 cm, particle size 5 μm, Thermo Fischer Scientific) and separated on an in-house packed analytical column (75 μm × 30 cm, particle size 3 μm, Reprosil-Pur C18, Dr. Maisch) using a linear gradient (solvent A; 0.2% formic acid in water and solvent B; 80% acetonitrile, 0.2% formic acid in water) from 7% to 35% of solvent B over 45 min followed by an increase to 100% solvent B in 5 min and finally 100% solvent B for 10 min, at a flow rate of 300 nL/min. MS1 scans were performed at 120,000 resolution, m/z range 600–2,000, the most abundant double or multiply charged precursors from the MS1 scans were selected with a duty cycle of 3 s, isolated with a 3 Da window, fragmented with higher-energy collision induced dissociation (HCD) at 30% normalized collision energy (NCE), m/z range 100–2,000, and then two times at 40% NCE, m/z range 100–2,000 and m/z 300–2,000. The maximum injection time was 118 ms and MS2 spectra were detected in the Orbitrap at 30,000 resolution. Dynamic exclusion was enabled with 10 ppm tolerance and 10 s duration. For selected samples, MS3 was performed on the m/z 657.23 [Neu5AcHexHexNAc]+ and m/z 698.26 [Neu5AcHexNAcHexNAc]+ ions using HCD 20% NCE at both the MS2 and the MS3 steps, and with detection in the orbitrap and the ion trap, respectively.
Glycoproteomic data analysis
The LC-MS/MS raw files were analyzed with the Byonic software (Protein metrics) using a modified list of the glycan modifications “182 human N-glycans” encompassing 217 glycoforms and “6 most common O-glycans”, such that e.g. glycoforms with a (Hex)4(HexNAc)5 core structure, differing from the normal (Hex)5(HexNAc)4 complex biantennary core, were added. Also, Man-6P glycoforms ranging from (Hex)3(HexNAc)2 to (Hex)9(HexNAc)2 with 1–2 phosphate groups and 0–2 additional HexNAc residues were used in separate Byonic searches. Further Byonic search criteria: the FASTA database was A. platyrhynchos (Organism ID 8840; 27,089 sequences, accessed 15 Jan 2019); A. fuligula (Organism ID 219594; 24,034 sequences, accessed 26 Aug 2021); and G. gallus (Organism ID 9031, 27,535 sequences, accessed 25 Feb 2019); C-terminal cleavage allowed after Lys and Arg; accuracy for MS1 was 5 ppm and for MS2 it was 20 ppm; static modification was methylthio on Cys (+45.9877 u) and variable modification, apart from glycans, was Met oxidation (+15.9949 u). A Byonic score cut off >300 was used for glycopeptide matches and Neu5Ac containing identities were manually verified with the following inclusion criteria 1) presence of the correct peptide+HexNAc ion with respect to the precursor mass and identified glycan mass; 2) presence of the Neu5Ac oxonium ions m/z 274.09 and m/z 292.10; and 3) for hits including fucose (dHex), presence of peptide+HexNAc+dHex ion for the identification of a core fucose, and/or presence of m/z 512.20 ion (HexHexNAcdHex) for identification of Fuc on the antennae. Extracted ion chromatograms were traced at diagnostic MS2 ions including peptide+HexNAc ions, chosen to identify additional glycoforms sharing the same peptide, and saccharide oxonium ions, for instance m/z 274.09 for tracing Neu5Ac; m/z 290.09 for tracing Neu5Gc; m/z 316.10 for tracing O-Ac Neu5Ac; and m/z 495.18 for tracing Neu5AcHexNAc.
The Neu5Acα2-3/Neu5Acα2-6 isomeric structures of glycopeptide hits were determined according to (Pett et al. 2018) Briefly, the relative LacNAc to Neu5Ac (L/N) oxonium ion ratio (m/z 204 + m/z 366) / (m/z 274 + m/z 292) were calculated at an NCE of 40% for N-glycopeptide hits carrying (Neu5Ac)2(Hex)5(HexNAc)4(dHex)0-1 complex biantennary structures; and for O-glycopeptide hits carrying (Neu5Ac)1-2HexHexNAc structures. The ratio was then multiplied with the stoichiometric compensation factor n(NeuAc) / n(HexNAc) to obtain the normalized ratio Ln/Nn. For the MS3 based methodology, the MS3 spectrum of the MS2 generated m/z 657.23 ion was manually inspected. When the (m/z 204 + m/z 366) ion intensities were larger than the (m/z 274 + m/z 292) intensities, the linkage structure was considered as Neu5Acα2-6 and when lower the linkage structure was considered as Neu5Acα2-3. For the MS3 experiments, Byonic matches with scores <300 were also considered, provided that the same identity was present with a score >300 in the corresponding MS2 experiment. The samples were analyzed in accordance with the MIRAGE and MIAPE guidelines for glycomic and proteomic analysis (Taylor et al. 2007; Kolarich et al. 2013; Struwe et al. 2016).
Bioinformatic analysis and visualization
To identify the proteins to which the glycans including Man-phosphate were attached, a sequence similarity search of the peptides was made using the Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1990). The similarity search was made using BLASTp v. 2.9.0+, using the blastp-short option with default settings, against the protein database from UniProt (release 2023_2, (UniProt-Consortium 2019) for each species. An additional peptide search against the UniProt database for each species (release 2023_2) was made for uncharacterised peptides with at least 7 amino acids to which the N-glycans were attached. To investigate where the identified N-glycans and the NeuAc isomers were located, the subcellular location of the glycoproteins to which they were attached was determined using information from the UniProt Knowledgebase.
To evaluate whether the identified N-glycans, sialylated N-glycans and glycoproteins were shared between or unique among the bird species and tissue types in our study, data was converted to a presence-absence matrix and UpSet plots were generated using the UpSetR v. 1.4.0 (Lex et al. 2014; Lex and Gehlenborg 2014) package in R v. 4.2.2 (RCoreTeam 2022). To visualize what glycoproteins and sialylated N-glycans were detected in mallard, tufted duck and chicken tissues, heatmaps were generated using the pheatmap v. 1.0.12 package (Kolde 2019) and the ComplexHeatmap v. 2.12.1 (Gu et al. 2016) in R.
Results
Glycoproteomics reveals candidate IAV receptors in bird tissues
To study how the expression of sialylated glycans potentially serving as receptors for IAV infection differ between avian species, we performed glycoproteomic analysis on tissue specimens of mallards, chickens, and tufted ducks. Epithelial cells were extracted from tissue samples collected from putative AIV target organs comprising trachea, lung, ileum, and colon from healthy, uninfected individuals. After trypsin digestion, hydrophilic interaction liquid chromatography (HILIC) was used to enrich glycopeptides, which were then subjected to LC-MS/MS analysis and identified using the Byonic software.
We initially focused on complex type Neu5Ac containing N-glycopeptides where we were able to discriminate between the Neu5Acα2-3- and Neu5Acα2-6-linkage isomers (Fig. 1). We characterized 243 such N-glycopeptides from 122 glycoproteins across the three species. Among these, we identified 42 structurally different sialylated N-glycans including bi-, tri- and tetra-antennary structures (Supplementary Table S1). For the sialylated N-glycans, non-fucosylated structures were found as well as structures fucosylated on the core GlcNAc or on one or more of the antennae, and in some cases also containing a bisecting GlcNAc (Supplementary Table S1). The N-glycan antennae were usually extended with one (or rarely two) GalGlcNAc disaccharides but occasionally also with HexNAcHexNAc disaccharides, probably composed of GalNAcGlcNAc (LacdiNAc), on one of the antennae. All protein names and peptide sequences are provided in Supplementary Tables S2–S4 for the mallard, tufted duck and chicken tissues, respectively. We could also characterize 121 O-glycopeptides from 87 glycoproteins with complete information on the Neu5Acα2-3-/ Neu5Acα2-6-linkage status. The HILIC enrichment strategy was developed to enrich for N-glycopeptides (Parker et al. 2013). However, by manual fragmentation analysis of the LC-MS/MS data we also identified fragmentation patterns consistent with mono- and disialo core 1 O-glycopeptides and consequently we also performed an O-glycoproteomic Byonic analysis (Fig. 2 and Supplementary Tables S2–S4). However, to our knowledge, it is not known how well the relatively short O-glycans become enriched using HILIC, but it is important to stress that we were not able to identify larger and more complex O-glycans that should be retained better in the HILIC purification.
Fig. 1.
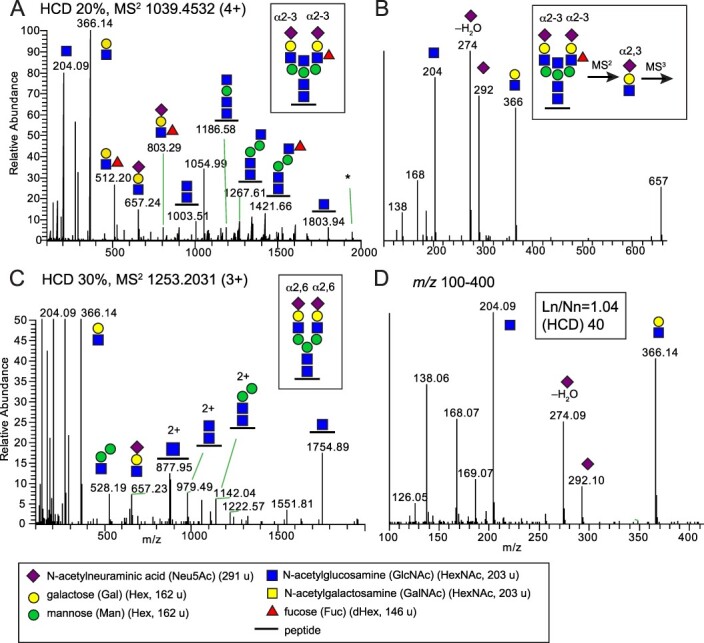
Glycoproteomic analysis of sialylated and fucosylated N-glycopeptides found in mallard. A) HCD-MS2 of a disialylated complex biantennary N-glycopeptide including a bisecting GlcNAc and a Lewis x fucose (boxed structure) from epithelial cell adhesion molecule (A0A8B9TTX1_ANAPL) with the sequence R.LNVSIDNEVVQLEK.A. The asterisk marks the presence of an ion corresponding to [peptide + GlcNAc + Fuc] possibly due to fucose migration or to co-elution of two glycoforms B) MS3 of the m/z 657 ion provides identification of the Neu5Acα2-3Gal linkages on both antennas (Pett et al. 2018). C) HCD-MS2 spectrum of a disialylated complex biantennary N-glycopeptide (boxed structure) from signal recognition particle receptor subunit beta (R0JEC8_ANAPL) and D) the m/z 100–400 region of the expanded MS2 spectrum shows the presence of several oxonium ions used to identify the Neu5Acα2-6-linkage of the sialic acids. Similar glycan structures were also detected in chickens and tufted ducks (Supplementary Table S1). Glycan symbols are according to the SNFG format (Varki et al. 2015).
Fig. 2.
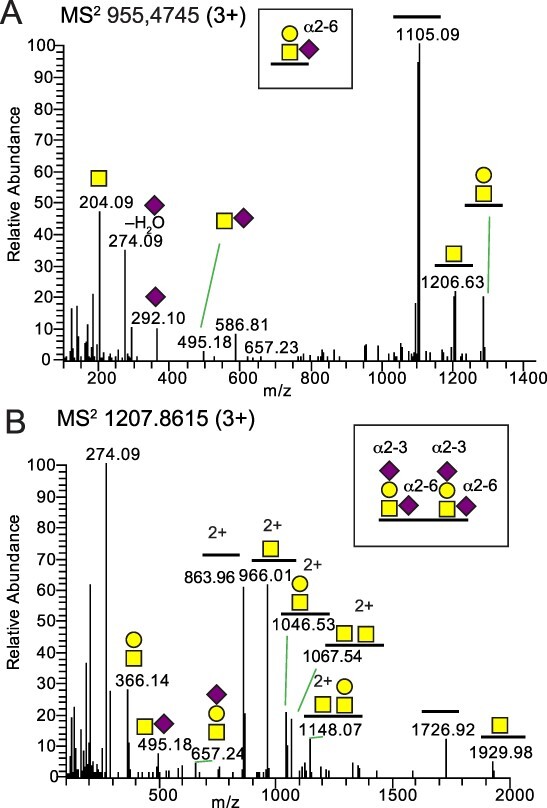
Glycoproteomic analysis of sialylated core 1 O-glycopeptides from mallard samples. A) HCD-MS2 of a Neu5Acα2-6 sialylated core 1 O-glycopeptide from fibrinogen beta chain (A0A8B9SHX9_ANAPL) with the R.ETAPTLRPVAPPISGTGYQPR.P amino acid sequence. B) HCD-MS2 of two Neu5Acα2-3 and Neu5Acα2-6 disialylated core 1 O-glycans of a glycopeptide from dystroglycan 1 (A0A493T5Q5_ANAPL) with the R.VISEATPTLAAGKDPEK.S sequence. Glycan symbols are according to the SNFG format (Varki et al. 2015).
Of particular interest, the recently identified putative IAV receptor structures, Man-6P N-glycopeptides (Byrd-Leotis et al. 2019a; Byrd-Leotis et al. 2019b) were identified in all tissues of the three bird species (Fig. 3 and Supplementary Tables S2–S4). In total, we characterized 469 such N-glycopeptides from 153 glycoproteins. These structures were enriched in ileum and colon and many of them were found on predicted lysosomal enzymes, including e.g. carboxypeptidase and aminopeptidase. Furthermore, despite previously published in vitro data showing binding of chicken-origin AIV isolates to sulfated sialylated glycopeptides (Gambaryan et al. 2005; Gambaryan et al. 2008), no such structures were identified in any of the samples. Also, we could not find the Sia structure, Neu5Gc in any of the tissues, and we could not find O-acetyl Neu5Ac in any of the tissues. In the following paragraphs we will present the results from the different bird species one by one as well as a short illustration of the glycoproteomic data analysis for mallard tissues.
Fig. 3.
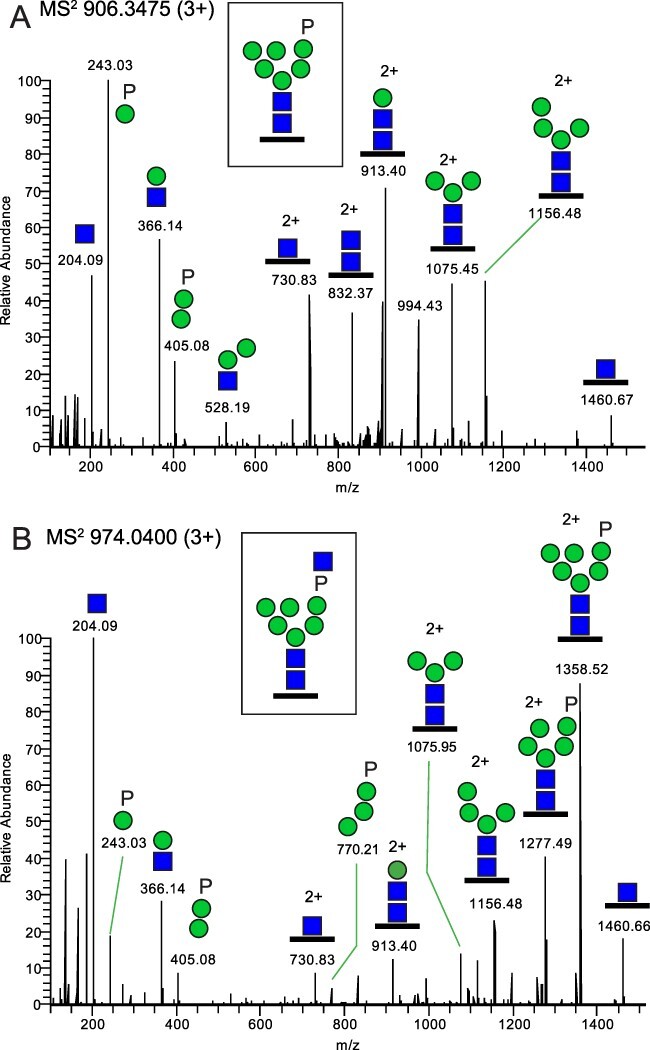
Identification of Man-6P N-glycopeptides. The example is taken from Man-6P glycopeptides, originating from chicken aminopeptidase (F1NB92_CHICK) with the R.ENHTVVSSNDR.A sequence, and the MS2 spectra show A) the (phosphate)1(Hex)6(HexNAc)2 glycoform and B) the (phosphate)1(Hex)6(HexNAc)3 glycoform. Glycan symbols are according to the SNFG format (Varki et al. 2015).
The mallard tissues
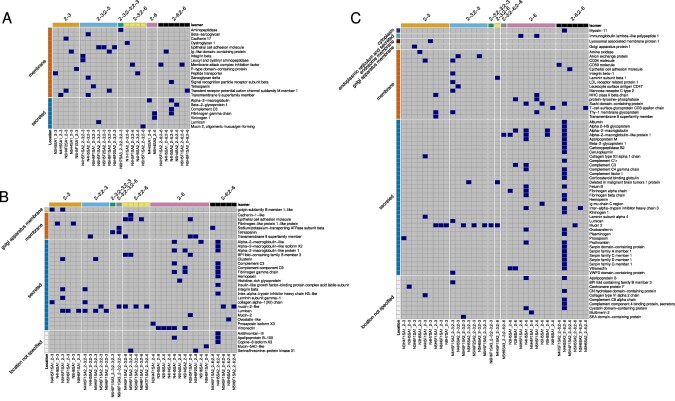
Blasting the glycosylated peptide sequences against the UniProt database (release 2023_2 (UniProt-Consortium 2019) revealed that N-glycans terminating with α2-3-linked Neu5Ac were found mainly on glycopeptides of membrane proteins from all mallard tissues sampled (Figs. 4 and 5A and Supplementary Table S1). In colon and ileum, α2-3-linked Neu5Ac were exclusively found on membrane proteins. N-glycans terminating with α2-6-linked Neu5Ac were instead found mainly on secreted glycoproteins but also on some typical membrane proteins (Figs. 4 and 5A and Supplementary Table S1). In total, 35 unique sialylated N-glycopeptides from 23 proteins were identified from the four tissues (Fig. 5A, Supplementary Table S2).
Fig. 4.
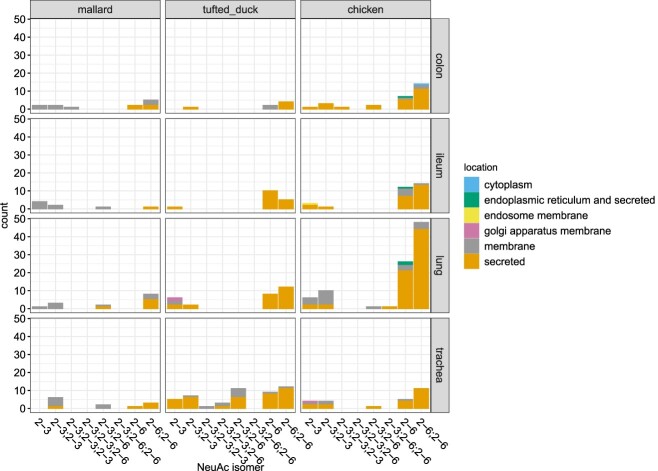
Overview of Neu5Ac modified N-glycopeptides from glycoproteins in different tissues in mallard, tufted duck and chicken. The figure displays the number and identity of all Neu5Ac isomers located on membrane or secreted glycoproteins for all species and tissue types. Orange color indicates Neu5Ac expressed on secreted glycoproteins, gray indicates expression on membrane bound glycoproteins, purple indicates expression on glycoproteins bound to the golgi apparatus membrane, yellow—glycoproteins bound to the endosome membrane, green—glycoproteinsfound in the endoplasmatic reticulum and secreted and blue indicates proteins found in the cytoplasm. The numbers on the X-axis denote the linkage between the Neu5Ac and the penultimate glycan on single or multi-antennary glycopeptides.
Fig. 5.
Glycan structure and protein information on avian sialylated N-glycopeptides. The heat maps display glycan structure including linkage conformation on terminal Sia and protein information for all N-linked glycopeptides detected in A) mallards, B) tufted ducks and C) chickens. The top X-axis indicates the linkage conformation on the Sia linkage and the bottom X-axis indicates the glycan structure including the linkage position. The right Y-axis indicates the name of the glycoprotein and the left Y-axis indicates whether the glycoprotein is membrane bound (orange), secreted (blue), located to the Golgi apparatus membrane (yellow), endosome membrane (brown), endoplasmic reticulum and secreted (green), cytoplasm (dark blue) or if glycoprotein location was not specified (white). Heatmaps were generated using the pheatmap v. 1.0.12 package (Kolde 2019) and the ComplexHeatmap v. 2.12.1 (Gu et al. 2016) in R.
As an illustration to the general workflow used to disentangle the glycan structures in this study, we provide the detailed results from the analysis of a tryptic glycopeptide, with the sequence R.LNVSIDNEVVQLEK.A, obtained from the epithelial cell adhesion molecule (A0A8B9TTX1_ANAPL) from the mallard ileum and colon samples (Fig. 1A). The MS/MS (MS2) spectrum, obtained at a relatively low normalized collision energy (NCE 20%), gave abundant glycosidic fragmentation into saccharide derived oxonium B-ions and a stepwise decomposition of the glycan while still attached to the intact peptide providing the Y-ions, which were all used to characterize the glycan structure. The MS2 derived ions at m/z 512.20 and m/z 803.29 demonstrated that the Fuc residue was attached to either of the antenna (as opposed to attachment to the core GlcNAc) and in line with a sialylated Lewis x-type fucose. Also, a manual analysis of N-glycopeptides sharing the same peptide showed the presence of a difucosylated glycoform (Supplementary Fig. S1) further demonstrating that one Fuc is attached to one of the antennae (see discussion regarding possible fucose migrations below).
Additionally, the ion at m/z 1186.58 showed that a HexNAc was attached to the innermost Man residue in line with a bisecting GlcNAc. The intense MS2 ion at m/z 657.24 (Fig. 1A) was selected for further fragmentation (MS3) providing the HexNAc oxonium ions at m/z 204 and m/z 366; and the Neu5Ac oxonium ions at m/z 274 and m/z 292. The relative intensities of ions at m/z 204 and 366 vs those of m/z 274 and 292 were then used to distinguish between α2-3- and α2-6-linked Neu5Ac (Pett et al. 2018). Since the sum of the m/z intensities for the Neu5Ac oxonium ions were higher than that of the HexNAc oxonium ions, this result showed that the Neu5Ac was linked α2-3- to the penultimate Gal residue. In summary, this bisected bi-antennary and di-fucosylated glycopeptide had two α2-3-linked Neu5Ac residues and an overall structure as shown boxed in Fig. 1A (Structure 21 in Supplementary Table S1).
The same oxonium ions, derived through MS2 at NCEs of 30% and 40%, of N-glycopeptides with (Neu5Ac)2(Hex)5(HexNAc)4Fuc0-1 and (Neu5Ac)3(Hex)6(HexNAc)5Fuc0-1 compositions, thus carrying one Neu5Ac per antenna but no bisecting GlcNAc, were also used to distinguish between α2-3- and α2-6-linked Neu5Ac. For instance, a disialo complex biantennary N-glycopeptide from Signal recognition particle receptor subunit beta (R0JEC8_ANAPL, Structure 10 in Supplementary Table S1) was identified in mallard lung and colon (Fig. 1C). The m/z 100–400 region of the expanded MS2 spectrum (Fig. 1D) showed the presence of the HexNAc and Neu5Ac oxonium ions, and their relative intensities were used to identify the Neu5Acα2-6 linkages of the sialic acids (Pett et al. 2018).
The numbers of N-glycopeptide identities terminated with Neu5Acα2-3Gal or Neu5Acα2-6Gal were similar for all the mallard tissues studied except for ileum where there was a dominance of Neu5Acα2-3Gal (Fig. 4 and Supplementary Table S1). Disialo glycoforms carrying one Neu5Acα2-3Gal and one Neu5Acα2-6Gal terminus were detected in a few cases. Importantly, complex sialylated N-glycans including fucosylated and sialylated Lewis structures were detected in all tissue samples from mallards (Supplementary Table S1).
The O-glycopeptide analyses of mallard samples identified 11 glycopeptides from 11 proteins (Supplementary Table S2) carrying both mono- and disialo core 1 glycans as exemplified by the MS2 analyses of glycopeptides from fibrinogen beta chain (A0A8B9SHX9_ANAPL) and dystroglycan 1 (A0A493T5Q5_ANAPL) (Fig. 2A and B, respectively). For the Neu5AcHexHexNAc composition, Neu5Acα2-6GalNAc linkage could in this case also be determined based on the low intensity of the m/z 274.09 and m/z 292.10 ions compared to the m/z 204.09 and m/z 366.14 ions (Pett et al. 2018) and the additional presence of a disaccharide ion at m/z 495.18 (Fig. 2A). When Neu5Acα2-3Gal was present (with or without an additional Neu5Acα2-6GalNAc) the relative intensities of the m/z 274.09 and m/z 292.10 ions increased characteristically (Fig. 2B).
The O-glycopeptides detected in mallards had core 1 structures (Galβ1-3GalNAcα1-O-Ser/Thr) with Neu5Acα2-6-linked to the GalNAc residue for secreted glycoproteins and Neu5Acα2-3-linked to the Gal, or both to the Gal and to the GalNAc residues, for membrane glycoproteins (Fig. 2). In general, there was a dominance of sialylated N-glycans compared to O-glycans in most tissues except for the lungs that had similar number of identities for both types of glycans.
The tufted duck tissues
Also in tufted ducks, glycopeptides of both membrane and secreted proteins terminated with Neu5Acα2-3Gal or Neu5Acα2-6Gal residues were identified although membrane glycoproteins were less frequently detected in this species (Figs. 4 and 5B and Supplementary Table S1). In total, 77 unique N-glycopeptides comprising 33 proteins; and 61 O-glycopeptides comprising 43 proteins were identified (Supplementary Table S3). Approximately equal ratios of the number of sialylated N- vs. O-glycan identities were detected in all tissues except for ileum that had dominance of N-glycan structures. Fucosylated and sialylated Lewis structures were detected in trachea and lung of the tufted duck. In trachea, there was also evidence of Neu5Acα2-3Gal terminated structures with elongated antennae containing poly-LacNAc (LacNAc)2 disaccharides (Structures 34 and 36, Supplementary Table S1). The Neu5Acα2-3Gal epitope was detected in all tissues, with an accentuation in trachea and lung (Fig. 4). Neu5Acα2-6Gal was detected in all tissues and in contrast to the mallards, membrane proteins in the tufted duck colon only carried Neu5Acα2-6Gal. N-glycan biantennary structures with both Neu5Acα2-3Gal and Neu5Acα2-6Gal termini were only found in trachea of this species.
The chicken tissues
Also in chickens, glycopeptides originating from both membrane and secreted glycoproteins terminating with Neu5Acα2-3Gal and/or Neu5Acα2-6Gal were identified (Figs. 4 and 5C and Supplementary Table S1) although the secreted glycoproteins dominated also in this species. In total, 131 unique N-glycopeptides comprising 66 proteins; and 49 O-glycopeptides comprising 33 proteins were identified (Supplementary Table S4). Secreted glycoproteins were preferentially found in the lung tissue with Neu5Acα2-6Gal termini on one or both antennas. Fucosylated and sialylated Lewis glycans were preferentially found in trachea on monosialylated glycans and in colon preferentially on disialylated glycans (Supplementary Table S1). Neu5Acα2-3Gal structures were found in all tissue samples from chicken on both biantennary and triantennary glycans, with or without a bisecting GlcNAc residue. A similar tissue distribution was found for Neu5Acα2-6Gal structures, although they were relatively more enriched in the lung samples (Fig. 4).
Tissues of all species compared
Comparing expressed glycopeptides terminating with α2-3- or α2-6-linked Neu5Ac in different organs from the three species revealed both differences and similarities (Fig. 4, Supplementary Table S1). Focusing only on the Neu5Ac isomers, glycopeptides containing a single α2-3- linked Neu5Ac were found in all tissues in all species except in tufted duck colon and mallard trachea. Glycopeptides containing a single α2-6- linked Neu5Ac were found in all tissues in all species except in mallard lung and ileum. Biantennary glycans containing two α2-3- or two α2-6-linked Neu5Ac were found in all tissues except tufted duck ileum that did not express biantennary glycans with two α2-3-linked Neu5Ac. Biantennary structures with combined α2-3 and α2-6-linked Neu5Ac and triantennary sialylated structures were less common, but over-represented in tufted duck trachea (Fig. 4). In general, α2-6-linked Neu5Ac residues were more commonly detected on secreted glycoproteins than on membrane glycoproteins in all three species. In mallards, there was a clear pattern that α2-3-linked Neu5Ac were expressed on membrane proteins and α2-6-linked Neu5Ac were mainly expressed on secreted proteins in all tissues. However, in tufted ducks or chickens this pattern was not as obvious and could only partly be seen in the lung of these species. A few sialylated N-glycans were found to be present in several tissues in all three species (Fig. 6 and Supplementary Table S6). However, only one; the biantennary di-α2-6 sialylated structure N4H5SA2_2-6; 2-6 was found in all tissues in all species (Supplementary Table S6). This structure was also found on the largest number of different proteins, of which the majority are secreted (4 different proteins in mallards, 12 in tufted ducks and 33 in chickens (Fig. 5)). Shared and unique N-glycopeptides in the three species are displayed in (Supplementary Figure S3 and Supplementary Table S7). The glycoproteins identified in the glycoproteomic approach were often specific to each species and of all glycoproteins identified, only 3 were found in all three species. These were identified as Transmembrane 9 super family member, Complement factor C3 and Lumican (Supplementary Fig. S4 and Supplementary Table S8).
Fig. 6.
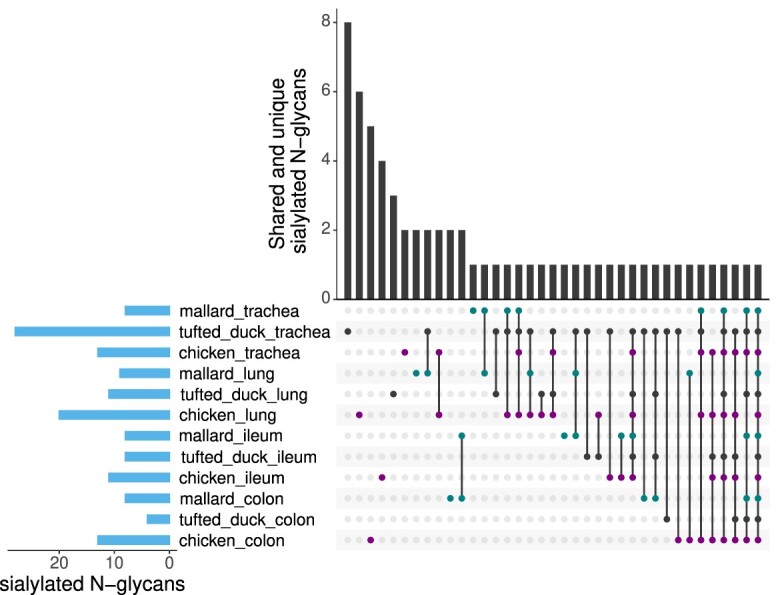
Shared and unique avian sialylated N-glycans. The UpSet plot indicates the intersection of sialylated N-glycans detected in trachea, lung, ileum and colon of mallard, tufted duck and chicken. Vertical bars visualize the number of shared and unique sialylated N-glycans to each species and tissue type. Horizontal bars represent the total number of sialylated N-glycans found in each tissue and species. The nature of a given intersection is indicated by the dots below the bar plot, with mallard shown in green, tufted duck in black and chicken in purple. For instance, the eight sialylated N-glycans in the first column were only detected in trachea of tufted duck, while the single sialylated N-glycan in the last column was detected in all tissues of all species. For information on which specific sialylated N-glycans that were shared or unique between species and tissue types, see Supplementary Table S6.
In summary, sialylated N-glycoproteins were identified in all the four studied tissues from all the three bird species. The total number of sialylated glycopeptide spectral matches that were found, irrespective of their Neu5Acα2-3/Neu5Acα2-6 status, are displayed for the individual birds and tissues in Supplementary Fig. S2A. Overall, the relative distribution of sialylation ranged from 1%–42% of all the glycoforms identified in each tissue (Supplementary Fig. S2A). A Byonic cut-off score of >300 was used, which produced false discovery rates of 0.42% for the complex type N-glycopeptides across the bird species and tissues, demonstrating a good reliability for the glycopeptide identities (Supplementary Table S5). Further, for sialylated N-glycopeptides, where the Neu5Ac α2-3 / α2-6 status could be determined, the fragmentation spectra were manually investigated to define the glycan structures according to the verification criteria presented in the materials and methods section.
Identification of oligomannose 6-phosphorylated (Man-6P) N-glycopeptides
Through further inspection of the LC-MS/MS data, we noted the presence of intense reoccurring ions at m/z 243.03 and m/z 405.08 corresponding to the masses of [Hex+phosphate+H]+ and [(Hex)2+phosphate+H]+, respectively (Fig. 3A). This would be in line with Man-6P N-glycopeptides (Čaval et al. 2019) and is of particular interest as such structures recently were proposed to serve as receptors for IAVs (Byrd-Leotis et al. 2019a; Byrd-Leotis et al. 2019b). Thus, the compositions (Hex)3-10(HexNAc)2(phosphate)1-2 were included in follow-up Byonic analyses. In addition, (Hex)5-10(HexNAc)3-4(phosphate)1-2 glycoforms were also observed and included in the analysis (Fig. 3B). The observed MS2 fragmentation patterns, including a particularly intense HexNAc ion at m/z 204.09 (Fig. 3B), in the absence of other prominent B-ions (suggestive of an additional HexNAc), is in line with a relatively labile phosphate-bridged terminal GlcNAc. Similar to the situation for the sialylated glycans, we identified Man-6P N-glycans in all tissues from all three bird species. In total, 132, 183 and 154 unique Man-6P N-glycopeptides were identified from 46, 53 and 54 glycoproteins from mallard, tufted duck and chicken, respectively (Supplementary Fig. S5 and Supplementary Tables S2–S4). Although several proteins carrying Man-6P N-glycan modifications in these birds were not annotated in UniProt as to their subcellular localizations, many of their human analogs are indeed annotated as endosomal/lysosomal proteins. This fits very well with the established receptor mediated sorting function of this particular glycan modification (Dahms et al. 2022) and may be of importance for the viral entry and endosomal escape mechanisms of AIV (Byrd-Leotis et al. 2019b). The total number of Man-6P glycopeptide identities in the samples of individual birds are displayed in Supplementary Fig. S2B which also illustrates a relative distribution of these Man-6P glycoforms vs all glycoforms varying between 1 and 11%.
Discussion
Species-specific differences in the glycan structures serving as receptors for IAV have been proposed as important barriers for interspecies transmission. Specifically, the receptor tropism for α2-3- vs. α2-6-linked Neu5Ac has been considered a hallmark for bird adapted vs. human adapted IAVs (Rogers and Paulson 1983; Rogers and D'Souza 1989; Ito et al. 1998). Glycomics and glycoproteomics experiments have earlier been performed in humans and pigs, providing extensive descriptions of expressed putative influenza A virus receptors in these animals (Walther et al. 2013; Byrd-Leotis et al. 2014; Byrd-Leotis et al. 2019b). In this study, we present site-specific information on N-glycans of glycoproteins obtained from epithelial cells of trachea, lung, ileum and colon of mallards, tufted ducks, and chickens, and discuss our results in relation to previous work.
Mallards are regarded as a key reservoir for AIV, as they carry almost all known AIV subtypes (Jourdain et al. 2010; Latorre-Margalef et al. 2014) and show limited signs of disease even when infected with some HPAIV strains (Pantin-Jackwood et al. 2016). In contrast, HPAIV cause significant morbidity and mortality in other bird species including genetically related ducks, such as tufted ducks (Keawcharoen et al. 2008; Kleyheeg et al. 2017), and in chickens (Wiethoelter et al. 2015). Although both oligomannose, hybrid and complex type glycan structures were identified, special focus was given to sialylated structures (Supplementary Table S1). Not only the number of Sia residues but also their identities and their linkage positions were analyzed by tandem mass spectrometry, using diagnostic oxonium ions obtained through higher-energy collision dissociation (HCD) at a lower energy setting (Fig. 1A). This information is critical since the more complex the structures become, the more glycan isoforms are theoretically possible to conclude from precursor ions, with the same mass.
A well-known complication with positive mode ionization and collisional activation of underivatized glycans and glycopeptides is that intra-molecular migration of fucose, and other monosaccharides, may obscure the MS/MS analysis (Wuhrer et al. 2011; Rath et al. 2018; Lettow et al. 2023). However, the presence of significant ions at m/z 512.20 [Gal(Fuc)GlcNAc] and m/z 803.29 [Neu5AcGal(Fuc)GlcNAc] having >10% relative abundance vs. m/z 366.14 [HexHexNAc] using the lower energy setting (NCE 20) make the identification of sialyl-Lewis x-like structures confident for these samples (Fig. 1A and Supplementary Fig. S1). On the other hand, weaker ion peaks of [peptide+GlcNAc+Fuc] at <5% relative abundance vs. [peptide+GlcNAc] were occasionally visible together with prominent m/z 512.20 and m/z 803.29 ion peaks also when the identified glycopeptide only included one Fuc residue. This may be due to fucose migration from the antenna to the core but can also be due to co-eluting isobaric glycoforms. In line with the latter possibility, we do not detect any presence of [peptide+GlcNAc+Fuc] ions for multiply fucosylated N-glycopeptides using similar LC-MS/MS settings (unpublished data). Furthermore, for several identified monofucosylated N-glycopeptides from lumican found in the present study, a core Fuc was clearly present but ions at m/z 512.20 and m/z 803.29 were also present at <5% relative abundances vs. m/z 366.14 and m/z 657.23, and their minor presence were discarded as Fuc migrations (Supplementary Fig. S6). These results suggest that Fuc migration from the core to the antenna indeed is present at these MS/MS settings but only at minor relative abundances.
As we have not performed glycomics on released glycans from glycoproteins or from glycolipids, it should be noted that the glycan structures reported only include peptide-bound glycans and that our quantitative estimates sometimes are based on a relatively low number of identities. However, and more importantly, our glycopeptide analysis strategy allowed us to structurally characterize glycans in a site-specific manner and thereby differentiate between membrane-bound and secreted glycoproteins which is essential for the interpretation of their role as putative receptors for IAV, and this direct information would not have become available using glycomics. Indeed, our quantitative analysis was conducted based on the relative numbers of glycopeptide spectral matches for both the identified sialic acid and the Man-P containing N-glycopeptides versus all glycopeptides identified in each sample (Supplementary Fig. S2).
In the glycoproteomic analysis, we detected N-glycans containing not only Neu5Acα2-3Gal but also Neu5Acα2-6Gal in all avian tissues sampled. In contrast to earlier studies, suggesting that avian glycosylation produce shorter, less complex glycans compared to human glycosylation (Bewley 2008; Walther et al. 2013), we found many di-, tri- and tetraantennary sialylated glycans as well as some extended, poly-LacNAc (Lac-NAc)2 sialylated glycan structures in the avian tissues analyzed (Supplementary Table S1). In studies of the human lung glycome, extended sialylated N-glycan structures have been identified that contained long poly-LacNAc structures (Jia et al. 2020). However, to our knowledge such structures have not previously been reported in birds. In a shotgun glycan micro array approach, (Byrd-Leotis et al. 2019b) it was demonstrated that the most abundantly bound receptors from the human lung, recognized by influenza viruses from humans, pigs and birds, was mainly constituted of bi- or tri-antennary sialylated and sometimes fucosylated glycans with α2-3- and/or α2-6-linked sialic acids. Hence, these are structures similar to those we found in the birds of this study and similar to what others have found in pig respiratory tract tissues (Byrd-Leotis et al. 2014).
Sulfated 3’SLN and sulfated S-Lex have been suggested as main receptors for H7 AIVs, H5 AIVs isolated from chickens, and gull-specific H13 and H16 viruses (Gambaryan et al. 2005; Gambaryan et al. 2008; Gambaryan et al. 2012; Gambaryan et al. 2018). Furthermore, it has been proposed that acquisition of binding ability to sulfated and/or fucosylated structures (in contrast to linear Neu5Acα2-3Gal containing receptors) represents an adaptation of duck viruses enabling them to cross the species barrier and readily infect chickens (Hiono et al. 2014; Kikutani et al. 2020; Kobayashi et al. 2022). However, in our study we could not identify sulfated sialylated glycopeptides in any of the investigated avian samples (including chickens), challenging the biological relevance of these structures as major AIV receptors. In contrast, as described above we did find fucosylated sialylated structures in all organs of all birds, including mallards. Hence, our studies do not corroborate that sulfated and/or fucosylated structures represent an important barrier for transmission of AIV between ducks and chickens. Instead, they are in line with our previous findings that mallard originating AIVs, of several subtypes, bind well to fucosylated sialylated structures (Verhagen et al. 2021). It should be noted however, that in this study we have only looked at glycoproteins and it might be possible that sulfated sialylated structures are expressed on glycolipids in these bird species.
Tropism for N-glycolylneuraminic acid (Neu5Gc) in wild type AIVs is extremely rare and this Sia structure has not been found in birds (Broszeit et al. 2019). Indeed, Neu5Gc was not detected in our glycoproteomic analysis in any of the avian species. Also, the presence of O-acetyl Neu5Ac was not detected. This hydrolysis-sensitive modification may be lost during the mild alkaline conditions used, although it is unlikely in this case since the Sia structure was abundantly detected in another study using similar preparation conditions (Mirgorodskaya et al. 2022). Although influenza A viruses have been shown to bind O-acetyl Neu5Ac and this Sia isoform has been found in many animal species, its role in the pathogenesis and zoonotic ecology of IAV is poorly understood (Higa et al. 1985; Park 2019).
However, we identified Man-6P N-glycopeptides in all three bird species. This is of considerable interest as IAVs (of avian, human, and swine origin) were recently described to bind human lung-derived phosphorylated non-sialylated glycans (particularly Man-6P containing glycans) in a Sia-independent manner (Byrd-Leotis et al. 2019b). We identified these N-glycopeptides on predicted lysosomal enzymes, including e.g. carboxypeptidase and aminopeptidase. It can be speculated that IAV binding to phosphorylated glycans takes place during a later phase than the cell membrane binding and entry, facilitating the virus uncoating process in endosomes by targeting lysosomal enzymes (Tian et al. 2019).
In mallards, we could observe a clear pattern in all tissues where α2-3-linked Neu5Ac were expressed on membrane proteins whereas α2-6-linked Neu5Ac were expressed mainly on secreted proteins. A similar pattern could only be observed in the chicken lung and to less extent in the tufted duck lung, whereas in the tufted duck colon α2-6-linked Neu5Ac was the only Sia found of membrane proteins. With the hypothesis in mind, that membrane bound Sia facilitate IAV infection and secreted Sia inhibits infection by serving as decoys for the virus (Olofsson et al. 2005), this observation could possibly explain why avian IAV infection in mallards take place both in the respiratory and intestinal tract, whereas in chickens IAV infection is more restricted to the respiratory tract and that in tufted ducks IAV infection is generally rare (Webster et al. 1978; Ellström et al. 2008; Lu and Zhao 2015; Bergervoet et al. 2019; Roy Chowdhury et al. 2019; Kye et al. 2021; Naguib et al. 2023). However, more detailed characterization of all expressed Sia in the three species together with infection experiments are needed to verify this hypothesis. For N-glycopeptides obtained from secreted glycoproteins Neu5Acα2-6Gal was the dominating glycoform in all three species. This is in line with what is known from humans and some domestic animals from which serum has, through its inhibitory capacity, been used as a surrogate marker for AIV receptor specificity (Ryan-Poirier et al. 1998).
Taken together, the results not only indicate great similarities in IAV receptor expression between the three bird species, but also demonstrate that avian glycosylation and the expression of glycan structures in avian tissues is more similar to that seen in mammalian tissues than what was previously anticipated. There are presently no systematic studies on host glycosylation response to influenza virus infection, neither acutely nor lately post infection, but Heindel et al. (Heindel et al. 2020) recently reported on changes in glycosylation in lung tissue of H1N1 infected ferrets. Although there was a large variation between individual animals as to glycophenotypes and infection kinetics, the authors could conclude that upon day 8 d.p.i. the level of oligomannose structures correlated with severity of the lung disease. Long term effects (>14 days) were not studied. In our analysis of glycoprotein expression in different tissues, we did not study experimentally infected animals. However, whether and how the glycoproteins of birds, even on an individual basis, would change in response to an earlier or an on-going AIV-infection should be the focus of future studies.
Conclusion
In conclusion, our glycoproteomic analysis of the avian respiratory and intestinal tracts clearly demonstrated that birds do produce complex α2-3- and α2-6-linked Neu5Ac N-glycans including α2-3-linked sialyl Lewis structures, as well as both N- and O- glycans terminated with both α2-3- and α2-6-linked Neu5Ac. This is contrary to the current dogma, whereby complex Neu5Ac containing structures are believed to be mammalian specific, and suggests that available receptors in mammals and birds are more closely related than previously thought. Avian influenza viruses have the capacity to attach to both α2-3- and α2-6-linked Neu5Ac (Rogers and Paulson 1983; Eriksson et al. 2018; Verhagen et al. 2021) and the affinity seems to partly depend on the glycan structure penultimate to the terminal Sia and the complexity of the glycan structure (Gambaryan et al. 2005; Stevens et al. 2006; Wang et al. 2013; Gambaryan et al. 2018). Furthermore, a recent study suggests that α2-6-linked sialoglycans can contribute to binding and cell entry of IAV with tropism for α2-3-linked Sia and vice versa for IAV with tropism for α2-6-linked Sia via hetero-multivalent interactions (Liu et al. 2022). Our findings in this study adds to the complexity of receptor incompatibility as a barrier for interspecies transmission of IAV and suggest that such transmission cannot only be explained by the presence or absence of specific glycan structures or the α2-3- v.s. α2-6-linkage of terminal sialic acids.
Supplementary Material
Acknowledgments
We acknowledge Dr. Roy Francis (NBIS, Science for Life Laboratory) for helpful input on bioinformatics. We are grateful of Inga-Britt and Arne Lundbergs Forskningsstiftelse for the donation of the Orbitrap Fusion Tribrid MS instrument.
Contributor Information
Jonas Nilsson, Department of Laboratory Medicine, University of Gothenburg, Sahlgrenska University Hospital, Vita Stråket 12, Gothenburg SE-413 45, Sweden; Laboratory of Clinical Chemistry, Sahlgrenska University Hospital, Bruna Stråket 16, Gothenburg SE-413 45, Sweden; Proteomics Core Facility, University of Gothenburg, Sahlgrenska Academy, Medicinaregatan 9E, Gothenburg SE-405 30, Sweden.
Per Eriksson, Zoonosis Science Center, Department of Medical Sciences, Husargatan 3, Uppsala University, Uppsala, SE-75185, Sweden.
Mahmoud M Naguib, Zoonosis Science Center, Department of Medical Biochemistry and Microbiology, Husargatan 3, Uppsala University, Uppsala, SE-75237, Sweden.
Elinor Jax, Department of Migration, Max Planck Institute of Animal Behavior, Am Obstberg 1, Radolfzell, Baden-Württemberg DE-78315, Germany.
Carina Sihlbom, Proteomics Core Facility, University of Gothenburg, Sahlgrenska Academy, Medicinaregatan 9E, Gothenburg SE-405 30, Sweden.
Britt-Marie Olsson, Proteomics Core Facility, University of Gothenburg, Sahlgrenska Academy, Medicinaregatan 9E, Gothenburg SE-405 30, Sweden.
Åke Lundkvist, Zoonosis Science Center, Department of Medical Biochemistry and Microbiology, Husargatan 3, Uppsala University, Uppsala, SE-75237, Sweden.
Björn Olsen, Zoonosis Science Center, Department of Medical Sciences, Husargatan 3, Uppsala University, Uppsala, SE-75185, Sweden.
Josef D Järhult, Zoonosis Science Center, Department of Medical Sciences, Husargatan 3, Uppsala University, Uppsala, SE-75185, Sweden.
Göran Larson, Department of Laboratory Medicine, University of Gothenburg, Sahlgrenska University Hospital, Vita Stråket 12, Gothenburg SE-413 45, Sweden; Laboratory of Clinical Chemistry, Sahlgrenska University Hospital, Bruna Stråket 16, Gothenburg SE-413 45, Sweden.
Patrik Ellström, Zoonosis Science Center, Department of Medical Sciences, Husargatan 3, Uppsala University, Uppsala, SE-75185, Sweden.
Author contributions
Patrik Ellström, Josef D. Järhult, Mahmoud M. Naguib, Per Eriksson, Jonas Nilsson, Göran Larson (Conceptualization), Mahmoud M. Naguib, Per Eriksson, Patrik Ellström, Josef D. Järhult (Animal experiment), Jonas Nilsson, Per Eriksson, Elinor Jax, Carina Sihlbom, Britt-Marie Olsson, Göran Larson (Glycoproteomic analysis), Per Eriksson, Jonas Nilsson, Elinor Jax (Data analysis and visualization), Per Eriksson, Mahmoud M. Naguib, Patrik Ellström, Jonas Nilsson, Göran Larson (Writing—original draft), and Mahmoud M. Naguib, Per Eriksson, Patrik Ellström, Elinor Jax, Josef D. Järhult, Åke Lundkvist, Björn Olsen, Jonas Nilsson, Göran Larson (Writing—review & editing)
CRediT author statement
Jonas Nilsson (Conceptualization [equal], Data curation [lead], Formal analysis [lead], Methodology [lead], Resources [equal], Software [equal], Validation [equal], Visualization [equal], Writing—original draft [equal], Writing—review & editing [equal]), Per Eriksson (Data curation [equal], Formal analysis [equal], Methodology [equal], Validation [equal], Visualization [equal], Writing—original draft [equal]), Mahmoud M. Naguib (Investigation [equal], Methodology [equal], Writing—original draft [equal]), Elinor Jax (Formal analysis [equal], Investigation [equal], Visualization [equal], Writing—review & editing [supporting]), Carina Sihlbom (Investigation [equal], Methodology [equal]), Britt-Marie Olsson (Investigation [equal], Methodology [equal]), Åke Lundkvist (Resources [equal], Supervision [equal]), Björn Olsen (Conceptualization [equal], Resources, Supervision), Josef D. Järhult (Investigation [equal], Resources [equal]), Goran Larson (Conceptualization [equal], Data curation [equal], Methodology [equal], Resources [equal], Supervision [equal], Validation [equal], Writing—original draft [equal], Writing—review & editing [equal]), Patrik Ellström (Conceptualization [lead], Data curation [equal], Investigation [equal], Methodology [equal], Resources [equal], Supervision [equal], Validation [equal], Writing—original draft [equal], Writing—review & editing [lead]).
Funding
Funding for this study was obtained from the Swedish Research Council (2016-02606 to JJ, 2016-02596 to BO, and 2017-00955 to GL). The glycoproteomic analysis was performed at the Proteomics Core Facility, Sahlgrenska academy, Gothenburg University, with financial support from SciLifeLab and BioMS. The Zoonosis Science Center is supported by the SciLifeLab PLP-1 ZSC grant - National core facility for Pandemic Preparedness.
Conflict of interest statement: We declare no conflict of interest for any of the authors.
Data availability
The authors declare that all data supporting the findings of this study are available within the article, including its supplementary information files, or are available from the authors upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2019) partner repository with the dataset identifier PXD036263.
Reviewer account details:
Username: reviewer_pxd036263@ebi.ac.uk
Password: CM60Uki5.
References
- Alqazlan N, Emam M, Nagy É, Bridle B, Sargolzaei M, Sharif S. Transcriptomics of chicken cecal tonsils and intestine after infection with low pathogenic avian influenza virus H9N2. Sci Rep. 2021:11(1):20462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990:215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Arike L, Seiman A, van der Post S, Rodriguez Piñeiro AM, Ermund A, Schütte A, Bäckhed F, Johansson MEV, Hansson GC. Protein turnover in epithelial cells and mucus along the gastrointestinal tract is coordinated by the spatial location and microbiota. Cell Rep. 2020:30(4):1077–1087.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergervoet SA, Germeraad EA, Alders M, Roose MM, Engelsma MY, Heutink R, Bouwstra R, Fouchier RAM, Beerens N. Susceptibility of chickens to low pathogenic avian influenza (LPAI) viruses of wild bird- and poultry-associated subtypes. Viruses. 2019:11(11):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley CA. Illuminating the switch in influenza viruses. Nat Biotechnol. 2008:26(1):60–62. [DOI] [PubMed] [Google Scholar]
- Blix FG, Gottschalk A, Klenk E. Proposed nomenclature in the field of neuraminic and sialic acids. Nature. 1957:179(4569):1088. [DOI] [PubMed] [Google Scholar]
- Bröjer C, Ågren EO, Uhlhorn H, Bernodt K, Mörner T, Désirée, Jansson S, Mattsson R, Zohari S, Thorén P, et al. Pathology of natural highly pathogenic avian influenza H5N1 infection in wild tufted ducks (Aythya fuligula). J Vet Diagn Investig. 2009:21(5):579–587. [DOI] [PubMed] [Google Scholar]
- Broszeit F, Tzarum N, Zhu X, Nemanichvili N, Eggink D, Leenders T, Li Z, Liu L, Wolfert MA, Papanikolaou A, et al. N-Glycolylneuraminic acid as a receptor for influenza a viruses. Cell Rep. 2019:27(11):3284–3294.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd-Leotis L, Liu R, Bradley KC, Lasanajak Y, Cummings SF, Song X, Heimburg-Molinaro J, Galloway SE, Culhane MR, Smith DF, et al. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc Natl Acad Sci U S A. 2014:111(22):E2241–E2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd-Leotis L, Gao C, Jia N, Mehta AY, Trost J, Cummings SF, Heimburg-Molinaro J, Cummings RD, Steinhauer DA. Antigenic pressure on H3N2 influenza virus drift strains imposes constraints on binding to Sialylated receptors but not phosphorylated glycans. J Virol. 2019a:93(22):e01178–e01119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd-Leotis L, Jia N, Dutta S, Trost JF, Gao C, Cummings SF, Braulke T, Müller-Loennies S, Heimburg-Molinaro J, Steinhauer DA, et al. Influenza binds phosphorylated glycans from human lung. Sci Adv. 2019b:5(2):eaav2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čaval T, Zhu J, Tian W, Remmelzwaal S, Yang Z, Clausen H, Heck AJR. Targeted analysis of lysosomal directed proteins and their sites of mannose-6-phosphate modification. Mol Cell Proteomics. 2019:18(1):16–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa T, Chaves AJ, Valle R, Darji A, van Riel D, Kuiken T, Majo N, Ramis A. Distribution patterns of influenza virus receptors and viral attachment patterns in the respiratory and intestinal tracts of seven avian species. Vet Res. 2012:43(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahms N, Braulke T, Varki A. P-type lectins. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, et al., editors. Essentials of Glycobiology [Internet]. 4th ed. Cold Spring Harbor Laboratory Press (NY); 2022. [Google Scholar]
- Ellström P, Latorre-Margalef N, Griekspoor P, Waldenström J, Olofsson J, Wahlgren J, Olsen B. Sampling for low-pathogenic avian influenza a virus in wild mallard ducks: oropharyngeal versus cloacal swabbing. Vaccine. 2008:26(35):4414–4416. [DOI] [PubMed] [Google Scholar]
- Ellström P, Jourdain E, Gunnarsson O, Waldenstrom J, Olsen B. The "human influenza receptor" Neu5Ac alpha 2,6Gal is expressed among different taxa of wild birds. Arch Virol. 2009:154(9):1533–1537. [DOI] [PubMed] [Google Scholar]
- Eriksson P, Lindskog C, Engholm E, Blixt O, Waldenstrom J, Munster V, Lundkvist A, Olsen B, Jourdain E, Ellstrom P. Characterization of avian influenza virus attachment patterns to human and pig tissues. Sci Rep. 2018:8(1):12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca M, Stallknecht DE, Howerth EW. Expression and distribution of sialic acid influenza virus receptors in wild birds. Avian Pathol. 2013:42(1):60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambaryan A, Yamnikova S, Lvov D, Tuzikov A, Chinarev A, Pazynina G, Webster R, Matrosovich M, Bovin N. Receptor specificity of influenza viruses from birds and mammals: new data on involvement of the inner fragments of the carbohydrate chain. Virology. 2005:334(2):276–283. [DOI] [PubMed] [Google Scholar]
- Gambaryan AS, Tuzikov AB, Pazynina GV, Desheva JA, Bovin NV, Matrosovich MN, Klimov AI. 6-sulfo sialyl Lewis X is the common receptor determinant recognized by H5, H6, H7 and H9 influenza viruses of terrestrial poultry. Virol J. 2008:5(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambaryan AS, Matrosovich TY, Philipp J, Munster VJ, Fouchier RA, Cattoli G, Capua I, Krauss SL, Webster RG, Banks J, et al. Receptor-binding profiles of H7 subtype influenza viruses in different host species. J Virol. 2012:86(8):4370–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambaryan AS, Matrosovich TY, Boravleva EY, Lomakina NF, Yamnikova SS, Tuzikov AB, Pazynina GV, Bovin NV, Fouchier RAM, Klenk HD, et al. Receptor-binding properties of influenza viruses isolated from gulls. Virology. 2018:522:37–45. [DOI] [PubMed] [Google Scholar]
- Gottschalk A. Carbohydrate residue of a urine mucoprotein inhibiting influenza virus haemagglutination. Nature. 1952:170(4329):662–663. [DOI] [PubMed] [Google Scholar]
- Gottschalk A. The influenza virus enzyme and its mucoprotein substrate. Yale J Biol Med. 1954:26(5):352–364. [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016:32(18):2847–2849. [DOI] [PubMed] [Google Scholar]
- Heindel DW, Koppolu S, Zhang Y, Kasper B, Meche L, Vaiana CA, Bissel SJ, Carter CE, Kelvin AA, Elaish M, et al. Glycomic analysis of host response reveals high mannose as a key mediator of influenza severity. Proc Natl Acad Sci U S A. 2020:117(43):26926–26935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa HH, Rogers GN, Paulson JC. Influenza virus hemagglutinins differentiate between receptor determinants bearing N-acetyl-, N-glycollyl-, and N,O-diacetylneuraminic acids. Virology. 1985:144(1):279–282. [DOI] [PubMed] [Google Scholar]
- Hiono T, Okamatsu M, Nishihara S, Takase-Yoden S, Sakoda Y, Kida H. A chicken influenza virus recognizes fucosylated α2,3 sialoglycan receptors on the epithelial cells lining upper respiratory tracts of chickens. Virology. 2014:456-457:131–138. [DOI] [PubMed] [Google Scholar]
- Horimoto T, Kawaoka Y. Pandemic threat posed by avian influenza a viruses. Clin Microbiol Rev. 2001:14(1):129–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Couceiro JNSS, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, et al. Molecular basis for the generation in pigs of influenza a viruses with pandemic potential. J Virol. 1998:72(9):7367–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Byrd-Leotis L, Matsumoto Y, Gao C, Wein AN, Lobby JL, Kohlmeier JE, Steinhauer DA, Cummings RD. The human lung glycome reveals novel glycan ligands for influenza a virus. Sci Rep. 2020:10(1):5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain E, Gunnarsson G, Wahlgren J, Latorre-Margalef N, Bröjer C, Sahlin S, Svensson L, Waldenström J, Lundkvist A, Olsen B. Influenza virus in a natural host, the mallard: experimental infection data. PLoS One. 2010:5(1):e8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain E, van Riel D, Munster VJ, Kuiken T, Waldenström J, Olsen B, Ellström P. The pattern of influenza virus attachment varies among wild bird species. PLoS One. 2011:6(9):e24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JM, Veguilla V, Belser JA, Maines TR, Van Hoeven N, Pappas C, Hancock K, Tumpey TM. The public health impact of avian influenza viruses. Poult Sci. 2009:88(4):872–879. [DOI] [PubMed] [Google Scholar]
- Keawcharoen J, van Riel D, van Amerongen G, Bestebroer T, Beyer WE, van Lavieren R, Osterhaus AD, Fouchier RA, Kuiken T. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1). Emerg Infect Dis. 2008:14(4):600–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikutani Y, Okamatsu M, Nishihara S, Takase-Yoden S, Hiono T, de Vries RP, McBride R, Matsuno K, Kida H, Sakoda Y. E190V substitution of H6 hemagglutinin is one of key factors for binding to sulfated sialylated glycan receptor and infection to chickens. Microbiol Immunol. 2020:64(4):304–312. [DOI] [PubMed] [Google Scholar]
- Kleyheeg E, Slaterus R, Bodewes R, Rijks JM, Spierenburg MAH, Beerens N, Kelder L, Poen MJ, Stegeman JA, Fouchier RAM, et al. Deaths among wild birds during highly pathogenic avian influenza a(H5N8) virus outbreak, the Netherlands. Emerg Infect Dis. 2017:23(12):2050–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D, Hiono T, Ichii O, Nishihara S, Takase-Yoden S, Yamamoto K, Kawashima H, Isoda N, Sakoda Y. Turkeys possess diverse Siaα2-3Gal glycans that facilitate their dual susceptibility to avian influenza viruses isolated from ducks and chickens. Virus Res. 2022:315:198771. [DOI] [PubMed] [Google Scholar]
- Kolarich D, Rapp E, Struwe WB, Haslam SM, Zaia J, McBride R, Agravat S, Campbell MP, Kato M, Ranzinger R, et al. The minimum information required for a glycomics experiment (MIRAGE) project: improving the standards for reporting mass-spectrometry-based glycoanalytic data. Mol Cell Proteomics. 2013:12(4):991–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. pheatmap: pretty heatmaps. 2019. R package version 1.0.12. https://cran.r-project.org/package=pheatmap.
- Kreuder Johnson C, Hitchens PL, Smiley Evans T, Goldstein T, Thomas K, Clements A, Joly DO, Wolfe ND, Daszak P, Karesh WB, et al. Spillover and pandemic properties of zoonotic viruses with high host plasticity. Sci Rep. 2015:5(1):14830–14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kye SJ, Park MJ, Kim NY, Lee YN, Heo GB, Baek YK, Shin JI, Lee MH, Lee YJ. Pathogenicity of H9N2 low pathogenic avian influenza viruses of different lineages isolated from live bird markets tested in three animal models: SPF chickens, Korean native chickens, and ducks. Poult Sci. 2021:100(9):101318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre-Margalef N, Tolf C, Grosbois V, Avril A, Bengtsson D, Wille M, Osterhaus AD, Fouchier RA, Olsen B, Waldenström J. Long-term variation in influenza a virus prevalence and subtype diversity in migratory mallards in northern Europe. Proc Biol Sci. 2014:281:20140098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettow M, Greis K, Mucha E, Lambeth TR, Yaman M, Kontodimas V, Manz C, Hoffmann W, Meijer G, Julian RR, et al. Decoding the fucose migration product during mass-spectrometric analysis of blood group epitopes. Angew Chem Int Ed Engl. 2023:62(24):e202302883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A, Gehlenborg N. Sets and intersections. Nat Methods. 2014:11(8):779–779. [Google Scholar]
- Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph. 2014:20(12):1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Han C, Wang X, Lin J, Ma M, Shu Y, Zhou J, Yang H, Liang Q, Guo C, et al. Influenza a virus receptors in the respiratory and intestinal tracts of pigeons. Avian Pathol. 2009:38(4):263–266. [DOI] [PubMed] [Google Scholar]
- Liu M, Huang LZX, Smits AA, Büll C, Narimatsu Y, van Kuppeveld FJM, Clausen H, de Haan CAM, de Vries E. Human-type sialic acid receptors contribute to avian influenza a virus binding and entry by hetero-multivalent interactions. Nat Commun. 2022:13(1):4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Zhao Q. Highly pathogenic avian influenza. Radiol Infect Dis. 2015:1:157–189. [Google Scholar]
- Mirgorodskaya E, Dransart E, Shafaq-Zadah M, Roderer D, Sihlbom C, Leffler H, Johannes L. Site-specific N-glycan profiles of α(5) β(1) integrin from rat liver. Biol Cell. 2022:114(6):160–176. [DOI] [PubMed] [Google Scholar]
- Mundt E, Gay L, Jones L, Saavedra G, Tompkins SM, Tripp RA. Replication and pathogenesis associated with H5N1, H5N2, and H5N3 low-pathogenic avian influenza virus infection in chickens and ducks. Arch Virol. 2009:154(8):1241–1248. [DOI] [PubMed] [Google Scholar]
- Naguib MM, Verhagen JH, Samy A, Eriksson P, Fife M, Lundkvist Å, Ellström P, Järhult JD. Avian influenza viruses at the wild-domestic bird interface in Egypt. Infect Ecol Epidemiol. 2019:9(1):1575687–1575687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib MM, Eriksson P, Jax E, Wille M, Lindskog C, Bröjer C, Krambrich J, Waldenström J, Kraus RHS, Larson G, et al. A comparison of host responses to infection with wild-type avian influenza viruses in chickens and tufted ducks. Microbiol Spectr. 2023:11(4):e0258622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson S, Kumlin U, Dimock K, Arnberg N. Avian influenza and sialic acid receptors: more than meets the eye? Lancet Infect Dis. 2005:5(3):184–188. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood MJ, Costa-Hurtado M, Shepherd E, DeJesus E, Smith D, Spackman E, Kapczynski DR, Suarez DL, Stallknecht DE, Swayne DE. Pathogenicity and transmission of H5 and H7 highly pathogenic avian influenza viruses in mallards. J Virol. 2016:90(21):9967–9982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SS. Post-glycosylation modification of sialic acid and its role in virus pathogenesis. Vaccines (Basel). 2019:7(4):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker BL, Thaysen-Andersen M, Solis N, Scott NE, Larsen MR, Graham ME, Packer NH, Cordwell SJ. Site-specific glycan-peptide analysis for determination of N-glycoproteome heterogeneity. J Proteome Res. 2013:12(12):5791–5800. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019:47(D1):D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pett C, Nasir W, Sihlbom C, Olsson BM, Caixeta V, Schorlemer M, Zahedi RP, Larson G, Nilsson J, Westerlind U. Effective assignment of α2,3/α2,6-sialic acid isomers by LC-MS/MS-based glycoproteomics. Angew Chem Int Ed Engl. 2018:57(30):9320–9324. [DOI] [PubMed] [Google Scholar]
- Rath CB, Schirmeister F, Figl R, Seeberger PH, Schäffer C, Kolarich D. Flagellin glycoproteomics of the periodontitis associated pathogen Selenomonas sputigena reveals previously not described O-glycans and rhamnose fragment rearrangement occurring on the glycopeptides. Mol Cell Proteomics. 2018:17(4):721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RCoreTeam . R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2022. https://www.R-project.org/. [Google Scholar]
- Rogers GN, D'Souza BL. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 1989:173(1):317–322. [DOI] [PubMed] [Google Scholar]
- Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983:127(2):361–373. [DOI] [PubMed] [Google Scholar]
- Roy Chowdhury I, Yeddula SGR, Kim SH. Pathogenicity and transmissibility of north American H7 low pathogenic avian influenza viruses in chickens and turkeys. Viruses. 2019:11(2):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan-Poirier K, Suzuki Y, Bean WJ, Kobasa D, Takada A, Ito T, Kawaoka Y. Changes in H3 influenza a virus receptor specificity during replication in humans. Virus Res. 1998:56(2):169–176. [DOI] [PubMed] [Google Scholar]
- Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006:440(7083):435–436. [DOI] [PubMed] [Google Scholar]
- Short KR, Richard M, Verhagen JH, van Riel D, Schrauwen EJA, van den Brand JMA, Mänz B, Bodewes R, Herfst S. One health, multiple challenges: the inter-species transmission of influenza A virus. One Health. 2015:1:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slemons RD, Swayne DE. Replication of a waterfowl-origin influenza virus in the kidney and intestine of chickens. Avian Dis. 1990:34(2):277–284. [PubMed] [Google Scholar]
- Stevens J, Blixt O, Paulson JC, Wilson IA. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat Rev Microbiol. 2006:4(11):857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe WB, Agravat S, Aoki-Kinoshita KF, Campbell MP, Costello CE, Dell A, Ten F, Haslam SM, Karlsson NG, Khoo KH, et al. The minimum information required for a glycomics experiment (MIRAGE) project: sample preparation guidelines for reliable reporting of glycomics datasets. Glycobiology. 2016:26(9):907–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K. The critical interspecies transmission barrier at the animal−human Interface. Trop Med Infect Dis. 2019:4(2):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK Jr, Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW, et al. The minimum information about a proteomics experiment (MIAPE). Nat Biotechnol. 2007:25(8):887–893. [DOI] [PubMed] [Google Scholar]
- Tian W, Ye Z, Wang S, Schulz MA, Van Coillie J, Sun L, Chen YH, Narimatsu Y, Hansen L, Kristensen C, et al. The glycosylation design space for recombinant lysosomal replacement enzymes produced in CHO cells. Nat Commun. 2019:10(1):1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt-Consortium . UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019:47(D1):D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, et al. Symbol nomenclature for graphical representations of glycans. Glycobiology. 2015:25(12):1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen JH, Eriksson P, Leijten L, Blixt O, Olsen B, Waldenström J, Ellström P, Kuiken T. Host range of influenza a virus H1 to H16 in Eurasian ducks based on tissue and receptor binding studies. J Virol. 2021:95(6):e01873–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T, Karamanska R, Chan RW, Chan MC, Jia N, Air G, Hopton C, Wong MP, Dell A, Malik Peiris JS, et al. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 2013:9(3):e1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chinoy ZS, Ambre SG, Peng W, McBride R, de Vries RP, Glushka J, Paulson JC, Boons GJ. A general strategy for the chemoenzymatic synthesis of asymmetrically branched N-glycans. Science. 2013:341(6144):379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Yakhno M, Hinshaw VS, Bean WJ, Copal Murti K. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology. 1978:84(2):268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiethoelter AK, Beltran-Alcrudo D, Kock R, Mor SM. Global trends in infectious diseases at the wildlife-livestock interface. Proc Natl Acad Sci U S A. 2015:112(31):9662–9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille M, Brojer C, Lundkvist A, Jarhult JD. Alternate routes of influenza a virus infection in mallard (Anas platyrhynchos). Vet Res. 2018:49(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009:6(5):359–362. [DOI] [PubMed] [Google Scholar]
- Wuhrer M, Deelder AM, van der Burgt YE. Mass spectrometric glycan rearrangements. Mass Spectrom Rev. 2011:30(4):664–680. [DOI] [PubMed] [Google Scholar]
- Yen HL, Webster RG. Pandemic influenza as a current threat. Curr Top Microbiol Immunol. 2009:333:3–24. [DOI] [PubMed] [Google Scholar]
- Yu JE, Yoon H, Lee HJ, Lee JH, Chang BJ, Song CS, Nahm S-S. Expression patterns of influenza virus receptors in the respiratory tracts of four species of poultry. J Vet Sci. 2011:12(1):7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Schorlemer M, Gomez Toledo A, Pett C, Sihlbom C, Larson G, Westerlind U, Nilsson J. Distinctive MS/MS fragmentation pathways of glycopeptide-generated oxonium ions provide evidence of the glycan structure. Chemistry. 2016:22(3):1114–1124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article, including its supplementary information files, or are available from the authors upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2019) partner repository with the dataset identifier PXD036263.
Reviewer account details:
Username: reviewer_pxd036263@ebi.ac.uk
Password: CM60Uki5.