Abstract
In this report we demonstrate that human immunodeficiency virus type 1 (HIV-1) minus-strand transfer, assayed in vitro and in endogenous reactions, is greatly inhibited by actinomycin D. Previously we showed that HIV-1 nucleocapsid (NC) protein (a nucleic acid chaperone catalyzing nucleic acid rearrangements which lead to more thermodynamically stable conformations) dramatically stimulates HIV-1 minus-strand transfer by preventing TAR-dependent self-priming from minus-strand strong-stop DNA [(−) SSDNA]. Despite this potent activity, the addition of NC to in vitro reactions with actinomycin D results in only a modest increase in the 50% inhibitory concentration (IC50) for the drug. PCR analysis of HIV-1 endogenous reactions indicates that minus-strand transfer is inhibited by the drug with an IC50 similar to that observed when NC is present in the in vitro system. Taken together, these results demonstrate that NC cannot overcome the inhibitory effect of actinomycin D on minus-strand transfer. Other experiments reveal that at actinomycin D concentrations which severely curtail minus-strand transfer, neither the synthesis of (−) SSDNA nor RNase H degradation of donor RNA is affected; however, the annealing of (−) SSDNA to acceptor RNA is significantly reduced. Thus, inhibition of the annealing reaction is responsible for actinomycin D-mediated inhibition of strand transfer. Since NC (but not reverse transcriptase) is required for efficient annealing, we conclude that actinomycin D inhibits minus-strand transfer by blocking the nucleic acid chaperone activity of NC. Our findings also suggest that actinomycin D, already approved for treatment of certain tumors, might be useful in combination therapy for AIDS.
Actinomycin D (Act D), a drug which binds to double- (reference 58 and references therein) and single-stranded (60, 71) DNA, has been known for many years to inhibit DNA-dependent DNA and RNA synthesis (reviewed in reference 58). For retrovirologists, use of Act D and knowledge of its inhibitory activities proved to be essential for early studies on the mechanisms involved in virus replication and assembly. Thus, the seminal observation that production of Rous sarcoma virus (RSV) particles early in infection is sensitive to Act D (3, 65, 70) initially led to the conclusion that retroviruses replicate via a DNA intermediate which is integrated into host DNA (provirus hypothesis [66; reviewed in reference 67]) and ultimately, to the discovery of reverse transcriptase (RT) (5, 68).
In other studies, it was shown that Act D treatment of retrovirus-infected cells results in a rapid shutdown of viral RNA synthesis (3, 6, 18, 66). Subsequent work indicated that despite the absence of ongoing RNA synthesis, noninfectious murine leukemia virus (MuLV) particles (termed Act D virions [24]), which are deficient in genomic RNA (42) but which contain the appropriate amounts of all of the viral proteins (24, 34, 43) and the select population of host tRNAs (44), continue to be produced for at least 8 to 12 h after the addition of the drug (42, 50, 54). These results demonstrated that genomic RNA is not required for MuLV assembly (42, 43) and that viral mRNAs can function for many hours after the cessation of viral RNA synthesis (43, 50, 54).
Act D has also been important for elucidation of the events which occur during the reverse transcription of genomic RNA. From experiments performed with detergent-treated RSV (48) or MuLV (47) particles (i.e., endogenous RT assays), it became clear that Act D blocks the conversion of a single-stranded form of viral DNA to a double-stranded DNA product. In later work on endogenous MuLV reverse transcription, Rothenberg et al. (61) found that with 100 μg of Act D per ml, the final 600 nucleotides (nt) in minus-strand DNA are not made. Under these conditions, the largest minus-strand DNA molecule is 8.2 kb and plus-strand strong-stop DNA [(+) SSDNA] is not detected; in the absence of the drug, full-length double-stranded DNA (8.8 kb) is synthesized (49, 61). All of these studies were consistent with the idea that the DNA-dependent step in viral DNA synthesis, i.e., synthesis of plus-strand DNA, is the primary target of the drug.
In contrast to the results with MuLV, Novak et al. (53) showed that the addition of 100 μg of Act D per ml to endogenous reaction mixtures with RSV leads to the accumulation of minus-strand strong-stop DNA [(−) SSDNA] and drastically inhibits the elongation of this product. These investigators also reported that at this high concentration of Act D, there is a 50% reduction in the amount of (−) SSDNA which hybridizes to virion RNA (8). It was concluded that nucleic acid hybridization is a necessary step for elongation of (−) SSDNA, in agreement with the model proposed by Gilboa et al. (25). Later work has confirmed this conclusion, and it is now established that the annealing of the R sequence at the 3′ end of viral RNA to the complementary sequence at the 3′ end of (−) SSDNA is a prerequisite for minus-strand transfer and subsequent elongation of minus-strand DNA (reference 64 and references therein).
In a more recent study on the effect of several RT inhibitors on endogenous HIV-1 reverse transcription, it was reported that in the presence of 20 μg of Act D per ml, the smear of 32P-labeled DNA products seen on an agarose gel was less intense and smaller than that observed in the absence of the drug (14). In this work, the effect of Act D was attributed to inhibition of plus-strand DNA synthesis.
We became interested in the effect of Act D on human immunodeficiency virus type 1 (HIV-1) reverse transcription during the course of studies with a reconstituted system (referred to here and in reference 28 as an in vitro system) which mimics the events in HIV-1 minus-strand transfer (see Fig. 1 in reference 28): In this system, the donor and acceptor RNA templates contain all of the R sequence and portions of U5 or U3, respectively; the primer is a short DNA oligonucleotide complementary to terminal U5 sequences in the donor. Our initial results demonstrated that HIV-1 nucleocapsid (NC) protein stimulates both the rate and extent of minus-strand transfer by preventing self-priming from (−) SSDNA (28). (Self-priming has also been observed by others [41, 46] in studies on (−) SSDNA synthesis.) In the minus-strand transfer system, we showed that self-priming results from conversion of fold-back structures formed by (−) SSDNA to a heterogeneous class of dead-end products termed SP DNAs (28). As a consequence, (−) SSDNA is unavailable for annealing to the acceptor. Mutational analysis indicated that self-priming is correlated with the presence of a large stem-loop containing minus-strand TAR sequences at the 3′ terminus of (−) SSDNA (28). We concluded that NC, a nucleic acid chaperone (32, 58a) catalyzing structural rearrangements of nucleic acids which result in more thermodynamically stable conformations (7, 13, 17, 20, 28, 33, 36, 39, 51, 69, 73, 75), exerts its effect by transiently destabilizing this complementary TAR structure.
The present study stemmed from a search for a drug which could specifically target a unique step in HIV-1 DNA synthesis. Here, we report that Act D selectively inhibits HIV-1 minus-strand transfer. Thus, we find that DNA-dependent DNA synthesis catalyzed by RT is significantly inhibited only at very high concentrations of Act D, whereas HIV-1 minus-strand transfer is highly sensitive to the drug. It is especially noteworthy that this sensitivity to Act D is manifested even in the presence of HIV-1 NC, which normally stimulates strand transfer (2, 16, 28, 40, 55, 75; also, see above). Experiments designed to elucidate the mechanism of inhibition of Act D indicate that it has no effect on RNA-dependent DNA synthesis of (−) SSDNA and little or no effect on RNase H degradation of donor RNA sequences but severely inhibits the annealing reaction. These results suggest that Act D, which is already being used in the treatment of certain tumors (19, 45), might also have an application in combination anti-HIV therapy.
MATERIALS AND METHODS
Materials.
Act D was obtained from Sigma. A PCR kit was purchased from Life Sciences Technology Inc. (Gaithersburg, Md.). [α-32P]dCTP (3,000 Ci/mmol) was purchased from Amersham Life Science Inc. The sources for all other materials are as specified by Guo et al. (28).
Preparation of donor and acceptor RNA templates.
Subclones of HIV-1 proviral clone NL4.3 (1), pDR5′ and pDR3′ (28), which were used to make the plasmids for transcription of the donor and acceptor RNA templates, were a gift from Robert Gorelick, SAIC-Frederick, NCI-Frederick Research and Development Center, Frederick, Md. Details on the procedures used to generate the 131-nt donor and 148-nt acceptor RNA templates containing sequences from nt 454 to 584 and from nt 9475 to 9622, respectively, (numbered according to the nucleotide positions in NL4.3 [1]) are given by Guo et al. (28).
RT assays. (i) In vitro strand transfer assay.
A detailed description of the in vitro strand transfer assay using a 32P-labeled 20-nt DNA primer with donor and acceptor RNA templates is given by Guo et al. (28). Where specified, HIV-1 NC was added to the annealed donor-primer hybrid together with acceptor RNA (28). In reaction mixtures containing Act D, the drug was added together with MgCl2 and the four deoxynucleoside triphosphates. Incubation was for 30 min at 37°C. Termination of reactions and analysis of DNA products by polyacrylamide gel electrophoresis were performed as described previously (28). The radioactivity in DNA products was quantified by using a PhosphorImager (Molecular Dynamics) and ImageQuant software. Calculations were performed as described by Guo et al. (28).
(ii) Assay of DNA-dependent DNA synthesis.
A 32P-labeled 20-nt DNA primer (JL239; 5′-CCC TTT TAG TCA GTG TGG AA-3′; nt 604 to 623 in NL4.3 [1]), was extended on a 50-nt DNA oligonucleotide template (JL279; 5′-GTC CCT GTT CGG GCG CCA CTG CTA GAG ATT TTC CAC ACT GAC TAA AAG GG-3′; complementary to nt 604 to 653 in NL4.3 [1]). DNA-dependent DNA synthesis was carried out under the same conditions as those used in the in vitro strand transfer assay (28; also, see above), except that no acceptor template was present. The final concentrations of template and RT were each 10 nM; the final concentration of the 20-nt DNA primer was 20 nM. Reactions (final volume, 20 μl) were initiated with MgCl2, the four deoxynucleoside triphosphates and Act D, where indicated, and the reaction mixtures were incubated for 30 min at 37°C.
(iii) HIV-1 endogenous RT assay.
The assay of endogenous reverse transcription in purified HIV-1 MN virions was performed as described by Guo et al. (28), except that the samples were analyzed without treatment with RNase ONE prior to electrophoresis. Act D was added as indicated. DNA products were detected by incorporation of 32P from [α-32P]dCTP and were analyzed in 6% sequencing gels. Where indicated, PCR analysis (see below) was also performed.
(iv) RNase H cleavage assay.
Reactions were performed under the conditions of the in vitro strand transfer assay as described by Guo et al. (28), except that the donor RNA was labeled with 32P at its 5′ end (∼105 cpm/0.2 pmol) (29) and the 20-nt DNA primer was unlabeled. Act D was added as indicated (see above). Samples were subjected to electrophoresis in a 12.5% sequencing gel. Radioactive cleavage products were quantified as described in subparagraph i.
PCR analysis of DNA products synthesized in endogenous RT reactions.
A 22.5-μl portion of the PCR mixture, i.e., the buffer and Taq polymerase supplied in the PCR kit, was combined with 1 μl of DNA products from an endogenous reaction and 1.5 μl of a solution containing the forward and reverse primers (final concentration, 0.24 μM each) in a final volume of 25 μl. The forward primers in each set were 5′ end labeled with 32P. Amplification of the DNA was carried out for 30 cycles as follows: 94°C, 1 min; 53°C, 1 min; and 72°C, 2 min. A 10-μl aliquot of the PCR reaction mixture was added to 4 μl of STOP solution from a Sequenase kit. The resulting mixture was heated at 90°C for 5 min, and a 2.5-μl aliquot was then loaded onto a 6% sequencing gel. A sequencing ladder was used to identify products with the expected sizes. Specific PCR products were quantified by PhosphorImager analysis, as described above.
The primers used to detect total minus-strand DNA contained sequences in (−) SSDNA. A 123-bp product was generated with a forward primer (JL308; nt 487 to 510; 5′-AGC TCT CTG GCT AAC TAG GGA ACC-3′) and a reverse primer (JL309; nt 609 to 586; 5′-AAA AGG ATC TGA GGG ATC TCT AGC-3′). A second set of primers was designed to detect only minus-strand DNA which had been transferred and elongated. A 159-bp product was generated with a forward primer (JL314; nt 8825 to 8847; 5′-GTC AAA ACG TGT GAC TGG ATG GC-3′) and a reverse primer (JL315; nt 8983 to 8961; 5′-GCA CAA TCA GCA TTG GTA GCT GC-3′). The nucleotide sequences refer to the positions in HIV-1 MN (30) (accession no., M17449).
To determine the sensitivity of the PCR, the DNA products from an endogenous RT reaction with HIV-1 MN virions were used as a template for PCR with unlabeled primers JL309 and JL314 (see above). The resulting DNA was 890 bp and contained the region covered by both sets of the primers described above (JL308 and JL309 and JL314 and JL315). After purification and quantification, the 890-bp fragment was used as a template for PCR with each set of primers; again, the forward primers in each set were 5′ end labeled. Serial dilutions of the template fragment were made to estimate the amount of label in the PCR products as a function of the number of DNA copies. The DNA products at each of the template dilutions were analyzed on a 6% sequencing gel.
Annealing assay.
Standard annealing reaction mixtures contained Tris-HCl (pH 8.0) and KCl at final concentrations of 50 and 75 mM, respectively, 0.2 pmol of unlabeled acceptor RNA, 0.2 pmol of 32P-labeled 131-nt synthetic (−) SSDNA (JL249) (106 to 2 × 106 cpm) (28), and Act D and NC (0.4 μM), as indicated, in a final volume of 20 μl; reaction mixtures were scaled up as needed. Annealing was performed at 37°C, and at the specified times, a 10-μl aliquot of the reaction mixture was mixed with 10 μl of 2× loading buffer (20 mM Tris-HCl [pH 8.0], 25 mM EDTA, 25% glycerol, 2% sodium dodecyl sulfate, and 0.01% bromphenol blue). For reactions without NC, a 5-μl portion of this mixture was applied to a 6% native polyacrylamide gel (6% acrylamide–0.32% bisacrylamide). For reactions with NC, a 10-μl portion was applied to a 5% agarose gel composed of 4% NuSieve 3:1 high-gel-strength agarose and 1% NuSieve GTG low-melting-temperature agarose. After electrophoresis, the gels were dried and radioactivity in the RNA-DNA hybrid was quantified with a PhosphorImager.
Recombinant NC.
The procedures for the preparation of recombinant HIV-1 NC are given in references 9 and 73.
RESULTS
Act D inhibits minus-strand transfer in vitro.
The fact that Act D has been shown to inhibit reverse transcription of avian and murine retroviruses (see above) and the recent finding that Act D can bind single-stranded DNA (60, 71) as well as RNA-DNA hybrids (63) led us to ask whether Act D might affect the transfer of (−) SSDNA to acceptor RNA in an HIV-1 system. As a first approach to this question, we measured the effect of the drug on HIV-1 DNA-dependent DNA synthesis and on in vitro minus-strand transfer (Fig. 1). In each case, we show gel data (IA and IIA) as well as the results of quantitative PhosphorImager analysis (IB and IIB).
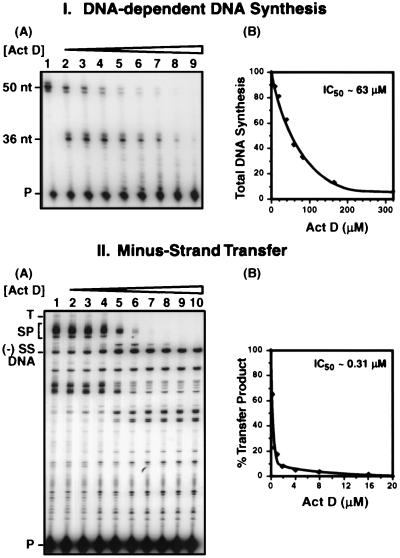
FIG. 1.
Selective inhibition of strand transfer by Act D. Reactions and analysis of DNA products were performed as described in Materials and Methods. (I) DNA-dependent DNA synthesis. (A) Gel analysis. The concentrations of Act D were as follows: lane 1, no drug; lane 2, 5 μM; lane 3, 10 μM; lane 4, 20 μM; lane 5, 40 μM; lane 6, 60 μM; lane 7, 80 μM; lane 8, 160 μM; and lane 9, 320 μM. The positions of the 36- and 50-nt products and the primer (P) are shown on the left. (B) Quantitative PhosphorImager analysis of gel data. The total amount of DNA synthesis (with the control value set at 100%) was plotted as a function of Act D concentration. (II) Minus-strand transfer. (A) Gel analysis. The concentrations of Act D used were as follows: lane 1, no drug; lane 2, 0.25 μM; lane 3, 0.5 μM; lane 4, 1 μM; lane 5, 2 μM; lane 6, 4 μM; lane 7, 8 μM; lane 8, 12 μM; lane 9, 16 μM; and lane 10, 20 μM. The positions of (−) SSDNA, the transfer product (T), SP DNAs (SP) (28), and primer are shown on the left. (B) Quantitative PhosphorImager analysis of gel data. To compare the amount of transfer product made in control and Act D-containing reaction mixtures, the percent transfer product represented in total DNA products was quantified for each reaction (with the control value set at 100%) and was plotted against the concentration of Act D.
The effect of adding increasing concentrations of Act D on plus-strand DNA synthesis was examined with reaction mixtures containing a 50-nt minus-strand DNA template and a 20-nt DNA primer (Fig. 1, panels IA and IB). In the absence of Act D (IA, lane 1), only the 50-nt plus-strand product was detectable, whereas in the presence of the drug (IA, lanes 2 to 9), a prominent pause product (36 nt) was observed at a G-C rich region in the template. This result is consistent with findings of Rill and Hecker (60), who assayed DNA-dependent primer extension catalyzed by several different DNA polymerases, including HIV-1 RT, in reaction mixtures containing Act D. They found that pause sites observed in the presence of Act D rarely corresponded to those generated in the absence of the drug. The pausing seen here (IA) presumably results from the preferred binding of Act D (11, 37, 52, 60, 62, 72; also, see reference 58 and references therein) to a 5′-GC-3′ sequence near the 5′ terminus of the 50-nt DNA template and the subsequent inability of RT to efficiently elongate nascent DNA from this site.
Inspection of the gel indicates that total DNA synthesis was not significantly inhibited at the lowest Act D concentrations tested (5 to 20 μM; Fig. 1, panel IA, lanes 2 to 4). In contrast, a more marked reduction in full-length DNA synthesis and the presence of additional pause products were observed with higher concentrations of the drug (40 to 80 μM; IA, lanes 5 to 7); total DNA synthesis ranged from ∼40 to 70% of the control, which was set at 100% (IB). At the highest concentrations of Act D tested (160 μM [IA, lane 8] and 320 μM [IA, lane 9]), the 50-nt full-length product was essentially undetectable and there was also a marked decrease in the amount of the 36-nt DNA and smaller pause products; in this case, total DNA synthesis was inhibited by ∼90% or more (IB).
To determine whether Act D is able to inhibit minus-strand transfer, increasing concentrations of Act D, from 0.25 to 20 μM, were added to in vitro strand transfer reaction mixtures (Fig. 1, panels IIA and IIB), as described in Materials and Methods. Although the actual amount of transfer product synthesized in the control reaction without the drug (IIA, lane 1) is fairly small, the product was detected in a highly reproducible manner (28). (In our system [28], minus-strand transfer is efficient only in the presence of NC [see below].) Remarkably, at an Act D concentration of 0.5 μM (IIA, lane 3), the amount of transfer product was only ∼20% (IIB) of the amount observed in the absence of the drug (IIA, lane 1; IIB). At concentrations of 2 μM or higher (IIA, lanes 5 to 10), strand transfer was severely inhibited and the amount of transfer product was less than 5% of the control value (IIB). Note, however, that synthesis of (−) SSDNA was not reduced by the drug. Indeed, as less of the transfer product was made, there was actually an increase in the total amount of (−) SSDNA (IIA; compare lanes 2 to 4 with lanes 5 to 10). Synthesis of SP DNAs was also inhibited by Act D (28) but was somewhat less sensitive to Act D than synthesis of the transfer product (IIA) (see below).
A comparison of the data obtained for DNA-dependent DNA synthesis and minus-strand transfer shows a striking difference in the sensitivities of these two reactions to Act D. Thus, the 50% inhibitory concentrations (IC50) for Act D in the DNA-templated and minus-strand transfer reactions were ∼63 (IB) and ∼0.31 μM (IIB), respectively.
Act D inhibits NC-catalyzed minus-strand transfer in vitro.
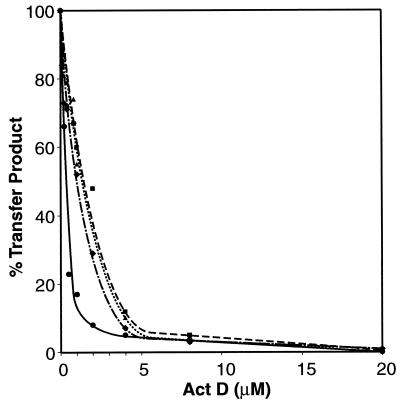
In retrovirus particles, genomic RNA is packaged in the virion core in association with NC, protease, RT, integrase, and primer tRNA (12, 13, 22). In view of previous work demonstrating that NC markedly stimulates minus-strand transfer (2, 16, 28, 40, 55, 75), it was of interest to determine whether NC could prevent or substantially reduce inhibition by Act D.
To examine this question, increasing amounts of Act D were added to reaction mixtures containing three different concentrations of NC: 0.4, 0.8, and 1.6 μM, corresponding to 7, 3.5, and 1.75 nt per NC molecule, respectively (Fig. 2). Data for the reaction without NC are taken from Fig. 1, panel IIB. As shown in Fig. 2, the addition of NC had a small but detectable effect on the extent of strand transfer inhibition by Act D. With 0.4 μM NC, the IC50 for Act D was ∼1.1 μM; with 0.8 and 1.6 μM NC, the IC50 was ∼1.4 to 1.5 μM. These values are 3- and 5-fold greater, respectively, than the IC50 in the absence of NC (∼0.3 μM; IIB) but are still at least 40-fold lower than the IC50 for DNA-dependent DNA synthesis (∼63 μM; IB). Thus, the presence of NC does not significantly affect the inhibition of minus-strand transfer by Act D.
FIG. 2.
Effect of Act D on NC-catalyzed minus-strand transfer. Reactions were carried out in the presence of NC and Act D, as indicated, according to the procedures detailed in Materials and Methods. The data shown for reactions without NC were taken from Fig. 1, panel IIB. The percent transfer product represented in total DNA products was quantified with a PhosphorImager and was plotted against the concentration of Act D as described in the legend to Fig. 1, panel IIB. Symbols: circles, solid line, no NC; diamonds, dot-dash line, 0.4 μM NC; triangles, dotted line, 0.8 μM NC; and squares, dashed line, 1.6 μM NC.
Inhibition of endogenous HIV-1 reverse transcription by Act D.
An alternate approach to testing the effect of Act D on NC-catalyzed minus-strand transfer is to determine whether Act D can inhibit this reaction during endogenous reverse transcription. The endogenous RT assay uses detergent-treated HIV-1 particles which contain NC (as well as the other viral proteins) and is more likely to mimic conditions present during virus infection than a purely in vitro assay.
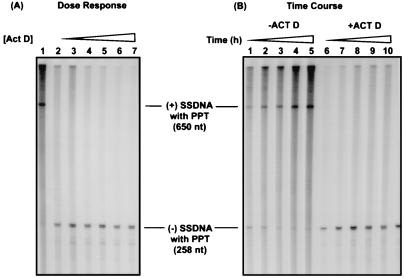
As shown in Fig. 3A, in 6-h endogenous reactions, every concentration of Act D tested, from 1 (lane 2) to 80 μM (lane 7), completely inhibited (+) SSDNA synthesis. In addition, (−) SSDNA accumulated in reaction mixtures containing Act D (lanes 2 to 7), whereas in the absence of the drug, very little (−) SSDNA was detected at 6 h (lane 1; also see Fig. 3B, lane 5, and reference 28).
FIG. 3.
Effect of Act D on endogenous HIV-1 reverse transcription. Reaction mixtures were incubated for 6 h with increasing concentrations of Act D (A) or in the absence or presence of Act D at 5 μM for increasing times (B), as described in Materials and Methods. Samples were analyzed on a 6% sequencing gel to visualize (−) and (+) SSDNA products still attached to the tRNA or PPT RNA primers, respectively. (A) Dose response. Lane 1, no drug; lanes 2 to 7, Act D at 1, 5, 10, 20, 40, and 80 μM, respectively. (B) Time course. Lanes 1 to 5, no Act D; lanes 6 to 10, 5 μM Act D. The incubation times were 0.5 (lanes 1 and 6), 1 (lanes 2 and 7), 2 (lanes 3 and 8), 4 (lanes 4 and 9), and 6 (lanes 5 and 10) h. The sizes of the products were verified by running a sequencing ladder generated with MP18 DNA, which is included in the Sequenase kit.
An investigation of the time course of endogenous DNA synthesis over a 6-h incubation in the absence of Act D revealed that (−) SSDNA gradually disappeared, while (+) SSDNA accumulated (Fig. 3B, lanes 1 to 5) (28). In contrast, in the presence of 5 μM Act D, (+) SSDNA could not be detected at any of the time points, i.e., from 30 min to 6 h (lanes 6 to 10), although there was significant accumulation of (−) SSDNA (lanes 6 to 10). These findings demonstrate that while Act D inhibits the synthesis of (+) SSDNA, it does not reduce the synthesis of (−) SSDNA, in agreement with the results of the in vitro assay (Fig. 1, panel IIA).
Synthesis of (+) SSDNA is the consequence of the following series of events. During minus-strand elongation, RT makes a copy of the polypurine tract (PPT) sequence in viral RNA. Once the PPT-containing RNA-DNA hybrid is formed, the RNase H activity of RT cleaves the substrate at its 3′ terminus to generate the plus-strand primer and synthesis of (+) SSDNA is initiated (57; reviewed in reference 10). Thus, if minus-strand transfer were blocked, this could account for our inability to detect (+) SSDNA in Act D-treated endogenous reaction mixtures.
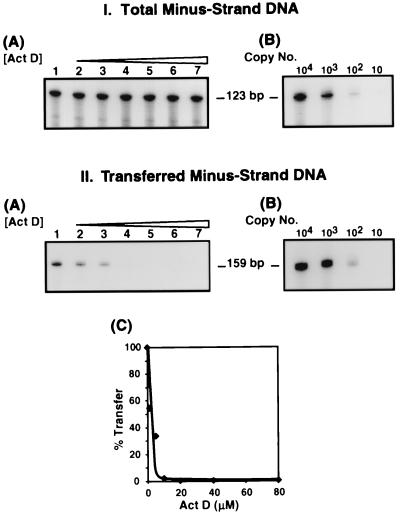
To test this hypothesis, we performed PCR analysis (Fig. 4) of the DNA products formed in the endogenous reverse transcription reactions shown in Fig. 3A (dose response). Two sets of primers were used: One set contained sequences in (−) SSDNA and could therefore detect (−) SSDNA as well as elongated minus-strand DNA present in the sample; the PCR product was 123 bp (Fig. 4, panel IA). The other set contained sequences in minus-strand DNA upstream of the PPT and could detect only minus-strand DNA that had been transferred and elongated; the PCR product in this case was 159 bp (IIA). To increase the sensitivity of the assay, the forward primer in each set was labeled at its 5′ end with 32P. The data indicate that with both primer sets, ∼102 copies of the DNA template could be detected (IB and IIB).
FIG. 4.
Detection of total and transferred minus-strand DNA synthesized during HIV-1 endogenous reverse transcription. The DNA products made in the reactions shown in Fig. 3A (Act D dose response) were stored for approximately 6 months, which allowed the original radioactivity to decay. The DNAs were then amplified by PCR with two sets of PCR primers (one primer of each set was labeled at its 5′ end with 32P), as described in Materials and Methods. Shown are gel analyses of the 123- and 159-bp PCR products (IA and IIA, respectively) and dilution analyses of known amounts of the 890-bp template (expressed as copy number) to indicate the sensitivity of the assay (IB and IIB). (IIC) PhosphorImager analysis. The amount of the 159-bp PCR product generated from the reaction without drug was set at 100%. The percent transfer relative to the control was plotted against the concentration of Act D. For panels IA and IIA, the designation of lanes 1 to 7 is the same as that given in the legend for Fig. 3A.
The gel data indicate that the total amounts of minus-strand DNA present in the control and Act D-treated endogenous reaction mixtures were fairly similar (Fig. 4, panel IA). In contrast, transferred minus-strand DNA was present in the control reaction mixture (IIA, lane 1) and to a lesser extent in reaction mixtures with 1 or 5 μM Act D (IIA, lanes 2 and 3; IIC) but was virtually undetectable in reaction mixtures containing Act D at concentrations of 10 μM or higher (IIA, lanes 4 to 7; IIC). Quantification of the PCR product (IIC) also showed that at 1 μM Act D, minus-strand transfer was inhibited by approximately 45%. These results indicate that minus-strand transfer is targeted by Act D during endogenous reverse transcription.
Although the PCR data should be viewed as only semiquantitative, it is of interest that the concentration of Act D which results in ∼50% inhibition of endogenous minus-strand transfer (estimated at between 1 and 2 μM) is consistent with the IC50 for Act D derived from the in vitro reaction mixtures containing NC (Fig. 2). Taken together, the results of Fig. 2 and 4 demonstrate that despite the dramatic stimulatory effect of NC on HIV-1 minus-strand transfer normally observed in our system (28), NC is unable to oppose the inhibitory activity of Act D.
Low concentrations of Act D do not inhibit RNase H cleavage during minus-strand transfer.
The results presented thus far demonstrate that Act D inhibits minus-strand transfer in the in vitro and endogenous assay systems. There are three reactions required for minus-strand transfer (64): (i) synthesis of (−) SSDNA; (ii) RNase H cleavage of donor RNA sequences; and (iii) annealing of (−) SSDNA to acceptor RNA, the actual transfer step. We have already shown that Act D does not inhibit the first reaction (Fig. 1, panel IIA; Fig. 3). The question then arises as to whether reactions ii and/or iii are targeted by the drug.
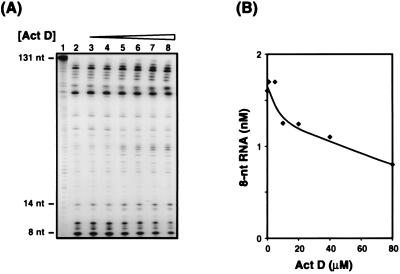
To investigate the effect of Act D on RNase H cleavage of the donor (reaction ii), minus-strand transfer was assayed as described in Materials and Methods, except that donor RNA was labeled at its 5′ end with 32P and an unlabeled DNA oligonucleotide primer was used (Fig. 5). Gel analysis of a reaction mixture incubated without RT or Act D indicates the position of the labeled 131-nt RNA (Fig. 5A, lane 1). Incubation with RT in the absence of the drug generated many cleavage fragments, including the terminal products of 14 and 8 nt (lane 2). The 8-nt band results from the 3′-OH-independent (i.e., polymerase-independent) (23, 27, 56) mode of RNase H cleavage which frees (−) SSDNA from the donor and which is required for subsequent annealing of (−) SSDNA to the acceptor (reviewed in references 10 and 64). The addition of Act D at concentrations of 1 and 5 μM (lanes 3 and 4) had no effect on the appearance of the 14- and 8-nt products; a small inhibitory effect was observed at higher concentrations of the drug, from 10 to 80 μM (lanes 5 to 8). Quantitative PhosphorImager analysis of the gel data showed that at the highest concentration of Act D tested (80 μM), the 8-nt RNA was reduced by only 50% (Fig. 5B). Similar results were obtained when increasing concentrations of Act D were added to reaction mixtures containing 0.8 μM NC (data not shown).
FIG. 5.
Effect of Act D on RNase H activity in the absence of NC. In vitro strand transfer reaction mixtures were incubated with donor RNA labeled at its 5′ end with 32P and increasing concentrations of Act D, as described in Materials and Methods. (A) Gel analysis. (B) Quantitative PhosphorImager analysis. The amount of the 8-nt RNA cleavage product was plotted against the concentration of Act D. Lanes: 2, no drug; lanes 3 to 8, Act D at 1, 5, 10, 20, 40, and 80 μM, respectively. Lane 1 is a control showing the position of the uncleaved labeled donor RNA from a reaction without RT and Act D.
Since low concentrations of Act D have no effect on RNase H cleavage of donor RNA, we conclude that inhibition of this reaction cannot account for the dramatic reduction of minus-strand transfer observed in the dose response experiments shown above (Fig. 1, 2, and 4).
Act D inhibits the annealing of (−) SSDNA and acceptor RNA.
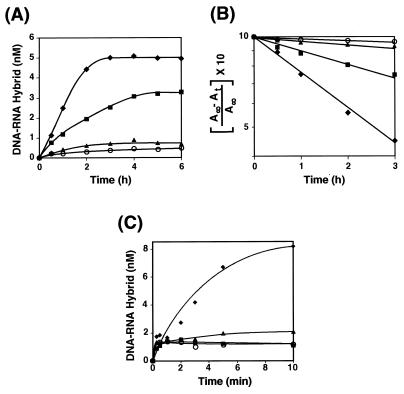
The effect of Act D on the annealing step in minus-strand transfer was assayed in the absence or presence of NC protein with a 131-nt synthetic (−) SSDNA oligonucleotide labeled with 32P at its 5′ end and unlabeled, 148-nt acceptor RNA, at the standard concentrations for the in vitro assay (Fig. 6). Under conditions where NC was not present, annealing was measured over a 6-h period; however, in the absence of the drug, the amount of hybrid formed did not appreciably increase after 3 h (Fig. 6A). With an Act D concentration of 0.5 μM, both the rate and extent of annealing were inhibited by approximately twofold. At concentrations of 1 and 8 μM, very little annealing was observed.
FIG. 6.
Effect of Act D on the kinetics of annealing of (−) SSDNA and acceptor RNA in the absence or presence of NC protein. In the absence of NC (A and B), the annealing reaction was carried out with Act D (as indicated), as described in Materials and Methods. Aliquots were removed for gel analysis at 0.5, 1, 2, 3, 4, 5, and 6 h. In panel C, reaction mixtures were preincubated with Act D (as indicated) for 10 min at 37°C prior to the addition of NC. Aliquots were removed for gel analysis at 0.25, 0.5, 1, 2, 3, 5, and 10 min. (A) Quantitative PhosphorImager analysis of annealing in the absence of NC. The concentration of the DNA-RNA hybrid was plotted against the time of incubation. (B) Kinetic plot of the PhosphorImager data. The data from panel A were plotted as a semilogarithmic plot of the concentration of hybrid formed with time (35). A∞ and At are the concentrations of hybrid formed at infinite time and at the indicated time, respectively. A∞ is a theoretical value; At was determined by PhosphorImager analysis of the gel data. (C) Quantitative PhosphorImager analysis of annealing in the presence of 0.4 μM NC. The concentration of the DNA-RNA hybrid was plotted against the time of incubation. Since the reactions performed in the presence of Act D and NC showed little or no progression to final hybridization after the first 15 to 30 s, the data did not lend themselves to a kinetic analysis. Symbols for panels A and B: diamonds, no drug; squares, 0.5 μM Act D; triangles, 1 μM Act D; circles, 8 μM Act D. Symbols for panel C: diamonds, no drug; squares, 1 μM Act D; triangles, 2 μM Act D; circles, 16 μM Act D.
A semilogarithmic plot of the concentration of the RNA-DNA hybrid formed as a function of time is shown in Fig. 6B; A∞ and At are the concentrations of the hybrid at infinite time and at the experimental time, respectively (35). These data are consistent with pseudo-first-order kinetics: a fast step in which an RNA-DNA complex is formed and a slow, rate-limiting step in which the complex is rearranged so that a linear hybrid molecule is formed. Act D appears to be inhibiting the slow step (discussed in more detail below).
We have also analyzed annealing in the presence of 0.4 μM NC and several concentrations of Act D (Fig. 6C). In agreement with You and McHenry (75), NC accelerated the rate of annealing in the reaction mixture without the drug and hybrid formation was almost complete by 10 min. The addition of NC to reaction mixtures preincubated for 10 min with 1, 2, and 16 μM Act D resulted in an initial burst of annealing at levels which were essentially the same in the control and all Act D-containing reaction mixtures; however, after 15 to 30 s, no further annealing was observed in the reaction mixtures containing the drug.
These findings demonstrate that inhibition of minus-strand transfer by Act D results from a marked reduction in annealing between (−) SSDNA and acceptor RNA. In accord with the observations on in vitro and endogenous minus-strand transfer (Fig. 2 and 4), NC is unable to prevent the inhibitory effect of Act D on the annealing reaction.
DISCUSSION
The present study demonstrates that Act D selectively inhibits HIV-1 minus-strand transfer in endogenous and in vitro RT assays. We find that DNA-dependent DNA synthesis catalyzed by RT is not strongly inhibited by Act D (IC50, ∼63 μM), whereas minus-strand transfer is considerably more sensitive to the drug (IC50, ∼0.3 μM (Fig. 1). Moreover, an analysis of the products synthesized in the endogenous reaction (Fig. 3 and 4) suggests that the primary target of Act D during HIV-1 reverse transcription is the minus-strand transfer step and not synthesis of (+) SSDNA. In addition to the effect on minus-strand transfer, relatively low concentrations of Act D also inhibit synthesis of SP DNAs (Fig. 1, panel IIA), the products of self-priming from (−) SSDNA (28). In view of the ability of Act D to inhibit the annealing reaction in minus-strand transfer (Fig. 6; also, see below), it seems likely that Act D also blocks the self-annealing step (28) required for DNA-dependent self-priming.
As discussed above, NC has been shown to dramatically stimulate both the rate and extent of HIV-1 minus-strand transfer (2, 16, 28, 40, 55, 75) by acting as a nucleic acid chaperone (32, 58a), which prevents TAR-dependent self-priming from (−) SSDNA (28) and which facilitates the annealing reaction (75). Despite this very potent activity of NC, there is only a modest increase in the IC50 for Act D in the in vitro reaction mixtures containing NC (Fig. 2). Similar results are obtained in endogenous RT assays with HIV-1 virions (Fig. 4), where the ratio of nucleotides in the two RNA templates to NC molecules is approximately 7 nt per NC molecule (17, 31, 38, 74) (equivalent to 0.4 μM in our in vitro system). Taken together, these results lead us to conclude that NC does not significantly interfere with Act D inhibition of minus-strand transfer.
The in vitro and endogenous RT assays demonstrate that synthesis of (−) SSDNA, i.e., the first product of reverse transcription, is not reduced in the presence of the drug and that, in fact, an accumulation of this intermediate is observed (Fig. 1 and 3). In addition, at concentrations which significantly reduce minus-strand transfer (1 to 5 μM), Act D has no effect on RNase H-catalyzed degradation of donor RNA (Fig. 5). The addition of NC gives similar results and, in agreement with the data of Kim et al. (40), does not affect the nature of the RNase H cleavage pattern in either the presence or absence of Act D (data not shown).
In contrast, Act D strongly inhibits the annealing step (Fig. 6), indicating that the inhibitory activity of the drug on minus-strand transfer can be accounted for by its effect on annealing. Thus, at a concentration of 0.5 μM, Act D already causes a significant reduction in the rate and extent of hybridization; more severe inhibition is observed with 1 and 8 μM Act D (Fig. 6A). The data from the semilogarithmic plot (Fig. 6B) suggest that the reaction is following pseudo-first-order kinetics, in accord with the observations of You and McHenry (75). One may envision a fast equilibrium step, i.e., formation of a transient complex containing both (−) SSDNA and acceptor, with each reactant still retaining its stem-loop structure, followed by a slow step in which the complex is rearranged to yield a linear molecule. The kinetic data suggest that Act D is affecting the slow step and is preventing the conversion of the transient complex to a fully annealed, functional RNA-DNA hybrid. This interpretation is strengthened by the finding that Act D can bind to an RNA-DNA hybrid (63).
Although the true nature of the complex containing Act D, acceptor RNA, and (−) SSDNA is not known, it is clear that it is unaffected by the addition of NC. Thus, NC is unable to reverse the Act D effect on annealing (Fig. 6C), despite the fact that in the absence of the drug, its nucleic acid chaperone activity dramatically increases the rate of annealing of (−) SSDNA and acceptor (Fig. 6C) (75). In addition, NC does not appear to be competing with Act D for the same sites. A large component of NC binding may be in an ionic mode (i.e., via electrostatic interactions with the phosphate backbone of the nucleic acid [15, 21]). In contrast, Act D is known to bind in a sequence-dependent manner. It intercalates into DNA at preferred 5′-GC-3′ sites (11, 26, 37; see also reference 60 and references therein) or at nonclassical binding sites such as TGGGT (4) and dissociates very slowly from these sites (11, 52; see also reference 60 and references therein). It should also be pointed out that while efficient annealing of (−) SSDNA and acceptor RNA requires NC (Fig. 6C) (40a, 75), the annealing reaction itself occurs in the absence of RT (Fig. 6) (8, 40a, 75). Thus, a major conclusion of this work is that Act D inhibits the minus-strand transfer step in reverse transcription but not the activity of RT.
The ability of Act D to bind to an RNA-DNA hybrid (63) raises the additional question of why the drug inhibits annealing but not the activity of RNase H. Annealing requires the formation of hydrogen bonds between complementary base sequences, and a drug which intercalates at specific sequences might be expected to inhibit this process. In contrast, RNase H cleavage involves interaction with the phosphate backbone of the RNA and in general is sequence independent, e.g., RT has been shown to degrade RNA in RNA-DNA hybrids having widely varying sequences (reviewed in reference 10). Finally, Act D is most likely binding directly to the DNA moiety in the RNA-DNA hybrid: interaction of the 2-amino group of the phenoxazone ring of Act D and the 2′-hydroxyl group of RNA results in steric hindrance which prevents Act D from intercalating into an RNA molecule (63).
The present work is in accord with early studies on retrovirus replication demonstrating that Act D also inhibits endogenous reverse transcription of the murine (47, 61) and avian (8, 48, 53) retroviruses. However, the HIV-1 minus-strand transfer reaction appears to be more sensitive to Act D than those of the murine and avian retroviruses. In an effort to clarify this observation, we considered the possibility that Act D might intercalate with greater efficiency into HIV-1 (−) SSDNA due to the long double-stranded stem in the complementary TAR structure at the 3′ terminus of the DNA. Several experiments were performed with mutant templates in which the TAR stem-loops at the ends of the acceptor and donor RNAs were destabilized by deletion of 16 nt (28). At low concentrations of Act D (0.25 to 2 μM), the level of inhibition of minus-strand transfer was the same or slightly lower in reaction mixtures containing the mutant templates. However, as the concentration of Act D was increased (4 μM), no further inhibition was observed; rather, the amount of transfer product formed in mutant reaction mixtures reached a plateau level (∼10 to 20% of that seen in the absence of the drug). In wild-type reaction mixtures, the amount of transfer product continued to decrease with the increase in Act D concentration, and in accord with the data of Fig. 1, panel IIB, the level of inhibition of strand transfer was 95 to 99% (data not shown). These results suggest that the TAR structure may only partially contribute to the inhibitory effect of Act D on HIV-1 minus-strand transfer. Sequence comparisons indicate that the numbers of G-C steps are similar in the RU5 regions of the three classes of retroviruses; however, there is not enough information available to know how these sites would rank with respect to preferred Act D binding. Other factors, such as possible differences in the nucleic acid binding affinities of the respective NC proteins or differences in the Act D-induced conformational changes (37, 52, 60) of the (−) SSDNAs, could also affect the response to the drug.
In summary, we have found that HIV-1 minus-strand transfer, a reaction which occurs in an efficient manner only in the presence of NC, is highly sensitive to Act D. Detailed analysis of this finding indicates that inhibition of the annealing step is responsible for the inhibitory activity of the drug. The observation that Act D interferes with NC function during a key step in reverse transcription is significant. To our knowledge, this is the first example of a drug which blocks the ability of NC to function as a nucleic acid chaperone. Although Act D has relatively high toxicity, it is used in treatment of two different tumors: Wilms’ tumor (19) and gestational choriocarcinoma (45). Our results raise the possibility that Act D might also be useful as part of a therapeutic regimen for AIDS patients in combination with protease and RT inhibitors and possibly agents which target the cysteine residues in the zinc fingers of NC (59).
ACKNOWLEDGMENTS
We thank Robert Gorelick for his generous gift of HIV-1 NL4.3 subclones and Bradley Kane for graciously providing us with HIV-1 NC protein. We are also indebted to Robert Crouch for helpful discussion and to Alan Rein and Robert Gorelick for critical reading of the manuscript.
This work was supported in part by the National Institutes of Health Intramural AIDS Targeted Antiviral Program.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J-L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bader J P. The role of deoxyribonucleic acid in the synthesis of Rous sarcoma virus. Virology. 1964;22:462–468. doi: 10.1016/0042-6822(64)90067-4. [DOI] [PubMed] [Google Scholar]
- 4.Bailey S A, Graves D E, Rill R. Binding of actinomycin D to the T(G)nT motif of double-stranded DNA: determination of the guanine requirement in nonclassical, non-GpC binding sites. Biochemistry. 1994;33:11493–11500. doi: 10.1021/bi00204a011. [DOI] [PubMed] [Google Scholar]
- 5.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 6.Bases R E, King A S. Inhibition of Rauscher murine leukemia virus growth in vitro by actinomycin D. Virology. 1967;32:175–183. doi: 10.1016/0042-6822(67)90268-1. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand E L, Rossi J J. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994;13:2904–2912. doi: 10.1002/j.1460-2075.1994.tb06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunte T, Novak U, Friedrich R, Moelling K. Effect of actinomycin D on nucleic acid hybridization: the cause of erroneous DNA elongation during DNA synthesis of RNA tumor viruses in vitro. Biochim Biophys Acta. 1980;610:241–247. doi: 10.1016/0005-2787(80)90006-4. [DOI] [PubMed] [Google Scholar]
- 9.Busch L K. Production of HIV-1 (MN) nucleocapsid protein (p7) by recombinant DNA technology. M.S. thesis. Frederick, Md: Hood College; 1994. [Google Scholar]
- 10.Champoux J J. Roles of ribonuclease H in reverse transcription. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 103–117. [Google Scholar]
- 11.Chen F-M. Binding specificities of actinomycin D to self-complementary tetranucleotide sequences-XGCY- Biochemistry. 1988;27:6393–6397. doi: 10.1021/bi00417a030. [DOI] [PubMed] [Google Scholar]
- 12.Chen M-J, Garon C F, Papas T S. Native ribonucleoprotein is an efficient transcriptional complex of avian myeloblastosis virus. Proc Natl Acad Sci USA. 1980;77:1296–1300. doi: 10.1073/pnas.77.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darlix J-L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 14.Debyser Z, Vandamme A-M, Pauwels R, Baba M, Desmyter J, De Clercq E. Kinetics of inhibition of endogenous human immunodeficiency virus type 1 reverse transcription by 2′,3′-dideoxynucleoside 5′-triphosphate, tetrahydroimidazo-[4,5,1-jk][1,4]-benzodiazepin-2(1H)-thione, and 1-[(2-hydroxyethoxy) methyl]-6-(phenylthio)thymine derivatives. J Biol Chem. 1992;267:11769–11776. [PubMed] [Google Scholar]
- 15.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 16.DeStefano J J. Human immunodeficiency virus nucleocapsid protein stimulates strand transfer from internal regions of heteropolymeric RNA templates. Arch Virol. 1995;140:1775–1789. doi: 10.1007/BF01384341. [DOI] [PubMed] [Google Scholar]
- 17.Dib-Hajj F, Khan R, Giedroc D P. Retroviral nucleocapsid proteins possess potent nucleic acid strand renaturation activity. Protein Sci. 1993;2:231–243. doi: 10.1002/pro.5560020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duesberg P H, Robinson W S. Inhibition of mouse leukemia virus (MLV) replication by actinomycin D. Virology. 1967;31:742–746. doi: 10.1016/0042-6822(67)90211-5. [DOI] [PubMed] [Google Scholar]
- 19.Farber S. Chemotherapy in the treatment of leukemia and Wilms’ tumor. JAMA. 1966;198:826–836. [PubMed] [Google Scholar]
- 20.Feng Y-X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher R J, Rein A, Fivash M, Urbaneja M A, Casas-Finet J R, Medaglia M, Henderson L E. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleissner E, Tress E. Isolation of a ribonucleoprotein structure from oncornaviruses. J Virol. 1973;12:1612–1615. doi: 10.1128/jvi.12.6.1612-1615.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furfine E S, Reardon J E. Reverse transcriptase · RNase H from the human immunodeficiency virus. Relationship of the DNA polymerase and RNA hydrolysis activities. J Biol Chem. 1991;266:406–412. [PubMed] [Google Scholar]
- 24.Gerwin B I, Levin J G. Interactions of murine leukemia virus core components: characterization of reverse transcriptase packaged in the absence of 70S genomic RNA. J Virol. 1977;24:478–488. doi: 10.1128/jvi.24.2.478-488.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilboa E, Mitra S W, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 26.Goodisman J, Rehfuss R, Ward B, Dabrowiak J C. Site-specific binding constants for actinomycin D on DNA determined from footprinting studies. Biochemistry. 1992;31:1046–1058. doi: 10.1021/bi00119a013. [DOI] [PubMed] [Google Scholar]
- 27.Gopalakrishnan V, Peliska J A, Benkovic S J. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc Natl Acad Sci USA. 1992;89:10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo J, Wu W, Yuan Z Y, Post K, Crouch R J, Levin J G. Defects in primer-template binding, processive DNA synthesis, and RNase H activity associated with chimeric reverse transcriptases having the murine leukemia virus polymerase domain joined to Escherichia coli RNase H. Biochemistry. 1995;34:5018–5029. doi: 10.1021/bi00015a013. [DOI] [PubMed] [Google Scholar]
- 30.Gurgo C, Guo H-G, Franchini G, Aldovini A, Collalti E, Farrell K, Wong-Staal F, Gallo R C, Reitz M S., Jr Envelope sequences of two new United States HIV-1 isolates. Virology. 1988;164:531–536. doi: 10.1016/0042-6822(88)90568-5. [DOI] [PubMed] [Google Scholar]
- 31.Henderson L E, Bowers M A, Sowder II R C, Serabyn S A, Johnson D G, Bess J W, Jr, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 33.Herschlag D, Khosla M, Tsuchihashi Z, Karpel R L. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamjoom G A, Naso R B, Arlinghaus R B. Selective decrease in the rate of cleavage of an intracellular precursor to Rauscher leukemia virus p30 by treatment of infected cells with actinomycin D. J Virol. 1976;19:1054–1072. doi: 10.1128/jvi.19.3.1054-1072.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jencks W P. Catalysis in chemistry and enzymology. New York, N.Y: McGraw-Hill; 1969. [Google Scholar]
- 36.Ji X, Klarmann G J, Preston B D. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry. 1996;35:132–143. doi: 10.1021/bi951707e. [DOI] [PubMed] [Google Scholar]
- 37.Kamitori S, Takusagawa F. Crystal structure of the 2:1 complex between d(GAAGCTTC) and the anticancer drug actinomycin D. J Mol Biol. 1992;225:445–456. doi: 10.1016/0022-2836(92)90931-9. [DOI] [PubMed] [Google Scholar]
- 38.Karpel R L, Henderson L E, Oroszlan S. Interactions of retroviral structural proteins with single-stranded nucleic acids. J Biol Chem. 1987;262:4961–4967. [PubMed] [Google Scholar]
- 39.Khan R, Giedroc D P. Recombinant human immunodeficiency virus type I nucleocapsid (NCp7) protein unwinds tRNA. J Biol Chem. 1992;267:6689–6695. [PubMed] [Google Scholar]
- 40.Kim J K, Palaniappan C, Wu W, Fay P J, Bambara R A. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J Biol Chem. 1997;272:16769–16777. doi: 10.1074/jbc.272.27.16769. [DOI] [PubMed] [Google Scholar]
- 40a.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix J-L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lapadat-Tapolsky M, Gabus C, Rau M, Darlix J-L. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- 42.Levin J G, Grimley P M, Ramseur J M, Berezesky I K. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J Virol. 1974;14:152–161. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levin J G, Rosenak M J. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc Natl Acad Sci USA. 1976;73:1154–1158. doi: 10.1073/pnas.73.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levin J G, Seidman J G. Selective packaging of host tRNAs by murine leukemia virus particles does not require genomic RNA. J Virol. 1979;29:328–335. doi: 10.1128/jvi.29.1.328-335.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis J L., Jr Chemotherapy of gestational choriocarcinoma. Cancer. 1972;30:1517–1521. doi: 10.1002/1097-0142(197212)30:6<1517::aid-cncr2820300616>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Quan Y, Arts E J, Li Z, Preston B D, de Rocquigny H, Roques B P, Darlix J-L, Kleiman L, Parniak M A, Wainberg M A. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA3Lys in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manly K F, Smoler D F, Bromfeld E, Baltimore D. Forms of deoxyribonucleic acid produced by virions of the ribonucleic acid tumor viruses. J Virol. 1971;7:106–111. doi: 10.1128/jvi.7.1.106-111.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonnell J P, Garapin A-C, Levinson W E, Quintrell N, Fanshier L, Bishop J M. DNA polymerases of Rous sarcoma virus: delineation of two reactions with actinomycin. Nature. 1970;228:433–435. doi: 10.1038/228433a0. [DOI] [PubMed] [Google Scholar]
- 49.Messer L I, Currey K M, O’Neill B J, Maizel J V, Jr, Levin J G, Gerwin B I. Functional analysis of reverse transcription by a frameshift pol mutant of murine leukemia virus. Virology. 1985;146:146–152. doi: 10.1016/0042-6822(85)90062-5. [DOI] [PubMed] [Google Scholar]
- 50.Messer L I, Levin J G, Chattopadhyay S K. Metabolism of viral RNA in murine leukemia virus-infected cells: evidence for differential stability of viral message and virion precursor RNA. J Virol. 1981;40:683–690. doi: 10.1128/jvi.40.3.683-690.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Müller G, Strack B, Dannull J, Sproat B S, Surovoy A, Jung G, Moelling K. Amino acid requirements of the nucleocapsid protein of HIV-1 for increasing catalytic activity of a Ki-ras ribozyme in vitro. J Mol Biol. 1994;242:422–429. doi: 10.1006/jmbi.1994.1592. [DOI] [PubMed] [Google Scholar]
- 52.Müller W, Crothers D M. Studies on the binding of actinomycin and related compounds to DNA. J Mol Biol. 1968;35:251–290. doi: 10.1016/s0022-2836(68)80024-5. [DOI] [PubMed] [Google Scholar]
- 53.Novak U, Friedrich R, Moelling K. Elongation of DNA complementary to the 5′ end of the avian sarcoma virus genome by the virion-associated RNA-dependent DNA polymerase. J Virol. 1979;30:438–452. doi: 10.1128/jvi.30.2.438-452.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paskind M P, Weinberg R A, Baltimore D. Dependence of Moloney murine leukemia virus production on cell growth. Virology. 1975;67:242–248. doi: 10.1016/0042-6822(75)90421-3. [DOI] [PubMed] [Google Scholar]
- 55.Peliska J A, Balasubramanian S, Giedroc D P, Benkovic S J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 56.Post K, Guo J, Kalman E, Uchida T, Crouch R J, Levin J G. A large deletion in the connection subdomain of murine leukemia virus reverse transcriptase or replacement of the RNase H domain with Escherichia coli RNase H results in altered polymerase and RNase H activities. Biochemistry. 1993;32:5508–5517. doi: 10.1021/bi00072a004. [DOI] [PubMed] [Google Scholar]
- 57.Powell M D, Levin J G. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J Virol. 1996;70:5288–5296. doi: 10.1128/jvi.70.8.5288-5296.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reich E, Goldberg I H. Actinomycin and nucleic acid function. Prog Nucleic Acid Res Mol Biol. 1964;3:183–234. doi: 10.1016/s0079-6603(08)60742-4. [DOI] [PubMed] [Google Scholar]
- 58a.Rein, A., L. E. Henderson, and J. G. Levin. Nucleic acid chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci., in press. [DOI] [PubMed]
- 59.Rice W G, Supko J G, Malspeis L, Buckheit R W, Jr, Clanton D, Bu M, Graham L, Schaeffer C A, Turpin J A, Domagala J, Gogliotti R, Bader J P, Halliday S M, Coren L, Sowder II R C, Arthur L O, Henderson L E. Inhibitors of HIV nucleocapsid protein zinc fingers as candidates for the treatment of AIDS. Science. 1995;270:1194–1197. doi: 10.1126/science.270.5239.1194. [DOI] [PubMed] [Google Scholar]
- 60.Rill R L, Hecker K H. Sequence-specific actinomycin D binding to single-stranded DNA inhibits HIV reverse transcriptase and other polymerases. Biochemistry. 1996;35:3525–3533. doi: 10.1021/bi9530797. [DOI] [PubMed] [Google Scholar]
- 61.Rothenberg E, Smotkin D, Baltimore D, Weinberg R A. In vitro synthesis of infectious DNA of murine leukaemia virus. Nature. 1977;269:122–126. doi: 10.1038/269122a0. [DOI] [PubMed] [Google Scholar]
- 62.Sobell H M, Jain S C. Stereochemistry of actinomycin binding to DNA II. Detailed molecular model of actinomycin-DNA complex and its implications. J Mol Biol. 1972;68:21–34. doi: 10.1016/0022-2836(72)90259-8. [DOI] [PubMed] [Google Scholar]
- 63.Takusagawa F, Takusagawa K T, Carlson R G, Weaver R F. Selectivity of F8-actinomycin D for RNA:DNA hybrids and its anti-leukemia activity. Bioorg Med Chem. 1997;5:1197–1207. doi: 10.1016/s0968-0896(97)00062-x. [DOI] [PubMed] [Google Scholar]
- 64.Telesnitsky A, Goff S P. Strong-stop strand transfer during reverse transcription. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 49–83. [Google Scholar]
- 65.Temin H M. The effects of actinomycin D on growth of Rous sarcoma virus in vitro. Virology. 1963;20:577–582. doi: 10.1016/0042-6822(63)90282-4. [DOI] [PubMed] [Google Scholar]
- 66.Temin H M. The participation of DNA in Rous sarcoma virus production. Virology. 1964;23:486–494. doi: 10.1016/0042-6822(64)90232-6. [DOI] [PubMed] [Google Scholar]
- 67.Temin H M. The DNA provirus hypothesis. The establishment and implications of RNA-directed DNA synthesis. Science. 1976;192:1075–1080. doi: 10.1126/science.58444. [DOI] [PubMed] [Google Scholar]
- 68.Temin H M, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 69.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vigier P, Goldé A. Effects of actinomycin D and of mitomycin C on the development of Rous sarcoma virus. Virology. 1964;23:511–519. doi: 10.1016/0042-6822(64)90235-1. [DOI] [PubMed] [Google Scholar]
- 71.Wadkins R M, Jovin T M. Actinomycin D and 7-aminoactinomycin D binding to single-stranded DNA. Biochemistry. 1991;30:9469–9478. doi: 10.1021/bi00103a012. [DOI] [PubMed] [Google Scholar]
- 72.Wells R D, Larson J E. Studies on the binding of actinomycin D to DNA and DNA model polymers. J Mol Biol. 1970;49:319–342. doi: 10.1016/0022-2836(70)90248-2. [DOI] [PubMed] [Google Scholar]
- 73.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.You J C, McHenry C S. HIV nucleocapsid protein. Expression in Escherichia coli, purification, and characterization. J Biol Chem. 1993;268:16519–16527. [PubMed] [Google Scholar]
- 75.You J C, McHenry C S. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J Biol Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]