Abstract
Vitamin B6 plays a crucial role in cellular metabolism and stress response, making it an essential component for growth in all known organisms. However, achieving efficient biosynthesis of vitamin B6 faces the challenge of maintaining a balanced distribution of metabolic flux between growth and production. In this study, our focus is on addressing this challenge through the engineering of phosphoserine aminotransferase (SerC) to resolve its redundancy and promiscuity. The enzyme SerC was semi-designed and screened based on sequences and predicted kcat values, respectively. Mutants and heterologous proteins showing potential were then fine-tuned to optimize the production of vitamin B6. The resulting strain enhances the production of vitamin B6, indicating that different fluxes are distributed to the biosynthesis pathway of serine and vitamin B6. This study presents a promising strategy to address the challenge posed by multifunctional enzymes, with significant implications for enhancing biochemical production through engineering processes.
Keywords: Vitamin B6, Phosphoserine aminotransferase SerC, Protein engineering, Multifunctional enzymes, Metabolic flux distribution
1. Introduction
Vitamin B6 is produced via the de novo biosynthetic pathway in various bacteria, fungi, and higher plants, excluding animals and humans [[1], [2], [3]]. The commercial form is pyridoxine (PN), which is synthesized through a Diels-Alder chemical reaction at high temperature after the formation of an oxazole intermediate. Raw materials such as oxalic acid, benzene, hydrochloric acid, liquid alkali and phosphorus oxychloride were used in this process [[4], [5], [6]]. Considering that some of the raw materials utilized in these processes are derived from fossil fuels, the depletion of fossil fuel reserves has led to a growing interest in exploring eco-friendly and sustainable alternative for the production of bulk chemicals. This includes the utilization of bio-based or renewable feedstocks as substitutes [7,8]. Indeed, these eco-friendly and sustainable processes for producing bulk chemicals using bio-based or renewable feedstocks offer several advantages. They have gained significant attention due to their potential to reduce dependence on fossil fuels, minimize environmental impact, and promote sustainability. Therefore, the establishment of a biosynthesis process for pyridoxine (PN) holds significant research importance in the field [9].
Aminotransferases also known as transaminases, are a group of enzymes with the capability to transfer an amino group, typically in a reversible manner, from a carbonyl or aldehyde carbon to α-keto acid to produce an amino acid. This enzymatic process is an essential part of amino acid metabolism in living organisms [10,11]. Aminotransferases rely on the presence of a cofactor called pyridoxal 5′-phosphate (PLP), which serves as a carrier for the amino group during the enzymatic transfer process [[12], [13], [14], [15]]. The amino group is often donated by glutamate, resulting in the formation of a pyridoxamine intermediate [16,17]. The pyridoxamine then donates the amine to the carbonyl carbon of an α-keto acid or an aldehyde [16,18]. Phosphoserine aminotransferase (SerC) from Escherichia coli (E. coli), known as a redundant and promiscuous enzyme, which participate in biosynthesis of serine and PLP, both the biosynthesis of serine and the catalytic activity of SerC itself require PLP as a cofactor to carry out the aminotransferase reaction [11,[19], [20], [21], [22], [23], [24]]. Zhang et al. changed the substrate specificity of SerC from l‐phosphoserine to l‐homoserine using a computation‐based rational approach in producing 1,3‐propanediol (1,3‐PDO) [20]. As reported in the study, the enzyme activity of the best mutant SerC(R42W/R77W) is successfully improved by 4.2‐fold compared to the wild type (WT) when using l‐homoserine as the substrate. However, it is important to note that the mutant's activity towards the natural substrate l-phosphoserine was completely deactivated. This trade-off in substrate specificity highlights the specificity of enzyme-substrate interactions and the challenges in engineering enzymes for targeted transformations. Thus, the rational design of SerC is important to improve the titer of special chemical compound.
In the vitamin B6 biosynthetic pathway, SerC catalyzed the 2-oxo-3-hydroxy-4-phosphobutanoate (OHPB) to 4-phosphonooxy-l-threonine (4HTP), this intermediate, 4HTP, is considered to be a toxic substance within the pathway [9,25,26]. In a previous study, parallel pathway engineering was performed to decouple PN and PLP production by introducing a heterologous PLP synthesis pathway [9]. The direct overexpression of SerC has been utilized for enhanced microbial pyridoxine production, which is realized by increasing the metabolic flux and providing alpha-ketoglutarate for potential PdxB multiple turnovers [27,28]. However, it did not change the flow of SerC to different metabolic pathways. Additionally, potential serine accumulation has been reported to hamper a number of different cellular processes, such as cell division, peptidoglycan synthesis, homoserine dehydrogenase (ThrA) involved in branched chain amino acid biosynthesis, etc [[29], [30], [31]]. Therefore, it is crucial to undertake rational design of SerC by modifying its substrate affinity to enhance pyridoxine yield and improve cell viability.
In this study, we firstly designed the serC by multiple sequence alignment and mining to achieve better performance for PN production. The residues or heterologous sequences were identified by molecular dynamics (MD) simulations and decomposition of binding free energy. Then, the expression of SerC was regulated by induced expression, changed CDS sequences, and to increase the copies to fit the modules with pathway. Amino acid analysis revealed a preferential metabolic flux towards the vitamin B6 synthesis pathway rather than the serine pathway. This study successfully tackles the challenges of carbon flux distribution through enzyme engineering, providing valuable insights for the engineering of multifunctional enzymes.
2. Material and methods
2.1. Construction of strains and plasmids
E. coli DH5α was used to construct and preserve plasmids. The E. coli strains utilized and modified in this work are derived from E. coli (MG1655). The strains, plasmids and primers used in this study are listed in Tables S1, S2, and S3, respectively. In general, all plasmids used in this study were based on p15ASI and pRSFDuet-1 according to the previous study [9]. DNA manipulations were performed according to the standard protocols. The primers used in this study were synthesized by GEMEWIZ biotechnology Co., Ltd (Suzhou, China). All recombinant plasmid were assembled by ClonExpress® MultiS One Step Cloning Kit (Vazyme Biotech Co., Ltd; catalog number C113-01). The mutated primers were designed using CE Design software from Vazyme Biotech Co., Ltd [32]. The sequences of heterologous proteins were displayed in Table S4. The coding sequence were codon-optimization according to E. coli expression system codon preferences, which synthesized by GENEWIZ biotechnology Co., Ltd (Suzhou, China).
Site-specific mutations were generated by polymerase chain reaction (PCR) [Takara PrimeSTAR Max DNA Polymerase (catalog number R045Q), direct and reverse primers 10 μM, template plasmid <100 ng] with plasmid pRSFDuet-1-serC used as the template. The PCR products were subjected to treatment with FD DpnI at 37 °C in a heat block for 5 min to eliminate the intact template plasmids, before being transformed into competent DH5α E. coli. Transformation assays used competent cell preparation kit (Generay Biotechnology Co., Ltd; catalog number GK6032).
2.2. Media and fermentation condition
Luria-Bertani (LB) medium (10 g/L NaCl, 5 g/L yeast extract and 10 g/L tryptone) was used to enrich cells for plasmid extraction. SerC variants were screened and cultured in 5 mL seed medium (10 g/L glycerol, 10 g/L Bacto peptone, 5 g/L yeast extract, 5 g/L NaCl, pH adjusted to 6.5) test tubes for 12 h. The bacterial culture was transferred into 30 mL of FM1.4 medium in 250 mL shake flask (15 g/L glycerol, 1 g/L glucose, 5 g/L yeast extract, 5 g/L Bacto peptone, 5 g/L NaCl, 200 mg/L MgSO4·7H2O, 10 mg/L MnSO4·5H2O, 10 mg/L FeSO4·7H2O, 100 mM Na2HPO4, pH 6.5) to an initial OD600 = 0.1 and cultured at 37 °C, 200 rpm for 48 h. All cultivations were performed in duplicates or triplicates. When necessary, kanamycin and chloramphenicol were added to the culture media at final concentrations of 50 and 34 μg/mL, respectively.
2.3. Analytical methods to monitor fermentation
The cell density (OD600) was measured by Hybrid Multi-Mode Reader (Synergy Neo2, Bio Tek, USA). Residual glycerol was determined using On-line biochemical analyzer (M101, Siemen, China).
The fermentation mixture was centrifuged at 10,625 × g, and the resulting supernatant was analyzed using high-performance liquid chromatography (HPLC) with an UltiMate™ 3000 equipped system with an FLD-3400 detector. Before being injected into the HPLC system, the sample was filtered through a membrane filter with a pore-size of 0.22 μm to remove impurities. The resulting samples were analyzed using fluorescence detection. The excitation and emission wavelengths were set to 293 nm and 395 nm, respectively. The Octadecylsilyl (ODS) column (Cosmosil AR-Ⅱ; 250 by 4.6 mm, 5 μm particle size; Nacalai Tesque) was used for separation. Mobile phase A (33 mM phosphoric acid and 8 mM 1-octanesulfonic acid, adjust pH to 2.5 with KOH) and mobile phase B (acetonitrile: water = 4:1, v/v). The column temperature was 35 °C. The total flow rate was 0.8 mL/min and the gradient elution program is set to: mobile phase B from 0% to 1% in 0–5 min, 1%–19% in 5–10 min, 19%–28% in 10–20 min, 28%–63% in 20–25 min, 63%–0% in 25–27 min, 0% B keep 5 min. The data from fermentation was analyzed by the software GraphPad Prism (version 8).
2.4. Molecular dynamics simulation
The complex structures of Eco_SerC and its mutant variants were constructed based on the crystal structure 1BJO from the PDB database [33]. The structures of SerC from other sources were predicted using the Alphafold2 algorithm [34]. According to the proposed catalytic mechanism [35], SerC requires the essential coenzyme pyridoxal 5′-phosphate (PLP) for catalysis. First, an imine is formed by the reaction of an amino group with glutamate, and then this imine is transferred onto the substrate 3-phosphooxypyruvate/(R)-3-hydroxy-2-oxo-4-phosphooxybutanoate by forming an imine with the carbonyl group of the substrate. To comprehensively study potential factors influencing catalysis, particularly the co-occurrence of substrate and coenzyme binding, we conducted molecular dynamics simulations to explore the binding energy and binding mode. The complex structures were prepared using the Amber20 software package with ff14SB force field [36,37]. Initial constrained molecular dynamics simulations were performed for 20 ns, followed by three independent unconstrained 100 ns MD simulations to capture the dynamic changes of the system. Further details regarding the simulation methodology can be found in the Supporting Information.
2.5. Amino acid and organic acid analysis
The intracellular serine content was accurately quantified using an amino acid analyzer (Hitech, L8900) [38]. The appropriate amount of fermented samples was collected and centrifuged at 10,625 × g to obtain the bacterial cells. The cells were lysed using a solution 0.8 M HClO4 and 0.8 M K2CO3. The resulting supernatant was obtained by centrifuging at 10,625 × g, subsequently treated with a 1:1 ratio of 10% sulfosalicylic acid dihydrate to remove proteins. The mixture was further centrifuged at 10,625 × g for 15 min. The volume of supernatant was adjusted by adding an equal volume of 0.02 M hydrochloric acid. To remove impurities, a filter membrane with a pore size of 0.22 μm was used. Each sample was filtered for a duration of 2.5 h (with a net analysis time of 110 min). Data acquisition and analysis for amino acid analysis were performed using the post-column derivatization method on the amino acid analyzer.
Organic acid such as acetic acid and pyruvate were detected by HPLC system (Agilent 1260 Infinity Ⅱ) equipped with a 1260 Infinity Ⅱ VWD detector. Choose polystyrene divinylbenzene column (Aminex® HPX-87H; 300 × 7.8 mm, 9 μm particle size; Bio-Rad) as the separation system of HPLC system. The flow rate of mobile phase A (5 mM H2SO4) is set to 0.5 mL/min.
2.6. SDS-PAGE analysis
After 16 h of fermentation, 1 mL of the fermentation broth was collected and centrifuged at 1844 × g for 10 min at 4 °C to separate the supernatant. The pellets were re-suspended with 1 mL of pre-cooled Phosphate-Buffered Saline (PBS) solution, then centrifuged at 4000 rpm for 30 min at 4 °C to remove the supernatant. This process is repeated three times, the resuspension was then diluted to an optical density (OD) of 600 nm equal to 0.2 with PBS solution, the diluted resuspension was lysed with Ultrasonic Cell Disruptor (operating for 2 s, pausing for 3 s, 60 times). The cell lysate (1 mL) was centrifuged at 7378 × g and 4 °C for 10 min, the supernatant was separated from the sediment. The supernatant (40 μL) was taken and mixed with 10 μL of 5 × Protein SDS- PAGE loading Buffer, and then heated to 100 °C for 20 min. The samples were added to the protein gel and separated at 130 V for nearly 40 min.
2.7. Sequence analysis and phylogenetic tree construction
The protein sequences of SerC were extracted from the BRENDA database based on EC number 2.6.1.52. Multiple sequence alignment and comparative analysis of the protein were conducted using the command-line version ClustalW2 [39]. Phylogenetic trees were constructed using the data from the alignments in MEGA11 software and viewed in ITOL [40,41]. Protein structure analysis and figure design were performed using PYMOL 2.1 [42].
Sequence alignment was performed using the ClustalW2 program and plotted with the ESPript program [43].
3. Results and discussion
3.1. Analysis of SerC protein by multiple sequence alignment to design sites mutation
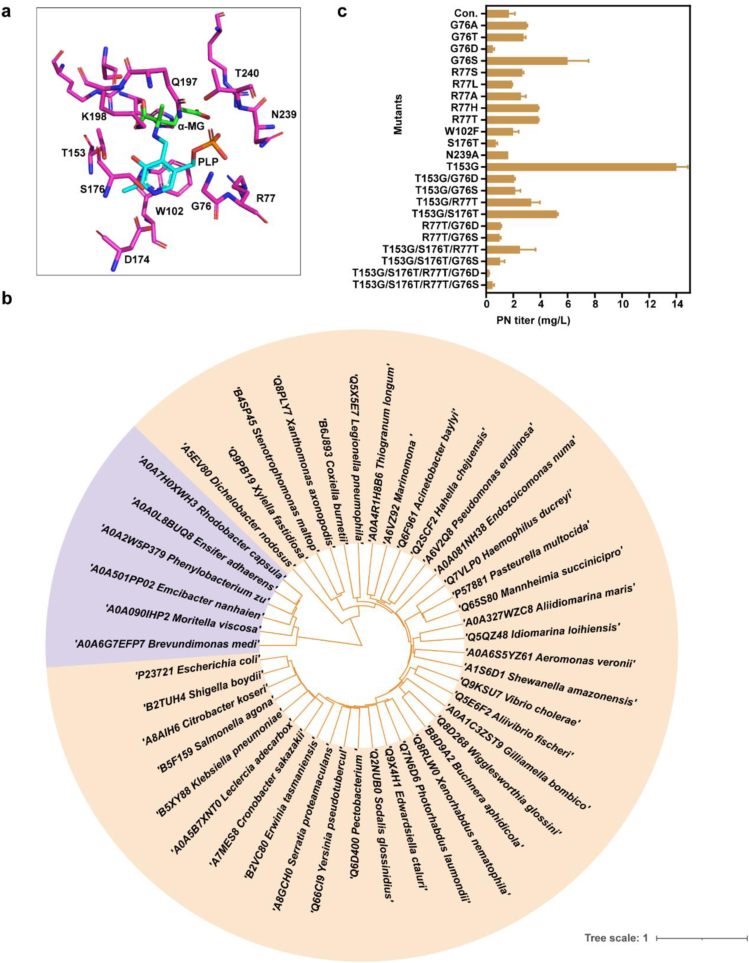
Serine is ubiquitously synthesized in all living organisms from the glycolysis intermediate 3-phosphoglycerate (3PG), including the phosphoserine aminotransferase SerC catalyzed 3-phosphohydroxylpyruvate (3PHP) and glutamate into 3-phosphoserine and α-ketoglutarate (α-KG), the second step in the phosphorylated pathway of serine biosynthesis [44]. SerC is also OHPB aminotransferase, participated in the third step of DXP (1-deoxy-d-xylulose-5-phosphate synthase)-dependent pathway to synthesize vitamin B6. The DXP-dependent vitamin B6 pathway has been revealed that it is only present in γ-proteobacteria or the α-proteobacteria by an in-silico analysis [45]. The orthologs of SerC (EC 2.6.1.52) were identified from the BRENDA enzyme database (www.brenda-enzymes.org) and restricted to mainly γ-proteobacteria or the α-proteobacteria to do multiple sequence alignment.
Crystal structure of phosphoserine aminotransferase SerC from E. coli has been solved (PDB: 1BJO) at 2.3 Å resolution [19]. The complete dimer of SerC is essential for activity for its active sites situated at the subunit interface. The region responsible for binding substrate molecules such as PLP and α-methyl-l-glutamate (α-MG) includes the following residues: G76, R77, W102, T153, D174, S176, Q197, K198, T240, and N239 (Fig. 1a). In this study, our analysis and mutation efforts were primarily focused on this specific 10 sites [19,46]. To identify mutations of key residues to better produce pyridoxine, multiple sequence alignment of amino acid sequences of closest SerC homologs that restricted in γ-proteobacteria and the α-proteobacteria were performed by ENDscript (Fig. 1b). Secondary structure of SerC was analyzed by ENDscript 3.0 indicated 9 α-helices and 16 β-sheets (Fig. S1). The conservation of certain sites was observed among the active sites, specifically D174, Q197, K198, and T240. These sites demonstrated a high degree of similarity across various species. However, some species such as Edwardsiella ctaluri, Citrobacter koseri, Xanthomonas axonopodis, Coxiella burnetiid, Legionella pneumophila, Haemophilus ducreyi showed in Fig. 1b exhibited variations in other sites, including G76, R77, W102, T153, S176, and N239. In order to boost the activity of the native serC gene from E. coli and improve pyridoxine production, we conducted mutation experiments aimed at identifying efficient mutations. Thirteen transformants were constructed through site-directed mutagenesis and subsequently tested for PN titer (Fig. 1c). Compared with the WT, the titer of SerC (T153G) overexpression was increased by more than 7-fold, and G76S increased 2.6-fold. To further enhance the performance of SerC, we conducted double‐site to fourth-site mutagenesis based on shared mutation sites across various bacterial strains, with particular emphasis on sites harboring the T153G mutation site. Among them, the mutants with the combination of residues of T153G and S176T increased the PN titer, although still lower than that of T153G mutants. The results showed that the mutation sites T153G, G76S, and T153G/S176T were preferable for PN production.
Fig. 1.
Mutation sites of SerC based on sequence. (a) The active sites of SerC protein from E. coli (PDB: 1BJO). (b) Phylogenetic tree constructed on the basis of SerC homologs chosen from γ-proteobacteria (orange) and the α-proteobacteria (purple). (c) The PN titer of SerC overexpression mutants. Data are presented as mean values ± SD from two independent biological replicates (n = 2).
To elucidate the underlying mechanisms behind the altered production yield of these mutants, we conducted molecular dynamics (MD) simulations on several variants: T153G (the variant with the highest yield), G76D (the variant with the lowest yield), G76S (a variant with improved yield compared to G76D), as well as the mutant T153G/S176T, which exhibited the highest yield among the combination mutants. The MD simulations revealed that the G76D mutation significantly reduced the binding affinity towards the substrate PLP_GLU (PLP-binding glutamic acid), as well as the substrates PLP_OHPB and PLP_3PHP (Table 1). Particularly noteworthy was the approximately 50% decrease in binding affinity towards PLP_OHPB and PLP_3PHP (PLP_OHPB binding energy increased from −200.90 to −77.15 kcal/mol, and PLP_3PHP binding energy increased from −176.20 to −95.83 kcal/mol). As a result, these mutants likely impact a portion of the metabolic flux that bypasses SerC enzyme, resulting in a decreased production yield in the vitamin B6 pathway.
Table 1.
The binding energy (kcal/mol) of native SerC from E. coli and its mutants to multiple substrates.
| mutations | PLP_GLUa | PLP_OHPBb | PLP_3PHPc | PLP_ OHPB-PLP_3PHP |
|---|---|---|---|---|
| WT | −130.04 | −200.90 | −176.20 | −24.70 |
| G76D | −125.74 | −77.15 | −95.83 | 18.68 |
| G76S | −173.35 | −150.78 | −190.66 | 39.88 |
| T153G | −174.59 | −191.78 | −157.08 | −34.70 |
| T153G_S176T | −170.05 | −158.72 | −173.50 | 14.78 |
PLP covalent with GLU.
PLP covalent with OHPB.
PLP covalent with 3PHP.
Conversely, the G76S mutation at the same position exhibited a significant decrease in binding energy towards the PLP_GLU substrate (approximately 43.4 kcal/mol) with less affecting the PLP_OHPB substrate. This finding provides a possible explanation for the higher productivity of G76S compared to the wild type and G76D. Additionally, both T153G and T153G/S176T demonstrated advantages over WT in terms of their binding to the substrate PLP_GLU. The T153G mutation has a decrease of 44.6 kcal/mol in binding energy towards PLP_OHPB, accompanied by no significant change in PLP_GLU. Notably, the T153G mutation increased the binding energy to PLP_3PHP from −176.20 to −157.08 kcal/mol, thereby reducing the binding energy difference for OHPB and PLP_3PHP from −24.70 to −34.70 kcal/mol. This particular modification in binding affinity has increased the selectivity of SerC towards the vitamin B6 pathway's OHPB substrate. Consequently, it enhanced the overall metabolic flux through SerC while simultaneously improving the competitive advantage of the OHPB substrate in the vitamin B6 pathway, resulting in a substantial increase in the production yield of T153G. It can be seen that the ability of substrate recruitment is a key factor in improving yield. To further explore SerC enzymes with inherent advantages in vitamin B6 pathway substrate metabolism, we conducted further investigations using heterologous SerC.
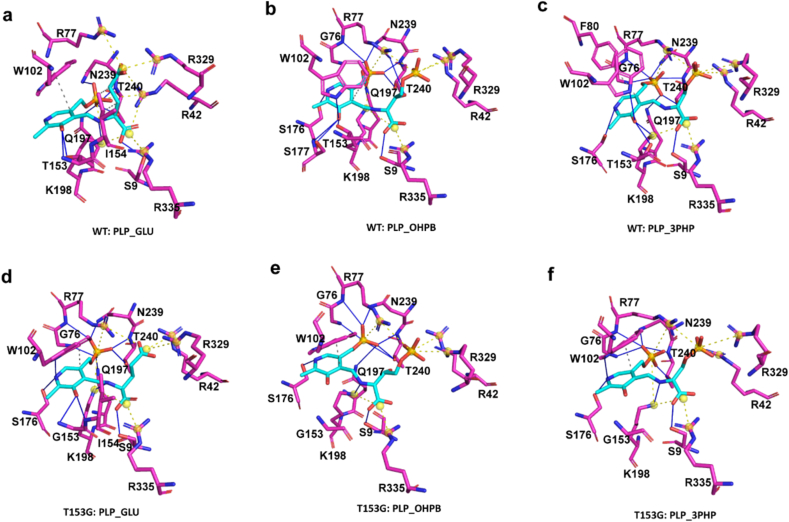
A comprehensive analysis was conducted to investigate the distinct binding characteristics between the high-yield mutant T153G and the WT protein. Using Protein-Ligand Interaction Profiler (PLIP) analysis, we analyzed the binding complexes of individual substrates with the protein, as depicted in Fig. 2a–f. Comparing the PLP_GLU complexes of T153G and WT, we observed that the T153G mutant displayed an additional 4 hydrogen bond interactions (residues S178, W102, G76, and R77) as shown in Fig. 2a and d, and Table S5. These interactions significantly favored the binding of the GLU substrate. Conversely, the T153G mutation led to the loss of 3 hydrogen bond interactions (W102, T153, and S177) with the PLP_OHPB substrate, as illustrated in Fig. 2b and e, and Table S5. The mutation introduced a novel hydrophobic interaction (W102) in the PLP_3PHP complex; however, it also resulted in the loss of 2 hydrogen bonds (T153 and N197) and one hydrophobic interaction (F78), as depicted in Fig. 2c and f, and Table S5. These alterations in the binding profile potentially enhance the recruitment of GLU and OHPB by the T153G mutant variant.
Fig. 2.
Interactions networks of different substrates with the WT and T153G enzymes. (a), (b), and (c) represent the interaction networks of WT with substrates PLP_GLU, PLP_OHPB, and PLP_3PHP, respectively. (d), (e), and (f) depict the interaction networks of T153G with substrates PLP_GLU, PLP_OHPB, and PLP_3PHP, respectively. Ligands are shown in cyan and protein residues in magenta. Hydrogen bonds (solid blue line), hydrophobic contacts (dashed gray line), and salt bridges (dashed yellow line with yellow balls) are detected between ligands and targets.
3.2. Mining beneficial SerC by GotEnzymes database
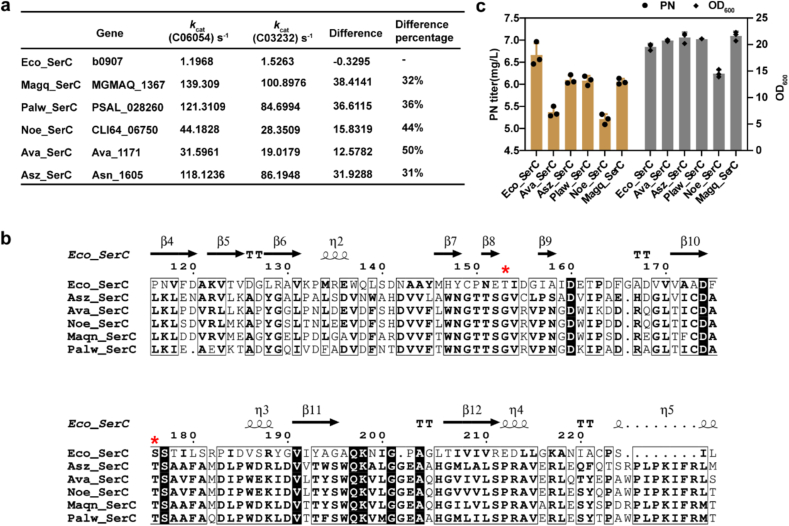
The BRENDA enzyme database has collected enzyme parameters, which are experimentally determined, much less than the numbers in GotEnzymes (https://metabolicatlas.org/gotenzymes). Based on the predicted parameters mainly enzyme turnover number kcat by the GotEnzymes database, it would guide enzyme selection and design from a large size of the data. From the database, we extracted the SerC enzyme information covering 6171 species available by submit the EC 2.6.1.52. The compound C06054 (OHPB) was the target molecule, while the compound C03232 (3PHP) was regarded as the competitive substrate. The following limits were applied: one is the relatively large kcat number, the other is the difference between the two and its percentage in the kcat number of the target compound. We selected five SerC homologous genes (Fig. 3a) for codon optimization and artificial gene synthesis. By multiple sequence alignment, these five sequences shared similar mutations at T153 and S176 sites (Fig. 3b). We overexpressed these 5 genes in the E. coli, the results showed that single overexpression of the heterologous genes was no better than the native one (Fig. 3c). The results may be due to the fact that the poor expression of foreign genes in E. coli from the results of the protein expression levels by SDS-PAGE (Fig. S2). The results showed that the expression of heterologous protein is weak other than Ava_SerC, which is consistent with our hypothesis.
Fig. 3.
Screening of heterologous SerC genes. (a) The heterologous genes mined from the GotEnzymes database based on kcat values. Magq, Magnetospira sp. QH-2; Palw, Pseudooceanicola algae; NOE, Nostoc sp. CENA543; Ava, Trichormus variabilis; Asz, Acetobacter senegalensis. (b) Multiple sequence alignment by the program ESPript (Easy Sequencing in PostScript). The red asterisk represented the position of 153 and 176 of native SerC from E. coli. (c)The PN titer and cell growth of heterologous SerC mutants. Data are presented as mean values ± SD from three independent biological replicates (n = 3), the circles or squares represent individual data points.
To investigate potential changes in substrate binding and selectivity, beyond the impact of expression levels alone, we performed MD simulations on five heterologous enzymes. The data revealed a significant decrease in the binding affinity of the heterologous enzymes towards all substrates (Table 2). Although, the NOE_SerC exhibiting the highest binding affinity to PLP_OHPB, displayed more than 50% increase compare with Eco_SerC (PLP_OHPB: increased from −200.90 to −70.07 kcal/mol). This shift in binding affinity has the potential to impact enzyme activity and, in conjunction with the reduction in expression levels, lead to a decrease in production yield. Of interest, despite the suboptimal expression levels and lower binding affinity, the heterologous enzymes demonstrated superior substrate selectivity within the vitamin B6 pathway compared to the native E. coli strain (the binding energy of PLP_OHPB - PLP_3PHP ranges from −26.43 to −58.07 kcal/mol). We hypothesized that when there is insufficient pulling and pushing forces upstream and downstream, the substrate's intrinsic binding affinity plays a dominant role. Due to the lower binding affinities of heterologous enzymes to vitamin B6 pathway substrates compared to Eco_SerC, this leads to inadequate flux and a subsequent decline in yield. However, when there is sufficient tension between the upstream and downstream processes, the competitive advantage of the vitamin B6 pathway becomes evident, providing more opportunities for efficient conversion of vitamin B6 substrates and potentially enhancing production yield. Consequently, these sourced SerC enzymes hold promise for enhancing vitamin B6 production by modulating metabolic flux diversion, following optimization of expression and upstream/downstream genes.
Table 2.
The binding energy (kcal/mol) of heterologous SerC to multiple substrates.
| species | PLP_GLUa | PLP_OHPBb | PLP_3PHPc | PLP_OHPB - PLP_3PHP |
|---|---|---|---|---|
| Eco | −130.04 | −200.90 | −176.20 | −24.70 |
| Asz | −43.34 | −88.60 | −62.17 | −26.43 |
| Ava | −50.19 | −58.90 | −24.45 | −34.45 |
| Magq | −53.44 | −67.59 | −31.68 | −35.91 |
| NOE | −37.53 | −70.07 | −12.00 | −58.07 |
| Palw | −26.76 | −56.72 | −13.09 | −43.63 |
PLP covalent with GLU.
PLP covalent with OHPB.
PLP covalent with 3PHP.
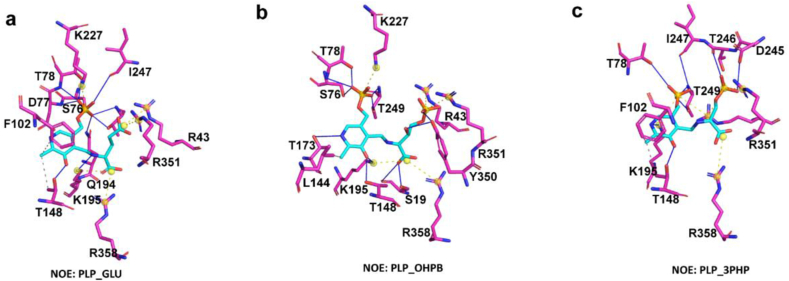
We further analyzed NOE_SerC, which exhibited the highest specificity differences. From Fig. 4a, b, and 4c, it is evident that each of the three substrates displayed distinct binding modes, particularly for PLP_3PHP. The substrate moiety in PLP_3PHP predominantly relied on residues R43, D245, T246, and R358 for stabilization, while the substrate moiety in PLP_OHPB was primarily stabilized by residues S19, R43, K195, Y350, R351, and R358. Notably, due to the presence of an additional hydroxyl group in the substrate moiety of PLP_OHPB compared to PLP_3PHP, Y350 may provide additional binding interactions and specificity, thereby further enhancing the selectivity of NOE_SerC towards to PLP_OHPB.
Fig. 4.
Interactions networks of different substrates with NOE_SerC. (a), (b), and (c) show the interaction networks of NOE_SerC with substrates PLP_GLU, PLP_OHPB, and PLP_3PHP, respectively. Ligands are shown in cyan and protein residues in magenta. Hydrogen bonds (solid blue line), hydrophobic contacts (dashed gray line), and salt bridges (dashed yellow line with yellow balls) are detected between ligands and targets.
3.3. Effective production of pyridoxine by fine-tuning the expression of SerC
In a previous study, we developed two plasmids carrying the entire enzymes of vitamin B6 pathway (Fig. S3) for the production of PN in an engineered E. coli strain [9]. One plasmid based on p15ASI backbone expressed the upstream genes of the biosynthetic pathway including epd from Glaciecola nitratireducens, native pdxB with codon-optimization, dxs from Ensifer meliloti, and native serC. The other plasmid based on pRSFDuet-1 backbone expressed the downstream genes of pdxA2 and pdxJ1, which were designed rationally based on the native enzyme from E. coli [9]. Notably, these plasmids were designed to achieve high PN yields without any modifications made to the SerC enzyme. To achieve a more balanced SerC expression level, we either replaced the serC gene on the plasmid or fine-tuned its expression, enabling us to allocate different metabolic fluxes to the serine and PN biosynthesis pathways.
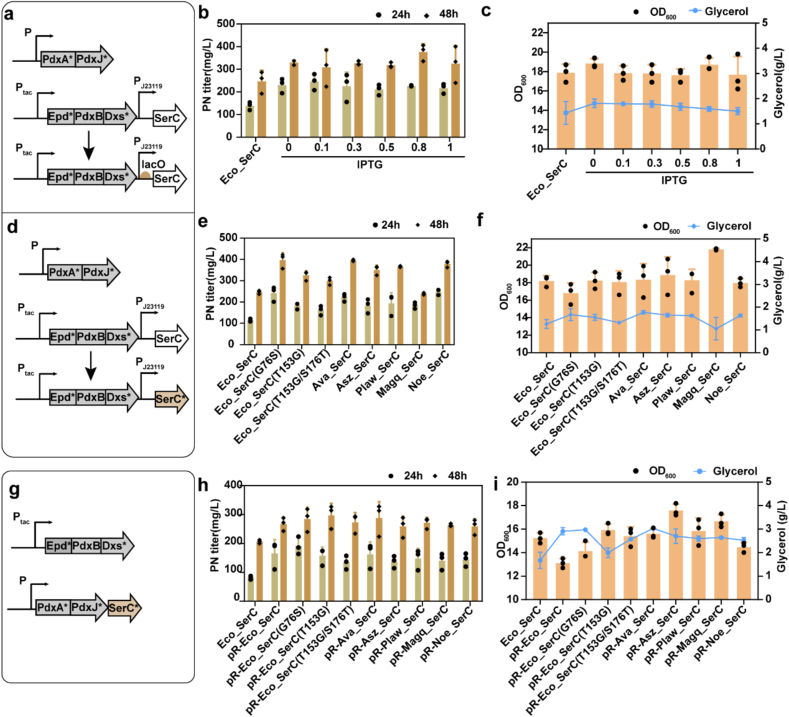
To fine-tune the expression of SerC to increase PN titer, we initially introduced the lac operator (lacO) sequences downstream of the J23119 promoter controlling SerC expression in the p15ASI plasmid, which contained a lacI gene (Fig. 5a). To induce the expression of SerC, we used IPTG (isopropyl-β-D-thiogalactoside), which is an effective inducer of protein expression in the concentration range of 100 μM to 1.0 mM. Therefore, we cultivated the E. coli mutant strain CK030 in FM1.4 medium supplemented with 0, 0.1, 0.3, 0.5, 0.8, and 1.0 mM of IPTG at OD600 = 0.6–0.8. After incubation for 24 and 48 h at 200 rpm, 37 °C, the PN titer was measured by HPLC. The results showed that the induced expression of SerC was better than the constitutive expression. However, the change of PN titer was not obviously different in the range of 0.1–1 mM IPTG with the maximum titer was 376.16 mg/L under the induction concentration of 0.8 mM IPTG (Fig. 5b). There were no significant differences observed in cell growth (OD600) between the mutant strains and the original strains, both of which utilized the constitutive expression promoter. Compared with the control strain, the titer increased by 52.6% and the glycerol consumption decreased by 3% (Fig. 5b and c). Although the influence of different IPTG concentrations on PN production was not significant, it indicated that optimization of SerC expression in time and intensity was necessary to improve PN production. It was speculated that the expression intensity of SerC is not the limitation of product improvement in the current strain.
Fig. 5.
Regulation of the expression of SerC based on the high-yield plasmid form. (a), (d), (g) Schematic diagram of the strategies for SerC regulation. (b), (e), (h) The PN titer of diferent mutants in 24 h and 48 h. (c), (f), (i) The cell growth (OD600) and residual glycerol. Eco_SerC, the native SerC of E. coli. Data are presented as mean values ± SD from three independent biological replicates (n = 3), the circles or squares represent individual data points.
To avoid inducer addition in the bioreactor and lower the fermentation cost, better mutants and homologs genes were further used to explore the constitutively expression by replacing the native serC gene. The native serC gene from E. coli was replaced by site-directed mutants, and the heterologous genes were introduced to screen for improved serC variants for PN titer (Fig. 5d). A PN production assay was conducted on the transformants containing the G76S, T153G, T153G/S176T mutations, as well as the other five heterologous genes. The shake flask results revealed that both the G76S mutation and the ava_serC gene exhibited higher performance compared to the other 6 mutant strains. The highest titer of PN produced by Eco_SerC (G76S) reached 241.09 mg/L at 24 h and 396.79 mg/L within 48 h, while the remaining glycerol were 1.67 g/L (Fig. 5e and f). The comparison between the PN yields at 24 h and 48 h indicated that the mutants exhibited a relatively stable production rate throughout cultivation. The cell growth was comparable among the mutants, indicating that any differences in capacity were due to variations in native serC mutans and heterologous genes.
From a proteomic analysis in the previous study, SerC expression was lower compared with other proteins in the vitamin B6 biosynthetic pathway [9]. As for the p15ASI was a low-copy-number vector (plasmid copy number 10–12), we transferred the placement of serC to the high copy plasmid pRSFDuet-1(copy number >100) containing the pdxA and pdxJ genes named pR-series (Fig. 5g). The PN titer of the resulting strains was tested and compared to the parental strain. As shown in Fig. 5h, the T153G mutation and the mutant with ava_serC expression produced PN 297.31 mg/L and 288.30 mg/L, respectively. The PN titers using pRSFDuet-1 expression were lower than p15ASI plasmid. It demonstrated that serC preferred to low-copy-number vector, while the over-expression of serC created a metabolic burden on the host by high-copy-number vector. This is consistent with the results that the growth (OD600) of the pR-series mutants is low, and then the residual glycerol was more than the expression of the low-copy plasmid (Fig. 5i).
The variations in product concentrations observed from native SerC mutations and heterologous protein expression could be attributed to the varying degrees of fitness cost imposed on the system compared to the control strain. These fitness costs may have affected the overall performance and productivity of the system, resulting in different product concentrations [47,48]. Microbial fitness and production rates are complex that are determined by multivariable factors, such as carbon metabolism and enzyme catalyzing efficiency [[49], [50], [51]]. The new fitness state could result from an alleviation of production load, or even site mutations in the enzyme from the match between the specialty and selection. This adaptation arises from the improved matching between the enzyme's selection and binding to the substrate [52]. In this study, the enhanced production of SerC mutation or heterologous proteins could be attributed to the different binding and catalyzing ability of OHPB and 3PHP. Mutation in serC gene (G76S, T153G or T153G/S176T) has previously successfully improved PN yield in the single overexpression assay. In silico analysis by molecular dynamics has shown that these mutants had lower binding free energy with glutamine. The improved binding ability between OHPB and 3PHP caused by G76S and T153G mutation as the major reason for titer improvement. It is interesting to note that the overexpression of the heterologous serC gene did not result in an improvement in PN yield in the single overexpression assay, despite low protein expression. This suggests that other factors in the metabolic pathway may be limiting PN production, and simply increasing the expression of the serC gene alone is not sufficient to enhance PN yield. However, the increase in PN titer observed in the combination plasmid can be attributed to the selectivity and competitive advantage of the vitamin B6 pathway over the serine pathway. Additionally, it was observed that there was a balanced distribution of metabolic flux between the growth requirement for serine and the production of PN. This suggests that the metabolic network is finely regulated to ensure a sufficient supply of serine for cell growth while also diverting a portion of the metabolic flux towards PN production. This balanced distribution is crucial for maintaining cellular homeostasis and optimizing PN yield.
3.4. Amino acids analysis to display the distribution of metabolic flux
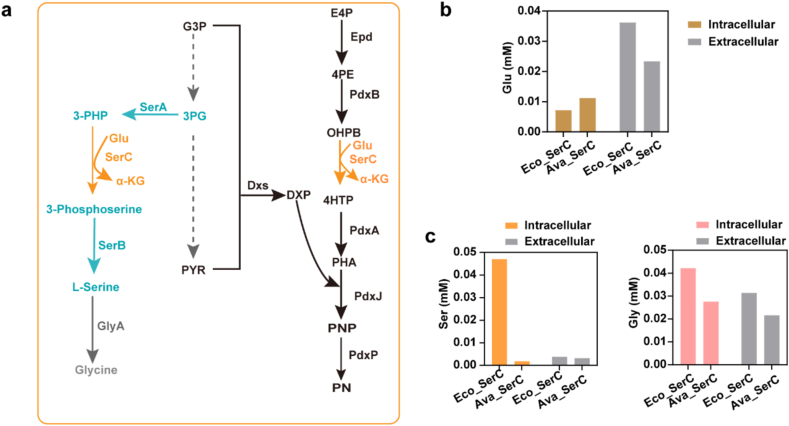
To better understand the metabolic flux states of serC engineering mutant, we measured the intracellular levels of serine and glutamate, as well as glycine which is the immediate product of the subsequent reaction in the biosynthesis of serine (Fig. 6a). The Eco_SerC (LL388 N) strain was used as the control, while the Ava_SerC (CK034) strain was used as the sample for amino acid analysis. The yield difference of PN between the CK034 strain and the control strain is approximately 200 mg/L. The availability of glutamate, which serves as a precursor in the biosynthetic process, was measured. The results showed that intracellular glutamate levels in CK034 were slightly higher compared to the control, while extracellular levels were slightly lower (Fig. 6b). According to a previous study, the absolute intracellular concentrations of glutamate in strains grown with glucose and glycerol ranged from 96 mM to 149 mM [17,53]. However, in the engineered strains, the glutamate levels were significantly reduced due to its depletion in the biosynthesis of vitamin B6, which aligns with the obtained results.
Fig. 6.
PN and serine biosynthetic pathways and the concentrations of specific amino acids. (a). The simplified metabolic pathways of serine and PN biosynthesis by engineered E. coli strain. Enzymes: Epd erythrose 4-phosphate dehydrogenase, PdxB 4-phosphoerythronate dehydrogenase, SerC 3-phosphserine aminotransferase, PdxA 4-phosphohydroxy-l-threonine dehydrogenase, PdxJ PNP synthase, Dxs 1-deoxyxylulose 5-phosphate synthase, PdxP PNP phosphatase, SerA D-3-phosphoglycerate dehydrogenase, SerB phosphoserine phosphatase, GlyA glycine hydroxymethyltransferase. Metabolites: E4P erythrose 4-phosphate, 4 PE 4-phosphoerythronate, OHPB 2-oxo-3-hydroxy-4-phosphobutanoate, 4HTP 4-phosphohydroxy-l-threonine, PHA 3-phosphohydroxy-1-aminoacetone, DXP 1-deoxy-d-xylulose 5-phosphate, G3P glyceraldehyde 3-phosphate, 3 PG 3-phosphoglycerate, PYR pyruvate, 3PHP 3-phosphohydroxylpyruvate, Glu glutamate, α-KG α-ketoglutarate. (b) and (c). The intracellular concentrations of glutamate (Glu), serine (Ser) and glycine (Gly). Eco_SerC was the control strain (LL388 N), and the Ava_SerC was the SerC engineered strain (CK034).
Additionally, another notable difference in the amino acid levels was observed in the concentrations of serine, which were consistent with our expectations (Fig. 6c). The intracellular and extracellular concentrations of serine in the CK034 strain were lower compared to the control strain (Fig. 6c). This decrease can be attributed to the fact that the SerC screened under the same culturing conditions exhibits an increased metabolic flux towards the PN production pathway, thereby reducing the flux into the serine biosynthesis pathway. However, in the control strain overexpression of the native SerC, the absolute intracellular concentration of serine (∼0.047 mM) did not show a significant increase, which was in the same order of magnitude compared to the previously reported about 0.068 mM in E. coli wild-type [53]. This could be attributed to the potential toxicity of serine to cells and the presence of feedback inhibition on the upstream pathway enzyme SerA. Consistent with the result of serine, the production of glycine catalyzed by one step of enzyme (GlyA) catalysis was also found to be consistent with the decrease in serine levels (Fig. 6c).
Furthermore, we measured the concentrations of organic acids, such as acetate and the precursor pyruvate, which can detectable in the fermentation supernatant. However, we observed no significant differences in their concentrations between the CK034 strain and the control strain (Fig. S4). The results suggest that engineering SerC did not significantly affect the levels of acetate and pyruvate. However, it effectively balanced the metabolic flux, leading to an enhancement in PN production.
4. Conclusions
In this study, the enzyme SerC was designed using sequence analysis and screened by predicted kcat values. Some mutants with potential and heterologous proteins were further optimized to improve the production of vitamin B6. MD analysis revealed that the substrate's intrinsic binding affinity and selectivity of SerC play a crucial role in determining the metabolic flux distribution. This finding highlights the significance of substrate interactions in influencing the direction of metabolic pathways and provides valuable insights into the regulation of metabolic fluxes. The fine-tuning assay, which involved regulating SerC expression through induced expression, modification of CDS sequences, and increasing copy numbers to fit the modules with the vitamin B6 pathway, showcased the engineered strain's ability to significantly enhance vitamin B6 production. This outcome suggests a redirection of metabolic fluxes towards both the serine biosynthesis pathway and the vitamin B6 biosynthesis pathway, contributing to the improved production of vitamin B6. This study showcases a promising strategy to overcome the challenges associated with multifunctional enzymes and holds significant potential for enhancing biochemical production through engineering processes. The findings suggest that optimizing enzyme performance and metabolic flux distribution can be instrumental in improving the production of valuable compounds.
CRediT authorship contribution statement
Kai Chen: Investigation, Methodology, Visualization, Writing – review & editing. Linxia Liu: Investigation, Methodology, Visualization, Funding acquisition, Writing – review & editing. Jinlong Li: Investigation, Methodology, Writing – original draft. Zhizhong Tian: Methodology, Visualization. Hongxing Jin: Supervision, Writing–review. Dawei Zhang: Conceptualization, Supervision, Funding acquisition, Resources, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.The author is an Editorial Board Member/Editor-in-Chief/Associate Editor/Guest Editor for [Journal name] and was not involved in the editorial review or the decision to publish this article.The authors declare the following financial interests/personal relationships which may be considered as potential competing interests.
Acknowledgements
This work was supported by the National Key R&D Program of China (2022YFC2106100), National Natural Science Foundation of China (22178372, 32200049), National Science Fund for Distinguished Young Scholars (22325807), Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-011, TSBICIP-CXRC-055) and Yellow River Delta Industry Leading Talents (DYRC20190212).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2024.03.005.
Contributor Information
Hongxing Jin, Email: jinhx87@126.com.
Dawei Zhang, Email: zhang_dw@tib.cas.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Samsatly J., Chamoun R., Gluck-Thaler E., Jabaji S. Genes of the de novo and salvage biosynthesis pathways of vitamin B6 are regulated under oxidative stress in the plant pathogen Rhizoctoniasolani. Front Microbiol. 2016;6 doi: 10.3389/fmicb.2015.01429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitzpatrick T.B., Amrhein N., Kappes B., Macheroux P., Tews I., Raschle T. Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem J. 2007;407(1):1–13. doi: 10.1042/BJ20070765. [DOI] [PubMed] [Google Scholar]
- 3.Tambasco-Studart M., Titiz O., Raschle T., Forster G., Amrhein N., Fitzpatrick T.B. Vitamin B6 biosynthesis in higher plants. Proc Natl Acad Sci U S A. 2005;102(38):13687–13692. doi: 10.1073/pnas.0506228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tasdelen M.A. Diels–Alder “click” reactions: recent applications in polymer and material science. Polym Chem. 2011;2(10):2133–2145. [Google Scholar]
- 5.Firestone R.A., Harris E.E., Reuter W. Synthesis of pyridozine by diels-alder reactions with 4-methyl-5-alkoxy oxazoles. Tetrahedron. 1967;23(2):943–955. [Google Scholar]
- 6.Osbond J.M. In: Harris R.S., Wool I.G., Loraine J.A., Marrian G.F., Thimann K.V., editors. vol. 22. Academic Press; 1964. Synthesis and Labeling of the vitamin B6 group; pp. 367–397. (Vitamins & hormones). [DOI] [PubMed] [Google Scholar]
- 7.Liu L., Zhao Z., Zhu R., Qin X. Can national environmental protection supervision and control have a lasting impact on corporate production efficiency? — an empirical study based on the multi-phase difference-in-difference model. Environ Sci Pollut Res. 2022;29(37):56136–56153. doi: 10.1007/s11356-022-19348-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ning P., Yang G., Hu L., Sun J., Shi L., Zhou Y., Wang Z., Yang J. Recent advances in the valorization of plant biomass. Biotechnol Biofuels. 2021;14(1):102. doi: 10.1186/s13068-021-01949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L., Li J., Gai Y., Tian Z., Wang Y., Wang T., Liu P., Yuan Q., Ma H., Lee S.Y., Zhang D. Protein engineering and iterative multimodule optimization for vitamin B6 production in Escherichia coli. Nat Commun. 2023;14(1):5304. doi: 10.1038/s41467-023-40928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villavicencio Kim J., Wu G.Y. Body building and aminotransferase elevations: a review. J Clin Transl Hepatol. 2020;8(2):161–167. doi: 10.14218/JCTH.2020.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lal P.B., Schneider B.L., Vu K., Reitzer L. The redundant aminotransferases in lysine and arginine synthesis and the extent of aminotransferase redundancy in Escherichia coli. Mol Microbiol. 2014;94(4):843–856. doi: 10.1111/mmi.12801. [DOI] [PubMed] [Google Scholar]
- 12.Percudani R., Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003;4(9):850–854. doi: 10.1038/sj.embor.embor914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.di Salvo M.L., Contestabile R., Safo M.K. Vitamin B(6) salvage enzymes: mechanism, structure and regulation. Biochim Biophys Acta. 2011;1814(11):1597–1608. doi: 10.1016/j.bbapap.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Koper K., Han S.-W., Pastor D.C., Yoshikuni Y., Maeda H.A. Evolutionary origin and functional diversification of aminotransferases. J Biol Chem. 2022;298(8) doi: 10.1016/j.jbc.2022.102122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cellini B., Zelante T., Dindo M., Bellet M.M., Renga G., Romani L., Costantini C. Pyridoxal 5'-phosphate-dependent enzymes at the crossroads of host-microbe tryptophan metabolism. Int J Mol Sci. 2020;21(16) doi: 10.3390/ijms21165823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werther T., Spinka M., Tittmann K., Schütz A., Golbik R., Mrestani-Klaus C., Hübner G., König S. Amino acids allosterically regulate the thiamine diphosphate-dependent α-keto acid decarboxylase from Mycobacterium tuberculosis. J Biol Chem. 2008;283(9):5344–5354. doi: 10.1074/jbc.M706569200. [DOI] [PubMed] [Google Scholar]
- 17.Walker M.C., van der Donk W.A. The many roles of glutamate in metabolism. J Ind Microbiol Biotechnol. 2016;43(2–3):419–430. doi: 10.1007/s10295-015-1665-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollmann F., Opperman D.J., Paul C.E. Biocatalytic reduction reactions from a chemist's perspective. Angew Chem Int Ed. 2021;60(11):5644–5665. doi: 10.1002/anie.202001876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hester G., Stark W., Moser M., Kallen J., Marković-Housley Z., Jansonius J.N. Crystal structure of phosphoserine aminotransferase from Escherichia coli at 2.3 Å resolution: comparison of the unligated enzyme and a complex with α-methyl-l-glutamate. J Mol Biol. 1999;286(3):829–850. doi: 10.1006/jmbi.1998.2506. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., Ma C., Dischert W., Soucaille P., Zeng A.P. Engineering of phosphoserine aminotransferase increases the conversion of l-homoserine to 4-hydroxy-2-ketobutyrate in a glycerol-independent pathway of 1,3-propanediol production from glucose. J Biotechnol. 2019;14(9) doi: 10.1002/biot.201900003. [DOI] [PubMed] [Google Scholar]
- 21.Shimizu S., Dempsey W.B. 3-hydroxypyruvate substitutes for pyridoxine in serC mutants of Escherichia coli K-12. J Bacteriol. 1978;134(3):944–949. doi: 10.1128/jb.134.3.944-949.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakai A., Kita M., Katsuragi T., Tani Y. serC Is involved in vitamin B6 biosynthesis in Escherichia coli but not in Bacillus subtilis. J Boisci Bioeng. 2002;93(3):334–337. doi: 10.1263/jbb.93.334. [DOI] [PubMed] [Google Scholar]
- 23.Rogerson D.T., Sachdeva A., Wang K., Haq T., Kazlauskaite A., Hancock S.M., Huguenin-Dezot N., Muqit M.M.K., Fry A.M., Bayliss R., Chin J.W. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat Chem Biol. 2015;11(7):496–503. doi: 10.1038/nchembio.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metcalf W.W., Zhang J.K., Shi X., Wolfe R.S. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J Bacteriol. 1996;178(19):5797–5802. doi: 10.1128/jb.178.19.5797-5802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parra M., Stahl S., Hellmann H. Vitamin B₆ and its role in cell metabolism and physiology. Cells. 2018;7(7) doi: 10.3390/cells7070084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mooney S., Leuendorf J.E., Hendrickson C., Hellmann H. Vitamin B6: a long known compound of surprising complexity. Molecules. 2009;14(1):329–351. doi: 10.3390/molecules14010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montaño López J., Duran L., Avalos J.L. Physiological limitations and opportunities in microbial metabolic engineering. Nat Rev Microbiol. 2022;20(1):35–48. doi: 10.1038/s41579-021-00600-0. [DOI] [PubMed] [Google Scholar]
- 28.Choi K.R., Lee S.Y. Systems metabolic engineering of microorganisms for food and cosmetics production. Nat Rev Microbiol. 2023;1(11):832–857. [Google Scholar]
- 29.Zhang X., Newman E. Deficiency in l‐serine deaminase results in abnormal growth and cell division of Escherichia coli K‐12. Mol Microbiol. 2008;69(4):870–881. doi: 10.1111/j.1365-2958.2008.06315.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X., El-Hajj Z.W., Newman E. Deficiency in L-serine deaminase interferes with one-carbon metabolism and cell wall synthesis in Escherichia coli K-12. J Bacteriol. 2010;192(20):5515–5525. doi: 10.1128/JB.00748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mundhada H., Seoane J.M., Schneider K., Koza A., Christensen H.B., Klein T., Phaneuf P.V., Herrgard M., Feist A.M., Nielsen A.T. Increased production of L-serine in Escherichia coli through adaptive laboratory evolution. Metab Eng. 2017;39:141–150. doi: 10.1016/j.ymben.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Rehati A., Abuduaini B., Liang Z., Chen D., He F. Identification of heat shock protein family A member 5 (HSPA5) targets involved in nonalcoholic fatty liver disease. Gene Immun. 2023;24(3):124–129. doi: 10.1038/s41435-023-00205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J.M., Dutta S., Feng Z., Ghosh S., Goodsell D.S., Green R.K., Guranović V., Guzenko D., Hudson B.P., Kalro T., Liang Y., Lowe R., Namkoong H., Peisach E., Periskova I., Prlić A., Randle C., Rose A., Rose P., Sala R., Sekharan M., Shao C., Tan L., Tao Y.-P., Valasatava Y., Voigt M., Westbrook J., Woo J., Yang H., Young J., Zhuravleva M., Zardecki C. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47(D1):D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchesani F., Zangelmi E., Murtas G., Costanzi E., Ullah R., Peracchi A., Bruno S., Pollegioni L., Mozzarelli A., Storici P., Campanini B. L‐serine biosynthesis in the human central nervous system: structure and function of phosphoserine aminotransferase. Protein Sci. 2023;32(4) doi: 10.1002/pro.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Case K B D.A., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham T.E., III, Cruzeiro V.W.D., Darden T.A., Duke R.E., Giambasu G., Gilson M.K., Gohlke H., Goetz A.W., Harris R., Izadi S., Izmailov S.A., Kasavajhala K., Kovalenko A., Krasny R., Kurtzman T., Lee T.S., LeGrand S., Li P., Lin C., Liu J., Luchko T., Luo R., Man V., Merz K.M., Miao Y., Mikhailovskii O., Monard G., Nguyen H., Onufriev A., Pan F., Pantano S., Qi R., Roe D.R., Roitberg A., Sagui C., Schott-Verdugo S., Shen J., Simmerling C.L., Skrynnikov N.R., Smith J., Swails J., Walker R.C., Wang J., Wilson L., Wolf R.M., Wu X., Xiong Y., Xue Y., York D.M., Kollman P.A. University of California; San Francisco: 2020. Amber 2020. [Google Scholar]
- 37.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. Ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theor Comput. 2015;11(8):3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox P.A., Metcalf J.S. Traditional food items in Ogimi, Okinawa: l-Serine content and the potential for neuroprotection. Curr Nutr Rep. 2017;6(1):24–31. doi: 10.1007/s13668-017-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I., Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1) doi: 10.1093/nar/gkab301. W293-w96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mooers B.H.M., Brown M.E. Templates for writing PyMOL scripts. Protein Sci. 2020;30(1):262–269. doi: 10.1002/pro.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gouet P., Courcelle E., Stuart D.I., Métoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15(4):305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 44.Singh R.K., Kumar D., Gourinath S. Phosphoserine aminotransferase has conserved active site from microbes to higher eukaryotes with minor deviations. Protein Pept Lett. 2021;28(9):996–1008. doi: 10.2174/0929866528666210215140231. [DOI] [PubMed] [Google Scholar]
- 45.Tian S., Wang D., Yang L., Zhang Z., Liu Y. A systematic review of 1-deoxy-D-xylulose-5-phosphate synthase in terpenoid biosynthesis in plants. Plant Growth Regul. 2022;96(2):221–235. [Google Scholar]
- 46.Mukherjee T., Hanes J., Tews I., Ealick S.E., Begley T.P. Pyridoxal phosphate: biosynthesis and catabolism. Biochim Biophys Acta. 2011;1814(11):1585–1596. doi: 10.1016/j.bbapap.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 47.Al-Hawash A.B., Zhang X., Ma F. Strategies of codon optimization for high-level heterologous protein expression in microbial expression systems. Gene Rep. 2017;9:46–53. [Google Scholar]
- 48.Pouresmaeil M., Azizi-Dargahlou S. Factors involved in heterologous expression of proteins in E. coli host. Arch Microbiol. 2023;205(5):212. doi: 10.1007/s00203-023-03541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khlebnikov A., Risa O., Skaug T., Carrier T.A., Keasling J.D. Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture. J Bacteriol. 2000;182(24):7029–7034. doi: 10.1128/jb.182.24.7029-7034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Downs D.M. Understanding microbial metabolism. Annu Rev Microbiol. 2006;60(1):533–559. doi: 10.1146/annurev.micro.60.080805.142308. [DOI] [PubMed] [Google Scholar]
- 51.Edwards H., Xu P. Unstructured kinetic models to simulate an arabinose switch that decouples cell growth from metabolite production. Synth Syst Biotechnol. 2020;5(3):222–229. doi: 10.1016/j.synbio.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su D., Kosciuk T., Yang M., Price I.R., Lin H. Binding affinity determines substrate specificity and enables discovery of substrates for N-myristoyltransferases. ACS Catal. 2021;11(24):14877–14883. doi: 10.1021/acscatal.1c03330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5(8):593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.