Abstract
This study introduces a series of novel Alkyl thio-1,2,4-triazole (4a-p) and mercapto-1,2,4-triazole (3a-d) compounds derived from nalidixic acid. The synthesis was streamlined, involving interactions between nalidixic acid hydrazide and various isothiocyanates to yield cyclic and alkyl(aryl) sulfide compounds, characterized using 1H NMR, 13C NMR, IR, and elemental analysis. Antioxidant capabilities were quantified through DPPH and ABTS assays, highlighting significant potential, especially for compound 3d, which demonstrated an ABTS IC50 value of 0.397 μM, on par with ascorbic acid (IC50 = 0.87 μM). Antibacterial efficacy was established through MIC assessments against a broad spectrum of Gram-positive and Gram-negative bacteria, including Candida albicans. Compounds 3b, 4e, 4h, 4j, 4i, 4m, and 4o showed broad-spectrum activity, with 4k and 4m exhibiting pronounced potency against E. coli. Molecular docking studies validated the antibacterial potential, with compounds 4f and 4h showing high binding affinities (docking scores of −9.8 and −9.6 kcal/mol, respectively), indicating robust interactions with the bacterial enzyme targets. These scores underscore the compounds' mechanistic basis for their antibacterial action and support their therapeutic promise. Furthermore, compounds 3b, 4i, and 4m, identified through drug-likeness and toxicity predictions, were highlighted for their favorable profiles, suggesting their suitability for oral antibiotic therapies. This comprehensive study, blending synthetic, in vitro, and in silico approaches, emphasizes the triazole derivatives' potential as future candidates for antibiotic and antioxidant applications, particularly spotlighting compounds 3b, 4i, and 4m due to their promising efficacy and safety profiles.
Keywords: Antioxidant, Antimicrobial, Isothiocyanate, Alkyl halide, Nalidixic acid, In silico
1. Introduction
A recent literature survey has revealed that 1,2,4-triazoles and their fused heterocyclic derivatives demonstrate a broad range of biological activities. A large number of clinically significant medications now contain the 1,2,4-triazole core, including letrozole, anastrozole, and voriconazole (antifungal), as well as the antivirals posaconazole, ribavirin, rizatriptan, alprazolam, and trazodone [[1], [2], [3], [4], [5], [6]].
The abundance of biological activities of 1,2,4-triazole derivatives, such as their antifungal [7,8], antitubercular [9], antioxidant [10], anticancer [11,12], anti-inflammatory [13], analgesic [14], antidiabetic [15], anticonvulsant [16], and anxiolytic activity [17], has recently piqued the interest of researchers. Triazole-based pharmacophores have supplanted the formerly common imidazole pharmacophore in systemically active azoles with regard to their antifungal activity because of lower toxicity, higher bioavailability, greater fungal cytochrome p450 selectivity, and a reduced effect on the production of human sterols [18].
The first synthetic quinolone derivative for the treatment of urinary tract infections, nalidixic acid (1,8-naphthyridine derivative), was introduced in 1963 [19]. It inhibits DNA synthesis and hastens bacterial mortality by promoting the breakdown of bacterial DNA in DNA gyrase enzyme complexes [20]. It is very resistant to most types of pseudomonas and extremely powerful against gram-negative bacteria, especially Escherichia coli [21,22]. By combining different pharmacophores into hybrid compounds, it is possible to produce medications with intriguing biological characteristics [21,22].
Because nalidixic acid causes a number of adverse effects, such as rash, itchy skin, blurred or double vision, light-halo effects, alterations in color vision, nausea, vomiting, and diarrhea. Additionally, nalidixic acid may result in hyperglycemia and seizures [23].
In silico methods like molecular docking and ADMET predictions are crucial in the early stages of drug discovery [24,25]. These computational approaches allow for quick and cost-effective screening of compounds, predicting how they might interact with targets (molecular docking) and their possible effects in the human body (ADMET profiles). This helps in identifying promising drug candidates by understanding their potential efficacy and safety before costly and time-consuming lab tests [24,25]. This study utilizes these techniques to advance the development of new triazole derivatives, demonstrating their value in finding effective treatments for antimicrobial and antioxidant needs more efficiently.
The urgent need for new treatments against these challenges highlights the importance of focusing on the antimicrobial and antioxidant properties of triazole derivatives. Chosen for their potential to offer new therapeutic strategies, these compounds are evaluated for their ability to bypass existing resistance mechanisms, a critical aspect in the fight against antimicrobial resistance (AMR). The selection of targets for molecular docking, based on their roles in AMR, aims to identify compounds capable of effectively interfering with these pathways [26]. This strategic approach, supported by recent findings, suggests that triazole derivatives could significantly contribute to developing new agents to combat AMR and oxidative stress, marking a significant step forward in addressing global health challenges [27].
This study aimed to synthesize novel mercapto- and alkylthio-1,2,4-triazole derivatives from nalidixic acid, assessing their potential as promising antibacterial and antioxidant compounds. Our objectives included evaluating their antibacterial efficacy against gram-positive and gram-negative bacteria, determining their antioxidant activity via DPPH and ABTS assays, and understanding their molecular interactions and safety profiles through molecular docking and ADMET predictions.
2. Experimental
2.1. Materials and equipment
Following the instructions in the literature, nalidixic acid hydrazides, 3a and 4a were produced [28,29], The experiment made use of readily accessible chemicals and solvents that were not further purified before use, including methanol, ethanol, DMSO, diethyl ether, chloroform, and nalidixic acid. Uncorrected melting points were found using an electrothermal-digital melting point equipment. Bruker 400 MHZ and 100 MHZ [1H NMR and 13C NMR] spectrometers were used to analyze the spectra in deuterated chloroform (CDCl3). Bruker's alpha FTIR was used to record IR spectra. elemental analyses of the elements (C, H and N) were carried out on Euro EA elemental analyzer 300.
2.1.1. General procedure for preparation of 2-(1-ethyl-7-methyl-4-oxo-1,4-dihydro-1,8-naphthyridine-3-carbonyl)-N-methyl(aryl)hydrazine-1-carbothioamide 2 a-d [12,24]
A dropwise addition of alkyl or aryl isothiocyanate RNCS (0.0143 mol) was added to a 3 g, 0.0155 mol solution of nalidixic acid hydrazide 1 in 50 mL of ethanol. The mixture was stirred at room temperature for around 24 h. Filtration was used to remove the resulting white precipitate or yellow precipitate from the methanol solution.
2.1.2. General procedure for preparation of 1-ethyl-3-(5-mercapto-4-methyl(aryl)-4H-1,2,4-triazol-3-yl)-7-methyl-1,8-naphthyridin-4(1H)-one 3a-d [12,24]
Compound 2a-d (0.0013 mol) was heated for almost 6 h at reflux in a solution of aqueous potassium hydroxide (0.0143 mol). Hydrochloric acid (HCl) was used to acidify the solution once, Filtration was used to get the white precipitate, which was then recrystallized from ethanol.
2.1.3. General procedure for the preparation of 1-ethyl-3-(5-(ethyl(substitutedbenzyl)thio)-4-methyl(aryl)-4H-1,2,4-triazol-3-yl)-7-methyl-1,8-naphthyridin-4(1H)-one 4a-p [12,24]
Potassium hydroxide (0.0143 mol), compound 3a-d (0.013 mol), ethanol (10 mL), and alkyl halide R'X (0.013 mol) were heated over reflux for approximately 4 h. Water was added after the solution had cooled, which led to the formation of a precipitate and the recrystallization of the chloroform/ether.
2.2. Evaluation of antioxidant activity
Compounds 3a-d were assessed for their antioxidant capacities using the DPPH and ABTS radical scavenging assays according to the procedures specified in the literature [30,31], with minor adjustments. All trials were performed in triplicate.
2.2.1. DPPH radical scavenging assay
DPPH (100 μM) and various concentrations of each tested compound (10 μM, 20 μM, 75 μM, 150 μM, and 600 μM) were prepared in DMSO 2 mL of each concentration was combined with 2 mL of the DPPH solution, and the resulting mixtures were incubated in darkness for 30 min. Following the incubation period, the absorbance of each solution was measured at a wavelength of 517 nm.
The inhibition percentage (I%) values were calculated using equation (1).
| (1) |
Where AS is the absorbance in the presence of the sample and AB is the absorbance of the blank.
2.2.2. ABTS radical scavenging assay
A solution of potassium persulfate (2.5 mM) was combined with ABTS (7.5 mM), leading to the generation of the ABTS•+ radical cation. This mixture was then left overnight at 4 °C, protected from light exposure. Five different concentrations of each tested compound (10 μM, 20 μM, 75 μM, 150 μM, and 600 μM) were prepared, The ABTS•+ solution was first diluted to achieve an absorbance of 0.75 at 734 nm. Next, 1 mL of each concentration of the tested compounds and 3 mL of the ABTS•+ solutions were mixed, and the resulting solutions were left to stand for 10 min. After that, the absorbance of each solution was measured at 734 nm.
The inhibition percentage (I%) values were calculated using equation (2).
| (2) |
Where AS is the absorbance in the presence of the sample and AB is the absorbance of the blank.
2.3. General procedure for the antimicrobial activity
Isolates of bacteria and Candida albicans were acquired from Yarmouk University's Central Laboratories for this study. The methicillin-resistant Staphylococcus aureus, Salmonella enteritidis, Enterococcus faecalis, Proteus mirabilis, Escherichia coli, and Klebsiella pneumonia clinical isolates were cultured in Muller Broth media for 24 h at 37 °C. Candida albicans was cultured on Sabouraud dextrose agar (SDA). By determining the minimal inhibitory concentration (MIC, g/mL) using the Hannan-described micro-broth dilution method, the biological activity of the substances was ascertained [32]. The chemicals' DMSO stock solutions were created in accordance with CLSI recommendations [33]. Standard sterile 96-well flat bottom micro-titer plates were used for the in vitro MIC. Each row of the layout included positive and negative control wells and spanned a concentration range of the individual chemicals under research from 500 to 0.5 g/mL 40 mL of the chosen compounds were added to each well at the proper concentration, and 40 mL of DMSO solvent was added to the control well. The next step was to add 150 mL of Muller Hinton media and 10 mL of the bacterial culture standardized using 0.5 McFarland turbidity standards to each well.
About 5.0 × 107 CFU/mL of bacteria were present in the inoculum at the end. Plates were sealed and incubated for 24 h at 37 °C in an atmosphere. With the aid of an ELIZA UV–vis spectrometer, microtiter plates were read. The MIC was determined to be the lowest concentration that had an optical density lower than the control.
2.4. Molecular docking
2.4.1. Protein structure preparation
Protein targets for further analysis were selected, including E. faecalis FabH (PDB ID: 3IL5) complexed with the ligand 2-((4-bromo 3[(diethylamino)sulfonyl]benzoyl)amino)benzoic acid [34], S. aureus FtsA protein (PDB ID: 3WQT) [35], KPC-2 beta-lactamase bound to hydrolyzed cefotaxime (PDB ID: 5UJ3) [36], and E. coli beta-Glucuronidase (PDB ID: 4JHZ) bound with a potent inhibitor termed as 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1 -yl]-N^[(ls,2S,SS)-Z,S dimethoxycyclohexyl]acetamide [37]. These structures were acquired from the Protein Data Bank [38] in PDB format on September 8th, 2023. Each complex's co-crystalized ligand was used as a reference drug for comparative purposes. The selection of protein targets for further analysis was based on their potential roles in resistance and pathogenic survival.
To facilitate molecular docking studies, the 3D structures of the protein targets underwent preprocessing procedures. Heteroatoms and non-essential water molecules were removed using Biovia Discovery Studio Visualizer [39]. The resulting structures were saved in PDB format. Additionally, any missing amino acids in the target structures were integrated using the YASARA web-server tool [[40], [41], [42]]. Titratable amino acid groups' ionization states at pH 7.0 were calculated through the use of H++ web-server tool [[41], [42], [43]]. Finally, AutoDock Tools version 1.5.6 tools transformed these outputs into PDBQT format by adding polar hydrogen atoms and Kollman charges [44].
2.4.2. Ligand preparation
The tested compounds (3b, 4f, 4h, 4i, 4j, 4k, 4m, 4n and 4o) were drawn using the ChemDraw JS web page (https://chemdrawdirect.perkinelmer.cloud/js/sample/index.html, accessed on September 10, 2023) and saved in the structural data file format. To achieve optimal stability for each ligand conformation, energy minimization was performed by applying the Universal Force Field (UFF) with a Conjugate Gradient (CG) optimization algorithm consisting of 1000 steps. The open-source Babel software was utilized for this optimization process and resulted in structures being saved in PDB format. Following that step with AutoDock tools version1.5.6., Gasteiger charges were incorporated into the ligands which allowed them to be uploaded as PDBQT files ready for molecular docking simulations.
2.4.3. Molecular docking preparation
Molecular docking analyses were conducted to assess the binding mode pattern, binding conformation, and various types of interactions between the active derivatives and the active site in proteins. The AutoDock 4.2 Release 4.2.6 program was applied for these docking calculations [45]. To optimize the binding poses of compounds, a Lamarckian genetic algorithm was employed while keeping ligands flexible and macromolecules rigid. The grid box size used for all proteins encompassed dimensions of 40 × 40 × 40 along the X, Y, and Z axes. Coordinates for the active binding sites during dockings were determined based on potential ligand-binding domain regions extracted from available crystal structures obtained through research efforts. To ensure consistent calculations, the maximum number of evaluations was set to 25,000,000 and the number of runs was adjusted to 100. All other parameters were kept at their default values. The binding energies and properties of various compounds were analyzed after docking in order to assess their potential as therapeutic agents. To validate the accuracy of our molecular docking simulations, we implemented a redocking procedure. This method entails removing the co-crystallized ligand from its complex with the target protein and repositioning it into the active site. The effectiveness of the docking software was verified by comparing the orientation of the ligand before and after redocking. An essential part of this validation involves calculating the Root Mean Square Deviation (RMSD) between the two ligand positions. An RMSD value below 2 Å is deemed acceptable [41,[46], [47], [48]], indicating a minimal deviation between the predicted and actual ligand configurations. This criterion ensures our docking approach can reliably replicate known ligand interactions, thereby enhancing the credibility of our simulation outcomes. Molecular interactions between ligands and proteins were visualized and analyzed in both 2D and 3D using BIOVIA Discovery Studio Visualizer.
In addressing the conformational flexibility of the compounds under investigation, our study utilized AutoDock 4.2 software, employing the Lamarckian genetic algorithm for conformational search and docking simulations. This approach allowed for a comprehensive exploration of potential conformations by generating a diverse set of possible structures for each compound. The conformational search was performed considering different environmental contexts (such as in solvent and vacuum) to approximate various physiological conditions [49]. Special attention was given to the energy penalties associated with transitions from the most stable conformer to energetically less stable ones. We recognized that the energy cost of conformational changes could be mitigated by the favorable interactions within the protein's active site, potentially enhancing binding affinity. The selection process for the conformers used in the docking studies was thus informed by this balance, ensuring that the impact of conformational diversity on the binding process was accurately accounted for. This meticulous approach aims to reflect the true dynamic nature of the molecular interactions and their critical role in determining the biological activity of the investigated compounds.
2.5. ADMET prediction
In this investigation, we utilized computational tools analysis likely SwissADME [50] and AdmetSAR 2.0 [51] online webservers tools for the prediction of ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) properties. This enabled us to assess the potential suitability of various compounds (3b, 4f, 4h, 4i, 4j, 4k, 4m, 4n, and 4o) as prospective candidates for drug development. The selection of specific ADMET properties was based on their well-established significance in relation to oral bioavailability including factors such as oral absorption rate and blood-brain barrier penetration capacity. We employed amoxicillin, a commercially available antibacterial agent that has undergone clinical use, to serve as a reference compound during our analysis.
To utilize SwissADME and AdmetSAR 2.0, the procedure involved uploading the compounds of interest in SMILES format to the web server, selecting the desired ADMET properties for prediction, and retrieving predicted values with corresponding confidence levels. The predictions were generated through various algorithms and models offered by SwissADME and AdmetSAR 2.0 such as multiple linear regression, decision trees, and artificial neural networks. In addition, to verify the accuracy of the predictions, we examined how well the anticipated ADMET properties of the compounds being tested aligned with known values for amoxicillin. Furthermore, a computational analysis was conducted on toxicity and drug-like characteristics using AdmetSAR 2.0. These predicted outcomes were then utilized to assess these particular compounds as potential lead candidates and identify any areas that may require further improvement in their composition.
3. Results and discussions
3.1. Synthesis and characterization
The synthetic route of Alkyl thio-1,2,4-triazole 4a-p and mercapto-1,2,4-triazole 3a-d from nalidixic acid hydrazide is summarized in Fig. 1. All characteristic data for the newly synthesized compound 3a-d and 4a-p are presented in Table 1 and Table S1.
Fig. 1.
Synthesis routs of 1-ethyl-7-methyl-3-(5-mercapto-4-subtituted-4H-1,2,4-triazol-3-yl)- 1,8-naphthyridin-4(1H)-one 3a-d and 1-ethyl-7-methyl-3-(5-substitutedthio-4-substituted-4H-1,2,4-triazol-3-yl)-1,8-naphthyridin-4(1H)-one 4a-p.
Table 1.
Molecular formula and M. Mass(g/mole), melting points, colors, yields and elemental analysis of compounds 3(a–d) and 4(a–p).
| Compound No. | Formula & M. mass (g/mol) |
Yield % | M.p (oC) | Color | Elemental analysis % |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Calculated |
Found |
|||||||||
| C | H | N | C | H | N | |||||
| 3a | C14H15N5OS (301.37) | 85.7 | 305–309 | Yellow | 55.80 | 5.02 | 23.24 | 55.72 | 4.91 | 23.17 |
| 3b | C19H17N5OS (363.44) | 81.0 | 313–318 | White | 62.79 | 4.71 | 19.27 | 62.71 | 4.68 | 19.14 |
| 3c | C20H19N5OS (377.47) | 94.7 | 310–314 | White | 63.64 | 5.07 | 18.55 | 63.57 | 4.97 | 18.51 |
| 3d | C19H16FN5OS (381.11) | 89.8 | 266–269 | White | 59.83 | 4.23 | 18.36 | 59.79 | 4.17 | 18.31 |
| 4a | C22H23N5OS (405.52) | 55 | 188–191 | Yellow | 65.16 | 5.72 | 17.27 | 65.05 | 5.62 | 17.15 |
| 4b | C21H20FN5OS (409.48) | 31.9 | 195–200 | yellow | 61.60 | 4.92 | 17.10 | 61.54 | 4.87 | 17.03 |
| 4c | C22H20BrN5O2S (498.40) | 52.2 | 220–223 | white | 53.02 | 4.04 | 14.05 | 52.98 | 4.01 | 16.99 |
| 4d | C16H19N5OS (329.42) | 27.3 | 192–194 | yellow | 58.34 | 5.81 | 21.26 | 58.29 | 5.77 | 21.19 |
| 4e | C21H20N6O3S (436.49) | 39.6 | 229–234 | Brown | 57.79 | 4.62 | 19.25 | 57.73 | 4.58 | 21.17 |
| 4f | C27H25N5OS (467.59) | 21.40 | 219–223 | White | 69.35 | 5.39 | 14.98 | 69.31 | 5.37 | 14.96 |
| 4g | C26H22FN5OS (471.55) | 67.5 | 209–211 | yellow | 66.22 | 4.70 | 14.85 | 66.18 | 4.65 | 14.80 |
| 4h | C27H22BrN5O2S (560.47) | 67.5 | 100–105 | yellow | 57.86 | 3.96 | 12.50 | 57.81 | 3.92 | 12.47 |
| 4i | C21H21N5OS (391.49) | 62.8 | 223–228 | white | 64.43 | 5.41 | 17.89 | 64.38 | 5.37 | 17.85 |
| 4j | C26H22N6O3S (498.56). | 45.5 | 166–170 | yellow | 62.64 | 4.45 | 16.86 | 62.59 | 4.41 | 16.85 |
| 4k | C28H27N5OS (481.62) | 52.9 | 177–180 | white | 69.83 | 5.65 | 14.54 | 69.78 | 5.63 | 14.51 |
| 4l | C27H24FN5OS (485.58) | 70.0 | 201–203 | white | 66.79 | 4.98 | 14.42 | 66.74 | 4.97 | 14.38 |
| 4m | C22H23N5OS (405.52) | 69.7 | 174–178 | light yellow | 65.16 | 5.72 | 17.27 | 65.11 | 5.69 | 17.25 |
| 4n | C27H24FN5OS (485.58) | 27.5 | 212–217 | light yellow | 66.79 | 4.98 | 14.40 | 66.76 | 4.96 | 14.37 |
| 4o | C26H21F2N5OS (489.54) | 41.2 | 182–187 | yellow | 63.79 | 4.32 | 14.31 | 63.75 | 4.28 | 14.27 |
| 4p | C21H20FN5OS (409.48) | 55.8 | 116–120 | white | 61.60 | 4.92 | 17.10 | 61.56 | 4.88 | 17.07 |
M.p = melting point; M. mass = Molar mass.
Components 3a-d through d's 1H NMR spectra revealed a singlet at 14.74–11.53 ppm that matched the SH proton. Aromatic protons were identified as the source of signals in the range 8.93–7.05 ppm. Other substituent protons, such as those from ethyl and methyl groups, resonate in the predicted places. The compounds 3a-d 13C NMR spectra support the proposed structures. Signals in the 148.3–169.1 ppm range are attributed to the triazole moiety's C-3 and C-5, while those in the 127.6–129.8 ppm range are attributed to the N–C in the aryl group. The resonant sites of the methyl and ethyl substituents were as expected. Compounds 3a-d infrared's spectra exhibit moderate absorption bands at 1619–1627 cm−1 for the (C O) functional group, 1440–1499 cm−1 for the (C N) functional group, and 1254–1289 cm−1 for the (C–N) bond.
Compounds 4a-p 1H NMR's spectra revealed a singlet at 4.38–4.91 ppm that corresponds to the –SCH2- protons. Aromatic protons were given credit for the signals in the range of 8.65 ppm to 6.95 ppm. The compounds 4a-p 13C NMR's spectra are consistent with the postulated structures. The triazole moiety's C-3 and C-5 signals have a range of 150.0–153.4 ppm. SCH2 carbons is allocated to signals in the ppm range of 26.8 ppm–41.3 ppm. At the anticipated chemical changes, other carbons resonant. Organic compounds 4a–p infrared p's spectra exhibit moderate absorption bands in the ranges of 1681–1630 cm−1 for the (C O) functional group, 1426–1507 cm−1 for the (C N) group, and 1340–1372 cm−1 for the (C–N) bond.
3.2. Determination of antioxidant activities of the newly synthesized compounds
Free radicals, which are a byproduct of oxidation events, can start a chain reaction. It is commonly known that a cell's damage or death can result from a free radical chain reaction. Antioxidants inhibit these chain reactions and limit further oxidative processes through eliminating the free radical intermediates [[52], [53], [54], [55]]. Antioxidants are in charge of organisms' defense mechanisms against diseases brought on by the onslaught of free radicals. Utilizing DPPH radical and ABTS tests, the antioxidant properties of some recently synthesized compounds, specifically 3a-d and 4a-p, were investigated. Spectrophotometric techniques were used to monitor the reaction. Few substances demonstrated strong antioxidant action.
None of the compounds 4a–p showed any appreciable scavenging ability. The results demonstrated that the ineffectiveness of compounds (4a-p negligible) in neutralizing free radicals was due to their inability to release hydrogen atoms or electrons for the radicals. The (SH) group found in the structures of compounds 3a–d may have contributed to the compounds' noticeably stronger hydrogen or electron donating properties. Notably, even at low concentrations, the antioxidant potential could be seen (10 μM, 20 μM, 75 μM, 150 μM and 600 μM).
3.2.1. DPPH radical scavenging activity: 2, 2-diphenyl-1-picrylhydrazyl radical
By using the DPPH method, the inhibitory effects of compounds 3a-d and 4a-p on free radicals were examined, and the results were compared to ascorbic acid. By measuring the reduction in DPPH absorbance at = 517 nm, the values of scavenging activity of the synthesized compounds 3a-d and 4a-p at concentrations (10 μM, 20 μM, 75 μM, 150 μM, and 600 μM) were calculated. Table 2 and Fig. 2 present the outcomes derived from the assessment of compounds 3a-d using the DPPH scavenging activity method. The outcomes demonstrate that the scavenging activity is reliant on the concentration of the compounds. Notably, at the highest concentration (600 μM), compound 3b exhibited the most substantial inhibition percentage (78.2%), followed by compound 3c (74.3%), compound 3d (55.6%), and finally, compound 3a (52.7%). However, it is noteworthy that ascorbic acid displayed superior scavenging activity compared to all compounds across all concentrations. According to the IC50 values presented in Table 2, compound 3b had the lowest IC50 (48.7), meaning that it has the highest antioxidant activity, followed by compound 3c (240), compound 3d (406), and compound 3a (700). The outcomes demonstrated that the concentration-dependent DPPH radical scavenging activities. As a result, the tested compounds' scavenging activity increased with increasing concentrations.
Table 2.
Inhibition percentage (I%) and IC50 of DPPH and ABTS radicals of compounds 3a-d, and the control ascorbic acid.
| Conc. Comd. |
DPPH |
ABTS |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 μM | 20 μM | 75 μM | 150 μM | 600 μM | IC50 | 10 μM | 20 μM | 75 μM | 150 μM | 600 μM | IC50 | |
| 3a | 10.95 | 15.87 | 22.38 | 32.5 | 52.6 | 700 | 54.55 | 70.62 | 78.33 | 87.97 | 96.97 | 3.96 |
| 3b | 30.95 | 39.04 | 49.57 | 71.63 | 78.22 | 48.7 | 33.8 | 59.81 | 76.88 | 87.74 | 95.78 | 15.93 |
| 3c | 10.7 | 14.99 | 25.55 | 31.88 | 74.6 | 240 | 46.43 | 63.05 | 90.51 | 95.93 | 98.19 | 7.43 |
| 3d | 4.93 | 11.33 | 27.08 | 37.97 | 55.56 | 406 | 68.87 | 80.34 | 95.55 | 97.85 | 99.14 | 0.397 |
| Ascorbic acid | 37.89 | 69.72 | 94.89 | 95.49 | 96.39 | 7.97 | 60.34 | 83.99 | 96.23 | 98.87 | 99.1 | 0.87 |
Fig. 2.
DPPH and ABTS radical scavenging percentage at various concentrations of compounds (3a-d) at room temperature (30 min for DPPH and 10 min for ABTS).
3.2.2. Scavenging activity of ABTS radical
The results obtained from ABTS scavenging activity of compounds 3a-d are shown in Table 2 and Fig. 2. The results show that the scavenging activity was concentration-dependent. At the highest concentration (600 μM), compound 3d had the highest inhibition percentage (99.14%), followed by compound 3c (98.19%), compound 20a (92.19%), compound 3a (96.97%), and lastly compound 3b (95.78%). However, compound 3d exhibited scavenging activity as similar as ascorbic acid but a higher scavenging activity than all the compounds at all concentrations. The compounds antioxidant activities are compared based on their IC50 value; lower IC50 value implies higher scavenging activity. Based on the data in Table 2, compound 3d had the highest scavenging activity with IC50 value of (0.397), then compound 3a (3.96), compound 3c (7.43), and compound 3b (15.93).
3.3. Antimicrobial activity
Compounds 3a-d and 4a-p's minimum inhibitory concentrations (MIC) against Gram-negative and Gram-positive bacteria were identified. A positive control was amoxicillin. Compounds 4a, 4g, 4i, and 4k were inactive against Enterococcus, whereas other compounds with MICs in the range of 16–32 g/mL were moderately active against Enterococcus. The synthesized compounds 3b, 4f, 4h, 4j, and 4o were moderately effective against some Gram-positive bacteria (Enterococcus). Other synthesized compounds with MICs of 32–64 g/mL were only moderately effective against Staphylococcus aureus, whereas compound 4n was effective. Except for 4e and 4f, all newly created compounds were revealed to be moderately active against Methicillin Resistant Staphylococcus aureus with MICs in the range of 128–256 g/mL, which were discovered to be inactive against Methicillin Resistant Staphylococcus aureus as shown in Table 3.
Table 3.
MIC of compounds 3a-d and 4a-p against gram-negative and gram-positive bacteria.
| Extract ID |
Gram positive |
Gram negative |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Comd No. | MRSA | MIC μg/mL | En |
MIC μg/mL |
Sa | MIC μg/mL | K |
MIC μg/mL |
Se | MIC μg/mL | E. coli | MIC μg/mL |
| 3a | ++ | 256 | ++ | 32 | N | N | ++ | 128 | ++ | 64 | ++ | 128 |
| 3b | ++ | 256 | +++ | 16 | ++ | 64 | ++ | 128 | ++ | 64 | ++ | 128 |
| 3c | ++ | 256 | ++ | 64 | ++ | 64 | ++ | 128 | ++ | 128 | N | N |
| 3d | ++ | 256 | ++ | 32 | ++ | 64 | ++ | 128 | ++ | 128 | N | N |
| 4a | ++ | 256 | N | N | ++ | 32 | ++ | 128 | ++ | 128 | ++ | 128 |
| 4b | ++ | 256 | ++ | 32 | ++ | 32 | ++ | 128 | ++ | 128 | N | N |
| 4c | ++ | 128 | ++ | 32 | ++ | 32 | ++ | 128 | ++ | 128 | N | N |
| 4d | ++ | 128 | ++ | 32 | N | N | ++ | 128 | ++ | 128 | ++ | 128 |
| 4e | N | N | ++ | 32 | ++ | 64 | ++ | 128 | ++ | 128 | N | 128 |
| 4f | N | N | +++ | 16 | ++ | 64 | ++ | 128 | ++ | 128 | N | N |
| 4g | ++ | 128 | N | N | ++ | 64 | N | N | ++ | 128 | ++ | 128 |
| 4h | ++ | 128 | +++ | 16 | ++ | 64 | ++ | 128 | ++ | 128 | ++ | 128 |
| 4i | ++ | 128 | N | N | ++ | 64 | +++ | 16 | ++ | 128 | ++ | 128 |
| 4j | ++ | 128 | +++ | 16 | ++ | 64 | ++ | 128 | ++ | 128 | N | N |
| 4k | ++ | 128 | N | N | ++ | 64 | +++ | 16 | ++ | 128 | ++++ | 8 |
| 4l | ++ | 128 | ++ | 32 | ++ | 64 | N | N | ++ | 128 | ++ | 128 |
| 4m | ++ | 128 | ++ | 32 | ++ | 64 | +++ | 16 | ++ | 128 | ++ | 128 |
| 4n | ++ | 128 | ++ | 32 | +++ | 16 | +++ | 16 | ++ | 128 | ++++ | 8 |
| 4o | ++ | 128 | +++ | 16 | ++ | 64 | ++ | 128 | ++ | 128 | +++ | 16 |
| 4p | ++ | 128 | ++ | 32 | ++ | 64 | ++ | 128 | ++ | 128 | ++ | 128 |
|
DMSO -ve control |
N | N | N | N | N | N | N | N | N | N | N | N |
|
Amoxicillin +ve control |
+++ | 16 | +++ | 16 | +++ | 16 | +++ | 16 | +++ | 16 | +++ | 32 |
Se=Salmonella enteritidis; Sa = Staphylococcus aureus; MRSA = Methicillin-resistant Staphylococcus aureus. Ec = Escherichia coli, Pm=Proteus mirabilis, En = Enterococcus faecalis, K=Klebsiella pneumonia; N, not detected, DMSO is a negative control, Amoxicillin is a positive control Key to interpretation: >16 mm (+++++): very active. 10–15 mm (+++): active; less active; 6–10 mm (++): moderately active; 1–5 mm (+): inactive; (N): No inhibition zone.
While 4o shown less activity, compounds 4k and 4n were quite effective against Gram-negative bacteria like E. Coli. Other substances were discovered to have just fair effectiveness against E. Coli. The compounds 3c, 3d, 4b, 4c, 4e, 4f, and 4j have a moderate to high efficacy against E. Coli, the current results are better than that reported in literature [56] with MICs between 8 and 128 g/mL. Compounds 4i, 4k, 4m, and 4n were effective against klebsiella pneumoniae, while other compounds were somewhat effective, with the exception of 4f and 4i, which had an inactive MIC in the range of 16–128 g/mL. The study findings are similar to that reported [57,58]. With MICs between 64 and 128 g/mL, all the new compounds showed promising moderately antibacterial activities which are better than many reported works [57,58].
3.4. Molecular docking simulations
In the endeavor to develop and design novel antibacterial agents, molecular docking assumes a pivotal role as a methodological cornerstone [59]. It enables the elucidation of the intricate binding interactions between synthesized compounds and their respective target proteins, thereby furnishing valuable insights into their binding affinities [25,60]. In this study, we conducted a series of molecular docking simulations to investigate the binding profiles of the most potent antibacterial compounds against both gram-positive strains (3b, 4f, 4h, 4j, 4n, and 4o) and gram-negative strains (4i, 4k, 4n, and 4o). These simulations were performed within the active binding sites of the respective bacterial proteins, allowing us the opportunity to determine the binding mode pattern, binding conformation, and different types of interaction of the active derivatives with the active site in proteins. Based on the observed biological activity, none of the synthesized compounds exhibited minimum inhibitory concentrations against Methicillin-resistant Staphylococcus aureus (MRSA) of the Gram-positive type or Salmonella enteritidis (Se) of the Gram-negative type. Therefore, this study proceeded to conduct molecular docking simulations to investigate the potential of the synthesized compounds against other bacterial strains. Specifically, the study focused on the main proteins or enzymes associated with Enterococcus faecalis, Staphylococcus aureus, Klebsiella pneumonia, and E. coli. The ensuing sections elaborate on these docking results.
3.4.1. Molecular docking with Enterococcus faecalis (E. faecalis)
In the pursuit of discovering antibacterial therapies against E. faecalis, the specific focus was directed toward the FabH (β-ketoacyl-acyl carrier protein synthase III) enzyme. FabH, a unique bacterial enzyme, plays a crucial role in initiating fatty acid biosynthesis [34,61]. Inhibition of FabH may directly lead to the disruption of bacterial fatty acid synthesis, thereby inhibiting the growth and survival of pathogenic bacteria [62]. The aim of the present study was to investigate the binding patterns and affinities of compounds 3b, 4f, 4h, 4j, and 4o when interacting with the FabH enzyme E. faecalis. The study was conducted to gain insight into the potential use of these compounds as inhibitors of the FabH enzyme, which is a promising target for the development of new antibiotics. For this purpose, the co-crystalized ligand (2-((4-bromo-3-[(diethylamino)sulfonyl] benzoyl)amino)benzoic acid) from the crystal structure (PDB ID:3IL5) [34] was used as a control in the docking experiments to compare and evaluate the binding affinity and interaction of potential inhibitors with the structure of E. faecalis FabH.
To validate the docking process parameters, the co-crystallized ligand, 2-((4-bromo-3 [(diethylamino)sulfonyl]benzoyl)amino)benzoic acid, underwent re-docking into the active binding site of the E. faecalis FabH enzyme structure (PDB ID: 3IL5) using AutoDock 4.2 software. The resulting RMSD value for the co-crystallized ligand was determined to be 1.61 Å (Fig. S1). This value falls well within the acceptable range for utilizing docking as a predictive tool for assessing the binding affinity of small molecules [41,[46], [47], [48]]. These findings not only affirm the reliability of the docking process parameters but also underscore the utility of docking as a robust tool for predicting the binding affinity of the tested compounds (3b, 4f, 4h, 4j, and 4o) against the FabH enzyme.
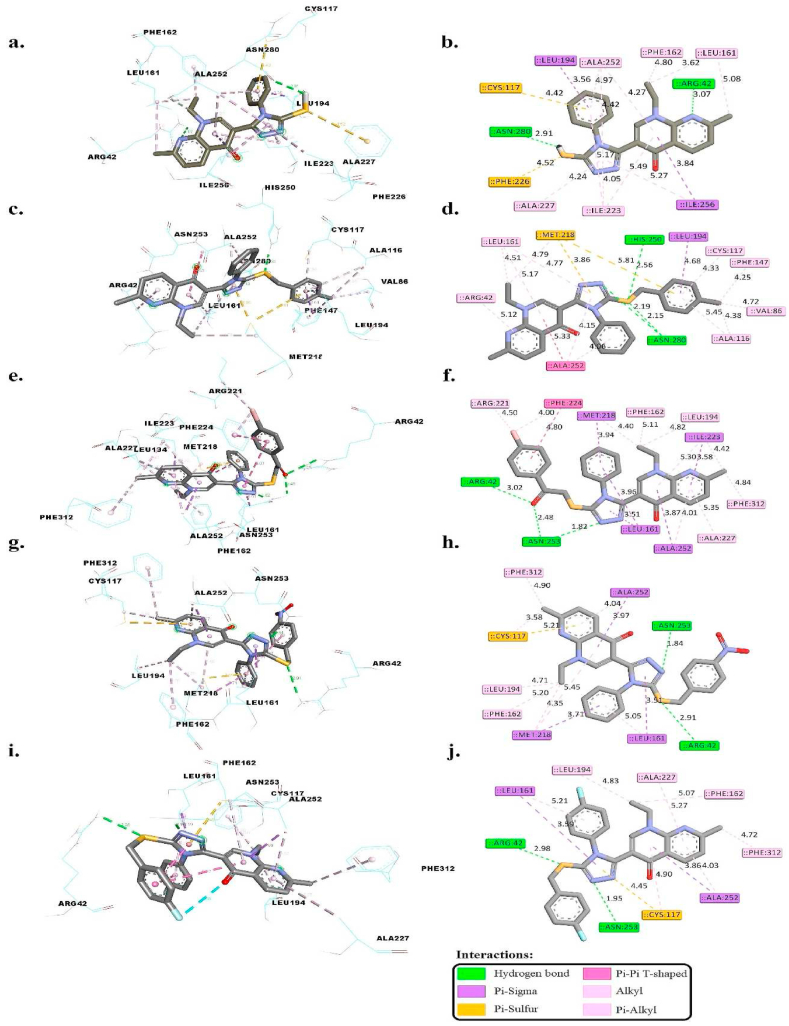
Table 4 and Fig. 3(a–j) present a detailed overview of the binding energies and molecular interactions within the E. faecalis FabH enzyme's active site, featuring compounds 3b, 4f, 4h, 4j, and 4o. Notably, these compounds demonstrated more negative binding energies than the control, indicating stronger interactions. The co-crystallized ligand, with a docking score of −8.45 kcal/mol, formed hydrogen bonds with ARG42 (2.36 Å) and ASN280 (2.99 Å), suggesting a tight and specific interaction that likely contributes to its strong binding affinity and robust inhibition of the enzyme's activity.
Table 4.
Docking scores (*ΔGbind (kcal/mol)) and molecular interactions analysis between 3b, 4f, 4h, 4j, and 4o and the co-crystallized ligand 2-((4-bromo-3-[(diethylamino)sulfonyl]benzoyl)amino)benzoic acid within the active binding site of the E. faecalis FabH protein (3IL5.PDB) using AutoDock 4.2.
| Compounds | *ΔGbind (kcal/mol) | Hydrogen Bond Interactions |
Pi-Sigma | Pi-Sulfur | Hydrophobic Interaction | |
|---|---|---|---|---|---|---|
| Residues | Distances (Å) | |||||
| 3b | −9.36 | ARG42 and ASN280 | 3.07 and 2.91 | LEU194 and ILE256 | – | LEU161, PHE162, ILE223, ALA227, and ALA252 |
| 4f | −10.12 | HIS250, ASN280, and ASN280 | 2.56, 2.15, and 2.19 | LEU194 | MET218 | ARG42, VAL86, ALA116, CYS117, PHE147, LEU161, and ALA252 |
| 4h | −10.87 | ARG42, ASN253, and ASN253 | 3.02, 1.82, and 2.48 | LEU161, MET218, ILE22, and ALA252 | – | PHE162, LEU194, ARG221, ALA227, PHE312 |
| 4j | −9.97 | ARG42 and ASN253 | 2.91 and 1.84 | LEU161, MET218, and ALA252 | CYS117 | LEU161, PHE162, LEU194, MET218, ALA252, and PHE312 |
| 4o | −9.72 | ARG42 and ASN253 | 2.98 and 1.95 | LEU161 and ALA252 | CYS117 | CYS117, PHE162, LEU194, ALA227, ALA252, and PHE312 |
| Co-crystalized ligand | −8.45 | ARG42 and ASN280 | 2.36 and 2.99 | – | CYS117 | LEU161, ARG221, ILE223, PHE224, ALA227, ALA252, and PHE312 |
Fig. 3.
3D and 2D interactions of 3b (a & b), 4f (c & d), 4h (e & f), 4j (g & h), and 4o (i & j) within the active binding site of the E. faecalis FabH enzyme. These models were generated using the Discovery Studio visualizer.
Compound 3b, showing a docking score of −9.36 kcal/mol, formed hydrogen bonds with ARG42 (3.07 Å) and ASN280 (2.91 Å). The slightly longer distances, compared to the co-crystallized ligand, might influence the binding orientation and effectiveness, potentially leading to a distinct inhibition mechanism or altered enzyme specificity.
Compound 4f, with the highest binding affinity at −10.12 kcal/mol, established hydrogen bonds at distances of 2.56 Å (HIS250), 2.15 Å, and 2.19 Å (ASN280). These optimal distances are indicative of efficient enzyme-ligand interactions, correlating with its high inhibitory potency against the FabH enzyme.
Compound 4h, displaying a docking score of −10.87 kcal/mol, engaged in hydrogen bonds with ARG42 (3.02 Å) and ASN253 (1.82 Å and 2.48 Å). The shorter bond with ASN253, in particular, suggests a strong and specific interaction that may contribute to its notable inhibitory activity, highlighting the role of precise molecular alignment in achieving effective enzyme inhibition.
Compound 4j, with a docking score of −9.97 kcal/mol, formed hydrogen bonds with ARG42 (2.91 Å) and ASN253 (1.84 Å), with the distances likely facilitating a stable interaction conducive to significant biological activity, particularly in disrupting the enzymatic function of FabH.
Lastly, compound 4o, featuring a docking score of −9.72 kcal/mol, established hydrogen bonds with ARG42 (2.98 Å) and ASN253 (1.95 Å). These interactions, especially the closer engagement with ASN253, may underline its efficiency in binding to and inhibiting the FabH enzyme, emphasizing the critical role of hydrogen bonding in mediating biological outcomes.
In general, this comprehensive analysis highlights the compelling binding affinities of all tested compounds (3b, 4f, 4h, 4j, and 4o) within the active binding site of the E. faecalis FabH enzyme. These interactions, encompassing hydrogen bonds, pi-sigma, pi-sulfur interactions, and hydrophobic contacts, collectively contribute to their binding stability. Notably, compounds 4f, 4h, 4j, and 4o display enhanced pi-sigma interactions, further accentuating their binding potential. These findings underscore the promising prospects of these compounds as potential inhibitors against E. faecalis FabH, warranting further exploration for their therapeutic applications.
3.4.2. Molecular docking with Staphylococcus aureus (S. aureus)
Among all the synthesized compounds, only compound 4n exhibited a minimum inhibitory concentration (MIC) comparable to that of the control amoxicillin when evaluated against the gram-positive bacterium S. aureus, as outlined in Table 4. It is noteworthy that the FtsA protein holds a pivotal role in bacterial cell division by serving as the anchor for the FtsZ protein within the cytoplasmic membrane [35]. This study builds upon the foundational research conducted by James et al., in 2015, wherein the essentiality of the FtsA protein in S. aureus, contrasted with its nonessential status in higher organisms, was proposed [63]. The identification of proteins deemed essential in bacteria yet nonessential in higher organisms presents an enticing avenue for the development of innovative antibiotics [64]. To delve deeper into comprehending the mode of action and the degree of affinity associated with compound 4n, we conducted a molecular docking investigation. This test was specifically designed to explore the potential binding interactions of compound 4n within the active binding site of the FtsA protein in S. aureus.
In the realm of structural biology, the determination of a protein's crystal structure holds paramount importance for comprehending its functionality and potential interactions with other molecules. Attaining the three-dimensional arrangement of atoms within a protein provides researchers with valuable insights into its mechanism of action and enables the identification of potential targets for drug development. Nonetheless, there are instances where the availability of crystal structures for a specific protein remains restricted. An illustrative case can be found in the protein FtsA sourced from S. aureus, for which only two crystal structures presently exist within the Protein Data Bank. These structures, bearing PDB codes 3WQT and 3WQU, offer resolutions of 2.2 Å and 2.8 Å, respectively. It is noteworthy, however, that neither of these crystal structures features a co-crystallized ligand within the active binding site. In light of this constraint and with the aim of delving deeper into the potential interactions of FtsA, the crystal structure boasting the highest resolution, namely 3WQT, was selected for meticulous examination in the context of this study.
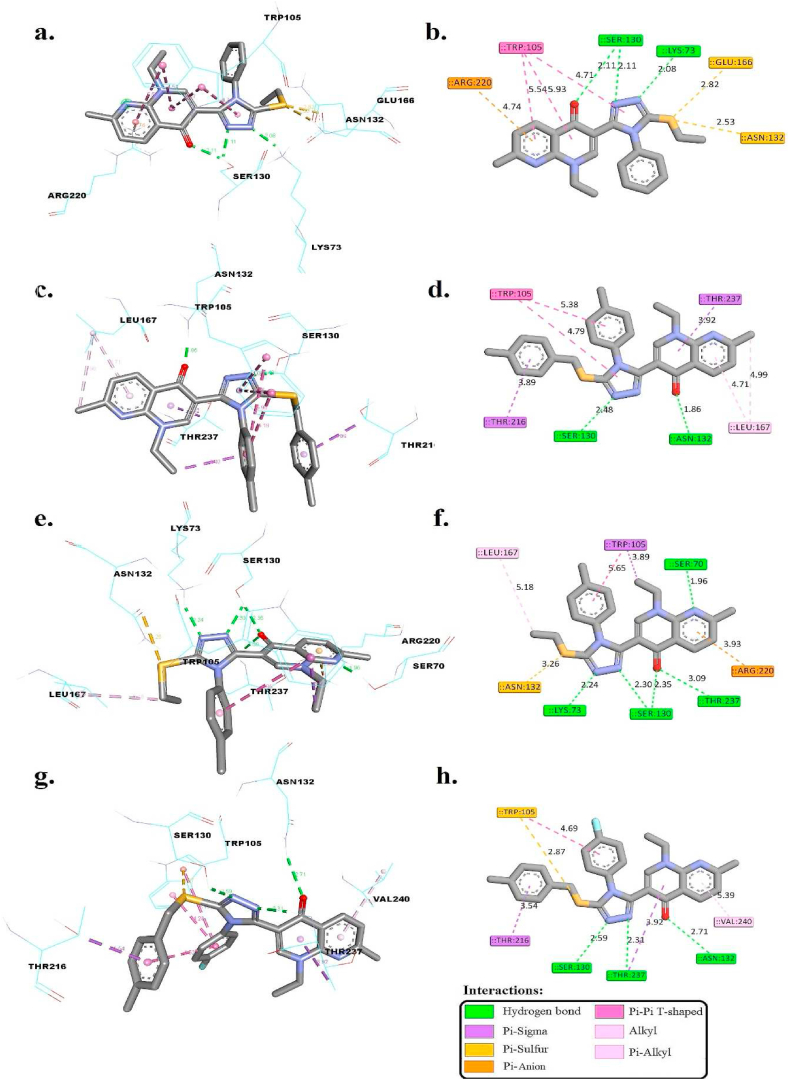
Compound 4n exhibits a remarkable affinity for the FtsA binding site, with a docking score of approximately −9.98 kcal/mol, indicative of its strong potential as a protein inhibitor. This affinity is substantiated by an in-depth analysis of molecular interactions, as illustrated in Fig. 4, where compound 4n is shown to effectively occupy the active binding site. The formation of four hydrogen bonds with SER14 (1.99 Å), SER15 (1.96 Å), LYS17 (2.49 Å), and LYS254 (2.19 Å) suggests a precise and stable engagement with the FtsA protein. These distances are particularly telling of the compound's ability to align closely with key active site residues, enhancing its inhibitory action.
Fig. 4.
3D and 2D interaction of 4n within the active binding site of FtsA protein in S. aureus.
The pi-sigma interaction with GLY208 and the pi-cation interaction with LYS17 further contribute to the compound's binding efficiency, allowing for additional specificity in its interaction with the protein. Moreover, the hydrophobic interactions with VAL211 and GLY325, along with a pi-sulfur interaction involving TYR37, complement the hydrogen bonds by stabilizing the compound within the binding site and potentially affecting the protein's conformational flexibility.
These intricate molecular interactions highlight that compound 4n presents itself in an advantageous conformation and shows potential as an inhibitor for the FtsA protein.
3.4.3. Molecular docking with Klebsiella pneumonia (K. pneumonia)
K. pneumoniae is a significant multidrug-resistant pathogen that affects humans and is a major source of hospital infections associated with high morbidity and mortality due to limited treatment options [65]. The pathogenicity of this microorganism is intricately linked to an ensemble of pivotal enzymes, among which the enzyme K. pneumoniae carbapenemase (KPC) especially KPC-2 hold particular prominence, given their recognized pivotal role in host cell invasion [66]. Consequently, the exploration of KPC-2 enzyme as prospective therapeutic targets holds promise in mitigating this pathogenic menace.
Within the ambit of biological assays conducted, compounds 4i, 4k, 4m, and 4n exhibited noteworthy MIC values against K. pneumoniae. In light of these findings, this study endeavors to employ molecular docking simulations, to elucidate and predict the optimal binding poses of these compounds concerning their interactions with the KPC-2 enzyme of K. pneumoniae. This endeavor seeks to shed light on potential molecular interactions that could pave the way for innovative therapeutic interventions against this pathogen.
In this research, we employed the crystal structure of KPC-2 beta-lactamase complexed with hydrolyzed cefotaxime (PDB ID: 5UJ3) as a model to investigate the interactions and binding mechanisms of compounds 4i, 4k, 4m, and 4n within the enzyme's active binding site. Our primary objective was to enhance our comprehension of these binding modes by comparing them with the control ligand, cefotaxime, originally identified in the native crystal structure.
In order to assess the performance and reliability of the docking process parameters, a thorough validation was conducted using AutoDock 4.2 software. The validation process involved the re-docking of the co-crystallized ligand, cefotaxime, into the active binding site of the enzyme K. pneumoniae e carbapenemase 2 (KPC-2) (PDB ID 5UJ3). This validation step was crucial in determining the accuracy and feasibility of using docking as a predictive tool for evaluating the binding affinity of small molecules.
The re-docking process resulted in an RMSD value of 1.29 Å, indicating that the docked conformation of the ligand closely resembles its native conformation in the crystal structure (Fig. S2). This RMSD value of 1.29 Å falls well within the acceptable range, as supported by previous literature. Previous studies have indicated that a successful docking process should yield an RMSD value of less than 2.0 Å when compared to the experimental crystal structure [42,[46], [47], [48],67]. These findings not only validate the accuracy of AutoDock 4.2 as a suitable docking tool but also highlight the robustness of docking as a predictive method for assessing the binding affinity of tested compounds (4i, 4k, 4m, and 4n) against KPC-2.
Table 5 details docking scores (*ΔGbind) and molecular interactions for compounds 4i, 4k, 4m, and 4n (Fig. 5(a–h)), alongside the co-crystallized ligand cefotaxime, within the K. pneumoniae carbapenemase 2 (KPC-2) active site (PDB ID 5UJ3), utilizing AutoDock 4.2. Cefotaxime, the co-crystallized ligand, showcases a high binding affinity (*ΔGbind of −10.19 kcal/mol), engaging in hydrogen bonds with SER70, SER130, and THR237, at distances between 2.42 Å and 3.20 Å, indicative of its strong interaction and potential for effective enzyme inhibition.
Table 5.
Docking scores (*ΔGbind in kcal/mol) and a molecular interaction analysis were performed between 4i, 4k, 4m, and 4n and the co-crystallized ligand (cefotaxime) within the active binding site of the enzyme K. pneumoniae carbapenemase 2 (KPC-2) (PDB ID 5UJ3) using AutoDock 4.2.
| Compounds | *ΔGbind (kcal/mol) | Hydrogen Bond Interactions |
Pi-Sigma | Pi-Sulfur | Pi-Cation | Ionic bond | Hydrophobic Interaction | |
|---|---|---|---|---|---|---|---|---|
| Residues | Distances (Å) | |||||||
| 4i | −8.31 | LYS73, SER130, and SER130 | 2.08, 2.11, and 2.11 | – | ASN132 and GLU166 | ARG220 | – | TRP105 |
| 4k | −8.12 | SER130 and ASN132 | 2.48 and 1.86 | THR216 and THR237 | – | – | – | TRP105 and LEU167 |
| 4m | −10.67 | SER70, LYS73, SER130, SER130, THR237 | 1.96, 2.24, 2.30, 2.35, and 3.09 | TRP105 | ASN132 | ARG220 | – | TRP105 and LEU167 |
| 4n | −8.97 | SER130, ASN132, and THR237 | 2.59, 2.71, and 2.31 | THR216 and THR237 | TRP105 | – | – | TRP105 and VAL240 |
| Co-crystalized ligand | −10.19 | SER70, SER130, SER130, THR23, THR23, and THR237 | 2.96, 3.06, 2.42, 2.68, 2.52, and 3.20 | – | TRP105 | – | LYS73, ARG220, and LYS234 | TRP105 |
Fig. 5.
3D and 2D interactions of 4i (a & b), 4k (c & d), 4m (e & f), and 4n (g & h) within the active binding site of the enzyme K. pneumoniae carbapenemase 2 (KPC-2).
Compound 4i, with a *ΔGbind of −8.31 kcal/mol, forms hydrogen bonds at distances of 2.08 Å and 2.11 Å with LYS73 and SER130, suggesting a significant binding efficacy that could translate into notable inhibitory activity against KPC-2. The pi-sulfur and pi-cation interactions, particularly with ASN132 and ARG220, further signify its potential as a strong inhibitor by stabilizing the compound within the binding site.
Compound 4k, displaying a *ΔGbind of −8.12 kcal/mol, establishes hydrogen bonds with SER130 and ASN132 at 2.48 Å and 1.86 Å, respectively. The absence of pi-sulfur or pi-cation interactions, contrasted with pi-sigma interactions with THR216 and THR237, points to a unique binding mode that may influence its inhibitory mechanism against KPC-2.
Compound 4m, exhibiting a *ΔGbind of −10.67 kcal/mol, forms hydrogen bonds across a range of 1.96 Å to 3.09 Å with SER70, LYS73, and SER130. These interactions, along with pi-sigma with TRP105 and pi-cation with ARG220, highlight its strong affinity and potential for effective inhibition, possibly exceeding that of cefotaxime due to its diversified interaction profile.
Lastly, compound 4n displayed a strong binding affinity with a *ΔGbind of −8.97 kcal/mol. It formed hydrogen bonds with SER130, ASN132, and THR237 at distances of 2.59 Å, 2.71 Å, and 2.31 Å, respectively. Additionally, 4n formed pi-sigma interactions with THR216 and THR237, pi-sulfur interactions with TRP105, and exhibited hydrophobic interactions with TRP105 and VAL240. Although it did not form ionic bonds or pi-cation interactions, these interactions collectively contributed to its stability within the binding site.
In comparison to the co-crystallized ligand, all tested compounds (4i, 4k, 4m, and 4n) displayed robust binding affinities, as reflected in their negative *ΔGbind values. They formed hydrogen bonds with key residues and, in some cases, additional interactions such as pi-sigma, pi-sulfur, pi-cation, and hydrophobic interactions. These findings underscore the potential of these compounds as inhibitors against the enzyme K. pneumoniae carbapenemase 2 (KPC-2), warranting further investigation for their therapeutic potential.
3.4.4. Molecular docking with Escherichia coli (E. coli)
E. coli, a Gram-negative bacterium known for its complex characteristics and resistance to drugs, has attracted significant attention in the field of science [68]. The enzyme β-glucuronidase plays a critical role in determining the viability of pathogenic E. coli by indicating inhibition of their survival mechanisms [69]. This versatile enzyme is involved in various metabolic processes within E. coli, breaking down sugar conjugates into glucuronides to produce ATP [69]. In addition to its metabolic functions, the activity of β-glucuronidase enables enterobacteria to thrive in carbohydrate-deficient environments like the urinary tract [70]. In light of recent investigations, there is an emerging theory connecting E. coli's β-glucuronidase to the onset of colon cancer [71]. In consideration of this biological intricacy and its potential health implications, our study focuses on evaluating compounds 4k, 4n, and 4o. These compounds are subjected to thorough molecular docking analyses with the objective of identifying their affinity as inhibitors against this crucial enzyme. The motivation behind this undertaking stems from notable observations regarding their significant minimum inhibitory concentrations during in vitro studies (see Table 3), which presents a hopeful avenue for further investigation in the field of antimicrobial research.
For this study, we chose to examine the structure of E. coli beta-Glucuronidase (PDB ID: 4JHZ) bound with a novel and potent inhibitor known as 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5-dimethoxycyclohexyl]acetamide from the protein data bank. Our aim was to conduct molecular docking investigations on tested compounds (4k, 4n, and 4o) using this target and compare their binding affinity and molecular interactions with those observed in the presence of the potent inhibitor (control).
The re-docking procedure resulted in an RMSD value of 1.09 Å, suggesting that the docked pose of the ligand closely resembles its native conformation observed in the crystal structure. This value falls within the acceptable range, as documented in previous studies. Literature suggests that a successful docking process should generate an RMSD value lower than 2.0 Å when compared to the experimental crystal structure [42,[46], [47], [48],67]. These findings not only validate AutoDock 4.2 as a reliable docking tool but also demonstrate the effectiveness of docking as a predictive method for evaluating the binding affinity of tested compounds (4k, 4n, and 4o) against E. coli beta-Glucuronidase enzyme.
The analysis of molecular docking scores (*ΔGbind in kcal/mol) and subsequent examination of molecular interactions for compounds 4k, 4n, and 4o are presented in Table 6 and Fig. 6(a–f). These calculations were performed using AutoDock 4.2 to analyze the active binding site of E. coli beta-Glucuronidase (PDB ID: 4JHZ) with the co-crystallized ligand, namely 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5dimethoxycyclohexyl]acetamide.
Table 6.
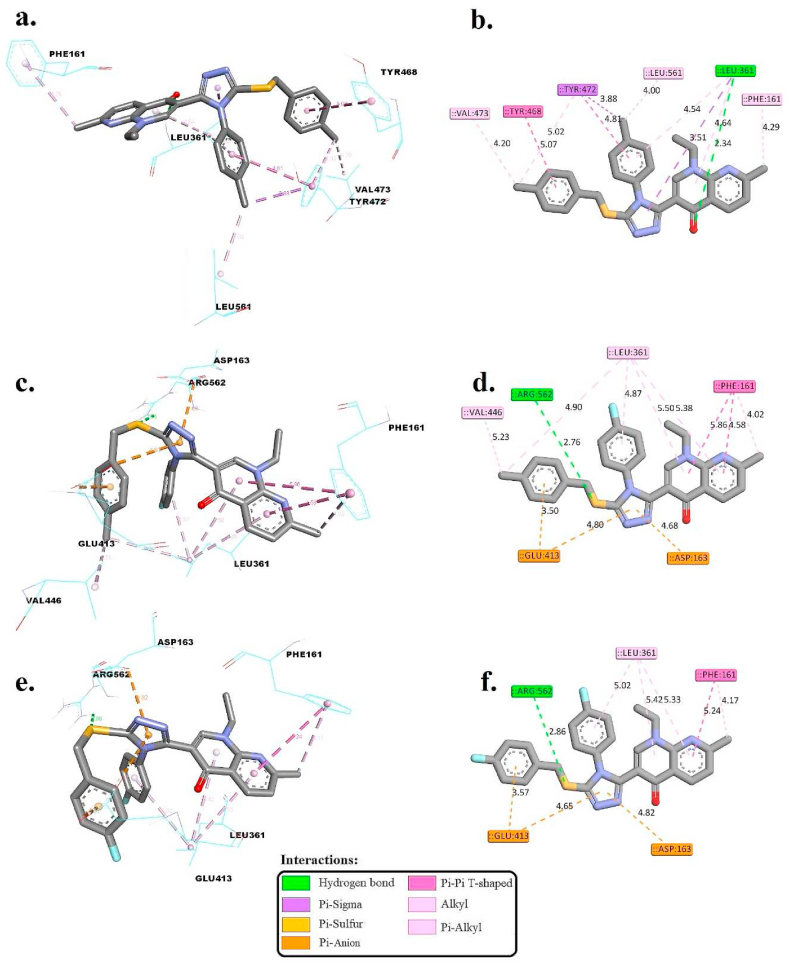
Docking scores (*ΔGbind (kcal/mol)) and molecular interactions analysis between 3k, 4n, and 4o and the co-crystallized ligand 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5 dimethoxycyclohexyl]acetamide within the active binding site of the E. coli beta-Glucuronidase (4JHZ.PDB) using AutoDock 4.2.
| Compounds | *ΔGbind (kcal/mol) | Hydrogen Bond Interactions |
Pi-Sigma | Pi-Anion | Hydrophobic Interaction | |
|---|---|---|---|---|---|---|
| Residues | Distances (Å) | |||||
| 4k | −8.69 | LEU361 | 2.34 | LEU361 and TYR472 | – | PHE161, TYR468, TYR472, VAL473, and LEU561 |
| 4n | −8.92 | ARG562 | 2.91 and 1.84 | – | ASP163 and GLU413 | PHE161, LEU361, and VAL446 |
| 4o | −7.98 | ARG562 | 2.86 | – | ASP163 and GLU413 | PHE161 and LEU361 |
| Co-crystalized ligand | −7.53 | GLU413 | 2.13 | TYR472 | ASP163 | LEU361 and TYR472 |
Fig. 6.
3D and 2D interactions of 4k (a & b), 4n (c & d), and 4o (e & f) within the active binding site of the enzyme E. coli beta-Glucuronidase.
The co-crystallized ligand demonstrated a significant binding affinity (*ΔGbind of −7.53 kcal/mol), primarily through a hydrogen bond with GLU413 (2.13 Å) as shown in Fig. S3. This interaction, alongside pi-sigma interactions with TYR472 and pi-anion bonds with ASP163, underlines a strategic engagement with the active site. The hydrophobic interactions with LEU361 and TYR472 contribute to the ligand's stability, suggesting a strong and specific inhibition mechanism.
Compound 4k, with a *ΔGbind of −8.69 kcal/mol, showcases an enhanced affinity, forming a hydrogen bond with LEU361 (2.34 Å). The pi-sigma interactions with LEU361 and TYR472, coupled with an extensive network of hydrophobic interactions including PHE161 and TYR468, highlight its potential for effective binding and inhibition. The detailed interaction profile of 4k suggests a possibly more potent inhibitory effect against the target enzyme than the co-crystallized ligand, attributed to its diverse and robust interaction pattern.
Turning our attention to compound 4n, it exhibited a strong binding affinity (*ΔGbind of −8.92 kcal/mol), with ARG562 forming crucial hydrogen bonds (2.91 Å and 1.84 Å). The compound's interaction is further enriched by hydrophobic bonds with PHE161 and LEU361, and pi-anion interactions with ASP163 and GLU413, enhancing its stability within the binding site. These interactions indicate a high potential for 4n to act as a potent inhibitor, with its diverse bonding possibly translating to significant biological activity.
Lastly, compound 4o, displaying a *ΔGbind of −7.98 kcal/mol, maintains hydrogen bonds with ARG562 (2.86 Å) and engages in similar hydrophobic and pi-anion interactions as 4n, with ASP163 and GLU413. This pattern of interactions, though slightly lesser in binding energy, suggests a comparable mechanism of action, underscoring the nuanced balance of interactions that contribute to binding affinity and potential inhibitory capacity.
In comparison to the co-crystallized ligand, all tested compounds (4k, 4n, and 4o) presented compelling binding affinities with negative *ΔGbind values. These interactions encompassed hydrogen bonds, pi-sigma and pi-anion interactions, and hydrophobic bonds, collectively underscoring their potential as inhibitors against the E. coli beta-Glucuronidase. These findings hold promise for further exploration in antimicrobial research and therapeutic development.
3.5. ADMET prediction
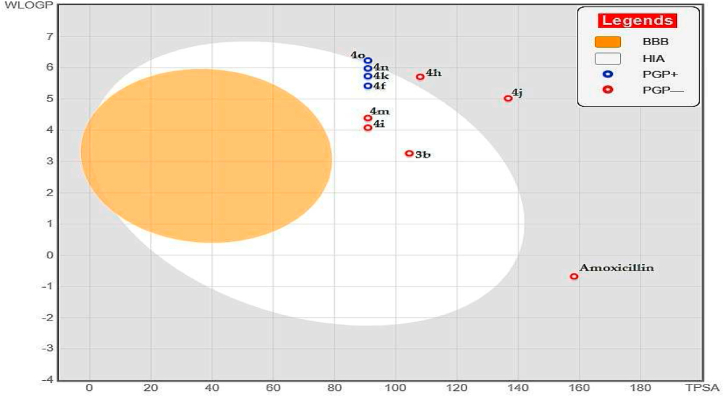
In the insightful depiction provided by Fig. 7, the 'boiled-egg' plot offers a visual representation that predicts pharmacokinetic attributes [50], with a specific focus on compounds 3b, 4i, 4j, and 4 m. These compounds are strategically positioned within the yellow and white regions, indicative of their potential to penetrate the blood-brain barrier (BBB) and be absorbed in the human intestine (HIA), respectively. The graphical layout, where the x-axis represents the topological polar surface area (TPSA) and the y-axis reflects the lipophilicity (WLOGP), is crucial for evaluating drug-likeness and absorption profiles. Compounds within the yellow region are suggestive of higher BBB permeability, while the white space aligns with favorable HIA potential [72]. Notably, compounds 3b, 4i, 4j, and 4m comply with Lipinski's rule of five, indicating a high likelihood of oral bioavailability, similar to the well-established oral drug, amoxicillin, with no apparent violations to detract from their potential for oral administration.
Fig. 7.
Pharmacokinetic profile egg-boiled shape model for 3b, 4f, 4h, 4i, 4j, 4k, 4m, 4n, 4o, and amoxicillin, generated using the SwissADME web server.
Additionally, the plot differentiates compounds as either P-glycoprotein (Pgp) substrates or non-substrates, marked by Pgp+ and Pgp-labels, respectively. This distinction is pivotal for assessing clinical outcomes, as Pgp substrates might necessitate increased dosages or the use of Pgp inhibitors to attain effective central nervous system (CNS) concentrations, whereas non-substrates could benefit from better CNS retention [73].
Remarkably, these compounds (3b, 4i, 4j, and 4m) are designated as non-substrates of P-glycoprotein (Pgp-), highlighted on the plot with specific labels. This status as non-substrates presents a significant advantage for CNS retention, as they are likely not to be actively effluxed by Pgp, suggesting enhanced and sustained CNS exposure. This could translate into a reduced need for higher dosages or co-administration of Pgp inhibitors, potentially streamlining therapeutic regimens and minimizing drug-drug interactions for CNS treatments.
However, it is imperative to approach these graphical predictions with due diligence, recognizing that the real-world clinical impact depends on corroborating these findings with experimental evidence. This includes permeability studies in Pgp-expressing cellular models and in vivo pharmacokinetic assessments to confirm the ability of these compounds to evade Pgp-mediated efflux [74]. The preliminary predictions offered by the 'boiled-egg' plot are promising, yet they mark only the beginning of a comprehensive evaluation process. The therapeutic viability of these compounds must be further supported by detailed toxicological and efficacy evaluations.
As we advance, the development of compounds with optimal pharmacokinetic properties for oral administration will rely on a holistic approach that seamlessly marries computational predictions with empirical data. The toxicity profiles for the selected compounds (3b, 4i, 4j, and 4m) along with amoxicillin have been meticulously assessed using AdmetSAR 2.0, with the findings detailed in Table 7. This critical analysis will inform the next steps in their development, ensuring a thorough understanding of their safety and potential as oral therapeutic agents.
Table 7.
Predicted the toxicity profiles of compounds (3b, 4i, 4j, and 4m) along with amoxicillin using AdmetSAR 2.0.
| Property | Model NaPDBme | Predicted Value |
||||
|---|---|---|---|---|---|---|
| 3b | 4i | 4j | 4 m | Amoxicillin | ||
| Toxicity | H-HT (Human Hepatotoxicity) | – | – | + | – | – |
| AMES (Ames Mutagenicity) | – | – | + | – | – | |
| Carcinogenicity | – | – | – | – | – | |
Table 7 provides a comprehensive insight into the predicted toxicity profiles of compounds 3b, 4i, 4j, and 4m, in comparison to the widely used antibiotic amoxicillin. The evaluation was performed using AdmetSAR 2.0, which offers a valuable assessment of potential adverse effects associated with these compounds.
For human hepatotoxicity (H-HT), compounds 3b, 4i, and 4m show a promising safety profile, as they are predicted as non-toxic (−). On the other hand, compound 4j, while indicating a potential risk (+), still remains within a manageable range. In terms of Ames Mutagenicity, compounds 3b, 4i, and 4m exhibit a non-mutagenic prediction (−), further enhancing their safety profile. Conversely, compound 4j suggests a mutagenic potential (+), which warrants careful consideration. Regarding carcinogenicity, none of the tested compounds (3b, 4i, 4j, and 4m) show indications of carcinogenicity, as all are predicted as non-carcinogenic (−).
Notably, compounds 3b, 4i, and 4m emerge as promising candidates for further investigation and development. They not only exhibit favorable antibacterial activity, as indicated by their MIC values, but also demonstrate a safe toxicity profile, making them suitable for potential oral administration. These findings highlight the potential of compounds 3b, 4i, and 4m as lead compounds in the development of novel antibiotics with enhanced safety and efficacy profiles.
4. Conclusion
In this study, we synthesized a novel series of 1,2,4-triazole derivatives and substituted sulfide-4-triazole compounds inspired by nalidixic acid, which were subjected to thorough spectroscopic and elemental analyses. Our comparative analysis indicates that these compounds show promising efficacy against gram-positive bacteria, compared to the benchmark antibiotic, amoxicillin, though their effectiveness against gram-negative bacteria was less pronounced. Additionally, compounds 3a through d displayed significant antioxidant activity, suggesting their potential as dual-purpose therapeutic agents. The antibacterial efficacy observed aligns with other contemporary studies on triazole derivatives, reaffirming the utility of such compounds in the search for new antibiotics. However, the varied responses from gram-negative bacteria highlight the challenge of bacterial membrane permeability and the need for structural optimization in future research. Predictive toxicity assessments indicated that compounds 3b, 4i, and 4m possess safety profiles that support their potential for oral administration, aligning with the low toxicity findings from similar studies. This work's integrated approach, combining synthetic chemistry, biological assays, and computational predictions, offers a comprehensive view of the therapeutic potential of these compounds. Despite these promising results, the study acknowledges its limitations, including the variable activity against gram-negative bacteria and the preliminary stage of the toxicity assessments. Future efforts will focus on modifying these compounds to broaden their antibacterial spectrum and conducting in vivo studies to better understand their pharmacological properties. While the path to clinical application is complex and requires further investigation, we remain optimistic about the potential of these compounds to contribute to the ongoing battle against antibiotic resistance.
Funding
This work was supported by the Researchers supporting project number (RSP2024R70), King Saud University, Riyadh, Saudi Arabia.
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Ibrahim Mhaidat: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. Sojoud Banidomi: Methodology. Fadel Wedian: Writing – review & editing, Writing – original draft, Funding acquisition, Conceptualization. Rahaf Badarneh: Methodology. Hasan Tashtoush: Writing – review & editing, Formal analysis. Waleed Almomani: Writing – review & editing, Investigation. Ghassab M. Al-Mazaideh: Writing – review & editing, Writing – original draft, Methodology, Funding acquisition. Naiyf S. Alharbi: Writing – review & editing, Funding acquisition. Muthu Thiruvengadam: Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28204.
Contributor Information
Ibrahim Mhaidat, Email: ibrahim.m@yu.edu.jo.
Sojoud Banidomi, Email: Sojoodbanidomi@yahoo.com.
Fadel Wedian, Email: alwedian@yu.edu.jo.
Rahaf Badarneh, Email: rahafriyad97@gmail.com.
Hasan Tashtoush, Email: htashtoush2005@yu.edu.jo.
Waleed Almomani, Email: waleed.momani@yu.edu.jo.
Ghassab M. Al-Mazaideh, Email: gmazideh@ttu.edu.jo.
Naiyf S. Alharbi, Email: nalharbi1@ksu.edu.sa.
Muthu Thiruvengadam, Email: thiruv30@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Haber J. Present status and perspectives on antimycotics with systemic effects. Cas. Lek. Cesk. 2001;140:596–604. [PubMed] [Google Scholar]
- 2.Heindel N.D., Reid J.R. 4‐Amino‐3‐mercapto‐4H‐1, 2, 4‐triazoles and propargyl aldehydes: a new route to 3‐R‐8‐aryl‐1, 2, 4‐triazolo [3, 4‐b]‐1, 3, 4‐thiadiazepines. J. Heterocycl. Chem. 1980;17:1087–1088. [Google Scholar]
- 3.Holla B., Kalluraya B., Sridhar K., Drake E., Thomas L., Bhandary K., Levine M. Synthesis, structural characterization, crystallographic analysis and antibacterial properties of some nitrofuryl triazolo [3, 4-b]-1, 3, 4-thiadiazines. Eur. J. Med. Chem. 1994;29:301–308. [Google Scholar]
- 4.Raslan M., Khalil M. Heterocyclic synthesis containing bridgehead nitrogen atom: synthesis of 3‐[(2H)‐2‐oxobenzo [b] pyran‐3‐yl]‐s‐triazolo [3, 4‐b]‐1, 3, 4‐thiadiazine and thiazole derivatives. Heteroat. Chem.: An International Journal of Main Group Elements. 2003;14:114–120. [Google Scholar]
- 5.Murty M., Ram K.R., Rao B.R., Rao R.V., Katiki M.R., Rao J.V., Pamanji R., Velatooru L. Synthesis, characterization, and anticancer studies of S and N alkyl piperazine-substituted positional isomers of 1, 2, 4-triazole derivatives. Med. Chem. Res. 2014;23:1661–1671. [Google Scholar]
- 6.Sahu N., Sahu J.K., Kaushik A. A review on ‘triazoles’: their chemistry and pharmacological potentials. Current Research in Pharmaceutical Sciences. 2013:108–113. [Google Scholar]
- 7.Karaca Gençer H., Acar Çevik U., Levent S., Sağlık B.N., Korkut B., Özkay Y., Ilgın S., Öztürk Y. New benzimidazole-1, 2, 4-triazole hybrid compounds: synthesis, anticandidal activity and cytotoxicity evaluation. Molecules. 2017;22:507. doi: 10.3390/molecules22040507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Appna N.R., Nagiri R.K., Korupolu R.B., Kanugala S., Chityal G.K., Thipparapu G., Banda N. Design and synthesis of novel 4-hydrazone functionalized/1, 2, 4-triazole fused pyrido [2, 3-d] pyrimidine derivatives, their evaluation for antifungal activity and docking studies. Med. Chem. Res. 2019;28:1509–1528. [Google Scholar]
- 9.Rode N.D., Sonawane A.D., Nawale L., Khedkar V.M., Joshi R.A., Likhite A.P., Sarkar D., Joshi R.R. vol. 90. Chemical Biology & Drug Design; 2017. pp. 1206–1214. (Synthesis, Biological Evaluation, and Molecular Docking Studies of Novel 3‐aryl‐5‐(alkyl‐thio)‐1H‐1, 2, 4‐triazoles Derivatives Targeting Mycobacterium tuberculosis). [DOI] [PubMed] [Google Scholar]
- 10.Peng Z., Wang G., Zeng Q.-H., Li Y., Wu Y., Liu H., Wang J.J., Zhao Y. Synthesis, antioxidant and anti-tyrosinase activity of 1, 2, 4-triazole hydrazones as antibrowning agents. Food Chem. 2021;341 doi: 10.1016/j.foodchem.2020.128265. [DOI] [PubMed] [Google Scholar]
- 11.Grytsai O., Valiashko O., Penco-Campillo M., Dufies M., Hagege A., Demange L., Martial S., Pagès G., Ronco C., Benhida R. Synthesis and biological evaluation of 3-amino-1, 2, 4-triazole derivatives as potential anticancer compounds. Bioorg. Chem. 2020;104 doi: 10.1016/j.bioorg.2020.104271. [DOI] [PubMed] [Google Scholar]
- 12.Shkoor M., Tashtoush H., Al-Talib M., Mhaidat I., Al-Hiari Y., Kasabri V., Alalawi S. Synthesis and antiproliferative and antilipolytic activities of a series of 1, 3-and 1, 4-Bis [5-(R-sulfanyl)-1, 2, 4-triazol-3-yl) benzenes. Russ. J. Org. Chem. 2021;57:1141–1151. [Google Scholar]
- 13.Li S.-M., Tsai S.-E., Chiang C.-Y., Chung C.-Y., Chuang T.-J., Tseng C.-C., Jiang W.-P., Huang G.-J., Lin C.-Y., Yang Y.-C. New methyl 5-(halomethyl)-1-aryl-1H-1, 2, 4-triazole-3-carboxylates as selective COX-2 inhibitors and anti-inflammatory agents: design, synthesis, biological evaluation, and docking study. Bioorg. Chem. 2020;104 doi: 10.1016/j.bioorg.2020.104333. [DOI] [PubMed] [Google Scholar]
- 14.Khanage S.G., Raju A., Mohite P.B., Pandhare R.B. Analgesic activity of some 1, 2, 4-triazole heterocycles clubbed with pyrazole, tetrazole, isoxazole and pyrimidine. Adv. Pharmaceut. Bull. 2013;3:13. doi: 10.5681/apb.2013.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hichri F., Omri A., Hossan A.S.M., Ben Jannet H. Alpha-glucosidase and amylase inhibitory effects of Eruca vesicaria subsp. longirostris essential oils: synthesis of new 1, 2, 4-triazole-thiol derivatives and 1, 3, 4-thiadiazole with potential inhibitory activity. Pharmaceut. Biol. 2019;57:564–570. doi: 10.1080/13880209.2019.1642363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaproń B., Łuszczki J.J., Siwek A., Karcz T., Nowak G., Zagaja M., Andres-Mach M., Stasiłowicz A., Cielecka-Piontek J., Kocki J. Preclinical evaluation of 1, 2, 4-triazole-based compounds targeting voltage-gated sodium channels (VGSCs) as promising anticonvulsant drug candidates. Bioorg. Chem. 2020;94 doi: 10.1016/j.bioorg.2019.103355. [DOI] [PubMed] [Google Scholar]
- 17.Navidpour L., Shabani S., Heidari A., Bashiri M., Ebrahim-Habibi A., Shahhosseini S., Shafaroodi H., Tabatabai S.A., Toolabi M. 5-[Aryloxypyridyl (or nitrophenyl)]-4H-1, 2, 4-triazoles as novel flexible benzodiazepine analogues: synthesis, receptor binding affinity and lipophilicity-dependent anti-seizure onset of action. Bioorg. Chem. 2021;106 doi: 10.1016/j.bioorg.2020.104504. [DOI] [PubMed] [Google Scholar]
- 18.Ostrosky-Zeichner L., Casadevall A., Galgiani J.N., Odds F.C., Rex J.H. An insight into the antifungal pipeline: selected new molecules and beyond. Nat. Rev. Drug Discov. 2010;9:719–727. doi: 10.1038/nrd3074. [DOI] [PubMed] [Google Scholar]
- 19.Grover G., Kini S.G. Synthesis and evaluation of new quinazolone derivatives of nalidixic acid as potential antibacterial and antifungal agents. Eur. J. Med. Chem. 2006;41:256–262. doi: 10.1016/j.ejmech.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Uddin T.M., Chakraborty A.J., Khusro A., Zidan B.R.M., Mitra S., Emran T.B., Dhama K., Ripon M.K.H., Gajdács M., Sahibzada M.U.K. Antibiotic resistance in microbes: history, mechanisms, therapeutic strategies and future prospects. Journal of infection and public health. 2021;14:1750–1766. doi: 10.1016/j.jiph.2021.10.020. [DOI] [PubMed] [Google Scholar]
- 21.Woodford N., Turton J.F., Livermore D.M. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 2011;35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 22.Hancock R.E. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin. Infect. Dis. 1998;27:S93–S99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- 23.Fraser A.G., Harrower A.D. Convulsions and hyperglycaemia associated with nalidixic acid. Br. Med. J. 1977;2:1518. doi: 10.1136/bmj.2.6101.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kar S., Roy K., Leszczynski J. In silico tools and software to predict ADMET of new drug candidates. In in silico methods for predicting drug toxicity; Springer. 2022:85–115. doi: 10.1007/978-1-0716-1960-5_4. [DOI] [PubMed] [Google Scholar]
- 25.Agu P., Afiukwa C., Orji O., Ezeh E., Ofoke I., Ogbu C., Ugwuja E., Aja P. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023;13 doi: 10.1038/s41598-023-40160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shriram V., Khare T., Bhagwat R., Shukla R., Kumar V. Inhibiting bacterial drug efflux pumps via phyto-therapeutics to combat threatening antimicrobial resistance. Front. Microbiol. 2018;9:2990. doi: 10.3389/fmicb.2018.02990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Upadhyay H.C. Coumarin-1, 2, 3-triazole hybrid molecules: an emerging scaffold for combating drug resistance. Curr. Top. Med. Chem. 2021;21:737–752. doi: 10.2174/1568026621666210303145759. [DOI] [PubMed] [Google Scholar]
- 28.Gupta A.K., Prachand S., Patel A., Jain S. Synthesis of some 4-amino-5-(substituted-phenyl)-4H-[1, 2, 4] triazole-3-thiol derivatives and antifungal activity. Int J Pharm Life Sci. 2012;3:1848–1857. [Google Scholar]
- 29.Wedian F., Mhaidat I., Braik N., Al-Mazaideh G. A corrosion inhibitor for aluminum by novel synthesized triazole compounds in basic medium. Int. J. Corros. Scale Inhib. 2022;11:364–381. [Google Scholar]
- 30.Kim B.-T., Chun J.-C., Hwang K.-J. Synthesis of dihydroxylated chalcone derivatives with diverse substitution patterns and their radical scavenging ability toward DPPH free radicals. Bull. Kor. Chem. Soc. 2008;29:1125–1130. [Google Scholar]
- 31.van der Werf R., Marcic C., Khalil A., Sigrist S., Marchioni E. ABTS radical scavenging capacity in green and roasted coffee extracts. LWT--Food Sci. Technol. 2014;58:77–85. [Google Scholar]
- 32.Hannan P. Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. Veterinary research. 2000;31:373–395. doi: 10.1051/vetres:2000100. [DOI] [PubMed] [Google Scholar]
- 33.Wayne P. Clinical and Laboratory Standards Institute (CLSI) performance standards for antimicrobial disk diffusion susceptibility tests 19th ed. approved standard. CLSI document M100-S19. 2009;29:M100–S121. [Google Scholar]
- 34.Gajiwala K.S., Margosiak S., Lu J., Cortez J., Su Y., Nie Z., Appelt K. Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme. FEBS Lett. 2009;583:2939–2946. doi: 10.1016/j.febslet.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Fujita J., Maeda Y., Nagao C., Tsuchiya Y., Miyazaki Y., Hirose M., Mizohata E., Matsumoto Y., Inoue T., Mizuguchi K. Crystal structure of FtsA from Staphylococcus aureus. FEBS Lett. 2014;588:1879–1885. doi: 10.1016/j.febslet.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 36.Pemberton O.A., Zhang X., Chen Y. Molecular basis of substrate recognition and product release by the Klebsiella pneumoniae carbapenemase (KPC-2) J. Med. Chem. 2017;60:3525–3530. doi: 10.1021/acs.jmedchem.7b00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts A.B., Wallace B.D., Venkatesh M.K., Mani S., Redinbo M.R. Molecular insights into microbial β-glucuronidase inhibition to abrogate CPT-11 toxicity. Mol. Pharmacol. 2013;84:208–217. doi: 10.1124/mol.113.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westbrook J., Feng Z., Chen L., Yang H., Berman H.M. The protein data bank and structural genomics. Nucleic acids research. 2003;31:489–491. doi: 10.1093/nar/gkg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biovia D.S. San Diego; CA, USA: 2017. Discovery Studio Visualizer; p. 936. [Google Scholar]
- 40.Land H., Humble M.S. YASARA: a tool to obtain structural guidance in biocatalytic investigations. Protein Eng.: methods and protocols. 2018:43–67. doi: 10.1007/978-1-4939-7366-8_4. [DOI] [PubMed] [Google Scholar]
- 41.Shalayel M.H.F., Al-Mazaideh G.M., Alanezi A.A., Almuqati A.F., Alotaibi M. Diosgenin and monohydroxy spirostanol from prunus amygdalus var amara seeds as potential suppressors of EGFR and HER2 tyrosine kinases: a computational approach. Pharmaceuticals. 2023;16:704. doi: 10.3390/ph16050704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdelbagi M.E., Al-Mazaideh G.M., Ahmed A.E., Al-Rimawi F., Ayyal Salman H., Almutairi A., Abuilaiwi F.A., Wedian F. Exploring securigera securidaca seeds as a source of potential CDK1 inhibitors: identification of hippeastrine and naringenin as promising hit candidates. Processes. 2023;11:1478. [Google Scholar]
- 43.Gordon J.C., Myers J.B., Folta T., Shoja V., Heath L.S., Onufriev A. H++: a server for estimating p K as and adding missing hydrogens to macromolecules. Nucleic acids research. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norgan A.P., Coffman P.K., Kocher J.-P.A., Katzmann D.J., Sosa C.P. Multilevel parallelization of AutoDock 4.2. J. Cheminf. 2011;3:1–9. doi: 10.1186/1758-2946-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh A., Singh J.V., Rana A., Bhagat K., Gulati H.K., Kumar R., Salwan R., Bhagat K., Kaur G., Singh N. Monocarbonyl curcumin-based molecular hybrids as potent antibacterial agents. ACS Omega. 2019;4:11673–11684. doi: 10.1021/acsomega.9b01109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh A., Kaur K., Kaur H., Mohana P., Arora S., Bedi N., Chadha R., Bedi P.M.S. Design, synthesis and biological evaluation of isatin-benzotriazole hybrids as new class of anti-Candida agents. J. Mol. Struct. 2023;1274 [Google Scholar]
- 48.Singh A., Kaur K., Mohana P., Singh K., Sharma A., Prajapati J., Goswami D., Khosla N., Kaur U., Kaur R., Kaur R., Rana A., Kour S., Ohri P., Arora S., Chadha R., Singh Bedi P.M. The development of thymol-isatin hybrids as broad-spectrum antibacterial agents with potent anti-MRSA activity. RSC Med. Chem. 2023;15(1):234–253. doi: 10.1039/d3md00580a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazaridis T., Karplus M. Effective energy function for proteins in solution. Proteins: Struct., Funct., Bioinf. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 50.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7 doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35:1067–1069. doi: 10.1093/bioinformatics/bty707. [DOI] [PubMed] [Google Scholar]
- 52.Ames B.N., Shigenaga M.K., Hagen T.M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sies H. Oxidative stress: oxidants and antioxidants. Exp. Physiol.: Translation and Integration. 1997;82:291–295. doi: 10.1113/expphysiol.1997.sp004024. [DOI] [PubMed] [Google Scholar]
- 54.Alam M.N., Bristi N.J., Rafiquzzaman M. Review on in vivo and in vitro methods evaluation of antioxidant activity. Saudi Pharmaceut. J. 2013;21:143–152. doi: 10.1016/j.jsps.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reische D., Lillard D., Eitenmiller R., Akoh C., Min D. Food lipids: chemistry, nutrition, and biotechnology. 15. Antioxidants. 2008;409 [Google Scholar]
- 56.Gumrukcuoglu N., Imran M., Iqbal I. Synthesis, characterization and antimicrobial activity of some novel 4-amino-5- phenyl-4H-1,2,4-triazole-3-thiol derivatives. İstanbul Journal of Pharmacy. 2023;53 doi: 10.26650/IstanbulJPharm.2023.130108. 294–230. [DOI] [Google Scholar]
- 57.Radwan A.A., Alanazi F.K., Al-Agamy M.H. 1,3,4-Thiadiazole and 1,2,4-triazole-3(4H)-thione bearing salicylate moiety: synthesis and evaluation as anti-Candida albicans. Brazilian Journal of Pharmaceutical Sciences. 2017;53 doi: 10.1590/s2175-97902017000115239. [DOI] [Google Scholar]
- 58.Singh R., Kashaw S.K., Mishra V.K., Mishra M., Rajoriya V., Kashaw V. Design and synthesis of new bioactive 1,2,4-triazoles, potential antitubercular and antimicrobial agents. Indian J Pharm Sci. 2018;80:36–45. doi: 10.4172/pharmaceutical-sciences.1000328. [DOI] [Google Scholar]
- 59.Ma D.-L., Chan D.S.-H., Leung C.-H. Molecular docking for virtual screening of natural product databases. Chem. Sci. 2011;2:1656–1665. [Google Scholar]
- 60.Alonso H., Bliznyuk A.A., Gready J.E. Combining docking and molecular dynamic simulations in drug design. Med. Res. Rev. 2006;26:531–568. doi: 10.1002/med.20067. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Y., Yang Y.-S., Song X.-D., Lu L., Zhu H.-L. Study of Schiff-base-derived with dioxygenated rings and nitrogen heterocycle as potential β-ketoacyl-acyl carrier protein synthase III (FabH) inhibitors. Chem. Pharm. Bull. 2017;65:178–185. doi: 10.1248/cpb.c16-00772. [DOI] [PubMed] [Google Scholar]
- 62.Belete T.M. Novel targets to develop new antibacterial agents and novel alternatives to antibacterial agents. Human Microbiome Journal. 2019;11 [Google Scholar]
- 63.James O.C., Francis U.U., Tajudeen O., Jude C. In silico identification of putative drug targets in methicillin resistant Staphylococcus aureus: a subtractive genomic approach. Int J Comput Bioinfo In Silico Model. 2015;4:585–591. [Google Scholar]
- 64.Mura A., Fadda D., Perez A.J., Danforth M.L., Musu D., Rico A.I., Krupka M., Denapaite D., Tsui H.-C.T., Winkler M.E. Roles of the essential protein FtsA in cell growth and division in Streptococcus pneumoniae. J. Bacteriol. 2017;199 doi: 10.1128/jb.00608-00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mahapatra S.R., Dey J., Raj T.K., Kumar V., Ghosh M., Verma K.K., Kaur T., Kesawat M.S., Misra N., Suar M. The potential of plant-derived secondary metabolites as novel drug candidates against Klebsiella pneumoniae: molecular docking and simulation investigation. South Afr. J. Bot. 2022;149:789–797. [Google Scholar]
- 66.Arnold R.S., Thom K.A., Sharma S., Phillips M., Johnson J.K., Morgan D.J. Emergence of Klebsiella pneumoniae carbapenemase (KPC)-producing bacteria. South. Med. J. 2011;104:40. doi: 10.1097/SMJ.0b013e3181fd7d5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elhady S.S., Abdelhameed R.F., Malatani R.T., Alahdal A.M., Bogari H.A., Almalki A.J., Mohammad K.A., Ahmed S.A., Khedr A.I., Darwish K.M. Molecular docking and dynamics simulation study of hyrtios erectus isolated scalarane sesterterpenes as potential SARS-CoV-2 dual target inhibitors. Biology. 2021;10:389. doi: 10.3390/biology10050389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stojiljkovic I., Bäumler A.J., Hantke K. Fur regulon in gram-negative bacteria: identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 69.Wang P., Jia Y., Wu R., Chen Z., Yan R. Human gut bacterial β-glucuronidase inhibition: an emerging approach to manage medication therapy. Biochem. Pharmacol. 2021;190 doi: 10.1016/j.bcp.2021.114566. [DOI] [PubMed] [Google Scholar]
- 70.Kovács Z., Glover L., Reidy F., MacSharry J., Saldova R. Novel diagnostic options for endometriosis–Based on the glycome and microbiome. J. Adv. Res. 2021;33:167–181. doi: 10.1016/j.jare.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Louis P., Hold G.L., Flint H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 72.Khamouli S., Belaidi S., Ouassaf M., Lanez T., Belaaouad S., Chtita S. Multi-combined 3D-QSAR, docking molecular and ADMET prediction of 5-azaindazole derivatives as LRRK2 tyrosine kinase inhibitors. J. Biomol. Struct. Dyn. 2022;40:1285–1298. doi: 10.1080/07391102.2020.1824815. [DOI] [PubMed] [Google Scholar]
- 73.O'Brien F.E. University College Cork; 2013. P-Glycoprotein Inhibition as a Strategy to Increase Drug Delivery across the Blood-Brain Barrier: Focus on Antidepressants. [Google Scholar]
- 74.Zahra R., Furqan M., Ullah R., Mithani A., Saleem R.S.Z., Faisal A. A cell-based high-throughput screen identifies inhibitors that overcome P-glycoprotein (Pgp)-mediated multidrug resistance. PLoS One. 2020;15 doi: 10.1371/journal.pone.0233993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van De Waterbeemd H., Gifford E. ADMET in silico modelling: towards prediction paradise? Nat. Rev. Drug Discov. 2003;2:192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 76.Xiong G., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., Yin M., Zeng X., Wu C., Lu A. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49:W5–W14. doi: 10.1093/nar/gkab255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.