Summary
Gametogenesis drives the maturation of germ cell precursors into functional gametes, facilitated by interactions with the niche environment. However, the molecular mechanisms, especially in invertebrates, remain incompletely understood. In this study, the gonadal microenvironment and gametogenic processes in the Pacific oyster, a model for diffuse gonadal organization and periodic gametogenesis, are investigated. We combine single-nucleus RNA-seq and bulk RNA-seq to analyze gonadal microenvironments in oysters. Twenty-three male and nineteen female gonadal cell clusters are identified, revealing four male and three female germ cell types, alongside follicular cells in females and Sertoli/Leydig cells in males. The NOTCH and BMP (bone morphogenetic protein) signaling pathways play a significant role in the male germline niche, suggesting similarities with mammalian germ cell microenvironment. This study offers valuable insights into germ cell developmental transitions and microenvironmental characteristics.
Subject areas: Wildlife reproduction, Developmental biology, Aquatic biology, Transcriptomics
Graphical abstract

Highlights
-
•
Single-nucleus sequencing provides insights into the cell types in the oyster gonad
-
•
Diverse germ cells identified, revealing oyster gametogenesis nuances
-
•
NRG, VISFATIN, and SEMA6 pathways aid niche-to-germ communication
-
•
NOTCH and BMP pathways play vital roles in male germline niche
Wildlife reproduction; Developmental biology; Aquatic biology; Transcriptomics
Introduction
Germ cells play a pivotal role in animal reproduction, genetic heredity, and evolution. In sexually reproducing animals, they are the exclusive progenitors of eggs and sperm and undergo several major fate decisions, including mitosis-meiosis, sperm-oocyte, and apoptosis-survival decisions.1 The strict regulation of germ cell development is essential for the development of all multicellular organisms.2 As such, the molecular and systemic mechanisms underlying the unique activities involved in germ cell development are of great interest to biologists, resulting in an enormous number of studies on the subject.
Studies in model organisms have provided significant insight into the molecular mechanisms that govern germline development. The intrinsic balance of gene expression in germ stem cells between renewal and differentiation is critical for maintaining fertility. This process is particularly important in the niches where gametes are encased by supportive somatic cells, creating units of reproduction. Studies of the gonad niche in mammals have well mapped out a thriving microenvironmental kingdom, where different cell types play specific roles in gametogenesis.3 In the testis, Sertoli cells provide structural, nutritive, and hormone-regulatory support to germ cells, while Leydig cells generate sex hormones.4 In the ovary, granulosa cells produce estradiol-17 beta (E2) for oocyte growth and maturation. The NOTCH and BMP signaling pathways are reciprocally expressed between fetal germ cells (FGCs) and associated gonadal niche cells.5 In Drosophila, nurse cells support oocyte growth by generating piRNAs and aid in the establishment of oocyte polarity.6
Although significant progress has been made in dissecting the molecular mechanisms of germline development in model organisms, the vast majority of invertebrates which constitute 99% of all animal life on earth, remain understudied.7 Invertebrates present excellent opportunities for germ cell research due to their remarkable reproductive diversity and the structural heterogeneity of their gonads. A comprehensive understanding of the molecular mechanisms governing gametogenesis development in diverse invertebrate species could provide valuable insights into the evolution of germ cells and the complex process of germ cell differentiation.
Gonadal tissue in some marine invertebrates, including bivalves such as oysters and clams, may appear deceptively simple. The gonads of these species consist of diffuse and episodic tissues that encircle the digestive gland and lack sexual dimorphism, which poses a great challenge for germ cell studies. Additionally, unlike mammals, gonadal formation and gamete maturation in these species occur periodically. During the initial stage of oyster gametogenesis, small clusters of self-renewing stem cells develop in conjunctive tissue.8 It remains unclear whether an alternative gonadal microenvironment accounts for the unique gonadal organization and periodic gametogenesis observed in these invertebrates. Transcriptomic analysis is commonly used in studies of the molecular mechanisms of germ cell development. However, bulk RNA-seq requires defining tissues a priori by morphology or marker gene expression, which is problematic in diffuse tissues like those found in some bivalves.9
To address this issue, we performed extensive single-nucleus RNA sequencing (snRNA-seq) characterization of Pacific oyster male and female gonadal cells. Our aim was to create a transcriptional cell atlas of all cell types in the gonad, effectively delineating cell heterogeneity and mapping the germline cells and gonadal niche interactions at the cellular level. Our work provides a valuable resource that supplements earlier transcriptome investigations by offering a detailed perspective on germ cell lineage development and establishing the germ cell niche in the most abundant gonadal somatic cell populations.
Results
The gene expression landscapes of Pacific oyster germline cells and somatic cells in the gonad
In this study, we conducted an unbiased snRNA-seq analysis on Pacific oyster gonad cells at stage I (Figures 1A and S1A). In this stage, gonadal tubules were scattered in the connective tissue. Spermatogonia and oogonia were distinctly observable at the margins of the inner walls of the gonadal tubules, signifying the commencement of development and formation of oocytes and spermatocytes (Figure S1A). After QC (quality control) filtering, we identified a total of 6,452 male cells and 7,231 female cells, which were classified into 23 and 19 distinct clusters, respectively. The cell clusters were visualized using t-distributed stochastic neighbor embedding (t-SNE) projection (Figure 1B). On average, we identified median 622 and 656 expressed genes per cell in each female and male gonad cell, respectively (Table S1).
Figure 1.
Global patterns of single-cell expression profiles and identification of cell types
(A) Workflow for single-nucleus RNA-seq of the oyster gonad.
(B) Two-dimensional t-SNE representation of female (left) and male (right) gonad cells analyzed in this study.
(C and D) Expression patterns of germline cell and niche cell marker genes exhibited on t-SNE plots; a gradient of gray and purple indicates low to high expression levels.
We performed functional enrichment analysis on differentially expressed genes (DEGs) to gain deeper insights into oyster germline cell development and the regulatory interactions between germ cells and their surrounding niche cells. Our findings revealed that significant enrichment of signaling pathways, including ribosome, base excision repair, and RNA transport in female clusters 3, 8, and 16, as well as male clusters 0, 1, 2, 4, and 15. Additionally, we observed the expression of well-established germline cell markers of C. gigas, including Ddx4 (Vasa) and Piwil1, as well as vertebrate markers such as TDRD1 and Boll (Dazl), within female clusters 3, 8, 16, and male clusters 0, 1, 2, 4, and 15 (Figures 1C and 1D, and Table S2).10,11,12 The presence of TDRD1 and Boll (Dazl) in germline cells was confirmed by in situ hybridization (ISH) (Figures S1B and S1C). High expression levels of TDRD1 and Boll (Dazl) in female and male gonads were further substantiated by bulk RNA-seq data (Figures S1D and S1E). Notably, Boll (Dazl) showed the highest expression in the adductor muscle (Figure S1E). We assessed and presented the correlation between the two cell clusters, and observed that female clusters 3, 8, and 16 exhibited similar gene expression patterns, suggesting they may represent the same cell type. Similarly, male clusters 0, 1, 2, 4, and 15 might also belong to the same cell type (Figures S2A and S2B). Therefore, female clusters 3, 8, and 16, as well as male clusters 0, 1, 2, 4, and 15, were categorized as germline cells.
Furthermore, we analyzed the gonadal somatic cells of female and male. Clusters 1, 4, 13, 20, and 21 in female were determined as follicular cells based on the expression pattern of female estrogenic yolk syntheses related and somatic cell-specific markers, such as Foxl2, Vg, TGF-β-like, and esr1 (Figure 1C).13,14,15,16 Clusters 3 and 10 in male were determined as Sertoli cells, exhibiting clear expression of TGF-β-like, esr1, CPT1A, Cpt1a, Arhgap18, CORO2A, Ggt1, lmbr1l, and Vwa5a (Figures 1D and S2C).5,13,17 We assigned cluster 12 in male as Leydig cells, characterized by the expression of testosterone production-related gene lhx9 and secretin receptors PTH1R and VIPR1 (Figures 1D and S2C).18,19 Male clusters 5, 6 and female clusters 5, 15 were identified as vesicular connective tissue cells (VCT-cells) as these cluster-specific genes, such as apolpp, LRP1, Soat1, and PCK1, are associated with lipid metabolism and glycogen synthesis (Figures S2D and S2E). The ISH of apolpp further supported our classification (Figure S2F). The expression of the gonadotropin-releasing hormone II receptor GNRHR was primarily observed in male cluster 13 and female cluster 12 (Figures S2D and S2E), indicating their characterization as putative nerve-like cells,20 but further experimental validation is needed to confirm this classification definitively. Additionally, we observed that a small subset of gonad cell types showed high expression of several genes, such as mlc-3, Mp20, MYL9, and Act42A (Figures S2D and S2E). These cells (cluster 2 and cluster 14 of female, cluster 9 of male) were labeled as myoepithelial cells, which may play a crucial role in facilitating germ cell migration and ovulation in oyster gonads. Boll (Dazl) was upregulated in myoepithelial cells (cluster 2 of female and cluster 9 of male), suggesting its potential utility as a marker gene for both germline and myoepithelial cells (Figure 1E; Table S2).
Subcluster-specific expression profiles of germline cells
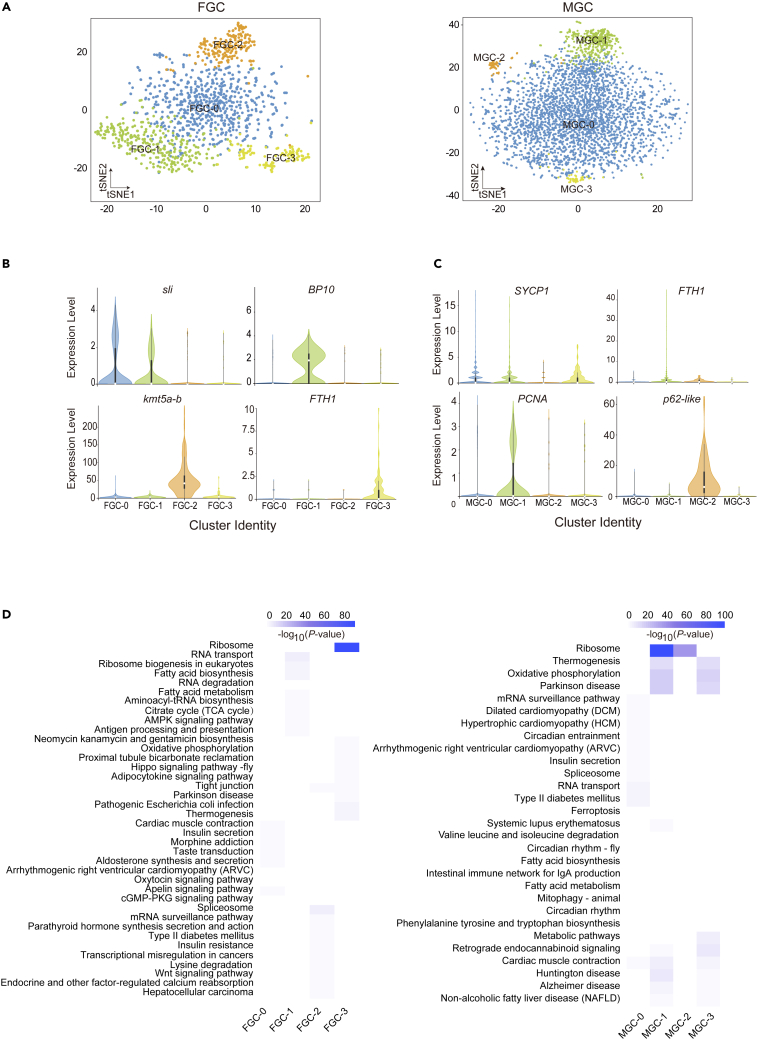
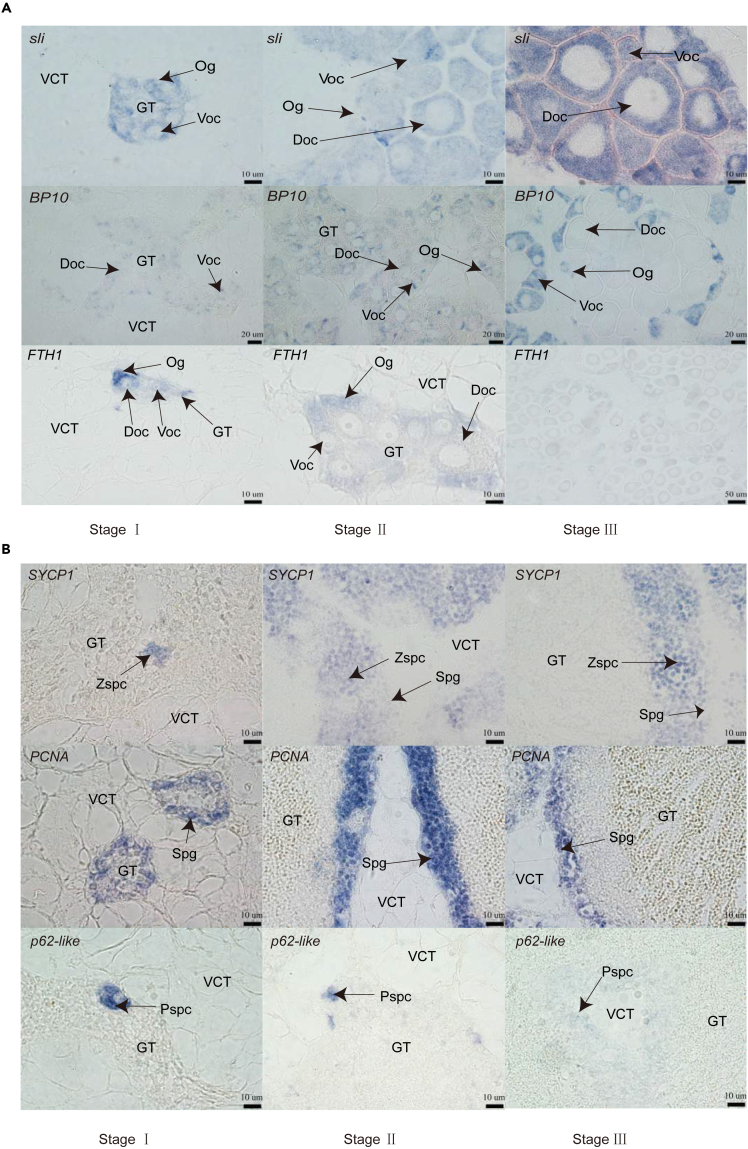
To characterize the heterogeneity within germline cells of oysters, we conducted further investigation into their sub-clustering. The clustering analysis revealed four distinct subclusters for both female and male germline cells, denoted as FGC-0 to FGC-3 and MGC-0 to MGC-3, respectively (Figure 2A). The identification of germline subgroups was based on the enrichment of recognized markers and DEGs, with experimental validation of germline-specific genes. The FTH1 showed widespread expression in FGC-3 and MGC-1 subtypes (Figures 2B and 2C).21 Moreover, the cell proliferation marker gene PCNA exhibited higher abundance in FGC-3 and MGC-1 compared to other germline subclusters, leading us to classify them as oogonia and spermatogonia (Figures 2C and S3A).22 ISH for PCNA and FTH1 confirmed its expression in oogonia and spermatogonia, located in the inner layer of the gonadal ducts (Figures 3A, 3B, and S3B). Subsequently, we determined that FGC-1 represented vitellogenic oocytes, characterized by the specific expression of genes related to lipid uptakes and lipogeneses, such as Lrp2, FASN-like, FASN, and Pnliprp2 (Figure S3A).23,24,25 Notably, ISH for BP-10, the marker gene of FGC-1, confirmed the expression in vitellogenic oocytes (Figures 2B and 3A). Based on the expression patterns of ZAN, ZAN-like1, ZAN-like2, JAG2-like, and sli, FGC-0 and FGC-2 were identified as developing oocytes (Figures 2B, 3A, and S3A).26,27,28 The cells within MGC-0 were classified as meiotic zygotene spermatocytes, exhibiting high expression levels of synaptonemal complex protein SYCP1 and meiotic recombination protein Prdm9 (Figures 2C, S3C, and S3D).29 Notably, MGC-2 was identified as meiotic pachytene spermatocytes, as evidenced by the clear expression of p62-like (Figures 2C and 3B).30 MGC-3 comprised 57 cells, identified as migration phase spermatocytes, characterized by the expression of genes related to aerobic respiration energy, such as ND1, ND2, and others (Figure S3C).
Figure 2.
Germline cell subclusters and their expression profiles
(A) Two-dimensional t-SNE plot displaying female and male germline cells.
(B and C) Violin plots showing the relative expression levels (TPM) of marker genes in each germline subclusters of female and male.
(D) Results of the KEGG enrichment test (p value < 0.05) demonstrating the enriched terms linked to upregulated genes in each of the four germline subclusters of female and male.
Figure 3.
Marker gene expression pattern in germline subcluster
(A) Localization of sli, BP10, and FTH1 with antisense probe in female gonads.
(B) Localization of SYCP1, PCNA, and p62-like with antisense probe in male gonads.
Accession numbers: sli, GeneBank: XM_011435634.3, BP10, GeneBank: XM_034446675.1, FTH1, GeneBank: NM_001305338.1, SYCP1, GeneBank: XM_034478449.1, PCNA, GeneBank: XM_011447436.3, p62-like, GeneBank: XM_034472977.1. Annotations: Og, oogonia; Oc, oocyte; Voc, vitellogenic oocyte; Doc, developing oocyte; Spg, spermatogonia; Zspc, zygotene spermatocyte; Pspc, pachytene spermatocyte. Scale bars for (A, top): 10 μm, (A, center): 20 μm, (A, bottom): 10 μm and 50 μm, (B): 10 μm.
A total of 759 and 601 differentially expressed unigenes were obtained in the female and male germline subclusters, respectively (Table S3). The marker genes associated with oogonia, spermatogonia, and pachytene spermatocytes were significantly enriched in Kyoto Encyclopedia of Genes and Genomes (KEGG) terms, such as ribosome pathway and tight junction (Figure 2D). Additionally, genes related to vitellogenic oocytes showed enrichment in the fatty acid metabolism pathway, while genes expressed in migrating spermatocytes were enriched in the oxidative phosphorylation and thermogenesis categories (Figure 2D).
To further investigate the genes involved in gametogenesis, we utilized bulk RNA-seq data from gonad development. When comparing stage 0 and stage I with differently expressed genes in germline cells (Table S3), we identified eight genes, including SYCP2L, that were uniquely expressed in developing oocytes (FGC-0). Notably, the upregulated genes in stage 0 were not entirely identical to those in germline cells. In stage I compared to stage III, three oogonia-specific genes, namely mrsA, SOD3-like, and GLIPR2, showed increased expression, while stage III exhibited higher expression of vitellogenic oocyte-specific genes (Atf7ip, uncharacterized LOC105334018, Incenp, and H1-8-like) and a developing oocyte-specific gene (DCLRE1A) (Figure S3E; Table S3). In male, zygotene spermatocyte-specific genes (uncharacterized LOC105348138 and uncharacterized LOC117689247) and pachytene spermatocyte-specific genes (uncharacterized LOC105329656 and uncharacterized LOC117689247) were upregulated in stage I when comparing stage 0 and stage I (Table S3). Surprisingly, we observed multiple overlapping marker genes between male stage I and stage III (Table S3). Of note, the meiotic spermatocyte-specific gene SYCP1 was included in the DEGs of male stage III, whereas the spermatogonia-specific maker gene FTH1 was listed in differently expressed genes of male stage I (Figure S3F; Table S3).
Cell lineage reconstruction reveals the fate of germline cells
To gain insights into the fate of germline cells, we performed cell lineage reconstruction by positioning 1,273 female germline cells and 4,328 male germline cells along a pseudotemporal axis. In the female germ cell lineage, we observed two distinct waves of oogenesis at branch points 1 and 2. Notably, the first wave of oogenesis (branch 1) occurred prior to the vitellogenesis stage, whereas the second wave of oogenesis (branch 2) occurred during the development of developing oocytes (Figure 4A). In the male germ cell lineage, pachytene and zygotene spermatocytes were situated in the middle of the pseudotemporal axis, while migrating spermatocytes were located toward the end (Figure 4B).
Figure 4.
Dynamic gene expression patterns of oyster germline cells
(A and B) Developmental pseudotime trajectory of germ cells, plotted in two dimensions using monocle. Different colors represent different cell types. The top of the figure shows the degree of differentiation of cell types in the pseudotime trajectory, with darker colors indicating a lower degree of differentiation.
(C and D) Heatmap of gene expression ordered by pseudotime, showing differential expression of genes across the germline cell types. Enriched GO terms (p value < 0.05) associated with biological processes are shown on the right.
Additionally, we selected 521 male and 384 female ordering genes that exhibited dynamic expression along pseudotime to investigate the characteristics of various stages of germline cell development. Hierarchical clustering analysis of the ordered genes identified five distinct groups with different expression patterns in female and male, respectively. Furthermore, we conducted Gene Ontology (GO) analyses on the DEGs within each cluster. In female, the first and second groups, consisting of 155 and 21 genes, respectively, were enriched for various metabolic processes, including sugar and energy metabolism, as well as gene expression. The fourth and fifth groups in female, comprising 128 and 71 genes, respectively, exhibited significant enrichment in planar cell polarity and microtubule-based processes (Figure 4C). In the male germline trajectory, the first and second clusters consisted of 239 and 23 ordering genes, respectively, which were mainly enriched in GO terms such as gene expression, mRNA catabolism, and meiosis Ⅰ (Figure 4D). Additionally, the genes in the fifth cluster of the male germline trajectory showed significant enrichment in ATP generation, which corresponded to the identification of the migrating spermatocytes (Figure 4D).
Transcription profiles of niche cells in the gonads
To validate the specific expression of TGF-β-like in niche cells, we performed ISH in the gonads. Interestingly, we observed that TGF-β-like positive niche cells were initially dispersed within the stage I gonadal ducts and subsequently migrated to the inner wall of the gonadal ducts during stage Ⅱ and stage III (Figures S4A and S4B). Additionally, unsupervised hierarchical clustering analysis revealed that female clusters 1 and 13 were closely related, suggesting potential functional similarities, while female clusters 4, 20, and 21 exhibited greater heterogeneity (Figures 1C and S4C). Likewise, male clusters 3 and 10 displayed much more similarity (Figure S4D).
We then conducted differential gene expression and GO analysis for these cells. By analyzing the profiles of DEGs, we identified 387, 318, 311, 250, and 345 genes that were explicitly upregulated in male clusters 1, 4, 13, 20, and 21, respectively (Figure 5A). GO analysis revealed that genes specific to female cluster 1 were enriched in terms such as mRNA-containing ribonucleoprotein complex export from the nucleus, dopamine secretion, and regulation of hormone secretion, suggesting that female cluster 1 may play a role in dopamine and hormones secretion (Figure 5B). Moreover, we observed elevated expression of genes evolved in spindle location, meiotic cell cycle, cell cycle process, and M phase in female clusters 4 and 13, indicating active cytokinesis activity or readiness for oocyte division in certain follicular cells (Figure 5B). On the other hand, female clusters 20 and 21 were enriched in ion transport and catabolic process (Figure 5B).
Figure 5.
Dynamic gene expression patterns of oyster gonadal niche cells
(A and C) Heatmap showing differentially expressed genes in female and male gonadal niche cells. The heatmap color key indicated the relative expression levels, ranging from low (blue) to high (yellow).
(B and D) Enriched GO terms (biological processes) for the differentially expressed genes in female and male gonadal niche cells. The top significantly enriched GO terms (p value < 0.05) are shown on the right.
Next, we investigated the characteristics of Sertoli cells and Leydig cells in males (Figures 1C and S4D). Noticeably, 383 genes specific to male cluster 3 were enriched in processes such as aromatic amino acid family metabolic process and regulation of hormone secretion (Figures 5C and 5D). In contrast, cluster 10 exhibited enrichment in terms such as morphogenesis of a branching epithelium, epithelial tube morphogenesis, and establishment of endothelial barrier, suggesting that Sertoli cells form tubular structures that enclose germ cells (Figure 5D). Additionally, the GO term, cellular lipid metabolic process, was highly enriched in the male Leydig cell cluster, suggesting that cells in cluster 12 may undergo lipid metabolism (Figure 5D).
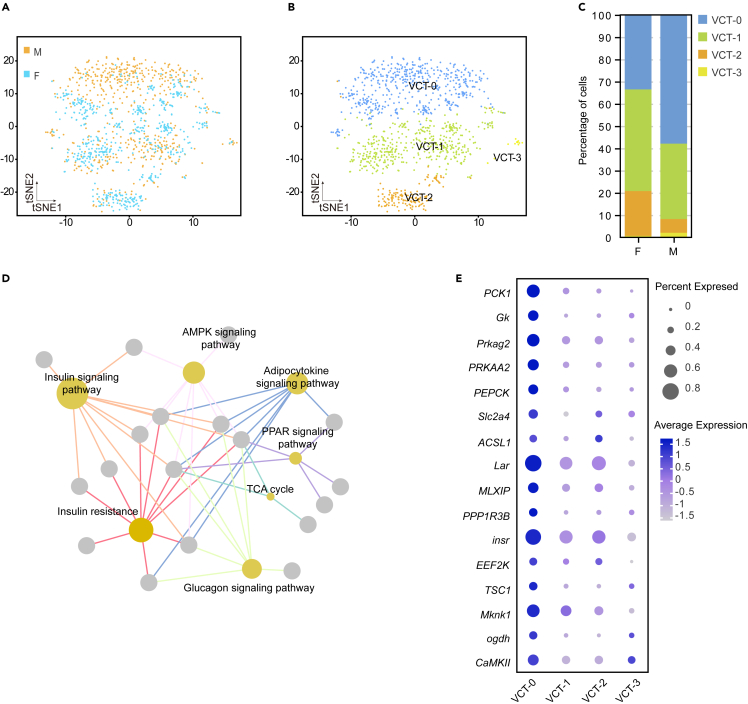
Oyster VCT-cells are involved in both lipid production and glycogen storage processes
The previous histological study has demonstrated that oyster VCT-cells possess the ability to store lipid droplets and glycogen, unlike scallops.31 To further investigate this finding at the molecular functional level, we performed an unsupervised hierarchical clustering analysis of female clusters 5 and 15, as well as male clusters 5 and 6, which correspond to VCT-cells (Figure 1C). We identified four distinct types of VCT-cells in both female and male cells, denoted as VCT-0 to VCT-3 (Figures 6A and 6B). The VCT-0 subpopulation was found to consist of 195 and 309 cells in female and male, respectively, accounting for 33.51% and 57.87% of the total VCT cells (Figure 6C). We conducted a differential gene expression analysis and KEGG analysis for these VCT-cells. The results showed that 305 genes exhibited high expression in VCT-0 and were significantly enriched in the glucagon signaling pathway, adipocytokine signaling pathway, PPAR signaling pathway, AMPK signaling pathway, and insulin-related pathways (Figure 6D). To further dissect the role of VCT-0 in glucose and lipid metabolism, we focused on essential genes involved in energy-related pathways. Several genes, such as PCK1, PEPCK, Slc2a4, PRKAA2, Prkag2, Lar, and insr were simultaneously active in the aforementioned pathways and displayed high expressions in VCT-0 (Figure 6E). Moreover, functional enrichment analysis of upregulated genes in VCT-1 to 3 subgroups revealed that VCT-1 and VCT-2 were enriched in KEGG terms related to regulating the actin cytoskeleton and adherence genes, whereas VCT-3 was enriched in the ribosome pathway (Table S4). Further, we identified 168 upregulated genes in female VCT-cells and 290 upregulated genes in male VCT-cells (Figure S5A). KEGG analysis indicated that the upregulated genes in females were significantly associated with focal adhesion and ECM-receptor interaction, while those in males were significantly enriched in terms related to lipid and glycogen metabolic processes (Figure S5B). DEGs related to cell adhesion and interaction-related in female VCT-cells included MATN1, ARHGAP5, act-2, ITGB1, COL6A3, act-2b, IGTA4, Vinc, and BIRC2 (Figure S5A). Male VCT-cells exhibited upregulation of lipid and glycogen-related genes, including SCD-1, SCD-2, JAK2, Lar, Gk, Prkag2, PCK1, PRKAA2, and PPP1R3B (Figure S5A).
Figure 6.
Oyster VCT cells performed both the lipid production and glycogen storage processes
(A and B) Two-dimensional t-SNE plots of the VCT cells.
(C) The taxonomic stacking chart illustrates the percentage of VCT cell subpopulations in female and male.
(D) The KEGG enrichment analysis shows the significantly enriched pathways of the metabolism process (p value < 0.05) in VCT-0.
(E) Dot plot analysis of the expression levels and the percentage of cells expressing metabolism-related genes in VCT-0.
Signaling pathway features in oyster germline cells and gonadal niche cells
To investigate cell-cell communication in oyster germline cells and gonadal niche cells, we used the CellChat software to analyze ligand-receptor interactions. This analysis was performed separately for male and female samples based on the results of germline re-clustering. Figure 7A illustrates the cell type present in the oyster gonad interaction network, along with the number of potential ligand-receptor pairs identified. In females, we identified 26 pairs of ligand-receptor relationships associated with 14 pathways, while in males, we found 37 ligand-receptor relationships connected to 19 pathways.
Figure 7.
Signaling pathway features in oyster germline cells and gonadal niche cell development
(A) Number of ligand-receptor pairs between any pair of two cell populations. The edge width is proportional to the indicated number of ligand-receptor pairs.
(B) Cell-cell communication patterns in female and male. The networks show the CellChat inferred latent patterns connecting cell groups sharing similar signaling pathways. The thickness of the water flow represents the relative contribution of the cell group or signaling pathway to a latent pattern, outgoing patterns for secreting cells.
(C) The inferred NRG, VISFATIN, and SEMA6 signaling networks. Edge width represents the communication probability.
(D) The dot plot of ligand-receptors in the NRG, VISFATIN, and SEMA6 signaling pathways of female.
(E) The dot plot of ligand-receptors in the NRG, VISFATIN, and SEMA6 signaling pathways of male.
We further examined the cell types involved in sending or receiving signals and identified three distinct patterns for both male and female germline niches (Figure 7B). Female germline cells were implicated in all three patterns of receiving signaling, while male germline cells were only involved in pattern one (Figure 7B). The niche cell, a necessary component of the germline cell microenvironment, primarily participated in patterns one and three of female outgoing signaling. The main pathways associated with this pattern included NCAM, LAMININ, NRG, SEMA5, SEMA6, VISFATIN, BMP, and EPHB. On the male outgoing end, the niche cell was connected to patterns two and three, with the main pathways being NRG, HSPG, VISFATIN, SEMA6, SELE, and NGL.
We identified NRG, VISFATIN, and SEMA6 as vital niche signaling pathways in both female and male samples (Figure 7C). We further investigated the ligand-receptor interactions within these pathways. In our results, the receptors in NRG, erbb4a, and erbb4b, were expressed in oogonia (FGC-3), vitellogenic oocyte (FGC-1), and developing oocyte (FGC-0) in female, as well as in four germline subclusters in male. Follicular cells (female clusters 1 and 13) and Sertoli cells (male cluster 3) expressed ligand nrg2b to transmit the signal to germline cells (Figures 7D and 7E). The VISFATIN signaling pathway included the ligand nampta, which interacted with the receptors insra and insrb. Developing oocytes (FGC-0, FGC-2) and male germline cells expressed receptors insra and insrb to receive signals from follicular cells (female cluster 1) and Sertoli cells (male cluster 10), which expressed the ligand nampta (Figures 7D and 7E). The SEMA6 signaling pathway consisted of the ligand sema6dL and its receptor plxna1a. Our study showed that plxna1a was expressed in female germline cells, zygotene spermatocytes (MGC-0), and pachytene spermatocytes (MGC-2), while sema6dl was expressed in follicular cells (female clusters 1, 13, and 20), Sertoli cells (male clusters 3 and 10), and Leydig cells (male cluster 12) (Figures 7D and 7E).
Additionally, we found that some signaling pathways, such as NOTCH and BMP, played important roles in the communication between male gonadal niche cells and germ cells, consistent with previous studies using single-cell transcriptomes of gonadal cells (Figure S6A).5 In our results, male Sertoli cells (male cluster 3) and germline cells expressed the ligands for the NOTCH signaling pathway (jagged1b, dll4, and dld), whereas Sertoli cells (male cluster 10) expressed its receptor, notch1b, to receive the signal (Figure S6B). Moreover, Sertoli cells (male clusters 3 and 10) specifically expressed bmp6, the ligand for the BMP signaling pathway, while male germline cells expressed bmpr1aa_acvr2aa to respond to the signal (Figure S6C). Interestingly, male Sertoli cells (male clusters 3 and 10) expressed receptors bmpr1aa_bmpr2b to receive ligand bmp6 from germline cells (Figure S6C). However, in females, the predicted communication pathways (BMP, BMP10) did not apply to the signal exchange between germ cells and follicle cells (Figure S6D).
Discussion
Altogether, our snRNA-seq profiling of oyster gonad tissues exposes the intricate transcriptional dynamics of germline cell development and the coordinated and reciprocal expression of several signaling pathways between germ cells and their gonadal niche cells. Our study provides a comprehensive characterization of the oyster gonadal cell profile and lays the foundation for a molecular atlas of invertebrate gonad development.
The sn-RNA transcriptome atlas: A platform for profiling gene expression dynamics during molluscan gametogenesis
The gametogenesis process in the Pacific oyster is divided into four stages. In stage 0, the gonads are in a resting phase and the sex of the individual cannot be distinguished. Stage I is characterized by gonial mitosis, which involves the division of germ cells within the gonadal tubules. During stage II, the gonads undergo further development, and in females, this is accompanied by vitellogenesis, the process of yolk accumulation in oocytes. In males, stage II is marked by the presence of the entire germinal lineage in males. In stage III, the gonads are fully developed and primarily contain either mature oocytes or spermatozoa. By analyzing the expression profiles of marker genes and integrating information from the gonadal bulk transcriptome, we precisely identified the various germ cells. In females, we identified three subpopulations of germline cells: oogonia, vitellogenic oocytes, and developing oocytes. In males, we identified four subpopulations of germline cells: spermatogonia, meiotic zygotene spermatocytes, meiotic pachytene spermatocytes, and migration phase spermatocytes.
Significantly, the study identified and experimentally verified FTH1, a gene involved in iron storage, was one of the marker genes for oogonia and spermatogonia, indicating that iron storage starts early in gametogenesis.32,33 Moreover, the gene sli, known to be essential in cell migration and cell communication, was primarily expressed in vitellogenic oocytes and developing oocytes.28 Therefore, it is plausible that the oocyte relies on niche cells to complete the assembly of yolk material before migrating inward to complete maturation. Interestingly, in the bulk transcriptome of stage Ⅰ and stage 0 gonads, we observed some marker genes of developing oocytes, zygotene spermatocytes, and pachytene spermatocytes were upregulated in stage Ⅰ, suggesting that stage I gonads contain a diverse range of germ cell subpopulations.
Notably, our study revealed that upregulated genes of oogonia and spermatogonia subclusters, including RPL37A, RPS26, and RPS5, were significantly enriched in the ribosome-related KEGG pathway. The presence of germplasm-related structures and the “germplasm granule formation complex” in spermatogonia and oogonia subclusters was consistent with previous single cell RNA-seq findings in vertebrates such as human and buffalo.5,34,35,36 Furthermore, we observed that the oogonia cluster displayed enrichment of genes related to the tight junction pathway, suggesting that these cells adhere to the niche cells, ensuring their differentiation. In contrast, the vitellogenic oocyte and developing oocyte clusters exhibited the expression of genes involved in fatty acid metabolic processes and aldosterone synthesis, which are known to be crucial for oocyte development.37 The genes specifically expressed in meiotic zygotene spermatocytes showed an enrichment associated with RNA transport and the mRNA surveillance pathway, suggesting active gene expression during meiosis. In single cell RNA-seq studies characterizing human and Drosophila spermatogenesis, it was discovered that male germ cell migration and spermatid elongation heavily rely on oxidative phosphorylation and mitochondrial ATP generation.5,38 We also discovered that migration phase spermatocytes possessed upregulated ATP synthesis-related genes. This may be connected to the fact that in early gametogenic gonads, a small number of spermatocytes migrating toward the gonadal tubules are accompanied by the process of spermatocyte formation.
The pseudotemporal analysis revealed distinct positioning of oogonia, spermatogonia, developing oocytes, and vitellogenic oocytes along the development axis. Oogonia and spermatogonia were located at the beginning of the axis, indicating their early development stages. The vitellogenic oocyte branch differentiated prior to the developing oocyte branch, suggesting a sequential progression in oogenesis. The initial wave of oogenesis likely involves the accumulation of vitellin, which is transported by follicular cells to the oocyte for processing into vitellogenin. The second wave of oogenesis contributes to oocyte maturation, as evidenced by an increase in oocyte size.39 The GO enrichment analysis highlighted key processes involved in oogenesis, including germplasm accumulation and energy storage, as well as the trafficking of mRNA-protein complexes within the oocyte, consistent with previous studies.40 In male germline cells, similar to the early stages of oogenesis, spermatogenesis also undergoes an initial stage of germplasm construction followed by the meiosis process.
Taken together, these findings not only confirm the presence of distinct subclusters of germline cells in our dataset but also provide valuable insights into the diversity and plasticity of molluscan germ cells, even within the same gonad.
The sn-RNA transcriptome atlas: A platform for excavating of gonadal somatic cells functions in Molluscs
In this study, five clusters (female clusters 1, 4, 13, 20, and 21) were classified as female follicular cells. Two clusters (male clusters 3 and 10) were identified as male Sertoli cells and male cluster 12 was proposed as male Leydig cells. By performing ISH of TGF-β-like, we observed that some niche cells migrate to the interior edge of the gonadal tubule as the gonads developed. The migration path of niche cells resembled that of germ cells within the gonadal tubule. The adhesion between niche cells and early germ cells suggested potential signal communication between niche cells and oogonia and spermatogonia. According to the results of GO analysis, we found that the follicular cells in female oysters have functions related to hormone synthesis and secretion, as well as the preparation of substances required for the division of female germ cells. This is similar to the role of follicular cells in humans.5 Moreover, the follicular cells exhibited ion transport functions, which we speculate are involved in maintaining the osmotic pressure of the gonadal niches. Our results revealed that Sertoli cells in oysters not only secrete hormones but also play a role in maintaining the morphology of gonadal tubules, which is inconsistent with that observed in mammal.5
Additionally, clusters 5 and 15 in female, and clusters 5 and 6 in male were determined as VCT-cells. Furthermore, we analyzed the expression patterns of VCT-cells. We observed that VCT-0 cells are predominantly associated with major glycogen metabolism processes in eukaryotes, serving as the primary sources of energy.41 Interestingly, we identified specific expressions of Slc2a4, PCK1, PEPCK, Gk, and ACSL1 in VCT-0 cells. Slc2a4 plays a pivotal role in mediating bidirectional glucose transport, while PCK1 and PEPCK are crucial flux-generating enzymes catalyzing the rate-limiting step of gluconeogenesis.42,43 Furthermore, studies on human brown fat biopsies have shown that PPAR and adipocytokine-related genes, such as Gk and ACSL1, play a role in selecting adipocyte proteins that enhance cellular fatty acid import and contribute to brown fat thermogenic capability.44,45 We found distinct expression peaks of Lar and insr specifically in VCT-0 cells. Previous research has demonstrated that Lar co-immunoprecipitates with insr and modulates insulin receptor signaling by dephosphorylating the regulatory phosphoryl-tyrosine residues.46 Interestingly, we observed different enrichment results in VCT-1 to VCT-3, implying that oyster VCT-cells may possess heterogeneity in maintaining gonadal morphology and gonadal development. As a class of cell cluster with both lipid and glycogen metabolic functions, the proportion of VCT-0 cells was shown to be greater in males than in females. More interestingly, genes concerning lipid and glycogen metabolic processes were upregulated in male VCT cells while genes concerning cell adhesion were up-regulated in female VCT cells. This implied that the meiotic process of germ cells in males might require more energy from VCT cells.
The sn-RNA transcriptome atlas: A platform for profiling signal pathways between germline cells and their somatic niche
In this study, we successfully identified and characterized the specific cell types involved in gametogenesis, namely the follicular cells in females, and Sertoli cells and Leydig cells in males. These somatic niche cells play crucial roles in regulating cell differentiation, nutrients supply, and physical adherence of the extracellular matrix during gametogenesis. Notably, we identified three information flows from niche cells to germ cells, namely NRG, SEMA6, and VISFATIN, shared between both sexes. Neuregulins have been shown to be essential for initiating spermatogonia meiosis.47 It is of particular interest that Sertoli cells express the ligand (nrg2b), while male germline cells express the receptor (erbb4a and erbb4b). This further supports the crucial role of Sertoli cells as positive regulators in initiating spermatogonia meiosis and maintaining spermatocyte meiosis. Additionally, semaphorin protein ligand sema6dl has been proven to facilitate the gonadotropin hormone-releasing system.48 Our findings highlighted that female germline cells, zygotene spermatocytes, and pachytene spermatocytes express the receptor plxna1a, underscoring the significant influence of gonadal hormones in the oyster’s gonadal microenvironment. Previous research has demonstrated that the ligand nampta of VISFATIN is an adipokine that inhibits granulosa cells from producing progesterone.49 Therefore, we speculate that follicular cells and Sertoli cells play a balancing role in ensuring the successful development of germ cells. Of particular interest, our finding indicated a similar expression pattern of NOTCH and BMP signaling pathways components in male oysters compared to humans.5 Sertoli cells (male cluster 10) expressed receptors in the NOTCH pathway to receive signals from male germline cells, while germ cells in male received signals from Sertoli cells (male clusters 3 and 10) through the BMP pathway. These findings suggest that the microenvironmental characteristics of molluscan germ cells share certain similarities with mammals while also exhibiting distinct features.
Our study offers crucial perspectives on the essential characteristics of invertebrate germ cells during their highly orchestrated mitotic, meiotic, and gametogenesis processes in vivo. Furthermore, we have established a robust platform for evaluating the identity and characteristics of germ cells that have undergone in vitro differentiation from pluripotent stem cells. The reciprocal interaction between the niche and germ cell signaling pathways uncovered in this study can provide vital information for enhancing the efficiency of germ cell differentiation in vitro. Our comprehensive roadmap of germ cell growth in vivo under physiological conditions can contribute to the analysis of issues pertaining to germ cells, including triploid infertility.
Limitations of the study
While our snRNA-seq analysis has provided insights into cell heterogeneity and elucidated the interactions between germline cells and gonadal niche in the Pacific oyster at the cellular level, it is subject to several limitations. Firstly, a significant portion of the marker genes employed for defining cell types, particularly non-germline cells, were extrapolated from vertebrate studies. Although we conducted a thorough examination of their homology, further experimental validation is warranted. Secondly, our study did not specifically identify undifferentiated germ cells, hence limiting our ability to provide a comprehensive elucidation of sex determination and differentiation. Future investigations could delve into the potential transcriptional regulation within the gonadal microenvironment to further elucidate the processes of sex determination and differentiation in oysters.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Digoxigenin-AP, Fab fragment | Roche | Cat# 11093274910; RRID: AB_2734716 |
| Biological samples | ||
| Diploid male and female Pacific oyster | Tianheng Island, Jimo City, Shandong Province, China | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Tween-20 | Sangon Biotech | Cat# 9005-64-5 |
| Deionized Formamide | LookChem | Cat# 75-12-7 |
| Triethanolamine | Sangon Biotech | Cat# 102-71-6 |
| 20×SSC Buffer, pH 7.0 | Phygene | PH1859 |
| Maleic Acid | Solarbio | Cat# M8050 |
| Proteinase K | Merck KGaA | Cat# 1245680100 |
| RNase Inhibitor | Beyotime | Cat# R0102-10KU |
| Dextran Sulphate | BBI | Cat# A600160-0050 |
| 50X Denhardt's Solution | Sangon Biotech | B548209-0050 |
| Yeast tRNA | Solarbio | Cat# 9014-25-9 |
| TRIzol reagent | Invitrogen | N/A |
| Critical commercial assays | ||
| Chromium Next GEM Single Cell 3’ Reagent Kits v3.1 | 10X Genomics | N/A |
| NBT/BCIP Stock Solution | Roche | Cat# 11681451001 |
| Gel Extraction Kit | Omega | D2500-02 |
| Deposited data | ||
| Oyster gonad transcriptome | Yue et al.50 | SRA: SRP112367 |
| Oyster tissue transcriptome | Zhang et al.51 | GEO: GSE31012 |
| snRNA-seq | This paper | SRA: SRR24954538 SRA: SRR24954539 |
| Oligonucleotides | ||
| See Table S5 for ISH | N/A | N/A |
| Software and algorithms | ||
| Cell Ranger v3.1.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| Seurat v3.1.1 | Butler et al., 201852 | https://satijalab.org/seurat/ |
| Monocle2 v2.28.0 | Qiu et al., 201753 | https://cole-trapnell-lab.github.io/monocle-release/docs/#introduction |
| CellChat v1.6.1 | Jin et al., 202154 | http://www.cellchat.org/ |
| Fastp v0.22.0 | Chen et al., 201855 | https://github.com/OpenGene/fastp/blob/master/README.md |
| Hisat2 v2.2.1 | Kim et al., 201556 | https://daehwankimlab.github.io/hisat2/ |
| Samtools v1.6 | Li et al., 200957 | http://samtools.sourceforge.net/ |
| clusterProfile | Yu et al., 201258 | https://guangchuangyu.github.io/software/clusterProfiler/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hong Yu (hongyu@ouc.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
The snRNA-seq data have been deposited in the GenBank SRA database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The bulk RNA-seq data for different gonadal development stages and different tissues of oysters were downloaded from the GenBank SRA database and GEO database (listed in the key resources table).
-
•
This article contains no original code.
-
•
Any additional information required to reanalyze the data reported in this study will be provided upon request to the lead contact.
Experimental model and study participant details
Animals
Two-year-old adult Pacific oysters were obtained monthly from Tianheng Island, Shandong, China. Male and female diploid oysters at stage Ⅰ were used in snRNA-seq and stage Ⅰ-Ⅲ were used in in suit hybridization.
Method details
Tissue processing and histological analysis
Two-year-old adult diploid Pacific oysters were collected monthly from March 2021 and July 2021 from Tianheng Island, Shandong, China. For each oyster, the gonad was sampled and stored at -80°C for RNA extraction. Simultaneously, the gonad was subjected to fixation in Bouin’s fluid and 4% paraformaldehyde overnight for histological analysis. The dehydrated gonad was embedded in paraffin and sectioned serially at thickness of five micrometers. Histologically staining was performed on the sections, and digital images were captured using an Olympus BX53 microscope (Olympus, Japan). The gonad development stage and sex of the oysters were identified by histological methods.
Isolation of nuclei from the oyster gonad
Oyster gonad tissues were minced to 1 mm3 and stored at -80°C prior to nuclei extraction. Following histological confirmation, the nuclei from male and female gonadal cells at stage Ⅰ were isolated and resuspended in 2 mL of phosphate-buffered saline containing 0.01% bovine serum albumin. The concentration of nuclei was determined using Thermofisher Countess II FL Automatic Cell Counter and was adjusted as needed to achieve the ideal range for loading on the 10X Chromium chip.
Construction of libraries and generation of cDNA on the 10X genomics platform
10× genomics-based libraries were generated following the manufacturer recommended protocol. Briefly, the re-suspended nuclei were loaded into the 10X Chromium system using the Single Cell 3′ Reagent Kit v3 to obtain nanoliter-scale Gel Bead-In-Emulsions (GEMs). Primers containing an Illumina® R1 sequence (read one sequencing primer), a 16 nt 10x Barcode, a ten nt Unique Molecular Identifier (UMI), and a poly-dT primer sequence were mixed with GEMs during incubation, which was performed on PCR instrument (Bio-Rad C1000 Touch). Silane magnetic beads were used to remove the biochemical reagents and primers. Then, the cDNA was amplified with 11 PCR cycles and purified with SPRlselect (Beckman Coulter B23318). Illumina bridge amplification was carried out using the final libraries that contained the P5 and P7 primers after primers had been added. The libraries were pooled and sequenced on the illumine 10 × Genomics Chromium platform. The sequencing output was subjected to quality testing with DNA 1000 assay Kit (Agilent Technologies).
Total RNA extraction
Gonadal total RNA was extracted using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. RNA concentration was measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific, USA). The integrity and purity of RNA were assessed by running the samples on 1% agarose gels. RNA samples that met the purity criteria (A260/A230 and A260/ A280 > 1.8) were selected for cDNA library preparation.
In Suit Hybridization (ISH)
The gonad tissue was embedded in paraffin and sectioned at a thickness of 5 μm. The RNA in situ hybridization (ISH) protocol from a prior study were followed with slight modifications.14 The key resources table and Table S5 contains information about the probes used.
snRNA-seq analysis pipeline
Cell Ranger (v.3.1.0) was used to convert the raw BCL files into FASTQ files, followed by quality control of the sequences and alignment to the C. gigas reference genome (GCF_902806645.1) with default parameters (10x Genomes). Downstream analysis was primarily performed using Seurat (v.3.1.1).52 In brief, gene expression matrices were trimmed based on quality metrics (> 500 and < 4000 genes, < 20 % mitochondrial reads, < 8000 UMI, and genes expressed in at least three cells). Potential batch effects were assessed using PCA visualization of the first three dimensions, and outliers were disregarded. Following QC filtering, expression data for 6,502 male gonad cells and 7,273 female gonad cells with median gene counts of 622 and 656, respectively, were obtained. Clusters were visualized using t-distributed Stochastic Neighbor Embedding (t-SNE).
Oyster tissue transcriptome data processing
Transcriptomic data of different stages of gonadal development and different tissues in the Pacific oyster were downloaded (accession number: SRA: SRP112367, GEO: GSE31012).50,51 The raw sequence reads were subjected to quality control using Fastp (v.0.22.0) (https://github.com/OpenGene/fastp), and low-quality reading were removed.50 The reads were then mapped to the reference genome using HISAT2 (v.2.2.1),59 and StringTie was used to calculate the read count and transcripts per million (TPM) values. Differential expression analysis was carried out using DESeq2, with a significance threshold of | log2 (fold change) | ≥1 and a false discovery rate (FDR) < 0.05 to identify significantly differentially expressed genes.
Quantification and statistical analysis
Wilcoxon rank-sum test was used to compare each gene's expression level to that of the other cells in a given cluster.60 Genes with at least 1.28-fold or 2-fold higher expression levels in the target cluster, expressed in at least 25% of the target cluster's cells, and had a P-value of less than 0.01 or 0.05 were considered significantly upregulated. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were subsequently conducted.61,62 The trajectory reconstruction program Monocle2 was used to analyze the kinetics of gene expression during the differentiation of germline cells on a set of highly variable genes.53
Acknowledgments
This study was supported by grants from National Natural Science Foundation of China (42276111) and Science Foundation of Shandong Province (ZR2022MC171).
Author contributions
The study was designed by H.Y and H.W. H.W helped in the sample collection. H.Y and H.W conducted the majority of the experimental work and data analysis. H.W drafted the original draft preparation, while H.Y and Q.L assisted with the review and editing. H.Y. and Q.L helped support the study. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 14, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109499.
Contributor Information
Hong Yu, Email: hongyu@ouc.edu.cn.
Qi Li, Email: qili66@ouc.edu.cn.
Supplemental information
References
- 1.Saffman E.E., Lasko P. Germline development in vertebrates and invertebrates. Cell. Mol. Life Sci. 1999;55:1141–1163. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ewen-Campen B., Schwager E.E., Extavour C.G.M. The molecular machinery of germ line specification. Mol. Reprod. Dev. 2010;77:3–18. doi: 10.1002/mrd.21091. [DOI] [PubMed] [Google Scholar]
- 3.Yang L., Ng H.-H. The making of an ovarian niche. Science. 2021;373:282–283. doi: 10.1126/science.abj8347. [DOI] [PubMed] [Google Scholar]
- 4.Wen Q., Cheng C.Y., Liu Y.-X. Development, function and fate of fetal Leydig cells. Semin. Cell Dev. Biol. 2016;59:89–98. doi: 10.1016/j.semcdb.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., Fan X., Wu X., Guo H., Wang X., et al. Single-Cell RNA-Seq Analysis Maps Development of Human Germline Cells and Gonadal Niche Interactions. Cell Stem Cell. 2017;20:858–873.e4. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Wang C., Lin H. Roles of piRNAs in transposon and pseudogene regulation of germline mRNAs and lncRNAs. Genome Biol. 2021;22:27. doi: 10.1186/s13059-020-02221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckelbarger K.J., Hodgson A.N. Invertebrate oogenesis - a review and synthesis: comparative ovarian morphology, accessory cell function and the origins of yolk precursors. Invertebr. Reprod. Dev. 2021;65:71–140. doi: 10.1080/07924259.2021.1927861. [DOI] [Google Scholar]
- 8.Fabioux C., Huvet A., Lelong C., Robert R., Pouvreau S., Daniel J.Y., Minguant C., Le Pennec M. Oyster vasa-like gene as a marker of the germline cell development in Crassostrea gigas. Biochem. Biophys. Res. Commun. 2004;320:592–598. doi: 10.1016/j.bbrc.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 9.La H., Yoo H., Lee E.J., Thang N.X., Choi H.J., Oh J., Park J.H., Hong K. Insights from the Applications of Single-Cell Transcriptomic Analysis in Germ Cell Development and Reproductive Medicine. Int. J. Mol. Sci. 2021;22:823. doi: 10.3390/ijms22020823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabioux C., Pouvreau S., Le Roux F., Huvet A. The oyster vasa-like gene: a specific marker of the germline in Crassostrea gigas. Biochem. Biophys. Res. Commun. 2004;315:897–904. doi: 10.1016/j.bbrc.2004.01.145. [DOI] [PubMed] [Google Scholar]
- 11.Geijsen N., Horoschak M., Kim K., Gribnau J., Eggan K., Daley G.Q. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature. 2004;427:148–154. doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- 12.Rui X., Qi L., Hong Y. Expression pattern of Piwi-like gene implies the potential role in germline development in the Pacific oyster Crossosrea gigas. Aquac. Rep. 2020;18 doi: 10.1016/j.aqrep.2020.100486. [DOI] [Google Scholar]
- 13.Matsumoto T., Nakamura A.M., Mori K., Akiyama I., Hirose H., Takahashi Y. Oyster estrogen receptor: cDNA cloning and immunolocalization. Gen. Comp. Endocrinol. 2007;151:195–201. doi: 10.1016/j.ygcen.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Yue C., Li Q., Yu H. Variance in expression and localization of sex-related genes CgDsx, CgBHMG1 and CgFoxl2 during diploid and triploid Pacific oyster Crassostrea gigas gonad differentiation. Gene. 2021;790 doi: 10.1016/j.gene.2021.145692. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto T., Nakamura A.M., Mori K., Kayano T. Molecular Characterization of a cDNA Encoding Putative Vitellogenin from the Pacific Oyster Crassostrea gigas. Zool. Sci. 2003;20:37–42. doi: 10.2108/zsj.20.37. [DOI] [PubMed] [Google Scholar]
- 16.Fleury E., Fabioux C., Lelong C., Favrel P., Huvet A. Characterization of a gonad-specific transforming growth factor-β superfamily member differentially expressed during the reproductive cycle of the oyster Crassostrea gigas. Gene. 2008;410:187–196. doi: 10.1016/j.gene.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 17.Huvet A., Fleury E., Corporeau C., Quillien V., Daniel J.Y., Riviere G., Boudry P., Fabioux C. In Vivo RNA Interference of a Gonad-Specific Transforming Growth Factor-β in the Pacific Oyster Crassostrea gigas. Mar. Biotechnol. 2012;14:402–410. doi: 10.1007/s10126-011-9421-4. [DOI] [PubMed] [Google Scholar]
- 18.Chu J.Y.S., Yung W.H., Chow B.K.C. Secretin: A Pleiotrophic Hormone. Ann. N. Y. Acad. Sci. 2006;1070:27–50. doi: 10.1196/annals.1317.013. [DOI] [PubMed] [Google Scholar]
- 19.Hu F., Zhu Q., Sun B., Cui C., Li C., Zhang L. Smad ubiquitylation regulatory factor 1 promotes LIM-homeobox gene 9 degradation and represses testosterone production in Leydig cells. Faseb. J. 2018;32:4627–4640. doi: 10.1096/fj.201701480R. [DOI] [PubMed] [Google Scholar]
- 20.Alavi S., Nagasawa K., Takahashi K., Osada M. 2017. Structure-Function of Serotonin in Bivalve Molluscs. [DOI] [Google Scholar]
- 21.Huan P., Liu G., Wang H., Liu B. Multiple ferritin subunit genes of the Pacific oyster Crassostrea gigas and their distinct expression patterns during early development. Gene. 2014;546:80–88. doi: 10.1016/j.gene.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 22.Franco A., Jouaux A., Mathieu M., Sourdaine P., Lelong C., Kellner K., Heude Berthelin C. Proliferating cell nuclear antigen in gonad and associated storage tissue of the Pacific oyster Crassostrea gigas: seasonal immunodetection and expression in laser microdissected tissues. Cell Tissue Res. 2010;340:201–210. doi: 10.1007/s00441-009-0923-6. [DOI] [PubMed] [Google Scholar]
- 23.Endo T., Todo T., Lokman P.M., Kudo H., Ijiri S., Adachi S., Yamauchi K. Androgens and Very Low Density Lipoprotein Are Essential for the Growth of Previtellogenic Oocytes from Japanese Eel, Anguilla japonica. Biol. Reprod. 2011;84:816–825. doi: 10.1095/biolreprod.110.087163. [DOI] [PubMed] [Google Scholar]
- 24.Tang H., Han M. Fatty Acids Regulate Germline Sex Determination through ACS-4-Dependent Myristoylation. Cell. 2017;169:457–469.e13. doi: 10.1016/j.cell.2017.03.049. [DOI] [PubMed] [Google Scholar]
- 25.Kim K.S., Kim B.K., Kim H.J., Yoo M.S., Mykles D.L., Kim H.-W. Pancreatic lipase-related protein (PY-PLRP) highly expressed in the vitellogenic ovary of the scallop, Patinopecten yessoensis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008;151:52–58. doi: 10.1016/j.cbpb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Tardif S., Wilson M.D., Wagner R., Hunt P., Gertsenstein M., Nagy A., Lobe C., Koop B.F., Hardy D.M. Zonadhesin Is Essential for Species Specificity of Sperm Adhesion to the Egg Zona Pellucida. J. Biol. Chem. 2010;285:24863–24870. doi: 10.1074/jbc.M110.123125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGinnis L.K., Limback S.D., Albertini D.F. Signaling Modalities During Oogenesis in Mammals. Curr. Top. Dev. Biol. 2013;102:227–242. doi: 10.1016/B978-0-12-416024-8.00008-8. [DOI] [PubMed] [Google Scholar]
- 28.Dickinson R.E., Duncan W.C. The SLIT/ROBO pathway: a regulator of cell function with implications for the reproductive system. Reprod. Camb. Engl. 2010;139:697–704. doi: 10.1530/REP-10-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soh Y.Q.S., Junker J.P., Gill M.E., Mueller J.L., van Oudenaarden A., Page D.C. A Gene Regulatory Program for Meiotic Prophase in the Fetal Ovary. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chien M.-L., Lai J.-H., Lin T.-F., Yang W.-S., Juang Y.-L. NUP62 is required for the maintenance of the spindle assembly checkpoint and chromosomal stability. Int. J. Biochem. Cell Biol. 2020;128 doi: 10.1016/j.biocel.2020.105843. [DOI] [PubMed] [Google Scholar]
- 31.Kalachev A.V., Yurchenko O.V. Autophagy in nutrient storage cells of the Pacific oyster, Crassostrea gigas. Tissue Cell. 2019;61:30–34. doi: 10.1016/j.tice.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Jin S., Fu H., Jiang S., Xiong Y., Qiao H., Zhang W., Gong Y., Wu Y. RNA Interference Analysis Reveals the Positive Regulatory Role of Ferritin in Testis Development in the Oriental River Prawn, Macrobrachium nipponense. Front. Physiol. 2022;13 doi: 10.3389/fphys.2022.805861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Missirlis F. Regulation and biological function of metal ions in Drosophila. Curr. Opin. Insect Sci. 2021;47:18–24. doi: 10.1016/j.cois.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Bontems F., Stein A., Marlow F., Lyautey J., Gupta T., Mullins M.C., Dosch R. Bucky Ball Organizes Germ Plasm Assembly in Zebrafish. Curr. Biol. 2009;19:414–422. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 35.Reunov A., Alexandrova Y., Reunova Y., Komkova A., Milani L. Germ plasm provides clues on meiosis: the concerted action of germ plasm granules and mitochondria in gametogenesis of the clam Ruditapes philippinarum. Zygote. 2019;27:25–35. doi: 10.1017/S0967199418000588. [DOI] [PubMed] [Google Scholar]
- 36.Huang L., Zhang J., Zhang P., Huang X., Yang W., Liu R., Sun Q., Lu Y., Zhang M., Fu Q. Single-cell RNA sequencing uncovers dynamic roadmap and cell-cell communication during buffalo spermatogenesis. iScience. 2023;26 doi: 10.1016/j.isci.2022.105733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seydoux G., Braun R.E. Pathway to Totipotency: Lessons from Germ Cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 38.Yu J., Li Z., Fu Y., Sun F., Chen X., Huang Q., He L., Yu H., Ji L., Cheng X., et al. Single-cell RNA-sequencing reveals the transcriptional landscape of ND-42 mediated spermatid elongation via mitochondrial derivative maintenance in Drosophila testes. Redox Biol. 2023;62 doi: 10.1016/j.redox.2023.102671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung E.-Y. Ultrastructural Studies Of Oogenesis and Sexual Maturation In Female Chlamys (azumapecten) farreri farreri (Jones & Preston, 1904) (Pteriomorphia: Pectinidae) On The Western Coast Of KoreA. Malacologia. 2008;50:279–292. doi: 10.4002/0076-2997-50.1.279. [DOI] [Google Scholar]
- 40.Becalska A.N., Gavis E.R. Lighting up mRNA localization in Drosophila oogenesis. Development. 2009;136:2493–2503. doi: 10.1242/dev.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Qin C., Yang L., Lu R., Zhao X., Nie G. A comparative genomics study of carbohydrate/glucose metabolic genes: from fish to mammals. BMC Genom. 2018;19:246–314. doi: 10.1186/s12864-018-4647-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunten P., Belunis C., Crowther R., Hollfelder K., Kammlott U., Levin W., Michel H., Ramsey G.B., Swain A., Weber D., Wertheimer S.J. Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site11Edited by I. J. Mol. Biol. 2002;316:257–264. doi: 10.1006/jmbi.2001.5364. [DOI] [PubMed] [Google Scholar]
- 43.Zhao F.-Q., Keating A.F. Functional Properties and Genomics of Glucose Transporters. Curr. Genom. 2007;8:113–128. doi: 10.2174/138920207780368187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lasar D., Rosenwald M., Kiehlmann E., Balaz M., Tall B., Opitz L., Lidell M.E., Zamboni N., Krznar P., Sun W., et al. Peroxisome Proliferator Activated Receptor Gamma Controls Mature Brown Adipocyte Inducibility through Glycerol Kinase. Cell Rep. 2018;22:760–773. doi: 10.1016/j.celrep.2017.12.067. [DOI] [PubMed] [Google Scholar]
- 45.Richards M.R., Harp J.D., Ory D.S., Schaffer J.E. Fatty acid transport protein 1 and long-chain acyl coenzyme A synthetase 1 interact in adipocytes. J. Lipid Res. 2006;47:665–672. doi: 10.1194/jlr.M500514-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Sevillano J., Sánchez-Alonso M.G., Pizarro-Delgado J., Ramos-Álvarez M.D.P. Role of Receptor Protein Tyrosine Phosphatases (RPTPs) in Insulin Signaling and Secretion. Int. J. Mol. Sci. 2021;22:5812. doi: 10.3390/ijms22115812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J., Eto K., Honmyou A., Nakao K., Kiyonari H., Abé S.i. Neuregulins are essential for spermatogonial proliferation and meiotic initiation in neonatal mouse testis. Development. 2011;138:3159–3168. doi: 10.1242/dev.062380. [DOI] [PubMed] [Google Scholar]
- 48.Messina A., Giacobini P. Semaphorin Signaling in the Development and Function of the Gonadotropin Hormone-Releasing Hormone System. Front. Endocrinol. 2013;4 doi: 10.3389/fendo.2013.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diot M., Reverchon M., Ramé C., Baumard Y., Dupont J. Expression and effect of NAMPT (visfatin) on progesterone secretion in hen granulosa cells. Reproduction. 2015;150:53–63. doi: 10.1530/REP-15-0021. [DOI] [PubMed] [Google Scholar]
- 50.Yue C., Li Q., Yu H. Gonad Transcriptome Analysis of the Pacific Oyster Crassostrea gigas Identifies Potential Genes Regulating the Sex Determination and Differentiation Process. Mar. Biotechnol. 2018;20:206–219. doi: 10.1007/s10126-018-9798-4. [DOI] [PubMed] [Google Scholar]
- 51.Zhang G., Fang X., Guo X., Li L., Luo R., Xu F., Yang P., Zhang L., Wang X., Qi H., et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490:49–54. doi: 10.1038/nature11413. [DOI] [PubMed] [Google Scholar]
- 52.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.-H., Myung P., Plikus M.V., Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Camp J.G., Sekine K., Gerber T., Loeffler-Wirth H., Binder H., Gac M., Kanton S., Kageyama J., Damm G., Seehofer D., et al. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–538. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- 61.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The snRNA-seq data have been deposited in the GenBank SRA database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The bulk RNA-seq data for different gonadal development stages and different tissues of oysters were downloaded from the GenBank SRA database and GEO database (listed in the key resources table).
-
•
This article contains no original code.
-
•
Any additional information required to reanalyze the data reported in this study will be provided upon request to the lead contact.