Abstract
The present work aims to synthesize four series of phenothiazine incorporation Mannich bases. Therefore, 10-methyl-10H-phenothiazine-3-sulfonamide (4) which was subjected to react with some secondary amines and formaldehyde to give the Mannich bases 5a-f, and 6–13. Compound 13 was then subjected to react with some secondary amines and formaldehyde to give the corresponding Mannich bases 14a-f. In total, twenty-two new compounds were synthesized and evaluated for in vitro growth inhibition activity against P. aeruginosa, E. coli, and S. aureus. Among the tested compounds, compounds 3, 5a, 5c, 6, 12, 13, 14d, and 14e exhibited good activity with a MIC value (12.5 μg/mL), compounds 5b, 10, 11, 14a, and 14c exhibited strong activity against the growth of S. aureus with a MIC value (6.25 μg/mL), and compound 14b superior against S. aureus with a MIC value (3.125 μg/mL) compared to drug reference ciprofloxacin with MIC value (2 μg/mL). The molecular docking investigation revealed the presence of many derivatives with high binding affinities and distinct interaction patterns with the target protein. Derivatives 14a-e emerged as the most promising possibilities, displaying the greatest binding energies and a varied variety of interaction types, including hydrogen bonding and pi interactions, over different distances, with derivative 14b exhibiting the highest binding energy at S = −8.3093 kcal/mol. These derivatives displayed superior binding affinities and various interaction mechanisms with the target protein, suggesting that they have great promise as lead compounds for future development into therapeutic medicines.
Keywords: Phenothiazine, N-Mannich bases, Molecular docking, Antimicrobial agent
Highlights
-
•
Synthesis of 3-phenothiazinesulfonamide (4).
-
•
Study the reaction of compound 4 with secondary amines and formaldehyde
-
•
Reaction of phenothiazine sulfonamide 5a with tetrahydrocarbazole and formaldehyde
-
•
Reaction of sulfamoyl phenyl phenothiazine sulfonamide 13 with secondary amines and formaldehyde.
-
•
Study the antimicrobial efficacy of the newly made compounds.
-
•
Some of newly compounds showed promising antimicrobial activity and their molecular docking was investigated.
1. Introduction
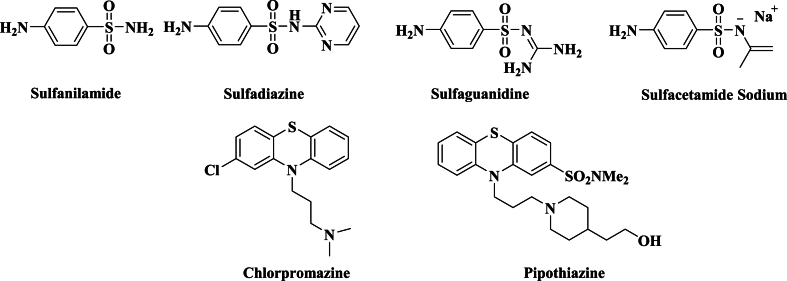
Formulating and synthesizing compounds with high medical properties is one of the main goals of organic and medicinal chemistry. The scientific community faces a significant dilemma because of the quick emergence of resistance to currently available antimicrobial medications. The synthesis of novel antimicrobial drugs with strong efficacy against drug-resistant microbes is therefore urgently needed [1]. Because the amino group can be readily converted into a variety of other functionalities, the chemistry of the amino alkylation of aromatic substrates by the Mannich reaction is of great interest for the synthesis and modification of biologically active compounds having physical [2], chemical importance [3], as well as physiological properties [4,5]. A sensible way to introduce a basic aminoalkyl chain into a variety of medications and substances is by the Mannich reaction [6]. Additionally, a significant degree of pharmacological activity of different Mannich bases for antibacterial, analgesic, anti-inflammatory, and anaesthetic properties as well as intermediates in drug production. Mannich bases have gained importance due to their application in antibacterial activity and other applications are in agrochemicals such as plant growth regulators. Moreover, N-bridged heterocyclic derivatives show important antibacterial activity. The amino alkylation of aromatic substrates by the Mannich reaction is of considerable importance for the synthesis and modification of biologically active compounds [2]. Mannich bases have several biological activities such as antimicrobial and anticancer. Morpholine derivatives were reported to possess antimicrobial, anti-inflammatory, and central nervous system activities. Therefore, bearing in mind the above observation, we were led to synthesize and test the antimicrobial activity of a new series of Mannich base derivatives [[7], [8], [9]]. A review of the literature in this area turned up several publications on the antibacterial action of N-Mannich bases. The combination of Pregnenolone and Carbamazepine exhibits reduced efficacy against S. aureus and E. coli [10]. Furthermore, sulphonamide is widely recognized for its antibacterial [[11], [12], [13]], anti-tubercular [14], anti-inflammatory [15], and carbonic inhibitory [16] properties. Strong antibacterial properties and lower toxicity than the parent sulphonamide are reported for the Mannich bases integrated with sulphonamides [17] (Fig. 1).
Fig. 1.
Some marketed sulfonamide and phenothiazine drugs.
However, phenothiazine derivatives have long been identified as biologically active therapeutic possibilities and a TB patient has been successfully treated with the phenothiazine drug chlorpromazine (CPZ) [18]. This biological effect is attributed to the presence of chlorine, the phenothiazine ring, and the alkylamine group (Fig. 1).
In addition, phenothiazines were the first drugs to be effectively utilized in the treatment of psychosis; nowadays, they are well-known for their therapeutic applications as antipsychotic, antiemetic, antihypertensive, and antihistaminic drugs [19]. Phenothiazines have demonstrated their ability to function as human cholinesterase inhibitors in recent years [20], and they have frequently been described as multidrug resistance (MDR) reversal medicines [21]. Since they have been utilized for more than 50 years, neuroleptic phenothiazines have been prepared in several ways that have been documented. These methods are primarily found in pharmaceutical company patents, such as Pipothiazine [22] which attributed its biological activity to the presence of phenothiazine, alkyl amine, and SO2 moieties (Fig. 1).
There are well-known techniques for synthesizing phenothiazines. [2-[(2-Chlorophenyl) thio]phenyl]amine derivatives can be used to synthesize phenothiazines with substituents in positions 2, 3, and 4 [21]. The reaction of 2-aminobenzenethiol with derivatives of 2-chloro-1-nitrobenzene, followed by acetylation and Smiles rearrangement, is a widely utilized technique for the synthesis of 2-substituted phenothiazines [23,24].
The Mannich reaction is a crucial stage in the synthesis of several bioactive compounds and is frequently employed in the production of secondary and tertiary amine derivatives [[25], [26], [27]]. N-Mannich bases that are generated from NH-heterocycles and related compounds exhibit a wide range of potential pharmacological effects, including neuroprotection, anticancer, antifungal, anti-HIV, antitubercular, and antibacterial properties [[28], [29], [30], [31], [32], [33], [34]].
2. Results and discussion
2.1. Characterization of new synthesizes
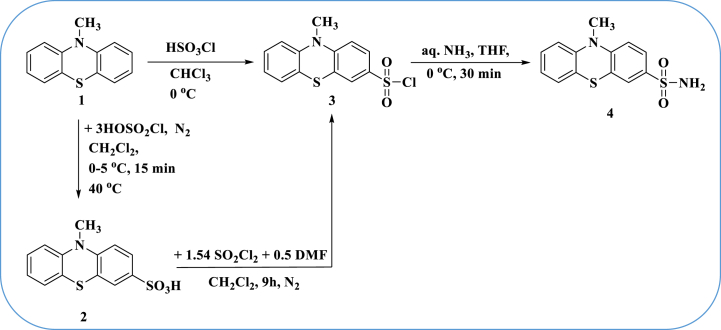
The intended 10H-phenothiazine-2-sulfonyl chloride was not produced when phenothiazine was sulfonated with chlorosulfonic acid. The literature claims make polysubstitution and easy sulfur atom oxidation make the electrophilic aromatic substitution of the phenothiazine ring challenging. This led us to develop a different strategy that starts with a suitably substituted phenothiazine.
To enhance the poor reactivity of phenothiazine toward chlorosulfonation, compound 3 was prepared to have N-methyl group. Finding this alkyl group in the aromatic ring parallel to the sulfur atom was the aim. Keeping this criterion in mind, our synthetic path toward structure 3 resembles that of Warburton et al. [35] in certain ways.
Compound 4 had two singlet signals in its 1H NMR spectrum, corresponding to NH2 and CH3 protons at δ 3.07 and 6.94 ppm, respectively. The NH2 function group was identified in the IR spectra as the source of an absorption band at 3450 cm−1 (Scheme 1).
Scheme 1.
Synthesis of 3-chlorosulfonyl and 3-sulfonamide phenothiazine derivatives 3 and 4.
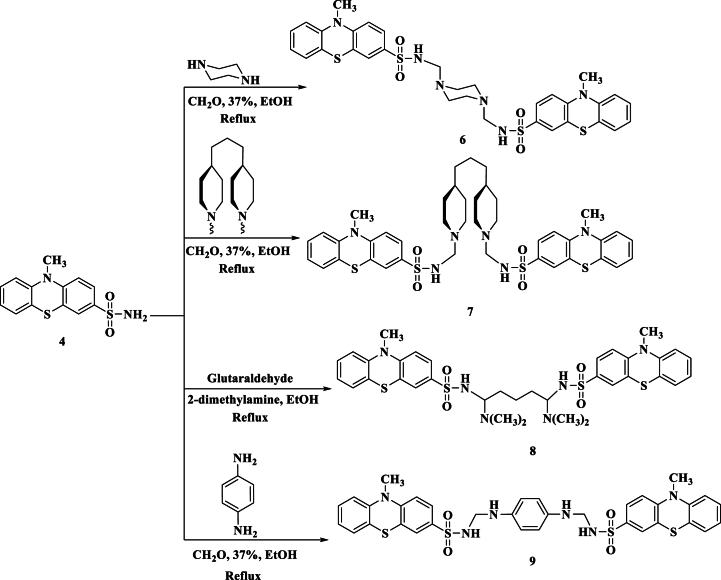
Following up on the Mannich-bases reported work [36], the current work focuses on attempting to broaden the scope of the Mannich reaction with phenothiazine derivatives. This includes synthesizing new N-Mannich bases and bis(Mannich bases), which may have uses in medicine. In the present study, phenothiazine 4 was treated with some secondary amines namely (piperidine, morpholine, diethanol amine, dimethyl amine, diethyl amine, and tetrahydrocarbazole) in the presence of formaldehyde to give Mannich bases 5a-f. In general, 1H NMR of compounds 5a-f showed a characteristic singlet signal of CH2 protons around δ 4.00–4.50 ppm. The IR spectra, in general, displayed an absorption band around 3150 cm−1 due to the NH function group (Scheme 2). (c.f. supplementary file).
Scheme 2.
Reaction of phenothiazine sulfonamide 4 with secondary amine and formaldehyde.
In the course of this study, the synthesis of bis(Mannich bases) 6–9 has been achieved by treating 2 mol compound 4 with piperazine/formaldehyde, 4,4′-trimethyelenedipiperidine (TMDP)/formaldehyde, glutaraldehyde/dimethylamine, and p-phenylenediamine/formaldehyde, respectively. The structure of compound 6 was confirmed based on its analytical and spectral data. The 1H NMR spectrum of compound 6 showed an interesting singlet signal at δ 2.27 ppm due to four CH2 protons of the piperazine ring, while its IR spectrum showed an absorption band at 3160 cm−1 for the NH group. On the other hand, compound 7 showed an absorption band of NH at 3155 cm−1, and 1H NMR showed a multiplet signal at δ 1.42–1.62 ppm corresponding to seven CH2 + 2CH protons. Additionally, compound 8 showed in its 1H NMR multiplet signal at δ 1.30 ppm due to CH–CH2–CH2–CH2–CH protons, quartet signal at δ 1.55 ppm due to CH–CH2–CH2–CH2–CH protons, and triplet signal at δ 3.80 ppm due to CH–CH2–CH2–CH2–CH protons. Finally, the bis base 9 showed in its IR spectrum absorption band at 3152 cm−1 due to the NH absorption band. The 1H NMR spectrum showed a characteristic singlet signal at δ 6.60 ppm corresponding to four aromatic protons of the AB system (Scheme 3).
Scheme 3.
Synthesis of bis-Mannich bases 6–9.
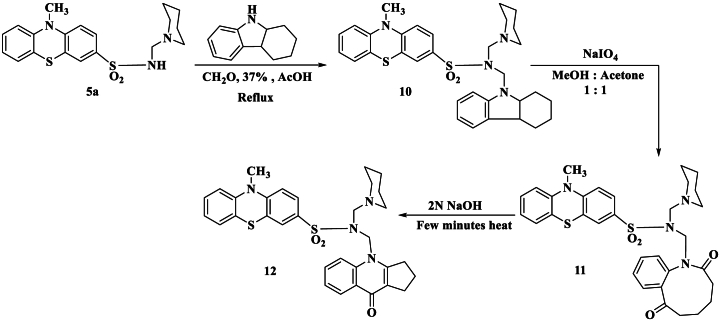
Considerable attention has been devoted to the tetrahydrocarbazole derivatives as anticonvulsant and antihybooxant activity [37,38]. In view of this and an extension of the presence study, we reported here the reaction of phenothiazine derivatives (5a) with tetrahydrocarbazole and formaldehyde to give N-((1,2,3,4,4a,9a-hexahydro-9H-carbazol-9-yl)methyl)-10-methyl-N-(piperidin-1-ylmethyl)-10H-phenothiazine-3-sulfonamide (10). Compound 10 showed a molecular ion peak at m/z = 574 (M+), corresponding to the molecular weight of the formula C32H38N4O2S2. The 1H NMR displayed new signals at δ 1.20–1.80 ppm as multiplet related to the tetrahydrocarbazole ring. The periodate oxidation of the tetrahydrocarbazole moiety in compound 10 is of particular interest because it provides convenient access to the formation of the functionalized hexahydrobenzo[b]azonine system N-((2,7-dioxo-2,3,4,5,6,7-hexahydro-1H-benzo[b]azonin-1-yl)methyl)-10-methyl-N-(piperidin-1-ylmethyl)-10H-phenothiazine-3-sulfonamide (11). Synthesis of compound 11 is of particular interest, because of azonine core is present in vinblastine and vincristine alkaloids [39,40] which possess significant antitumor activity and have been widely used clinically. In addition, some azonine and benzoazonine have been studied with interest centred on their potential pharmaceutical activity [[41], [42], [43], [44]]. In continuation compound 11 has been used as a precursor to 10-methyl-N-((9-oxo-1,2,3,9-tetrahydro-4H-cyclopenta[b]quinolin-4-yl)methyl)-N-(piperidin-1-ylmethyl)-10H-phenothiazine-3-sulfonamide (12), which was obtained on treating compound 11 with dil. sodium hydroxide solution. The tendency of 11 to cyclize readily under the mild basic condition to give compound 12 is in line with an earlier report by Witkop et al. [45] (Scheme 4).
Scheme 4.
Reaction of phenothiazine sulfonamide 5a with tetrahydrocarbazole and formaldehyde: synthesis of benzoazonine derivative 11 and related compound.
Moreover, compound 3 reacted with sulfanilamide to give the corresponding 10-methyl-N-(4-sulfamoylphenyl)-10H-phenothiazine-2-sulfonamide (13). The 1H NMR spectrum showed three singlet signals at δ 2.97, 6.84, and 10.10 ppm corresponding to CH3, NH2, and NH protons, while its mass spectrum displayed m/z = 447 as exact mass. Compound 13 was subjected to react with some secondary amines in the presence of formaldehyde to give N-Mannich bases (14a-f) (Scheme 5). The structure of compounds 14a-f was characterized by analytical and spectroscopical data. (c.f. supplementary data)
Scheme 5.
Reaction of sulfamoyl phenyl phenothiazine sulfonamide 13 with secondary amines and formaldehyde.
2.2. Antimicrobial evaluation
In vitro antibacterial activity and Structure-activity relationship (SAR):
Using the agar dilution method to determine the lowest inhibitory concentration, all the resulting compounds were evaluated for their in vitro antibacterial activity against strains of P. aeruginosa, E. coli, and S. aureus [46]. The commercial antibiotic ciprofloxacin was used as the reference drug for comparison. Compared to the other tested strains, most of the compounds showed superior inhibitory efficacy against the S. aureus strain (Table 1).
Table 1.
Antimicrobial activity of the newly synthesized compounds.
| MIC (μg/mL) | |||
|---|---|---|---|
| Gram Positive bacteria |
Gram Negative bacteria |
||
| Compound No. | S. aureus | E. coli | P. aeruginosa |
| 3 | 12.5 | 50 | 50 |
| 4 | 25 | 50 | 100 |
| 5a | 12.5 | 25 | 50 |
| 5b | 6.25 | 12.5 | 25 |
| 5c | 12.5 | 25 | 50 |
| 5d | 25 | 50 | 50 |
| 5e | 25 | 50 | 50 |
| 5f | 50 | 100 | 100 |
| 6 | 12.5 | 25 | 50 |
| 7 | 25 | 25 | 50 |
| 8 | 100 | 100 | 100 |
| 9 | 50 | 50 | 100 |
| 10 | 6.25 | 25 | 50 |
| 11 | 6.25 | 12.5 | 50 |
| 12 | 12.5 | 25 | 100 |
| 13 | 12.5 | 25 | 50 |
| 14a | 6.25 | 12.5 | 50 |
| 14b | 3.125 | 6.25 | 25 |
| 14c | 6.25 | 12.5 | 25 |
| 14d | 12.5 | 25 | 50 |
| 14e | 12.5 | 25 | 50 |
| 14f | 25 | 50 | 50 |
| Ciprofloxacin | 2 | 2 | 4 |
Interestingly, compounds with an electron-withdrawing group and a chlorine atom exhibited outstanding inhibitory activity, indicating that the chlorine substituent plays a substantial role in the compound's antibacterial action. Compound 3's MIC value, however, indicates that it has a better inhibitory effect against S. aureus (MIC 12.5 μg/mL) than it does against the other examined bacterial strains. When comparing the results of Mannich bases 14a-f with their counterparts Mannich bases 5a-f, it was found that the sulfamoyl phenyl group in Mannich bases 14a-f increased the effectiveness of the compounds as antibacterial agents, as sulfonamides are known to interfere with bacterial folic acid synthesis. However, the specific activity against Gram-positive or Gram-negative bacteria can depend on various factors, including the overall structure, charge, and lipophilicity of the compounds. The results in Table 1 showed antibacterial activity against S. aureus due to the presence of morpholinomethyl group (compounds 14b and/or 5b) (MIC 3.125, and/or 6.25 μg/mL) which enhance antibacterial activity compared to piperidinomethyl group in 14a and/or 5a (6.25, and/or 12.5 μg/mL).
Compounds 6–9 showed moderate to weak activity against the tested strains (MIC 12.5–100 μg/mL). The antibacterial activity of a compound can depend on various factors, including its size, structure, and ability to penetrate bacterial cells. Generally, larger compounds may face challenges in entering bacterial cells, but their antimicrobial activity can still be influenced by other factors. This result is based on the assumption that the more flexible and shorter linkers in compounds 6 and 7 may facilitate better interaction with bacterial cells compared to the longer and less flexible linkers in compounds 8 and 9.
When comparing the antibacterial activity of compounds 10 and 11 with the biological effect of compound 5a as an antibacterial agent against S. aureus strain, it was found that the process of replacing NH in compound 5a with methyl tetrahydrocarbazol and methyl benzoazonine increased the biological effect significantly. It is noteworthy that, all compounds carrying electron-withdrawing groups are active. Depending on this fact, compound 11 (MIC 6.25 μg/mL) which contains two carbonyl groups is more active than compounds 10 and 12. Compound 11 contains hexahydrobenzoazonine ring, which is more structurally complex and might have a different mode of action compared to the other compounds, potentially contributing to higher activity. Compound 10 has a hexahydrocarbazolylmethyl group, which is also complex but less so than the hexahydrobenzoazonin ring in compound 11.
2.3. Molecular docking
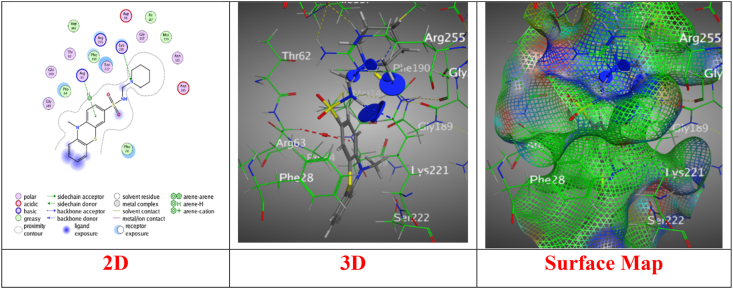
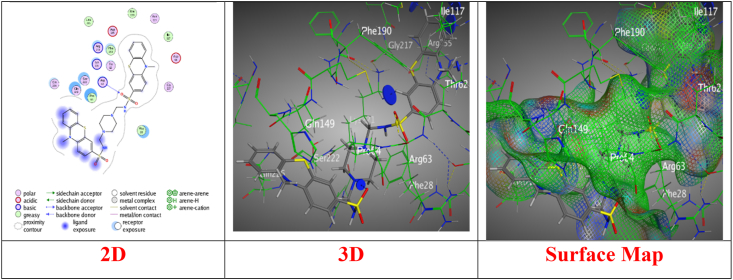
In sight the molecular docking stimulation for the synthesized derivatives with good antibacterial activities with the DHPS (dihydropteroate synthase) protein structure (PBD: 3TZF) to establish the most preferred mode of interaction (Table S1) [47]. Derivative 3 displayed binding energy score (S) = −6.0013 kcal/mol, indicating moderate affinity, RMSD = 1.0146, suggesting a reliable docking pose, and hydrogen acceptor bonds with Arg255 and Thr62, crucial for binding through intermolecular distance 3.49 Å and 2.89 Å, respectively, enhancing interaction stability (Fig. 2). Meanwhile, derivative 5a revealed a good energy score (S) = −7.6491 kcal/mol, showing strong affinity, RMSD = 1.6136, indicating a stable conformation upon binding, over N16, engages Lys221 via hydrogen bonding, while the benzene ring forms pi-H interactions with Arg63, suggesting a robust binding mechanism (Fig. 3). Though derivative 5b showed a binding Energy score (S) with −6.1538 kcal/mol, reflecting moderate to high affinity, showcases a mix of hydrogen and both of pi-H and H-pi interactions, indicating versatile binding capabilities (Fig. 4). Likewise, derivative 5c presented binding energy score (S) = −6.9959 kcal/mol, highlighting strong affinity, RMSD = 1.5995, suggesting a stable interaction, over hydrogen bonding and pi-H interactions, indicating a complex interaction pattern with the target (Fig. 5). However, derivative 6 recorded a binding energy score (S) = −7.1426 kcal/mol, indicative of strong affinity, through a singular hydrogen acceptor bond with Arg63, showing specific but potentially narrow interaction over an intermolecular distance of 3.26 Å, within the ideal range for hydrogen bonding, suggesting effective but focused interaction (Fig. 6). Nevertheless, derivative 11 displayed binding energy score (S) = −6.6470 kcal/mol, showing good affinity through RMSD = 1.9820, indicating a stable but slightly variable binding mode, and forms a hydrogen bond with Arg 63, similar to Derivative 6, suggesting a targeted but potentially limited interaction scope (Fig. 7). Derivative 12 exhibited a potent energy score (S) = −8.0267 kcal/mol, one of the highest affinities observed, indicating excellent potential as an inhibitor through RMSD = 1.7753, suggesting a reliable docking pose, and A strong hydrogen bond with Arg 63, emphasizing a potent and specific interaction over intermolecular distance 3.38 Å. Moreover, derivative 13 showed energy score (S) = −7.9849 kcal/mol, showcasing high affinity comes from RMSD = 1.2861, reflecting a precise and stable binding conformation. Features a combination of hydrogen donor/acceptor and pi interactions, suggesting multifaceted binding, through distances ranging from 2.83 Å to 4.46 Å, enabling a strong and diverse set of interactions (Fig. 7). Furthermore, Derivatives 14a-e demonstrated amazing binding energy score (S) ranging from −8.2036 to −7.7650 kcal/mol, all showing very high affinity with RMSD Values indicating stable and reliable docking poses over a rich diversity of interaction types, including hydrogen bonding (donor/acceptor) and pi interactions, indicative of strong, specific, and span intermolecular distances from 2.90 Å to 4.58 Å which are optimal for the interactions, suggesting these derivatives have potent inhibitory capabilities through varied binding modes (Figs. S1–S5; supplementary file).
Fig. 2.
Interaction images between derivative 3 with (PDB: 3tzf).
Fig. 3.
Interaction images between derivative 5a with (PDB: 3tzf).
Fig. 4.
Interaction images between derivative 5b with (PDB: 3tzf).
Fig. 5.
Interaction images between derivative 5c with (PDB: 3tzf).
Fig. 6.
Interaction images between derivative 6 with (PDB: 3tzf).
Fig. 7.
Interaction images between derivative 13 with (PDB: 3tzf).
3. Experiment
3.1. Synthesis of 10-methyl-10H-phenothiazine (1)
It was prepared in accordance with the reported work [48].
3.2. Synthesis of 10-methyl-10H-phenothiazine-3-sulfonic acid (2)
It was prepared in accordance with the reported method [49].
3.3. Synthesis of 10-methyl-10H-phenothiazine-3-sulfonyl chloride (3) and 10-methyl-10H-phenothiazine-3-sulfonamide (4)
The procedure of their preparation and analysis were displayed in the supplementary file.
3.4. Synthesis of mannich bases (5a-f)
3.4.1. General procedure
In absolute ethanol (30 mL) containing formaldehyde 37% (0.1 g, 3 mmol), a mixture of compound 4 (0.87 g, 3 mmol) and the appropriate secondary amines (3 mmol) namely ''piperidine, morpholine, diethanolamine, dimethyl amine, diethyl amine, and tetrahydrocarbazole'' was heated for 30 min. Each mixture was heated at 70–75 °C for 7–8 h, depending upon the type of amine. The reaction mixture was left to stand at room temperature overnight. The obtained solid material was filtered off and crystallized from ethanol to give compounds 5a-f.
3.5. 10-Methyl-N-(piperidin-1-ylmethyl)-10H-phenothiazine-3-sulfonamide (5a)
Yield, 74%; m.p. = 247–249 °C; IR (KBr): νmax, cm−1: 3150 (NH), 1610 (C N), 1342, 1171 (SO2); 1H NMR (DMSO‑d6) δ ppm: 1.60 (m, 6H, 3CH2), 2.25 (m, 4H, 2CH2), 3.28 (s, 3H, CH3), 4.25 (s, 2H, CH2), 6.99–7.16 (m, 4H, Ar–H), 7.45 (d, 1H, Ar–H), 7.55 (s, 1H, Ar–H), 7.69 (d, 1H, Ar–H), 7.81 (s, 1H, NH). MS (m/z, %): 389 (M+, 56). Anal. Calced for C19H23N3O2S2 (389.53): C, 58.59; H, 5.95; N, 10.79%. Found: C, 58.52; H, 5.87; N, 10.72%.
3.6. 10-Methyl-N-(morpholinomethyl)-10H-phenothiazine-3-sulfonamide (5b)
Yield, 68%; m.p. = 189–191 °C; IR (KBr): νmax, cm−1: 3155 (NH), 1615 (C N), 1339, 1169 (SO2); 13C NMR (DMSO‑d6) δ ppm: 36.5, 53.4 (2C), 64.9, 66.8 (2C), 115.0, 118.2, 120.0, 121.8, 123.7, 126.7, 128.2 (3C), 129.6, 144.4, 148.2. MS (m/z, %): 391 (M+, 52). Anal. Calced for C18H21N3O3S2 (391.50): C, 55.22; H, 5.41; N, 10.73%. Found: C, 55.15; H, 5.35; N, 10.66%.
3.7. N-((bis(2-hydroxyethyl)amino)methyl)-10-methyl-10H-phenothiazine-3-sulfonamide (5c)
Yield, 71%; m.p. = 177–179 °C; IR (KBr): νmax, cm−1: 3450 (OH), 3145 (NH), 1609 (C N), 1345, 1175 (SO2); 1H NMR (DMSO‑d6) δ ppm: 2.30 (t, 4H, 2CH2), 3.31 (s, 3H, CH3), 3.69 (t, 4H, 2CH2), 4.22 (s, 2H, 2OH), 4.40 (s, 2H, CH2), 7.09–7.25 (m, 4H, Ar–H), 7.55 (d, 1H, Ar–H), 7.65 (s, 1H, Ar–H), 7.79 (d, 1H, Ar–H), 7.92 (s, 1H, NH). MS (m/z, %): 409 (M+, 42). Anal. Calced for C18H23N3O4S2 (409.52): C, 52.79; H, 5.66; N, 10.26%. Found: C, 52.71; H, 5.55; N, 10.19%.
3.8. N-((dimethylamino)methyl)-10-methyl-10H-phenothiazine-3-sulfonamide (5d)
Yield, 68%; m.p. = 157–159 °C; IR (KBr): νmax, cm−1: 3144 (NH), 1600 (C N), 1342, 1171 (SO2); 1H NMR (DMSO‑d6) δ ppm: 2.20 (s, 6H, 2CH3), 3.19 (s, 3H, CH3), 4.47 (s, 2H, CH2), 6.95–7.12 (m, 4H, Ar–H), 7.41 (d, 1H, Ar–H), 7.52 (s, 1H, Ar–H), 7.64 (d, 1H, Ar–H), 7.80 (s, 1H, NH). MS (m/z, %): 349 (M+, 61). Anal. Calced for C16H19N3O2S2 (349.47): C, 54.99; H, 5.48; N, 12.02%. Found: C, 54.93; H, 5.40; N, 11.93%.
3.9. N-((diethylamino)methyl)-10-methyl-10H-phenothiazine-3-sulfonamide (5e)
Yield, 77%; m.p. = 171–173 °C; IR (KBr): νmax, cm−1: 3150 (NH), 1610 (C N), 1342, 1171 (SO2); 1H NMR (DMSO‑d6) δ ppm: 1.10 (t, 6H, 2CH3), 2.84 (q, 4H, 2CH2), 3.21 (s, 3H, CH3), 4.37 (s, 2H, CH2), 7.00–7.20 (m, 4H, Ar–H), 7.49 (d, 1H, Ar–H), 7.60 (s, 1H, Ar–H), 7.70 (d, 1H, Ar–H), 7.83 (s, 1H, NH). MS (m/z, %): 377 (M+, 66). Anal. Calced for C18H23N3O2S2 (377.52): C, 57.27; H, 6.14; N, 11.13%. Found: C, 57.19; H, 6.05; N, 11.08%.
3.10. N-((1,2,3,4,4a,9a-hexahydro-9H-carbazol-9-yl)methyl)-10-methyl-10H-phenothiazine-3-sulfonamide (5f)
Yield, 78%; m.p. = 215–217 °C; IR (KBr): νmax, cm−1: 3140 (NH), 1605 (C N), 1335, 1166 (SO2); 13C NMR (DMSO‑d6) δ ppm: 25.9 (2C), 29.0, 32.0, 36.0, 44.5, 60.1, 64.0, 109.9, 114.4, 117.0 (2C), 120.1, 121.9 (2C), 123.8, 125.7 (2C), 128.7 (3C), 130.5, 138.0, 144.0, 146.4, 150.2. MS (m/z, %): 477 (M+, 45). Anal. Calced for C26H27N3O2S2 (477.64): C, 65.38; H, 5.70; N, 8.80%. Found: C, 65.31; H, 5.61; N, 8.73%.
3.11. Synthesis of N,N'-(piperazine-1,4-diylbis(methylene))bis(10-methyl-10H-phenothiazine-3-sulfonamide) (6)
In 30 mL of ethanol containing 37% formalin (0.18 g, 6 mmol), compound 4 (1.74 g, 6 mmol) and piperazine (0.25 g, 3 mmol) were reacted and heated for 45 min. After 10 h of stirring at standard temperature (25 °C), such a mixture was allowed to stand overnight. Compound 6 was obtained by filtering the isolated product and purifying it chromatographically with an ethyl acetate-diethyl ether (2:1) eluent.
Yield, 81%; m.p. = 251–253 °C; IR (KBr): νmax, cm−1: 3160 (NH), 1611 (C N), 1342, 1171 (SO2); 1H NMR (DMSO‑d6) δ ppm: 2.27 (s, 8H, 4CH2), 3.10 (s, 6H, 2CH3), 4.05 (s, 4H, 2CH2), 7.17–7.40 (m, 8H, Ar–H), 7.58 (d, 2H, Ar–H), 7.70 (s, 2H, Ar–H), 7.82 (d, 2H, Ar–H), 7.95 (s, 2H, 2NH); 13C NMR (DMSO‑d6) δ ppm: 35.9 (2C), 52.4 (4C), 63.9 (2C), 116.1 (2C), 119.8 (2C), 121.5 (2C), 122.9 (2C), 123.9 (2C), 128.0 (2C), 129.9 (6C), 131.0 (2C), 145.9 (2C), 149.8 (2C). MS (m/z, %): 694 (M+, 40). Anal. Calced for C32H34N6O4S4 (694.90): C, 55.31; H, 4.93; N, 12.09%. Found: C, 55.25; H, 4.85; N, 12.00%.
3.12. Synthesis of N,N'-((propane-1,3-diylbis(piperidine-4,1-diyl))bis(methylene))bis(10-methyl-10H-phenothiazine-3-sulfonamide) (7)
In ethanol (30 mL) containing 37% formalin (0.18 g, 6 mmol), compound 4 (1.74 g, 6 mmol) and 4,4′-trimethylenedipiperidine (0.63 g, 3 mmol) were heated for 30 min. After 10 h of stirring at 25 °C, such a mixture was allowed to stand overnight. Compound 7 was obtained by filtration and the isolated solid was purified chromatographically with an eluent of ethyl acetate – diethyl ether (2:1).
Yield, 73%; m.p. = 266–268 °C; IR (KBr): νmax, cm−1: 3155 (NH), 1616 (C N), 1343, 1174 (SO2); 1H NMR (DMSO‑d6) δ ppm: 1.42–1.62 (m, 16H, 7CH2 + 2CH), 2.22–2.38 (m 8H, 4CH2), 3.25 (s, 6H, 2CH3), 4.11 (s, 4H, 2CH2), 7.23–7.49 (m, 8H, Ar–H), 7.66 (d, 2H, Ar–H), 7.78 (s, 2H, Ar–H), 7.89 (d, 2H, Ar–H), 8.00 (s, 2H, 2NH). MS (m/z, %): 819 (M+, 38). Anal. Calced for C41H50N6O4S4 (819.13): C, 60.12; H, 6.15; N, 10.26%. Found: C, 60.02; H, 6.09; N, 10.17%.
3.13. Synthesis of N,N'-(1,5-bis(dimethylamino)pentane-1,5-diyl)bis(10-methyl-10H-phenothiazine-3-sulfonamide) (8)
Compound 4 (1.74 g, 6 mmol), glutaraldehyde (0.27 g, 3 mmol), and dimethyl amine (0.27 g, 6 mmol) were dissolved in 30 mL of ethanol and heated in a water bath for 2 h. After 10 h of stirring at 25 °C, such a mixture was allowed to stand overnight. Compound 8 was obtained by filtration and the isolated product was purified chromatographically with an n-hexane-ethyl acetate (6:1) eluent.
Yield, 71%; m.p. = 218–220 °C; IR (KBr): νmax, cm−1: 3120 (NH), 1600 (C N), 1340, 1174 (SO2); 1H NMR (DMSO‑d6) δ ppm: 1.30 (m, 2H, CH–CH2–CH2–CH2–CH), 1.55 (q, 4H, CH–CH2–CH2–CH2–CH), 2.15 (s, 12H, 4CH3), 3.15 (s, 6H, 2CH3), 3.80 (t, 2H, CH–CH2–CH2–CH2–CH), 6.78–7.00 (m, 8H, Ar–H), 7.19 (d, 2H, Ar–H), 7.30 (s, 2H, Ar–H), 7.40 (d, 2H, Ar–H), 7.51 (s, 2H, 2NH); 13C NMR (DMSO‑d6) δ ppm: 14.5, 31.0 (2C), 35.5 (2C), 42.0 (4C), 82.5 (2C), 115.6 (2C), 118.8 (2C), 120.1 (2C), 122.0 (2C), 124.0 (2C), 127.0 (2C), 128.7 (6C), 129.9 (2C), 144.9 (2C), 148.6 (2C). MS (m/z, %): 739 (M+, 46). Anal. Calced for C35H42N6O4S4 (739.00): C, 56.89; H, 5.73; N, 11.37%. Found: C, 56.82; H, 5.66; N, 11.31%.
3.14. Synthesis of N,N'-((1,4-phenylenebis(azanediyl))bis(methylene))bis(10-methyl-10H-phenothiazine-3-sulfonamide) (9)
After being refluxed for 1 h in 30 mL of ethanol, compound 4 (1.74 g, 6 mmol), formaldehyde 37% (0.2 g, 6 mmol), and p-phenylenediamine (0.35 g, 3 mmol) were combined. After 20 h of stirring at 25 °C, such a mixture was allowed to stand overnight. Compound 9 was obtained by filtration and the isolated product was purified chromatographically with an eluent of n-hexane - diethyl ether (6:1).
Yield, 76%; m.p. = 273–275 °C; IR (KBr): νmax, cm−1: 3152 (NH), 1612 (C N), 1342, 1171 (SO2); 1H NMR (DMSO‑d6) δ ppm: 3.20 (s, 6H, 2CH3), 5.05 (s, 4H, 2CH2), 5.91 (s, 2H, 2NH), 6.60 (s, 4H, A-H), 7.18–7.41 (m, 8H, Ar–H), 7.58 (d, 2H, Ar–H), 7.70 (s, 2H, Ar–H), 7.81 (d, 2H, Ar–H), 7.93 (s, 2H, 2NH). MS (m/z, %): 716 (M+, 50). Anal. Calced for C34H32N6O4S4 (716.91): C, 56.96; H, 4.50; N, 11.72%. Found: C, 56.90; H, 4.42; N, 11.66%.
3.15. Synthesis of N-((1,2,3,4,4a,9a-hexahydro-9H-carbazol-9-yl)methyl)-10-methyl-N-(piperidin-1-ylmethyl)-10H-phenothiazine-3-sulfonamide (10)
A 30-min water bath heating process was used to combine compound 5a (0.39 g, 1 mmol), formaldehyde 37% (0.03 g, 1 mmol), and 1,2,3,4-tetrahydrocarbazole (0.17 g, 1 mmol) in glacial acetic acid (15 mL). After being diluted with water, such a mixture was left to stand at 25 °C. Compound 10 was obtained by crystallizing the isolated product with ethanol to purify it.
Yield, 74%; m.p. = 191–193 °C; IR (KBr): νmax, cm−1: 1610 (C N), 1339, 1171 (SO2); 13C NMR (DMSO‑d6) δ ppm: 23.0, 25.0 (2C), 25.1 (2C), 29.4, 31.5, 36.3, 44.8, 54.3 (2C), 60.9, 69.0 (2C), 110.7, 115.8, 118.0 (2C), 120.6, 123.1 (3C), 125.0, 126.5, 129.8 (3C), 131.5, 139.0, 145.1, 148.0, 151.5. MS (m/z, %): 574 (M+, 54). Anal. Calced for C32H38N4O2S2 (574.80): C, 66.87; H, 6.66; N, 9.75%. Found: C, 62.80; H, 6.58; N, 9.69%.
3.16. Synthesis of N-((2,7-dioxo-2,3,4,5,6,7-hexahydro-1H-benzo[b]azonin-1-yl)methyl)-10-methyl-N-(piperidin-1-ylmethyl)-10H-phenothiazine-3-sulfonamide (11)
A solution of sodium periodate (0.44 g, 2 mmol) in water (5 mL) was mixed with 10 (0.57 g, 1 mmol) in a mixture of methanol and acetone (60 mL, 1:1). Compound 11 crystallized from methanol after being stirred at 25 °C overnight. The solvent was then extracted under decreased pressure, and the result was repeatedly washed with water (3 x 10 mL).
Yield, 64%; m.p. = 216–218 °C; IR (KBr): νmax, cm−1: 1608 (C N), 1342, 1174 (SO2); 13C NMR (DMSO‑d6) δ ppm: 22.0 (2C), 25.7 (2C), 27.0, 36.8, 37.8, 42.1, 55.1 (2C), 64.0, 68.0, 116.0 (2C), 118.8, 123.8 (2C), 125.4 (2C), 127.0 (4C), 130.2 (2C), 132.0, 135.9, 139.7, 145.8, 148.2, 171.1, 194.0. MS (m/z, %): 604 (M+, 60). Anal. Calced for C32H36N4O4S2 (604.78): C, 63.55; H, 6.00; N, 9.26%. Found: C, 63.49; H, 5.91; N, 9.18%.
3.17. Synthesis of 10-methyl-N-((9-oxo-1,2,3,9-tetrahydro-4H-cyclopenta[b]quinolin-4-yl) methyl)-N-(piperidin-1-ylmethyl)-10H-phenothiazine-3-sulfonamide (12)
Compound 11 (0.60 g, 1 mmol) was dissolved in a cold 2 N sodium hydroxide solution (15 mL). After a few seconds of heat, the yellow solution became colorless. Compound 12 was produced by neutralizing ethyl acetate, drying it, and crystallizing the white powder.
Yield, 71%; m.p. = 200–202 °C; IR (KBr): νmax, cm−1: 1616 (C N), 1340, 1176 (SO2); 1H NMR (DMSO‑d6) δ ppm: 1.40 (m, 6H, 3CH2), 1.95 (m, 2H, CH2), 2.25 (t, 2H, CH2), 2.31 (t, 4H, 2CH2), 2.75 (t, 2H, CH2), 3.14 (s, 3H, CH3), 4.45 (s, 2H, CH2), 4.99 (s, 2H, CH2), 7.15–7.80 (m, 11H, Ar–H). MS (m/z, %): 586 (M+, 48). Anal. Calced for C32H34N4O3S2 (586.77): C, 65.50; H, 5.84; N, 9.55%. Found: C, 65.45; H, 5.79; N, 9.45%.
3.18. Synthesis of 10-methyl-N-(4-sulfamoylphenyl)-10H-phenothiazine-3-sulfonamide (13)
Compound 3 (0.93 g, 3 mmol) was portion-wise added at 0 °C to a solution of sulphanilamide (0.25 g, 3 mmol) in THF (30 mL) and stirred for 40 min at the same temperature. Lower pressure was used to remove the excess solvent. Compound 13 was obtained by dissolving the crude solid material in 30 mL of ethyl acetate, followed by 100 mL of water, 30 mL of diluted HCl, 50 mL of water, and 50 mL of saturated sodium chloride solution. The solid material was then dried over anhydrous sodium sulfate, filtered, and evaporated under reduced pressure.
Yield, 80%; m.p. = 240–242 °C; IR (KBr): νmax, cm−1: 3410 (NH2), 3120 (NH), 1610 (C N), 1342, 1171 (SO2); 1H NMR (DMSO‑d6) δ ppm: 2.97 (s, 3H, CH3), 6.84 (s, 2H, NH2), 7.25–8.08 (m, 11H, Ar–H), 10.10 (s, 1H, NH). MS (m/z, %): 447 (M+, 55). Anal. Calced for C19H17N3O4S3 (447.54): C, 50.99; H, 3.83; N, 9.39%. Found: C, 50.91; H, 3.75; N, 9.33%.
3.19. Synthesis of mannich bases (14a-f)
They were prepared according to the previously mentioned procedure for the synthesis of compounds 5a-f.
3.20. 10-Methyl-N-(4-(N-(piperidin-1-ylmethyl)sulfamoyl)phenyl)-10H-phenothiazine-3-sulfonamide (14a)
Yield, 68%; m.p. = 210–212 °C; IR (KBr): νmax, cm−1: 3130 (NH), 1620 (C N), 1350, 1168 (SO2); 1H NMR (DMSO‑d6) δ ppm: 1.55 (t, 6H, 3CH2), 2.25 (t, 4H, 2CH2), 3.21 (s, 3H, CH3), 4.20 (s, 2H, CH2), 7.11 (s, 1H, NH), 7.28–7.70 (m, 11H, Ar–H), 10.15 (s, 1H, NH); 13C NMR (DMSO‑d6) δ ppm: 24.0, 25.5 (2C), 37.0, 54.5 (2C), 64.6, 115.7, 118.9 (3C), 120.2, 122.1, 123.9, 127.1, 128.8 (3C), 130.0 (3C), 134.9, 141.0, 145.0, 148.7. MS (m/z, %): 544 (M+, 50). Anal. Calced for C25H28N4O4S3 (544.70): C, 55.13; H, 5.18; N, 10.29%. Found: C, 55.05; H, 5.09; N, 10.21%.
3.21. 10-Methyl-N-(4-(N-(morpholinomethyl)sulfamoyl)phenyl)-10H-phenothiazine-3-sulfonamide (14b)
Yield, 77%; m.p. = 231–233 °C; IR (KBr): νmax, cm−1: 3125 (NH), 1617 (C N), 1340, 1176 (SO2); 1H NMR (DMSO‑d6) δ ppm: 2.33 (t, 4H, 2CH2), 3.26 (s, 3H, CH3), 3.65 (t, 4H, 2CH2), 4.15 (s, 2H, CH2), 7.05 (s, 1H, NH), 7.11–7.65 (m, 11H, Ar–H), 10.22 (s, 1H, NH); 13C NMR (DMSO‑d6) δ ppm: 37.2, 53.0 (2C), 64.2, 66.6 (2C), 116.5, 119.9 (3C), 121.5, 123.3, 125.1, 128.1, 130.0 (3C), 131.1 (3C), 136.0, 142.0, 146.1, 150.0. MS (m/z, %): 546 (M+, 42). Anal. Calced for C24H26N4O5S3 (546.68): C, 52.73; H, 4.79; N, 10.25%. Found: C, 52.65; H, 4.70; N, 10.16%.
3.22. N-(4-(N-((bis(2-hydroxyethyl)amino)methyl)sulfamoyl)phenyl)-10-methyl-10H-phenothiazine-3-sulfonamide (14c)
Yield, 73%; m.p. = 218–220 °C; IR (KBr): νmax, cm−1: 3430 (OH), 3130 (NH), 1605 (C N), 1350, 1166 (SO2); 1H NMR (DMSO‑d6) δ ppm: 2.80 (t, 4H, 2CH2), 3.28 (s, 3H, CH3), 3.59 (t, 4H, 2CH2), 4.25 (s, 2H, 2OH), 4.44 (s, 2H, CH2), 7.21 (s, 1H, NH), 7.40–7.84 (m, 11H, Ar–H), 10.40 (s, 1H, NH). MS (m/z, %): 564 (M+, 53). Anal. Calced for C24H28N4O6S3 (564.69): C, 51.05; H, 5.00; N, 9.92%. Found: C, 50.99; H, 4.93; N, 9.85%.
3.23. N-(4-(N-((dimethylamino)methyl)sulfamoyl)phenyl)-10-methyl-10H-phenothiazine-3-sulfonamide (14d)
Yield, 77%; m.p. = 197–199 °C; IR (KBr): νmax, cm−1: 3110 (NH), 1613 (C N), 1338, 1175 (SO2); 13C NMR (DMSO‑d6) δ ppm: 36.3, 44.9 (2C), 69.0, 117.0, 120.1 (3C), 121.9, 123.8, 125.5, 128.6, 130.4 (3C), 131.8 (3C), 136.1, 142.2, 146.6, 150.1. MS (m/z, %): 504 (M+, 66). Anal. Calced for C22H24N4O4S3 (504.64): C, 52.36; H, 4.79; N, 11.10%. Found: C, 52.29; H, 4.71; N, 11.00%.
3.24. N-(4-(N-((diethylamino)methyl)sulfamoyl)phenyl)-10-methyl-10H-phenothiazine-3-sulfonamide (14e)
Yield, 71%; m.p. = 216–218 °C; IR (KBr): νmax, cm−1: 3115 (NH), 1614 (C N), 1344, 1177 (SO2); MS (m/z, %): 532 (M+, 64). Anal. Calced for C24H28N4O4S3 (532.69): C, 54.11; H, 5.30; N, 10.52%. Found: C, 54.05; H, 5.24; N, 10.45%.
3.25. N-(4-(N-((1,2,3,4,4a,9a-hexahydro-9H-carbazol-9-yl)methyl)sulfamoyl)phenyl)-10-methyl-10H-phenothiazine-3-sulfonamide (14f)
Yield, 73%; m.p. = 187–189 °C; IR (KBr): νmax, cm−1: 3111 (NH), 1611 (C N), 1343, 1173 (SO2); 1H NMR (DMSO‑d6) δ ppm: 1.00–1.90 (m, 10H, 4CH2 + 2CH), 3.29 (s, 3H, CH3), 5.35 (s, 2H, CH2), 7.00 (s, 1H, NH), 7.17–7.59 (m, 15H, Ar–H), 10.30 (s, 1H, NH); 13C NMR (DMSO‑d6) δ ppm: 25.8 (2C), 29.5, 31.9, 35.9, 44.9, 60.5, 65.0, 110.0, 114.8, 117.5 (4C), 120.3, 122.0 (2C), 124.0, 125.9 (2C), 129.0 (3C), 130.6 (3C), 134.9, 138.2, 141.0, 144.1, 150.7 (2C). MS (m/z, %): 632 (M+, 48). Anal. Calced for C32H32N4O4S3 (632.81): C, 60.74; H, 5.10; N, 8.85%. Found: C, 60.67; H, 5.01; N, 8.78%.
3.25.1. Molecular docking
the synthesized derivatives with good antibacterial activities were created using the software version (MOE.2019.01). The ligands were subjected to computational docking with the DHPS (dihydropteroate synthase) protein structure (PBD: 3TZF) to establish the most preferred mode of interaction [47].
4. Conclusion
In conclusion, it was synthesized four series of phenothiazine derivatives linked to Mannich bases by mixing two active pharmacophores, phenothiazine, and alkyl amines. All the newly synthesized compounds were evaluated for in vitro growth inhibition activity against P. aeruginosa, E. coli, and S. aureus. Eight compounds showed good antibacterial activity with a MIC value of 12.5 μg/mL. The bis Mannich bases 6–9 showed moderate to weak activity against the tested strains with a MIC value of 12.5–100 μg/mL indicating that larger compounds may face challenges in entering bacterial cells, but their antimicrobial activity can still be influenced by other factors. In general, compounds 14a-f exhibited significant activity due to the presence of sulfonamoyl phenyl group which may enhance the antibacterial activity. These results were further supported by molecular docking studies, which showed the presence of many derivatives with high binding affinities and distinct interaction patterns with the target protein.
Ethical approval
Not applicable.
Consent to participate
All authors participated directly in the current research work.
Consent to publish
The authors agree to publish the article under the Creative Commons Attribution License.
Availability of data and materials
All relevant data are within the manuscript and available from the corresponding author upon request.
CRediT authorship contribution statement
Ahmed M. Abdula: Resources, Methodology, Data curation, Conceptualization. Ahmad Fawzi Qarah: Validation, Software, Resources, Methodology. Kahdr Alatawi: Methodology, Investigation, Formal analysis, Data curation. Jihan Qurban: Writing – original draft, Software, Methodology, Investigation. Matokah M. Abualnaja: Writing – original draft, Validation, Software, Methodology. Hanadi A. Katuah: Writing – original draft, Resources, Methodology, Formal analysis. Nashwa M. El-Metwaly: Writing – review & editing, Supervision, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28573.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Malhotra M., Sanduja M., Samad A., Deep A. J. Serb. Chem. Soc. 2011;76:1–15. [Google Scholar]
- 2.Tramontini M., Angiolini L. Tetrahedron. 1990;46:1791. [Google Scholar]
- 3.Tramontini M., Angiolini L., Ghedini N. Polymer. 1998;29:771. [Google Scholar]
- 4.Sriram D., Yogeeswari P., Reddy P. Bioorg Med Chem Lett. 2006;16:2113–2116. doi: 10.1016/j.bmcl.2006.01.064. [DOI] [PubMed] [Google Scholar]
- 5.Ali A.M., Shaharynar M. Bioorg Med Chem Lett. 2007;17:3314–3316. doi: 10.1016/j.bmcl.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Tramontini M., Angiolini L. CRC Press; Boca Raton: 1994. Mannich Base:Chemistry and Uses; pp. 7–20. [Google Scholar]
- 7.a) Blanton C.D., Nobles W.L. J Pharm Sci. 1962;51:878.5. [Google Scholar]; b) Holla B.S., Shivananda M.K., Shenoy M.S., Antony G. Farmaco. 1998;53:531–535. doi: 10.1016/s0014-827x(98)00058-5. [DOI] [PubMed] [Google Scholar]; c) Sarangapani M., Reddy V.M. Indian J. Pharm. Sci. 1994;56:174–177. [Google Scholar]; d) Mannich C., Krosche W. Arch. Pharm. (Weinheim) 1912;250:647–667. [Google Scholar]
- 8.a) Jesudason P.E., Sridhar S.K., PadmaMalar E.J., Shanmugapandiyan P., Inayathullah M., Arul V., Selvaraj D., Jayakumar R. Eur. J. Med. Chem. 2009;44:2307–2312. doi: 10.1016/j.ejmech.2008.03.043. [DOI] [PubMed] [Google Scholar]; b) Turan-Zitouni G., Kaplancıklı Z.A., Yıldız M.T., Chevallet P., Kaya D. Eur. J. Med. Chem. 2005;40:607–613. doi: 10.1016/j.ejmech.2005.01.007. [DOI] [PubMed] [Google Scholar]; c) Edic-Saric M., Maysinger D., Movrin M., Dvorzak I. Chemotherapy. 1980;26:263–267. doi: 10.1159/000237915. [DOI] [PubMed] [Google Scholar]; d) Borenstein M.R., Doukas P.H. J. Pharm. Sci. 1987;76:300–302. doi: 10.1002/jps.2600760407. [DOI] [PubMed] [Google Scholar]
- 9.a) Joshi S., Bilgayan P., Pathak A., Chil J. Chem. Soc. 2012;58:3. [Google Scholar]; b) Tramontini M. 1973. Synthesis; pp. 703–775. [Google Scholar]; c) Thompson B.B. J. Pharm. Sci. 1968;57:715–733. doi: 10.1002/jps.2600570501. [DOI] [PubMed] [Google Scholar]; d) Cummings T.F., Shelton J.R. J. Org. Chem. 1960;25:419–423. [Google Scholar]
- 10.Valverde L.F., Cedillo F.D., Ramos M.L., Cervera E.G., Cruz R.T., Moo J.R. Int. J. Pharma. Sci. Rev. Res. Oct- 2010;51(4):1. [Google Scholar]
- 11.Joshi S., Khosla N., Tiwari P. Bioorg. Med. Chem. 2003;12:571. doi: 10.1016/j.bmc.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Almajan G.L., Barbusceanu S.P., Almajan E.R., Draghici C., Saramet G. Eur. J. Med. Chem. 2009;44:3083–3089. doi: 10.1016/j.ejmech.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Bayrak H., Demirbas A., Karaoglu S.A., Demirbas N. Eur. J. Med. Chem. 2009;44:1057–1066. doi: 10.1016/j.ejmech.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 14.Nishihara H. J. Biol. Chem. 1953;40:579. [Google Scholar]
- 15.Li J.J., Anderson Q.D., Burton E.G., Cogburn J.N., Collins J.T., Garland D.J., Gregory S.A., Huang H.C., Isakson P.C., Koboldt C.M., Logush E.W., Norton M.B., Perkns W.E., Reinherd E.J., Seibert K., Veenhuizen A.W., Zang Y., Reitz D.B. J. Med. Chem. 1995;38:4570. doi: 10.1021/jm00022a023. [DOI] [PubMed] [Google Scholar]
- 16.Supuran C.T., Scozzafava A., Jurca B.C., Ilies M.A. Eur. J. Med. Chem. 1998;33:83. [Google Scholar]
- 17.Joshi S., Manikpuri A., Tiwari P. Bioorg Med Chem Lett. 2006;17:645. doi: 10.1016/j.bmcl.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Hollister L.E., Eikenberry D.T., Raffel S. Am. Rev. Respir. Dis. 1960;81:562–566. doi: 10.1164/arrd.1960.81.4.562. [DOI] [PubMed] [Google Scholar]
- 19.Mosnaim A., Ranade V. Am J. Ther. 2006;13:261. doi: 10.1097/01.mjt.0000212897.20458.63. [DOI] [PubMed] [Google Scholar]
- 20.Daversh S., McDonald R.S., Penwell A., Conrad S., Darvesh K.V., Mataija D., Gomez G., Caines A., Walsh R., Martin E. Biorg. Med. Chem. 2005;13:211. doi: 10.1016/j.bmc.2004.09.059. [DOI] [PubMed] [Google Scholar]
- 21.(a) Schmidt M., Teitge M., Castillo M.E., Brandt T., Dobner B., Langner A. Arch. Pharm. (Weinheim) 2008;341:624. doi: 10.1002/ardp.200800115. [DOI] [PubMed] [Google Scholar]; (b) Bissi A., Meli M., Gobbi S., Rampa A., Tolomeo M., Dusonchet L. Bioorg. Med. Chem. 2008;16:6474. doi: 10.1016/j.bmc.2008.05.040. [DOI] [PubMed] [Google Scholar]
- 22.Jacob R.M. Robert, J. G. Ger. Pat. DE 1117584, 1961. Chem. Abstr. 1962;57 [Google Scholar]
- 23.Yale H.L. J. Am. Chem. Soc. Mass Spectrom. 1955;77:2270. [Google Scholar]
- 24.Evans W.J., Smiles S. J. Chem. Soc. 1935;181:1263. [Google Scholar]
- 25.Subramaniapillai S.G. J. Chem. Sci. 2013;125:467. [Google Scholar]
- 26.Roman G. Eur. J. Med. Chem. 2015;89:743. doi: 10.1016/j.ejmech.2014.10.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimmock J.R., Kumar P. Curr. Med. Chem. 1997;4:1. [Google Scholar]
- 28.Bogdanov A.V., Vazykhova A.M., Khasiyatullina N.R., Krivolapov D.B., Dobrynin A.B., Voloshina A.D., Mironov V.F., V. F. Chem Heterocycl Comp. 2016;52:25. [Google Scholar]
- 29.George S., Chkraborty R., Parameswaran M., Rajan A., Ravi T.K. J. Heterocycl. Chem. 2015;52:211. [Google Scholar]
- 30.Guo H. Eur. J. Med. Chem. 2019;164:678. doi: 10.1016/j.ejmech.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 31.Shen S., Sun X., Liu Y., Chen B., Jin R., Ma H. J. Heterocycl. Chem. 2015;52:1296. [Google Scholar]
- 32.Bal T.R., Anand B., Yogeeswari P., Sriram D. Bioorg Med Chem Lett. 2005;15:4451. doi: 10.1016/j.bmcl.2005.07.046. [DOI] [PubMed] [Google Scholar]
- 33.Aboul‐Fadl T., Bin‐Jubair F.A.S. Int. J. Res. Pharm. Sci. 2010;1:113. [Google Scholar]
- 34.Chen G., Ning Y., Zhao W., Zhang Y., Zhang Y., Hao X., Wang Y., Mu S. Lett. Drug Des. Discov. 2016;13:395. [Google Scholar]
- 35.Farrington K.J., Warburton W.K.J. Aut. Chem. 1987;10:502. [Google Scholar]
- 36.Afsah E.M., Fadda A.A., Bondock S., Hammouda M.M. Synthesis and Some Reactions of Functionalized Benzo (b) azonines and Bi (benzo (b) azonines) ChemInform. 2009;40(34):i. [Google Scholar]
- 37.Yan Y., Li G., Wang F., Mao W. Huadong Huagong Xueyuan Xuebao. 1992;18(192) CA 118, 127985k. [Google Scholar]
- 38.Khan S.A., Siddiqui A.A., Bhatt S. Asian J. Chem. 2002;14(14):417. [Google Scholar]
- 39.Neuss N., Gorman M., Svoboda G.H., Maciak G., Beer C.T. J. Amer. Chem. Soc. 1959;81:4754. [Google Scholar]
- 40.Neuss N., Gorman M., Boaz H.E., Cone N.J. J. Amer. Chem. Soc. 1962;84:1509. [Google Scholar]
- 41.Klayman D.L., Scovill J.P., Bartosevich J.F., Mason C.J. J. Med. Chem. 1979;22:1367. doi: 10.1021/jm00193a020. [DOI] [PubMed] [Google Scholar]
- 42.Clark C.R., Halfpenny P.R., Hill R.G., Horwell D.C., Hughes J., Jarvis T.C., Rees D.C., Schofield D. J. Med. Chem. 1988;31:831. doi: 10.1021/jm00399a025. [DOI] [PubMed] [Google Scholar]
- 43.Thorsett E.D., Harris E.E., Aster S.D., Peterson E.R., Snyder J.P., Springer J.P., Hirshfield J., Tristram E.W., Patchett A.A., Ulm E.U., Vassil T.C. J. Med. Chem. 1986;29:251. doi: 10.1021/jm00152a014. [DOI] [PubMed] [Google Scholar]
- 44.Elison C., Lien E.J., Zinger A.P., Hussain M., Tong G.L., Golden M. J. Pharm. Sci. 1971;60:1058. doi: 10.1002/jps.2600600712. [DOI] [PubMed] [Google Scholar]
- 45.Witkop B., Rosenblum M. J. Amer. Chem. Soc. 1951;73:2641. [Google Scholar]
- 46.a) Schwalbe R., Steele-Moore L., Goodwin A.C. Crc Press; 2007. Antimicrobial Susceptibility Testing Protocols. [Google Scholar]; b) Abbood A.F., Abdula A.M., Mohsen G.L., Baqi Y. Al-Mustansiriyah Journal of Science. 2021;32(4):26–32. [Google Scholar]; c) Khan A.K. Al-Mustansiriyah Journal of Science. 2017;28(3):122–133. [Google Scholar]; d) Sultan M.I., Abdula A.M., Faeq R.I., Radi M.F. Al-Mustansiriyah Journal of Science. 2021;32(3):8–14. [Google Scholar]
- 47.Mondal S., Mandal S.M., Mondal T.K., Sinha C. Spectroscopic characterization, antimicrobial activity, DFT computation and docking studies of sulfonamide Schiff bases. J. Mol. Struct. 2017;1127:557–567. [Google Scholar]
- 48.Venkatesan K., Satyanarayana V.S.V., Mohanapriya K., Khora S.S., Sivakumar A. Ultrasound-mediated synthesis of phenothiazine derivatives and their in vitro antibacterial and antioxidant studies. Res. Chem. Intermed. 2015;41:595–607. [Google Scholar]
- 49.Emese Gal. Synthesis and characterization of some new heterocyclic aromatic derivatives, precursors for materials with nonlinear optical properties. "BABEŞ-BOLYAI" University CLUJ-NAPOCA, Faculty of Chemistry and Chemical Engineering, Organic Chemistry Department. 2010 Thesis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the manuscript and available from the corresponding author upon request.