Abstract
Mitochondria participate in varieties of cellular events. It is widely accepted that human mitochondrial genome encodes 13 proteins, 2 rRNAs, and 22 tRNAs. Gene variation derived from human nuclear genome cannot completely explain mitochondrial diseases. The advent of high-throughput sequencing coupled with novel bioinformatic analyses decode the complexity of mitochondria-derived transcripts. Recently, circular RNAs (circRNAs) from both human mitochondrial genome and nuclear genome have been found to be located at mitochondria. Studies about the roles and molecular mechanisms underlying trafficking of the nucleus encoded circRNAs to mitochondria and mitochondria encoded circRNAs to the nucleus or cytoplasm in mammals are only beginning to emerge. These circRNAs have been associated with a variety of diseases, especially cancers. Here, we discuss the emerging field of mitochondria-located circRNAs by reviewing their identification, expression patterns, regulatory roles, and functional mechanisms. Mitochondria-located circRNAs have regulatory roles in cellular physiology and pathology. We also highlight future perspectives and challenges in studying mitochondria-located circRNAs, as well as their potential biomedical applications.
Keywords: mitochondria, mitochondrial genome, noncoding RNAs, circular RNAs, nuclear genome
Introduction
Mitochondria are ancient organelles and are the power factory of cells, producing adenosine triphosphate (ATP) via the electron-transport chain (ETC) and oxidative phosphorylation system (OXPHOS). Mitochondria are involved in many biological functions, such as ATP transport, cell cycle, cell signaling, development, and neuronal function1, 2. Different from other subcellular organelles, such as Golgi, lysosomes, and endosomes, mitochondria have their own genome. Human mitochondrial genome (mitochondrial DNA, mtDNA) is a double-stranded, 16 569 base pairs (bp) long circular DNA, which is lack of introns and resides within the mitochondrial matrix. Human mtDNA is present in most cells and is conventionally considered to encode 13 protein subunits of the OXPHOS, 2 rRNAs, and 22 tRNAs3 (Figure 1). Noncoding RNAs transcribed from the mtDNA are located at the cytosol, nucleus, and plasma4. In per diploid cell, the mtDNA copy number is maintained at approximately 1000 to 10 000 copies5. The division of mitochondrial genetic information between the nucleus and the mitochondria occurred with gene transfer events during evolution6. Gene variation derived from human nuclear genome (nuDNA) cannot completely explain mitochondrial diseases in maternally inherited diabetes and deafness7, aging8, renal disease9, cardiomyopathies10, inflammation and immunity11, and cancers12. This may be resulted from the difference between human mtDNA and nuDNA. Human mtDNA differs from human nuDNA in many aspects, such as non-Mendelian genetics13, transcriptional machinery14, repair pathways15, and the polyploid nature of the genome16. Human mtDNAs are packaged into nucleoids. Nucleoids are chromosome-like organellar nuclei17, that exhibit a discrete macromolecular assembly that determines mitochondrial genetics and dysfunction18 and cardiac homeostasis19.
Figure 1.
Human mitochondrial DNA composition. Heavy strand in red and light strand in blue. The rRNAs, mRNAs, and tRNAs were separately labeled in black, white, and purple. HSP and LSP are the promoters of heavy and light strands seperately. The validated mecciRNAs were marked in black. The figure was generated by using BioRender (https://app.biorender.com/).
Noncoding RNAs, mainly including long noncoding RNAs (lncRNA), microRNAs (miRNAs), and circular RNAs (circRNAs), play important roles in physiology and pathology20, 21. CircRNAs are a class of single-stranded noncoding RNAs that are formed in a circular conformation via non-canonical splicing or back-splicing event22. CircRNAs were first reported in Sendai virus in 1976, and reported as by-products of abnormal splicing23, 24. The advent of high-throughput sequencing makes circRNAs shine25-27. Recently, the roles of circRNAs are deeply explored in cancers28, immune responses and immune diseases29, brain development and central nervous system diseases30, and cardiovascular system31. Mechanically, circRNAs can regulate transcription, splicing and chromatin interactions, act as miRNA decoys, sequester proteins27 and translate proteins, and function as protein scaffolds32. CircRNAs are mainly located in the cytoplasm and nucleus25, while they are also found in mitochondria33. It was briefly reported that 118 mitochondria-located circRNAs were derived from a human cell line HepG233. To classify circRNAs related with the mitochondria clearly, we adopt the term “mecciRNAs” for mitochondrial genome encoded circRNAs which was previously used by Ren BB34, and the term “mt-circRNAs” for mitochondria-located circRNAs which was previously used by Liang HX4. CircRNAs encoded by nuclear genome were termed “nuc-circRNA”. Mt-circRNAs include both a part of mecciRNAs and a part of nuc-circRNAs. Therefore, circRNAs related with the mitochondria can be classified into three types based on their location and genome origin: circRNAs encoded by mitochondrial genome and located at mitochondria, circRNAs encoded by mitochondrial genome and located at cytoplasm or secreted out of the cells, and circRNAs encoded by nuclear genome and located at mitochondria.
The question arises as to why nuclear genome encoded the same circRNAs are located at mitochondria, despite their crucial roles when located at the cytoplasm and nucleus. Since the discovery of mecciRNAs, their biological roles remain an enigma. The roles of mitochondria-located mecciRNAs are beyond our understanding too. A deeper understanding of mitochondria-located mecciRNAs will bring a new direction. In this review, we highlight the identification, expression patterns, regulatory roles, and functional mechanisms of mt-circRNAs, as well as their potential biomedical applications.
Historical perspective of mitochondria
Over the past 130 years, the discovery of mitochondrial genome and its roles has gone through the discovery of genetic functions, the discovery of mitochondria DNA, and the discovery and implication of mitochondrial genome. In the 1890s, Richard Altmann first proposed that mitochondria are organelles of eukaryotes, and speculated that mitochondria have genetic autonomy35. In Nass MM and Nass S' work, they found mitochondria contain DNA by electron microscope36. In the 1960s, the existence of DNA in mitochondria was confirmed and widely accepted. In 1976, Trembath MK and his collogues first completed mapping the genetic and physical map of yeast mtDNA37. The human mtDNA sequence currently in use is modified from the portrait of mitochondrial genome by Grivell LA38. The studies conducted by Wallace DC's team had opened up a new field of medical research——mitochondrial disease39. In 2016, the advance of mitochondrial replacement therapy (MRT) made it possible that mother with a defective mtDNA could give birth to a healthy child40. The studies of circRNA blossomed in recent ten years. However, the discovery of circRNA located at mitochondria is just the beginning.
Mitochondrial function and disfunction
The most famous role of mitochondria is “powerhouse” of cell41, 42. The OXPHOS system is composed of several multi-protein complexes, which constitute of the electron transport chain within the extensive inner membrane of the mitochondria43. Mitochondrial ATP generation is intimately linked through the function of the ETC, and thus efficient measurement of ETC function can provide insight into mechanisms of physiology and disease. A consequence of electron transfer is the generation of reactive oxygen species (ROS)44. There are two main antioxidant systems in the mitochondrial matrix, the glutathione and thioredoxin/peroxiredoxin systems, which regulate the concentration and redox species in the organelle45. The overproduction of ROS can promote cancer development by inducing genomic instability, modifying gene expression, participating in signaling pathways, and leading to mtDNA and nuDNA mutate46.
The other main function of mitochondria is Ca2+ dynamics. The uptake of Ca2+ by mitochondria was first observed by Slater EC and his colleagues47. There are three pathways of Ca2+ entry into mitochondrial matrix: mitochondrial calcium uniporter (MCU)48, the “rapid mode” (RaM) mechanism49, and the mitochondrial ryanodine receptor (RyR)50. Ca2+ were excluded mainly by the mitochondrial Na+/ Ca2+/Li+ exchanger (NCLX) exchange for sodium (NaC1)51 or lithium (LiC1)52 or H+53. However, the capacity for Ca2+ of mitochondria is finite. The overload of Ca2+ may lead to produce reactive oxygen species and ultimately lead to cell death54.
Mitochondria also have metabolism and synthesis roles, such as amino acid55 and ascorbate56. GOT1 can consume aspartate to transfer electrons into mitochondria, however, upon ETC inhibition, it reverses to generate aspartate in the cytosol. Aspartate supplementation or overexpression of an aspartate transporter allows cells without ETC activity to proliferate55. Mitochondria can regenerate ascorbic acid from its oxidized forms, which may help to maintain the vitamin both in mitochondria and in the cytoplasm. This recycling of ascorbic acid is mitochondrial complex III depended57.
Cross-talk between mitochondria and the nucleus
The realization of mitochondrial roles relies on the genetic information both in mtDNA and nuDNA. Proteins that participate in mitochondrial transcription are nuDNA encoded14. Proteins encoded by human nuDNA were imported into mitochondria through several pathways, the presequence pathway, the carrier protein pathway, the redox-regulated import pathway, and the β-barrel pathway58. The presequence pathway directs proteins to matrix and inner membrane, the carrier protein pathway directs proteins to the inner membrane, the redox-regulated import pathway directs proteins to intermembrane space, and the β-barrel pathway directs proteins to the outer membrane58. TOM, TIM23, TIM22, and SAM complexes are embedded in the outer and inner membranes of mitochondria, and participate in protein transport into mitochondria59, 60. The import of proteins was finely regulated61. It is reported that mecciRNAs facilitate the mitochondria entry of nuclear-encoded proteins by serving as molecular chaperones in the folding of imported proteins62.
MiRNAs are located at cytosol, nucleus63, and exosomes64. Besides, miRNAs were found in human mitochondria65, 66. Blanchette M and his collogues developed a computational tool, miRdup, which can predict the location of miRNAs67. Other RNA were also imported into mitochondria, such as tRNA68 and pre-miRNAs69. The import of nuclear DNA into mitochondria was reviewed by Konstantinov YM70 and Verechshagina NA71. The import process includes: before cross-membrane translocation, translocation across the mitochondrial outer membrane, and translocation across the mitochondrial inner membrane72. Though, it is suggested that PNPASE mediates the import of most lncRNAs and miRNA-378 into mitochondrial matrix, the mitochondrial translocation mechanisms of human RNAs and DNA are largely unclear73, 74.
Mitochondria also export RNAs75 and mtDNA76. The miRNAs encoded by mtDNA seem to be located within mitochondria77. It is reported that the export of double-stranded RNAs (dsRNAs) into the cytosol is in a PNPASE-dependent manner78. The export of mtDNA is related with mitochondrial permeability transition pore (mPTP) and the outer membrane pore formed by VDAC oligomerization76.
Isolation and identification of nuclear-encoded and mitochondria-encoded circRNAs
CircRNAs arising from linear precursor RNAs and the 5′ and 3′ ends were covalently ligated, therefore the traditionally used biochemical and computational approaches for linear RNA studies cannot perfectly fit for the studies of circRNAs. CircRNAs are resistant to degradation by exonucleases, so the 3'-5' exonuclease RNase R is used to improve their circularity as well as enrichment. Xiao MS and his colleagues improved this standard procedure, thus improving the purification efficiency of RNase R79. For the annotation of nuclear-encoded circRNA, the pipeline for predicting circRNAs from ribominus sequencing data was applied as mentioned in this work80. The start and end positions of circRNAs in the chromosome can be predicted, but the full-length sequences of circRNAs cannot be identified. Hossain MT and his colleagues solved this problem, they presented an R package FcircSEC (Full Length circRNA Sequence Extraction and Classification), which can extract the full-length circRNA sequences based on gene annotation and the output of any circRNA prediction tools81. To acquire more information on mecciRNAs, the enrichment of mecciRNAs is needed. Liu X and his colleagues developed a method for mitochondrial RNA isolation to enrich mecciRNAs, so that more information of mecciRNAs could be acquired from RNA-seq, and provide a brief description of the computational method for mecciRNA identification82. To identify circRNAs more reliably, the detection tool constantly advancing83. It is well known that mRNAs have isogenous. It has been reported that alternative back-splicing also occurs in circRNAs, similar to linear RNAs, generating isogenous circRNAs (ISO circRNAs)33, 84. A novel algorithm, CIRI-long, is proposed for circRNA characterization and isoform quantification, also identified including 156 mecciRNAs with GT/AG signals85.
There are many datasets which can predict circRNAs and their roles or have collected many information of circRNAs which have been validated, such as circBank (http://www.circbank.cn/help.html)86, CIRCpedia v2 (http://yang-laboratory.com/circpedia/)87, circBase (http://www.circbase.org/)88, CSCD (http://gb.whu.edu.cn/CSCD/)89, circRNADb (http://reprod.njmu.edu.cn/cgi-bin/circrnadb/circRNADb.php)90, CircNet 2.0 (http://circnet.mbc.nctu.edu.tw/)91. But there are different naming systems of circRNAs among them, which may lead to confusion of the same circRNAs. But, the annotation of mecciRNA is limited. Until now, there don't have a specific database for mecciRNAs prediction and function annotation.
Mitochondria-located circRNAs derived from nuclear genome and their roles
Recently, the mechanisms of circRNA biogenesis have been fully elucidated25. Exons, introns, and a combination of both can generate circRNAs, which are respectively named ecircRNAs, ciRNAs, and eiciRNAs25. The circularization can be mediated by RNA-binding proteins (RBPs) and spliceosome92, 93, and reverse complementary motifs rely on intron-pairing and lariat-driven circularization94, 95. After the formation of circRNAs, commonly they are distributed to nucleus and cytoplasm or secreted out of the cells, but recent studies have found that they are also distributed in mitochondria33. Maybe this is resulted from the limited proteins or RNAs encoded by mtDNA3. Proteins and RNAs derived from mtDNA are limited. To sustain the homeostasis and duplication of mitochondria, proteins and RNAs encoded by the nuDNA are transported into mitochondria61. However, less is known about the biological role and molecular mechanism underlying import of circRNA into human mitochondria in contrast with that of proteins, DNA, and miRNAs.
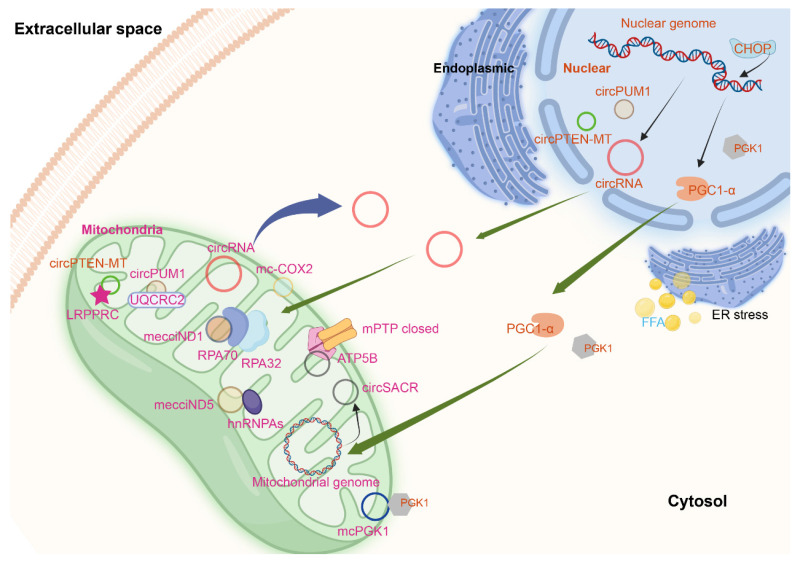
CircPUM1 is a circRNA generated from the PUM1 gene on human chromosome 1. And multiple confocal assays found that circPUM1 and UQCRC2 co-localized in the mitochondria2 (Figure 2). CircPUM1 is positively correlated with HIF1α accumulation under CoCl2-induced intracellular hypoxic-like condition in esophageal squamous cell carcinoma (ESCC) cell lines. Mechanically, circPUM1 acts as a scaffold for the interaction between UQCRC1 and UQCRC2, and circPUM1 depletion induces dysfunction of the mitochondrial complex III and the cleavage of caspase3, thus circPUM1 plays a critical role in maintaining the stability of mitochondrial complex III to enhance oxidative phosphorylation for ATP production of ESCC cells2. CircSamd4 was supposed as a biomarker for predicting vascular calcification96. CircSamd4 was also found to be mitochondria-located97 (Figure 2). CircSamd4 can induce the mitochondrial translocation of the Vcp protein, resulting in the downregulation of Vdac1 and preventing the open of mitochondrial permeability transition pore, thus reducing oxidative stress generation and maintaining mitochondrial dynamics97. CircPTEN-MT is another circRNA encoded by nuDNA and located at mitochondria98.
Figure 2.
Mitochondria-located circRNAs derived from nuclear and mitochondrial genome. The figure was generated by using BioRender (https://app.biorender.com/).
There are many circRNAs located outside of mitochondria but can regulate the function of mitochondria, such as circHIPK399, circ-CBFB100, circ_0004463101, and circFAM160A2102. However, it is not reported whether these circRNAs were imported into mitochondria. Exogenous mRNAs, antireplicative RNAs, and single-stranded DNAs were imported into mitochondria103-105, which occurs without the use of any carriers58. The import of proteins into mitochondria is supervised through mitochondrial protein quality control system106, however, it is not reported whether the import of circRNAs was supervised or not. Because of the presence of double mitochondrial membranes and the lack of studies about mitochondrial specific circRNA delivery systems, the transfer of nuDNA encoded circRNAs to mitochondria is largely unknown. It is difficult to decode the roles of mitochondria-located circRNAs encoded by nuclear genome now and the regulation of circRNAs import into mitochondria needs further investigation.
Mitochondria-located circRNAs derived from mitochondrial genome and their roles
The existing hypotheses of circularization mechanisms of circRNAs are mainly based on the studies of nuclear genome derived circRNAs, but the mechanism of mecciRNAs back-splicing has not been proposed until now. Our understanding of the circularization mechanism of mecciRNAs lags behind our knowledge of nuc-circRNAs. MecciRNAs were encoded by both the light and heavy strands of the mtDNA, and the heavy strand of mtDNA encoded the majority62. In human HEK cell line, mitochondrial mRNA fragments can be circularized107. An earlier study found that in the cells without mitochondria (rho0 MEF cells), no mecciRNAs were identified, whereas mecciRNAs were found in wild-type MEF cells62. Despite located at mitochondria, mecciRNAs were also present outside of the mitochondria62. In recent years, it has been found that hundreds of mecciRNAs are critical for the adaption of mitochondria to physiological conditions and diseases62. The discovery of mecciRNAs may shine a novel light on the communication between mitochondria and the nucleus.
In the plasma samples from chronic lymphocytic leukemia (CLL) patients, 51 circRNAs were remarkably and abnormally expressed. Among the 28 upregulated circRNAs, the top four circRNAs (hsa_circ_0089763, hsa_circ_0008882, hsa_circ_0002363, and hsa_circ_0089762) were all mecciRNAs. Hsa_circ_0089762 is derived by back-splicing from gene COX2, which is located at mtDNA, so termed mc-COX2108 (Figure 2). Mc-COX2 is less stable than ciRS-7 and circRPL15 (both circRNAs were derived from nuclear genome), but is much more stable than linear RNAs108, 109. Mc-COX2 was highly expressed in CLL patients compared with age- and sex-matched healthy persons, and mc-COX2high CLL patients had a worse overall survival (OS) compared with the mc-COX2low group108. Functionally, knock down mc-COX2 by siRNAs resulted in a decrease of mitochondrial membrane potential and ATP production, while the proliferation and apoptosis of CLL cells were also regulated108. The separate use of crbonyl cyanide 3-chlorophenylhydrazone (CCCP), doxycycline and metformin, can dramatically downregulate the expression of mc-COX2, while the combination of siRNAs against mc-COX2 with CCCP, doxycycline or metformin enhanced the anti-leukemic activity of these drugs108.
CircRNA SCAR/has-circ-0089762 (Figure 2), one of the four circRNAs (has-circ-0089736, has-circ-0089762, has-circ-0089763 and has-circ-0008882) derived from mitochondria, was found in liver fibroblasts from patients with nonalcoholic steatohepatitis (NASH). CircRNA SCAR is associated with steatosis-to-NASH progression109. Nuclear genome derived circRNAs are produced through a back-splicing mechanism mostly from repetitive elements, such as ALU elements and short intronic repeats (∼30- to 40-nt)110. However, different from this, the biogenesis of circRNA SCAR is regulated by hnRNPM, which was verified by siRNAs against hnRNPM and CLIP-seq data. Silencing RNase L increased the circRNA SCAR level in poly(I:C)-treated fibroblasts109, 111. In vitro, circRNA SCAR inhibits mitochondrial ROS (mROS) output and fibroblast activation. PGC-1α mediates circRNA SCAR binding to ATP5B and shuts down mPTP by blocking CypD-mPTP interaction. Furthermore, the effect of PGC-1α is inhibited by lipid overload through ER stress-induced CHOP. Targeting circRNA SCAR in vivo alleviates high fat diet-induced cirrhosis and insulin resistance. CircRNA SCAR is one of the molecular components that participate in ER-nucleus-mitochondria-cytosol communication pathway which drives lipid-mediated inflammation109.
MecciND1 and mecciND5 are mecciRNAs encoded by the mitochondrial genes ND1 and ND5 respectively, and they are located at both mitochondria and cytosol. RPA70 and RPA32 proteins interact with mecciND1 through TOM40, and the overexpression of mecciND1 results in a higher protein level of RPA70 and RPA32 in the mitochondria, but doesn't change the overall protein level of RPA70 and RPA32. HnRNPA1, hnRNPA2B1, and hnRNPA3 interact with mecciND5. The overexpression of mecciND5 results in a higher protein and mRNA level of hnRNPA1, hnRNPA2B1, and hnRNPA3 in the mitochondria, but the levels of all three hnRNPA proteins and mRNAs were much less affected. These results indicated that mecciRNAs promoted mitochondrial importation of specific protein partners. In hepatocellular carcinoma, the expression of mecciND1 and mecciND5 were upregulated. The use of UV and hydrogen peroxide increased mecciND1 levels, so as to RPA70 and RPA32 protein levels in mitochondria62 (Figure 2).
Another circRNA encoded by mitochondrial ND5 gene is circMTND5 (chrM: 14068-14413+)112. CircMTND5 sponge MIR6812 and colocalize in mitochondria, alleviating renal mitochondrial injury and kidney fibrosis112 (Figure 2). McPGK1 (mitochondrial circRNA for translocating phosphoglycerate kinase 1) is highly expressed in liver tumour-initiating cells (TICs). Its overexpression can drive liver TIC self-renewal113. Mitochondria-located circRNAs are listed in Table 1.
Table 1.
Mitochondria-located circRNAs
| CircRNA name | Derived genome | Validation | Diseases | Ref. |
|---|---|---|---|---|
| circPUM1 | Nuclear | Yes | Esophageal squamous cell carcinoma | 2 |
| circSmad4 | Nuclear | Yes | Myocardial infarction | 97 |
| circPTEN-MT | Nuclear | Yes | Hepatocellular carcinoma | 98 |
| mecciND1, mecciND5 | Mitochondrial | Yes, Yes | Hepatocellular carcinoma | 62 |
| hsa_circ_0089763, hsa_circ_0008882, hsa_circ_0002363, hsa_circ_0089762 | Mitochondrial | No, No, No, Yes |
Chronic lymphocytic leukemia | 108 |
| has-circ-0089761, has-circ-0089762, has-circ-0089763, has-circ-0008882 | Mitochondrial | No, Yes, Yes, Yes |
Nonalcoholic steatohepatitis | 109 |
| circMTND5 | Mitochondrial | Yes | Lupus nephritis | 112 |
| mcPGK1 | Mitochondrial | Yes | Liver cancer | 113 |
Applications and future aspirations of mt-circRNAs
Mt-circRNA can regulate the energy metabolism of mitochondria. It is reported that the level of PTEN is related with mitochondrial energy metabolism in cell lines114. The PTEN expression cells have a lower ATP content and higher ADP/ATP ratio, higher AMPK activating-phosphorylation evoking energy impairment, higher OXPHOS complexes and PGC1α-Sirt3-p53 protein abundance114. CircPTEN-MT is a circRNA encoded by exons 3, 4, and 5 of PTEN, which is localized at mitochondria and physically associated with leucine-rich pentatricopeptide repeat-containing protein (LRPPRC)98. The downregulation by siRNAs against circPTEN-MT can decrease the mRNA level of the mitochondrial complex Ι subunit and reduce mitochondrial membrane potential and ATP production98. It seems that PTEN and circPTEN-MT have an opposite role in regulating mitochondrial energy metabolism. However, these two studies were carried out in different labs and different disease models. The relative expression level and dynamics of PTEN and circPTEN-MT have not been elucidated. Whether PTEN and circPTEN-MT counteract with each other to balance the energy metabolism of mitochondria needs further study. It is reported that mcPGK1 regulates metabolic reprogramming by inhibiting mitochondrial OXPHOS and promoting glycolysis113. Those studies indicate that targeting mt-circRNAs has the potential to regulate the energy metabolism of mitochondria and further a potential to regulate the chemoresistance of cancers.
Mt-circRNA can assist the transport of proteins to mitochondria. TOM, TIM23, TIM22, and SAM complexes are embedded in the outer and inner membranes of mitochondria, and participate in protein transport into mitochondria59, 60. It is reported that mecciRNAs facilitate the mitochondrial entry of nuclear-encoded proteins by serving as molecular chaperones in the folding of imported proteins62. McPGK1 can promote PGK1 mitochondrial import via TOM40 interactions at outer mitochondria membrane113. The most studied mechanism of circRNA is miRNA sponges and protein scaffolds, however, whether circRNAs can sponge or carry drugs or small-molecule inhibitors to assist the import of them to mitochondria has not been reported. It is worth to devote our efforts to figure out the mystery.
Discussion
Mitochondrial dysfunction is related with a series of diseases, such as neurodegenerative diseases115, cancer115, diabetic kidney disease116, cardiovascular diseases117, and rare diseases118. Mitochondrial genome is in small size but highly utilized. The advent of high-throughput sequencing has expanded our understanding of the known complexity of mitochondrial transcriptome. Besides the generally known mRNAs, rRNAs, and tRNAs, mtDNA also encodes a variety of noncoding RNAs such as lncRNAs, sncRNAs, dsRNAs, and circRNAs with diverse regulatory functions. So far, studies of circRNAs encoded by nuDNA have been performed widely, while the roles of mecciRNAs were less understood. MecciRNAs may reside in or shuttle out of the mitochondria and contribute to the nucleus-mitochondria communication, thus posing a difficulty in functional study, such as mecciND1 and mecciND562. Despite the high utilization efficiency of mtDNA, the homeostasis and duplication of mitochondria is resort to proteins and noncoding RNAs encoded by the nuclear genome. CircPUM1 is mitochondria-located circRNA derived from nuDNA. CircPUM1 plays important roles in mitochondria.
Recent studies have brought hints of circRNAs involved in maintaining mitochondrial function, indicating that the circRNAs located at mitochondria are of importance. Nevertheless, many aspects of circRNAs in mitochondria remain unsolved. Firstly, the import processing of nuclear genome encoded circRNAs into mitochondria is not clear. MtDNA encodes limited genes3, not only the transcription of mtDNA needs the assistance of nuDNA encoded proteins but also the processing of mitochondrial RNA (mtRNA) needs119. NuDNA encoded proteins designated to mitochondria were imported into mitochondrial membrane and mitochondrial matrix by several pathways58. CircRNA is the new star of research, the majority of circRNAs encoded by nuclear genome are located at cytoplasm and nucleus25. However, recent studies found that in mitochondria there are nuc-circRNAs. But, none of them elucidate the import processing of nuc-circRNAs into mitochondria. Are there any carriers or chaperonins or portholes? Are the import pathways generally applicable or specialized? We need further studies in this field. Secondly, the mechanism of the dynamic regulation of mt-circRNAs has not been identified. CircRNAs are both 5' and 3' end lacking, so the major RNA decay pathways which are often initiated via exoribonucleases are unserviceable to decay circRNAs120. Endoribonucleases are enzymes that can cleave RNA without a free 5' or 3' end121. It is reported that circRNAs can be degraded by endoribonuclease with the binding of miRNAs and subsequent Ago2122. Upon poly(I:C) stimulation or viral infection, circRNAs are globally degraded by endoribonuclease RNase L123. Leung AKLand his collogues found that UPF1 and G3BP1 regulate highly-structured circRNAs124. GW182 regulate a subset of circRNAs degradation in Drosophila, three homologs of GW182-TNRC6A, TNRC6B, and TNRC6C in human control degradation of human circRNAs similarly125. These circRNAs decay pathways have not been verified in mitochondria, and it is unknown if there are other mechanisms that can dynamically regulate mt-circRNAs. Deeper investigations are needed to elucidate these questions. Thirdly, it is suggested that nuDNA encoded spliceosomes can mediate the splicing of mtRNA111, but it is unknown whether they are participated in regulating the splice of mecciRNAs. There are other questions to be solved, such as the modification of mt-circRNAs, the roles of the identified mecciRNAs, and the functions of mecciRNAs outside mitochondria. Besides, there is a lack of standard nomenclature for circRNAs, which may result in ambiguity in different circRNAs. Such as circRNA SCAR108 and mc-COX2109 were both named has-circ-0089762. Because of the organelle double membranes, there are still no efficient methods to directly modify the mtDNA in vivo. There are some circRNAs that are not located in mitochondria, but can interfere the function of mitochondria, such as circ_0004463101, circEZH2126, and circFoxO3127. The future focuses on isolation, identification, verification, and modification of mecciRNAs will enhance our understanding about the characters and roles of mecciRNAs.
The expression of mt-circRNAs is correlated with mitochondrial function. The aberrant expression of diverse mt-circRNAs has been observed in various cancer cell types, as evidenced by multiple studies. The precise contribution of mt-circRNAs in different cancer development and its clinical implications remain to be elucidated. Further study on the mechanism and clinical significance of mt-circRNAs is conducive to the discovery of new targeted drugs and clinical markers for mitochondria-related diseases, especially cancer.
We are just arrived at the gate of mt-circRNAs, the indoor world landscape of mt-circRNA is far beyond our understanding now. Much work is needed to unveil the circRNA world in mitochondria. Our future studies should concentrate on but not be limited to: the import mechanism of nuc-circRNA into mitochondria and their roles, the genesis of mecciRNA and their function in mitochondria, the dynamics regulation, and the clinical significance of mt-circRNAs.
Acknowledgments
Funding
This work was supported by grants from No.71 General Support from China Postdoctoral Science Foundation (2022M713840).
Data availability statement
Data sharing not available to this article as no datasets were generated or analyzed during the current study.
Author contributions
Jun Zhao conceived this review and critically revised the manuscript, Donghong Liu did major work of writing the manuscript. Xinyu Zhou and Yida He made contributions to writing the manuscript. All authors gave final approval of the version to be published.
Abbreviations
- ATP
adenosine triphosphate
- ETC
electron-transport chain
- OXPHOS
oxidative phosphorylation system
- mtDNA
mitochondrial genome
- nuDNA
nuclear genome
- lncRNA
long noncoding RNAs
- miRNAs
microRNAs
- circRNAs
circular RNAs
- mecciRNAs
circRNAs transcribed from the mitochondrial genome
- mt-circRNA
mitochondria-located circRNAs
- nuc-circRNA
circRNAs transcribed from the nuclear genome
- ROS
reactive oxygen species
- MCU
mitochondrial calcium uniporter
- RyR
mitochondrial ryanodine receptor
- NCLX
Na+/ Ca2+/Li+ exchanger
- dsRNAs
double-stranded RNAs
- mPTP
mitochondrial permeability transition pore
- FcircSEC
full length circRNA sequence extraction and classification
- ER
endoplasmic reticulum
- ISO circRNAs
isogenous circular RNAs
- RBPs
RNA-binding proteins
- ESCC
esophageal squamous cell carcinoma
- CLL
chronic lymphocytic leukemia
- CCCP
crbonyl cyanide 3-chlorophenylhydrazone
- SCAR
Steatohepatitis-associated circRNA ATP5B regulator
- NASH
nonalcoholic steatohepatitis
- mROS
mitochondrial ROS
- CHOP
C/EBP homologous protein
- TICs
tumour-initiating cells
- LRPPRC
leucine-rich pentatricopeptide repeat-containing protein
- mtRNA
mitochondrial RNA
References
- 1.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 2.Gong W, Xu J, Wang Y, Min Q, Chen X, Zhang W. et al. Nuclear genome-derived circular RNA circPUM1 localizes in mitochondria and regulates oxidative phosphorylation in esophageal squamous cell carcinoma. Signal Transduct Target Ther. 2022;7:40–52. doi: 10.1038/s41392-021-00865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J. et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 4.Liang H, Liu J, Su S, Zhao Q. Mitochondrial noncoding RNAs: new wine in an old bottle. RNA Biol. 2021;18:2168–2182. doi: 10.1080/15476286.2021.1935572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrera-Paez JD, Moraes CT. Mitochondrial genome engineering coming-of-age. Trends Genet. 2022;38:869–880. doi: 10.1016/j.tig.2022.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson SG, Karlberg O, Canback B, Kurland CG. On the origin of mitochondria: a genomics perspective. Philos Trans R Soc Lond B Biol Sci. 2003;358:165–177. doi: 10.1098/rstb.2002.1193. discussion 177-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nesbitt V, Pitceathly RD, Turnbull DM, Taylor RW, Sweeney MG, Mudanohwo EE. et al. The UK MRC mitochondrial disease patient cohort study: clinical phenotypes associated with the m.3243A>G mutation-implications for diagnosis and management. J Neurol Neurosurg Psychiatry. 2013;84:936–938. doi: 10.1136/jnnp-2012-303528. [DOI] [PubMed] [Google Scholar]
- 8.Leuthner TC, Meyer JN. Mitochondrial DNA nutagenesis: feature of and biomarker for environmental exposures and aging. Curr Environ Health Rep. 2021;8:294–308. doi: 10.1007/s40572-021-00329-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govers LP, Toka HR, Hariri A, Walsh SB, Bockenhauer D. Mitochondrial DNA mutations in renal disease: an overview. Pediatr Nephrol. 2021;36:9–17. doi: 10.1007/s00467-019-04404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohammed S, Bahitham W, Chan A, Chiu B, Bamforth F, Sergi C. Mitochondrial DNA related cardiomyopathies. Front Biosci (Elite Ed) 2012;4:1706–1716. doi: 10.2741/491. [DOI] [PubMed] [Google Scholar]
- 11.Riley JS, Tait SW. Mitochondrial DNA in inflammation and immunity. EMBO Rep. 2020;21:e49799. doi: 10.15252/embr.201949799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopinski PK, Singh LN, Zhang S, Lott MT, Wallace DC. Mitochondrial DNA variation and cancer. Nat Rev Cancer. 2021;21:431–445. doi: 10.1038/s41568-021-00358-w. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz M, Vissing J. Paternal inheritance of mitochondrial DNA. N Engl J Med. 2002;347:576–580. doi: 10.1056/NEJMoa020350. [DOI] [PubMed] [Google Scholar]
- 14.Basu U, Bostwick AM, Das K, Dittenhafer-Reed KE, Patel SS. Structure, mechanism, and regulation of mitochondrial DNA transcription initiation. J Biol Chem. 2020;295:18406–18425. doi: 10.1074/jbc.REV120.011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen NB, Rasmussen M, Rasmussen LJ. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion. 2005;5:89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 16.St John J. The control of mtDNA replication during differentiation and development. Biochim Biophys Acta. 2014;1840:1345–1354. doi: 10.1016/j.bbagen.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 17.Bogenhagen DF. Mitochondrial DNA nucleoid structure. Biochim Biophys Acta. 2012;1819:914–920. doi: 10.1016/j.bbagrm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Gilkerson RW. Mitochondrial DNA nucleoids determine mitochondrial genetics and dysfunction. Int J Biochem Cell Biol. 2009;41:1899–1906. doi: 10.1016/j.biocel.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Feng Y, Huang W, Paul C, Liu X, Sadayappan S, Wang Y. et al. Mitochondrial nucleoid in cardiac homeostasis: bidirectional signaling of mitochondria and nucleus in cardiac diseases. Basic Res Cardiol. 2021;116:49–74. doi: 10.1007/s00395-021-00889-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hombach S, Kretz M. Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17. doi: 10.1007/978-3-319-42059-2_1. [DOI] [PubMed] [Google Scholar]
- 21.Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19:1310–1335. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 24.Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8:547–555. doi: 10.1016/0092-8674(76)90223-3. [DOI] [PubMed] [Google Scholar]
- 25.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 26.Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19:172–190. doi: 10.1186/s12943-020-01286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6:319–336. doi: 10.1016/j.trecan.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q. et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9:588–607. doi: 10.7150/thno.29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehta SL, Dempsey RJ, Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol. 2020;186:101746–101782. doi: 10.1016/j.pneurobio.2020.101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. 2019;16:503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 32.He L, Man C, Xiang S, Yao L, Wang X, Fan Y. Circular RNAs' cap-independent translation protein and its roles in carcinomas. Mol Cancer. 2021;20:119–129. doi: 10.1186/s12943-021-01417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Zhang X, Li C, Yue L, Ding N, Riordan T. et al. Circular RNA profiling provides insights into their subcellular distribution and molecular characteristics in HepG2 cells. RNA Biol. 2019;16:220–232. doi: 10.1080/15476286.2019.1565284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren B, Guan MX, Zhou T, Cai X, Shan G. Emerging functions of mitochondria-encoded noncoding RNAs. Trends Genet. 2023;39:125–139. doi: 10.1016/j.tig.2022.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Altmann R. Die Elementarorganismen und ihre Beziehungen zu den Zellen. Zweite vermehrte Auflage (The elementary organisms and their relationships to thecells. Second Extended Edition). Leipzig: Verlag Von Veit and Company, 1894.
- 36.Nass MM, Nass S. Intramitochondrial fibers with DNA characteristics. I. Fixation and electron stainingreactions. J Cell Biol. 1963;19:593–611. doi: 10.1083/jcb.19.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linnane AW, Lukins HB, Molloy PL, Nagley P, Rytka J, Sriprakash KS. et al. Biogenesis of mitochondria: molecular mapping of the mitochondrial genome of yeast. Proc Natl Acad Sci U S A. 1976;73:2082–2085. doi: 10.1073/pnas.73.6.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borst P, Grivell LA. Small is beautiful-portrait of a mitochondrial genome. Nature. 1981;290:443–444. doi: 10.1038/290443a0. [DOI] [PubMed] [Google Scholar]
- 39.Brown MD, Trounce IA, Jun AS, Allen JC, Wallace DC. Functional analysis of lymphoblast and cybrid mitochondria containing the 3460, 11778, or 14484 Leber's hereditary optic neuropathy mitochondrial DNA mutation. J Biol Chem. 2000;275:39831–39836. doi: 10.1074/jbc.M006476200. [DOI] [PubMed] [Google Scholar]
- 40.Hyslop LA, Blakeley P, Craven L, Richardson J, Fogarty NM, Fragouli E. et al. Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature. 2016;534:383–386. doi: 10.1038/nature18303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willis EJ. The powerhouse of the cell. Ultrastruct Pathol. 1992;16:iii–vi. doi: 10.3109/01913129209061353. [DOI] [PubMed] [Google Scholar]
- 42.Siekevitz P. Powerhouse of the cell. Scientific American. 1957;197:131–144. [Google Scholar]
- 43.Zhao RZ, Jiang S, Zhang L, Yu ZB. Mitochondrial electron transport chain, ROS generation and uncoupling (Review) Int J Mol Med. 2019;44:3–15. doi: 10.3892/ijmm.2019.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen PK. Antimycin-insensitive oxidation of succinate and reduced nicotinamide-adenine dinucleotide in electron-transport particles. II. Steroid effects. Biochim Biophys Acta. 1966;122:167–174. doi: 10.1016/0926-6593(66)90058-0. [DOI] [PubMed] [Google Scholar]
- 45.Nolfi-Donegan D, Braganza A, Shiva S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020;37:101674–101682. doi: 10.1016/j.redox.2020.101674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Karakhanova S, Hartwig W, D'Haese JG, Philippov PP, Werner J. et al. Mitochondria and mitochondrial ROS in Cancer: novel targets for anticancer therapy. J Cell Physiol. 2016;231:2570–2581. doi: 10.1002/jcp.25349. [DOI] [PubMed] [Google Scholar]
- 47.Slater EC, Cleland KW. The effect of calcium on the respiratory and phosphorylative activities of heart-muscle sarcosomes. Biochem J. 1953;55:566–590. doi: 10.1042/bj0550566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholls DG. Mitochondria and calcium signaling. Cell Calcium. 2005;38:311–317. doi: 10.1016/j.ceca.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 49.Sparagna GC, Gunter KK, Sheu SS, Gunter TE. Mitochondrial calcium uptake from physiological-type pulses of calcium. A description of the rapid uptake mode. J Biol Chem. 1995;270:27510–27515. doi: 10.1074/jbc.270.46.27510. [DOI] [PubMed] [Google Scholar]
- 50.Beutner G, Sharma VK, Giovannucci DR, Yule DI, Sheu SS. Identification of a ryanodine receptor in rat heart mitochondria. J Biol Chem. 2001;276:21482–21488. doi: 10.1074/jbc.M101486200. [DOI] [PubMed] [Google Scholar]
- 51.Carafoli E, Tiozzo R, Lugli G, Crovetti F, Kratzing C. The release of calcium from heart mitochondria by sodium. J Mol Cell Cardiol. 1974;6:361–371. doi: 10.1016/0022-2828(74)90077-7. [DOI] [PubMed] [Google Scholar]
- 52.Crompton M, Capano M, Carafoli E. The sodium-induced efflux of calcium from heart mitochondria. European Journal of Biochemistry. 2008;69:453–462. [Google Scholar]
- 53.Pozzan T, Bragadin M, Azzone GF. Disequilibrium between steady-state Ca2+ accumulation ratio and membrane potential in mitochondria. Pathway and role of Ca2+ efflux. Biochemistry. 1977;16:5618–5625. doi: 10.1021/bi00644a036. [DOI] [PubMed] [Google Scholar]
- 54.Dridi H, Santulli G, Bahlouli L, Miotto MC, Weninger G, Marks AR. Mitochondrial calcium overload plays a causal role in oxidative stress in the failing heart. Biomolecules. 2023;13:1409–1477. doi: 10.3390/biom13091409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.May JM, Li L, Qu ZC, Cobb CE. Mitochondrial recycling of ascorbic acid as a mechanism for regenerating cellular ascorbate. Biofactors. 2007;30:35–48. doi: 10.1002/biof.5520300105. [DOI] [PubMed] [Google Scholar]
- 57.Li X, Cobb CE, May JM. Mitochondrial recycling of ascorbic acid from dehydroascorbic acid: dependence on the electron transport chain. Arch Biochem Biophys. 2002;403:103–110. doi: 10.1016/S0003-9861(02)00205-9. [DOI] [PubMed] [Google Scholar]
- 58.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 60.Mokranjac D, Neupert W. Thirty years of protein translocation into mitochondria: unexpectedly complex and still puzzling. Biochim Biophys Acta. 2009;1793:33–41. doi: 10.1016/j.bbamcr.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 61.Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab. 2014;19:357–372. doi: 10.1016/j.cmet.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 62.Liu X, Wang X, Li J, Hu S, Deng Y, Yin H. et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci China Life Sci. 2020;63:1429–1449. doi: 10.1007/s11427-020-1631-9. [DOI] [PubMed] [Google Scholar]
- 63.Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P. et al. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3' trailers. PLoS One. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mori MA, Ludwig RG, Garcia-Martin R, Brandao BB, Kahn CR. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 2019;30:656–673. doi: 10.1016/j.cmet.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Erturk E, Enes Onur O, Akgun O, Tuna G, Yildiz Y, Ari F. Mitochondrial miRNAs (MitomiRs): their potential roles in breast and other cancers. Mitochondrion. 2022;66:74–81. doi: 10.1016/j.mito.2022.08.002. [DOI] [PubMed] [Google Scholar]
- 66.Li P, Jiao J, Gao G, Prabhakar BS. Control of mitochondrial activity by miRNAs. J Cell Biochem. 2012;113:1104–1110. doi: 10.1002/jcb.24004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leclercq M, Diallo AB, Blanchette M. Computational prediction of the localization of microRNAs within their pre-miRNA. Nucleic Acids Res. 2013;41:7200–7211. doi: 10.1093/nar/gkt466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rubio MA, Rinehart JJ, Krett B, Duvezin-Caubet S, Reichert AS, Soll D. et al. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc Natl Acad Sci U S A. 2008;105:9186–9191. doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X. Pre-microRNA and mature microRNA in human mitochondria. PLoS One. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Konstantinov YM, Dietrich A, Weber-Lotfi F, Ibrahim N, Klimenko ES, Tarasenko VI. et al. DNA import into mitochondria. Biochemistry (Mosc) 2016;81:1044–1056. doi: 10.1134/S0006297916100035. [DOI] [PubMed] [Google Scholar]
- 71.Verechshagina NA, Konstantinov YM, Kamenski PA, Mazunin IO. Import of proteins and nucleic acids into mitochondria. Biochemistry (Mosc) 2018;83:643–661. doi: 10.1134/S0006297918060032. [DOI] [PubMed] [Google Scholar]
- 72.Huang J, Wu S, Wang P, Wang G. Non-coding RNA regulated cross-talk between mitochondria and other cellular compartments. Front Cell Dev Biol. 2021;9:688523–688533. doi: 10.3389/fcell.2021.688523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM. et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shepherd DL, Hathaway QA, Pinti MV, Nichols CE, Durr AJ, Sreekumar S. et al. Exploring the mitochondrial microRNA import pathway through Polynucleotide Phosphorylase (PNPase) J Mol Cell Cardiol. 2017;110:15–25. doi: 10.1016/j.yjmcc.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Landerer E, Villegas J, Burzio VA, Oliveira L, Villota C, Lopez C. et al. Nuclear localization of the mitochondrial ncRNAs in normal and cancer cells. Cell Oncol (Dordr) 2011;34:297–305. doi: 10.1007/s13402-011-0018-8. [DOI] [PubMed] [Google Scholar]
- 76.Kim J, Gupta R, Blanco LP, Yang S, Shteinfer-Kuzmine A, Wang K. et al. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science. 2019;366:1531–1536. doi: 10.1126/science.aav4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ro S, Ma HY, Park C, Ortogero N, Song R, Hennig GW. et al. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 2013;23:759–774. doi: 10.1038/cr.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rotig A. et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature. 2018;560:238–242. doi: 10.1038/s41586-018-0363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiao MS, Wilusz JE. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3' ends. Nucleic Acids Res. 2019;47:8755–8769. doi: 10.1093/nar/gkz576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 81.Hossain MT, Peng Y, Feng S, Wei Y. FcircSEC: an R package for full length circRNA sequence extraction and classification. Int J Genomics. 2020;2020:9084901–9084911. doi: 10.1155/2020/9084901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu X, Yang Y, Shan G. Identification and detection of mecciRNAs. Methods. 2021;196:147–152. doi: 10.1016/j.ymeth.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 83.Gao Y, Zhang J, Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform. 2018;19:803–810. doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- 84.Dang Y, Yan L, Hu B, Fan X, Ren Y, Li R. et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17:130. doi: 10.1186/s13059-016-0991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang J, Hou L, Zuo Z, Ji P, Zhang X, Xue Y. et al. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat Biotechnol. 2021;39:836–845. doi: 10.1038/s41587-021-00842-6. [DOI] [PubMed] [Google Scholar]
- 86.Liu M, Wang Q, Shen J, Yang BB, Ding X. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019;16:899–905. doi: 10.1080/15476286.2019.1600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dong R, Ma XK, Li GW, Yang L. CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteomics Bioinformatics. 2018;16:226–233. doi: 10.1016/j.gpb.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F. et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46:D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen Y, Yao L, Tang Y, Jhong JH, Wan J, Chang J. et al. CircNet 2.0: an updated database for exploring circular RNA regulatory networks in cancers. Nucleic Acids Res. 2022;50:D93–D101. doi: 10.1093/nar/gkab1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2020;98:87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 93.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 95.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 96.Zhou Y, Liu Y, Xuan S, Jin T, Chen K, Wu Z. et al. CircSamd4: a novel biomarker for predicting vascular calcification. J Clin Lab Anal. 2022;36:e24156. doi: 10.1002/jcla.24156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng H, Huang S, Wei G, Sun Y, Li C, Si X. et al. CircRNA Samd4 induces cardiac repair after myocardial infarction by blocking mitochondria-derived ROS output. Mol Ther. 2022;30:3477–3498. doi: 10.1016/j.ymthe.2022.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ruan D, Xu J, Liu Y, Luo J, Zhao X, Li Y. et al. CircPTEN-MT from PTEN regulates mitochondrial energy metabolism. J Genet Genomics. 2024 doi: 10.1016/j.jgg.2023.12.011. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 99.Chen G, Shan X, Li L, Dong L, Huang G, Tao H. circHIPK3 regulates apoptosis and mitochondrial dysfunction induced by ischemic stroke in mice by sponging miR-148b-3p via CDK5R1/SIRT1. Exp Neurol. 2022;355:114115. doi: 10.1016/j.expneurol.2022.114115. [DOI] [PubMed] [Google Scholar]
- 100.Wang Z, Zhao Y, Sun R, Sun Y, Liu D, Lin M. et al. circ-CBFB upregulates p66Shc to perturb mitochondrial dynamics in APAP-induced liver injury. Cell Death Dis. 2020;11:953–967. doi: 10.1038/s41419-020-03160-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu S, Deng H, He H, Xu R, Wang Y, Zhu X. et al. The circ_0004463/miR-380-3p/FOXO1 axis modulates mitochondrial respiration and bladder cancer cell apoptosis. Cell Cycle. 2020;19:3563–3580. doi: 10.1080/15384101.2020.1852746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bao J, Lin C, Zhou X, Ma D, Ge L, Xu K. et al. circFAM160A2 promotes mitochondrial stabilization and apoptosis reduction in osteoarthritis chondrocytes by targeting miR-505-3p and SIRT3. Oxid Med Cell Longev. 2021;2021:5712280. doi: 10.1155/2021/5712280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang G, Shimada E, Zhang J, Hong JS, Smith GM, Teitell MA. et al. Correcting human mitochondrial mutations with targeted RNA import. Proc Natl Acad Sci U S A. 2012;109:4840–4845. doi: 10.1073/pnas.1116792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Loutre R, Heckel AM, Jeandard D, Tarassov I, Entelis N. Anti-replicative recombinant 5S rRNA molecules can modulate the mtDNA heteroplasmy in a glucose-dependent manner. PLoS One. 2018;13:e0199258. doi: 10.1371/journal.pone.0199258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang B, Lv X, Wang Y, Wang Z, Liu Q, Lu B. et al. CRISPR/Cas9-mediated mutagenesis at microhomologous regions of human mitochondrial genome. Sci China Life Sci. 2021;64:1463–1472. doi: 10.1007/s11427-020-1819-8. [DOI] [PubMed] [Google Scholar]
- 106.Jadiya P, Tomar D. Mitochondrial protein quality control mechanisms. Genes (Basel) 2020;11:563–585. doi: 10.3390/genes11050563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mance LG, Mawla I, Shell SM, Cahoon AB. Mitochondrial mRNA fragments are circularized in a human HEK cell line. Mitochondrion. 2020;51:1–6. doi: 10.1016/j.mito.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 108.Wu Z, Sun H, Wang C, Liu W, Liu M, Zhu Y. et al. Mitochondrial genome-derived circRNA mc-COX2 functions as an oncogene in chronic lymphocytic leukemia. Mol Ther Nucleic Acids. 2020;20:801–811. doi: 10.1016/j.omtn.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao Q, Liu J, Deng H, Ma R, Liao JY, Liang H. et al. Targeting mitochondria-located circRNA SCAR alleviates NASH viareducing mROS output. Cell. 2020;183:76–93. doi: 10.1016/j.cell.2020.08.009. e22. [DOI] [PubMed] [Google Scholar]
- 110.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 111.Herai RH, Negraes PD, Muotri AR. Evidence of nuclei-encoded spliceosome mediating splicing of mitochondrial RNA. Hum Mol Genet. 2017;26:2472–2479. doi: 10.1093/hmg/ddx142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luan J, Jiao C, Ma C, Zhang Y, Hao X, Zhou G. et al. circMTND5 participates in renal mitochondrial injury and fibrosis by sponging MIR6812 in lupus nephritis. Oxid Med Cell Longev. 2022;2022:2769487. doi: 10.1155/2022/2769487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen Z, He Q, Lu T, Wu J, Shi G, He L. et al. mcPGK1-dependent mitochondrial import of PGK1 promotes metabolic reprogramming and self-renewal of liver TICs. Nat Commun. 2023;14:1121–1136. doi: 10.1038/s41467-023-36651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Comelli M, Pretis I, Buso A, Mavelli I. Mitochondrial energy metabolism and signalling in human glioblastoma cell lines with different PTEN gene status. J Bioenerg Biomembr. 2018;50:33–52. doi: 10.1007/s10863-017-9737-5. [DOI] [PubMed] [Google Scholar]
- 115.Chan DC. Mitochondrial dynamics and its involvement in disease. Annu Rev Pathol. 2020;15:235–259. doi: 10.1146/annurev-pathmechdis-012419-032711. [DOI] [PubMed] [Google Scholar]
- 116.Wei PZ, Szeto CC. Mitochondrial dysfunction in diabetic kidney disease. Clin Chim Acta. 2019;496:108–116. doi: 10.1016/j.cca.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 117.Chistiakov DA, Shkurat TP, Melnichenko AA, Grechko AV, Orekhov AN. The role of mitochondrial dysfunction in cardiovascular disease: a brief review. Ann Med. 2018;50:121–127. doi: 10.1080/07853890.2017.1417631. [DOI] [PubMed] [Google Scholar]
- 118.Dard L, Blanchard W, Hubert C, Lacombe D, Rossignol R. Mitochondrial functions and rare diseases. Mol Aspects Med. 2020;71:100842. doi: 10.1016/j.mam.2019.100842. [DOI] [PubMed] [Google Scholar]
- 119.Jedynak-Slyvka M, Jabczynska A, Szczesny RJ. Human Mitochondrial RNA Processing and Modifications: Overview. Int J Mol Sci. 2021;22:7999–8021. doi: 10.3390/ijms22157999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Labno A, Tomecki R, Dziembowski A. Cytoplasmic RNA decay pathways - Enzymes and mechanisms. Biochim Biophys Acta. 2016;1863:3125–3147. doi: 10.1016/j.bbamcr.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 121.Li WM, Barnes T, Lee CH. Endoribonucleases-enzymes gaining spotlight in mRNA metabolism. FEBS J. 2010;277:627–641. doi: 10.1111/j.1742-4658.2009.07488.x. [DOI] [PubMed] [Google Scholar]
- 122.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880. doi: 10.1016/j.cell.2019.03.046. e21. [DOI] [PubMed] [Google Scholar]
- 124.Fischer JW, Busa VF, Shao Y, Leung AKL. Structure-mediated RNA Decay by UPF1 and G3BP1. Mol Cell. 2020;78:70–84. doi: 10.1016/j.molcel.2020.01.021. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jia R, Xiao MS, Li Z, Shan G, Huang C. Defining an evolutionarily conserved role of GW182 in circular RNA degradation. Cell Discov. 2019;5:45. doi: 10.1038/s41421-019-0113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao X, Ma X, Guo J, Mi M, Wang K, Zhang C. et al. Circular RNA circEZH2 suppresses transmissible gastroenteritis coronavirus-induced opening of mitochondrial permeability transition pore via targeting miR-22 in IPEC-J2. Int J Biol Sci. 2019;15:2051–2064. doi: 10.7150/ijbs.36532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lin SP, Hu J, Wei JX, Ye S, Bu J, Xu W. et al. Silencing of circFoxO3 protects HT22 cells from glutamate-induced oxidative injury via regulating the mitochondrial apoptosis pathway. Cell Mol Neurobiol. 2020;40:1231–1242. doi: 10.1007/s10571-020-00817-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not available to this article as no datasets were generated or analyzed during the current study.