Abstract
The hepatitis C virus (HCV) nonstructural 3 protein (NS3) contains at least two domains associated with multiple enzymatic activities; a serine protease activity resides in the N-terminal one-third of the protein, whereas RNA helicase activity and RNA-stimulated nucleoside triphosphatase activity are associated with the C-terminal portion. To study the possible mutual influence of these enzymatic activities, a full-length NS3 polypeptide of 67 kDa was expressed as a nonfusion protein in Escherichia coli, purified to homogeneity, and shown to retain all three enzymatic activities. The protease activity of the full-length NS3 was strongly dependent on the activation by a synthetic peptide spanning the central hydrophobic core of the NS4A cofactor. Once complexed with the NS4A-derived peptide, the full-length NS3 protein and the isolated N-terminal protease domain cleaved synthetic peptide substrates with comparable efficiency. We show that, as in the case of the isolated protease domain, the protease activity of full-length NS3 undergoes inhibition by the N-terminal cleavage products of substrate peptides corresponding to the NS4A-NS4B and NS5A-NS5B. We have also characterized and quantified the NS3 ATPase, RNA helicase, and RNA-binding activities under optimized reaction conditions. Compared with the isolated N-terminal and C-terminal domains, recombinant full-length NS3 did not show significant differences in the three enzymatic activities analyzed in independent in vitro assays. We have further explored the possible interdependence of the NS3 N-terminal and C-terminal domains by analyzing the effect of polynucleotides on the modulation of all NS3 enzymatic functions. Our results demonstrated that the observed inhibition of the NS3 proteolytic activity by single-stranded RNA is mediated by direct interaction with the protease domain rather than with the helicase RNA-binding domain.
Hepatitis C virus (HCV) is a member of the Flaviviridae and is now recognized as the major cause of both parenterally transmitted and community-acquired non-A, non-B hepatitis (30). Chronic HCV infection, which develops in more than half of afflicted individuals, has also been linked to the development of liver cirrhosis and of hepatocellular carcinoma (6). Although the rate of new infections has been significantly reduced as a result of the introduction of reliable blood tests, it has been estimated that at least 1% of the world’s population is affected by the desease (1). So far, no efficient therapy and no vaccine is available.
HCV was identified by molecular cloning in 1989 (8). The viral genome is a 9.5-kb, positive-sense single-stranded RNA (ssRNA) molecule that contains a single open reading frame encoding a polyprotein of 3,010 to 3,030 amino acids (9, 19, 35, 61). The ORF is flanked by 5′ and 3′ untranslated regions (15, 41, 62, 67, 68). The HCV polyprotein undergoes proteolytic processing by both host signal peptidases and viral proteases (3, 12, 17, 18, 26–28, 42, 43, 51, 53, 65), giving rise to at least 10 mature proteins, which are encoded on the viral RNA in the following order: NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH. C, E1, and E2 are believed to be viralstructural proteins, whereas the role of p7 has not been established. The remaining viral proteins (NS2 to NS5B) are believed to be nonstructural proteins, i.e., components of the viral replication machinery.
Whereas the structural HCV proteins arise through the action of cellular proteinases, two viral enzymes are required for the maturation of the nonstructural region of the polyprotein. The NS2-NS3 junction is cleaved by a zinc-dependent autoproteinase composed of NS2 and the N-terminal one-third of the NS3 protein (17, 27). The C-terminal remainder of the HCV polyprotein is further processed by the serine protease contained within the NS3 protein to give rise to mature NS3 (67 kDa), NS4A (6 kDa), NS4B (26 kDa), NS5A (56 to 58 kDa), and NS5B (65 kDa) proteins (3, 11, 18, 46, 65). The location of the sites cleaved by the NS3 protease within the HCV polyprotein has been obtained by N-terminal sequencing of the mature NS4A, NS4B, NS5A, and NS5B proteins. Based on a comparative analysis of the sequences flanking the cleaved peptide bonds, it has been possible to derive the following consensus sequence for the NS3-dependent cleavage site: Asp/GluXaa4Cys/Thr-Ser/Ala (18). The catalytic domain of the NS3 protease has been mapped to the N-terminal 180-amino-acid region of NS3, which contains a characteristic serine protease catalytic triad (2, 14, 24, 40, 56, 63). Although the N-terminal serine protease domain of NS3 shows enzymatic activity on its own, NS4A is a protease cofactor essential for efficient proteolytic processing. The degree of NS4A activation depends on the location of the cleavage site (2, 13, 43, 64). A 14-amino-acid, hydrophobic region of NS4A has been identified as being sufficient for the stimulation of the NS3 protease (7, 39, 44, 52, 66). In addition to the N-terminal protease domain, the C-terminal two-thirds of the NS3 protein contains conserved sequence motifs which are the hallmark of RNA helicases (16, 32). Various forms of recombinant proteins containing the C-terminal domain of NS3 have been shown to possess RNA-stimulated nucleoside triphosphatase (NTPase) (22, 49, 59) and RNA helicase (20, 31, 36, 49, 60) activities. The minimal requirement for both these activities lies in the C-terminal 465 amino acids of NS3 (36). The NS3 helicase can unwind double-stranded RNA (dsRNA) as well as dsDNA and RNA-DNA heteroduplexes in the 3′-to-5′ direction by using any nucleoside triphosphate (NTP) or dNTP as the energy source (20, 21, 60). Mutations of the conserved residues in the ATPase and helicase motifs severely impair both functions (25, 37).
The three-dimensional structures of the NS3 protease domain, both alone (45) and in complex with NS4A-derived peptides designed to include the essential NS3-binding region (38, 69), were recently determined by X-ray crystallography. Analysis of the three-dimensional structures has revealed a chymotrypsin-like fold and a structural zinc-binding site. The crystal structure of the C-terminal, 451-residue helicase domain of NS3 has also been determined (70), revealing two RecA-like domains containing the helicase motifs and a third, C-terminal domain that is unique to the HCV enzyme.
To date, the study of the HCV protease and helicase enzymatic activities on the two separate domains has proved to be a useful approach to dissect the multiple functions of the full-length protein. The major and obvious limitation is that the structural and catalytic properties of the NS3 protein may not be accurately represented by studies on independent domains. In particular, information on the possible mutual influence of the various enzymatic activities requires the characterization of a more physiologically relevant protein. To date, these kinds of studies have been hampered by the difficulties in obtaining sufficient amounts of soluble and pure full-length NS3 enzyme. Several reports (29, 47) have recently focused on the protease and RNA helicase activities associated with HCV full-length NS3-NS4A complexes partially purified at relative low yields from eukaryotic cells.
Here we describe the overexpression of a native form of full-length NS3 protein in E. coli. We report the procedure used to purify it to homogeneity in milligrams amounts and the characterization of its enzymatic properties. We have characterized the serine protease activity of the full-length NS3 protein to compare it with that of the isolated protease domain. We have also analyzed and quantified NS3 ATPase, RNA helicase, and RNA-binding activities under optimized reaction conditions. Finally, we have explored the possible interdependence of the two domains by analyzing the effect of polynucleotides and protease inhibitors on the modulation of all NS3 enzymatic functions.
MATERIALS AND METHODS
Expression and purification of the NS3 protein from bacteria.
The NS3 protease domain from HCV BK (amino acids 1027 to 1207) was expressed in Escherichia coli and purified as previously described (56). A cDNA fragment encoding the full-length (FL) NS3 polypeptide (amino acids 1027 to 1657 of the HCV BK polyprotein) was obtained by PCR and cloned downstream of the T7 promoter of the pT7-7 vector, in frame with the first ATG of the protein of gene 10 of the T7 phage. The resulting construct, pT7 NS3 FL, was sequenced with an Applied Biosystems model 373 DNA sequencer. The NS3 protein was expressed in E. coli BL21 (DE3) (58) by a method modified from the one described in reference 50. A 1-liter liquid culture derived from a single transformed bacterial colony was grown at 37°C to an absorbance at 600 nm of 0.8 in M9 modified minimal medium (5 g of glucose per liter, 1 g of ammonium sulfate per liter, 100 mM potassium phosphate [pH 7], 5 μM biotin, 7 μM thiamine, 0.5% Casamino Acids, 0.5 mM MgSO4, 0.5 mM CaCl2, 13 μM FeSO4, 50 mg of ampicillin per liter). It was then cooled to 18°C, made up 100 μM ZnCl2, and induced with 600 μM IPTG (isopropyl-β-d-thiogalactopyranoside) for 22 h at 18°C. All subsequent operations were performed at 4°C unless otherwise indicated. Cells were harvested and then disrupted with a Microfluidizer (model 110-S) in lysis buffer (25 mM HEPES [pH 7.6], 1 mM EDTA, 20% glycerol, 0.3 M NaCl, 0.1% n-octyl-β-d-glucopyranoside [Calbiochem], 3 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, Complete [Boehringer] protease inhibitor mixture). The insoluble material was pelleted at 27,000 × g for 30 min in a Sorvall SS34 rotor. The clarified supernatant, containing about 90% of the recombinant protein, was filtered through DEAE-Sepharose Fast Flow resin (Pharmacia) preequilibrated in lysis buffer. This sample was concentrated by 50% ammonium sulfate precipitation and dialyzed against lysis buffer containing 0.1 M NaCl, and NS3 was purified by fast protein liquid chromatography (Pharmacia), as follows. The dialyzed sample was loaded onto a 10-ml High Trap heparin-Sepharose column (Pharmacia) and eluted with a 0.1 to 1 M NaCl linear gradient in a buffer containing 25 mM HEPES [pH 7.6], 1 mM EDTA, 20% glycerol, 0.1% n-octyl-β-d-glucopyranoside, and 3 mM DTT (buffer A). The protein peak was detected in fractions containing approximately 0.35 M NaCl, which were then pooled, dialyzed against buffer A containing 0.2 M NaCl, and loaded onto a 2-ml poly(U)-Sepharose affinity column (Pharmacia). After a wash with 5 column volumes of the same buffer, the NS3 protein was eluted in a >95% pure form with buffer A—1 M NaCl. Protein stocks were quantified by amino acid analysis and stored at −80°C at 5 to 20 μM in buffer A–50% glycerol–0.5 M NaCl after being subjected to shock-freezing in liquid nitrogen. Control experiments had shown that this freezing procedure does not affect the NS3 protease and helicase specific activity. NS3 was >95% pure as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). N-terminal sequencing of the purified protein was performed on an Applied Biosystems model 470A gas phase sequencer.
Gel filtration chromatography.
The native molecular weight of NS3 protein purified from bacteria was determined on a Pharmacia Superdex 200 HR 10/30 column in a buffer containing 25 mM HEPES (pH 7.6), 1 mM EDTA, 10% glycerol, 0.3 M NaCl, 0.1% n-octyl-β-d-glucopyranoside, and 3 mM DTT. The flow rate was 0.3 ml/min, and 0.6-ml fractions were collected and analyzed by Western blotting. Blue dextran (2,000 kDa), aldolase (153 kDa), bovine serum albumin (67 kDa), and RNase A (13.7 kDa) were obtained from Pharmacia and used as molecular mass standards.
Peptides and HPLC protease assays.
The peptide substrate derived from the NS4A-NS4B cleavage sequence (acetylated [Ac]-DEMEEC-ASHLPYK-NH2) was purchased from Peptides International. All the other peptides were synthesized by solid-phase synthesis based on Fmoc/t-Bu chemistry, as described previously (5, 54). The identity of the peptides was determined by mass spectrometry and amino acid analysis. The concentration of stock peptide aliquots, prepared in buffered aqueous solutions and kept at −80°C until use, was determined by quantitative amino acid analysis performed on HCl-hydrolyzed samples.
If not specified differently, cleavage assays were performed in 57 μl of 50 mM Tris-HCl (pH 7.5)–2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS; Calbiochem)–50% glycerol–30 mM DTT to which 3 μl of NS4A-NS4B peptide substrate was added. As the protease cofactor, a peptide spanning the central hydrophobic core (residues 21 to 34) of the NS4A protein carrying a three-lysine solubilizing tag at the N terminus was used (5). The NS4A-derived peptide Pep4AK (KKKGSVVIVGRIILSGR-NH2) was preincubated for 15 min at 23°C with 20 nM NS3, and the reactions were started by addition of the substrate. Incubation times at 23°C were chosen to obtain <10% conversion. Reactions were stopped by the addition of 40 μl of 1% trifluoroacetic acid. Cleavage of peptide substrates was determined by high-pressure liquid chromatography (HPLC) with a Merck-Hitachi chromatograph equipped with an autosampler, as described previously (5, 56). Cleavage products were quantified by integration of chromatograms with respect to appropriate standards. Initial rates of cleavage were determined on samples with <10% substrate conversion. Kinetic parameters were calculated from nonlinear least-squares fit of initial rates as a function of substrate concentration with the aid of a Kaleidagraph software, assuming Michaelis-Menten kinetics.
The dissociation constant of the NS3-Pep4AK complex was calculated from measurements of the rate of proteolysis (V0) by nonlinear least-squares fit to the equation: V = V0 + (Vmax[Pep4AK])/(Kd + [Pep4AK]) (5).
The 50% inhibitory concentrations (IC50) of the peptides Ac-DEMEEC-OH and Ac-EDVVCC-OH and of ssRNA inhibitors were calculated from protease assay experiments performed in the presence of increasing concentrations of inhibitor. In the ssRNA inhibition experiments, either an 18-mer oligouridylic acid [oligo(U)18] (Genset) or polyuridylic acid [poly(U)] (Pharmacia) was used, and in both cases the inhibitor concentrations were expressed as UMP concentrations due to the size heterogeneity of ssRNA molecules in the poly(U) sample. The IC50s were obtained by multiparameter logistic fitting of the experimental data.
Protease assays on in vitro-translated substrates.
In vitro translation of the HCV proteins NS4A-NS4B (residues 1649 to 1964) and NS5A-NS5B ΔC51 (residues 1965 to 2470) was described previously (55).
In vitro transcription was performed with T7 RNA polymerase (Stratagene). The transcripts were translated for 1 h at 30°C in the presence of [35S]methionine (1,175 Ci/mmol; Dupont NEN) with an RNA-dependent rabbit reticulocyte lysate (Promega). Aliquots of purified NS3 were added to the translated protein substrates in the absence or presence of 15 μM Pep4AK, and the mixtures were incubated for 60 min at 30°C. Cleavage of radiolabelled precursors was assessed by SDS-PAGE followed by autoradiography.
NTPase activity assay.
NTPase activity was directly determined by monitoring [γ-32P]ATP hydrolysis by thin-layer chromatography. Protein titration assays were carried out by incubating 1.25 to 80 nM NS3 FL protein for 30 min at 37°C under standard conditions: 25 mM morpholinepropanesulfonic acid (MOPS)-NaOH (pH 7)–2.5 mM DTT–2.5 U of RNasin (Promega)–100 μg of bovine serum albumin (BSA) per ml–3 mM MgCl2–1 mM ATP–2 μCi of [γ-32P]ATP (6,000 Ci/mmol, 10 mCi/ml; Dupont NEN) with or without 0.1 mM poly(U) (see above) in a final volume of 10 μl. Poly(U) titration experiments were performed by incubating 2 nM enzyme in the presence of increasing concentrations of poly(U) (0.78 to 100 μM UMP). Portions (0.5 μl) of each reaction mixture were spotted onto polyethyleneimine-cellulose sheets and developed by ascending chromatography in 150 mM LiCl–150 mM formic acid (pH 3.0). The cellulose sheets were dried, and released [32P]phosphoric acid was quantified with a PhosphorImager by volumetric integration with ImageQuant software.
Helicase and RNA-binding assays.
The partially double-stranded RNA substrate is schematically described in Fig. 7A. The substrate was obtained by annealing the two complementary RNA oligonucleotides, 5′-AGAGAGAGAGGUUGAGAGAGAGAGAGUUUGAGAGAGAGAG-3′ (40-mer, template strand) and 5′-CAAACUCUCUCUCUCUCAACAAAAAA-3′ (26-mer, release strand), synthesized by Genset and purified on a 20% polyacrylamide–7 M urea denaturing gel. The release strand was 5′-end labelled with [γ-32P]ATP by using T4 polynucleotide kinase (Pharmacia) before the annealing reaction. The two RNA oligonucleotides were combined at a molar ratio of 3:1 (template/release), and annealing was performed by denaturation for 5 min at 80°C followed by slow renaturation at 23°C in 20 mM Tris-HCl (pH 8)–0.5 M NaCl–1 mM EDTA. The partial duplex RNA was purified on a G50-80 Sephadex spun column and stored at −20°C in H2O containing 0.25 U of RNasin (Promega) per μl.
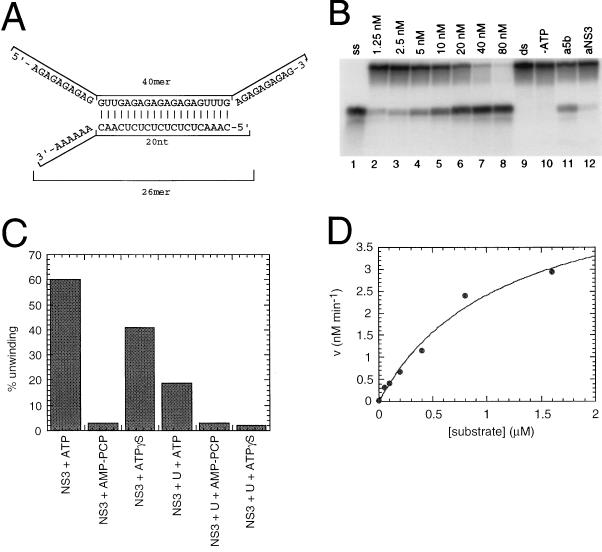
FIG. 7.
Helicase activity of full-length NS3 protein. (A) The partially dsRNA substrate used in the helicase assay. (B) An NS3 stock solution was diluted in 25 mM MOPS (pH 7)–50% glycerol–1 mg of BSA per ml to yield a series of diluted solutions. Portions (2 μl) of these solutions were incubated with 1 nM 32P-labelled partial duplex RNA substrate under standard reaction conditions, as described in Materials and Methods. Lanes: 1, sample heated in 4% formamide to yield the release strand ssRNA marker; 2 to 8, increasing concentrations of full-length NS3 protein; 9, assay performed in the absence of added NS3 protein; 10, assay performed in the absence of ATP; 11 and 12, 3 μl of protein A-Sepharose-purified antisera against NS5B and NS3, respectively, were preincubated with 10 nM NS3 protein in the reaction mixture for 15 min at 23°C, before ATP addition. (C) ATP, β,γ-methylene-ATP (AMP-PCP), or ATP(γ)S (each at 5 mM) was added after preincubation of NS3 (40 nM) with 2 nM [32P]dsRNA substrate in the absence or presence of 30 nM oligo(U)18 ssRNA. (D) The enzyme (20 nM) was incubated with increasing concentrations of unlabeled dsRNA substrate (0.019 to 0.599 μM) and with 1 nM of [32P]dsRNA. Up to 0.05 μM substrate, conversion was higher than 20%. From the 0.05 to 1.6 μM substrate data points, initial rates were calculated and plotted against substrate concentration. Assuming Michaelis-Menten kinetics, a kcat/Km of 3746 s−1 M−1 and a Km of at least 1.16 μM were determined.
Unless otherwise specified, the NS3 helicase activity assay was performed in 20 μl reaction volume containing 25 mM MOPS-NaOH (pH 7), 2.5 mM DTT, 2.5 U of RNasin, 100 μg of BSA per ml, 3 mM MgCl2, 1.25 to 80 nM NS3 protein, and 1 nM 32P-labelled partial duplex RNA substrate. After preincubation for 15 min at 23°C, 3 mM ATP was added to start the helicase reaction. This was carried out at 37°C for 30 min and then stopped by adding 5 μl of termination buffer (0.1 M Tris [pH 7.5], 20 mM EDTA, 0.5% SDS, 0.1% Nonidet P-40, 0.1% bromophenol blue, 0.1% xylene cyanol, 25% glycerol). Aliquots (8 μl) were analyzed on a native 8% polyacrylamide gel containing 0.5× Tris-borate-EDTA. Strand separation was visualized by autoradiography, and the efficiency of the helicase reaction was calculated by quantification of the radioactivity with a PhosphorImager and ImageQuant software. The percent unwinding was calculated as the ratio of the radioactivity associated with the release strand and total radioactivity associated with both the unwound substrate and the released strand.
Gel retardation reaction mixtures (20 μl) contained 25 mM MOPS-NaOH (pH 7.0), 2.5 mM DTT, 2.5 U of RNasin, 100 μg of BSA per ml, 3 mM MgCl2, 5 to 80 nM NS3 protein, and 1 nM 5′-32P-labelled 26-mer ssRNA oligonucleotide corresponding to the release strand. After being incubated for 30 min at 37°C in the absence or presence of 5 mM ATP, the samples were adjusted to 5% glycerol and were electrophoresed in a native 6% polyacrylamide gel containing 0.25× Tris-borate-EDTA.
RESULTS
Expression and purification of the full-length NS3 protein.
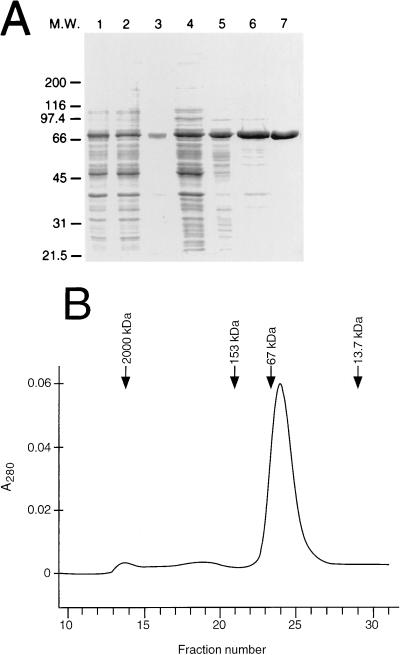
For the production of large amounts of FL NS3 protein (amino acids 1027 to 1657), we devised a method involving expression and purification from E. coli. To this end, we chose a T7-based expression vector and decided not to rely on a fusion protein system to obtain an enzyme as close as possible to the native one. In preliminary experiments, we successfully expressed this protein in E. coli by using the standard Luria-Bertani medium for bacterial cultures and a temperature of 37°C for the IPTG induction of expression. Nevertheless, the enzyme produced by this method was quantitatively found in an insoluble form (data not shown). Therefore, we tried to push the system toward the expression of a more soluble protein and to minimize the formation of inclusion bodies, normally associated with the presence of large amounts of misfolded protein. To this end, transformed bacterial cultures were grown in a defined modified minimal medium and after exponential growth at 37°C to the desired optical density, induction with IPTG was carried out at 18°C for 22 h. Similar methods have been demonstrated to significantly improve the yields and to simplify the purification of several proteins (50). In addition, to provide a source of the structural zinc required for the proper folding of the enzyme (10, 57), 100 μM ZnCl2 was added just before induction to minimize misfolding. About 90% of NS3 (as shown in Fig. 1A and in Western blot experiments [data not shown]) was recovered in a soluble form upon disruption of cells in a glycerol- and 0.1% n-octyl-β-d-glucopyranoside-containing hypertonic buffer. Only glycerol was strictly required for solubility, whereas the presence of detergents was optional. After filtration through DEAE-Sepharose to separate nucleic acids and concentration by ammonium sulfate precipitation, NS3 was further purified by two subsequent chromatographic steps on High Trap heparin-Sepharose and poly(U)-Sepharose (Fig. 1A). The latter purification step was very effective due to the high affinity of the enzyme for ssRNA (see below). The purified enzyme was homogeneous, as judged by SDS-PAGE (Fig. 1A). Experimental sequence analysis revealed the removal of the N-terminal methionine and alanine residues, yielding the sequence P-I-T-A-Y-S-S-Q, analogous to what has already been observed in the purified NS3 protease domain (56). This method yielded about 8 mg of purified NS3 from 1 liter of culture at a concentration of 1.5 mg/ml.
FIG. 1.
Purification of the full-length NS3 protein from E. coli. (A) SDS-PAGE analysis of the purification steps. Lanes: 1, total extract; 2, soluble fraction; 3, inclusion bodies; 4, DEAE-Sepharose flowthrough; 5, 50% ammonium sulfate cut; 6, High Trap heparin-Sepharose; 7, poly(U)-Sepharose. M.W., molecular mass markers in kilodaltons. (B) Gel filtration analysis. A UV elution profile of a Superdex 200 analytical column is shown. Fractions of 0.6 ml were collected. The peaks in the chromatogram correspond to the monomeric and aggregated NS3 protein identified by Western blotting. The positions of the peaks corresponding to the molecular mass standard proteins are indicated by arrows. A280, absorbance at 280 nm.
To investigate the multimerization or aggregation state of NS3, 500 μg of a purified preparation was applied to a Pharmacia Superdex 200 gel filtration column. The elution profile was monitored by measuring UV absorbance at 280 nm (Fig. 1B), and the eluted fractions were analyzed by Western blotting with a specific anti-NS3 antiserum (data not shown). More than 95% of NS3 eluted in close proximity to the BSA (67-kDa) marker, in agreement with the predicted molecular mass of NS3 monomeric form (67 kDa). Only a small fraction of the protein eluted in the column void volume (≥ 2,000 kDa). These data indicate that the purification conditions used yielded a monomeric, nonaggregated protein.
Protease activity on in vitro-translated substrates.
To assay the trans-cleavage activity of the purified NS3 on HCV polyprotein precursors, we incubated the enzyme at 100 nM with 35S-labelled precursor proteins NS4A-NS4B and NS5A-NS5BΔC51, synthesized by in vitro translation from the appropriate RNAs. To assess the dependency of NS3 protease activity on the NS4A cofactor, the experiments were performed in the presence or absence of a large excess (15 μM) of an NS4A-derived peptide, spanning the central hydrophobic core (residues 21 to 34) of the NS4A protein (Pep4AK). As shown in Fig. 2, no cleavage was detectable on either protein substrate in the absence of the cofactor whereas processing was evident in the samples in which Pep4AK was added. Similar results were obtained when the full-length NS5A-NS5B precursor was used as the substrate (data not shown).
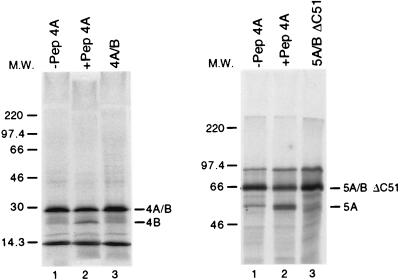
FIG. 2.
Full-length NS3 protease activity on in vitro-translated precursor substrates. NS4A-NS4B and NS5A-NS5BΔC51 precursor proteins were synthesized by in vitro translation of the corresponding RNAs in the presence of [35S]methionine, as described in Materials and Methods. An NS3 stock solution was diluted to 143 nM in 7 μl of 25 mM HEPES (pH 7.5)–1 mM EDTA–50% glycerol–3 mM DTT and preincubated in the absence or presence of 15 μM Pep4AK for 10 min at 23°C. Then 3 μl of the appropriate in vitro-translated precursor was added to the protein and the mixture was incubated for 1 h at 30°C. Reactions were terminated by the addition of 20 μl of SDS sample buffer, and 10 μl-aliquots were analyzed by SDS-PAGE followed by autoradiography. Lanes: 1, reactions in the absence of Pep4AK; 2, reactions in the presence of Pep4AK; 3, control samples in the absence of NS3 protein and in the presence of Pep4AK. Bands corresponding to the 5A/BΔC51 and 4AB substrates and to the 5A and 4B products are indicated. The complementary products (5BΔC51 and 4A, respectively) were not detected in the gel system used because of the small size. M.W., molecular mass markers in kilodaltons.
This stringent requirement for NS4A in NS4A-NS4B and NS5A-NS5B trans processing in vitro was not detected with the isolated NS3 protease domain in analogous experiments (55), but our finding is in agreement with the study of Hamatake et al. (23), who compared the trans-cleavage activity of purified FL NS3 and NS3-NS4A complex in the same type of assay.
Protease activity on synthetic peptide substrates.
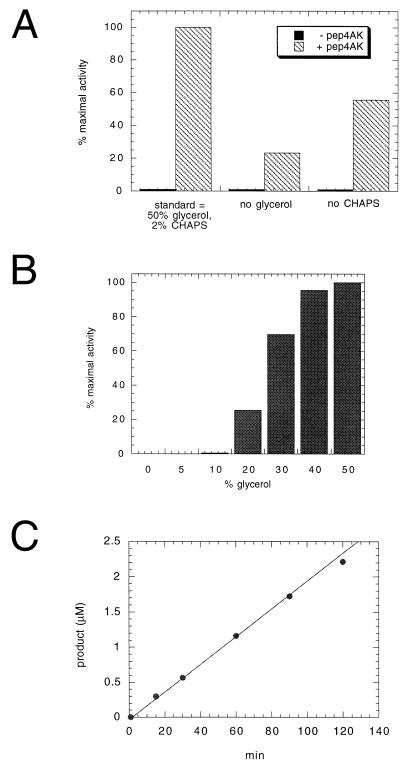
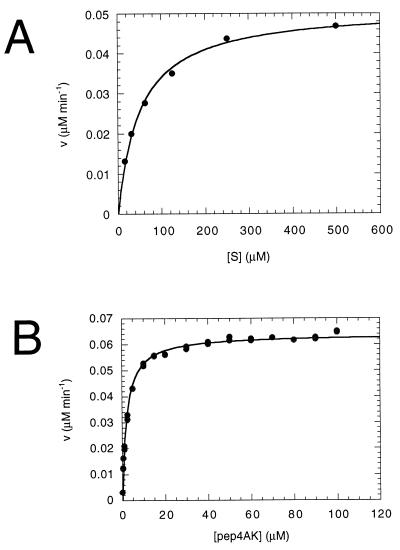
To determine whether the purified NS3 protein was enzymatically active on a synthetic peptide substrate, aliquots were incubated with the 13-mer peptide Ac-DEMEECASHLPYK-NH2, derived from the NS4A-NS4B cleavage site (5). The cleavage efficiency was strictly dependent on the presence of a saturating concentration of an NS4A-derived synthetic peptide (Pep4AK) (Fig. 3A). The NS4A-derived core peptide had been demonstrated to increase the cleavage efficiency of the isolated NS3 protease domain on similar NS4A-NS4B peptide substrates (5, 55, 56). In contrast to FL NS3, the isolated protease domain was previously shown to possess a significant level of basal activity when analyzed under the same experimental conditions (55, 56). The cleavage efficiency of the Pep4AK-activated enzyme was only partially dependent on the presence of detergents (e.g., 2% CHAPS) but was negatively affected by the omission of glycerol from the assay mixture (Fig. 3A). In the presence of the cofactor, a glycerol-dependent activity increase was observed, with a plateau value between 40 and 50% glycerol (Fig. 3B). We then analyzed the time course of the NS3-catalyzed cleavage of NS4A-NS4B peptide at a substrate concentration of 50 μM (Km [see Table 1]) and 20 nM enzyme. No significant loss of activity was observed for up to 2 h of incubation (Fig. 3C), indicating that the enzyme was substantially stable under the assay conditions used. A minor deviation from linearity was visible at the 2-h time point due to product inhibition, which starts to be significant at sufficiently high substrate conversion (see below).
FIG. 3.
Dependence of full-length NS3 protease activity on glycerol, CHAPS, Pep4AK, and time course of the proteolysis reaction. NS3 protein (20 nM) was incubated with 50 μM NS4A-NS4B peptide substrate in the absence or presence of 16 μM Pep4AK in 50 mM Tris (pH 7.5)–30 mM DTT. The reactions were stopped by addition of 1% trifluoroacetic acid, and cleavage products were analyzed and quantified by HPLC. (A) Standard optimized conditions included 50% glycerol and 2% CHAPS. The effect of omitting either CHAPS or glycerol was evaluated in the absence and presence of Pep4AK. Reactions were carried out at 23°C for 60 min. (B) CHAPS (2%) and increasing concentrations of glycerol in the presence of Pep4AK were analyzed at 23°C for 60 min. (C) Increasing incubation times under standard optimized conditions and in the presence of Pep4AK were analyzed.
TABLE 1.
Comparison between FL NS3 and N-terminal domain protease activities
| NS3 protein | Kcat (min−1) | Km (μM) | Kcat/Km (M−1 s−1) | Kd Pep4AK (μM) | Fold activation by Pep4AK |
|---|---|---|---|---|---|
| FL | 5.14 | 50.9 | 1,683 | 2 | 25 |
| N-terminal domain | 3.5 | 40 | 1,458 | 5 | 7 |
Kinetic analysis of NS3 protease activity.
We performed a kinetic analysis of the cleavage reaction of NS4A-NS4B peptide substrate under the optimized conditions, i.e., in 50 mM Tris (pH 7.5)–50% glycerol–2% CHAPS–30 mM DTT in the presence of the NS4A cofactor (Fig. 4A), and we found that Km was 50.9 μM and kcat was 5.14 min−1. These values are similar to those observed with the recombinant protease domain alone under the same reaction conditions (5). The Pep4AK dependence of the enzymatic activity was used to determine the dissociation constant of the NS3-Pep4AK complex. From the titration curve in Fig. 4B, we calculated a Kd of 2 μM, i.e., 2.5-fold lower than the Kd calculated for the NS3 protease domain-Pep4AK complex (5). Due to the very low basal proteolytic activity associated with the full-length NS3 protein, the maximum degree of activation induced by Pep4AK peptide was 25-fold, a value significantly higher than the 7-fold activation measured with the protease domain alone (56). Table 1 shows a comparison between the kinetic parameters obtained with FL NS3 and with the isolated protease domain. Similarly to the protease domain, both the affinity of FL NS3 for the NS4A-NS4B substrate and the dissociation constant for Pep4AK were negatively affected by lowering the glycerol concentration (Km > 100 μM and Kd = 8.8 μM in 15% glycerol [data not shown]).
FIG. 4.
Steady-state kinetic analysis of NS4A-NS4B cleavage by full-length NS3 and kinetic determination of the dissociation constant of the NS3-Pep4AK complex. Full-length NS3 (10 nM) was incubated under standard conditions at 23°C for 60 min. (A) Pep4AK (16 μM) and increasing concentrations of NS4A-NS4B peptide substrate were added. Six data points at substrate concentrations between 15 and 500 μM were used to calculate the kinetic parameters. Initial rates of cleavage were determined on samples with <10% substrate conversion. Kinetic parameters (Km = 50.9 μM, and kcat = 5.14 min−1) were calculated from a nonlinear least-squares fit of initial rates as a function of substrate concentration, assuming Michaelis-Menten kinetics. (B) NS4A-NS4B peptide substrate (50 μM) was added. NS4A peptide concentrations were varied between 0.2 and 100 μM, and 17 duplicate data points were determined. The dissociation constant of the NS3-Pep4AK complex (Kd = 2 μM) was calculated from a nonlinear least-squares fit to the equation V = V0 + (Vmax[Pep4AK]) / (Kd + [Pep4AK]).
Inhibition of NS3 protease activity.
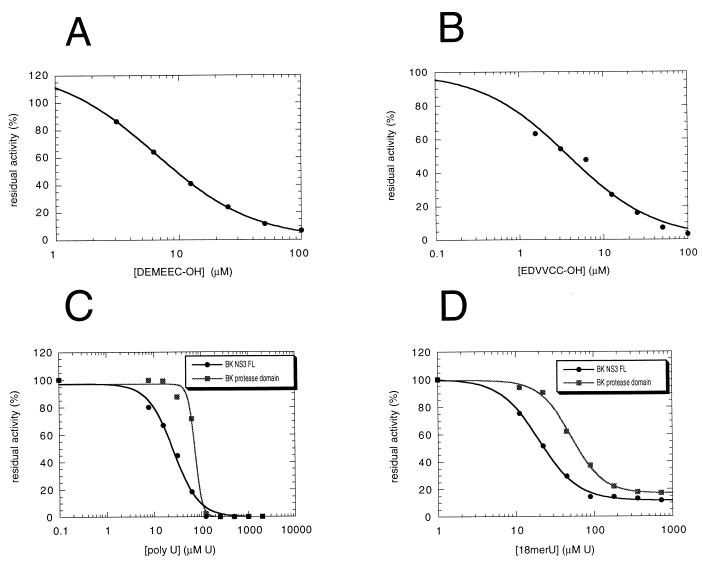
It has recently been reported that hexamer synthetic peptides derived from the P-side fragments of both the NS4A-NS4B (Ac-DEMEEC-OH) and the NS5A-NS5B (Ac-EDVVCC-OH) cleavage sites are micromolar inhibitors of the NS3 protease domain (54). On this basis, it has been suggested that the NS3 protease is subjected to significant product inhibition. To assess whether product inhibition also affects the full-length enzyme, we have performed inhibitor titration experiments on NS3 protease activity in the presence of Pep4AK cofactor with the NS4A-NS4B substrate (Fig. 5A and B). We have found that both product-derived peptides inhibited the NS3 activity with IC50s in the low micromolar range (6.4 μM for Ac-DEMEEC-OH and 3.8 μM for Ac-EDVVCC-OH).
FIG. 5.
Inhibition of NS3 protease activity by P6-P1 product peptides and by ssRNA. (A and B) Full-length NS3 (20 nM) trans-cleavage activity was assayed under standard conditions on 50 μM NS4A-NS4B peptide substrate at 23°C for 60 min after preincubation with 16 μM Pep4AK and in the presence of increasing concentrations of inhibitor. Seven data points at product peptide concentrations between 1.5 and 100 μM were used. (A) Titration of Ac-DEMEEC-OH. IC50 = 6.4 μM. (B) Titration of Ac-EDVVCC-OH. IC50 = 3.8 μM. (C and D) A standard cleavage assay was performed with either FL NS3 or the isolated protease domain (20 nM) in the presence of increasing concentrations of poly(U) or oligo(U)18. These were expressed as UMP (U) concentration because of the size heterogeneity of the RNA molecules contained in the poly(U) sample. (C) Poly(U) titration. IC50 = 25.4 μM UMP for FL NS3; IC50 = 73.7 μM UMP for the NS3 protease domain. (D) Oligo(U)18 titration. IC50 = 20 μM UMP for FL NS3; IC50 = 51.7 μM UMP for the protease domain.
Since RNA homopolymers are known to modulate the NTPase-helicase activity of both the isolated helicase domain (31, 49, 59, 60) and the full-length protein (29, 47), we were interested in exploring whether RNA could affect also the protease activity of our FL NS3 enzyme under the controlled reaction conditions established for the HPLC assay. To this end, we performed the standard NS3 cleavage assay in the presence of increasing concentrations of either poly(U) or an 18-mer oligo(U)18. Interestingly, both RNA molecules significantly inhibited FL NS3 protease with similar IC50s of 25.4 μM UMP for the poly(U) and 20 μM UMP for oligo(U)18 (Fig. 5C and D). Expressed in terms of RNA molecule concentration, the corresponding IC50s were 85 nM for poly(U), assuming an average length of about 300 nucleotides, and 1.1 μM for oligo(U)18. To investigate whether this RNA-mediated inhibitory effect was due to the binding of ssRNA to the helicase domain of NS3 protein, we performed similar RNA titration experiments on the isolated NS3 protease domain (Fig. 5C and D). Surprisingly, a strong inhibitory effect was evident also in this case, with IC50s only 2.5- to 3-fold higher than those measured with the full-length NS3 enzyme, i.e., 74 μM UMP for poly(U) and 51.7 μM UMP for oligo(U)18, respectively. This result suggests that the inhibition observed might be mediated by a different RNA-binding site located in NS3 protease domain. Interestingly, when 5 mM ATP was added to the poly(U) titration experiments (data not shown), a threefold decrease in the inhibition potency was observed for full-length NS3 (IC50 = 75 μM UMP) whereas the IC50 for the protease domain remained constant at 74 μM [UMP], suggesting that binding of RNA to the helicase region might participate in the inhibitory effect on the full-length enzyme.
NS3 ATPase activity.
Full-length NS3 displayed poly(U)-dependent ATPase activity, which was proportional to the concentration of enzyme added (Fig. 6A). In the linear range of the titration (up to 2 nM protein), NS3 hydrolyzed 17 fmol of ATP per fmol of enzyme in the absence of poly(U) and 136 fmol of ATP per fmol of enzyme in the presence of 0.1 mM poly(U) over the course of a 30-min reaction. The degree of stimulation induced by poly(U) was approximately eightfold. Poly(U) titration experiments (Fig. 6B) revealed that the concentration required to reach half-maximal ATPase activity was 0.5 μM UMP. This value was 50-fold lower than the IC50 (25.4 μM UMP) calculated for the inhibition of the NS3 protease activity. This would suggest that at polynucleotide concentrations which effectively stimulate the ATPase activity of NS3, its protease activity should not be affected.
FIG. 6.
RNA-stimulated ATPase activity of full-length NS3 protein. The ATPase activity was analyzed by incubating the NS3 enzyme for 30 min at 37°C under standard helicase conditions: 25 mM MOPS-NaOH (pH 7)–2.5 mM DTT–2.5 U of RNasin–100 μg of BSA per ml–3 mM MgCl2. Then 1 mM ATP hot-cold mix containing 2 μCi of [γ-32P]ATP was added in a final volume of 10 μl. (A) Protein titration assays were carried out by incubating 1.25 to 40 nM of NS3 enzyme with or without 0.1 mM poly(U) (UMP). Protein dilutions were as in Fig. 7. The initial rates were determined on samples with <30% substrate conversion. (B) RNA titration experiments were performed by incubating 2 nM enzyme in the presence of increasing concentrations of poly(U) (0.78 to 50 μM UMP). The dissociation constant of the NS3-poly(U) complex (Kd = 0.5 μM UMP) was calculated from the nonlinear least-squares fit to the equation V = V0 + (Vmax[U] / (Kd + [U]).
NS3 helicase activity.
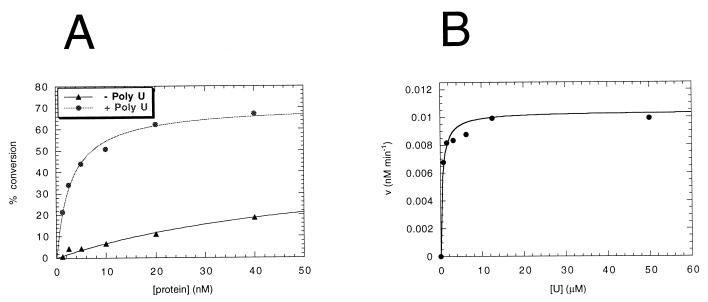
The full-length NS3 helicase activity was measured with the partially dsRNA substrate shown in Fig. 7A. Titration of NS3 under optimized reaction conditions showed that this enzyme displayed linear helicase activity up to 40 nM protein, where it reached a plateau at about 80% strand displacement (Fig. 7B). Similar results were obtained with the corresponding partially dsDNA substrate (data not shown). The addition of NS3-specific polyclonal antibodies (65) significantly reduced the strand displacement efficiency (Fig. 7B, lane 12), demonstrating that no contaminants in the NS3 preparation contributed to the unwinding activity shown in our experiments. In the absence of ATP, the RNA substrate was not affected by addition of the enzyme (lane 10). Moreover, substitution of ATP with a nonhydrolyzable ATP analog (β,γ-methylene-ATP) reduced unwinding to background levels (Fig. 7C). When the oligo(U)18 shown to affect the protease activity of NS3 was added to the helicase reaction mixture at a 15-fold molar excess relative to the dsRNA substrate, strand displacement was decreased to 33%. The same amount of oligo(U)18 inhibited the unwinding activity to background levels when ATP was replaced with the poorly hydrolyzed analog ATP(γ)S, which by itself reduced unwinding to 66% of the control value (Fig. 7C). Addition of the Ac-DEMEEC-OH product inhibitor to the strand-displacement reaction did not affect NS3 helicase activity at peptide concentrations up to 120 μM (data not shown).
To evaluate the catalytic efficiency of the unwinding activity associated with NS3, we performed a strand displacement experiment in which increasing concentrations of the dsRNA substrate (0.02 to 1.6 μM) were added to 20 nM enzyme (an enzyme concentration which showed 50% of unwinding on 1 nM substrate) in the assay. We analyzed substrate titration values from 0.05 to 1.6 μM (Fig. 7D), at which conversion was less than 20%. From the theoretical curve which fitted our experimental data, we could calculate a catalytic efficiency value (kcat/Km) of 3746 s−1 M−1 and a Michaelis constant (Km) of at least 1.16 μM. From the experiment in Fig. 7D, it is also possible to deduce that the amount of product generated is always in excess with respect to amount of the enzyme used in the assay. This observation implies that the enzyme is undergoing multiple dsRNA-unwinding cycles.
Analysis of the helicase reaction conditions.
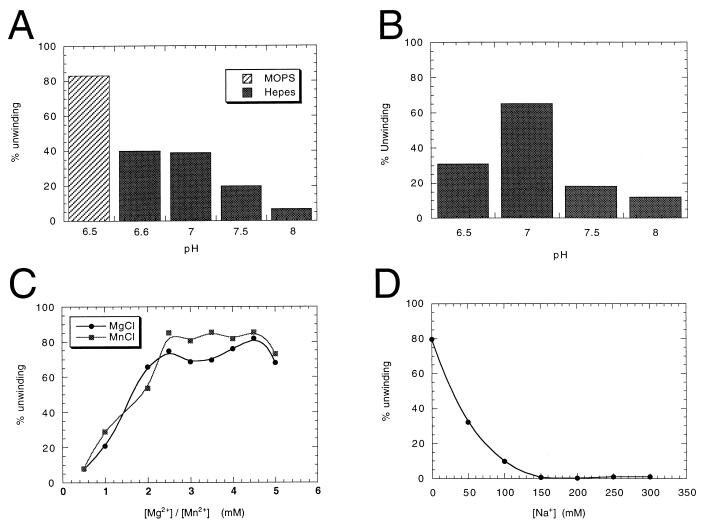
Since the reaction conditions used in different laboratories to assay the NS3 helicase activity are quite different (20, 29, 31, 47, 49, 59, 60), we determined the optimal buffer, pH, and divalent and monovalent cation concentrations for the maximal helicase activity of the NS3 enzyme purified in our laboratory (Fig. 8). First, we found that NS3 strand displacement efficiency was affected not only by the pH of the reaction, but also by the nature of the buffer used (Fig. 8A and B). MOPS was the optimal buffer among those we analyzed (Fig. 8A and data not shown), whereas HEPES (and Tris [data not shown]) reduced the activity at all pHs tested. With MOPS, the optimal pH for the reaction was 7.0 (Fig. 8B). We found that divalent cations, either Mg2+ or Mn2+, were strictly required and that optimal concentrations were at least 2.5 mM (Fig. 8C). On the other hand, the presence of increasing concentrations of monovalent cations dramatically decreased the level of helicase activity (Fig. 8D). In fact, the maximal activity was obtained in the absence of sodium ions. As described in Materials and Methods, the optimized values were used in all subsequent strand displacement experiments.
FIG. 8.
Optimization of helicase reaction conditions for the full-length NS3 protein. The activity was quantified by determining the percentage of unwinding on 1 nM dsRNA substrate by 40 nM enzyme, as described in Materials and Methods. (A) MOPS (pH 6.5) or HEPES (pH 6.6, 7, 7.5, and 8) (each at 25 mM) compared in reactions performed at 0 mM NaCl, 3 mM MgCl2, 3 mM ATP. (B) Determinations of optimal pH were performed in 25 mM MOPS (pH 6.5, 7, 7.5, and 8), as in panel A. (C) Mg2+ and Mn2+ titration was performed in 25 mM MOPS (pH 7) at 0 mM NaCl and 3 mM ATP. (D) Na+ titration was performed in 25 mM MOPS (pH 7)–3 mM MgCl2–3 mM ATP.
NS3 RNA-binding activity.
We analyzed the RNA-binding activity of full-length NS3 under helicase-optimized reaction conditions on the 5′-32P-labelled 26-mer ssRNA oligonucleotide corresponding to the release strand. The gel retardation experiment (Fig. 9) demonstrated that in the absence of ATP, NS3 is able to form a stable complex with the ssRNA probe in a concentration-dependent manner. Titration of NS3 RNA-binding activity paralleled titration of NS3 helicase activity (Fig. 7B), showing linearity up to 40 nM protein, where saturation of the free probe was observed. In the presence of 5 mM ATP, the formation of the complex was strongly reduced at all protein concentrations tested. This result suggests that under standard helicase reaction conditions, ATP is required to dissociate NS3 from RNA, implying that high-affinity binding to ssRNA is mediated by the RNA-binding domain in the helicase portion of the protein. Similar data were obtained with oligo(U)18 as the ssRNA probe (data not shown). NS3 protein was also able, in the absence of ATP, to form a stable complex on the partially dsRNA helicase substrate, which was completely dissociated by adding a 15-fold molar excess of oligo(U)18 (data not shown).
FIG. 9.
Binding activity of full-length NS3 on ssRNA. Increasing concentrations (5 to 80 nM) of NS3 protein were analyzed in binding reactions performed in helicase reaction mixtures (20 μl) containing 1 nM 5′-32P-labelled 26-mer ssRNA oligonucleotide corresponding to the release strand. After a 30-min incubation at 37°C in the absence (lanes 1 to 5) or presence (lanes 6 to 10) of 5 mM ATP, 6-μl aliquots were electrophoresed in a native 6% polyacrylamide gel containing 0.25× Tris-borate-EDTA. Lane 11 contains a control reaction mixture in the absence of NS3 protein and ATP.
DISCUSSION
We describe here a procedure for a large-scale production of the full-length NS3 protein from the 1b HCV genotype in E. coli and the characterization of its enzymatic activities. We have previously produced the NS3 protein in insect cells by using a baculovirus expression system (data not shown). Although the enzyme purified from eukaryotic cells possessed an efficient serine protease and NTPase/helicase activity in vitro, the total amount of pure NS3 protein obtained in this experiment was not satisfactory. Therefore, we devised a method of producing milligram amounts of the enzyme in a soluble form in bacteria. To this end, in contrast to previously published reports (25, 33), we successfully expressed NS3 as a nonfusion protein to obtain an enzyme as close as possible to the native viral enzyme. Indeed, our biochemical data demonstrated that the polypeptide obtained from prokaryotic cells was completely indistinguishable from that expressed in eukaryotic systems in terms of proper folding, oligomerization, and enzymatic activity.
Although it is likely that NS3 is stably associated with its NS4A cofactor in HCV-infected cells, we decided to produce the mature form of NS3 without NS4A for two reasons. First, we were interested in the possibility of investigating the effect of the NS4A cofactor on the protease activity of FL NS3 by using the same synthetic NS4A-derived core peptide which had been demonstrated to increase the cleavage efficiency of the isolated protease domain (5, 55, 56). This has allowed a direct comparison of the proteolytic properties of the full-length and truncated enzymes. Second, we also expressed and purified the NS3-NS4A native complex (unpublished data), and FL NS3 would provide a useful tool to study how all the enzymatic activities of NS3 are influenced by the presence of the full-length NS4A protein.
The synthesis of soluble protein in bacteria was crucially temperature dependent, with induction above 23°C leading quantitatively to the formation of insoluble aggregates. Protein induction at 18°C, together with the use of a defined modified minimal medium, significantly improved the yield of soluble protein obtained, by minimizing the amount of misfolded protein. The purification scheme described in this work led to several milligrams of >95% pure NS3 from 1 liter of culture, an amount suitable both for enzymological and structural studies and for screening of potential enzyme inhibitors.
Deletion experiments have shown that the helicase and protease domains of NS3 can work independently of each other, with the separate polypeptide chains expressing the respective activities (2, 14, 20, 24, 31, 36, 40, 49, 56, 60, 63). This has recently permitted the crystallization of the NS3 protease and helicase domains (38, 45, 69, 70). Despite the apparent independence of the two enzymatic activities, there is no evidence indicating that the corresponding two regions of the NS3 protein are cleaved during the virus life cycle. This could simply reflect economical packaging of essential viral replicative components, but it could also suggest that there is functional interdependence between the two domains. The analysis of the relationship between the two enzymatic activities in the context of the full-length NS3 protein could provide an insight into the physiological role of NS3 during HCV viral infection. Therefore, we have tried to ascertain whether the two domains have any functional overlaps or have any interdependent regulation by any cofactor. Our study demonstrated that the enzymatic activities associated with recombinant full-length NS3, namely, protease, ATPase, and helicase, do not differ significantly from those associated with the protein individual domains.
The catalytic efficiencies displayed by the full-length protein and by the isolated protease domain in a trans-cleavage assay under the same experimental conditions were quite comparable, due both to similar turnover numbers (kcat) and to similar affinities for the same NS4A-NS4B substrate (Km) (Table 1). Activity titration of FL NS3 with Pep4AK indicated an apparent dissociation constant in the low micromolar range, also comparable to that measured for the N-terminal protease domain alone. The trans-cleavage activity of the full-length NS3 protein was increased about 25-fold in the presence of an NS4A-derived peptide, a value significantly higher than the 7-fold activation measured with the isolated protease domain. This difference was due to the extremely low basal activity associated with the full-length protein compared with its N-terminal domain. The reason for this more stringent requirement for NS4AK in the trans processing of the NS4A-NS4B substrate is unknown. The NS4A cofactor might be more crucial in the stabilization of the active conformation of the full-length protein under our in vitro conditions. Structural studies are needed to clarify this point. On the other hand, titration of NS4AK peptide in the strand displacement assay caused a concentration-dependent inhibition of the helicase activity, with an IC50 of approximately 2 μM, consistent with the apparent dissociation constant measured in the protease activity assay (data not shown). This data might indicate that stabilization of the enzyme in the active protease conformation would affect its capacity to unwind dsRNA. This result would support a model in which NS3 could assume two alternative, mutually exclusive active conformations in the presence or absence of NS4A cofactor. Control experiments to demonstrate the specificity of this inhibitory effect are in progress, together with the study of the helicase activity associated with the native NS3-NS4A complex.
The isolated protease domain of NS3 is inhibited by synthetic peptides derived from the P6-P1 sequence of NS4A-NS4B and NS5A-NS5B cleavage sites. The N-terminal cleavage products were demonstrated to bind to the enzyme active site with low micromolar affinities, comparable to or higher than those of the corresponding substrates. Although the mechanism of this product inhibition has recently been clarified (54), the question whether this phenomenon has any physiological relevance is still open to debate. Hexapeptide product inhibitors based on the NS4A-NS4B and NS5A-NS5B cleavage sites were similarly found to efficiently inhibit the serine protease activity of full-length NS3. Interestingly, the same peptides did not affect the helicase activity of the protein at any of the concentrations tested. This result suggests that complete inhibition of the NS3 protease activity has no influence on its helicase activity.
Notably, poly(U) and oligo(U)18 RNA molecules significantly inhibited FL NS3 protease activity with similar potency. Our results are in contrast to those described in a recent study (47), which showed a poly(U)-mediated fivefold stimulation of the protease activity of a recombinant NS3-NS4A complex on an in vitro-translated NS5A-NS5B substrate. We have tried to reproduce this RNA-mediated activation on our full-length NS3 trans-cleavage activity by using both NS4A-NS4B and NS5A-NS5B in vitro-translated substrates and Pep4AK as a cofactor, but we did not see any effect of adding either poly(U) or oligo(U) at concentrations up to 2 mM UMP and 180 μM UMP, respectively (data not shown). This discrepancy might be explained by a different mode of interaction with RNA between the NS3 complexed with the synthetic peptide and the native NS3-NS4A complex used by Morgenstern et al. (47). Furthermore, the RNA-mediated inhibitory effect observed in this study did not occur through the binding of ssRNA to the helicase domain. Indeed, similar inhibitory effects were also evident on the isolated protease domain, with IC50s only 2.5- to 3-fold higher than those measured with the full-length NS3 enzyme. Our results suggest that the inhibition observed might be mediated by a different RNA-binding site, located in the NS3 protease domain. The mechanism of inhibition is unknown, but experiments are in progress to define the position of this RNA recognition site and its relationship to the active site and the substrate-binding site within the NS3 protease domain. One attractive hypothesis is that the RNA might interact with a positively charged surface in the proximity of the specificity pocket, possibly at a site which may be involved in the recognition of the acidic residues in the P6 region of protease substrates. The physiological significance of the observed RNA-protease domain interaction remains unknown. Interestingly, the recent resolution of the structure of two picornavirus 3C chymotrypsin-like proteases (hepatitis A virus and poliovirus) (4, 48) has revealed that a well-defined surface with a strongly charged electrostatic potential might participate in the recognition of the 5′ and 3′ untranslated regions of the RNA virus genome.
We found that the full-length NS3 ATPase-associated activity displayed a greater sensitivity to polynucleotide stimulation than was observed with the C-terminal helicase domain produced in E. coli (31, 49), although it was comparable to that shown by a NS3-NS4A complex purified from COS cells (47). This difference in poly(U) sensitivity therefore seems to represent a distinct feature of the full-length enzyme relative to the C-terminal helicase domain. The poly(U) concentration required to reach half-maximal NS3 ATPase activity was 50-fold lower than that required for the inhibition of half-maximal protease activity. Consistent with the protease inhibition data, this result would also support the existence of an independent RNA-binding site on the NS3 protease domain.
The helicase activity of our enzyme was evaluated by using a partially dsRNA synthetic oligosubstrate, which allowed a better quantification of results due to the reproducibility in the RNA oligonucleotide labelling and annealing procedure. Quantification of the strand displacement activity data provided a catalytic efficiency value (kcat/Km) of about 3,700 s−1 M−1, indicating a highly active enzyme preparation. Moreover, our data show that an NS3 helicase molecule can undergo multiple rounds of dsRNA unwinding. As expected, we observed that oligo(U)18 RNA had an inhibitory effect on NS3 helicase activity, probably resulting from a binding competition between the oligo(U)18 and the 3′ single-stranded region of the RNA substrate for the same binding site on NS3.
Our results, taken together, demonstrated that the ATPase-RNA helicase activity of full-length NS3 does not substantially differ from that of the C-terminal helicase domain analyzed by several groups (20, 22, 31, 36, 49, 59, 60) both in terms of optimal reaction conditions (pH, temperature, and divalent-cation dependence, inhibition by monovalent cations, etc.) and in terms of specific activity. Furthermore, we directly compared the efficiency of unwinding of full-length NS3 and that of a recombinant C-terminal helicase domain (a kind gift of G. Heilek) (25) under our standard assay conditions and found that the two enzymes possess similar strand displacement efficiencies, indicating that the N-terminal protease domain has little, if any, effect on the helicase activity (data not shown).
The ability of the HCV helicase to interact preferentially with the HCV genomic or antigenomic RNA has not been addressed in this study. We have shown that in the absence of ATP, NS3 protein binds ssRNA tightly with no particular sequence specificity, analogous to the results reported for the isolated helicase domain (20, 49). From these experiments, we could not evaluate whether the protease region does influence the ability of the full-length enzyme to selectively bind RNA. A recent report has indicated that the HCV helicase may bind preferentially to the poly(U) sequence near the 3′ end of the viral genome (34, 62). It will be of interest to assess whether full-length NS3 shows a selectively of binding to the 3′ ends of the HCV positive and negative strands, which are presumably the initiation sites for negative- and positive-strand RNA synthesis, respectively. Alternatively, protein-protein interaction could confer specificity of binding to the NS3 protein upon recruitment in the replication complex.
ACKNOWLEDGMENTS
We thank A. Pessi and E. Bianchi for peptide synthesis, P. Neuner for oligonucleotide synthesis, R. Petruzzelli for N-terminal sequence analysis, and M. Emili for artwork. We are grateful to G. Heilek for the gift of the recombinant NS3 helicase domain.
REFERENCES
- 1.Alter H J. To C or not to C: these are the questions. Blood. 1995;85:1681–1695. [PubMed] [Google Scholar]
- 2.Bartenschlager R, Ahlborn Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann E M, Mosimann S C, Chernaia M M, Malcolm B A, James M N. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J Virol. 1997;71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchi E, Urbani A, Biasiol G, Brunetti M, Pessi A, De Francesco R, Steinkühler C. Complex formation between the hepatitis C virus serine protease and a synthetic NS4A cofactor peptide. Biochemistry. 1997;36:7890–7897. doi: 10.1021/bi9631475. [DOI] [PubMed] [Google Scholar]
- 6.Bisceglie A M. Hepatitis C and hepatocellular carcinoma. Semin Liver Dis. 1995;15:64–69. doi: 10.1055/s-2007-1007263. [DOI] [PubMed] [Google Scholar]
- 7.Butkiewicz N J, Wendel M, Zhang R, Jubin R, Pichardo J, Smith E B, Hart A M, Ingram R, Durkin J, Mui P W, Murray M G, Ramanathan L, Dasmahapatra B. Enhancement of hepatitis C virus NS3 proteinase activity by association with NS4A-specific synthetic peptides: identification of sequence and critical residues of NS4A for the cofactor activity. Virology. 1996;225:328–338. doi: 10.1006/viro.1996.0607. [DOI] [PubMed] [Google Scholar]
- 8.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 9.Choo Q L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina Selby R, Barr P J, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Francesco R, Urbani A, Nardi M C, Tomei L, Steinkühler C, Tramontano A. A zinc binding site in viral serine proteinases. Biochemistry. 1996;35:13282–13287. doi: 10.1021/bi9616458. [DOI] [PubMed] [Google Scholar]
- 11.D’Souza E D, O’Sullivan E, Amphlett E M, Rowlands D J, Sangar D V, Clarke B E. Analysis of NS3-mediated processing of the hepatitis C virus non-structural region in vitro. J Gen Virol. 1994;75:3469–3476. doi: 10.1099/0022-1317-75-12-3469. [DOI] [PubMed] [Google Scholar]
- 12.Eckart M R, Selby M, Masiarz F, Lee C, Berger K, Crawford K, Kuo C, Kuo G, Houghton M, Choo Q L. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem Biophys Res Commun. 1993;192:399–406. doi: 10.1006/bbrc.1993.1429. [DOI] [PubMed] [Google Scholar]
- 13.Failla C, Tomei L, De Francesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994;68:3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Failla C, Tomei L, De Francesco R. An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J Virol. 1995;69:1769–1777. doi: 10.1128/jvi.69.3.1769-1777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukushi S, Katayama K, Kurihara C, Ishiyama N, Hoshino F-B, Ando T, Oya A. Complete 5′ noncoding region is necessary for the efficient internal initiation of hepatitis C virus RNA. Biochem Biophys Res Commun. 1994;199:425–432. doi: 10.1006/bbrc.1994.1246. [DOI] [PubMed] [Google Scholar]
- 16.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparison and structure-function relationship. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 17.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gwack Y, Kim D W, Han J H, Choe J. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem Biophys Res Commun. 1996;225:654–659. doi: 10.1006/bbrc.1996.1225. [DOI] [PubMed] [Google Scholar]
- 21.Gwack Y, Kim D W, Han J H, Choe J. DNA helicase activity of the hepatitis C virus nonstructural protein 3. Eur J Biochem. 1997;250:47–54. doi: 10.1111/j.1432-1033.1997.00047.x. [DOI] [PubMed] [Google Scholar]
- 22.Gwack Y, Wook D, Han J H, Choe J. NTPase activity of hepatitis C virus NS3 protein expressed in insect cells. Mol Cells. 1995;5:171–175. [Google Scholar]
- 23.Hamatake R, Wang H G, Butcher J A, Bifano M, Clark G, Hernandez D, Zhang S, Racela J, Standring D, Colonno R. Establishment of an in vitro assay to characterize hepatitis C virus NS3-4A protease trans-processing activity. Intervirology. 1996;39:249–258. doi: 10.1159/000150525. [DOI] [PubMed] [Google Scholar]
- 24.Han D S, Hahm B, Rho H-M, Jang S K. Identification of the protease domain in NS3 of hepatitis C virus. J Gen Virol. 1995;76:985–993. doi: 10.1099/0022-1317-76-4-985. [DOI] [PubMed] [Google Scholar]
- 25.Heilek G M, Peterson M G. A point mutation abolishes the helicase but not the nucleoside triphosphatase activity of hepatitis C virus NS3 protein. J Virol. 1997;71:6264–6266. doi: 10.1128/jvi.71.8.6264-6266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hijikata M, Mizushima H, Tanji Y, Komoda Y, Hirowatari Y, Akagi T, Kato N, Kimura K, Shimotono K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci USA. 1993;90:10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong Z, Ferrari E, Wright Minogue J, Chase R, Risano C, Seelig G, Lee C G, Kwong A D. Enzymatic characterization of hepatitis C virus NS3/4A complexes expressed in mammalian cells by using the herpes simplex virus amplicon system. J Virol. 1996;70:4261–4268. doi: 10.1128/jvi.70.7.4261-4268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1035–1058. [Google Scholar]
- 31.Jin L, Peterson D L. Expression, isolation, and characterization of the hepatitis C virus ATPase/RNA helicase. Arch Biochem Biophys. 1995;323:47–53. doi: 10.1006/abbi.1995.0008. [DOI] [PubMed] [Google Scholar]
- 32.Kadaré G, Haenni A-L. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kakiuchi N, Hijikata M, Komoda Y, Tanji Y, Hirowatari Y, Shimotohno K. Bacterial expression and analysis of cleavage activity of HCV serine proteinase using recombinant and synthetic substrate. Biochem Biophys Res Commun. 1995;210:1059–1065. doi: 10.1006/bbrc.1995.1764. [DOI] [PubMed] [Google Scholar]
- 34.Kanai A, Tanabe K, Kohara M. Poly(U) binding activity of hepatitis C virus NS3 protein, a putative RNA helicase. FEBS Lett. 1995;376:221–224. doi: 10.1016/0014-5793(95)01283-x. [DOI] [PubMed] [Google Scholar]
- 35.Kato M, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkosci S, Sugimura T, Shimotono K. Molecular cloning of human hepatitis C virus genome from Japanese patients with non-A non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 37.Kim D W, Kim J, Gwack Y, Han J H, Choe J. Mutational analysis of the hepatitis C virus RNA helicase. J Virol. 1997;71:9400–9409. doi: 10.1128/jvi.71.12.9400-9409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J L, Morgenstern K A, Lin C, Fox T, Dwyer M D, Landro J A, Chambers S P, Markland W, Lepre C A, O’Malley E T, Harbeson S L, Rice C M, Murcko M A, Caron P R, Thomson J A. Crystal structure of the hepatitis virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 39.Koch J O, Lohmann V, Herian U, Bartenschlager R. In vitro studies on the activation of the hepatitis C virus NS3 proteinase by the NS4A cofactor. Virology. 1996;221:54–66. doi: 10.1006/viro.1996.0352. [DOI] [PubMed] [Google Scholar]
- 40.Kolykhalov A A, Agapov E V, Rice C M. Specificity of the hepatitis C virus NS3 serine protease: effects of substitutions at the 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites on polyprotein processing. J Virol. 1994;68:7525–7533. doi: 10.1128/jvi.68.11.7525-7533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komoda Y, Hijikata M, Tanji Y, Hirowatari Y, Mizushima H, Kimura K, Shimotohno K. Processing of hepatitis C viral polyprotein in Escherichia coli. Gene. 1994;145:221–226. doi: 10.1016/0378-1119(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 43.Lin C, Pragai B M, Grakoui A, Xu J, Rice C M. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J Virol. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin C, Thomson J A, Rice C M. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J Virol. 1995;69:4373–4380. doi: 10.1128/jvi.69.7.4373-4380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love R A, Parge H E, Wickersham J A, Hostomsky Z, Habuka N, Moomaw E W, Adachi T, Homstomska Z. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 46.Manabe S, Fuke I, Tanishita O, Kaji C, Gomi Y, Yoshida S, Mori C, Takamizawa A, Yosida I, Okayama H. Production of nonstructural proteins of hepatitis C virus requires a putative viral protease encoded by NS3. Virology. 1994;198:636–644. doi: 10.1006/viro.1994.1075. [DOI] [PubMed] [Google Scholar]
- 47.Morgenstern K A, Landro J A, Hsiao K, Lin C, Gu Y, Su M S-S, Thompson J A. Polynucleotide modulation of the protease, nucleoside triphosphatase, and helicase activities of a hepatitis C virus NS3-NS4A complex isolated from transfected COS cells. J Virol. 1997;71:3767–3775. doi: 10.1128/jvi.71.5.3767-3775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mosimann S C, Cherney M M, Sia S, Plotch S, James M N. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J Mol Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 49.Preugschat F, Averett D R, Clarke B E, Porter D J T. A steady-state and pre-steady state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J Biol Chem. 1996;271:24449–24457. doi: 10.1074/jbc.271.40.24449. [DOI] [PubMed] [Google Scholar]
- 50.Pryor K D, Leiting B. High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expression Purif. 1998;10:309–319. doi: 10.1006/prep.1997.0759. [DOI] [PubMed] [Google Scholar]
- 51.Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimizu Y, Yamaji K, Masuho Y, Yokota T, Inoue H, Sudo K, Satoh S, Shimotohno K. Identification of the sequence of NS4A required for enhanced cleavage of the NS5A/5B site by hepatitis C virus NS3 protease. J Virol. 1996;70:127–132. doi: 10.1128/jvi.70.1.127-132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimotohno K, Tanji Y, Hirowatari Y, Komoda Y, Kato N, Hijikata M. Processing of the hepatitis C virus precursor protein. J Hepatol. 1995;22:87–92. [PubMed] [Google Scholar]
- 54.Steinkühler, C., G. Biasiol, M. Brunetti, A. Urbani, R. Cortese, A. Pessi, and R. De Francesco. Product inhibition of the hepatitis C virus NS3 protease. Biochemistry, in press. [DOI] [PubMed]
- 55.Steinkühler C, Tomei L, De Francesco R. In vitro activity of hepatitis C virus protease NS3 purified from recombinant baculovirus-infected Sf9 cells. J Biol Chem. 1996;271:6367–6373. doi: 10.1074/jbc.271.11.6367. [DOI] [PubMed] [Google Scholar]
- 56.Steinkühler C, Urbani A, Tomei L, Biasiol G, Sardana M, Bianchi E, Pessi A, De Francesco R. Activity of purified hepatitis C virus protease NS3 on peptide substrates. J Virol. 1996;70:6694–6700. doi: 10.1128/jvi.70.10.6694-6700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stempniak M, Homstomska Z, Nodes B R, Hostomsky Z. The NS3 proteinase domain of hepatitis C virus is a zinc-containing enzyme. J Virol. 1997;71:2881–2886. doi: 10.1128/jvi.71.4.2881-2886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of the T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1998;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 59.Suzich J A, Tamura J K, Palmer Hill F, Warrener P, Grakoui A, Rice C M, Feinstone S M, Collett M S. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tai C-L, Chi W-K, Chen D-S, Hwang L-H. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3) J Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka T, Kato N, Cho M J, Shimotohno K. A novel sequence found at the 3′ terminus of the hepatitis C virus genome. Biochem Biophys Res Commun. 1995;215:744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 63.Tanji Y, Hijikata M, Hirowatari Y, Shimotohno K. Hepatitis C virus polyprotein processing: kinetics and mutagenic analysis of serine proteinase-dependent cleavage. J Virol. 1994;68:8418–8422. doi: 10.1128/jvi.68.12.8418-8422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomei L, Failla C, Vitale R L, Bianchi E, De Francesco R. A central hydrophobic domain of the hepatitis C virus NS4A protein is necessary and sufficient for the activation of the NS3 protease. J Gen Virol. 1996;77:1065–1070. doi: 10.1099/0022-1317-77-5-1065. [DOI] [PubMed] [Google Scholar]
- 67.Tsukiyama-Koharak D, Iizukam N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan Y, Munshi S, Sardana V, Blue J, Johns B, Cole J, Steinkuheler C, Tomei L, De Francesco R, Kuo L, Chen Z. Complex of NS3 proteinase and NS4A peptide of BK strain hepatitis C virus: a 2.2 Å resolution structure in a hexagonal crystal form. Protein Sci. 1988;7:837–847. doi: 10.1002/pro.5560070402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao N, Hesson T, Cable M, Hong Z, Kwong A D, Le H V, Weber P C. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]