Abstract
The product of open reading frame 14 (orf14) of herpesvirus saimiri (HVS) exhibits significant homology with mouse mammary tumor virus superantigen. orf14 encodes a 50-kDa secreted glycoprotein, as shown previously (Z. Yao, E. Maraskovsky, M. K. Spriggs, J. I. Cohen, R. J. Armitage, and M. R. Alderson, J. Immunol. 156:3260–3266, 1996). orf14 expressed from recombinant baculovirus powerfully induces proliferation of CD4-positive cells originating from several different species. To study the role of orf14 in transformation, a mutant form of HVS (HVS Δorf14) was constructed with a deletion in the orf14 gene. The transforming potential of HVS Δorf14 was tested in cell culture and in common marmosets. Parental HVS subgroup C strain 488 immortalized common marmoset T lymphocytes in vitro to interleukin-2-independent growth, while the HVS Δorf14 mutant did not produce such a growth transformation. In addition, HVS Δorf14 was nononcogenic in common marmosets. In contrast to other nononcogenic HVS mutant viruses which were repeatedly isolated from peripheral blood mononuclear cells of infected marmosets for more than 5 months, HVS Δorf14 did not persist at a high level in vivo. These results demonstrate that orf14 of HVS is not required for replication but is required for transformation and for high-level persistence in vivo.
Superantigens interact with T lymphocytes largely on the basis of their specificity for particular variable β (Vβ) regions of the αβ T-cell receptor (TCR). All superantigens, regardless of their origin, have the ability to elicit a very powerful response from mature T cells, a characteristic that is responsible at least in part for their pathogenic effects (22). A virus-encoded superantigen was first identified from within the U3 region of the 3′ long terminal repeat of mouse mammary tumor virus (MMTV) (9). MMTV is a type B retrovirus that is responsible for the induction and transmission of mammary carcinomas in mice (19, 23). This viral superantigen is a type II membrane-anchored glycoprotein (6, 27, 28, 39). Expression of an endogenous superantigen leads to the deletion in vivo of immature T cells expressing particular TCR Vβ chains (1). Sequence comparisons of various superantigen genes have suggested that the specificity of the superantigen-TCR Vβ interaction may be determined by a polymorphic region at the carboxyl terminus of the superantigen molecule. A unique major histocompatibility complex (MHC) class II binding site has been shown to exist in the carboxyl-terminal region of the MMTV superantigen (31). During the stimulation phase, MMTV-infected B cells are activated by superantigen-reactive T cells. The activation, differentiation, and expansion of the infected B cells are key steps during the life cycle of MMTV, and the presentation of a functional superantigen is apparently required for establishment of a productive infection (21). In addition to MMTV superantigen, herpesvirus-associated superantigens have been suggested to be responsible for viral infection, latency, or pathogenicity (13, 14, 35, 38).
Herpesvirus saimiri (HVS) belongs to the gamma subfamily of herpesviruses (Gammaherpesvirinae) (18). Some of members of this group, e.g., Epstein-Barr virus, HVS, herpesvirus ateles, and herpesvirus sylvilagus, are capable of inducing lymphoproliferative disorders in natural or experimental hosts. Recently, Kaposi’s sarcoma-associated herpesvirus has been shown to have high sequence homology to HVS (8, 33). HVS naturally infects squirrel monkeys (Saimiri sciureus), a common primate species of South American rain forests, without any apparent disease association. Infection of marmosets, owl monkeys, and other species of New World primates results in rapidly progressing fulminant lymphomas, lymphosarcomas, and leukemias of T-cell origin (18, 24). Sequence divergence among HVS isolates is most extensive at the left end of the unique L-DNA of the viral genome and is the basis for classification of HVS into subgroups A, B, and C (29). Variation in this region is correlated with differences in the capacity of the viruses to immortalize T lymphocytes in vitro and to produce lymphomas in nonhuman primates (5, 10–12, 26, 32). Both subgroup A and subgroup C viruses immortalize common marmoset T lymphocytes to interleukin-2 (IL-2)-independent proliferation (12, 36). Highly oncogenic subgroup C strains also immortalize human, rabbit, and rhesus monkey lymphocytes and can produce fulminant lymphomas in rhesus monkeys as well as in New World primates (2–5, 7, 30).
DNA sequence analysis of the whole HVS genome has revealed an open reading frame which is homologous to the product of the superantigen open reading frame of MMTV: 43% identity and 60% similarity in restricted areas (37, 40). Yao et al. have recently shown that open reading frame 14 (orf14) of HVS strain S295C encodes a secreted, highly glycosylated protein (40). A chimeric orf14-Fc fusion protein was found to bind to heterodimeric MHC class II HLA-DR molecules (40). Finally, the supernatant from HVS orf14-transfected cells induced proliferation of human peripheral blood mononuclear cells (PBMC) (40). These results suggest that orf14 of HVS functions as an immunomodulator that may contribute to the immunopathology of HVS.
In the present studies, we characterized the orf14 gene product of HVS subgroup C strain 488 (C488) and assessed its contribution to infection by the virus. orf14 expressed from recombinant baculovirus powerfully induced proliferation of CD4+ cells of several origins. orf14 deletion virus (HVS Δorf14) was unable to immortalize common marmoset T lymphocytes in vitro, nor was it able to induce lymphomas in common marmosets. HVS Δorf14 did not persist at a high level in vivo. The results presented here demonstrate that HVS C488 orf14 is required not only for transformation but also for high-level persistence in vivo. This is the first demonstration of an HVS viral gene which contributes to productive infection in vivo.
MATERIALS AND METHODS
Cell culture and virus propagation.
Owl monkey kidney (OMK) cells (OMK 637) that had been cultivated in minimal essential medium supplemented with penicillin, streptomycin, l-glutamine, and 10% (vol/vol) heat-inactivated fetal bovine serum (GIBCO BRL, Grand Island, N.Y.) were used for the propagation of HVS strain C488. Low-passage OMK cells (<30 passages) were used for transfections. Culture of common marmoset lymphocytes in immortalization assays with HVS recombinants was performed in RPMI 1640 medium supplemented with penicillin, streptomycin, fungizone, l-glutamine, 20% (vol/vol) heat-inactivated fetal bovine serum, and 5 mg of beta-mercaptoethanol per liter. Sf9 cells were maintained at 27°C in Grace’s medium containing 10% fetal calf serum, yeastolate, and lactalbumin hydrolysate.
Virion DNA isolation.
HVS virion preparations were obtained from the media of infected OMK cells by removing cell debris by low-speed centrifugation followed by pelleting of the virus at 18,000 rpm for 2 h in an SS-34 rotor. To purify intact virion DNA, the virus was disrupted at 60°C for 2 h in lysis buffer containing 10 mM Tris (pH 8.5), 1 mM EDTA, 1% (vol/vol) Sarkosyl, and 0.1 mg of proteinase K per ml. Extraction of the aqueous solution first with an equal volume of phenol and then twice with chloroform was sufficient to purify the virion DNA for transfections. To prevent shearing, sterile cut pipette tips were used for manipulating virion DNA.
Construction of an orf14 deletion plasmid.
A deletion in plasmid pNEB193, containing orf14 within a 3.7-kbp XbaI fragment of HVS C488 virion DNA, was made by restriction enzyme digestion followed by cloning of the reporter cassette into each deletion. Deletion of nucleotides 27825 to 28725 of HVS C488 by KpnI/NheI digestion removed 900 bp of HVS DNA, retaining only the carboxyl-terminal 51 amino acids of the 240 total amino acids of orf14 (see Fig. 3). Since NheI and KpnI digestion deleted an additional noncoding sequence from nucleotides 28424 to 28725, the deleted fragment was replaced with PCR-amplified DNA containing KpnI and NheI at their respective ends for cloning. After that, a secreted engineered alkaline phosphatase (SEAP) expression cassette was inserted into the deleted regions in plasmid DNA, as previously described (15, 16).
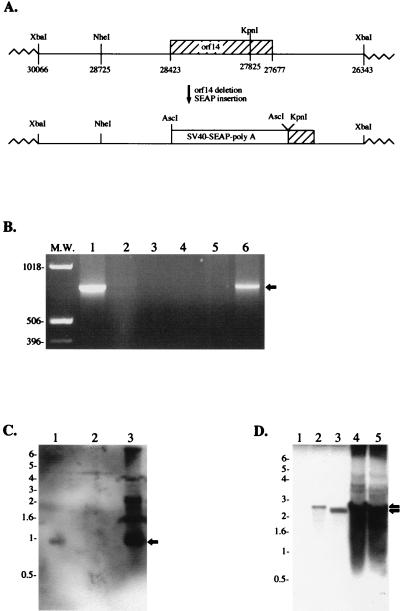
FIG. 3.
Absence of the orf14 gene in recombinant HVS Δorf14. (A) Schematic diagram of the deletion of orf14 and insertion of an SEAP expression cassette. (B) PCR amplification of orf14 DNA. One microliter of wild-type HVS (lane 1) or purified HVS Δorf14 (lane 2) was used directly for PCR analysis with specific primers to amplify the orf14 gene. A plasmid containing the orf14 gene was used as a positive control (lane 6). Lack of template virion DNA (lane 3), template plasmid DNA (lane 4), and primers (lane 5) were used as negative controls. PCR-amplified DNA (arrow) was expected to be 746 bp. (C) Southern blot analysis of the orf14 gene. Purified virion DNAs of wild-type HVS (lane 1), HVS Δorf14 (lane 2), and plasmid DNA containing an XbaI fragment (lane 3) were digested with NheI/KpnI restriction enzymes and separated on an agarose gel, followed by blotting with labeled orf14 probes. The orf14-positive band (arrow) was expected to be a 0.9-kb DNA fragment. (D) Southern blot analysis of the SEAP expression cassette. Purified virion DNAs of wild-type HVS (lane 1), HVS Δorf14 (lanes 2 and 3), and plasmid DNA containing an XbaI fragment with a deletion of orf14 and insertion of the SEAP expression cassette (lanes 4 and 5) were digested with NheI/KpnI restriction enzymes (lanes 1, 2, and 4) or with AscI restriction enzyme (lanes 3 and 5). A labeled SEAP probe was used for the hybridization. The SEAP-positive band (arrows) was expected to be a 2.8-kb DNA fragment for NheI/KpnI digestion and a 2.5-kb DNA fragment for AscI digestion.
Transfection and isolation of HVS recombinants and SEAP assay.
A reporter gene expression cassette containing the SEAP gene under the control of the simian virus 40 early promoter and enhancer was used as a selection marker for the identification of viral recombinants. HVS C488 recombinants with specific gene deletions were generated by mixed transfection of virion and cloned DNA and identification of recombinants which express SEAP activity, as described previously (15, 16). Recombinants expressing SEAP were isolated in pure form by repeated passage of limiting dilutions of virus stocks to OMK cell monolayers in 48-well tissue culture plates (Corning). SEAP production in individual wells containing cells showing cytopathic effects (CPE) was assessed with the Phospha-Light chemiluminescent assay (Tropix), performed in opaque 96-well microtiter plates with a MicroBeta scintillation counter (Wallac, Gaithersburg, Md.).
In vitro immortalization of common marmoset lymphocytes.
Assays of lymphocyte immortalization in vitro have been described previously (12). PBMC were isolated from 3-ml heparinized blood specimens from common marmosets (Callithrix jacchus) by centrifugation through lymphocyte separation medium (Organon Teknika Corp., Malvern, Pa.), followed by a wash in RPMI culture medium. PBMC from each animal were individually washed, resuspended in RPMI, and then distributed in 1-ml volumes containing approximately 106 cells into 12-well tissue culture plates. Cells were then infected at multiplicities of infection ranging from 1 to 5 with 1 ml of purified HVS viral stocks. Cells were maintained with RPMI 1640 growth medium changed every 3 to 4 days. Immortalization or cell death was assessed microscopically.
Experimental infection of common marmosets.
In vivo oncogenicity of the HVS C488 recombinants was assessed by experimental infection of common marmosets (C. jacchus). Marmosets were injected intramuscularly with 105 50% tissue culture infective doses of virus in a volume of 1 ml. Sera and blood cell pellets were collected and frozen at −70°C weekly during the first 4 weeks and every 2 weeks thereafter. Viral loads in PBMC specimens were assessed periodically by duplicate plating of 106 PBMC and serial threefold dilutions of PBMC on OMK cells in 24-well tissue culture plates. Selected culture supernatants from the PBMC viral load plates and selected sera were also tested for SEAP expression from recombinant HVS. Animals that became moribund were euthanized and received complete necropsies. Tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin.
Antibody responses against HVS virion proteins were assessed by an enzyme-linked immunosorbent assay (ELISA) to purified, lysed whole virus. Purified HVS was prepared from OMK cell lysates. Cell debris was removed by low-speed centrifugation and filtration through 0.45-μm-pore-size filters. Virus was then pelleted at 40,000 × g and resuspended in 1 ml of a solution containing 20 mM Tris, 100 mM NaCl, and 1 mM EDTA. Virus was then further purified by passage through a 10-ml Sepharose 4B column (15). Virion particles were collected in the void volume. Binding of antibodies in sera from infected marmosets to HVS was assayed by using plates coated with 20 μg of purified HVS per 96-well plate. Antibodies to HVS in diluted sera were detected by using alkaline phosphatase-conjugated protein A and measuring absorbance at 410 nm (15).
Antibody production.
An EcoRI-XhoI fragment containing the orf14 coding sequences from amino acid residues 34 to 249 (GST-orf14ΔN) was inserted into the EcoRI and XhoI sites of the expression vector pGEX-4T (Pharmacia LKB, Piscataway, N.J.). Glutathione S-transferase (GST) fusion protein expression and purification were performed essentially as described by Smith and Johnson (34). For fusion protein recovery with glutathione-Sepharose, bacterial cell pellets were frozen once, resuspended with 1/10 volume lysis buffer (1% Triton X-100 and 0.1% sarcosinate in phosphate-buffered saline [PBS]) containing protease inhibitors, and disrupted by sonication. After centrifugation to remove cell debris, supernatant fluids were mixed with pre-equilibrated glutathione-Sepharose for 30 min at 4°C. The beads were then washed three times with PBS and once with buffer (10 mM MgCl2, 1 mM dithiothreitol, 20 mM Tris [pH 7.0]). The purified recombinant GST-orf14ΔN protein was used to generate polyclonal antibody in New Zealand White rabbits.
Metabolic labeling and immunoprecipitation.
Sf9 insect cells infected with recombinant orf14 baculovirus or OMK cells infected with HVS at 50 to 60% CPE were rinsed three times with PBS, washed once with labeling medium, and then incubated overnight with 2 ml of the same medium containing 200 μCi of [35S]methionine and [35S]cysteine (New England Nuclear, Boston, Mass.). In all cases, cells were incubated in labeling medium for 30 min prior to addition of the radioisotopes. Cells were harvested and lysed with lysis buffer (0.3 M NaCl, 0.1% Nonidet P-40, 50 mM HEPES buffer [pH 8.0]) or RIPA buffer (0.15 M NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris [pH 7.5]) containing 0.1 mM Na2 VO3 and protease inhibitors (leupeptin, aprotinin, phenylmethylsulfonyl fluoride, and bestatin). Precleared cell lysates or supernatants were used for immunoprecipitation with rabbit anti-orf14 antibody. Immunoprecipitated proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and detected by autoradiography of the dried gel slabs.
Construction of recombinant baculovirus.
The EcoRI-XhoI fragment containing the orf14 gene was inserted into the EcoRI and XhoI sites of the baculovirus transfer vector pAcSG1 (PharMingen, San Diego, Calif.). Vector plasmids were cotransfected into Sf9 cells with linearized baculovirus DNA. Four days later, virus-containing supernatants were harvested. The recombinant baculovirus was amplified to obtain a high-titer stock solution. Sf9 cells infected with baculovirus were assayed for expression of recombinant protein by labeling with [35S]methionine. For routine production of recombinant proteins, 106 cells were infected with 0.2 ml of each baculovirus supernatant and lysed 48 h postinfection with lysis buffer, and the cleared cell lysates were used for immunoprecipitation or stimulation.
FACS analysis.
Cells (5 × 105) were washed with RPMI medium containing 10% fetal calf serum and incubated with fluorescein isothiocyanate-conjugated or phycoerythrin-conjugated monoclonal antibody for 30 min at 4°C. After a wash, each sample was fixed with 1% formalin solution and fluorescence-activated cell sorting (FACS) analysis was performed with a FACScan (Becton Dickinson Co., Mountainview, Calif.). Antibodies for CD2 (RPA-2.10; PharMingen), CD4 (RPA-T4; PharMingen), CD8 (RPA-T8; PharMingen), and CD20 (2H7; PharMingen) were purchased commercially.
Proliferation assays.
PBMC from healthy donors were purified with lymphocyte separation medium (Organon Teknika Corp.) and washed three times with RPMI with 20% fetal calf serum. In some cases, purified PBMC were mixed with CD4 or CD8 Dynabeads to deplete the CD4 or CD8 cell population, as recommended by the manufacturer (Dynal, Great Neck, N.Y.). Isolated PBMC were stimulated with 100-μg cell lysates of Sf9 cells infected with either wild-type baculovirus or recombinant orf14 baculovirus in 96-well round-bottom plates. After 7 days of culture, 1 μCi of [3H]thymidine per well was added, plates were cultured overnight before harvesting, and [3H]thymidine incorporation was determined.
In vitro translation.
One microgram of pBluescript-orf14 plasmid DNA was directly subjected to the TNT coupled reticulocyte lysate system from Promega (Madison, Wis.). In some cases, the reaction mixture was mixed with 2 μl of canine pancreatic microsomal membranes for 90 min. Radioactively labeled proteins were separated by SDS-PAGE and detected by autoradiography.
Southern blot analysis.
Purified virion DNA was digested overnight with NheI/KpnI or AscI restriction enzyme. Digested virion DNA was separated on a 1% agarose gel, transferred to a nitrocellulose membrane, and subjected to a hybridization reaction. A 0.6-kb DNA fragment that contained the deleted portion of orf14 and a 1.5-kb DNA fragment that contained the SEAP gene were labeled with a High Prime digoxigenin DNA labeling kit. Detection of DNA bands was performed with the protocol provided by the manufacturer (Boehringer Mannheim, Indianapolis, Ind.).
RESULTS
Identification of the HVS orf14 protein.
A recombinant baculovirus system was employed to express orf14 of HVS strain C488. The orf14 gene was cloned into a baculovirus vector, followed by transfection into Sf9 insect cells of defective, linearized, baculovirus virion DNA. Recombinant orf14 baculovirus was amplified in Sf9 insect cells. To demonstrate expression of orf14, we generated a rabbit polyclonal antibody against a purified bacterial GST-orf14ΔN fusion protein. The anti-orf14 antibody reacted specifically with a protein having an apparent molecular size of 50 kDa upon immunoprecipitation in Sf9 insect cells infected with recombinant orf14 baculovirus (Fig. 1B). No such protein was detected in control insect cells infected with wild-type baculovirus lacking the orf14 gene. While the molecular mass of orf14 predicted from the DNA sequence was 28 kDa, the protein migrated at 50 kDa in SDS-PAGE. The orf14 protein is predicted to have five potential sites for N-linked glycosylation (NX[S/T]). To demonstrate the posttranslational modification of orf14, RNA of orf14 was subjected to in vitro translation with or without canine pancreatic microsomal membranes. orf14 migrated at 35 kDa in SDS-PAGE before modification and at 50 kDa after modification (Fig. 1A). These results suggest that glycosylation of the protein most likely contributes to the slow migration of the orf14 protein in SDS-PAGE. These findings are consistent with previous analysis by Yao et al. (40).
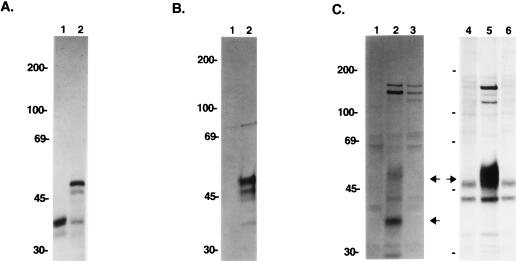
FIG. 1.
Expression of orf14. (A) In vitro translation of the orf14 gene. In vitro translation was performed with (lane 2) or without (lane 1) canine pancreatic microsomal membranes. (B) orf14 expression from the recombinant baculovirus. Insect cells were infected with wild-type baculovirus (lane 1) or recombinant orf14 baculovirus (lane 2), labeled with [35S]methionine, and subjected to immunoprecipitation with anti-orf14 antibody. (C) orf14 expression in wild-type and recombinant HVS. OMK cells were mock infected (lanes 1 and 4) and infected with wild-type HVS (lanes 2 and 5) or HVS Δorf14 (lanes 3 and 6). At 50 to 60% CPE, cells were labeled overnight with [35S]methionine and [35S]cysteine. Radioactive cell lysates (lanes 1 to 3) or culture media (lanes 4 to 6) were used for immunoprecipitation with an anti-orf14 antibody. Arrows indicate the orf14 proteins.
Induction of proliferation of primary T cells by HVS orf14.
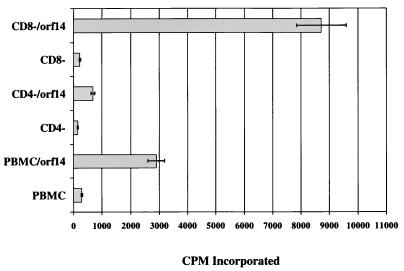
In order to investigate the ability of orf14 to induce T-cell proliferation, PBMC from humans, rhesus monkeys, HVS-negative squirrel monkeys, and common marmosets were cultured for 14 days with 100 μg of cell lysates of insect cells infected with mock virus, wild-type baculovirus or recombinant orf14 baculovirus. At day 14, the numbers of viable cells were evaluated with trypan blue stain. These measurements show that cell lysates containing orf14 induced a dramatic proliferation of PBMC, whereas control lysates of insect cells or lysates of insect cells infected with wild-type baculovirus had no effect (Table 1). A similar response was detected with PBMC of additional independent human, rhesus monkey, squirrel monkey, and common marmoset donors. To determine the cell type responsive to stimulation of orf14, PBMC were mixed with Dynabeads conjugated with anti-CD4 antibody to deplete CD4-positive T cells or with Dynabeads conjugated with anti-CD8 antibody to deplete CD8-positive T cells. Subsequently, these cells were cultured for 7 days with or without insect cell lysates containing orf14 under the same conditions as described above and assessed for proliferation by thymidine incorporation. CD8-depleted PBMC treated with orf14 showed threefold-higher stimulation than parental PBMC (Fig. 2). In contrast, CD4-depleted PBMC responded insignificantly to orf14 stimulation (Fig. 2). In addition, human PBMC stimulated for 7 days were examined for surface expression of CD4 and CD8. The proportion of CD4-positive T cells was significantly increased after stimulation; conversely, the proportion of CD8-positive T cells was decreased (data not shown). These results demonstrate that orf14 specifically stimulates CD4-positive T lymphocytes in culture.
TABLE 1.
Induction of proliferation of PBMC by HVS orf14
| PBMC type | Incubation conditiona | No. of cellsb |
|---|---|---|
| Human | Sf9 | <104 |
| Sf9/BV | <104 | |
| Sf9/orf14 | 5.2 × 106 | |
| Rhesus monkey | Sf9 | <104 |
| Sf9/BV | <104 | |
| Sf9/orf14 | 4.0 × 106 | |
| Squirrel monkeyc | Sf9 | <104 |
| Sf9/BV | <104 | |
| Sf9/orf14 | 6.2 × 106 | |
| Common marmoset | Sf9 | <104 |
| Sf9/BV | <104 | |
| Sf9/orf14 | 2.6 × 106 |
Sf9 insect cells were infected with wild-type baculovirus (Sf9/BV) or infected recombinant baculovirus expressing HVS orf14 (Sf9/orf14). After 3 days, 100 μg of Sf9, Sf9/BV, or Sf9/orf14 cell extracts were incubated with 5 × 105 PBMC for 14 days.
At day 14, cell numbers were counted by trypan blue staining. Values are means of two separate experiments.
PBMC from HVS-negative squirrel monkeys were used.
FIG. 2.
Proliferation response of human PBMC to orf14 protein. PBMC from healthy donors were mixed with Dynabeads conjugated with anti-CD4 antibody to deplete CD4-positive T cells (CD4−) or with Dynabeads conjugated with anti-CD8 antibody to deplete CD8-positive T cells (CD8−). Afterward, cells (105 cells/well) were cultured for 7 days with or without insect cell lysates containing orf14 and assessed for proliferation by thymidine incorporation. Means of two independent experiments are shown.
Isolation of orf14 deletion mutants.
Recombinant HVS containing a deletion in orf14 was generated by replacing selected portions of a 3.7-kbp XbaI-cloned HVS C488 virion DNA fragment with a SEAP expression cassette driven by the simian virus 40 early promoter (Fig. 3A). Deletion of nucleotides 27825 to 28725 of HVS C488 by KpnI/NheI digestion removed 900 bp of HVS DNA, leaving only the carboxyl-terminal 51 amino acids of the original 240 amino acids of orf14. The KpnI/NheI restriction enzyme digestion eliminated an additional 302 bp of 5′ noncoding sequence of orf14. This 302-bp fragment was reinserted by using PCR-amplified sequences. At this point, we introduced an AscI restriction enzyme site, allowing for insertion of a SEAP expression cassette into the deleted orf14 region in the plasmid DNA. DNA sequence analysis confirmed the desired deletion mutation within orf14 and the absence of other unwanted alterations in the fragment used. Following cotransfection of this plasmid with infectious wild-type virion DNA, recombinants expressing SEAP were isolated by repeated limiting dilution passage of virus in OMK cells.
After repeated purification by limiting dilution of virus with SEAP activity, the absence of orf14 was confirmed by PCR and Southern blot analysis. One microliter of isolated HVS Δorf14 virus was used directly for PCR analysis with specific primers to amplify the orf14 gene. Wild-type HVS virus and the plasmid containing the orf14 gene were used as controls. PCR analysis showed the absence of the orf14 gene in purified HVS Δorf14 recombinant virus (Fig. 3B). Additionally, we performed Southern blot analysis by using the 600-bp deleted orf14 fragment or the 1.5-kb SEAP gene as a probe. Purified virion DNAs were digested with NheI/KpnI or AscI restriction enzyme and separated on an agarose gel, followed by hybridization with labeled probes. The orf14 probe detected the expected 0.9-kb DNA band from wild-type HVS virion DNA but not from HVS Δorf14 virion DNA (Fig. 3C). Conversely, the SEAP probe reacted with the 2.5- and 2.8-kb fragments of the SEAP expression cassette gene from HVS Δorf14 virion DNA but not from wild-type HVS virion DNA (Fig. 3D). Finally, the loss of orf14 expression was examined by immunoprecipitation with an anti-orf14 antibody. OMK cells were infected with wild-type HVS or recombinant HVS Δorf14 deletion virus. When CPE progressed to 50 to 60%, cells were radioactively labeled overnight. Radioactive cell lysates were used for immunoprecipitation with anti-orf14 antibody. These experiments showed that the 35- and 50-kDa species of orf14 protein were not detected in cells infected with the orf14 deletion mutants, while they were readily detected in cells infected with wild-type HVS (Fig. 1C, lanes 2 and 3). Since orf14 has recently been shown to be a secreted protein (40), the labeled culture medium was also used for immunoprecipitation with anti-orf14 antibody. The glycosylated 50-kDa species of orf14 protein was strongly detected from the culture medium of wild-type HVS-infected cells, but it was not detected from the culture medium of HVS Δorf14-infected cells (Fig. 1C, lanes 5 and 6).
In vitro T-lymphocyte immortalization and in vivo lymphoma induction.
Common marmoset T lymphocytes are immortalized efficiently to IL-2 independent cell growth by infection with wild-type HVS C488 (12, 36). Therefore, an in vitro immortalization assay was used to test the transforming activity of HVS recombinants. Wild-type HVS was used as a positive control, and HVS ΔSTP was used as a negative control. HVS ΔSTP, which has a deletion in the STP gene, has been shown to be nononcogenic in in vitro immortalization and lymphoma induction in animals (15). Equivalent titers of HVS Δorf14, HVS ΔSTP, and wild-type HVS were added to aliquots of unstimulated PBMC from individual common marmosets. Five different common marmosets were used individually as donors for independent experiments. Wild-type HVS uniformly immortalized lymphocytes from all five of the common marmosets to IL-2-independent growth (Table 2). Deletion of orf14 resulted in loss of the ability to transform common marmoset T lymphocytes in vitro (Table 2). In addition, HVS ΔSTP did not produce growth transformation, as has been described previously (Table 2) (15). These results demonstrate that although orf14 is not required for replication, it is required for in vitro immortalization of primary marmoset lymphocytes.
TABLE 2.
In vitro immortalization and in vivo oncogenicity of HVS Δorf14
| Virus | No. of marmosets with positive result/no. tested for:
|
Survival (days) | ||
|---|---|---|---|---|
| In vitro immortalization | Virus recoverya | Lymphoma induction | ||
| None | 0/5 | |||
| HVS C488 | 5/5 | 3/3 | 3/3 | 17, 19, 20 |
| HVS ΔSTP | 0/5 | 2/2 | 0/2 | >370 |
| HVS Δorf14 | 0/5 | 2/2 | 0/2 | >370 |
Virus was recovered from PBMC of infected marmosets as described previously (15).
Experimental infections of common marmosets with virus deleted in orf14 demonstrated that this gene is also essential for induction of lymphomas in vivo. The three marmosets infected with wild-type HVS C488 died on days 17, 19, and 20 after inoculation. However, animals infected with the Δorf14 recombinants remained healthy over the 12 months of observation after experimental infection (Table 2). Animals infected with wild-type HVS C488 developed fulminant multicentric lymphomas consistent with the disease, as described previously (10). Thus, the orf14 gene is required not only for T-lymphocyte immortalization in vitro but also for lymphoma induction in marmosets.
Persistent infection by attenuated HVS Δorf14 deletion viruses in vivo.
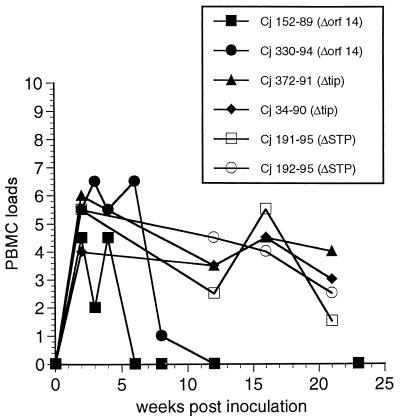
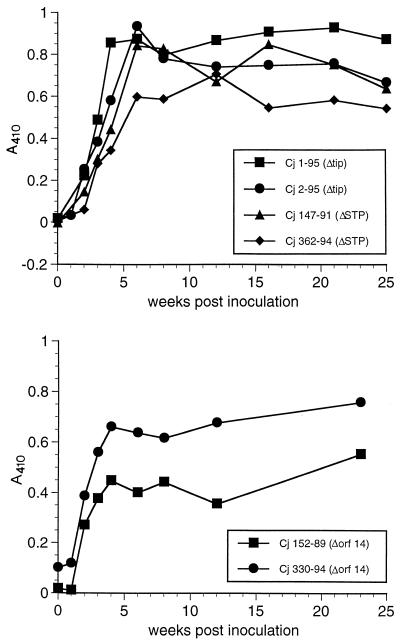
We have developed procedures for evaluating virus load in vivo by measuring the numbers of PBMC required to isolate HVS. This assay measures the number of infectious cells in PBMC. Recently, this method has been used for quantitating virus loads in marmosets infected with the nononcogenic HVS strains HVS ΔSTP and HVS ΔTip (15). We have shown that these nononcogenic viruses are readily recovered from PBMC of animals infected with the deletion viruses for at least 5 months (Fig. 4) (15). However, we were unable to isolate HVS Δorf14 from both infected marmosets beyond 4 to 8 weeks postinfection (Fig. 4). The number of infectious cells from marmoset 152-89, infected with HVS Δorf14, rapidly declined after 4 weeks postinoculation. Marmoset 330-94, infected with HVS Δorf14, showed a PBMC load equivalent to those from marmosets infected with HVS ΔSTP or HVS ΔTip at the acute phase of infection; however, viral loads dramatically declined thereafter (Fig. 4).
FIG. 4.
Cell-associated HVS viral loads. Cell-associated viral loads were measured in common marmoset PBMC collected following experimental infection with HVS ΔSTP, HVS ΔTip, and HVS Δorf14 recombinants. Values on the y axis indicate the number of PBMCs required to recover HVS, coded as follows: 0, >106 (i.e., virus was not recovered with 106 PBMC); 1, 106; 2, 333,333; 3, 111,111; 4, 37,037; 5, 12,345; 6, 4,115; 7, 1371; 8, 457. HVS ΔSTP and HVS ΔTip have been described previously (15).
Antibody responses against HVS were also measured by ELISA with plates coated with detergent-lysed, purified virions. The four marmosets infected with HVS ΔSTP and HVS ΔTip recombinant viruses showed strong antibody responses to HVS structural antigens, as reported previously (Fig. 5) (15). The two marmosets infected with HVS Δorf14 recombinant virus also showed persistent antibody responses, although they were slightly weaker than those of animals infected with HVS ΔSTP and HVS ΔTip (Fig. 5). These antibodies persisted at similar levels for the entire 24-week period of measurement (Fig. 5).
FIG. 5.
In vivo antibody responses against HVS virion proteins. Binding of antibodies in sera from infected marmosets to HVS was assayed by ELISA with plates coated with 20 μg of purified HVS per plate. Sera were tested at a 1:400 dilution. Alkaline phosphatase-conjugated protein A was used to detect antibodies bound to HVS. Results are expressed as absorbance at 410 nm.
DISCUSSION
The results described herein demonstrate that orf14 induces proliferation of CD4-positive T cells in culture in a manner similar to that observed previously for the MMTV superantigen (22). The properties of the deletion mutant indicate that the orf14 gene of HVS is required for T-lymphocyte transformation in vitro and lymphoma induction in vivo. The inability to isolate recombinant HVS Δorf14 from marmosets beyond 4 to 8 weeks after inoculation and the persistence of antibody responses suggest that the orf14 deletion virus may persist in vivo but at a very low level. Our results indicate that orf14 is even more important than STP and Tip in maintaining the levels of persisting virus in marmosets.
Since HVS contains other growth-promoting genes (STP, Tip, vIL-8 receptor, and v-cyclin, etc.), virus without orf14 may still be expected to retain some growth-promoting activity. Indeed, Knappe et al. (25) have reported that mutant viruses without the orf14/vsag gene are replication competent and fully capable of transforming human and marmoset T cells. The conditions used by Knappe et al. included repeated lectin stimulation, addition of IL-2 to the media, and use of feeder layers (25). Since the conditions used by our group are very different from those reported by Knappe et al., our results are not necessarily in disagreement. However, we have found that common marmoset PBMC can grow for prolonged periods in the presence of IL-2 and can even grow continuously under these conditions. Furthermore, the Δorf14 virus provided to us by Knappe et al. (25) did not produce cell growth transformation in our assay. Our results demonstrate that, unlike the parental virus, virus lacking orf14 does not cause lymphomas in common marmosets and does not cause continuous, IL-2-independent growth of common marmoset T lymphocytes.
To further confirm that the loss of transforming activity of our orf14 deletion virus was derived from the specific gene deletion and not from other unexpected alterations, the reading frame for the MMTV superantigen was recombined into the deleted region of HVS Δorf14. Replacement of orf14 with the MMTV superantigen gene fully reconstituted the in vitro immortalization ability of this virus (unpublished results). Thus, the loss of transforming activity in the deletion mutant virus was attributed solely to the specific orf14 gene deletion and not to any inadvertently selected alterations.
In addition to insight into the role of orf14 in viral oncogenesis, we describe here a greatly diminished level of viral persistence resulting from deletion of orf14. Upon MMTV infection, B lymphocytes express the viral superantigen at their surfaces in the context of MHC class II molecules. Presentation of the superantigen by B cells leads to stimulation and proliferation of T lymphocytes bearing a defined TCR Vβ chain. During the stimulation phase, superantigen-reactive T cells activate the infected B cells, which ultimately results in an increase in the number of MMTV-infected B cells. Thus, a functional superantigen is important for maintaining a high level of MMTV in exposed mice (21). Our results with HVS in monkeys suggest that orf14 may be similar to the MMTV superantigen in terms of the general features of its functional contribution. Certainly, orf14 is important for the induction of dramatic T-cell proliferation by HVS; without it the levels at which HVS is maintained become dramatically reduced.
Despite this similarity in sequence and functional contribution, HVS orf14 may be processed and presented differently from the MMTV superantigen. Yao et al. have demonstrated that orf14 binds to HLA-DR and that T-cell proliferation induced by orf14 is dependent on the presence of antigen-presenting cells (40). This suggests that orf14 protein secreted during HVS infection may efficiently bind to an antigen-presenting cell and thereby activate T cells. We have found that most HVS-transformed common marmoset T cells contain unusually high levels of expression of HLA-DR on their surfaces (17, 20). These observations suggest that orf14 secreted from HVS-transformed marmoset T cells may bind directly to HLA-DR on the surfaces of infected T cells, which could contribute to their proliferation. Additionally, several groups including ours have found that orf14 induces T-cell proliferation in a TCR Vβ-independent fashion (25). Thus, orf14 does not appear to possess all of the characteristics of a superantigen based upon a classic definition. However, the mode of action of orf14 is analogous in many respects to that of the classic superantigens.
ACKNOWLEDGMENTS
M. Duboise and J. Guo contributed equally to this work.
We thank H. Fickenscher for providing the orf14 deletion virus and J. Newton for preparing the manuscript.
This work was supported by Public Health Service grants CA31363 and AI38131 and by grant RR00168 from the Division of Research Resources.
REFERENCES
- 1.Acha-Orbea H, Shakhov A, Scarpellino N, L, Kolb E, Müller V, Vessaz-Shaw A, Fuchs R, Blöchlinger K, Rollini P, Billotte J, Sarafidou M, MacDonald H R, Diggelmann H. Clonal deletion of VB14-bearing T cells in mice transgenic for mammary tumour virus. Nature. 1991;350:207–211. doi: 10.1038/350207a0. [DOI] [PubMed] [Google Scholar]
- 2.Alexander L, Du Z, Rosenzweig M, Jung J U, Desrosiers R C. A role for natural simian immunodeficiency virus and human immunodeficiency virus type 1 Nef alleles in lymphocyte activation. J Virol. 1997;71:6094–6099. doi: 10.1128/jvi.71.8.6094-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berend K R, Jung J U, Boyle T J, DiMaio J M, Mungal S A, Desrosiers R C, Lyerly H K. Phenotypic and functional consequences of herpesvirus saimiri infection of human CD8+ cytotoxic T lymphocytes. J Virol. 1993;67:6317–6321. doi: 10.1128/jvi.67.10.6317-6321.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biesinger B, Müller-Fleckenstein I, Simmer B, Lang G, Wittmann S, Platzer E, Desrosiers R C, Fleckenstein B. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc Natl Acad Sci USA. 1992;89:3116–3119. doi: 10.1073/pnas.89.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biesinger B, Trimble J J, Desrosiers R C, Fleckenstein B. The divergence between two oncogenic herpesvirus saimiri strains in a genomic region related to the transforming phenotype. Virology. 1990;176:505–514. doi: 10.1016/0042-6822(90)90020-r. [DOI] [PubMed] [Google Scholar]
- 6.Brandt-Carlson C, Butel J S. Detection and characterization of a glycoprotein encoded by the mouse mammary tumor virus long terminal repeat gene. J Virol. 1991;65:6051–6060. doi: 10.1128/jvi.65.11.6051-6060.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bröker B M, Tsygankov A Y, Müller-Fleckenstein I, Guse A H, Chitaev N A, Biesinger B, Fleckenstein B, Emmrich F. Immortalization of human T cell clones by Herpesvirus saimiri. J Immunol. 1993;151:1184–1192. [PubMed] [Google Scholar]
- 8.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 9.Choi Y, Kappler J W, Marrack P. A superantigen encoded in the open reading frame of the 3′ long terminal repeat of mouse mammary tumour virus. Nature. 1991;350:203–206. doi: 10.1038/350203a0. [DOI] [PubMed] [Google Scholar]
- 10.Desrosiers R C, Bakker A, Kamine J, Falk L A, Hunt R D, King N W. A region of the herpesvirus saimiri genome required for oncogenicity. Science. 1985;228:184–187. doi: 10.1126/science.2983431. [DOI] [PubMed] [Google Scholar]
- 11.Desrosiers R C, Burghoff R L, Bakker A, Kamine J. Construction of replication-competent herpesvirus saimiri deletion mutants. J Virol. 1984;49:343–348. doi: 10.1128/jvi.49.2.343-348.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desrosiers R C, Silva D, Waldron L M, Letvin N L. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J Virol. 1986;57:701–705. doi: 10.1128/jvi.57.2.701-705.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobrescu D, Ursea B, Pope M, Asch A, Posnett D. Enhanced HIV-1 replication in V beta 12 T cells due to human cytomegalovirus in monocytes: evidence for a putative herpesvirus superantigen. Cell. 1995;82:753–763. doi: 10.1016/0092-8674(95)90472-7. [DOI] [PubMed] [Google Scholar]
- 14.Doherty P C, Tripp R A, Hamilton-Easton A M, Cardin R D, Woodland D L, Blackman M A. Tuning into immunological dissonance: an experimental model for infectious mononucleosis. Curr Opin Immunol. 1997;9:477–483. doi: 10.1016/s0952-7915(97)80098-2. [DOI] [PubMed] [Google Scholar]
- 15.Duboise S M, Guo J, Czajak S, Desrosiers R C, Jung J U. STP and Tip are essential for herpesvirus saimiri oncogenicity. J Virol. 1998;72:1308–1313. doi: 10.1128/jvi.72.2.1308-1313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duboise S M, Guo J, Desrosiers R C, Jung J U. Use of virion DNA as a cloning vector for the construction of mutant and recombinant herpesviruses. Proc Natl Acad Sci USA. 1996;93:11389–11394. doi: 10.1073/pnas.93.21.11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duboise S M, Lee H, Guo J, Choi J-K, Czajak S, Simon M, Desrosiers R C, Jung J U. Mutation of the Lck-binding motif of Tip enhances lymphoid cell activation by herpesvirus saimiri. J Virol. 1998;72:2607–2614. doi: 10.1128/jvi.72.4.2607-2614.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleckenstein B, Desrosiers R C. Herpesvirus saimiri and herpesvirus ateles. In: Roizman B, editor. The herpesviruses. Vol. 1. New York, N.Y: Plenum Publishing Corporation; 1982. pp. 253–332. [Google Scholar]
- 19.Golovkina T V, Chervonsky A, Dudley J P, Ross S R. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell. 1992;69:637–645. doi: 10.1016/0092-8674(92)90227-4. [DOI] [PubMed] [Google Scholar]
- 20.Guo J, Williams K, Duboise S M, Alexander L, Veazey R, Jung J U. Substitution of ras for the herpesvirus saimiri STP oncogene in lymphocyte transformation. J Virol. 1998;72:3698–3704. doi: 10.1128/jvi.72.5.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Held W, Waanders G A, Shakhov A, Scarpellino N, L, Acha-Orbea H, MacDonald H R. Superantigen-induced immune stimulation amplifies mouse mammary tumor virus infection and allows virus transmission. Cell. 1993;74:529–540. doi: 10.1016/0092-8674(93)80054-i. [DOI] [PubMed] [Google Scholar]
- 22.Herman A, Kappler J W, Marrack P, Pullen M. Superantigens: mechanisms of T-cell stimulation and role in immune responses. Annu Rev Immunol. 1991;9:745–772. doi: 10.1146/annurev.iy.09.040191.003525. [DOI] [PubMed] [Google Scholar]
- 23.Heston W E, Deringer M K, Andervont H B. Gene-milk agent relationship in mammary tumor development. J Natl Cancer Inst. 1945;5:289–307. [Google Scholar]
- 24.Jung J U, Desrosiers R C. Herpesvirus saimiri and ateles. In: Webster R, Granoff A, editors. Encyclopedia of virology. Philadelphia, Pa: Saunders Scientific Publications, Inc.; 1994. pp. 614–622. [Google Scholar]
- 25.Knappe A, Hiller C, Thurau M, Wittmann S, Hofmann H, Fleckenstein B, Fickenscher H. The superantigen-homologous viral immediate-early gene ie14/vsag in herpesvirus saimiri-transformed human T cells. J Virol. 1997;71:9124–9133. doi: 10.1128/jvi.71.12.9124-9133.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koomey J M, Mulder C, Burghoff R L, Fleckenstein B, Desrosiers R C. Deletion of DNA sequences in a nononcogenic variant of herpesvirus saimiri. J Virol. 1984;50:662–665. doi: 10.1128/jvi.50.2.662-665.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korman A J, Bourgarel P, Meo T, Rieckhof G E. The mouse mammary tumor virus long terminal repeat encodes a type II transmembrane glycoprotein. EMBO J. 1992;11:1901–1905. doi: 10.1002/j.1460-2075.1992.tb05242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krummenacher C, Diggelman H. The mouse mammary tumor virus long terminal repeat encodes a 47 kDa glycoprotein with a short half-life in mammalian cells. Mol Immunol. 1993;30:1151–1157. doi: 10.1016/0161-5890(93)90133-v. [DOI] [PubMed] [Google Scholar]
- 29.Medveczky P, Szomolayi E, Desrosiers R C, Mulder C. Classification of herpesvirus saimiri into three groups based on extreme variation in a DNA region required for oncogenicity. J Virol. 1984;52:938–944. doi: 10.1128/jvi.52.3.938-944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mittrücker H-W, Müller-Fleckenstein I, Fleckenstein B, Fleishcher B. CD2-mediated autocrine growth of herpes virus saimiri-transformed human T lymphocytes. J Exp Med. 1995;176:900–913. doi: 10.1084/jem.176.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mottershead D G, Hus P-N, Urban R G, Strominger J L, Huber B T. Direct binding of the Mtv7 superantigen (Mls-1) to soluble MHC class II molecules. Immunity. 1995;2:149–154. doi: 10.1016/s1074-7613(95)80027-1. [DOI] [PubMed] [Google Scholar]
- 32.Murthy S C S, Trimble J J, Desrosiers R C. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J Virol. 1989;63:3307–3314. doi: 10.1128/jvi.63.8.3307-3314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo J J, Bohenzxy R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi’s sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusion with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 35.Sutkowski N, Palkama T, Ciurli C, Sekaly R P, Thorley-Lawson D A, Huber B T. An Epstein-Barr virus-associated superantigen. J Exp Med. 1996;184:971–980. doi: 10.1084/jem.184.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szomolanyi E, Medveczky P, Mulder C. In vitro immortalization of marmoset cells with three subgroups of herpesvirus saimiri. J Virol. 1987;61:3485–3490. doi: 10.1128/jvi.61.11.3485-3490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson B J, Nicholas J. Superantigen function. Nature. 1991;351:530. doi: 10.1038/351530a0. [DOI] [PubMed] [Google Scholar]
- 38.Tripp R, Hamilton-Easton A, Cardin R, Nguyen P, Behm F, Woodland D, Doherty P, Blackman M. Pathogenesis of an infectious mononucleosis-like disease induced by a murine gamma-herpesvirus: role for a viral superantigen? J Exp Med. 1997;185:1641–1650. doi: 10.1084/jem.185.9.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winslow G M, Scherer M T, Kappler J W, Marrack P. Detection and biochemical characterization of the mouse mammary tumor virus 7 superantigen (Mls-1a) Cell. 1992;71:719–730. doi: 10.1016/0092-8674(92)90549-r. [DOI] [PubMed] [Google Scholar]
- 40.Yao Z, Maraskovsky E, Spriggs M K, Cohen J I, Armitage R J, Alderson M R. Herpesvirus saimiri open reading frame 14, a protein encoded by a T lymphotropic herpesvirus, binds to MHC class II molecules and stimulates T cell proliferation. J Immunol. 1996;156:3260–3266. [PubMed] [Google Scholar]