Abstract
Functional analysis of naturally occurring hepatitis B virus (HBV) mutations is crucial in understanding their impact on disease. We have recently identified two mutations in the HBV core promoter of an HBV strain associated with fulminant hepatitis leading to highly (15-fold) enhanced replication as a result of increased viral encapsidation of pregenomic RNA into the core particles (T. F. Baumert et al., J. Clin. Invest. 98:2268–2276, 1996). Functional studies in an encapsidation assay had demonstrated that the increase in encapsidation was largely independent of pregenomic RNA transcription. In this study, we define the molecular mechanism whereby the two core promoter mutations (C to T at nucleotide [nt] 1768 and T to A at nt 1770) result in enhanced viral encapsidation and replication. The effect of these mutations leading to increased encapsidation is mediated through enhanced core protein synthesis (15-fold) by the mutant virus. The marked increase in core protein synthesis is largely a result of posttranscriptional or translational effect of the mutations because the mutations resulted in only a twofold increase in pregenomic RNA transcription. In addition, this effect appears to be selective for core expression since reverse transcriptase-polymerase expression was increased only twofold. trans-complementation analyses of HBV replication demonstrated that enhanced replication occurred only when the mutations were provided together with the core protein in trans, confirming the functional association of the core promoter mutations and core protein expression. In addition, the effect of the mutations appears to be quantitatively dependent on the strain background to which the mutations were introduced. Our study suggests that the HBV core promoter regulates core protein expression at both transcriptional and posttranscriptional levels.
Hepatitis B virus (HBV) is a partially double stranded DNA virus that replicates through an RNA intermediate (for reviews, see references 8 and 39). The virally encoded reverse transcriptase polymerase (RT-Pol) is essential for this unique form of genome replication. Viral replication occurs exclusively in the core particle, which is assembled through complex interactions among pregenomic RNA, core protein, and polymerase. A well-defined cis-acting element in the pregenomic RNA (encapsidation signal ɛ) has been shown to mediate the interaction of pregenomic RNA with the encapsidation complex (14, 27, 29, 30). The capsid containing the replicative intermediate is then enveloped by HBV surface antigens (HBsAg) in a lipid bilayer. Although the molecular mechanism of HBV encapsidation and replication has been largely elucidated with the identification of various essential elements (11, 14, 27, 30, 37, 41), it is not clear whether any other sequences outside these elements may play a role in the replicative process. The core promoter contains multiple cis-acting elements with nuclear receptor binding sites (32) and regulates the transcription of 3.5-kb RNAs with heterogeneous 5′ ends (42, 44). There are two 3.5-kb RNA species, the precore and core RNAs, which direct the translation of HBV e and core antigens, respectively. The core RNA also functions as the pregenomic RNA.
We have recently identified two mutations in the core promoter of a viral strain associated with a fatal outbreak of fulminant hepatitis B (FH strain) (10, 19) resulting in markedly enhanced viral replication (2). These mutations comprised a C-to-T change at nucleotide (nt) 1768 as well as a T-to-A change at nt 1770 (MT5/6) in the HBV basal core promoter (nucleotide numbering according to reference 32). Functional characterization of these mutations in a tissue culture model had demonstrated that the phenotype of enhanced replication was the result of enhanced viral encapsidation of pregenomic RNA into HBV nucleocapsids (2). The core promoter mutations resulted in only minor changes of transcription of pregenomic RNA and precore RNAs (2). In contrast, encapsidation of pregenomic RNA into HBV core particles was increased 15-fold in the mutant strain compared to the wild-type strain (2). Although the identified core promoter mutations resulted in two amino acid changes of the overlapping HBX open reading frame (ORF), the mutated HBX protein was not responsible for the phenotype of enhanced encapsidation and replication (2). The aim of this study was to identify the molecular mechanism of enhanced encapsidation and replication induced by these mutations. The identification of this mechanism may have important implications in understanding the viral life cycle as well as in the pathogenesis of fulminant hepatitis associated with these mutations.
MATERIALS AND METHODS
Constructs.
Replication-competent constructs of wild-type adw (adwR9) and ayw (aywR9) and MT5/6 (MT5/6R9) mutant strains were described previously (2). These constructs contained a 1.2× genomic length of HBV. To generate a construct that is deficient in encapsidating pregenomic RNA, the HBV encapsidation signal was altered by introducing a single nucleotide change (G to A at nt 1882 without affecting the precore ORF) into the stem-loop (loop 3) of the pregenomic RNA encapsidation signal (29). The following primers were used to generate mutant HBV DNA by PCR mutagenesis (underlined nucleotides represent introduced mutations) (1): 5′ AAGCCTCCAAGCTATGCCTTGGGTGG 3′ (sense, nt 1868 to 1894) and 5′ CGAGGGAGTTCTTCTTCTAG 3′ (antisense outer primer, nt 2359 to 2339) as well as 5′ TCTCGGGGCCGCTTGGGGACTCTC (sense outer primer, nt 1464 to 1486) and 5′ ACCCAAGGCATAGCTTGGAGGC 3′ (antisense, nt 1892 to 1870). The PCR products were combined, reannealed, and amplified by a second PCR using the above-described outer primers. After subcloning of the PCR-generated fragments into PCR II vector (Invitrogen, San Diego, Calif.), an RsrII-BglII fragment (HBV nt 1525 to 1986) was subcloned into the replication-competent construct adwR9. To inactivate the core gene, a frameshift mutation was introduced into the core gene at nt 1986 by digesting adwR9 DNA with BglII, treatment with Klenow enzyme, and subsequent religation. To inactivate the polymerase gene, a BspEI-EcoRI (nt 2355 to 3200) fragment of wild-type adwR9 was replaced with the same fragment of HBV strain HBV5-15 (3). HBV 5-15 contains a naturally occurring missense mutation (A to C at nt 2798) in the polymerase gene terminating HBV replication. All constructs were analyzed structurally by sequencing and restriction digestion as well as functionally by demonstrating their inability to replicate in HuH-7 cells (Fig. 1B). The Altered Sites in vitro system (Promega, Madison, Wis.) was used to introduce HBV core promoter mutations and to eliminate the precore ORF. Various nucleotide changes (Table 1) at positions 1764, 1766, 1768, and 1770 were generated by using oligonucleotide 5′ AGGTTAANGNTNTNTGTATTAGGAG 3′ (sense, nt 1757 to 1781). Wild-type HBV adw DNA was used as the template for mutagenesis (plasmid pSelectHBVadw [2, 10]). The replication-competent mutant constructs were generated by subcloning an RsrII-BglII fragment (nt 1525 to 1986) of pSelectHBVMutant into adwR9. To eliminate the precore protein expression in the replication-competent R9 constructs, the precore protein start codon at nt 1816 was mutated to GTG in pSelectHBV, using the primer 5′ CACCAGCACCGTGCAACTTTTTCACC 3′ (sense, nt 1802 to 1827) to generate the pre-C−. The replication-competent constructs adwR9preC− and MT5/6preC− were constructed by ligating the FspI-BspEI fragment (nt 1802 to 2329) of pSelectHBVpreC− into adwR9 or MT5/6R9. Replication-competent aywR9 constructs containing MT5/6 (C to T at nt 1768 and A to T at nt 1770) and a stop codon in the precore ORF (G to A at nt 1896) were generated by PCR mutagenesis (QuickChange site-directed mutagenesis kit; Stratagene, La Jolla, Calif.), using mutant sense and antisense primers and HBV ayw cDNA (a generous gift from H. Schaller, University of Heidelberg, Heidelberg, Germany) as a template. To generate MT5/6 in HBV ayw (nt 1768 and 1770), we used primers 5′ GGAGGAGATTAGGTTAAAGGTTTATGTACTAGGAGG 3′ (sense, nt 1747 to 1782) and 5′ CCTCCTAGTACATAAACCTTTAACCTAATCTCCTCC 3′ (antisense, nt 1782 to 1747). To generate a precore stop codon mutation at nt 1896 in HBV ayw, the primers used were 5′ GGGTGGCTTTAGGGCATGGACATCG 3′ (sense, nt 1886 to 1910) and 5′ CGATGTCCATGCCCTAAAGCCACCC 3′ (antisense, nt 1910 to 1886). Subsequently, an RsrII-BglII fragment (HBV nt 1525 to 1986) of the PCR product containing either MT5/6 or the precore stop codon mutation was subcloned into the replication-competent aywR9 construct. All mutant constructs were sequenced to confirm the correct mutations and ensure absence of other mutations that could have been introduced during mutagenesis. Core expression constructs were generated by subcloning the ScaI-RsrII fragment (containing HBV nt 963 to 1526) of pGEM7HBXadw (2) and the RsrII-ApaI fragment (containing HBV nt 1526 to 2604) of wild-type or mutant (MT5/6) adwR9 (2) into the ScaI and ApaI sites of a modified pGEM7 plasmid (Promega) containing the simian virus 40 tIVS and poly(A) (subcloned as a HindIII-AflIII fragment from pSV-SPORT1 [Gibco, Gaithersburg, Md.]) at the 3′ end of its multiple cloning site. To analyze polymerase expression, the β-galactosidase gene (lacZ) was fused in frame with the polymerase at a BglII site (HBV nt 2403) 99 nt after its start codon. The BamHI fragment of pBSLacZ (24) containing the lacZ gene with its 3′ end blunt ended was subcloned into the BglII (nt 2403; 5′ end) and HpaI (nt 963; 3′ end) sites of the replication-competent constructs adwR9 and MT5/6R9 to generate constructs padwpolLacZ and pMT5/6polLacZ. Correct orientation of the inserted fragment was analyzed by restriction digestion and sequencing of the RT-Pol-LacZ junction. The control LacZ expression construct pCDLacZ was constructed by ligating a lacZ-cDNA fragment into the mammalian expression vector pcDNA1 (Invitrogen, Carlsbad, Calif.).
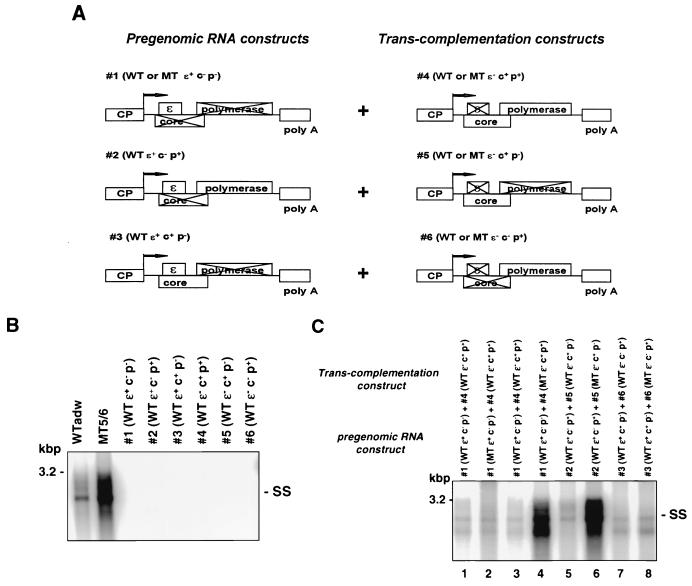
FIG. 1.
Mapping of the HBV genetic element targeted by MT5/6 to induce enhanced replication in a trans-complementation assay. (A) Constructs. For trans-complementation analysis, a construct generating pregenomic RNA (1, 2, or 3) was cotransfected with a trans-complementing protein expression construct (4, 5, or 6). CP, core promoter; c, core; p, polymerase; ɛ, RNA encapsidation signal; WT, wild type; MT, mutant. (B) Lack of replication of individual trans-complementation constructs. HuH-7 cells were transfected with replication-competent constructs adwR9 and MT5/6R9 or one of the knockout construct 1 to 6 (A). Four days posttransfection, viral replicative intermediates were analyzed by Southern blotting of core particle-associated viral DNA. SS, single-stranded DNA. (C) trans-complementation analysis. A pregenomic RNA-generating construct (1, 2, or 3 [A]) was cotransfected with a protein expression construct (4, 5, or 6 [A]) into HuH-7 cells. MT5/6 was provided either in cis in the RNA-generating construct (lane 2) or in trans together with core and/or polymerase ORF (lanes 4, 6, and 8). Four days posttransfection, replication was analyzed as described above. Transfection efficiencies as monitored by pTKGH cotransfection were similar among all samples. SS, single-stranded DNA.
TABLE 1.
Effect of naturally occurring and randomly designed promoter mutations on HBV replicationa
| Construct | Mutation
|
Replicationc | |
|---|---|---|---|
| Nucleotide position(s) | Position(s)b in mutated core promoter sequence | ||
| Mutants | |||
| MT5/6 | 1768, 1770 | TAAAGGTTTATGTATT | 15 |
| MT8 | 1764, 1766 | TAATGATCTTTGTATT | 1.5–2 |
| MT9 | 1770 | TAAAGGTCTGTGTATT | No change |
| MT10 | 1768, 1770 | TAAAGGTATGTGTATT | No change |
| MT11 | 1768, 1770 | TAAAGGTGTATGTATT | No change |
| MT12 | 1768 | TAAAGGTATTTGTATT | No change |
| MT13 | 1768, 1770 | TAAAGGTATATGTATT | No change |
| MT14 | 1772 | TAAAGGTCTTTATATT | No change |
| WT adw | TAAAGGTCTTTGTATT | 1 | |
Various mutations were introduced into the replication-competent construct adwR9 by oligonucleotide-directed mutagenesis. Four days after transfection of wild-type (WT) and mutant constructs into HuH-7 cells, viral replication was analyzed by Southern blotting of core particle-associated viral replicative intermediates as shown in Fig. 8A. The amount of viral replicative intermediates was quantified with a phosphor imager. Core promoter nucleotide positions are numbered according to reference 32 and MT8, represents mutations described in reference 34.
Underlined.
Shown as fold increase compared to replication of wild-type HBV adw (replicative intermediates of wild-type adw construct = 1).
Tissue culture and transfection.
Human hepatoma HuH-7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) plus 10% fetal bovine serum (Sigma, St. Louis, Mo.) at 37°C and 5% CO2. HuH-7 were grown to 70% confluence and transfected with DNA by a CaPO4 transfection kit (Calcium Phosphate Mammalian Cell Transfection kit; 5 Prime-3 Prime Inc., Boulder, Colo.). For analysis of transfection efficiency in all experiments, plasmid pTKGH containing the human growth hormone gene (driven by the thymidine kinase enhancer and promoter) was cotransfected with various HBV constructs. Typically, 15 μg of HBV construct was cotransfected with 1 μg of plasmid pTKGH (Nichols Institute Diagnostics, San Juan Capistrano, Calif.) into HuH-7 cells grown in 10-cm-diameter dishes. From each transfection experiment, medium was harvested and human growth hormone was measured with a radioimmunoassay from Nichols Institute Diagnostics.
Analysis of viral nucleic acids and HBsAg and HBeAg expression.
Three or four days after transfection, HuH-7 cells were harvested for viral RNA and DNA analysis. RNA was prepared by the guanidium isothiocyanate-acid-phenol method (1), analyzed by formaldehyde agarose gel electrophoresis (10 μg of RNA), and hybridized with an HBV-specific probe as described recently (2, 13). For primer extension analysis, an HBV primer (5′ TCTAAGGCTTCTCGATACAGAGCTG 3′) spanning nt 2006 to 2030 in the antisense orientation was end labeled with [γ-32P]ATP and then reacted with guanidium isothiocyanate-acid-phenol-purified HBV RNA by a standard protocol (1). Primer extension products were separated on a 8% polyacrylamide-urea gel and subjected to autoradiography (2). Viral replicative DNA intermediates associated with intracellular core particles were isolated by ultracentrifugation of cell lysate through a 30% sucrose cushion and then analyzed by Southern blot hybridization (2, 10). HBsAg and HBV e antigen (HBeAg) synthesis was analyzed in the culture medium of transfected HuH-7 cells by using commercially available radioimmunoassays (for HBsAg, Ausria II from Abbott, North Chicago, Ill.; for HBeAg, EBK from Sorin Biomedica, Saluggia, Italy).
Analysis of core expression and nucleocapsid assembly.
Three days after transfection of HuH-7 cells with replication-competent or core expression HBV constructs, the cells were lysed with lysis buffer containing 1% Nonidet P-40, 50 mM Tris (pH 7.4), 50 mM NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml, and 2 μg of leupeptin per ml. The cell lysate was cleared of cell debris and nuclei by low-speed centrifugation (15 min at 20,000 × g and 4°C). For immunoblotting, a fraction of the supernatant was subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (15% gel). After gel transfer to polyvinylidene difluoride membranes (Immobilon P; Millipore Corp., Bedford, Mass.), the blots were probed with anticore (dilution of 1:1,000) antibody (polyclonal rabbit antibody; generously provided by J. Ou, University of Southern California, Los Angeles) followed by horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) antibody (dilution of 1:4,000; Amersham Corp., Arlington Heights, Ill.) and subsequent chemiluminescence detection (ECL kit; Amersham). The analysis of core expression was reproduced by using a commercially available anticore antibody (DAKO Corp., Carpinteria, Calif.). To control for differences in sample processing and gel loading, the blot was reprobed with antiactin antibody (dilution of 1:2,000; Sigma) and analyzed as described above. For metabolic labeling of the core protein, HuH-7 cells (day 3 posttransfection) were starved for 2 h in methionine- and cysteine-free DMEM and then labeled for 15 min with 250 μCi of [35S]methionine and -cysteine (NEN Express labeling mix; New England Nuclear, Boston, Mass.). The cells were then washed with phosphate-buffered saline and lysed with a lysis buffer containing 1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 10 mM Tris (pH 8.0), 140 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml, and 2 μg of leupeptin per ml. The cell lysate was cleared of cell debris and nuclei by low-speed centrifugation (15 min at 20,000 × g and 4°C). Then 500 μl of the cleared supernatant was incubated with 1 μl of anticore antibody for 16 h at 4°C, followed by incubation with 50 μl of protein A-Sepharose 4B-CL beads (Pharmacia Biotech Inc., San Francisco, Calif.) for 1 h at room temperature with mixing. The beads were washed repeatedly, and the bound proteins were released and denatured by heating for 5 min at 95°C in SDS sample buffer. The immunoprecipitated proteins were subjected to electrophoresis on a 15% polyacrylamide gel. After gel drying, the immunoprecipitated proteins were analyzed by a phosphor imager (STORM; Molecular Dynamics, Sunnyvale, Calif.) and quantified by using the ImageQuant program (Molecular Dynamics). For analysis of nucleocapsid assembly lysates of 35S-labeled, transfected HuH-7 cells (labeling conditions, 1,000 μCi of [35S]methionine and -cysteine in DMEM containing unlabeled methionine and cysteine at 5% of the standard concentration; labeling time, 16 to 24 h) were lysed and subjected to sucrose gradient centrifugation (10 to 60% sucrose step gradient) as described previously (45). In brief, 700-μl aliquots of lysates were layered onto a 10 to 60% sucrose step gradient (700-μl steps of 10, 20, 30, 40, 50, and 60% sucrose in 50 mM Tris–100 mM NaCl [wt/wt], pH 7.4). After ultracentrifugation (SW55 rotor for 2:30 h at 40,000 rpm and 4°C), 10 fractions were collected from the top and the core protein was immunoprecipitated (45) from the sucrose fractions, using an anticore specific antibody (DAKO) and protein A-Sepharose 4B-CL beads. After extensive washing of the beads, the bound proteins were released and denatured by heating for 5 min at 95°C in SDS sample buffer. The immunoprecipitated core protein was then analyzed by SDS-PAGE and autoradiography.
Analysis of LacZ and polymerase-LacZ (Pol-LacZ) fusion protein expression.
Three days after transfection of HuH-7 cells with constructs pCDLacZ, padwpolLacZ, and pMT5/6polLacZ, the cells were lysed as described above. The lysate was subjected to SDS-PAGE (10% gel) and immunoblotting with a mouse monoclonal anti-β-galactosidase antibody (dilution of 1:1,000; Boehringer Mannheim, Indianapolis, Ind.) as described above.
RESULTS
Core promoter mutations result in enhanced replication when provided with the core ORF in trans.
To map the HBV genetic element mediating MT5/6-induced enhanced encapsidation, we systematically analyzed the HBV elements known to be required for encapsidation (encapsidation signal ɛ, core, and polymerase protein) and developed a trans-complementation assay as shown in Fig. 1A. Since the mutant constructs lacked one or more of the required elements for encapsidation, they did not exhibit viral encapsidation or replication when transfected individually (Fig. 1B). However, when a pregenomic RNA-generating construct (1, 2, or 3) was cotransfected with a core- or polymerase-expressing construct (4, 5, or 6), viral encapsidation and replication were restored by complementation in trans (Fig. 1C). By providing MT5/6 together with the various elements required for encapsidation, we were able to functionally map the HBV element affected by MT5/6.
When MT5/6 was provided in cis, modifying the transcription of pregenomic RNA (construct 1), replication was not affected to any major extent (Fig. 1C, lane 2 compared to lane 1), indicating that the phenotype of enhanced replication was largely independent of an effect of MT5/6 on the level of pregenomic RNA transcription. In contrast, MT5/6, when provided together with the core and polymerase proteins (construct 4) in trans, induced the phenotype of enhanced replication (Fig. 1C, lane 4 compared to lane 3). This experiment confirms our previous results of an encapsidation assay in which MT5/6 exerted a major transcription-independent effect on encapsidation when provided in trans (2). The next experiment was designed to discern the effect of MT5/6 on either the core or polymerase protein. Therefore, MT5/6 was provided in trans with either the core or the polymerase protein (Fig. 1A, construct 5 or 6). Analysis of viral replication demonstrated that MT5/6 was able to induce enhanced replication when provided together with core in trans (Fig. 1C, lane 6) but not when provided together with the polymerase (Fig. 1C, lane 8). These data suggest that MT5/6 exerts on core protein a functional effect resulting in enhanced viral encapsidation and replication.
Core promoter mutations led to enhanced core expression as a result of increased synthesis.
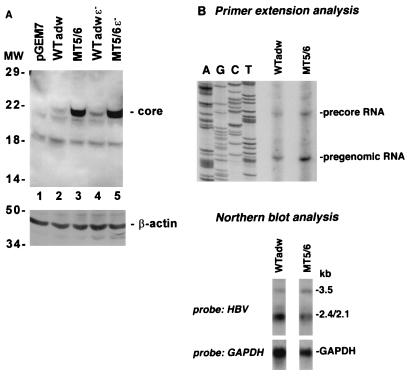
To analyze the effect of MT5/6 on core protein expression directly, wild-type and mutant replication-competent constructs were transfected into HuH-7 cells and the steady-state level of core protein expression was analyzed by immunoblotting of transfected cell lysates. MT5/6 led to a 15-fold increase in core protein expression compared to the wild type (Fig. 2A, lanes 2 and 3). The increase in core protein expression was independent of pregenomic RNA encapsidation, because elimination of the pregenomic RNA encapsidation signal (construct 4 [Fig. 1A]) did not affect the increase in the core protein level (Fig. 2A, lane 5). The increase in core protein expression was much greater than the twofold increase of pregenomic RNA (serving as the core mRNA) transcription induced by the mutations (Fig. 2B). Although it is possible that MT5/6 induced alternatively initiated pregenomic RNA resulting in increased core translation, the primer extension analysis did not reveal any unusual 5′ ends of the pregenomic RNA associated with MT5/6 (Fig. 2B, top).
FIG. 2.
(A) Core protein expression in wild-type and mutant virus. HuH-7 cells were transfected with replication-competent wild-type adw (WTadw) or mutant constructs, and core expression was analyzed by SDS-PAGE (15% gel) and immunoblotting of transfected cell lysates with an anticore antibody (top). The blot was stripped and reprobed with an antiactin antibody (bottom). Positions of molecular weight (MW) markers (in kilodaltons) are indicated on the left. (B) Transcription of wild-type and mutant viruses. HBV RNA was purified from HuH-7 cells transfected either with adwR9 or MT5/6R9 and analyzed by primer extension using an HBV-specific antisense primer (top) or Northern blotting using an HBV-specific probe (bottom). Transfection efficiencies as monitored by pTKGH cotransfection were similar among all samples.
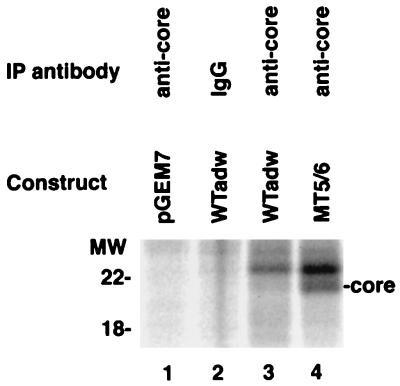
To distinguish between the possibilities of increased synthesis versus decreased turnover resulting in enhanced core expression, transfected HuH-7 cells were pulse-labeled with [35S]methionine and -cysteine and cell lysates were examined by immunoprecipitation with anticore antibodies. The replication-competent construct containing MT5/6 displayed a markedly increased synthesis of core protein (>15-fold) compared with the wild-type construct (Fig. 3). Subsequent chase in the presence of excess nonradioactive methionine and cysteine revealed little or no difference in the turnover rate of core protein between the two transfected cells (not shown).
FIG. 3.
Core protein synthesis in mutant and wild-type HBV. HuH-7 cells were transfected with either a wild-type adw (WTadw) or mutant replication-competent HBV DNA construct as indicated. Three days after transfection, HuH-7 cells were subjected to metabolic labeling. After pulse-labeling with [35S]methionine and -cysteine for 15 min, HuH-7 cells were lysed and subjected to immunoprecipitation (IP) with an core-specific antibody (anti-core) or nonimmune serum (IgG). Immunoprecipitated proteins were analyzed by SDS-PAGE and autoradiography. The identity of the core band was established by a parallel Western immunoblot with anticore antibodies (not shown); the band above the core band did not react with anticore antibodies and therefore probably represents a nonspecific protein from immunoprecipitation. Positions of molecular weight (MW) markers (in kilodaltons) are indicated on the left. Quantitation of the core protein with the ImageQuant program revealed a >15-fold-higher level of signal intensity in the mutant- than in wild-type-transfected cells.
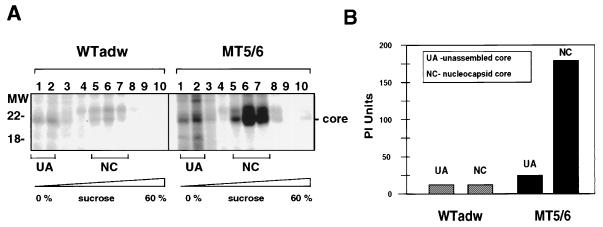
We next examined whether the increase in core protein expression is reflected in an increase in HBV nucleocapsid assembly. Lysates of 35S-labeled HuH-7 cells transfected with either wild-type or mutant constructs were subjected to sucrose velocity centrifugation, and nucleocapsid formation was analyzed by immunoprecipitation of the 35S-labeled core protein in high-density fractions of the sucrose gradient (Fig. 4A). The mutant construct demonstrated an approximately 15-fold increase of nucleocapsids (core protein in high-density sucrose fractions 5, 6, and 7 in Fig. 4A) compared to the wild type. The sedimentation coefficient of the nucleocapsids from mutant and wild-type strains was 120S to 130S (determined according to reference 22), similar to what was reported in the literature (6). This result is consistent with a much higher level of replicative intermediates in cells transfected with the MT5/6 construct.
FIG. 4.
Nucleocapsid assembly in mutant and wild-type HBV. HuH-7 cells were transfected with either the wild-type adw (WTadw; left) or mutant (MT5/6; right) replication-competent R9 construct. After labeling with [35S]methionine and -cysteine for 16 h, cells were lysed and the lysates were subjected to 10 to 60% sucrose velocity centrifugation. Ten fractions were collected from the top and analyzed for core by immunoprecipitation with an anticore antibody. The immunoprecipitated proteins were subjected to SDS-PAGE and autoradiography (A). Nucleocapsid-associated core sedimented to fractions 5 to 7, whereas unassembled core monomers and dimers did not sediment and remained in fractions 1 and 2. Positions of molecular weight (MW) markers (in kilodaltons) are indicated on the left. (B) Quantitation of unassembled and nucleocapsid-associated core protein in wild-type and mutant virus by phosphor imager (PI) analysis.
It is possible that a small increase in transcription can result in an exponential increase in protein level due to a limitation in cellular protein degradation. In particular, the capacity of core protein degradation may be near saturation at the wild-type level of core synthesis, and the substantial increase in steady-state level of core protein disproportional to the minor increase of transcription in mutant-transfected cells may represent this threshold phenomenon. To investigate whether the observed accumulation was secondary to this possibility under our experimental conditions, we transfected various amounts of wild-type and mutant constructs in a 2-log range into HuH-7 cells and analyzed core protein levels as described before. The fold increases in core expression were similar in all quantities of plasmids transfected (Fig. 5). These data indicate that the mutation-induced increase of core expression was not due to a threshold effect of core protein degradation.
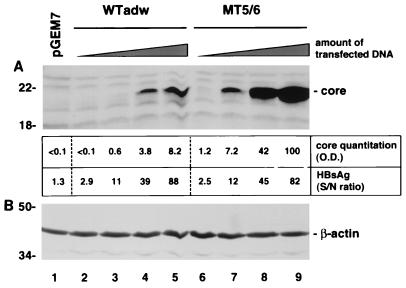
FIG. 5.
Increase in mutation-induced core expression is independent of the amount of transfected plasmid DNA. HuH-7 cells were transfected with 20 μg of the control plasmid pGEM7 (lane 1) or 1, 3, 10, and 20 μg of wild-type adwR9 (WTadw; lanes 2 to 5) or MT5/6R9 construct (lanes 6 to 9) together with 19, 17, 10, or 0 μg of pGEM7, respectively (total amount of transfected DNA was 20 μg in all experiments). Core expression was analyzed by SDS-PAGE (15% gel) and immunoblotting of HuH-7 cell lysates with an anticore antibody (A). Quantitation (optical density [O.D.]) of the core protein by using the ImageQuant program is shown below (top row). Transfection efficiencies of wild-type and mutant constructs were similar, as indicated by similar amounts of secreted HBsAg (bottom row). S/N, signal/noise. To demonstrate similar protein loading, the blot was stripped and reprobed with an antiactin antibody (B). Positions of molecular weight markers (in kilodaltons) are indicated on the left.
Effect of MT5/6 on HBV RT-Pol expression.
Since the core (pregenomic) RNA also codes for the RT-Pol, it is conceivable that MT5/6 could similarly increase the expression of RT-Pol as it did on the core protein. To study the effect of MT5/6 on HBV RT-Pol expression, lysates of transfected HuH-7 cells were subjected to immunoprecipitation and immunoblotting with RT-Pol-specific antibodies. However, none of the antibodies available to us detected RT-Pol expression in transfected HuH-7 cells (not shown). Therefore, RT-Pol was fused in frame with the β-galactosidase (lacZ) gene in the wild-type and mutant constructs (Fig. 6A). The resulting constructs, although no longer replication competent, should allow us to study the effect of MT5/6 on RT-Pol expression. After transfection of wild-type and mutant RT-Pol-LacZ constructs into HuH-7 cells, expression of the Pol-LacZ fusion protein was analyzed by SDS-PAGE and immunoblotting with an anti-LacZ antibody. MT5/6 increased the level of the RT-Pol-LacZ fusion protein only 1.5- to 2-fold (Fig. 6B; quantitation after correction for transfection efficiency and protein loading), indicating that MT5/6 affected RT-Pol expression to the same extent as would be expected from the twofold increase of the transcript. This result also confirmed the trans-complementation experiment in which MT5/6 together with the polymerase did not confer a high-replication phenotype (Fig. 1C, lanes 7 and 8).
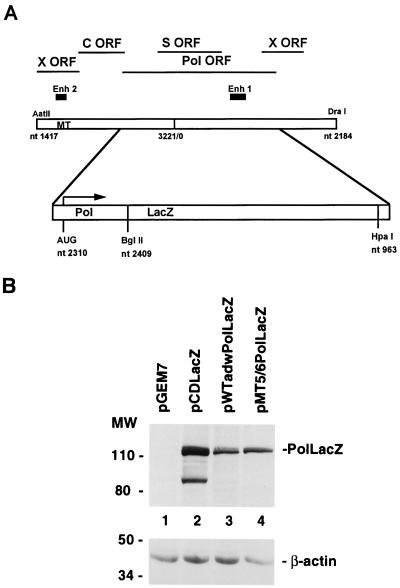
FIG. 6.
Effect of MT5/6 on Pol expression. (A) In the terminal redundant R9 construct, an RT-Pol-LacZ fusion gene was generated by exchanging a major part of the RT-Pol ORF with a cDNA fragment of lacZ. (B) After transfection of the adw or MT5/6 RT-Pol fusion construct into HuH-7 cells, expression of the RT-Pol-LacZ fusion protein was analyzed by SDS-PAGE (10% gel) and immunoblotting with an anti-LacZ antibody (top). Analysis of LacZ expression of plasmid pCDLacZ served as positive control. The lower band of the pCDLacZ lane probably represents a degradation product of the full-length LacZ protein. The blot was stripped and reprobed with an antiactin antibody (bottom). Positions of molecular weight (MW) markers (in kilodaltons) are indicated on the left. Transfection efficiencies as monitored by pTKGH cotransfection were similar among all samples.
Effect of MT5/6 on core expression is independent of HBV elements outside the core promoter and core gene.
To study whether any other virus-specific genetic elements outside the core promoter and core protein were required for the observed enhanced core expression and nucleocapsid assembly, we generated wild-type and mutant core expression constructs as illustrated in Fig. 7A. Transfection of these constructs into HuH-7 revealed enhanced core expression (Fig. 7B), core synthesis (Fig. 7C), and nucleocapsid assembly (Fig. 7D) similar to that seen with the replication-competent, full-length constructs (Fig. 2 to 4). Interestingly, the nucleocapsids generated by the core expression constructs sedimented slightly differently (nucleocapsid peak in fraction 5 of sucrose velocity gradient [Fig. 7B]) from the nucleocapsids generated from the replication-competent constructs (nucleocapsid peak in fraction 6 of sucrose velocity gradient [Fig. 4]). This difference is probably due to the lack of pregenomic RNA and RT-Pol in the core particles generated from the core expression constructs, resulting in a difference in biophysical property from the assembled nucleocapsids of the replication-competent constructs. Taken together, these data demonstrate that the mutation-induced enhanced nucleocapsid assembly and core protein expression are independent of other HBV genetic elements outside the core promoter and core gene sequences.
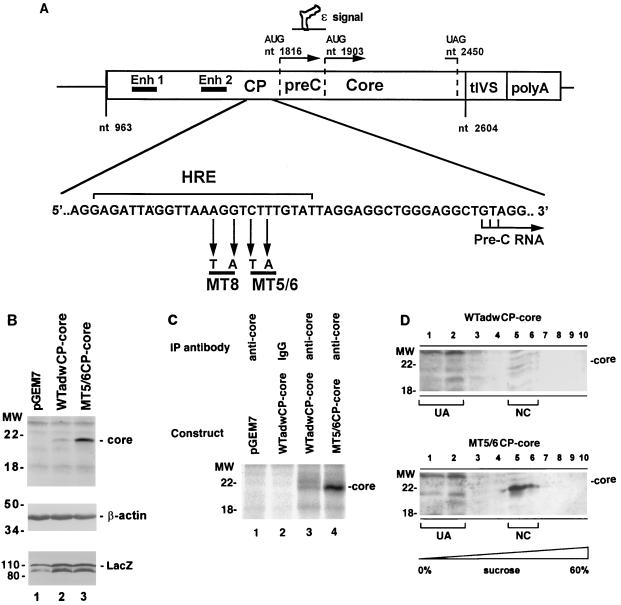
FIG. 7.
Effect of MT5/6 on core expression is independent of HBV elements outside the core promoter and core gene. (A) Core expression constructs. The core expression constructs contain coding sequences for only core promoter and core protein. Enhancers I and II (Enh 1 and Enh 2), encapsidation signal (ɛ), and core promoter (CP) are illustrated above. tIVS, small t-antigen intron. The sequence of HBV adw strain in the core promoter region is shown below; the hormone response element (HRE) (where various members of nuclear receptor superfamily bind), MT5/6, and MT8 are indicated. (B) Core protein expression. adwCP-core or MT5/6CP-core constructs were cotransfected with a LacZ expression construct (pCDLacZ) as a transfection control into HuH-7 cells. Three days after transfection, cells were lysed and core expression was analyzed by immunoblotting (upper panel). To demonstrate similar protein loading and transfection efficiency in the experiment, the blot was stripped and reprobed with antiactin and anti-LacZ antibodies, respectively (middle and lower panels). In panels B to D, positions of molecular weight (MW) markers (in kilodaltons) are indicated on the left. (C) Core protein synthesis. After labeling of cellular and viral proteins with [35S]methionine and -cysteine for 15 min, HuH-7 cells were lysed and subjected to immunoprecipitation (IP) with a core-specific antibody (anti-core) or nonimmune serum (IgG). Immunoprecipitated proteins were analyzed by SDS-PAGE and autoradiography. (D) Nucleocapsid assembly. Lysates of transfected HuH-7 cells were subjected to sucrose velocity centrifugation (10 to 60% step gradient); 10 fractions were collected from the top and analyzed for core protein by immunoblotting with an anticore antibody. Nucleocapsid-associated core (NC) sedimented to fractions 5 and 6, whereas unassembled core monomers and dimers (UA) did not sediment and remained in fractions 1 and 2.
The precore protein is not involved in the MT5/6-induced enhanced replication.
Since the precore protein has been suggested to interfere with viral encapsidation when expressed under the control of a strong promoter (17) and some core promoter mutations have been shown to result in a modest decrease in precore protein and HBeAg expression (4, 25), a decrease in precore protein expression may be responsible for the enhanced replication associated with core promoter mutants. To address this hypothesis, we eliminated the precore ORF in both wild-type and mutant constructs by mutating the precore start codon AUG to GUG at nt 1816. Mutating this codon should have no effect on the downstream encapsidation signal and DR1 sequences, precluding the possibility of any confounding effects on replication. One caveat of this approach is that the elimination of precore AUG may lead to initiation of core synthesis at the downstream core AUG, resulting in increased core expression and encapsidation of the precore mRNA (27). However, since the precore mRNA accounts for only a minor fraction (<1/3 [Fig. 2B]) of the total 3.5-kb RNA, this effect should have only a small impact on the overall core expression and encapsidation. Furthermore, this possibility should be independent of the effect of MT5/6 on core synthesis from the core mRNA. Elimination of the precore expression had no effect on the enhanced replication (Fig. 8A, lane 7) and core expression (Fig. 8B, lane 7) associated with MT5/6, although there was a minor increase of replication associated with the pre-C− mutant over wild type (Fig. 8A or B; compare lanes 2 and 3), perhaps reflecting the possibility discussed above. The successful elimination of precore expression was functionally confirmed by a complete absence of HBeAg synthesis (Fig. 8C). Similar HBsAg production (Fig. 8C) as well as similar levels of growth hormone levels expressed from the cotransfected plasmid pTKGH (data not shown) demonstrated that the transfection efficiencies were comparable in these experiments. These data exclude a role of the precore protein in MT5/6-induced enhanced replication and are consistent with previous studies showing that a naturally occurring mutation leading to a precore stop codon (G to A at nt 1896) did not significantly alter HBV replication in transfected cells (10, 40, 43). The lack of effect of precore elimination on MT5/6-induced enhanced replication is not surprising since MT5/6 did not result in a dramatic change of precore gene transcription (Fig. 2B and reference 2) or precore protein expression as indicated by HBeAg synthesis (Fig. 8C).
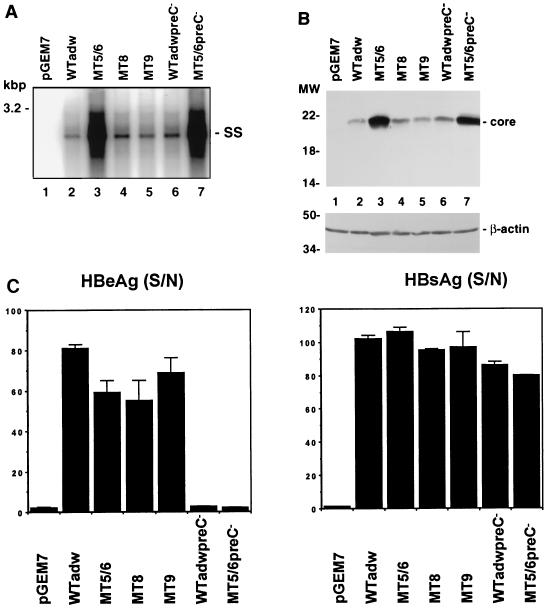
FIG. 8.
Effects of core promoter and precore mutations on viral replication and protein expression. Replication-competent constructs containing either wild-type adw (WTadw), precore (WTadwpreC− or MT5/6preC−, each containing a mutated precore start codon), or core promoter mutant sequences (Table 1) were transfected into HuH-7 cells. The transfected cells were analyzed for viral replication, core protein expression, and HBeAg and HBsAg synthesis. (A) Viral replication. Viral replicative intermediates were analyzed by Southern blotting of core particle-associated viral DNA. SS, single stranded DNA. (B) Core protein expression. Transfected cell lysates from the experiment shown in panel A were subjected to SDS-PAGE and immunoblotting with an anticore antibody. Positions of molecular weight (MW) markers (in kilodaltons) are indicated on the left. (C) HBeAg and HBsAg synthesis. HBeAg (left) and HBsAg (right) were analyzed from the medium of transfected HuH-7 cells by using radioimmunoassays. Transfection efficiencies as monitored by pTKGH cotransfection were similar among all samples. Results are shown as signal-to-noise ratio (S/N) of three independent experiments (average ± standard deviation).
Functional comparison of naturally occurring and randomly introduced core promoter mutants.
Recent studies have identified another cluster of mutations in the HBV core promoter (A to T at nt 1764 and G to A at nt 1766; designated MT8 in Table 1) associated with fulminant or severe hepatitis (12, 34). Functional studies in a tissue culture system have demonstrated that these mutations exhibited a high-replication phenotype (4, 25), although not as dramatic as that of MT5/6 (2). To directly compare MT5/6 and these mutations in the capacity to increase viral replication, we introduced MT8 into the replication-competent construct adwR9. Analysis of viral replication of the mutant constructs in HuH-7 cells demonstrated at most a 2-fold increase in replication for MT8, versus a 15-fold increase in replication for MT5/6 (Table 1; Fig. 8A). To further define HBV core promoter sequences involved in the regulation of encapsidation and replication, various other mutations were randomly introduced into the region of the core promoter of the replication-competent HBV construct adwR9. Analysis of viral replication of the mutant constructs in HuH-7 cells demonstrated that only the naturally occurring core promoter mutation in construct MT5/6 resulted in a substantially enhanced replication, whereas other mutations had little or no effect on replication (Table 1; Fig. 8A).
The effect of MT5/6 on HBV replication is quantitatively dependent on strain background.
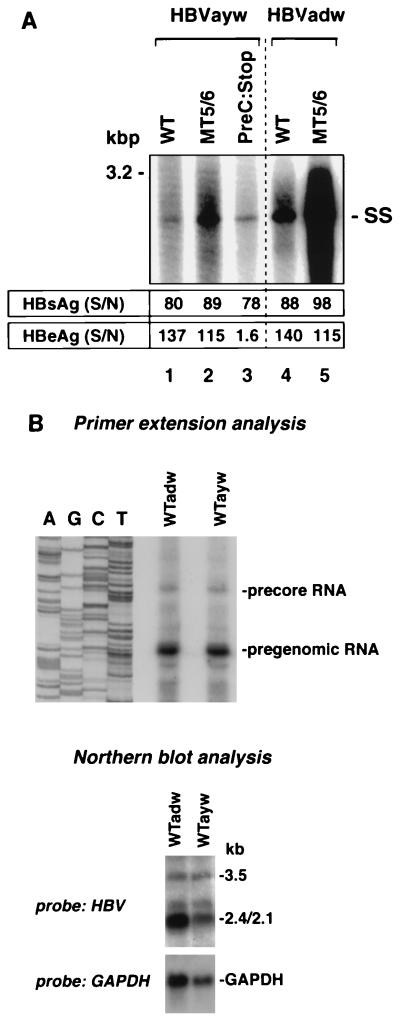
To study whether the observed MT5/6-induced enhanced encapsidation and replication also occur in an HBV wild-type strain other than adw, we introduced MT5/6 into a replication-competent construct of HBV strain ayw. This ayw strain was originally described by Galibert et al. (7). Compared to a 15-fold increase of replication in the adw background, MT5/6 led to a 2.5- to 5-fold increase in replication when introduced into the ayw strain (Fig. 9A). This finding demonstrates that the identified core promoter mutations result in enhanced replication independent of the strain background, though the effect on replication is quantitatively different between the two strains. In contrast, the introduction of a mutation (G to A at nt 1896) leading to a stop codon in the precore ORF and loss of HBeAg synthesis did not result in enhanced viral replication (Fig. 9A). Interestingly, HBV adw replicated ∼3- to 5-fold more efficiently than ayw although the levels of pregenomic and precore RNA transcripts (3.5-kb RNAs) were comparable, as indicated by primer extension and Northern blot analysis of HBV RNA (Fig. 9B).
FIG. 9.
Effects of core promoter and precore stop codon mutations on replication of different HBV strains. MT5/6 (C to T at nt 1768 and T to A at nt 1770) and precore stop codon mutation (G to A at nt 1896) were introduced into replication-competent constructs (HBVayw strain) as described in Materials and Methods. These constructs were transfected into HuH-7 cells, and viral replication, transcription, and antigen expression were analyzed as described in Materials and Methods. (A) Viral replication. WT, wild type; SS, single-stranded DNA. HBs and HBeAg levels are indicated as signal-to-noise ratio (S/N). (B) Transcription of adw and ayw constructs. HBV RNA was purified from HuH-7 cells transfected with either adwR9 or aywR9 and analyzed by primer extension using a HBV-specific antisense primer (upper panel) or Northern blotting using an HBV-specific probe (lower panel). Transfection efficiencies as monitored by pTKGH cotransfection were similar among all samples.
DISCUSSION
In this study, we have defined the molecular mechanism of enhanced viral replication and encapsidation induced by naturally occurring core promoter mutations (in construct MT5/6) isolated from patients with a fatal outbreak of fulminant hepatitis. The enhanced replication is apparently mediated through the effect of the core promoter mutations on core synthesis largely at the posttranscriptional or translational level, leading to enhanced encapsidation of pregenomic RNA. The enhanced core expression was the result of two different effects of the core promoter mutations. First, the core promoter mutations resulted in a minor (twofold) increase in pregenomic RNA transcription, as demonstrated by primer extension, RNase protection, and Northern blot analysis of HBV transcripts in HuH-7 cells transfected with replication-competent wild-type and mutant constructs (Fig. 2B and reference 2). Second, the twofold increase in pregenomic RNA transcription is accompanied by a much larger (15-fold) increase of core protein expression, which is paralleled by a similar increase in core protein synthesis in the metabolic labeling experiments (Fig. 3). Therefore, it is likely that the core promoter mutations exert a novel posttranscriptional or translational effect on core synthesis. Furthermore, this effect appears to be specific for core protein and not for RT-Pol (Fig. 6), which is translated from the same RNA. Both mechanisms, a minor increase in transcription and a substantial enhancement in translation (or other posttranscriptional processes), contributed to a 15-fold augmentation of core protein expression, nucleocapsid assembly, and replication.
Since the precore protein has been suggested to interfere with viral encapsidation (17), it is conceivable that a change in precore expression could alter encapsidation efficiency. Similarly, some core promoter mutations have been shown to result in a moderate decrease in precore protein and HBeAg expression, possibly explaining the phenotype of enhanced replication (4, 25). However, in our study the elimination of the precore ORF did not affect the phenotype of enhanced core expression and replication, essentially excluding a role of the precore protein in MT5/6-induced enhanced replication. This is consistent with a lack of effect of MT5/6 on precore gene transcription (2) and protein expression (HBeAg synthesis [Fig. 8C]). This finding has also been substantiated by several previous reports that the introduction of a mutation resulting in a precore stop codon (G to A in nt 1896) did not result in enhanced replication (10, 40, 43). In contrast, one report suggested that the precore stop codon mutation led to a high-replication phenotype (35). The explanation for this discrepancy is unclear but possibly includes laboratory strain differences or technical aspects of the experiments.
It is conceivable that elements outside the core promoter and gene are required for the effect of the core promoter mutations. However, several lines of evidence indicate that this is unlikely. First, the trans-complementation experiment demonstrated that enhanced replication occurred only when the core promoter mutations are provided together with the core protein (Fig. 1). Presence of the mutations in a core promoter construct directing pregenomic RNA synthesis had no effect on replication when core protein was provided in trans (Fig. 1). Second, the core promoter mutations did not appear to affect RT-Pol expression substantially other than a minor effect on transcription (Fig. 6). Third, analysis of the core expression constructs containing only the HBV enhancer and promoter elements driving the core ORF resulted in a similar increase in core synthesis associated with the mutations (Fig. 7). Fourth, our previous studies demonstrated that HBX or any possible antisense ORFs overlapping with the core promoter played no role in this effect (2).
By demonstrating that MT5/6 resulted in a posttranscriptional effect on core promoter activities, we have identified a novel function of the core promoter in the regulation of core protein expression. The molecular mechanism whereby MT5/6 induces the posttranscriptional or translational effect on core synthesis is not completely understood. It is interesting that the mutations are not part of the transcribed core RNA sequences (Fig. 7A) and therefore probably exert their main effect cotranscriptionally without affecting the transcriptional rate substantially. We reason that the mutations likely confer specific cis-acting sequence information to the core promoter, which is then translated into a functional effect. It is interesting that this core promoter region has been shown to contain sequence information (Fig. 7A) important for interaction with various members of nuclear receptor family (32) as well as in differential regulation of precore and pregenomic RNA transcription (42). These nuclear receptors appear to exert a differential effect on the transcription of these two forms of RNAs. Two other naturally occurring mutations (MT8 in this report) in this region have been shown to induce a selective decrease of precore RNA transcription (4). Although the sequence affected by MT5/6 is not part of the critical motifs important for interaction with these factors (42), it is situated immediately adjacent to them. Therefore, it is conceivable that MT5/6 can affect the differential bindings of these cellular factors. One of the effects of MT5/6 is evidenced by the twofold increase in the transcription of pregenomic RNA without affecting the precore RNA. In addition, we speculate that MT5/6 induces a specific change in the composition of transcription factors binding to this region in such a way that some of the factors may become complexed specifically with the transcript and, through some unknown mechanism, function to enhance translation of the RNA. The RNA polymerase II transcription complex has been shown to interact with splicing and polyadenylation factors to form an mRNA factory leading to coupled transcription, splicing, processing, and possibly transport of mRNAs (21, 26). In addition, there is evidence suggesting that nuclear processing and export of mRNA are closely and possibly physically linked to cytoplasmic translation (20). Therefore, transcription and translation may be tightly coupled, representing interaction of cellular factors with distinct functions in a highly interdependent manner.
Candidates for such a factor(s) could be some of the translation initiation factors such as eIF-4E or its associated factors, which bind to the 5′-terminal cap structure of most mRNAs to facilitate translation initiation in the cytoplasm (38). This factor has also been shown to be present in the nucleus (18) and probably plays an additional role in the transport of mRNA (33). However, such an effect of MT5/6 must be selective on the pregenomic RNA only, since the synthesis of precore protein is not similarly affected. This phenomenon can be explained by the possibility that MT5/6 confers the binding of such a factor only to the pregenomic RNA, or more plausibly that the pregenomic RNA is affected to a greater extent by this factor than the precore RNA. It is interesting that the encapsidation signal lies between the precore and core start codons, and it may function as a deterrant for translation of core protein from the pregenomic RNA, whose 5′ end lies upstream of the encapsidation signal. Such secondary structures have been shown to impede ribosomal binding, and eIF-4F, of which eIF-4E is a subunit, functions to relieve the translation inhibition of these structures (15). The ribosomal binding site (RBS; Kozak sequences) of the precore ORF, because of its position upstream of the encapsidation signal, may be more accessible to the translational machinery. In contrast, the RBS of the core ORF is part of the encapsidation signal (part of the stem structure) (27) and therefore may function less efficiently as a translation initiation signal. In addition, the RBS of the precore ORF (GCACCATG) conforms much better to the Kozak consensus sequence (CCACCATG) (16) than that of the core ORF (GGGGCATG), underscoring the possibility that translation of the core ORF is less efficient and therefore more sensitive to regulation by translational factors. The validity of this hypothesis awaits further experimentation.
Our data suggest that MT5/6 appears to induce less replication enhancement in the ayw than the adw strain. The difference in phenotypic expression of the mutations is probably due to sequence heterogeneity in this core promoter region, which may confer a qualitatively similar but quantitatively distinct response to the mutations. There are several sequence polymorphisms in this region distinctive for the adw and ayw strains, and whether they contribute to the observed difference in the impact of MT5/6 on replication is not known. Again this effect may be the result of posttranscriptional mechanism, since adw replicates ∼4- to 5-fold more efficiently than ayw but their levels of transcription are comparable (Fig. 9B and reference 2). Further experiments are necessary to resolve this issue.
Previous studies have identified core promoter mutations in various patient populations which share the phenotype of more aggressive liver disease (9, 12, 31, 34, 36). A common hallmark of several of these mutations is the phenotype of enhanced replication (4, 9, 25, 31). The mechanism of the enhanced replication in these strains is only partially understood. Two mutations in the enhancer II of the core promoter (indicated as MT8 in Table 1) have been shown to affect precore protein expression (4). This finding led the authors to conclude that the decrease in precore expression may have been responsible for the enhanced replication by derepressing nucleocapsid assembly, although no functional studies (effect of elimination of the precore ORF in the construct containing the mutation) were performed to verify this hypothesis (4, 25). Other mutations have been shown to alter the binding of nuclear transcription factors to the core promoter (9, 31). For some of these variants, the alteration of binding of transcription factors was associated with increased pregenomic RNA transcription and core and polymerase expression (9). Further studies are necessary to elucidate whether these naturally occurring core promoter mutations affect the same functional core promoter element as MT5/6 or whether different mechanisms apply to the individual mutations.
Core promoter mutations have been associated with more aggressive disease, including fulminant hepatitis (2, 9, 31, 34, 36). The MT5/6-induced increase in core protein expression and increased replication could potentially play an important role in the pathogenesis of fulminant hepatitis associated with this mutation. Since the core protein is a major target of the host immune response (5), the increase in core protein expression may render hepatocytes more vulnerable to host immune response and the enhanced replication may result in more widespread HBV infection in the liver. In addition, the precore stop codon mutation (G to A at nt 1896), which has been found in many cases of fulminant hepatitis B, results in the absence of HBeAg production, which, in turn, may direct a more Th1-like, proinflammatory response (23). A more vigorous and extensive immune response with enhanced viral replication may lead to massive liver injury and ultimately fulminant hepatic failure. Further studies in animal models such as woodchuck and chimpanzee (28) are necessary to ascertain whether the findings in the tissue culture system are applicable in vivo and whether the observed changes in replication and protein expression are indeed responsible for the more aggressive disease associated with these mutations. Functional analysis of hepatitis B virus mutants in vivo will be crucial in validating current concepts of virus-host interaction and the impact of these viral factors on HBV-induced disease.
ACKNOWLEDGMENTS
We thank Jim Ou (University of Southern California, Los Angeles) for providing the anticore antibody and Junying Yuan (Harvard Medical School, Boston, Mass.) for the gift of plasmid pBSlacZ. We also thank Jay Hoofnagle and Reed Wickner (NIDDK, National Institutes of Health, Bethesda, Md.) for helpful discussions.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1991. [Google Scholar]
- 2.Baumert T F, Rogers S A, Hasegawa K, Liang T J. Two core promoter mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J Clin Invest. 1996;98:2268–2276. doi: 10.1172/JCI119037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum H E, Galun E, Liang T J, von Weizsäcker F, Wands J R. Naturally occurring missense mutation in the polymerase gene terminating hepatitis B virus replication. J Virol. 1991;65:1836–1842. doi: 10.1128/jvi.65.4.1836-1842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckwold V E, Xu Z, Chen M, Yen T S B, Ou J-H. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J Virol. 1996;70:5845–5851. doi: 10.1128/jvi.70.9.5845-5851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chisari F V, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 6.Fields H A, Hollinger F B, Desmyter J, Melnick J L, Dreesman G R. Biochemical and biophysical characterization of hepatitis B core particles derived from Dane particles and infected hepatocytes. Intervirology. 1977;8:336–350. doi: 10.1159/000148909. [DOI] [PubMed] [Google Scholar]
- 7.Galibert F E, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequences of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 8.Ganem D, Varmus H E. The molecular biology of the hepatitis B virus. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 9.Günther S, Piwon N, Iwanska A, Schilling R, Meisel H, Will H. Type, prevalence, and significance of core promoter/enhancer II mutations in hepatitis B viruses from immunosuppressed patients with severe liver disease. J Virol. 1996;70:8318–8331. doi: 10.1128/jvi.70.12.8318-8331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasegawa K, Huang J, Rogers S A, Blum H E, Liang T J. Enhanced replication of a hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. J Virol. 1994;68:1651–1659. doi: 10.1128/jvi.68.3.1651-1659.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch R C, Lavine J E, Chang L-J, Varmus H E, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 12.Honda A, Ehata T, Ypkosuka O, Ohto M, Omata M. Mutations in enhancer 2/core promoter, X protein region of hepatitis B virus correlate with fatal fulminant hepatitis. Hepatology. 1994;20:792A. [Google Scholar]
- 13.Huang J, Liang T J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Junker-Niepmann M, Bartenschlager R, Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990;9:3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koromilas A E, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamberts C, Nassal M, Velhagen I, Zentgraf H, Schroeder C H. Precore-mediated inhibition of hepatitis B virus progeny DNA synthesis. J Virol. 1993;67:3756–3762. doi: 10.1128/jvi.67.7.3756-3762.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lejbkowicz F, Goyer C, Darveau A, Neron S, Lemieux R, Sienberg N. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc Natl Acad Sci USA. 1992;89:9612–9616. doi: 10.1073/pnas.89.20.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang T J, Hasegawa H, Rimon N, Wands J R, Ben-Porath E. A hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. N Engl J Med. 1991;324:1705–1709. doi: 10.1056/NEJM199106133242405. [DOI] [PubMed] [Google Scholar]
- 20.Maquat L E. When cells stop making sense: effect of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 21.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 22.McEwen C R. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal Biochem. 1967;20:114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- 23.Milich D R, Schödel F, Hughes J, Jones J E, Peterson D L. The hepatitis B virus core and e antigen elicit different Th cell subsets: antigen structure can affect Th cell phenotype. J Virol. 1997;71:2192–2201. doi: 10.1128/jvi.71.3.2192-2201.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Induction of apoptosis in fibroblasts by IL-1β-converting enzyme, a mammalian homolog of the C. elegans cell death gene ced-3. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 25.Moriyama K, Okamoto H, Tsuda F, Mayumi M. Reduced precore transcription and enhanced core-pregenome transcription of hepatitis B virus DNA after replacement of the precore-core promoter with sequences associated with e antigen-seronegative persistent infections. Virology. 1996;226:269–280. doi: 10.1006/viro.1996.0655. [DOI] [PubMed] [Google Scholar]
- 26.Mortillaro M J, Blencowe B J, Wei X, Nakayasu H, Du L, Warren S, Sharp P A, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nassal M, Junker-Niepmann M, Schaller H. Translational inactivation of RNA function: discrimination against a subset of genomic transcripts during HBV nucleocapsid assembly. Cell. 1990;63:1357–1363. doi: 10.1016/0092-8674(90)90431-d. [DOI] [PubMed] [Google Scholar]
- 28.Ogata N, Miller R H, Ishak K G, Purcell R H. The complete nucleotide sequence of a pre-core mutant of hepatitis B virus implicated in fulminant hepatitis and its biological characterization in chimpanzees. Virology. 1993;194:263–276. doi: 10.1006/viro.1993.1257. [DOI] [PubMed] [Google Scholar]
- 29.Pollack J R, Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993;67:3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollack J R, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pult I, Chouard T, Wieland S, Klemenz R, Yaniv M, Blum H E. A hepatitis B virus mutant with a new hepatocyte nuclear factor 1 binding site emerging in transplant-transmitted fulminant hepatitis B. Hepatology. 1997;25:1507–1515. doi: 10.1002/hep.510250633. [DOI] [PubMed] [Google Scholar]
- 32.Raney A K, Johnson J L, Palmer C N A, McLachlan A. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci USA. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato S, Suzuki K, Akahane Y, Akamatsu K, Akiyama K, Yunomura K, Tsuda F, Tanaka T, Okamoto H, Miyakawa Y, Mayumi M. Hepatitis B virus strains with mutations in the core promoter in patients with fulminant hepatitis. Ann Intern Med. 1995;122:241–248. doi: 10.7326/0003-4819-122-4-199502150-00001. [DOI] [PubMed] [Google Scholar]
- 35.Scaglioni P P, Melegari M, Wands J R. Biologic properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame. Virology. 1997;233:374–381. doi: 10.1006/viro.1997.8594. [DOI] [PubMed] [Google Scholar]
- 36.Sterneck M, Günther S, Sanantonia T, Fischer L, Rogiers X, Brölsch C E, Will H. The complete nucleotide sequence of hepatitis B virus in 9 patients with fulminant hepatitis B infection. Hepatology. 1996;24:300–306. doi: 10.1002/hep.510240203. [DOI] [PubMed] [Google Scholar]
- 37.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 38.Thach R E. Cap recap: the involvement of eIF-4F in regulating gene expression. Cell. 1992;68:177–180. doi: 10.1016/0092-8674(92)90461-k. [DOI] [PubMed] [Google Scholar]
- 39.Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317:489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- 40.Tong S-P, Li J-S, Vitvitski L, Trepo C. Replication capacities of natural and artificial precore stop codon mutants of hepatitis B virus: relevance of pregenome encapsidation signal. Virology. 1992;191:237–245. doi: 10.1016/0042-6822(92)90185-r. [DOI] [PubMed] [Google Scholar]
- 41.Will H, Reiser W, Weimer T, Pfaff E, Buescher M, Sprengel R, Cattaneo R, Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987;61:904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu X, Mertz J E. Differential regulation of the pre-C and pregenomic promoters of human hepatitis B virus by members of the nuclear receptor superfamily. J Virol. 1997;71:9366–9374. doi: 10.1128/jvi.71.12.9366-9374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan T T T, Faruqi A, Shih J W K, Shih C. The mechanism of natural occurrence of two closely linked HBV precore predominant mutations. Virology. 1995;211:144–156. doi: 10.1006/viro.1995.1387. [DOI] [PubMed] [Google Scholar]
- 44.Yuh C-H, Chang Y-L, Ting L-P. Transcriptional regulation of precore and pregenomic RNAs of hepatitis B virus. J Virol. 1992;66:4073–4084. doi: 10.1128/jvi.66.7.4073-4084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou S, Yang S Q, Standring D. Characterization of hepatitis B virus capsid particle assembly in Xenopus oocytes. J Virol. 1992;66:3086–3092. doi: 10.1128/jvi.66.5.3086-3092.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]