Abstract
This review focuses on technologies at the core of calcific aortic valve disease (CAVD) and drug target research advancement, including transcriptomics, proteomics, and molecular imaging. We examine how bulk RNA sequencing and single-cell RNA sequencing have engendered organismal genomes and transcriptomes, promoting the analysis of tissue gene expression profiles and cell subpopulations, respectively. We bring into focus how the field is also largely influenced by increasingly accessible proteome profiling techniques. In unison, global transcriptional and protein expression analyses allow for increased understanding of cellular behavior and pathogenic pathways under pathologic stimuli including stress, inflammation, low-density lipoprotein accumulation, increased calcium and phosphate levels, and vascular injury. We also look at how direct investigation of protein signatures paves the way for identification of targetable pathways for pharmacologic intervention. Here, we note that imaging techniques, once a clinical diagnostic tool for late-stage CAVD, have since been refined to address a clinical need to identify microcalcifications using positron emission tomography/computed tomography and even detect in vivo cellular events indicative of early stage CAVD and map the expression of identified proteins in animal models. Together, these techniques generate a holistic approach to CAVD investigation, with the potential to identify additional novel regulatory pathways.
Calcific aortic valve disease (CAVD) is a rapidly increasing disorder in developed countries. Disease severity ranges from aortic valve sclerosis, marked by leaflet thickening and calcification without major disruption to outflow dynamics, to aortic valve stenosis, marked by leaflet rigidity and flow obstruction. The latter is associated with an increased risk of cardiovascular mortality and morbidity.1 Mortality rates have increased from more than 50,000 cases in 1990 to roughly 125,000 cases in 2019.2 According to a meta-analysis and modeling study,3 the number of elderly patients (age, ≥75 years) with calcific aortic stenosis is expected to double by 2050 in the United States and Europe.4 Although the prevalence of CAVD is an increasing concern, treatment options remain restricted to two techniques for aortic valve replacement (AVR): surgical AVR (SAVR) and transcatheter AVR (TAVR).5, 6, 7 However, there is an increasing motivation to treat CAVD independent of such surgical measures, suggestive of pharmacologic treatments.8 For example, statins, via their lipid-lowering mechanisms, have been largely successful at reducing cardiovascular risk, however, to date, have shown no apparent benefit to mitigating aortic stenosis,9 which is indicative of a lack of comprehensive understanding of the underlying disease mechanisms. Strides to elucidate CAVD pathobiology and drivers prompt the implementation of innovative technologies, multi-omics, and network medicine.
Valvular calcification was first recorded in 1697 during an autopsy and, less than a decade later, became clinically associated with symptoms of dyspnea, angina, faintness, and aortic valve petrification.10 With the advent and implementation of clinical diagnostic tools such as radiography and fluoroscopy, autopsy was no longer the sole method of studying pathologic features of diseased valves.10,11 Such techniques now routinely prompt preoperative diagnoses of valvular calcification by visualizing calcified valves using roentgen film and orthodiagrams.12 By the mid-20th century, tomographic techniques showed that valvular calcification disturbs valve function by perturbing blood flow.13 The impact of these radiologic technologies have remained diagnostic rather than etiologic.10
Recent basic and clinical research using molecular imaging,14 multi-omics, and systems biology has provided insight into mechanisms that drive CAVD,15 resulting in its current classification as a disease driven by inflammatory cell infiltration, extracellular matrix formation, valve thickening, and remodeling, finally leading to aortic valve calcification and stenosis with leaflet rigidity and outflow obstruction (Figure 1).15, 16, 17, 18, 19, 20, 21, 22, 23 Disease progression leading to aortic stenosis can be broken down into three basic etiologic categories. First, rheumatic heart disease is the leading cause of aortic valve stenosis globally.24 Generally associated with overcrowding and inadequate access to health care, it is preceded by rheumatic fever and results in commissural fusion, fibrosis, and calcification.24,25 Second, congenital abnormal aortic valves (bicuspid and unicuspid valves) account for roughly half of all stenotic cases in developed countries.25, 26, 27 Notably, bicuspid valves develop aortic stenosis 10 to 20 years earlier than tricuspid valves,24 suggesting a significant role for hemodynamic flow patterns across the valve in pathogenesis.28 Third, calcification of normal aortic valves is suggested to have origins associated with inflammation, hyperlipidemia, aging, and poor economic lifestyle.9,26 In light of these distinct etiologies, comprehensive characterization of CAVD progression is necessary, thereby providing opportunities to identify therapeutic targets.
Figure 1.
Roadmap of advances in technology and understanding of disease pathophysiology. A PubMed query of “calcific aortic valve disease” yielded 5461 results from 1944 to 2023, showing the increasing trend of published research that paralleled the increased understanding of earlier stages of calcific aortic valve disease (CAVD). Within this 80-year period, the ability to analyze CAVD improved alongside development of novel techniques and groundbreaking technological advancements—tomography, RNA sequencing, near-infrared fluorescence, and proteomics to name a few—leading to the increase in discoveries made and benchmark findings published. This increase in novel technologies corresponds to a focal shift in the study of disease pathophysiology, with current technology and methodologies facilitating early stage CAVD investigation.5,10,14, 15, 16, 17, 18, 19, 20, 21 BAV, bicuspid aortic valve; TAV, tricuspid aortic valve. Images obtained from BioRender.com (Toronto, ON, Canada).
This review therefore focuses on technologies at the core of CAVD and drug target research advancement, including transcriptomics, proteomics, and molecular imaging. The authors of this review article examine how bulk RNA sequencing (RNA-seq) and single-cell RNA-seq (scRNA-seq) have engendered organismal genomes and transcriptomes, promoting the analysis of tissue gene expression profiles and cell subpopulations, respectively.29 They bring into focus how the field is also largely influenced by increasingly accessible proteome profiling techniques. In unison, global transcriptional and protein expression analyses (multi-omics) improve the understanding of cellular and molecular pathway responses to pathologic stimuli including stress, inflammation, low-density lipoprotein accumulation, increased calcium and phosphate levels, and vascular injury. The authors then examine how direct investigation of protein signatures paves the way for the identification of targetable pathways for pharmacologic intervention. Here, they note that imaging techniques, once a clinical diagnostic tool for late-stage CAVD marked by macrocalcifications, have since been refined to identify early stage microcalcifications. Positron emission tomography (PET) combined with computed tomography (CT) can now detect in vivo cellular events indicative of early stage CAVD15 and map the expression of identified proteins in animal models.16 Together, these techniques generate a holistic approach to CAVD investigation, with the potential to identify additional novel regulatory pathways.
Molecular Imaging
PET and CT
Various imaging modalities have become integral to many cardiovascular disease studies. For example, CT scans were among the earliest forms of imaging technology used to analyze CAVD.11 However, this technology, because of its low spatial sensitivity/resolution, is only capable of identifying late-stage, advanced calcification, not the subclinical microcalcifications that are proposed to precede the more readily detected macrocalcifications.30 On the other hand, a combination of CT paired with PET is able to detect microcalcifications.14
By using 18F-NaF, which binds to hydroxyapatite crystals, CT and PET scans can be used to identify vascular microcalcifications noninvasively in vivo, ex vivo, and in vitro.14,31,32 In the same light, 18F-fluorodeoxyglucose (18F-FDG) uptake is examined with PET to assess inflammation.14 Multiple studies confirm a relationship between 18F-FDG signal and atherosclerotic plaques, as an early indicator of CAVD, highlighting an overlap between macrophage-positive areas and 18F-FDG–positive areas.33, 34, 35 It has been suggested that 18F-FDG acts as a glucose analog that binds to glucose transport proteins to be taken up by the cell, mimicking the process by which macrophages and other inflammatory cells uptake glucose for cellular processes. The ability of these cells to incorporate higher rates of glucose than neighboring cell types makes 18F-FDG a suitable inflammatory target for PET imaging.34,36 Improvements in image quality, acquisition, and quantification make it more likely for clinical studies to be performed at the microcalcification stage.33
Near-Infrared Fluorescence
A more recent alternative to PET-CT scans is near-infrared fluorescence (NIRF) intravital molecular imaging. Commercially available NIRF dye, IRDye800CW (BroadPharm, San Diego, CA), can be paired with antibodies or nanobodies and injected into patients to be visualized as a real-time guiding target during procedures such as pediatric, oncological, or laparoscopic colorectal surgeries.32,37 Advantages of this procedure include the rapid clearance of the dye, low genotoxicity, high signal-to-background ratio, and deep tissue penetrance.38 In the context of cardiovascular disease–related surgeries, the use of such agents is not yet approved; instead, near-infrared autofluorescence imaging (coupled with optical coherence tomography) has been shown to identify high-risk plaques in patients undergoing catheterization procedures.39 Nonetheless, a NIRF calcium tracer technique can be used to visualize microcalcifications in explanted carotid artery plaques40; and the imaging modality has been used to characterize plaque morphology in animal models of atherosclerosis.41
NIRF imaging has also been used to study disease progression in atherosclerotic plaques, providing insight into aortic valve stenosis.42 Imaging probes target hydroxyapatite producing an osteogenic NIRF signal, which enables researchers to identify microcalcifications in vivo, ex vivo, and in vitro.15 Furthermore, intravenous co-injection of two different imaging probes that have excitation in different NIRF spectra allows simultaneous visualization of two different biological processes43 (eg, calcification and inflammation) (Figure 2).42
Figure 2.
Visualization of calcification ex vivo and in vivo using multiple imaging modalities. A: Co-injection of spectrally distinct fluorescent probes in apolipoprotein E (ApoE)–deficient mice visualizes a correlation between macrophages and osteogenesis in the root, arch, and abdominal aorta (ex vivo fluorescence reflectance imaging). B: Intravital near-infrared fluorescent molecular imaging was performed in ApoE-deficient mice using a calcified carotid artery to visualize the macrophages (macrophages, green) and calcium deposition (osteogenesis, red). The individual imaging signals were superimposed on one another (merged), showing the colocalization between macrophages and calcification in diseased tissue (in vivo near-infrared fluorescence molecular imaging). Scale bars: 3 mm (A); 250 μm (B). Adapted from Aikawa et al.42
Colocalization of the signals in atherosclerotic plaques has led to the hypothesis that macrophages themselves undergo the process of calcification through the release of calcifying extracellular vesicles, confirming the notion that calcification is an inflammatory disease.44 This hypothesis has been supported further through NIRF imaging by using a calcium tracer to identify the calcifying extracellular vesicles released from macrophages that cause the microcalcifications.45 Results from NIRF imaging were reinforced with gold standard histologic methods.46 Such corroborative stains often include, but are not limited to, hematoxylin and eosin, von Kossa, Oil Red O, Picrosirius red, and immunohistochemistry, which identify morphologic changes, calcification, lipid deposition, collagen stability, and protein localization, respectively.47, 48, 49, 50, 51 von Kossa substantiates calcification ex vivo. When paired with Oil Red O, the two stains validated a relationship between atherosclerosis and calcification in plaques identified with NIRF imaging.17,52 Immunofluorescence supports NIRF findings at the molecular level. Immunofluorescent staining against both macrophages (eg, anti-CD68 antibody) and calcification [eg, OsteoSense-680 fluorophore (PerkinElmer, Boston, MA)] in calcified mouse carotid arteries provided additional evidence to support the hypotheses that calcification is an inflammatory condition.42 NIRF and immunohistochemical techniques have also been applied to mouse valves, confirming the link between inflammation and mineralization in aortic valve leaflets.15
Because of the highly specific nature of the probes, NIRF imaging is especially useful when studying inflammation because it is less affected by the autofluorescence of surrounding tissues relative to other imaging modalities. This high specificity enables probes to monitor, over time, enzymes with extremely low abundances. For example, temporal analysis of the low-abundant enzyme, zymosan, showed zymosan-induced inflammation in murine models.53 This novel application has the potential to be useful in understanding the role of low-abundant molecules in CAVD.
NIRF imaging can visualize distinct cellular changes of early aortic valve disease in vivo, validating targets identified by transcriptomics and proteomics. In the aortic valves of hypercholesterolemic apolipoprotein E–deficient mice, macrophage accumulation, an established feature of CAVD,8 was detected by NIRF imaging.15 NIRF also was used to detect increased matrix metalloproteinases (MMP-1, -9, and -12) in CAVD, which could be used as a target in stenotic tissue.54 To confirm their role in CAVD, tracers bound to the activated MMPs were imaged at different stages of the disease. The MMP signal peaked before the valves were fully calcified, suggesting a role of MMPs in early CAVD progression and the potential for MMP imaging to predict CAVD.23
Limitations of NIRF imaging include a 24-hour time delay between injection of the agent and detection, suggesting a need for faster-acting agents,55,56 and signal attenuation by blood requiring a consensus on correction methods.57 However, molecular imaging in general is limited because it is not a method for identification of novel disease driver or disease biomarker molecules. Large-scale screening efforts to identify these molecules are undertaken with the assistance of multi-omics technologies.
Transcriptomics
Transcriptomics is the characterization of multiple gene-expressed RNAs from cells or tissues that in its infancy relied on microarray or fluorescently labeled nucleic acid hybridization technologies. More recently, transcriptomics has transitioned to unbiased RNA-seq methods.58 Bulk RNA-seq is any sequencing method that provides an average gene expression or abundance profile from a cell population or tissue. scRNA-seq, on the other hand, relies on isolation of individual cells for subsequent gene expression profiling. The latter therefore provides a view of the extent of cellular identity and heterogeneity in a cell population or tissue. Defining the extent of cellular heterogeneity may help identify disease driver cell subpopulations,59,60 which, in turn, can be targeted pharmacologically.61 A comprehensive review of the various RNA-seq platforms has been reported.62 The following paragraphs describe the application of this technology to uncover the molecular mechanisms at the root of CAVD.
Bulk RNA-Seq of Calcified Aortic Valve Leaflets
Within the past 10 years, a few studies have used RNA-seq to profile the transcriptomes of leaflets derived from patients with calcified tricuspid aortic valve (TAV) and/or bicuspid aortic valve (BAV) disease.20,21,63 BAV is a congenital heart disease that promotes early onset aortic stenosis and calcification in patients. Therefore, it has been hypothesized that the disease drivers of BAV and TAV are distinct. One study compared calcified TAV and BAV (n = 10 each) leaflets from patients who underwent aortic valve replacement; as controls, normal tricuspid aortic valves were used from patients (n = 10) who underwent orthotopic heart transplantation, but had normal aortic valves.21 Interestingly, only two genes were expressed differentially when comparing calcified TAV versus calcified BAV: IGF1 and RSPO2 were increased in calcified BAV. However, 462 and 329 genes were expressed differentially when calcified BAV and normal TAV, and calcified TAV and normal TAV were compared, respectively.21 An explanation for the limited number of differentially expressed transcripts between the calcified tissues is that they represent the end stage of each disease, which likely has more commonality than the early stages.21,64 This underwhelming result is not a drawback of RNA-seq itself, rather it is a limitation resulting from the inaccessibility of early stage disease samples. As shown further in the next paragraph, alternative strategies to analyze late-stage diseased samples have enabled inferences of early stage molecular signatures.
An alternative approach to RNA-seq alone to identify CAVD drivers from late-stage diseased samples is the implementation of genome-wide associations. RNA-seq–dependent expression of quantitative trait loci mapping (n = 233 human aortic valve samples) combined with genome-wide associations, termed transcriptome-wide association study, identified the gene PALMD as a candidate driver of CAVD, particularly when PALMD is underexpressed.65 This finding inspired a subsequent phenome-wide study (2359 cases and 350,060 controls) of PALMD as a possible therapeutic target, which, in turn, confirmed that aortic valve–specific, genetically deduced expression of PALMD is associated inversely with calcific aortic valve stenosis.66
scRNA-seq for the Identification of CAVD Driver Cell Subpopulations
scRNA-seq confronts the limitations of understanding aortic valve heterogeneity, which bulk RNA-seq is unable to accomplish on its own. Resolution at the single-cell level allows for transcriptome analysis of, and distinction between, cell subpopulations. scRNA-seq was introduced in 2009 to determine transcriptomes of single blastomeres and oocytes from mice.18 As a result of very recent scRNA-seq studies, it now has been established that aortic valve tissue is highly heterogeneous, consisting of distinct extracellular matrix layers with various cell subpopulations.16,59,60,67, 68, 69
One such study performed scRNA-seq analysis of aortic valve leaflets from patients with CAVD (n = 4) and patients whose valvular tissues were removed during corrective surgery for aortic dissection (n = 2).60 The combined patient scRNA-seq data analysis showed 12 candidate cell subpopulations that were ascribed to either valvular stromal, interstitial, or endothelial cells; or monocytes, macrophages, or lymphocytes.60 For example, based on differential transcripts, the valvular stromal cells were classified into five subpopulations, the interstitial cells were classified into three subpopulations, and the endothelial cells were classified into two subpopulations. This study also confirmed that 7 of the 12 cell populations were unique to, or more prevalent in, the CAVD leaflets, and that the monocyte cells were more prevalent in normal leaflets. Subsequent immunofluorescence or immunochemistry was used to localize the transcript products (proteins), which defined each of the seven CAVD-enriched cell populations in diseased leaflets such as the three stromal cell–derived populations that were defined by high expression of either metallothionein-1A (MT1A), peptidase inhibitor 3 (PI3), or the ribosomal subunit homolog protein (CMSS1). Immunohistochemical analysis showed that MT1A and CMSS1 were localized to the spongiosa and fibrosa layers, whereas the PI3 signal was increased in the vicinity of the endothelial layer of CAVD leaflets. Such integrational studies highlight the benefits of this multimodal approach (scRNA-seq to molecular imaging) to generate an understanding of the disease as a system and identify ways to treat it.60
A second study used a high-throughput screening–flow cytometry panel strategy to identify a potential subpopulation of progenitor/stem cell–like population of valvular cells that may drive the tissues calcification potential.59 These candidate disease-driver cells then were isolated from CAVD leaflets using fluorescence-activated cell sorting for subsequent cell culture and scRNA-seq analysis. The isolated disease-driver cells showed an increased potential to calcify in vitro when compared with unsorted valvular interstitial cells. Moreover, scRNA-seq showed that when these cells were cultured in normal versus osteogenic media, each culture condition induced differentiation into multiple yet distinct cell subpopulations. One of the osteogenic clusters contained high levels of a known procalcification enzyme, alkaline phosphatase, and also the newly described amine oxidase-A. Subsequent studies confirmed that loss of amine oxidase-A inhibited the calcification potential in vitro.59 This scRNA-seq analysis of a cell culture model for valvular calcification being mediated by a progenitor cell population provides means to glean insight into the early mechanism that drives disease progression, even from end-stage diseased tissue.
Proteomics
DNA is transcribed into RNA transcripts, which are translated into proteins.70 Proteins are metabolic and physiological drivers of all bodily functions as well as disease pathogenesis.71 They provide insight into disease mechanisms and can be interrogated on a large scale via proteomic techniques. Proteomic techniques range in complexity and level of data generation. The earlier chromatography and blotting techniques are useful for smaller sample sizes but are incapable of depicting protein expression levels. As more advanced technologies such as protein arrays, advanced gel methods, mass spectrometry, and some sequencing methods rose to the forefront, the ability to perform proteomic analysis on more complex samples with a higher degree of sensitivity arose with them.72,73 There are three main technologies that are used to conduct high-throughput proteomics: the aptamer platform,74 the proximity ligation assay,75 and mass spectrometry.73 However, unlike mass spectrometry, which is permissive to unbiased screening of previously uncharacterized proteins and post-translational modifications, the aptamer platform and the proximity ligation assay are detection and quantification panels of characterized proteins. Nonetheless, any of these technologies permit analysis of thousands of proteins simultaneously, increasing the potential for identifying targets for further research and drug therapy tremendously.76
Mass Spectrometry–Based Proteomics of CAVD Tissues
Proteome profiling of CAVD tissues has emerged only within the past 5 to 6 years, enabled by mass spectrometry–based proteomics. The first proteome of human aortic valves was published in 2018.16 Mass spectrometry was used to characterize the differential distribution of proteins across the three layers that make up the human aortic valve—the ventricularis, fibrosa, and spongiosa—to better understand their constituents’ roles in disease pathogenesis. Leaflets obtained from patients who underwent aortic valve replacement surgeries (diseased) or from autopsy (nondiseased) were compared. Their findings identified layer-enriched proteins, including apolipoprotein B in fibrosa, glial fibrillary acidic protein in spongiosa, and calponin-1 in ventricularis, which all were increased in the diseased compared with nondiseased sample leaflets.16 These and other identified proteins were validated with imaging technologies that, in part, confirmed glial fibrillary acidic protein as a novel marker of the spongiosa layer (Figure 3). This study also noted that in addition to apolipoprotein (Apo) B, other apolipoproteins, such as ApoA-I and ApoA-II, which are located primarily on high-density lipoprotein (HDL), and ApoC-III, which is located on all classes of lipoproteins (HDL, and very-low-density and low-density lipoproteins), were increased in calcified versus noncalcified samples, leading to the hypothesis that one or more of these apolipoproteins may contribute to the disease pathogenesis.
Figure 3.
Immunofluorescent verification of layer-specific proteins. Microlayer atlas of calcific aortic valve disease (CAVD) tissue. Immunofluorescence corroborates layer proteomic findings and visualizes the localization of apolipoprotein B (ApoB), proline-arginine rich end leucine rich repeat protein (PRELP), sulfatase 1 (SULF1), and procollagen C-endopeptidase enhancer 2 (PCOLCE2) to the fibrosa layer; glial fibrillary acidic protein (GFAP) to the spongiosa; and cysteine and glycine rich protein 1 (CSRP1), calponin-1 (CNN1), and alpha-2-glycoprotein 1, zinc-binding (AZGP1) to the ventricularis valvular interstitial cells. Scale bars = 100 μm. Adapted from Schlotter et al.16
In a follow-up study, 12 apolipoproteins were investigated in more detail.19 First, the investigators used a targeted proteomics strategy,73 entailing the use of mass spectrometry peptide standards, to precisely quantify and verify the subset of apolipoproteins (8 of 12) that were increased in calcified portions of aortic valve leaflets. ApoC-III was one such apolipoprotein that also was confirmed to co-localize with calcified regions within CAVD leaflets. The potential of ApoC-III to promote disease progression was shown in vitro, increasing calcification potential, compared with controls, when incubated with human valvular interstitial cells. Subsequent proteomic analysis showed that ApoC-III induced an enrichment of proteins involved in oxidative stress, inflammation, and removal of reactive oxygen species.19 This study showed the value in conducting disease tissue–sourced proteomics to identify CAVD drivers, but tissue acquisition is dependent on patients reaching stages of morbidity or mortality. Thus, there is an initiative to investigate, at a minimum, the circulatory system for potential prognostic molecules for CAVD development.
Proteomics of Plasma and Urine to Identify CAVD Biomarkers
Urine is a clinically relevant and stable medium for uncovering proteins associated with CAVD. Proteomic analysis of urine from patients with aortic valve stenosis and coronary artery disease identified von Willebrand factor and tissue inhibitor matrix metalloproteinase 1 as signature proteins of diseased samples relative to healthy controls.77 Furthermore, a strong positive correlation was found between von Willebrand factor and HDL-cholesterol and between tissue inhibitor matrix metalloproteinase 1 and HDL-cholesterol in patients with aortic valve stenosis.77 Subsequent confirmation of the strong association between serum HDL and CAVD progression strengthened the proposed relationship between known biomarkers, von Willebrand factor, and tissue inhibitor matrix metalloproteinase 1, and the progression of CAVD.78 This finding not only has implications for therapeutic targeting, but also suggests an influential role of urine for routine screening of early CAVD. As a noninvasive alternative for sample collection, it is particularly appealing for biomarker classification.
When it comes to stable and clinically relevant media on which to perform proteomic analysis, human plasma is the gold standard. Human plasma is a unique compartment for extensive biomarker discovery, drawing from proteins secreted from many organs and tissues throughout the body.79 The high throughput and widespread nature of plasma proteomics generates efficient yield of hundreds of proteins within thousands of samples: a large-scale performance that is rarely achieved using tissue samples.80 Applied to cardiovascular disease cohorts, this technology has verified the role of apolipoproteins, among others, in atherosclerosis and other agents of cardiovascular disease. Plasma proteomics also identified novel proteins such as low-density lipoprotein receptor–related protein 2 to be investigated further.81 With its increasing application among cardiovascular and other diseases, plasma proteomics is the next frontier for extracting information about CAVD biomarkers.73,81 Moving forward, it is important to develop standardized pipelines concerning the design and execution of the study, as well as how the resulting data are processed and analyzed.82 The Human Plasma Proteome Project is a platform that promotes this sort of systemization, while engendering a space to investigate collective challenges to proteomic studies.82,83
Proteomics at the Single-Cell Level
Proteomics provides valuable insight into the protein profiles of diseased tissue, shedding light on the protein interactions that contribute to CAVD pathophysiology. Combining proteomic and molecular imaging technologies, as previously mentioned, could serve as an efficient tool to visualize proteome spatial profiles. Until recently, single-cell proteomics or single-molecule proteomics were unexplored areas of translational research.84 Although there have been strides in performing single-cell proteomics, specifically by mass spectrometry, this analysis has been limited by the sensitivity of such high-resolution samples. Even when single-cell proteomic analysis is achieved, drawbacks remain with its low availability and its inability to provide high-throughput results.85 These limitations, in addition to the importance of understanding expression profiles when investigating CAVD, necessitate transcriptomic techniques. Transcriptional analysis not only resolves concerns surrounding proteomic analysis of high-throughput single-cell resolution, but also provides contextual information behind CAVD expression profiles.
Multi-Omics Applied to CAVD Research
We now understand that transcript levels do not directly translate to protein levels, leaving an imbalance between gene and protein profiles. The poor correlation between the multiple gene-expressed RNA and protein can be accounted for by various biological influences including the secondary structure of multiple gene-expressed RNAs, regulatory proteins, regulatory soluble RNAs, ribosomal availability, and the half-life of individual proteins.84,86 In the same study that conducted the first proteome profiling of normal and diseased aortic valve leaflets, it was shown that the accumulation of most apolipoproteins in diseased valves was independent of their transcript levels, supporting the notion that circulating lipoproteins are the likely source of their apolipoproteins.16,19
Integrative approaches also help draw the bridge between transcript or gene levels, and their protein product and localization within the body. Of late, amyloid protein deposition has been implicated in the development of calcification within the aortic valve.87 Two-dimensional gel electrophoresis has identified serum amyloid P–component up-regulation within calcific aortic valves.88 A deeper dive into the role of amyloid protein in CAVD progression was taken with a three-dimensional multi-omics approach that analyzed prior transcriptomics and proteomics studies. This approach found the gene for APP as a connector gene for three input molecules from this transcriptomic and proteomic data, even though the APP gene did not exist within the transcriptomic and proteomic data used to generate this three-dimensional network. Subsequent immunohistochemical staining localized APP exclusively to areas of calcification within extracted aortic valves, illustrating how the integration of techniques allows for a more comprehensive understanding of the interaction between transcripts and protein products within the scope of CAVD.89
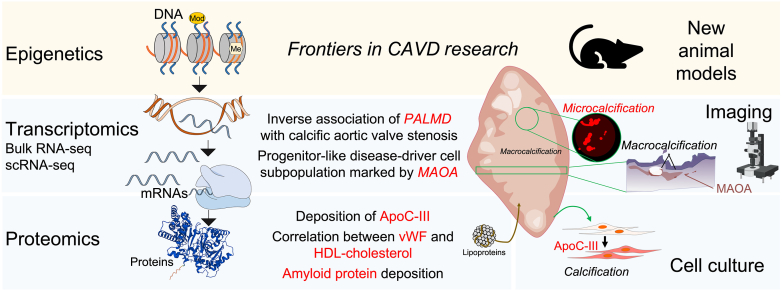
Frontiers in CAVD Research
Epigenetics
Epigenetics is the study of the factors, independent of mutations, that directly affect a gene's transcription activity, such as noncoding RNAs, DNA methylation, and heterochromatin components such as histones.90 The assay for transposase-accessible chromatin (ATAC-seq) is a fast, sensitive technique used to assess genome-wide chromatin accessibility by adding the viral enzyme, transposase, to DNA to tag open chromogen sites that can be mapped to known transcriptional start sites and used to infer transient gene expression.91 Only recently has this technique been applied to calcific aortic valves to examine disease development.92 Given the dynamic nature of CAVD, epigenetics most likely will be crucial in understanding the diversity in etiologic mechanisms of disease (Figure 4).92 As of now, epigenetics has been used to identify a role of Notch1 expression in valvular interstitial cell differentiation.93
Figure 4.
The multi-omics landscape for calcific aortic valve disease (CAVD). Molecular profiling enabled by the three -omics fields that encompass the central dogma of biology, DNA (epigenetics) to multiple gene–expressed RNA (mRNA) (transcriptomics) to protein (proteomics), and the integration of animal models, imaging technologies, and cell culture models to enable the understanding of CAVD. Molecules of interest, epigenetics, and the further development of animal models represent part of the frontiers for CAVD research. Most studies to date have relied on human aortic valves retrieved from valve-replacement surgeries. ApoC-III, apolipoprotein C-III; HDL, high-density lipoprotein; MAOA, monoamine oxidase A; PALMD, palmdelphin; RNA-seq, RNA sequencing; scRNA-seq, single-cell RNA sequencing; vWF, von Willebrand factor.
The Potential Impact of TAVR on CAVD Research
TAVR, a catheter-based, less-invasive alternative to SAVR, generates a potential obstacle surrounding sample (explanted valves) accessibility. As the advantages of TAVR are documented, therein lies a decreased dependence on SAVR for AVR procedures,94 and thus fewer explants for valvular research. Clinical cohort studies have shown that patients who underwent a TAVR procedure spend half the time in the hospital after surgery as compared to those who have undergone a SAVR procedure. Patients who underwent a TAVR procedure were also more commonly discharged to their home, compared with those who underwent a SAVR procedure who more commonly were discharged to a rehabilitation facility. It also was found that there was a lower hospital mortality rate after surgery among TAVR patients compared with SAVR patients.95 These benefits of TAVR procedures compared with SAVR procedures point to the anticipated increase in the use of these catheter-based procedures. As the acceptance of TAVR increases, it is increasingly important to maximize the information extracted from dwindling diseased tissue, thrusting human tissue studies into a race against time.64 The multimodal analytical tools mentioned earlier are not only highly effective techniques in discovering new insights in pathobiology, but also eliminate restrictions involving the inability to perform high-throughput, robust analysis. As these techniques become widespread, standardization of methods will allow for more effective use of tissue and ensure reproducible results.
Limitations to Current Animal Models
Apolipoprotein E–deficient mouse models, among other CAVD models in which diet and/or mutations in lipid metabolism (such as low-density lipoprotein receptor and apolipoproteins) induce dyslipidemia, are beneficial because they are used commonly to study multiple cardiovascular diseases and thus are readily accessible. Although such disease models may represent certain aspects of the disease, they assume dyslipidemia-dependent etiology that does not exhaustively represent the pathogenesis of any of the three basic etiologic categories of human CAVD: rheumatic heart disease, congenitally abnormal valvular disease, and atherosclerotic calcification of congenitally normal valves.9 The effectiveness of statins is the perfect example of the limitations of these mouse models. Statins, which have been proven by randomized trials, the Simvastatin and Ezetimibe in Aortic Stenosis trial and the Scottish Aortic Stenosis and Lipid Lowering Trial, Impact on Regression, only to be effective with dyslipidemia as the disease facilitator, do not act on all pathways of CAVD pathogenesis. When factors such as disease pathogenesis and stage are not controlled, they show no effect on aortic valve stenosis progression.9 This suggests that CAVD cases of differing etiologic origins likewise have differing underlying mechanisms that require suitable animal models for further investigation. Identifying such cost-effective animal models (Figure 4) will be beneficial for understanding the pathogenesis of atherosclerotic calcification of congenitally normal valves.
Conclusions
Research in the field of valvular calcification has advanced rapidly in recent years. The use of proteomics, transcriptomics, and molecular imaging (Figure 4) has unlocked new avenues of study that enhance the current understanding of CAVD pathobiology and, in conjunction with one another, become even more powerful. Although considerable progress has been made, there still are unanswered questions regarding the disease mechanisms. For example, how can the calcification potential of progenitor-like cells in the aortic valve be mitigated, and is amine oxidase-A a viable clinical target (Figure 4)? If the documented accumulation of apolipoproteins in the valve (Figure 4) is owing to a life-long deposition of lipoproteins and their constituents within the extracellular matrix, would lipid-lowering interventions be effective?
Multi-omics technologies are often used to discover disease drivers of late-stage CAVD tissue; however, much of the current research fails to investigate the origins of early stage drivers. As a result, therapeutic targeting remains possible only with late stages of the disease, and falls short of thwarting disease progression.96,97 To prevent CAVD, it is crucial to continue to examine disease pathophysiology at early stages, which will become increasingly accessible with technological advancement and with suitable animal and cell culture models. Addressing this issue requires coordination between clinical and basic research institutions to ensure the integrity of the discoveries being made via translational research.
Disclosure Statement
None declared.
Footnotes
Advances in Understanding Cardiovascular Disease Pathogenesis Theme Issue
Supported by NIH grants R01 HL136431, R01 HL141917, and R01 HL147095 (E.A.’s laboratory).
This article is part of a review series highlighting the novel insights provided by next-generation approaches to our understanding of cardiovascular disease pathogenesis and treatment.
References
- 1.Otto C.M., Lind B.K., Kitzman D.W., Gersh B.J., Siscovick D.S. Association of aortic-valve sclerosis with cardiovascular mortality and morbidity in the elderly. N Engl J Med. 1999;341:142–147. doi: 10.1056/NEJM199907153410302. [DOI] [PubMed] [Google Scholar]
- 2.Yu J., Wang Z., Bao Q., Lei S., You Y., Yin Z., Xie X. Global burden of calcific aortic valve disease and attributable risk factors from 1990 to 2019. Front Cardiovasc Med. 2022;9 doi: 10.3389/fcvm.2022.1003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osnabrugge R.L., Mylotte D., Head S.J., Van Mieghem N.M., Nkomo V.T., LeReun C.M., Bogers A.J., Piazza N., Kappetein A.P. Aortic stenosis in the elderly: disease prevalence and number of candidates for transcatheter aortic valve replacement: a meta-analysis and modeling study. J Am Coll Cardiol. 2013;62:1002–1012. doi: 10.1016/j.jacc.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Tsao C.W., Aday A.W., Almarzooq Z.I., Alonso A., Beaton A.Z., Bittencourt M.S., et al. Heart disease and stroke statistics-2022 update: a report from the American Heart Association. Circulation. 2022;145:e153–e639. doi: 10.1161/CIR.0000000000001052. [DOI] [PubMed] [Google Scholar]
- 5.Cribier A., Eltchaninoff H., Bash A., Borenstein N., Tron C., Bauer F., Derumeaux G., Anselme F., Laborde F., Leon M.B. Percutaneous transcatheter implantation of an aortic valve prosthesis for calcific aortic stenosis: first human case description. Circulation. 2002;106:3006–3008. doi: 10.1161/01.cir.0000047200.36165.b8. [DOI] [PubMed] [Google Scholar]
- 6.Falasa M., Holmes H.R., Neal D., Choi C.Y., Park K., Bavry A.A., Freeman K.A., Manning E.W., Stinson W.W., Jeng E.I. Outcome and cost comparisons between surgical and transcatheter aortic valve replacements. Innovations. 2022;17:482–490. doi: 10.1177/15569845221125474. [DOI] [PubMed] [Google Scholar]
- 7.Saito S., Sairenchi T., Hirota S., Niitsuma K., Yokoyama S., Kanno Y., Kanazawa Y., Tezuka M., Takei Y., Tsuchiya G., Konishi T., Shibasaki I., Ogata K., Monta O., Tsutsumi Y., Fukuda H. Prosthetic valve function after aortic valve replacement for severe aortic stenosis by transcatheter procedure versus surgery. J Cardiovasc Dev Dis. 2022;9:355. doi: 10.3390/jcdd9100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aikawa E., Libby P. A rock and a hard place: chiseling away at the multiple mechanisms of aortic stenosis. Circulation. 2017;135:1951–1955. doi: 10.1161/CIRCULATIONAHA.117.027776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermans H., Herijgers P., Holvoet P., Verbeken E., Meuris B., Flameng W., Herregods M.C. Statins for calcific aortic valve stenosis: into oblivion after SALTIRE and SEAS? An extensive review from bench to bedside. Curr Probl Cardiol. 2010;35:284–306. doi: 10.1016/j.cpcardiol.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Davies C.E., Steiner R.E. Calcified aortic valve; clinical and radiological features. Br Heart J. 1949;11:126–136. doi: 10.1136/hrt.11.2.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sparks J.V., Evans C. Radiography of calcification in cardiac valves during life. Br Med J. 1934;1:1028–1040.4. [PMC free article] [PubMed] [Google Scholar]
- 12.Sosman M.C. The position of the heart valves and their relation to the anterior chest wall in living subjects with abnormal hearts. Am Heart J. 1934;10:156–162. [Google Scholar]
- 13.Soloff L.A., Zatuchni J., Fisher H. Visualization of valvular and myocardial calcification by planigraphy. Circulation. 1954;9:367–370. doi: 10.1161/01.cir.9.3.367. [DOI] [PubMed] [Google Scholar]
- 14.Dweck M.R., Jones C., Joshi N.V., Fletcher A.M., Richardson H., White A., Marsden M., Pessotto R., Clark J.C., Wallace W.A., Salter D.M., McKillop G., van Beek E.J., Boon N.A., Rudd J.H., Newby D.E. Assessment of valvular calcification and inflammation by positron emission tomography in patients with aortic stenosis. Circulation. 2012;125:76–86. doi: 10.1161/CIRCULATIONAHA.111.051052. [DOI] [PubMed] [Google Scholar]
- 15.Aikawa E., Nahrendorf M., Sosnovik D., Lok V.M., Jaffer F.A., Aikawa M., Weissleder R. Multimodality molecular imaging identifies proteolytic and osteogenic activities in early aortic valve disease. Circulation. 2007;115:377–386. doi: 10.1161/CIRCULATIONAHA.106.654913. [DOI] [PubMed] [Google Scholar]
- 16.Schlotter F., Halu A., Goto S., Blaser M.C., Body S.C., Lee L.H., Higashi H., DeLaughter D.M., Hutcheson J.D., Vyas P., Pham T., Rogers M.A., Sharma A., Seidman C.E., Loscalzo J., Seidman J.G., Aikawa M., Singh S.A., Aikawa E. Spatiotemporal multi-omics mapping generates a molecular atlas of the aortic valve and reveals networks driving disease. Circulation. 2018;138:377–393. doi: 10.1161/CIRCULATIONAHA.117.032291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otto C.M., Kuusisto J., Reichenbach D.D., Gown A.M., O'Brien K.D. Characterization of the early lesion of 'degenerative' valvular aortic stenosis. Histological and immunohistochemical studies. Circulation. 1994;90:844–853. doi: 10.1161/01.cir.90.2.844. [DOI] [PubMed] [Google Scholar]
- 18.Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A., Lao K., Surani M.A. mRNA-seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 19.Schlotter F., de Freitas R.C.C., Rogers M.A., Blaser M.C., Wu P.J., Higashi H., Halu A., Iqbal F., Andraski A.B., Rodia C.N., Kuraoka S., Wen J.R., Creager M., Pham T., Hutcheson J.D., Body S.C., Kohan A.B., Sacks F.M., Aikawa M., Singh S.A., Aikawa E. ApoC-III is a novel inducer of calcification in human aortic valves. J Biol Chem. 2021;296 doi: 10.1074/jbc.RA120.015700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padang R., Bagnall R.D., Tsoutsman T., Bannon P.G., Semsarian C. Comparative transcriptome profiling in human bicuspid aortic valve disease using RNA sequencing. Physiol Genomics. 2015;47:75–87. doi: 10.1152/physiolgenomics.00115.2014. [DOI] [PubMed] [Google Scholar]
- 21.Guauque-Olarte S., Droit A., Tremblay-Marchand J., Gaudreault N., Kalavrouziotis D., Dagenais F., Seidman J.G., Body S.C., Pibarot P., Mathieu P., Bosse Y. RNA expression profile of calcified bicuspid, tricuspid, and normal human aortic valves by RNA sequencing. Physiol Genomics. 2016;48:749–761. doi: 10.1152/physiolgenomics.00041.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman R.V., Otto C.M. Spectrum of calcific aortic valve disease: pathogenesis, disease progression, and treatment strategies. Circulation. 2005;111:3316–3326. doi: 10.1161/CIRCULATIONAHA.104.486738. [DOI] [PubMed] [Google Scholar]
- 23.New S.E., Aikawa E. Molecular imaging insights into early inflammatory stages of arterial and aortic valve calcification. Circ Res. 2011;108:1381–1391. doi: 10.1161/CIRCRESAHA.110.234146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aluru J.S., Barsouk A., Saginala K., Rawla P., Barsouk A. Valvular heart disease epidemiology. Med Sci. 2022;10:32. doi: 10.3390/medsci10020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pomerance A. Pathogenesis of aortic stenosis and its relation to age. Br Heart J. 1972;34:569–574. doi: 10.1136/hrt.34.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts W.C., Ko J.M. Frequency by decades of unicuspid, bicuspid, and tricuspid aortic valves in adults having isolated aortic valve replacement for aortic stenosis, with or without associated aortic regurgitation. Circulation. 2005;111:920–925. doi: 10.1161/01.CIR.0000155623.48408.C5. [DOI] [PubMed] [Google Scholar]
- 27.Roberts W.C., Vowels T.J., Filardo G., Ko J.M., Mathur R.P., Shirani J. Natural history of unoperated aortic stenosis during a 50-year period of cardiac valve replacement. Am J Cardiol. 2013;112:541–553. doi: 10.1016/j.amjcard.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 28.Atkins S.K., Sucosky P. Etiology of bicuspid aortic valve disease: focus on hemodynamics. World J Cardiol. 2014;6:1227–1233. doi: 10.4330/wjc.v6.i12.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pimpalwar N., Czuba T., Smith M.L., Nilsson J., Gidlof O., Smith J.G. Methods for isolation and transcriptional profiling of individual cells from the human heart. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e05810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung J.J., Jadbabaie F., Sadeghi M.M. Molecular imaging of calcific aortic valve disease. J Nucl Cardiol. 2018;25:1148–1155. doi: 10.1007/s12350-017-1158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irkle A., Vesey A.T., Lewis D.Y., Skepper J.N., Bird J.L., Dweck M.R., Joshi F.R., Gallagher F.A., Warburton E.A., Bennett M.R., Brindle K.M., Newby D.E., Rudd J.H., Davenport A.P. Identifying active vascular microcalcification by (18)F-sodium fluoride positron emission tomography. Nat Commun. 2015;6:7495. doi: 10.1038/ncomms8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayami S., Matsuda K., Iwamoto H., Ueno M., Kawai M., Hirono S., Okada K., Miyazawa M., Tamura K., Mitani Y., Kitahata Y., Mizumoto Y., Yamaue H. Visualization and quantification of anastomotic perfusion in colorectal surgery using near-infrared fluorescence. Tech Coloproctol. 2019;23:973–980. doi: 10.1007/s10151-019-02089-5. [DOI] [PubMed] [Google Scholar]
- 33.Weiss R.M., Miller J.D., Heistad D.D. Fibrocalcific aortic valve disease: opportunity to understand disease mechanisms using mouse models. Circ Res. 2013;113:209–222. doi: 10.1161/CIRCRESAHA.113.300153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudd J.H., Narula J., Strauss H.W., Virmani R., Machac J., Klimas M., Tahara N., Fuster V., Warburton E.A., Fayad Z.A., Tawakol A.A. Imaging atherosclerotic plaque inflammation by fluorodeoxyglucose with positron emission tomography: ready for prime time? J Am Coll Cardiol. 2010;55:2527–2535. doi: 10.1016/j.jacc.2009.12.061. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z., Machac J., Helft G., Worthley S.G., Tang C., Zaman A.G., Rodriguez O.J., Buchsbaum M.S., Fuster V., Badimon J.J. Non-invasive imaging of atherosclerotic plaque macrophage in a rabbit model with F-18 FDG PET: a histopathological correlation. BMC Nucl Med. 2006;6:3. doi: 10.1186/1471-2385-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leppanen O., Bjornheden T., Evaldsson M., Boren J., Wiklund O., Levin M. ATP depletion in macrophages in the core of advanced rabbit atherosclerotic plaques in vivo. Atherosclerosis. 2006;188:323–330. doi: 10.1016/j.atherosclerosis.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 37.Esposito C., Settimi A., Del Conte F., Cerulo M., Coppola V., Farina A., Crocetto F., Ricciardi E., Esposito G., Escolino M. Image-guided pediatric surgery using indocyanine green (ICG) fluorescence in laparoscopic and robotic surgery. Front Pediatr. 2020;8:314. doi: 10.3389/fped.2020.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang E., Liu Q., Huang G., Liu J., Wei W. Engineering nanobodies for next-generation molecular imaging. Drug Discov Today. 2022;27:1622–1638. doi: 10.1016/j.drudis.2022.03.013. [DOI] [PubMed] [Google Scholar]
- 39.Ughi G.J., Verjans J., Fard A.M., Wang H., Osborn E., Hara T., Mauskapf A., Jaffer F.A., Tearney G.J. Dual modality intravascular optical coherence tomography (OCT) and near-infrared fluorescence (NIRF) imaging: a fully automated algorithm for the distance-calibration of NIRF signal intensity for quantitative molecular imaging. Int J Cardiovasc Imag. 2015;31:259–268. doi: 10.1007/s10554-014-0556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hutcheson J.D., Goettsch C., Bertazzo S., Maldonado N., Ruiz J.L., Goh W., Yabusaki K., Faits T., Bouten C., Franck G., Quillard T., Libby P., Aikawa M., Weinbaum S., Aikawa E. Genesis and growth of extracellular-vesicle-derived microcalcification in atherosclerotic plaques. Nat Mater. 2016;15:335–343. doi: 10.1038/nmat4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chowdhury M.M., Piao Z., Albaghdadi M.S., Coughlin P.A., Rudd J.H.F., Tearney G.J., Jaffer F.A. Intravascular fluorescence molecular imaging of atherosclerosis. Methods Mol Biol. 2022;2419:853–872. doi: 10.1007/978-1-0716-1924-7_52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aikawa E., Nahrendorf M., Figueiredo J.L., Swirski F.K., Shtatland T., Kohler R.H., Jaffer F.A., Aikawa M., Weissleder R. Osteogenesis associates with inflammation in early-stage atherosclerosis evaluated by molecular imaging in vivo. Circulation. 2007;116:2841–2850. doi: 10.1161/CIRCULATIONAHA.107.732867. [DOI] [PubMed] [Google Scholar]
- 43.Figueiredo J.L., Aikawa M., Zheng C., Aaron J., Lax L., Libby P., de Lima Filho J.L., Gruener S., Fingerle J., Haap W., Hartmann G., Aikawa E. Selective cathepsin S inhibition attenuates atherosclerosis in apolipoprotein E-deficient mice with chronic renal disease. Am J Pathol. 2015;185:1156–1166. doi: 10.1016/j.ajpath.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawakami R., Katsuki S., Travers R., Romero D.C., Becker-Greene D., Passos L.S.A., Higashi H., Blaser M.C., Sukhova G.K., Buttigieg J., Kopriva D., Schmidt A.M., Anderson D.G., Singh S.A., Cardoso L., Weinbaum S., Libby P., Aikawa M., Croce K., Aikawa E. S100A9-RAGE axis accelerates formation of macrophage-mediated extracellular vesicle microcalcification in diabetes mellitus. Arterioscler Thromb Vasc Biol. 2020;40:1838–1853. doi: 10.1161/ATVBAHA.118.314087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krahn K.N., Bouten C.V., van Tuijl S., van Zandvoort M.A., Merkx M. Fluorescently labeled collagen binding proteins allow specific visualization of collagen in tissues and live cell culture. Anal Biochem. 2006;350:177–185. doi: 10.1016/j.ab.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 46.Aikawa E., Blaser M.C. 2020 Jeffrey M. Hoeg award lecture: calcifying extracellular vesicles as building blocks of microcalcifications in cardiovascular disorders. Arterioscler Thromb Vasc Biol. 2021;41:117–127. doi: 10.1161/ATVBAHA.120.314704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feldman A.T., Wolfe D. Tissue processing and hematoxylin and eosin staining. Methods Mol Biol. 2014;1180:31–43. doi: 10.1007/978-1-4939-1050-2_3. [DOI] [PubMed] [Google Scholar]
- 48.Schneider M.R. Von Kossa and his staining technique. Histochem Cell Biol. 2021;156:523–526. doi: 10.1007/s00418-021-02051-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kinkel A.D., Fernyhough M.E., Helterline D.L., Vierck J.L., Oberg K.S., Vance T.J., Hausman G.J., Hill R.A., Dodson M.V. Oil red-O stains non-adipogenic cells: a precautionary note. Cytotechnology. 2004;46:49–56. doi: 10.1007/s10616-004-3903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lattouf R., Younes R., Lutomski D., Naaman N., Godeau G., Senni K., Changotade S. Picrosirius red staining: a useful tool to appraise collagen networks in normal and pathological tissues. J Histochem Cytochem. 2014;62:751–758. doi: 10.1369/0022155414545787. [DOI] [PubMed] [Google Scholar]
- 51.Sukswai N., Khoury J.D. Immunohistochemistry innovations for diagnosis and tissue-based biomarker detection. Curr Hematol Malig Rep. 2019;14:368–375. doi: 10.1007/s11899-019-00533-9. [DOI] [PubMed] [Google Scholar]
- 52.Chistiakov D.A., Melnichenko A.A., Myasoedova V.A., Grechko A.V., Orekhov A.N. Mechanisms of foam cell formation in atherosclerosis. J Mol Med (Berl) 2017;95:1153–1165. doi: 10.1007/s00109-017-1575-8. [DOI] [PubMed] [Google Scholar]
- 53.Caglic D., Globisch A., Kindermann M., Lim N.H., Jeske V., Juretschke H.P., Bartnik E., Weithmann K.U., Nagase H., Turk B., Wendt K.U. Functional in vivo imaging of cysteine cathepsin activity in murine model of inflammation. Bioorg Med Chem. 2011;19:1055–1061. doi: 10.1016/j.bmc.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 54.Bosse Y., Miqdad A., Fournier D., Pepin A., Pibarot P., Mathieu P. Refining molecular pathways leading to calcific aortic valve stenosis by studying gene expression profile of normal and calcified stenotic human aortic valves. Circ Cardiovasc Genet. 2009;2:489–498. doi: 10.1161/CIRCGENETICS.108.820795. [DOI] [PubMed] [Google Scholar]
- 55.Khraishah H., Jaffer F.A. Intravascular molecular imaging: near-infrared fluorescence as a new frontier. Front Cardiovasc Med. 2020;7 doi: 10.3389/fcvm.2020.587100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaffer F.A., Vinegoni C., John M.C., Aikawa E., Gold H.K., Finn A.V., Ntziachristos V., Libby P., Weissleder R. Real-time catheter molecular sensing of inflammation in proteolytically active atherosclerosis. Circulation. 2008;118:1802–1809. doi: 10.1161/CIRCULATIONAHA.108.785881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rauschendorfer P., Wissmeyer G., Jaffer F.A., Gorpas D., Ntziachristos V. Accounting for blood attenuation in intravascular near-infrared fluorescence-ultrasound imaging using a fluorophore-coated guidewire. J Biomed Opt. 2023;28 doi: 10.1117/1.JBO.28.4.046001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wirka R.C., Pjanic M., Quertermous T. Advances in transcriptomics: investigating cardiovascular disease at unprecedented resolution. Circ Res. 2018;122:1200–1220. doi: 10.1161/CIRCRESAHA.117.310910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Decano J.L., Iwamoto Y., Goto S., Lee J.Y., Matamalas J.T., Halu A., Blaser M., Lee L.H., Pieper B., Chelvanambi S., Silva-Nicolau J., Bartoli-Leonard F., Higashi H., Shibata H., Vyas P., Wang J., Gostjeva E., Body S.C., Singh S.A., Aikawa M., Aikawa E. A disease-driver population within interstitial cells of human calcific aortic valves identified via single-cell and proteomic profiling. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110685. [DOI] [PubMed] [Google Scholar]
- 60.Xu K., Xie S., Huang Y., Zhou T., Liu M., Zhu P., Wang C., Shi J., Li F., Sellke F.W., Dong N. Cell-type transcriptome atlas of human aortic valves reveal cell heterogeneity and endothelial to mesenchymal transition involved in calcific aortic valve disease. Arterioscler Thromb Vasc Biol. 2020;40:2910–2921. doi: 10.1161/ATVBAHA.120.314789. [DOI] [PubMed] [Google Scholar]
- 61.Decano J.L., Singh S.A., Gasparotto Bueno C., Ho Lee L., Halu A., Chelvanambi S., Matamalas J.T., Zhang H., Mlynarchik A.K., Qiao J., Sharma A., Mukai S., Wang J., Anderson D.G., Ozaki C.K., Libby P., Aikawa E., Aikawa M. Systems approach to discovery of therapeutic targets for vein graft disease: PPARalpha pivotally regulates metabolism, activation, and heterogeneity of macrophages and lesion development. Circulation. 2021;143:2454–2470. doi: 10.1161/CIRCULATIONAHA.119.043724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stark R., Grzelak M., Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20:631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 63.Kossar A.P., Anselmo W., Grau J.B., Liu Y., Small A., Carter S.L., Salvador L., Zhao L., Cvijic M.E., Li Z., Yarde M., Rioux N., Rader D.J., Levy R.J., Ferrari G. Circulating and tissue matricellular RNA and protein expression in calcific aortic valve disease. Physiol Genomics. 2020;52:191–199. doi: 10.1152/physiolgenomics.00104.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atkins S.K., Singh S.A., Aikawa E. Calcific aortic valve disease “omics” is timely, but are we looking too late? JACC Basic Transl Sci. 2020;5:1178–1180. doi: 10.1016/j.jacbts.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Theriault S., Gaudreault N., Lamontagne M., Rosa M., Boulanger M.C., Messika-Zeitoun D., Clavel M.A., Capoulade R., Dagenais F., Pibarot P., Mathieu P., Bosse Y. A transcriptome-wide association study identifies PALMD as a susceptibility gene for calcific aortic valve stenosis. Nat Commun. 2018;9:988. doi: 10.1038/s41467-018-03260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z., Gaudreault N., Arsenault B.J., Mathieu P., Bosse Y., Theriault S. Phenome-wide analyses establish a specific association between aortic valve PALMD expression and calcific aortic valve stenosis. Commun Biol. 2020;3:477. doi: 10.1038/s42003-020-01210-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iqbal F., Schlotter F., Becker-Greene D., Lupieri A., Goettsch C., Hutcheson J.D., Rogers M.A., Itoh S., Halu A., Lee L.H., Blaser M.C., Mlynarchik A.K., Hagita S., Kuraoka S., Chen H.Y., Engert J.C., Passos L.S.A., Jha P.K., Osborn E.A., Jaffer F.A., Body S.C., Robson S.C., Thanassoulis G., Aikawa M., Singh S.A., Sonawane A.R., Aikawa E. Sortilin enhances fibrosis and calcification in aortic valve disease by inducing interstitial cell heterogeneity. Eur Heart J. 2023;44:885–898. doi: 10.1093/eurheartj/ehac818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Majumdar U., Choudhury T.Z., Manivannan S., Ueyama Y., Basu M., Garg V. Single-cell RNA-sequencing analysis of aortic valve interstitial cells demonstrates the regulation of integrin signaling by nitric oxide. Front Cardiovasc Med. 2022;9 doi: 10.3389/fcvm.2022.742850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang K., Zheng Q., Liu X., Geng B., Dong N., Shi J. Identifying hub genes of calcific aortic valve disease and revealing the immune infiltration landscape based on multiple WGCNA and single-cell sequence analysis. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.1035285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 71.LaPelusa A., Kaushik R. StatPearls; Treasure Island, FL: 2022. Physiology, Proteins. [PubMed] [Google Scholar]
- 72.Aslam B., Basit M., Nisar M.A., Khurshid M., Rasool M.H. Proteomics: technologies and their applications. J Chromatogr Sci. 2017;55:182–196. doi: 10.1093/chromsci/bmw167. [DOI] [PubMed] [Google Scholar]
- 73.Singh S.A., Aikawa E., Aikawa M. Current trends and future perspectives of state-of-the-art proteomics technologies applied to cardiovascular disease research. Circ J. 2016;80:1674–1683. doi: 10.1253/circj.CJ-16-0499. [DOI] [PubMed] [Google Scholar]
- 74.Mehan M.R., Ostroff R., Wilcox S.K., Steele F., Schneider D., Jarvis T.C., Baird G.S., Gold L., Janjic N. Highly multiplexed proteomic platform for biomarker discovery, diagnostics, and therapeutics. Adv Exp Med Biol. 2013;735:283–300. doi: 10.1007/978-1-4614-4118-2_20. [DOI] [PubMed] [Google Scholar]
- 75.Gullberg M., Fredriksson S., Taussig M., Jarvius J., Gustafsdottir S., Landegren U. A sense of closeness: protein detection by proximity ligation. Curr Opin Biotechnol. 2003;14:82–86. doi: 10.1016/s0958-1669(02)00011-3. [DOI] [PubMed] [Google Scholar]
- 76.Leopold J.A., Loscalzo J. Emerging role of precision medicine in cardiovascular disease. Circ Res. 2018;122:1302–1315. doi: 10.1161/CIRCRESAHA.117.310782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perpetuo L., Barros A.S., Dalsuco J., Nogueira-Ferreira R., Resende-Goncalves P., Falcao-Pires I., Ferreira R., Leite-Moreira A., Trindade F., Vitorino R. Coronary artery disease and aortic valve stenosis: a urine proteomics study. Int J Mol Sci. 2022;23 doi: 10.3390/ijms232113579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olgun Kucuk H., Kucuk U., Demirtas C., Ozdemir M. Role of serum high density lipoprotein levels and functions in calcific aortic valve stenosis progression. Int J Clin Exp Med. 2015;8:22543–22549. [PMC free article] [PubMed] [Google Scholar]
- 79.Huang Z., Ma L., Huang C., Li Q., Nice E.C. Proteomic profiling of human plasma for cancer biomarker discovery. Proteomics. 2017;17 doi: 10.1002/pmic.201600240. [DOI] [PubMed] [Google Scholar]
- 80.Cominetti O., Nunez Galindo A., Corthesy J., Oller Moreno S., Irincheeva I., Valsesia A., Astrup A., Saris W.H., Hager J., Kussmann M., Dayon L. Proteomic biomarker discovery in 1000 human plasma samples with mass spectrometry. J Proteome Res. 2016;15:389–399. doi: 10.1021/acs.jproteome.5b00901. [DOI] [PubMed] [Google Scholar]
- 81.Lygirou V., Latosinska A., Makridakis M., Mullen W., Delles C., Schanstra J.P., Zoidakis J., Pieske B., Mischak H., Vlahou A. Plasma proteomic analysis reveals altered protein abundances in cardiovascular disease. J Transl Med. 2018;16:104. doi: 10.1186/s12967-018-1476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ignjatovic V., Geyer P.E., Palaniappan K.K., Chaaban J.E., Omenn G.S., Baker M.S., Deutsch E.W., Schwenk J.M. Mass spectrometry-based plasma proteomics: considerations from sample collection to achieving translational data. J Proteome Res. 2019;18:4085–4097. doi: 10.1021/acs.jproteome.9b00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vidal M., Chan D.W., Gerstein M., Mann M., Omenn G.S., Tagle D., Sechi S., Workshop P. The human proteome - a scientific opportunity for transforming diagnostics, therapeutics, and healthcare. Clin Proteonomics. 2012;9:6. doi: 10.1186/1559-0275-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alfaro J.A., Bohlander P., Dai M., Filius M., Howard C.J., van Kooten X.F., et al. The emerging landscape of single-molecule protein sequencing technologies. Nat Methods. 2021;18:604–617. doi: 10.1038/s41592-021-01143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mund A., Brunner A.D., Mann M. Unbiased spatial proteomics with single-cell resolution in tissues. Mol Cell. 2022;82:2335–2349. doi: 10.1016/j.molcel.2022.05.022. [DOI] [PubMed] [Google Scholar]
- 86.Maier T., Guell M., Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 87.Sud K., Narula N., Aikawa E., Arbustini E., Pibarot P., Merlini G., Rosenson R.S., Seshan S.V., Argulian E., Ahmadi A., Zhou F., Moreira A.L., Cote N., Tsimikas S., Fuster V., Gandy S., Bonow R.O., Gursky O., Narula J. The contribution of amyloid deposition in the aortic valve to calcification and aortic stenosis. Nat Rev Cardiol. 2023;20:418–428. doi: 10.1038/s41569-022-00818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weisell J., Ohukainen P., Napankangas J., Ohlmeier S., Bergmann U., Peltonen T., Taskinen P., Ruskoaho H., Rysa J. Heat shock protein 90 is downregulated in calcific aortic valve disease. BMC Cardiovasc Disord. 2019;19:306. doi: 10.1186/s12872-019-01294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heuschkel M.A., Skenteris N.T., Hutcheson J.D., van der Valk D.D., Bremer J., Goody P., Hjortnaes J., Jansen F., Bouten C.V.C., van den Bogaerdt A., Matic L., Marx N., Goettsch C. Integrative multi-omics analysis in calcific aortic valve disease reveals a link to the formation of amyloid-like deposits. Cells. 2020;9:2164. doi: 10.3390/cells9102164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cavalli G., Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 91.Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Menon V., Lincoln J. The genetic regulation of aortic valve development and calcific disease. Front Cardiovasc Med. 2018;5:162. doi: 10.3389/fcvm.2018.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou Y., Li J., Zhou K., Liao X., Zhou X., Shen K. The methylation of Notch1 promoter mediates the osteogenesis differentiation in human aortic valve interstitial cells through Wnt/beta-catenin signaling. J Cell Physiol. 2019;234:20366–20376. doi: 10.1002/jcp.28638. [DOI] [PubMed] [Google Scholar]
- 94.Gupta T., Khera S., Kolte D., Goel K., Kalra A., Villablanca P.A., Aronow H.D., Abbott J.D., Fonarow G.C., Taub C.C., Kleiman N.S., Weisz G., Inglessis I., Elmariah S., Rihal C.S., Garcia M.J., Bhatt D.L. Transcatheter versus surgical aortic valve replacement in patients with prior coronary artery bypass grafting: trends in utilization and propensity-matched analysis of in-hospital outcomes. Circ Cardiovasc Interv. 2018;11 doi: 10.1161/CIRCINTERVENTIONS.117.006179. [DOI] [PubMed] [Google Scholar]
- 95.Brennan J.M., Thomas L., Cohen D.J., Shahian D., Wang A., Mack M.J., Holmes D.R., Edwards F.H., Frankel N.Z., Baron S.J., Carroll J., Thourani V., Tuzcu E.M., Arnold S.V., Cohn R., Maser T., Schawe B., Strong S., Stickfort A., Patrick-Lake E., Graham F.L., Dai D., Li F., Matsouaka R.A., O'Brien S., Li F., Pencina M.J., Peterson E.D. Transcatheter versus surgical aortic valve replacement: propensity-matched comparison. J Am Coll Cardiol. 2017;70:439–450. doi: 10.1016/j.jacc.2017.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yamamoto K., Yamamoto H., Yoshida K., Kisanuki A., Hirano Y., Ohte N., Akasaka T., Takeuchi M., Nakatani S., Ohtani T., Sozu T., Masuyama T. Prognostic factors for progression of early- and late-stage calcific aortic valve disease in Japanese: the Japanese aortic stenosis study (JASS) retrospective analysis. Hypertens Res. 2010;33:269–274. doi: 10.1038/hr.2009.225. [DOI] [PubMed] [Google Scholar]
- 97.Blaser M.C., Kraler S., Luscher T.F., Aikawa E. Multi-omics approaches to define calcific aortic valve disease pathogenesis. Circ Res. 2021;128:1371–1397. doi: 10.1161/CIRCRESAHA.120.317979. [DOI] [PMC free article] [PubMed] [Google Scholar]