Abstract
Atherosclerosis is a chronic inflammatory disease of the arterial wall, characterized by the buildup of plaques with the accumulation and transformation of lipids, immune cells, vascular smooth muscle cells, and necrotic cell debris. Plaques with collagen-poor thin fibrous caps infiltrated by macrophages and lymphocytes are considered unstable because they are at the greatest risk of rupture and clinical events. However, the current histologic definition of plaque types may not fully capture the complex molecular nature of atherosclerotic plaque biology and the underlying mechanisms contributing to plaque progression, rupture, and erosion. The advances in omics technologies have changed the understanding of atherosclerosis plaque biology, offering new possibilities to improve risk prediction and discover novel therapeutic targets. Genomic studies have shed light on the genetic predisposition to atherosclerosis, and integrative genomic analyses expedite the translation of genomic discoveries. Transcriptomic, proteomic, metabolomic, and lipidomic studies have refined the understanding of the molecular signature of atherosclerotic plaques, aiding in data-driven hypothesis generation for mechanistic studies and offering new prospects for biomarker discovery. Furthermore, advancements in single-cell technologies and emerging spatial analysis techniques have unveiled the heterogeneity and plasticity of plaque cells. This review discusses key omics-based discoveries that have advanced the understanding of human atherosclerotic plaque biology, focusing on insights derived from omics profiling of human atherosclerotic vascular specimens.
Atherosclerosis is a chronic inflammatory disease of the arterial wall,1 characterized by the buildup of plaque with the accumulation and transformation of lipids, immune cells, vascular smooth muscle cells (SMCs), and necrotic cell debris.2 Atherosclerosis progression drives the thickening and hardening of arteries, reducing blood flow in the lumen. Lesions can undergo rupture or erosion.1,2 Plaque rupture is the most frequent cause of arterial thrombosis,3 resulting in most myocardial infarctions and strokes.1,2 Atherosclerosis is the root cause of atherosclerotic cardiovascular disease (ACVD), including coronary heart disease, cerebrovascular disease, peripheral artery disease, and aortic atherosclerotic disease.4 ACVD-related conditions remain the leading cause of morbidity and mortality globally.4,5 The current standard of care for ACVD involves reducing lipid levels and managing other risk factors such as hypertension, diabetes, obesity, and smoking. However, despite significant advancements in the past few decades, the therapeutic impact of these strategies on cardiovascular outcomes has reached a plateau.6 Discovering novel mechanisms and new therapeutic targets and identifying novel biomarkers for better risk stratification are essential for improving the management of ACVD.
Plaques with collagen-poor thin fibrous caps infiltrated by macrophages and lymphocytes are considered unstable because they are at the greatest risk of rupture and clinical events.7 The relative contribution of plaque erosion to acute arterial thrombus formation is on the rise.1 The current histologic definition of plaque types may not capture the complex molecular nature of atherosclerotic plaque biology and the underlying mechanisms that lead to the progression and rupture of plaques.8 In addition to histology- and imaging-based definitions of plaque vulnerability, understanding the molecular signature of vulnerable plaques represents an opportunity to uncover new mechanisms and therapeutic targets to address the residual risks of ACVD beyond the management of classic risk factors. The development of improved approaches to identifying patients with high-risk atherosclerotic plaques before the onset of symptoms and atherothrombotic events is an unmet need.
The growing application of “omics” approaches in the last two decades has marked an important turning point in atherosclerosis research. Omics tools, including genomics, epigenomics, transcriptomics, proteomics, metabolomics, and lipidomics, have provided a more comprehensive and unbiased view of the molecular changes underlying atherosclerotic plaque biology. These tools have fueled hypothesis generation and facilitated novel discoveries. Moreover, recent advancements in single-cell omics and spatial omics9 have opened up opportunities to characterize the heterogeneity and spatial information of cell subpopulations within atherosclerotic plaques. These breakthroughs in omics technologies have transformed atherosclerosis research, allowing for in-depth mechanistic studies and improving the identification of biomarkers and risk stratification.
Omics technologies and their applications in atherosclerosis research are rapidly evolving. The aim of the current review is to highlight key omics-based discoveries that have deepened understanding of human atherosclerotic plaque biology. Specifically, this review focuses on insights derived from omics profiling of human atherosclerotic vascular specimens. It is worth noting that this review does not provide a comprehensive coverage of all literature in omics-based atherosclerosis research, as each omics approach warrants independent reviews. Instead, it highlights landmark studies and the most recent advancements while directing readers to review articles authored by experts in specific areas. For each omics approach, each respective section first introduces the technology, then highlights how each omics approach works, and, when applicable, how the integrative multi-omics approach has contributed to a comprehensive understanding of the molecular signature of atherosclerotic plaques. Notably, the review emphasizes how identifying target genes through omics profiling has facilitated mechanistic studies and improved biomarker discovery and risk stratification. Furthermore, the review summarizes the emerging application of single-cell and spatial analyses in atherosclerosis research, highlighting the impact of these technical advancements on understanding the underlying mechanisms. Lastly, the knowledge gaps and potential directions for future research endeavors are discussed.
Genomics: Understanding the Genetic Spectrum Contributing to Atherosclerosis—From Germline Variation to Somatic Mutations
Improved technologies and declined costs of genotyping microarrays and next-generation sequencing have played a pivotal role in accelerating the understanding of how germline and somatic variations contribute to ACVD.
Genotyping microarrays capture the majority of common interindividual genetic variations and are widely used in common variant association studies [ie, genome-wide association studies (GWAS)]. Coronary artery disease (CAD) and myocardial infarction are prime examples of complex diseases with a polygenic genetic architecture and have been most comprehensively studied through approaches using GWAS. From the first CAD GWAS published in 200710, 11, 12 to the latest GWAS published in 2022, >300 loci have been identified as associated with CAD.13,14
The discovery of genetic loci has outpaced the elucidation of their mechanisms in CAD pathogenesis. Because the majority of risk variants reside in noncoding regions of the genome, identifying the causal genes and cell types for associated loci is essential for discovering genetics-guided therapeutic targets. Although functional assays are required to confirm the causality, integrative genomic pipelines leveraging genomic, transcriptomic, and epigenomic data sets can systematically prioritize the most likely candidates for functional validation. As reported by Mountjoy et al,15 using fine-mapped genetics and functional genomics data, including molecular quantitative trait loci such as gene expression, chromatin accessibility, and interaction data, a machine learning model was trained to prioritize causal genes for all published GWAS. Data can be queried via the Open Targets Genetics web portal, with new data sources continually incorporated. Because most CAD loci are not associated with traditional risk factors,16 determining the genetic contribution of vascular wall cells is the focus of functional follow-up of CAD GWAS. Indeed, integrative genomic pipelines using data sets from CAD-relevant tissues and cell types as well as from diseased samples allow for identifying more candidate genes than when using disease-unspecific data sets.17 The Stockholm-Tartu Atherosclerosis Reverse Networks Engineering Task (STARNET) data sets are, therefore, invaluable by providing gene expression quantitative trait locus data from nine cardiometabolic tissue/cell types from CAD patients and control subjects; these types include atherosclerotic aortic wall, non-atherosclerotic mammary artery, blood, liver, skeletal muscle, subcutaneous fat and visceral fat, macrophages, and lipid-loaded macrophages (foam cells). Leveraging the STARNET data sets, Hao et al18 prioritized 162 unique candidate causal CAD genes, which exert their effects from between one and up to seven CAD-relevant tissues/cell types. Using the STARNET data, Koplev et al19 also identified gene-regulatory coexpression networks contributing to CAD heritability and associated with clinical severity of CAD.
When incorporating single-cell transcriptomic and single-nucleus chromatin accessibility profiling (snATAC-seq) of human atherosclerotic plaque cells, gene expression and chromatin accessibility can be assigned to specific cell types, predicting the cell type of action for the loci. snATAC-seq profiling of 28,316 nuclei from 41 individuals for coronary arteries at various stages of CAD suggests that CAD variants mostly reside in SMC and macrophage peaks, highlighting the value of cell type–specific analysis.20 Another study using snATAC-seq of human carotid plaque cells (>7000 nuclei from three individuals) showed that CAD-associated genetic variants are particularly enriched in endothelial and SMC–specific open chromatin regions.21 These studies establish a framework for functional and integrative genomics studies, aiming to elucidate the candidate causal genes and genetic mechanisms of CAD inspired by GWAS findings.
In contrast to genotyping microarrays, whole-exome sequencing and whole-genome sequencing studies aim to capture all genetic variations. Population-based exome-sequencing studies using blood DNA estimated that somatic mutations in blood cells, a process termed clonal hematopoiesis (CH), are common and increase in prevalence with age.22 CH largely results from mutations in a very restricted set of genes associated with hematopoietic malignancies such as DNMT3A, TET2, ASXL1, JAK2, and TP53. Subsequent studies showed the association between CH and higher risks for ACVD.23 The underlying mechanisms of the association represent an active area of research. As recently reviewed by Tall and Fuster,24 the expansion of hematopoietic stem and progenitor cells, increased myelopoiesis, macrophage inflammation, and inflammasome activation contribute to the expansion of lesional inflammatory cells and lesion development; increased production of inflammatory cytokines such as IL-1β or IL-6; and amplify hematopoietic stem and progenitor cell proliferation and myelopoiesis. Understanding an individual’s CH status could guide individualized approaches to high-risk populations with targeted anti-inflammatory therapies. For example, most CH variants are associated with elevated levels of IL-6.25 Genetic IL-6 signaling deficiency attenuates cardiovascular risk in CH.26 Blockade of IL-6 signaling alleviates atherosclerosis in Tet2-deficient CH mice.27 Moving forward, completing the list of CH-related genes and understanding the underlying mechanisms are essential to identifying individuals with CH, evaluating risks of developing related diseases, and developing targeted treatment strategies.24
Genomic data also enable rare variant association analyses. Rare variant association studies have linked inactivating mutations in a handful of genes to the risks of CAD, primarily through effects on lipids or blood pressure.28 For example, inactive mutations in LDLR confer increased risks, whereas inactive mutations in PCSK9 and APOC3 confer decreased risks of CAD. A recent rare variant association analysis did not identify additional strong signals.13 The general implication is that the evidence supporting the theory that rare variation makes a substantial contribution to the genetic architecture of CAD is moderate; functional testing of promising variants is needed to establish pathogenicity.29
Taken together, genomic discoveries over the last two decades have had a fundamental impact on the understanding of the pathogenesis of ACVD. Integrative functional genomic studies leverage tissue- and disease-specific omics data sets, translate genomic discoveries into mechanistic knowledge, and guide the translational process from target identification to therapeutic application.
Epigenomics: Exploring Epigenetic Landscape and Mechanism-Based Therapeutic Targets
Epigenetic regulation is being recognized as an important factor in the pathogenesis of ACVD.30 Epigenomic approaches profile DNA methylation, chromatin accessibility, posttranslational histone modifications, and transcription factor binding. These modifications, including noncoding RNAs that serve as epigenetic regulators, modulate gene expression at both transcriptional and posttranscriptional levels without changing the DNA sequence.31
Epigenetic signature of blood cells is of interest for biomarker discovery in patients with atherosclerosis.30 Global epigenetic profiling data for human atherosclerotic vascular tissues, however, remain relatively limited. A recent review has summarized genome-wide DNA methylation studies conducted in human atherosclerotic vascular tissues, revealing various differentially methylated sites.32 The current section highlights notable contributions toward understanding the epigenetic landscape of human atherosclerotic plaque tissue at various pathologic stages. Zaina et al33 identified an epigenetic signature of 1858 atherosclerosis-specific methylated CpGs. Of these, 91% appeared hypermethylated in atherosclerotic aorta samples compared with donor-matched healthy counterparts (n = 15 individuals). A follow-up study revealed 1631 CpG loci in atherosclerotic plaques that exhibited increasing methylation as the lesions advance.34 Aavik et al35 reported a predominant hypomethylation of promoter sites in human femoral artery atherectomy samples (n = 22) compared with normal mammary artery samples (n = 9). Li et al36 profiled carotid plaques from six symptomatic and eight asymptomatic patients. The findings suggest hypomethylation as a feature of vulnerable plaques, identifying 107 hypomethylated genes with up-regulated expression through intersection with transcriptomic data (http://www.ncbi.nlm.nih.gov/geo; accession numbers GSE2882937 and GSE4157138). The interpretation of current epigenetic profiling studies faces challenges due to the relatively small sample sizes, heterogeneity in atherosclerotic plaque cell composition across different vascular beds, and individual variations in age-related changes in DNA methylation. Single-cell epigenomic analysis will address the challenges posed by the heterogeneity of cell types in atherosclerotic plaques. Moving beyond global profiling, the unmet challenge is to show a causal link between epigenetic signals and disease phenotypes. Nevertheless, CH frequently entails mutations in genes responsible for epigenetic modifications such as TET2, DNMT3A, and ASXL1.39 Preclinical models of atherosclerosis have indicated the potential of epigenetic drugs that target DNA methylation, histone acetylation, methylation, and enhancer regulation, as reviewed by Khyzha et al.40
Collectively, there is substantial evidence establishing the association between epigenetic changes and atherosclerosis. Epigenetic regulation plays an important role in the pathogenesis of ACVD. Epigenetic studies also facilitate the interpretation of genomic discoveries and contribute to the development of mechanism-based therapies targeting epigenetic modifications.
Transcriptomics: Providing the Molecular Signature of Human Atherosclerosis and Facilitating Data-Driven Hypothesis Generation
Transcriptomic profiling of human atherosclerotic vascular samples has revolutionized the understanding of the molecular signature of atherosclerosis. Microarrays and RNA-sequencing (RNA-seq) are the major techniques for transcriptomic profiling. Microarrays detect nucleic acids in a sample by hybridization to probes on microchips. Microarrays have been widely used for decades to analyze large numbers of samples in drug development and clinical research.41 RNA-seq (ie, transcriptomic profiling based on next-generation sequencing) brings a qualitative and quantitative improvement to transcriptome analysis.42,43
Table 137,38,44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54 summarizes publicly available transcriptomic data sets using microarrays or RNA-seq to profile human atherosclerotic vascular samples. All the data sets were submitted to the Gene Expression Omnibus (GEO) database by the scientific community to allow for analysis using GEO2R, an interactive web tool. GEO2R enables users to compare two or more groups of samples in a GEO Series to identify differentially expressed genes across experimental conditions. Results are presented as a table of genes ordered according to P value and a collection of graphic plots to help visualize differentially expressed genes. R script used to perform the calculation can be saved and used as a reference for reproducing the analysis.
Table 1.
Publicly Available Microarray and RNA-Seq Data of Human Atherosclerotic Plaque Samples in the Gene Expression Omnibus Repository (https://www.ncbi.nlm.nih.gov/geo)
| GSE | Sample | Platform | Reference |
|---|---|---|---|
| GSE21545 | 97 peripheral blood mononuclear cell samples and 126 carotid plaque samples with 97 overlapping samples (from symptomatic and asymptomatic patients) | Affymetrix Human Genome U133 Plus 2.0 Array | Folkersen et al44 |
| GSE23314 | Peripheral plaques (n = 101 samples from 67 individuals); LCM of smooth muscle cells and macrophages of carotid plaques (n = 3 samples from one individual) |
Agilent microarrays | Puig et al45 |
| GSE28829 | 13 early and 16 advanced carotid plaque samples | Affymetrix Human Genome U133 Plus 2.0 Array | Döring et al37 |
| GSE40231 | 278 transcriptional profiles of liver, skeletal muscle, and visceral fat (n = 66/tissue) and atherosclerotic and unaffected arterial wall (n = 40/tissue) isolated from CAD patients during coronary artery bypass surgery | Affymetrix Human Genome U133 Plus 2.0 Array | Hägg et al46 |
| GSE41571 | LCM of macrophage-rich regions from 5 ruptured and 6 stable carotid plaque samples | Affymetrix Human Genome U133 Plus 2.0 Array | Lee et al38 |
| GSE43292 | 68 specimens from 32 patients (paired intact tissue and atheroma plaque from carotid plaque samples) | Affymetrix GeneChip Human Gene 1.0 ST arrays | Ayari et al47 |
| GSE100927 | 31 healthy arteries (10 carotid arteries, 11 femoral arteries, and 10 infrapopliteal arteries) and 65 atherosclerotic arteries (27 carotid arteries, 25 femoral arteries, and 13 infrapopliteal arteries) | Agilent Human Gene Expression Microarrays | Steenman et al48 |
| GSE118481 | LCM of macrophage-enriched regions from human carotid plaque samples, including 16 nondiabetic (control) samples containing 6 asymptomatic and 10 symptomatic plaques, and 8 diabetic samples containing 6 asymptomatic and 2 symptomatic plaques | Illumina HumanHT-12 V4.0 expression beadchip | Chai et al49 |
| GSE125771 | 40 carotid plaque samples | Affymetrix HTA 2.0 arrays | Karlöf et al50 |
| GSE163154 | 16 non-IPH and 27 IPH from carotid plaque samples | Illumina HumanRef-8 v2.0 expression BeadChip | Jin et al51 |
| GSE120521 | RNA-seq on stable and unstable regions dissected from carotid plaque samples obtained at carotid endarterectomy in symptomatic patients (n = 4 individuals) | RNA-seq | Mahmoud et al52 |
| GSE198600 | 5 asymptomatic and 6 symptomatic carotid plaques | RNA-seq | Bazan et al53 |
| GSE226790 | 3 human normal arterial intimae and 3 advanced atherosclerotic plaques | RNA-seq | NA |
| SE236610 | 27 human coronary plaques in live patients. Of the 27 patients, 13 were confirmed to have stable CAD and 14 confirmed to have been performed on lesions causing acute coronary syndrome | RNA-seq | Widlansky et al54 |
CAD, coronary artery disease; IPH, intraplaque hemorrhage; LCM, laser capture microscopy; RNA-seq, RNA sequencing.
Summarizing all the published data sets in the current review would be impractical and potentially overwhelming for readers. Instead, the aim of this section was to highlight exemplary studies using large-scale transcriptomic data sets [eg, by the Biobank of Karolinska Endarterectomy (BiKE), Athero-Express, and STARNET]. These studies offer valuable insights into pathways and gene regulatory networks for hypothesis testing and mechanistic studies.19,55 This section also underscores the importance of biobanking initiatives in uncovering new biomarkers.
BiKE was established in 2002 for the prospective collection of atherosclerotic plaque tissue and blood samples from patients undergoing carotid endarterectomy, with a database of clinical parameters, including risk factors, medication, symptoms, time of surgery, preoperative imaging, and anthropometric and laboratory measurements.44 Microarray profiling was performed in atherosclerotic carotid artery lesions from patients undergoing endarterectomy surgery for stable (asymptomatic, n = 40) or unstable (symptomatic, n = 87) carotid stenosis. The data sets facilitated the prioritization and mechanistic studies of multiple coding genes and noncoding RNAs. They also allowed for the identification of gene expression signatures in relation to clinical parameters by multivariate analysis, contributing to the discovery of new biomarkers for improved risk stratification.56, 57, 58, 59, 60, 61, 62 For example, in the BiKE microarray data, the long noncoding MIAT was among the most significantly up-regulated noncoding RNAs in human carotid plaques compared with controls. Miat deletion in atherosclerosis-prone Apoe–/– mice exposed to the inducible plaque rupture model led to thinner fibrous caps and more unstable plaque phenotype.63 Proprotein convertase subtilisin/kexin type 6 (PCSK6) showed elevated expression in the symptomatic lesion and exhibited a positive correlation with genes linked to inflammation, matrix degradation, and mitogens.57 Pcsk6−/− mice exhibited a reduced intimal hyperplasia response after carotid ligation, supporting its role in vascular remodeling.64 Perisic et al65 performed pathway and network analyses on the BiKE microarray data, highlighting the involvement of hemoglobin metabolism, calcification, inflammation, apoptosis, and extracellular matrix pathways in plaque instability. Prediction modeling indicated a panel of 30 genes capable of distinguishing plaques from symptomatic versus asymptomatic patients. This model suggests additional novel candidate genes and pathways warranting further mechanistic studies.65
The Athero-Express biobank included >3500 patients with atherosclerosis; 2400 underwent carotid endarterectomy, and the remaining patients underwent other vascular surgeries.66,67 The database encompasses pathologic specifications, clinical characteristics, a 3-year follow-up, whole-transcriptome analyses (n = 700), whole-genome methylation profile (n = 700) and single nucleotide polymorphism data (n = 2000), and single-cell sequencing data (n = 45). The Athero-Express study can relate atherosclerotic plaque characteristics to systemic cardiovascular outcome (ie, the atherosclerotic plaque, excised during vascular surgery procedures, serves as a fingerprint of the vascular system) to determine the vulnerability for future vascular events.67 The most recent Athero-Express publication reported the transcriptomic profiles of 654 advanced human carotid plaques by RNA-seq that revealed five dominant plaque types based on unsupervised, transcriptome-driven clustering.68 The plaque phenotypes were correlated with clinical presentations, and the plaque type exhibiting the most severe clinical symptoms was linked to inflammatory and fibrotic cell lineages.
In summary, the transcriptomic profiling of human atherosclerotic vascular tissues offers valuable insights into the molecular signature of human atherosclerosis. It serves as a robust resource for identifying novel candidates for functional follow-up, expediting hypothesis generation, and facilitating mechanistic studies. Notably, the Athero-Express data also established that, compared with histopathologic data, incorporating transcriptomic information of the local atherosclerotic plaque provides an improved prediction model for local disease progression (ie, restenosis) and systemic cardiovascular outcome during follow-up.68 This progress underscores the potential for studies utilizing other omics approaches to further advance knowledge in atherosclerosis research.
Proteomics, Metabolomics and Lipidomics, and Multi-Omics: Holding Promise for Improving Risk Prediction
In addition to transcriptomic data providing a valuable overview of global gene expression, other omics approaches, including proteomics, metabolomics, and lipidomics, capture protein and metabolite signatures. Protein and metabolite profiles represent a phenotypic manifestation of the genomic, epigenomic, and transcriptomic changes and complement transcriptomic measurement.69 This section highlights studies conducted on human atheroma samples using proteomics, metabolomics, and lipidomics approaches. These data sets provide insights into the protein and metabolite signatures of human atherosclerosis and investigate whether tissue-based omics and multi-omics profiling at the protein and metabolite levels can shed light on mechanistic studies and improve risk prediction.
Proteomics-Based Studies
This section highlights four proteomics studies that compare different human vascular beds, advanced versus early stages, unstable versus stable plaques, and determine sex differences. Herrington et al70 performed tandem mass spectrometry analysis of coronary artery and abdominal aorta specimens from 100 autopsied young adults to establish a proteomic signature of human arteries with and without atherosclerosis as defined by using pathology grading. Both the atherosclerotic coronary artery and aorta samples present a proteomic profile consistent with tumor necrosis factor-α activation. However, the atherosclerotic coronary artery samples also display a proteomic pattern indicating reduced mitochondria mass, as well as inhibition of peroxisome proliferator–activated receptor alpha and gamma and insulin receptor–regulated proteins, a pattern not observed in the atherosclerotic aorta samples. These findings highlight significant anatomic distinctions in the proteome between atherosclerotic coronary and aortic tissues. Nehme et al71 performed a liquid chromatography with tandem mass spectrometry analysis in carotid endarterectomy samples dissected into early and advanced atherosclerotic lesion (≤ type II lesion and ≥ type IV lesion, respectively, based on Stary et al72,73) (n = 6 individuals). A total of 95 proteins were up-regulated and 117 proteins were down-regulated in advanced lesions compared with early lesions. The up-regulated proteins were associated with proatherogenic processes. In contrast, down-regulated proteins were involved in extracellular matrix organization and vascular smooth muscle cytoskeleton. When intersecting with transcriptomic data of 32 paired early and advanced lesions (http://www.ncbi.nlm.nih.gov/geo; accession number GSE4329247), a total of 19 genes (eg, C1QA, C4B, CTSB, F13A1, STAB1) were commonly up-regulated and 30 genes were down-regulated (eg, LMOD1, CNN1, NEXN, DSTN) at the mRNA and protein levels.
To gain insights into the mechanisms of human plaque rupture, Vaisar et al74 performed liquid chromatography with tandem mass spectrometry on ruptured and stable areas of human carotid plaques (n = 6 individuals) that identified 1161 unique proteins, with 150 proteins more abundant and 339 proteins less abundant in ruptured areas versus stable areas. Overrepresented pathways among the more abundant proteins in ruptured areas included those related to inflammation (eg, activation of immune response and complement activation), atherosclerosis, blood coagulation, and apoptotic cell clearance. Overrepresented pathways among less abundant proteins in the ruptured areas included extracellular matrix and basement membrane protein–related categories. The pathways identified based on proteomic patterns are overall consistent with those identified based on transcriptomic analyses.74 Theofilatos et al75 generated proteomics data of 219 carotid endarterectomy samples from 120 patients. The study examined the proteomic signature of the plaque core versus periphery and extracellular proteins. A signature of four key proteins (calponin, protein C, serpin H1, and versican) predicted future cardiovascular mortality. This large cohort also allowed for the exploration of sex differences in atherosclerosis, implicating the large-aggregating proteoglycans versican and aggrecan as being more abundant in female subjects.
Metabolomics and Lipidomics-Based Studies
Metabolomics measures small-molecule metabolites and have displayed applications in identifying metabolites and metabolomic signature associated with ACVD risks (eg, branched-chain amino acids, select unsaturated lipid species, trimethylamine-N-oxide).69 Lipidomics is a subfield of metabolomics that identifies and measures lipids. This section highlights a few studies using targeted metabolomics and lipidomics to profile human atherosclerotic plaque samples. Tomas et al76 collected 159 carotid plaques from patients undergoing endarterectomy and measured 165 different metabolites. A distinct metabolite signature is linked to symptoms, histologically assessed markers of vulnerability, and protein levels of inflammatory mediators and cytokines in plaque homogenates. The metabolite signature in high-risk plaques aligns with elevated glucose utilization, reduced fatty acid oxidation flux, and increased amino acid anaplerosis, a pattern consistent with activated T cells and pro-inflammatory macrophages. Edsfeldt et al77 analyzed 200 carotid plaques for six sphingolipids: glucosylceramide, lactosylceramide, ceramide, dihydroceramide, sphingomyelin, and sphingosine-1-phosphate. All six analyzed sphingolipids displayed elevated levels in plaques associated with symptoms and exhibited correlations with inflammatory cytokines in plaque homogenate. All sphingolipids, except sphingosine-1-phosphate, also correlated with histologic markers of plaque instability. Fredman et al78 examined lipid mediators in both vulnerable and stable regions of carotid plaque samples (n = 8 individuals). The analysis revealed a significant decrease in the levels of specialized pro-resolving mediators, particularly resolvin D1, and the ratio of these mediators to pro-inflammatory leukotriene in the vulnerable regions. These findings in human plaque sparked research interest in the mechanistic underpinnings of sphingolipids and specialized pro-resolving mediators in atherosclerosis, as reviewed by Choi et al79 and Doran.80
Multi-Omics Integration
Integrating multi-omics data can be a powerful approach. Matic et al81 leveraged transcriptomic and proteomic data to compare plaques with adjacent carotid arterial tissue. In addition, they analyzed proteomic data comparing local and peripheral plasma. BLVRB emerged as the only differentially expressed molecule when the results of these analyses were intersected. Substantial validation confirmed BLVRB as a biomarker for intraplaque hemorrhage and plaque instability in carotid atherosclerosis. Beyond intersecting multi-omics data, Jin et al51 applied Data Integration Analysis for Biomarker Discovery Using Latent Components (DIABLO),82 an integrative multivariate classification and feature selection method for multi-omics assays. The analysis identified a protein/gene–associated multi-omics model that effectively distinguishes stable, non-hemorrhaged lesions from vulnerable, hemorrhaged lesions with high predictive performance (area under the curve >0.95). The model improved the stratification of low- and high-risk carotid artery plaques in independent cohorts.51 Moreover, BLVRB was also within this multi-omics model, contributing to its predictive capabilities for intraplaque hemorrhage. This workflow serves as a showcase for biomarker discovery using multi-omics data. The application of proteomics and lipidomics in predicting the risk of ACVD has also been recently reviewed by Nurmohamed et al.83
In summary, proteomic, metabolomic, and lipidomic profiling offer additional layers of information, facilitating biomarker discovery for improved risk stratification and the identification of potential targets for pharmacotherapeutic intervention.
Single-Cell Analysis: Building an Integrative Atlas and Moving toward Mechanisms and Translation
Omics analyses of tissue samples at bulk levels have yielded important insights into human atherosclerotic plaque biology. Leveraging single-cell technology to examine cell subpopulations presents an important opportunity to gain insights into cellular and molecular heterogeneity, identify disease mechanisms, and exploit new therapeutic strategies (eg, by targeting specific cell subpopulations). The classic flow cytometry methods analyze single-cell information, usually for panels of no more than 12 to 18 markers. Two cytometry-based techniques, spectral flow cytometry and mass cytometry (cytometry by time of flight),84 enable the detection of ≥40 markers. The first single-cell genome-wide mRNA sequencing (scRNA-seq) method was reported in 2019.85 As the most widely used single-cell omics method, scRNA-seq can be paired with other single-cell omics approaches such as cellular indexing of transcriptomes and epitopes by sequencing86 and snATAC-seq.87 These technical advances have enabled high-dimensional profiling and analysis of individual cells’ transcriptome, proteome, and epigenome, with unprecedented resolution and throughput. The past decade has seen a revolutionary advance in single-cell technologies in atherosclerosis research. This section provides a concise overview of insights into plaque cell populations derived from single-cell analysis.
The application of scRNA-seq analysis has discovered not only disease-related cell-specific transcriptomic profiles but also novel subpopulations of cells that were once considered homogeneous. A number of scRNA-seq studies published in 2018 to 2019 provided the initial single-cell landscape of atherosclerotic plaque cells in mice88, 89, 90, 91 and humans92 with a focus on immune cells. In 2020, Zernecke et al93 published a meta-analysis of published scRNA-seq data to summarize immune cell profiles in atherosclerotic mouse aortas. These initial single-cell analyses largely focused on providing an overview of immune cell heterogeneity. Since then, single-cell analyses leveraged lineage tracing and fate mapping mice to understand plaque cell origin and phenotype properties. scRNA-seq was also performed for all plaque cells in human carotid arteries94, 95, 96 and coronary arteries.97 Mice with genetic modification of regulators of atherosclerosis (eg, TCF21, KLF4, AHR98) were profiled by scRNA-seq to gain insights into their causal regulatory roles in SMC heterogeneity and plasticity. In 2021, Conklin et al99 published a meta-analysis of scRNA-seq data using SMC-lineage tracing mice.94,96, 97, 98 Most recently, another meta-analysis was performed by Mosquera et al100 to integrate 22 public human coronary and carotid scRNA-seq libraries.94,97,101,102 In addition to cell type annotation and leveraging murine lineage-traced SMC gene modules to identify modulated SMC populations in human data, this integrative analysis included integration of data from GWAS for the identification of etiologic cell types, cell–cell communication analyses, pseudotime inference, and transcription factor activity predictions.100 To date, scRNA-seq has been increasingly incorporated in preclinical mouse studies, and efforts are ongoing to systematically profile human atherosclerotic plaque samples at single-cell levels.103
Other single-cell omics and multi-omics techniques are increasingly being used in atherosclerosis research. Single-cell epigenomic profiling (eg, snATAC-seq) has been performed to investigate cell type–specific regulatory elements to understand cell identity and interpret CAD-relevant genetic variation.20,21 Cellular indexing of transcriptomes and epitopes by sequencing that combined protein (40 antibodies) and transcript scRNA-seq provides higher resolution than mass cytometry or scRNA-seq without antibodies to resolve blood immune cell subsets,104 and it has been used to profile human plaque samples.92,103 Publicly available single-cell data sets of human plaque samples are summarized in Table 2.20,21,92,94,96,97,102,105
Table 2.
Publicly Available Single-Cell Data of Human Atherosclerotic Plaque Samples in the Gene Expression Omnibus Repository (https://www.ncbi.nlm.nih.gov/geo)
| GSE | Sample | Platform | Reference |
|---|---|---|---|
| GSE131778 | Human atherosclerotic coronary arteries, obtained from explanted hearts of patients undergoing heart transplantation (n = 4 patients) | scRNA-seq (10× Genomics) | Wirka et al97 |
| GSE150644 | Human atherosclerotic plaques obtained from 14 male and 4 female patients undergoing a primary carotid endarterectomy in the Athero-Express biobank study | SORT-seq CEL-Seq2 |
Alencar et al96 |
| GSE155512 | Human carotid artery plaques (n = 3 patients) | scRNA-seq (10× Genomics) | Pan et al94 |
| GSE159677 | Calcified atherosclerotic core plaques and patient-matched proximal adjacent portions of carotid artery (n = 3 patients) | scRNA-seq (10× Genomics) | Alsaigh et al102 |
| GSE224273 | Human carotid artery plaques scRNA-seq: n = 4 asymptomatic and 2 symptomatic; CITE-seq: n = 1 symptomatic |
scRNA-seq (10× Genomics) CITE-seq |
Fernandez et al92 |
| GSE234077 | CD45+-selected leukocytes from femoral (n = 9) and carotid (n = 4) plaques | scRNA-seq (10× Genomics) | Slysz et al105 |
| https://doi.org/10.6084/m9.figshare.14501985.v2 | Human carotid endarterectomy samples (n = 3 patients) | snATAC-seq | Örd et al21 |
| GSE175621 | Human coronary artery segments from 41 patients with varying stages of CAD | snATAC-seq | Turner et al20 |
CAD, coronary artery disease; CITE-seq, cellular indexing of transcriptomes and epitopes by sequencing; scRNA-seq, single-cell genome-wide mRNA sequencing; snATAC-seq, single-nucleus chromatin accessibility profiling.
In the era of widespread use of single-cell analyses, Eberhardt et al106 and de Winther et al107 recently reviewed how single-cell technologies offer new insights and translational opportunities in atherosclerosis research. Some key findings are highlighted here. First, single-cell atherosclerosis studies confirm that human populations share striking similarities with those identified in the murine models.107 Nonetheless, important differences in human and experimental atherosclerosis need to be defined. This is particularly important when interpreting data from experimental studies for application to human contexts. As reviewed by de Winther et al, macrophages account for the bulk of immune cells in the murine aorta (around 50% of CD45+ cells) but comprise 16% to 20% of total CD45+ live cells in human carotid endarterectomies. Analyses of mouse aortas identified a proliferating and an interferon-responsive macrophage cluster, which were not observed in current human scRNA-seq data. In addition, Dib et al108 identified TREM1highPLIN2high macrophages in human carotid plaques but not in mouse models of atherosclerosis. Moreover, T cells outnumber macrophages in human atherosclerotic lesions92 and show a pattern of clonal expansion of effector CD4+ T cells.109 As data sets continue to expand, a more comprehensive assessment can be achieved.
A second key finding is that single-cell analysis in conjunction with lineage tracing and fate mapping in mice reveal new knowledge regarding SMC phenotypic switching, implying that cell population annotation in humans cannot rely on traditional markers but should leverage data in SMC-specific, endothelial cell–specific, and macrophage-specific lineage tracing mice and other approaches. Third, single-cell analyses redefined the heterogeneity of endothelial cells and immune cells. For example, studies consistently identify inflammatory, resident-like, and Trem2high macrophage subpopulations in both mouse and human plaques.107,110 Trem2high plaque macrophages express lipid-processing genes and show a lack of inflammation signature.89,93 The role of macrophage Trem2 in atherosclerosis is an active area of research. Patterson et al111 show that Trem2 is essential for foamy macrophage survival and that myeloid-specific knockout of Trem2 resulted in attenuation of plaque progression in mice. Piollet et al112 found that Trem2 is essential for macrophage efferocytosis. Hematopoietic or global Trem2 deficiency increases necrotic core formation in early atherosclerosis in mice. Based on trajectory analysis of scRNA-seq data and in vitro cell culture experiments, Dib et al108 proposed that TREM2high plaque macrophages can give rise to TREM1highPLIN2high inflammatory foamy macrophages within human atherosclerotic plaques. The data highlight a novel role of TREM2high plaque macrophages. However, to conclusively establish cell fate, additional studies using lineage tracing methods in suitable model systems will be necessary.
A final key finding is that sex differences in atherosclerosis at the transcriptomic level have been increasingly recognized.55 Autopsy studies revealed that the plaque erosion pathology is more pronounced in younger, premenopausal women.113 scRNA-seq data identified sex-stratified gene regulatory networks from human carotid atherosclerotic plaques.114 A female-specific gene regulatory network has been identified, showing enrichment for genes involved in regulating endothelial-to-mesenchymal transition, a potential mechanism for plaque erosion.115
As the research community continues to construct the atherosclerosis single-cell atlas, a significant research focus will be placed on understanding the role of various subpopulations of plaque cells, their potential as targets for intervention, and the molecular mechanisms driving the sex differences in atherosclerotic phenotypes. The integration of novel single-cell omics and multi-omics techniques, along with conducting comprehensive studies involving larger patient cohorts, holds potential for further unlocking the capabilities of single-cell technologies to facilitate a deeper understanding of the cellular and molecular mechanisms underlying clinical outcomes.
Spatial Analysis: First Steps toward Building the Spatial Map of Atherosclerosis
Single-cell analyses rely on digested tissue and isolated cells or nuclei and thus lack spatial information. Spatial information is critical to understanding tissue homeostasis and disease (eg, by identifying the localization and cell–cell crosstalk of disease-driving cell populations for spatially localized disease mechanisms and examining expression profile in the context of tissue microenvironment).9,87 In this context, the emerging applications of spatial transcriptomics, proteomics, and metabolomics in profiling human atherosclerosis plaques are summarized in this section.
The first spatial transcriptomic technique that can target the entire transcriptome, introduced by Ståhl et al in 2016,116 was later adapted by 10× Genomics (Pleasanton, CA) as the Visium platform. Visium maps the whole transcriptome but currently lacks single-cell resolution. The Xenium platform, however, allows the characterization of hundreds of RNAs and multiplexed proteins in cells and tissues, achieving subcellular resolution. Moreover, the tissue section used in the process can be employed for histologic studies post-run for histopathologic characterization. Other spatial transcriptomics and multi-omics techniques were recently reviewed by Baysoy et al.87
Sun et al117 reported data using the Visium platform in human carotid plaques. First, RNA-seq was performed to define gene expression signatures in different regions of the plaque along the longitudinal blood flow direction (the proximal, the most stenotic, and distal regions) in the context that the proximal and most stenotic regions had more intraplaque hemorrhage and a higher histologic vulnerability index. Examining the prioritized top differentially expressed genes between the ruptured-prone region and the distal region, matrix metalloproteinase-9 expression displayed a correlation with T cells and macrophages, exhibiting a spatial pattern mainly expressed in locations near the rupture. Mendelian randomization analyses validated that higher levels of circulating matrix metalloproteinase-9 causally increase the risk of coronary atherosclerosis. This study serves as a demonstration of leveraging spatial transcriptomics and bulk RNA-seq to provide spatial insights into the mechanisms of plaque instability. The prioritization of matrix metalloproteinase-9 aligns with findings from an early proteomics study on human carotid plaque specimens. The study involved six symptomatic and six asymptomatic patients, identifying a four-biomarker signature (matrix metalloproteinase-9, S100A8/S100A9, cathepsin D, and galectin-3–binding protein) that improved risk prediction.118
The spatial proteomics field has also expanded with the development of technologies such as CODEX (Co-Detection by Indexing).119 Schneider et al120 performed Visium spatial transcriptomics and CODEX imaging with 51-multiplexed antibodies in human carotid plaques. In their study, spatial analyses were used to confirm the origin of near-infrared auto-photoacoustic signal and to establish that the combination of this imaging and ultrasound imaging can detect vulnerable carotid plaque. Indeed, spatial analyses established that the highest near-infrared auto-photoacoustic signal was spatially correlated with bilirubin and associated blood-based residue and inflammatory macrophage subpopulations bearing CD74, HLA-DR, CD14, and CD163 markers. Spatial analyses also highlight the inflammatory macrophages lining the lumen of vulnerable plaque.
Spatial metabolomics uses imaging mass spectrometry to detect metabolites, lipids, and other small molecules within the spatial context of tissues and cells that correlate with pathologic findings in situ.121 Matrix-assisted laser desorption/ionization mass spectrometry imaging and desorption electrospray ionization mass spectrometry imaging have been often used for spatial profiling of metabolites in human atherosclerosis samples. This section highlights recent spatial metabolomics studies that have provided insights into the spatial distribution of specific lipid classes in relation to in situ histopathologic features of plaque instability. These studies were conducted in human carotid arteries122, 123, 124 and coronary arteries.125 Li et al122 used desorption electrospray ionization mass spectrometry imaging to profile carotid plaque samples from 12 patients, identifying 55 metabolites. Pathway enrichment analysis showed that the sphingolipid signaling pathway and necroptosis pathway are enriched in lipid-rich regions. In addition, glycerophospholipid metabolism and the ether lipid metabolism pathway were mainly enriched in collagen-rich regions. Greco et al123 performed matrix-assisted laser desorption/ionization mass spectrometry imaging and identified that macrophage-rich regions in symptomatic lesions exhibited enrichment in sphingomyelins. Moreover, intimal SMCs in symptomatic plaques were found to be enriched in cholesterol and cholesteryl esters (n = 6 patients). Moerman et al124 conducted matrix-assisted laser desorption/ionization mass spectrometry imaging on human carotid plaques; their findings suggest a distinct colocalization between plaque features and specific lipid classes, as well as individual lipid species in high-risk atherosclerotic plaques. Seeley et al125 conducted matrix-assisted laser desorption/ionization mass spectrometry imaging on human coronary artery samples from nine unmatched individuals, classified based on the Stary classification scale,72,73 and further categorized into stable and unstable atheromas (n = 5 and 4, respectively). Among the 170 annotated metabolites, >60 differed between stable and unstable plaques. Acylcarnitines and acylglycines were increased in stable plaques, whereas tryptophan metabolites were enriched in unstable plaques. Spatial analysis in stable plaques revealed lactic acid in the necrotic core, with elevated pyruvic acid in the fibrous cap. In unstable plaques, 5-hydroxyindoleacetic acid was enriched in the fibrous cap. The study also integrated publicly available RNA-seq data set (http://www.ncbi.nlm.nih.gov/geo; accession number GSE12052152) , comparing stable versus unstable human atherosclerosis to identify the enrichment of relevant metabolic pathways.
To summarize, spatial transcriptomics and proteomics are emerging tools in atherosclerosis research. Spatial metabolomics analyses using imaging mass spectrometry have already provided insights into the spatial distribution of various metabolites within atherosclerosis plaques. The continuous development of advanced imaging mass spectrometry techniques, offering single-cell and subcellular spatial resolution,121 along with ongoing advancements in spatial omics technologies such as spatial assays for chromatin accessibility126 will accelerate the creation of a spatial atlas of human atherosclerosis.
Limitations and Challenges
To date, omics data at bulk levels have largely been derived from carotid endarterectomy samples. Expanding the data collection to diverse vascular beds holds promise for assessing both vascular bed–specific mechanisms and general mechanisms underlying atherosclerosis. A practical limitation also arises from the sample preparation process, in which a portion of the lesion is designated for histologic analysis, and adjacent plaque segments for omics data acquisition. Emerging technologies, such as spatial analysis, facilitate the simultaneous acquisition of omics and histology data from the same tissue section, offering a partial solution to this constraint. The scalability of spatial analysis, however, remains an ongoing challenge. Integration of bulk omics data with single-cell and spatial analyses has been applied to infer spatial information from bulk omics data.75
For scRNA-seq analysis, major challenges arise from dissociation methods that may result in the loss of fragile cells, alterations in gene expression, and loss of spatial resolution. Limitations of CITE analysis include the loss of surface epitopes due to enzymatic dissociation and the capture of only pre-selected surface targets. Single-nucleus RNA-seq has the capability to capture transcripts from cells vulnerable to dissociation. It also serves as an alternative method for cells >30 μm that are incompatible to droplet-based microfluidic devices. Nevertheless, scRNA-seq remains indispensable for obtaining a comprehensive transcriptional profile of both the nucleus and cytoplasm, particularly when cytoplasmic transcripts play a crucial role in the biological process being studied.127 Lastly, although various omics technologies enable high-resolution molecular snapshots of cells, the necessity for sample destruction precludes analyses over time. The current molecular event recording methods continues to face significant limitations that need to be addressed and adapted for atherosclerosis research to reconstruct a high-content biological history.
Summary and Future Directions
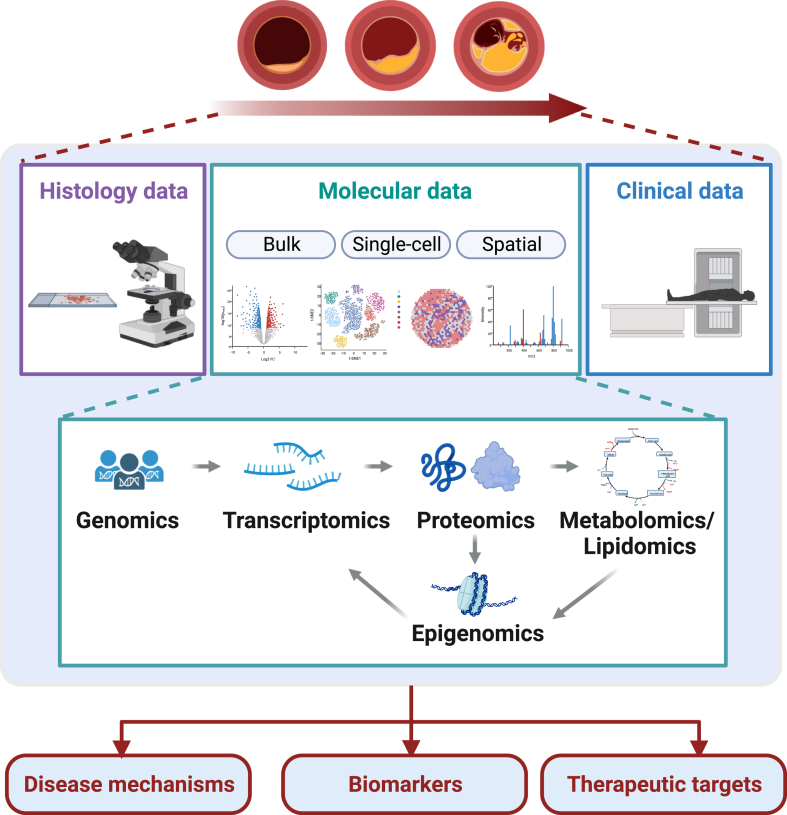
Omics technologies have revolutionized the understanding of atherosclerosis plaque biology, opening new avenues for enhancing risk prediction and identifying potential therapeutic targets. Genomic studies offer insights into the genetic predisposition to ACVD, and integrative genomic studies accelerate the translation of genomic discoveries. Transcriptomic, proteomic, metabolomic, and lipidomic studies further refine our understanding of the molecular signature of atherosclerotic plaques, guiding data-driven hypothesis generation for mechanistic studies and introducing novel avenues for biomarker discovery. Single-cell technologies and emerging spatial analysis unveil the heterogeneity and plasticity of plaque cells with spatial resolution. These advancements unlock the potential to target specific markers and disease-driving cell populations, paving the way for more precise and effective therapeutic interventions (Figure 1).
Figure 1.
Omics approaches unveiling the biology of human atherosclerotic plaques. This review covers the application of omics approaches, including genomics, epigenomics, transcriptomics, proteomics, and metabolomics/lipidomics, to unveil the underlying biology of human atherosclerotic plaques. Omics data define the molecular signature of human atherosclerotic plaques. Integration and analysis of data that spans molecular, histologic, and clinical traits provide insights into disease mechanisms, new biomarkers, and therapeutic targets.
Potential areas for future research have been discussed in the respective sections. This final section highlights three areas that hold relevance across all omics-based studies. First, omics data are crucial in prioritizing candidate causal genes supported by human genomic evidence, identifying differentially expressed molecules between conditions, and uncovering novel cell subpopulations. To advance further, it is crucial to expedite the validation of causal regulators and key drivers to develop mechanism-based therapeutic targets. Addressing this bottleneck can be achieved by enhancing prioritization strategies and optimizing the efficiency of validation workflows through the implementation of high-throughput screening assays. Second, efforts in biobanking have yielded significant discoveries.128 Previous reviews have highlighted the contributions of biobanking initiatives, such as the Athero-Express biobank66,67 and the Munich Vascular Biobank,61 in atherosclerosis research. Building large cohorts of well-characterized patients and integrating multi-omics and single-cell multi-omics analyses, along with histopathologic and clinical data, will enable comprehensive analysis across different parameters, including age, sex, ethnicity, pathologic features (eg, instability, rupture, erosion), and the evaluation of individual risks and therapeutic effects on specific pathways. The availability of rich data sets will also facilitate data-driven hypothesis generation. Lastly, alongside improved omics technologies and data analysis tools, open source tools and web portals for data access and visualization (eg, Open Targets Genetics,15 GEO2R, PlaqView129) will expedite scientific progress by facilitating comprehensive visualization and interpretation of results. In summary, advances in omics-based research are expected to continue to fuel scientific discoveries in atherosclerosis research.
Disclosure Statement
None declared.
Footnotes
Advances in Understanding Cardiovascular Disease Pathogenesis Theme Issue
Supported by NIHR00HL130574 (H.Z.), R01HL151611 (H.Z.), and R01HL168174 (H.Z.), and the Irving Scholars award through UL1TR001873 by the National Center for Advancing Translational Sciences and the NIH (H.Z.), the Russell Berrie Foundation Diabetes Scholar Program (X.W.), and the American Heart Association Postdoctoral Fellowship (21POST829654) (X.W.).
This article is part of a review series highlighting the novel insights provided by next-generation approaches to our understanding of cardiovascular disease pathogenesis and treatment.
References
- 1.Libby P. The changing landscape of atherosclerosis. Nature. 2021;592:524–533. doi: 10.1038/s41586-021-03392-8. [DOI] [PubMed] [Google Scholar]
- 2.Björkegren J.L.M., Lusis A.J. Atherosclerosis: recent developments. Cell. 2022;185:1630–1645. doi: 10.1016/j.cell.2022.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentzon J.F., Otsuka F., Virmani R., Falk E. Mechanisms of plaque formation and rupture. Circ Res. 2014;114:1852–1866. doi: 10.1161/CIRCRESAHA.114.302721. [DOI] [PubMed] [Google Scholar]
- 4.Arnett D.K., Blumenthal R.S., Albert M.A., Buroker A.B., Goldberger Z.D., Hahn E.J., Himmelfarb C.D., Khera A., Lloyd-Jones D., McEvoy J.W., Michos E.D., Miedema M.D., Muñoz D., Smith S.C., Jr., Virani S.S., Williams K.A., Sr., Yeboah J., Ziaeian B. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;140:e563–e595. doi: 10.1161/CIR.0000000000000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsao C.W., Aday A.W., Almarzooq Z.I., Anderson C.A.M., Arora P., Avery C.L., et al. American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee Heart Disease and Stroke Statistics–2023 update: a report from the American Heart Association. Circulation. 2023;147:e93–e621. doi: 10.1161/CIR.0000000000001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timmis A., Vardas P., Townsend N., Torbica A., Katus H., De Smedt D., Gale C.P., Maggioni A.P., Petersen S.E., Huculeci R., Kazakiewicz D., de Benito Rubio V., Ignatiuk B., Raisi-Estabragh Z., Pawlak A., Karagiannidis E., Treskes R., Gaita D., Beltrame J.F., McConnachie A., Bardinet I., Graham I., Flather M., Elliott P., Mossialos E.A., Weidinger F., Achenbach S., Atlas Writing Group, European Society of Cardiology European Society of Cardiology: cardiovascular disease statistics 2021. Eur Heart J. 2022;43:716–799. doi: 10.1093/eurheartj/ehab892. [DOI] [PubMed] [Google Scholar]
- 7.Yahagi K., Kolodgie F.D., Otsuka F., Finn A.V., Davis H.R., Joner M., Virmani R. Pathophysiology of native coronary, vein graft, and in-stent atherosclerosis. Nat Rev Cardiol. 2016;13:79–98. doi: 10.1038/nrcardio.2015.164. [DOI] [PubMed] [Google Scholar]
- 8.Bashore A.C., Zhu L.Y., Reilly M.P. A new era in understanding atherosclerotic plaques. Nat Cardiovasc Res. 2022;12:1127–1129. doi: 10.1038/s44161-022-00187-6. [DOI] [PubMed] [Google Scholar]
- 9.Vandereyken K., Sifrim A., Thienpont B., Voet T. Methods and applications for single-cell and spatial multi-omics. Nat Rev Genet. 2023;24:494–515. doi: 10.1038/s41576-023-00580-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McPherson R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., Boerwinkle E., Hobbs H.H., Cohen J.C. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samani N.J., Erdmann J., Hall A.S., Hengstenberg C., Mangino M., Mayer B., Dixon R.J., Meitinger T., Braund P., Wichmann H.-E., Barrett J.H., König I.R., Stevens S.E., Szymczak S., Tregouet D.-A., Iles M.M., Pahlke F., Pollard H., Lieb W., Cambien F., Fischer M., Ouwehand W., Blankenberg S., Balmforth A.J., Baessler A., Ball S.G., Strom T.M., Braenne I., Gieger C., Deloukas P., Tobin M.D., Ziegler A., Thompson J.R., Schunkert H., WTCC and the Cardiogenics Consortium Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aragam K.G., Jiang T., Goel A., Kanoni S., Wolford B.N., Atri D.S., et al. CARDIoGRAMplusC4D Consortium Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat Genet. 2022;54:1803–1815. doi: 10.1038/s41588-022-01233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tcheandjieu C., Zhu X., Hilliard A.T., Clarke S.L., Napolioni V., Ma S., et al. Large-scale genome-wide association study of coronary artery disease in genetically diverse populations. Nat Med. 2022;28:1679–1692. doi: 10.1038/s41591-022-01891-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mountjoy E., Schmidt E.M., Carmona M., Schwartzentruber J., Peat G., Miranda A., Fumis L., Hayhurst J., Buniello A., Karim M.A., Wright D., Hercules A., Papa E., Fauman E.B., Barrett J.C., Todd J.A., Ochoa D., Dunham I., Ghoussaini M. An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nat Genet. 2021;53:1527–1533. doi: 10.1038/s41588-021-00945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musunuru K., Kathiresan S. Genetics of common, complex coronary artery disease. Cell. 2019;177:132–145. doi: 10.1016/j.cell.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Wainberg M., Sinnott-Armstrong N., Mancuso N., Barbeira A.N., Knowles D.A., Golan D., Ermel R., Ruusalepp A., Quertermous T., Hao K., Björkegren J.L.M., Im H.K., Pasaniuc B., Rivas M.A., Kundaje A. Opportunities and challenges for transcriptome-wide association studies. Nat Genet. 2019;51:592–599. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao K., Ermel R., Sukhavasi K., Cheng H., Ma L., Li L., Amadori L., Koplev S., Franzén O., d’Escamard V., Chandel N., Wolhuter K., Bryce N.S., Venkata V.R.M., Miller C.L., Ruusalepp A., Schunkert H., Björkegren J.L.M., Kovacic J.C. Integrative prioritization of causal genes for coronary artery disease. Circ Genom Precis Med. 2022;15 doi: 10.1161/CIRCGEN.121.003365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koplev S., Seldin M., Sukhavasi K., Ermel R., Pang S., Zeng L., Bankier S., Di Narzo A., Cheng H., Meda V., Ma A., Talukdar H., Cohain A., Amadori L., Argmann C., Houten S.M., Franzén O., Mocci G., Meelu O.A., Ishikawa K., Whatling C., Jain A., Jain R.K., Gan L.-M., Giannarelli C., Roussos P., Hao K., Schunkert H., Michoel T., Ruusalepp A., Schadt E.E., Kovacic J.C., Lusis A.J., Björkegren J.L.M. A mechanistic framework for cardiometabolic and coronary artery diseases. Nat Cardiovasc Res. 2022;1:85–100. doi: 10.1038/s44161-021-00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner A.W., Hu S.S., Mosquera J.V., Ma W.F., Hodonsky C.J., Wong D., Auguste G., Song Y., Sol-Church K., Farber E., Kundu S., Kundaje A., Lopez N.G., Ma L., Ghosh S.K.B., Onengut-Gumuscu S., Ashley E.A., Quertermous T., Finn A.V., Leeper N.J., Kovacic J.C., Björkegren J.L.M., Zang C., Miller C.L. Single-nucleus chromatin accessibility profiling highlights regulatory mechanisms of coronary artery disease risk. Nat Genet. 2022;54:804–816. doi: 10.1038/s41588-022-01069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Örd T., Õunap K., Stolze L., Aherrahrou R., Nurminen V., Selvarajan I., Toropainen A., Lönnberg T., Aavik E., Ylä-Herttuala S., Civelek M., Romanoski C.E., Kaikkonen M.U. Single-cell epigenomics and functional fine-mapping of atherosclerosis GWAS loci. Circ Res. 2021;129:240–258. doi: 10.1161/CIRCRESAHA.121.318971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genovese G., Kähler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., Purcell S.M., Svantesson O., Landén M., Höglund M., Lehmann S., Gabriel S.B., Moran J.L., Lander E.S., Sullivan P.F., Sklar P., Grönberg H., Hultman C.M., McCarroll S.A. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaiswal S., Natarajan P., Silver A.J., Gibson C.J., Bick A.G., Shvartz E., McConkey M., Gupta N., Gabriel S., Ardissino D., Baber U., Mehran R., Fuster V., Danesh J., Frossard P., Saleheen D., Melander O., Sukhova G.K., Neuberg D., Libby P., Kathiresan S., Ebert B.L. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tall A.R., Fuster J.J. Clonal hematopoiesis in cardiovascular disease and therapeutic implications. Nat Cardiovasc Res. 2022;1:116–124. doi: 10.1038/s44161-021-00015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bick A.G., Weinstock J.S., Nandakumar S.K., Fulco C.P., Bao E.L., Zekavat S.M., et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 2020;586:763–768. doi: 10.1038/s41586-020-2819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bick A.G., Pirruccello J.P., Griffin G.K., Gupta N., Gabriel S., Saleheen D., Libby P., Kathiresan S., Natarajan P. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation. 2020;141:124–131. doi: 10.1161/CIRCULATIONAHA.119.044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu W., Yalcinkaya M., Maestre I.F., Olszewska M., Ampomah P.B., Heimlich J.B., Wang R., Vela P.S., Xiao T., Bick A.G., Levine R., Papapetrou E.P., Libby P., Tabas I., Wang N., Tall A.R. Blockade of IL-6 signaling alleviates atherosclerosis in Tet2-deficient clonal hematopoiesis. Nat Cardiovasc Res. 2023;2:572–586. doi: 10.1038/s44161-023-00281-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khera A.V., Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet. 2017;18:331–344. doi: 10.1038/nrg.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M., Lee-Kim V.S., Atri D.S., Elowe N.H., Yu J., Garvie C.W., Won H.-H., Hadaya J.E., MacDonald B.T., Trindade K., Melander O., Rader D.J., Natarajan P., Kathiresan S., Kaushik V.K., Khera A.V., Gupta R.M. Rare, damaging DNA variants in CORIN and risk of coronary artery disease: insights from functional genomics and large-scale sequencing analyses. Circ Genom Precis Med. 2021;14 doi: 10.1161/CIRCGEN.121.003399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costantino S., Paneni F. In: Prevention and Treatment of Atherosclerosis: Improving State-of-the-Art Management and Search for Novel Targets. von Eckardstein A., Binder C.J., editors. Springer; Cham, Switzerland: 2022. The epigenome in atherosclerosis; pp. 511–535. [PubMed] [Google Scholar]
- 31.Rizzacasa B., Amati F., Romeo F., Novelli G., Mehta J.L. Epigenetic modification in coronary atherosclerosis: JACC Review Topic of the Week. J Am Coll Cardiol. 2019;74:1352–1365. doi: 10.1016/j.jacc.2019.07.043. [DOI] [PubMed] [Google Scholar]
- 32.Aavik E., Babu M., Ylä-Herttuala S. DNA methylation processes in atheosclerotic plaque. Atherosclerosis. 2019;281:168–179. doi: 10.1016/j.atherosclerosis.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Zaina S., Heyn H., Carmona F.J., Varol N., Sayols S., Condom E., Ramírez-Ruz J., Gomez A., Gonçalves I., Moran S., Esteller M. DNA methylation map of human atherosclerosis. Circ Cardiovasc Genet. 2014;7:692–700. doi: 10.1161/CIRCGENETICS.113.000441. [DOI] [PubMed] [Google Scholar]
- 34.del Pilar Valencia-Morales M., Zaina S., Heyn H., Carmona F.J., Varol N., Sayols S., Condom E., Ramírez-Ruz J., Gomez A., Moran S., Lund G., Rodríguez-Ríos D., López-González G., Ramírez-Nava M., de la Rocha C., Sanchez-Flores A., Esteller M. The DNA methylation drift of the atherosclerotic aorta increases with lesion progression. BMC Med Genomics. 2015;8:7. doi: 10.1186/s12920-015-0085-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aavik E., Lumivuori H., Leppänen O., Wirth T., Häkkinen S.-K., Bräsen J.-H., Beschorner U., Zeller T., Braspenning M., van Criekinge W., Mäkinen K., Ylä-Herttuala S. Global DNA methylation analysis of human atherosclerotic plaques reveals extensive genomic hypomethylation and reactivation at imprinted locus 14q32 involving induction of a miRNA cluster. Eur Heart J. 2015;36:993–1000. doi: 10.1093/eurheartj/ehu437. [DOI] [PubMed] [Google Scholar]
- 36.Li J., Zhang X., Yang M., Yang H., Xu N., Fan X., Liu G., Jiang X., Fan J., Zhang L., Zhang H., Zhou Y., Li R., Gao S., Jin J., Jin Z., Zheng J., Tu Q., Ren J. DNA methylome profiling reveals epigenetic regulation of lipoprotein-associated phospholipase A2 in human vulnerable atherosclerotic plaque. Clin Epigenetics. 2021;13:161. doi: 10.1186/s13148-021-01152-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Döring Y., Manthey H.D., Drechsler M., Lievens D., Megens R.T.A., Soehnlein O., Busch M., Manca M., Koenen R.R., Pelisek J., Daemen M.J., Lutgens E., Zenke M., Binder C.J., Weber C., Zernecke A. Auto-antigenic protein-DNA complexes stimulate plasmacytoid dendritic cells to promote atherosclerosis. Circulation. 2012;125:1673–1683. doi: 10.1161/CIRCULATIONAHA.111.046755. [DOI] [PubMed] [Google Scholar]
- 38.Lee K., Santibanez-Koref M., Polvikoski T., Birchall D., Mendelow A.D., Keavney B. Increased expression of fatty acid binding protein 4 and leptin in resident macrophages characterises atherosclerotic plaque rupture. Atherosclerosis. 2013;226:74–81. doi: 10.1016/j.atherosclerosis.2012.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natarajan P. Genomic aging, clonal hematopoiesis, and cardiovascular disease. Arterioscler Thromb Vasc Biol. 2023;43:3–14. doi: 10.1161/ATVBAHA.122.318181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khyzha N., Alizada A., Wilson M.D., Fish J.E. Epigenetics of atherosclerosis: emerging mechanisms and methods. Trends Mol Med. 2017;23:332–347. doi: 10.1016/j.molmed.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Shendure J. The beginning of the end for microarrays? Nat Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 42.Cloonan N., Forrest A.R.R., Kolle G., Gardiner B.B.A., Faulkner G.J., Brown M.K., Taylor D.F., Steptoe A.L., Wani S., Bethel G., Robertson A.J., Perkins A.C., Bruce S.J., Lee C.C., Ranade S.S., Peckham H.E., Manning J.M., McKernan K.J., Grimmond S.M. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 43.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 44.Folkersen L., Persson J., Ekstrand J., Agardh H.E., Hansson G.K., Gabrielsen A., Hedin U., Paulsson-Berne G. Prediction of ischemic events on the basis of transcriptomic and genomic profiling in patients undergoing carotid endarterectomy. Mol Med. 2012;18:669–675. doi: 10.2119/molmed.2011.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puig O., Yuan J., Stepaniants S., Zieba R., Zycband E., Morris M., Coulter S., Yu X., Menke J., Woods J., Chen F., Ramey D.R., He X., O’Neill E.A., Hailman E., Johns D.G., Hubbard B.K., Yee Lum P., Wright S.D., Desouza M.M., Plump A., Reiser V. A gene expression signature that classifies human atherosclerotic plaque by relative inflammation status. Circ Cardiovasc Genet. 2011;4:595–604. doi: 10.1161/CIRCGENETICS.111.960773. [DOI] [PubMed] [Google Scholar]
- 46.Hägg S., Skogsberg J., Lundström J., Noori P., Nilsson R., Zhong H., Maleki S., Shang M.-M., Brinne B., Bradshaw M., Bajic V.B., Samnegård A., Silveira A., Kaplan L.M., Gigante B., Leander K., de Faire U., Rosfors S., Lockowandt U., Liska J., Konrad P., Takolander R., Franco-Cereceda A., Schadt E.E., Ivert T., Hamsten A., Tegnér J., Björkegren J. Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: the Stockholm Atherosclerosis Gene Expression (STAGE) study. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ayari H., Bricca G. Identification of two genes potentially associated in iron-heme homeostasis in human carotid plaque using microarray analysis. J Biosci. 2013;38:311–315. doi: 10.1007/s12038-013-9310-2. [DOI] [PubMed] [Google Scholar]
- 48.Steenman M., Espitia O., Maurel B., Guyomarch B., Heymann M.-F., Pistorius M.-A., Ory B., Heymann D., Houlgatte R., Gouëffic Y., Quillard T. Identification of genomic differences among peripheral arterial beds in atherosclerotic and healthy arteries. Sci Rep. 2018;8:3940. doi: 10.1038/s41598-018-22292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chai J.T., Ruparelia N., Goel A., Kyriakou T., Biasiolli L., Edgar L., Handa A., Farrall M., Watkins H., Choudhury R.P. Differential gene expression in macrophages from human atherosclerotic plaques shows convergence on pathways implicated by genome-wide association study risk variants. Arterioscler Thromb Vasc Biol. 2018;38:2718–2730. doi: 10.1161/ATVBAHA.118.311209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karlöf E., Seime T., Dias N., Lengquist M., Witasp A., Almqvist H., Kronqvist M., Gådin J.R., Odeberg J., Maegdefessel L., Stenvinkel P., Matic L.P., Hedin U. Correlation of computed tomography with carotid plaque transcriptomes associates calcification with lesion-stabilization. Atherosclerosis. 2019;288:175–185. doi: 10.1016/j.atherosclerosis.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Jin H., Goossens P., Juhasz P., Eijgelaar W., Manca M., Karel J.M.H., Smirnov E., Sikkink C.J.J.M., Mees B.M.E., Waring O., van Kuijk K., Fazzi G.E., Gijbels M.J.J., Kutmon M., Evelo C.T.A., Hedin U., Daemen M.J.A.P., Sluimer J.C., Matic L., Biessen E.A.L. Integrative multiomics analysis of human atherosclerosis reveals a serum response factor-driven network associated with intraplaque hemorrhage. Clin Transl Med. 2021;11:e458. doi: 10.1002/ctm2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahmoud A.D., Ballantyne M.D., Miscianinov V., Pinel K., Hung J., Scanlon J.P., Iyinikkel J., Kaczynski J., Tavares A.S., Bradshaw A.C., Mills N.L., Newby D.E., Caporali A., Gould G.W., George S.J., Ulitsky I., Sluimer J.C., Rodor J., Baker A.H. The human-specific and smooth muscle cell-enriched lncRNA SMILR promotes proliferation by regulating mitotic CENPF mRNA and drives cell-cycle progression which can be targeted to limit vascular remodeling. Circ Res. 2019;125:535–551. doi: 10.1161/CIRCRESAHA.119.314876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bazan H.A., Brooks A.J., Vongbunyong K., Tee C., Douglas H.F., Klingenberg N.C., Woods T.C. A pro-inflammatory and fibrous cap thinning transcriptome profile accompanies carotid plaque rupture leading to stroke. Sci Rep. 2022;12 doi: 10.1038/s41598-022-17546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Widlansky M.E., Liu Y., Tumusiime S., Hofeld B., Khan N., Aljadah M., Wang J., Anger A., Qiu Q., Therani B., Liu P., Liang M. Coronary plaque sampling reveals molecular insights into coronary artery disease. Circ Res. 2023;133:532–534. doi: 10.1161/CIRCRESAHA.123.323022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hartman R.J.G., Owsiany K., Ma L., Koplev S., Hao K., Slenders L., Civelek M., Mokry M., Kovacic J.C., Pasterkamp G., Owens G., Björkegren J.L.M., den Ruijter H.M. Sex-stratified gene regulatory networks reveal female key driver genes of atherosclerosis involved in smooth muscle cell phenotype switching. Circulation. 2021;143:713–726. doi: 10.1161/CIRCULATIONAHA.120.051231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salagianni M., Galani I.E., Lundberg A.M., Davos C.H., Varela A., Gavriil A., Lyytikäinen L.-P., Lehtimäki T., Sigala F., Folkersen L., Gorgoulis V., Lenglet S., Montecucco F., Mach F., Hedin U., Hansson G.K., Monaco C., Andreakos E. Toll-like receptor 7 protects from atherosclerosis by constraining “inflammatory” macrophage activation. Circulation. 2012;126:952–962. doi: 10.1161/CIRCULATIONAHA.111.067678. [DOI] [PubMed] [Google Scholar]
- 57.Perisic L., Hedin E., Razuvaev A., Lengquist M., Osterholm C., Folkersen L., Gillgren P., Paulsson-Berne G., Ponten F., Odeberg J., Hedin U. Profiling of atherosclerotic lesions by gene and tissue microarrays reveals PCSK6 as a novel protease in unstable carotid atherosclerosis. Arterioscler Thromb Vasc Biol. 2013;33:2432–2443. doi: 10.1161/ATVBAHA.113.301743. [DOI] [PubMed] [Google Scholar]
- 58.Kojima Y., Downing K., Kundu R., Miller C., Dewey F., Lancero H., Raaz U., Perisic L., Hedin U., Schadt E., Maegdefessel L., Quertermous T., Leeper N.J. Cyclin-dependent kinase inhibitor 2B regulates efferocytosis and atherosclerosis. J Clin Invest. 2014;124:1083–1097. doi: 10.1172/JCI70391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller C.L., Haas U., Diaz R., Leeper N.J., Kundu R.K., Patlolla B., Assimes T.L., Kaiser F.J., Perisic L., Hedin U., Maegdefessel L., Schunkert H., Erdmann J., Quertermous T., Sczakiel G. Coronary heart disease-associated variation in TCF21 disrupts a miR-224 binding site and miRNA-mediated regulation. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perisic Matic L., Rykaczewska U., Razuvaev A., Sabater-Lleal M., Lengquist M., Miller C.L., Ericsson I., Röhl S., Kronqvist M., Aldi S., Magné J., Paloschi V., Vesterlund M., Li Y., Jin H., Diez M.G., Roy J., Baldassarre D., Veglia F., Humphries S.E., de Faire U., Tremoli E., Odeberg J., Vukojević V., Lehtiö J., Maegdefessel L., Ehrenborg E., Paulsson-Berne G., Hansson G.K., Lindeman J.H.N., Eriksson P., Quertermous T., Hamsten A., Hedin U. Phenotypic modulation of smooth muscle cells in atherosclerosis is associated with downregulation of LMOD1, SYNPO2, PDLIM7, PLN, and SYNM. Arterioscler Thromb Vasc Biol. 2016;36:1947–1961. doi: 10.1161/ATVBAHA.116.307893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pelisek J., Hegenloh R., Bauer S., Metschl S., Pauli J., Glukha N., Busch A., Reutersberg B., Kallmayer M., Trenner M., Wendorff H., Tsantilas P., Schmid S., Knappich C., Schaeffer C., Stadlbauer T., Biro G., Wertern U., Meisner F., Stoklasa K., Menges A.-L., Radu O., Dallmann-Sieber S., Karlas A., Knipfer E., Reeps C., Zimmermann A., Maegdefessel L., Eckstein H.-H. Biobanking: objectives, requirements, and future challenges—experiences from the Munich Vascular Biobank. J Clin Med. 2019;8:251. doi: 10.3390/jcm8020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rykaczewska U., Zhao Q., Saliba-Gustafsson P., Lengquist M., Kronqvist M., Bergman O., Huang Z., Lund K., Waden K., Pons Vila Z., Caidahl K., Skogsberg J., Vukojevic V., Lindeman J.H.N., Roy J., Hansson G.K., Treuter E., Leeper N.J., Eriksson P., Ehrenborg E., Razuvaev A., Hedin U., Matic L. Plaque evaluation by ultrasound and transcriptomics reveals BCLAF1 as a regulator of smooth muscle cell lipid transdifferentiation in atherosclerosis. Arterioscler Thromb Vasc Biol. 2022;42:659–676. doi: 10.1161/ATVBAHA.121.317018. [DOI] [PubMed] [Google Scholar]
- 63.Fasolo F., Jin H., Winski G., Chernogubova E., Pauli J., Winter H., Li D.Y., Glukha N., Bauer S., Metschl S., Wu Z., Koschinsky M.L., Reilly M., Pelisek J., Kempf W., Eckstein H.-H., Soehnlein O., Matic L., Hedin U., Bäcklund A., Bergmark C., Paloschi V., Maegdefessel L. Long noncoding RNA MIAT controls advanced atherosclerotic lesion formation and plaque destabilization. Circulation. 2021;144:1567–1583. doi: 10.1161/CIRCULATIONAHA.120.052023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rykaczewska U., Suur B.E., Röhl S., Razuvaev A., Lengquist M., Sabater-Lleal M., van der Laan S.W., Miller C.L., Wirka R.C., Kronqvist M., Gonzalez Diez M., Vesterlund M., Gillgren P., Odeberg J., Lindeman J.H., Veglia F., Humphries S.E., de Faire U., Baldassarre D., Tremoli E., IMPROVE Study Group. Lehtiö J., Hansson G.K., Paulsson-Berne G., Pasterkamp G., Quertermous T., Hamsten A., Eriksson P., Hedin U., Matic L. PCSK6 is a key protease in the control of smooth muscle cell function in vascular remodeling. Circ Res. 2020;126:571–585. doi: 10.1161/CIRCRESAHA.119.316063. [DOI] [PubMed] [Google Scholar]
- 65.Perisic L., Aldi S., Sun Y., Folkersen L., Razuvaev A., Roy J., Lengquist M., Åkesson S., Wheelock C.E., Maegdefessel L., Gabrielsen A., Odeberg J., Hansson G.K., Paulsson-Berne G., Hedin U. Gene expression signatures, pathways and networks in carotid atherosclerosis. J Intern Med. 2016;279:293–308. doi: 10.1111/joim.12448. [DOI] [PubMed] [Google Scholar]
- 66.Verhoeven B.A.N., Velema E., Schoneveld A.H., de Vries J.P.P.M., de Bruin P., Seldenrijk C.A., de Kleijn D.P.V., Busser E., van der Graaf Y., Moll F., Pasterkamp G. Athero-express: differential atherosclerotic plaque expression of mRNA and protein in relation to cardiovascular events and patient characteristics. Rationale and design. Eur J Epidemiol. 2004;19:1127–1133. doi: 10.1007/s10564-004-2304-6. [DOI] [PubMed] [Google Scholar]
- 67.Hellings W.E., Moll F.L., de Kleijn D.P.V., Pasterkamp G. 10-Years experience with the Athero-Express study. Cardiovasc Diagn Ther. 2012;2:63–73. doi: 10.3978/j.issn.2223-3652.2012.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mokry M., Boltjes A., Slenders L., Bel-Bordes G., Cui K., Brouwer E., et al. Transcriptomic-based clustering of human atherosclerotic plaques identifies subgroups with different underlying biology and clinical presentation. Nat Cardiovasc Res. 2022;1:1140–1155. doi: 10.1038/s44161-022-00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng S., Shah S.H., Corwin E.J., Fiehn O., Fitzgerald R.L., Gerszten R.E., Illig T., Rhee E.P., Srinivas P.R., Wang T.J., Jain M., American Heart Association Council on Functional Genomics and Translational Biology. Council on Cardiovascular and Stroke Nursing. Council on Clinical Cardiology. Stroke Council Potential impact and study considerations of metabolomics in cardiovascular health and disease: a scientific statement from the American Heart Association. Circ Cardiovasc Genet. 2017;10 doi: 10.1161/HCG.0000000000000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herrington D.M., Mao C., Parker S.J., Fu Z., Yu G., Chen L., Venkatraman V., Fu Y., Wang Y., Howard T.D., Jun G., Zhao C.F., Liu Y., Saylor G., Spivia W.R., Athas G.B., Troxclair D., Hixson J.E., Vander Heide R.S., Wang Y., Van Eyk J.E. Proteomic architecture of human coronary and aortic atherosclerosis. Circulation. 2018;137:2741–2756. doi: 10.1161/CIRCULATIONAHA.118.034365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nehme A., Kobeissy F., Zhao J., Zhu R., Feugier P., Mechref Y., Zibara K. Functional pathways associated with human carotid atheroma: a proteomics analysis. Hypertens Res. 2019;42:362–373. doi: 10.1038/s41440-018-0192-4. [DOI] [PubMed] [Google Scholar]
- 72.Stary H.C., Chandler A.B., Dinsmore R.E., Fuster V., Glagov S., Insull W., Jr., Rosenfeld M.E., Schwartz C.J., Wagner W.D., Wissler R.W. A definition of advanced types of atherosclerotic lesions and a histological classification of atherosclerosis. A report from the Committee on Vascular Lesions of the Council on Arteriosclerosis, American Heart Association. Circulation. 1995;92:1355–1374. doi: 10.1161/01.cir.92.5.1355. [DOI] [PubMed] [Google Scholar]
- 73.Stary H.C. Natural history and histological classification of atherosclerotic lesions: an update. Arterioscler Thromb Vasc Biol. 2000;20:1177–1178. doi: 10.1161/01.atv.20.5.1177. [DOI] [PubMed] [Google Scholar]
- 74.Vaisar T., Hu J.H., Airhart N., Fox K., Heinecke J., Nicosia R.F., Kohler T., Potter Z.E., Simon G.M., Dix M.M., Cravatt B.F., Gharib S.A., Dichek D.A. Parallel murine and human plaque proteomics reveals pathways of plaque rupture. Circ Res. 2020;127:997–1022. doi: 10.1161/CIRCRESAHA.120.317295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Theofilatos K., Stojkovic S., Hasman M., van der Laan S.W., Baig F., Barallobre-Barreiro J., Schmidt L.E., Yin S., Yin X., Burnap S., Singh B., Popham J., Harkot O., Kampf S., Nackenhorst M.C., Strassl A., Loewe C., Demyanets S., Neumayer C., Bilban M., Hengstenberg C., Huber K., Pasterkamp G., Wojta J., Mayr M. Proteomic atlas of atherosclerosis: the contribution of proteoglycans to sex differences, plaque phenotypes, and outcomes. Circ Res. 2023;133:542–558. doi: 10.1161/CIRCRESAHA.123.322590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomas L., Edsfeldt A., Mollet I.G., Perisic Matic L., Prehn C., Adamski J., Paulsson-Berne G., Hedin U., Nilsson J., Bengtsson E., Gonçalves I., Björkbacka H. Altered metabolism distinguishes high-risk from stable carotid atherosclerotic plaques. Eur Heart J. 2018;39:2301–2310. doi: 10.1093/eurheartj/ehy124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edsfeldt A., Dunér P., Ståhlman M., Mollet I.G., Asciutto G., Grufman H., Nitulescu M., Persson A.F., Fisher R.M., Melander O., Orho-Melander M., Borén J., Nilsson J., Gonçalves I. Sphingolipids contribute to human atherosclerotic plaque inflammation. Arterioscler Thromb Vasc Biol. 2016;36:1132–1140. doi: 10.1161/ATVBAHA.116.305675. [DOI] [PubMed] [Google Scholar]
- 78.Fredman G., Hellmann J., Proto J.D., Kuriakose G., Colas R.A., Dorweiler B., Connolly E.S., Solomon R., Jones D.M., Heyer E.J., Spite M., Tabas I. An imbalance between specialized pro-resolving lipid mediators and pro-inflammatory leukotrienes promotes instability of atherosclerotic plaques. Nat Commun. 2016;7 doi: 10.1038/ncomms12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi R.H., Tatum S.M., Symons J.D., Summers S.A., Holland W.L. Ceramides and other sphingolipids as drivers of cardiovascular disease. Nat Rev Cardiol. 2021;18:701–711. doi: 10.1038/s41569-021-00536-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doran A.C. Inflammation resolution: implications for atherosclerosis. Circ Res. 2022;130:130–148. doi: 10.1161/CIRCRESAHA.121.319822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matic L.P., Jesus Iglesias M., Vesterlund M., Lengquist M., Hong M.-G., Saieed S., Sanchez-Rivera L., Berg M., Razuvaev A., Kronqvist M., Lund K., Caidahl K., Gillgren P., Pontén F., Uhlén M., Schwenk J.M., Hansson G.K., Paulsson-Berne G., Fagman E., Roy J., Hultgren R., Bergström G., Lehtiö J., Odeberg J., Hedin U. Novel multiomics profiling of human carotid atherosclerotic plaques and plasma reveals biliverdin reductase B as a marker of intraplaque hemorrhage. JACC Basic Transl Sci. 2018;3:464–480. doi: 10.1016/j.jacbts.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh A., Shannon C.P., Gautier B., Rohart F., Vacher M., Tebbutt S.J., Cao K.-A.L. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics. 2019;35:3055–3062. doi: 10.1093/bioinformatics/bty1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nurmohamed N.S., Kraaijenhof J.M., Mayr M., Nicholls S.J., Koenig W., Catapano A.L., Stroes E.S.G. Proteomics and lipidomics in atherosclerotic cardiovascular disease risk prediction. Eur Heart J. 2023;44:1594–1607. doi: 10.1093/eurheartj/ehad161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bandura D.R., Baranov V.I., Ornatsky O.I., Antonov A., Kinach R., Lou X., Pavlov S., Vorobiev S., Dick J.E., Tanner S.D. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 85.Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A., Lao K., Surani M.A. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 86.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baysoy A., Bai Z., Satija R., Fan R. The technological landscape and applications of single-cell multi-omics. Nat Rev Mol Cell Biol. 2023;24:695–713. doi: 10.1038/s41580-023-00615-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cochain C., Vafadarnejad E., Arampatzi P., Pelisek J., Winkels H., Ley K., Wolf D., Saliba A.-E., Zernecke A. Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res. 2018;122:1661–1674. doi: 10.1161/CIRCRESAHA.117.312509. [DOI] [PubMed] [Google Scholar]
- 89.Kim K., Shim D., Lee J.S., Zaitsev K., Williams J.W., Kim K.-W., Jang M.-Y., Seok Jang H., Yun T.J., Lee S.H., Yoon W.K., Prat A., Seidah N.G., Choi J., Lee S.-P., Yoon S.-H., Nam J.W., Seong J.K., Oh G.T., Randolph G.J., Artyomov M.N., Cheong C., Choi J.-H. Transcriptome analysis reveals nonfoamy rather than foamy plaque macrophages are proinflammatory in atherosclerotic murine models. Circ Res. 2018;123:1127–1142. doi: 10.1161/CIRCRESAHA.118.312804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Winkels H., Ehinger E., Vassallo M., Buscher K., Dinh H.Q., Kobiyama K., Hamers A.A.J., Cochain C., Vafadarnejad E., Saliba A.-E., Zernecke A., Pramod A.B., Ghosh A.K., Anto Michel N., Hoppe N., Hilgendorf I., Zirlik A., Hedrick C.C., Ley K., Wolf D. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass cytometry. Circ Res. 2018;122:1675–1688. doi: 10.1161/CIRCRESAHA.117.312513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin J.-D., Nishi H., Poles J., Niu X., Mccauley C., Rahman K., Brown E.J., Yeung S.T., Vozhilla N., Weinstock A., Ramsey S.A., Fisher E.A., Loke P. Single-cell analysis of fate-mapped macrophages reveals heterogeneity, including stem-like properties, during atherosclerosis progression and regression. JCI Insight. 2019;4 doi: 10.1172/jci.insight.124574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fernandez D.M., Rahman A.H., Fernandez N.F., Chudnovskiy A., Amir E.D., Amadori L., Khan N.S., Wong C.K., Shamailova R., Hill C.A., Wang Z., Remark R., Li J.R., Pina C., Faries C., Awad A.J., Moss N., Bjorkegren J.L.M., Kim-Schulze S., Gnjatic S., Ma’ayan A., Mocco J., Faries P., Merad M., Giannarelli C. Single-cell immune landscape of human atherosclerotic plaques. Nat Med. 2019;25:1576–1588. doi: 10.1038/s41591-019-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zernecke A., Winkels H., Cochain C., Williams J.W., Wolf D., Soehnlein O., Robbins C.S., Monaco C., Park I., McNamara C.A., Binder C.J., Cybulsky M.I., Scipione C.A., Hedrick C.C., Galkina E.V., Kyaw T., Ghosheh Y., Dinh H.Q., Ley K. Meta-analysis of leukocyte diversity in atherosclerotic mouse aortas. Circ Res. 2020;127:402–426. doi: 10.1161/CIRCRESAHA.120.316903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pan H., Xue C., Auerbach B.J., Fan J., Bashore A.C., Cui J., Yang D.Y., Trignano S.B., Liu W., Shi J., Ihuegbu C.O., Bush E.C., Worley J., Vlahos L., Laise P., Solomon R.A., Connolly E.S., Califano A., Sims P.A., Zhang H., Li M., Reilly M.P. Single-cell genomics reveals a novel cell state during smooth muscle cell phenotypic switching and potential therapeutic targets for atherosclerosis in mouse and human. Circulation. 2020;142:2060–2075. doi: 10.1161/CIRCULATIONAHA.120.048378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Depuydt M.A.C., Prange K.H.M., Slenders L., Örd T., Elbersen D., Boltjes A., de Jager S.C.A., Asselbergs F.W., de Borst G.J., Aavik E., Lönnberg T., Lutgens E., Glass C.K., den Ruijter H.M., Kaikkonen M.U., Bot I., Slütter B., van der Laan S.W., Yla-Herttuala S., Mokry M., Kuiper J., de Winther M.P.J., Pasterkamp G. Microanatomy of the human atherosclerotic plaque by single-cell transcriptomics. Circ Res. 2020;127:1437–1455. doi: 10.1161/CIRCRESAHA.120.316770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alencar G.F., Owsiany K.M., Karnewar S., Sukhavasi K., Mocci G., Nguyen A.T., Williams C.M., Shamsuzzaman S., Mokry M., Henderson C.A., Haskins R., Baylis R.A., Finn A.V., McNamara C.A., Zunder E.R., Venkata V., Pasterkamp G., Björkegren J., Bekiranov S., Owens G.K. Stem cell pluripotency genes Klf4 and Oct4 regulate complex SMC phenotypic changes critical in late-stage atherosclerotic lesion pathogenesis. Circulation. 2020;142:2045–2059. doi: 10.1161/CIRCULATIONAHA.120.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wirka R.C., Wagh D., Paik D.T., Pjanic M., Nguyen T., Miller C.L., Kundu R., Nagao M., Coller J., Koyano T.K., Fong R., Woo Y.J., Liu B., Montgomery S.B., Wu J.C., Zhu K., Chang R., Alamprese M., Tallquist M.D., Kim J.B., Quertermous T. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019;25:1280–1289. doi: 10.1038/s41591-019-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim J.B., Zhao Q., Nguyen T., Pjanic M., Cheng P., Wirka R., Travisano S., Nagao M., Kundu R., Quertermous T. Environment-sensing Aryl hydrocarbon receptor inhibits the chondrogenic fate of modulated smooth muscle cells in atherosclerotic lesions. Circulation. 2020;142:575–590. doi: 10.1161/CIRCULATIONAHA.120.045981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Conklin A.C., Nishi H., Schlamp F., Örd T., Õunap K., Kaikkonen M.U., Fisher E.A., Romanoski C.E. Meta-analysis of smooth muscle lineage transcriptomes in atherosclerosis and their relationships to in vitro models. Immunometabolism. 2021;3 doi: 10.20900/immunometab20210022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mosquera J.V., Auguste G., Wong D., Turner A.W., Hodonsky C.J., Alvarez-Yela A.C., Song Y., Cheng Q., Lino Cardenas C.L., Theofilatos K., Bos M., Kavousi M., Peyser P.A., Mayr M., Kovacic J.C., Björkegren J.L.M., Malhotra R., Stukenberg P.T., Finn A.V., van der Laan S.W., Zang C., Sheffield N.C., Miller C.L. Integrative single-cell meta-analysis reveals disease-relevant vascular cell states and markers in human atherosclerosis. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hu Z., Liu W., Hua X., Chen X., Chang Y., Hu Y., Xu Z., Song J. Single-cell transcriptomic atlas of different human cardiac arteries identifies cell types associated with vascular physiology. Arterioscler Thromb Vasc Biol. 2021;41:1408–1427. doi: 10.1161/ATVBAHA.120.315373. [DOI] [PubMed] [Google Scholar]
- 102.Alsaigh T., Evans D., Frankel D., Torkamani A. Decoding the transcriptome of calcified atherosclerotic plaque at single-cell resolution. Commun Biol. 2022;5:1084. doi: 10.1038/s42003-022-04056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bashore A.C., Yan H., Xue C., Zhu L.Y., Kim E., Mawson T., Coronel J., Chung A., Ho S., Ross L.S., Kissner M., Passegué E., Bauer R.C., Maegdefessel L., Li M., Reilly M.P. High-dimensional single-cell multimodal landscape of human carotid atherosclerosis. medRxiv. 2023 doi: 10.1101/2023.07.13.23292633. [Preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vallejo J., Saigusa R., Gulati R., Armstrong Suthahar S.S., Suryawanshi V., Alimadadi A., Durant C.P., Ghosheh Y., Roy P., Ehinger E., Pattarabanjird T., Hanna D.B., Landay A.L., Tracy R.P., Lazar J.M., Mack W.J., Weber K.M., Adimora A.A., Hodis H.N., Tien P.C., Ofotokun I., Heath S.L., Shemesh A., McNamara C.A., Lanier L.L., Hedrick C.C., Kaplan R.C., Ley K. Combined protein and transcript single-cell RNA sequencing in human peripheral blood mononuclear cells. BMC Biol. 2022;20:193. doi: 10.1186/s12915-022-01382-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Slysz J., Sinha A., DeBerge M., Singh S., Avgousti H., Lee I., Glinton K., Nagasaka R., Dalal P., Alexandria S., Wai C.M., Tellez R., Vescovo M., Sunderraj A., Wang X., Schipma M., Sisk R., Gulati R., Vallejo J., Saigusa R., Lloyd-Jones D.M., Lomasney J., Weinberg S., Ho K., Ley K., Giannarelli C., Thorp E.B., Feinstein M.J. Single-cell profiling reveals inflammatory polarization of human carotid versus femoral plaque leukocytes. JCI Insight. 2023;8 doi: 10.1172/jci.insight.171359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eberhardt N., Giannarelli C. How single-cell technologies have provided new insights into atherosclerosis. Arterioscler Thromb Vasc Biol. 2022;42:243–252. doi: 10.1161/ATVBAHA.121.315849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.de Winther M.P.J., Bäck M., Evans P., Gomez D., Goncalves I., Jørgensen H.F., Koenen R.R., Lutgens E., Norata G.D., Osto E., Dib L., Simons M., Stellos K., Ylä-Herttuala S., Winkels H., Bochaton-Piallat M.-L., Monaco C. Translational opportunities of single-cell biology in atherosclerosis. Eur Heart J. 2023;44:1216–1230. doi: 10.1093/eurheartj/ehac686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dib L., Koneva L.A., Edsfeldt A., Zurke Y.-X., Sun J., Nitulescu M., Attar M., Lutgens E., Schmidt S., Lindholm M.W., Choudhury R.P., Cassimjee I., Lee R., Handa A., Goncalves I., Sansom S.N., Monaco C. Lipid-associated macrophages transition to an inflammatory state in human atherosclerosis, increasing the risk of cerebrovascular complications. Nat Cardiovasc Res. 2023;2:656–672. doi: 10.1038/s44161-023-00295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Depuydt M.A.C., Schaftenaar F.H., Prange K.H.M., Boltjes A., Hemme E., Delfos L., de Mol J., de Jong M.J.M., Bernabé Kleijn M.N.A., Peeters J.A.H.M., Goncalves L., Wezel A., Smeets H.J., de Borst G.J., Foks A.C., Pasterkamp G., de Winther MPJ, Kuiper J., Bot I., Slütter B. Single-cell T cell receptor sequencing of paired human atherosclerotic plaques and blood reveals autoimmune-like features of expanded effector T cells. Nat Cardiovasc Res. 2023;2:112–125. doi: 10.1038/s44161-022-00208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zernecke A., Erhard F., Weinberger T., Schulz C., Ley K., Saliba A.-E., Cochain C. Integrated single-cell analysis based classification of vascular mononuclear phagocytes in mouse and human atherosclerosis. Cardiovasc Res. 2022;119:1676–1689. doi: 10.1093/cvr/cvac161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Patterson M., Firulyova M.M., Xu Y., Hillman H., Bishop C., Zhu A., Hickok G.H., Schrank P.R., Ronayne C.E., Caillot Z., Fredrickson G., Kennedy A.E., Acharya N., Neels J.G., Chinetti G., Revelo X., Stromnes I.M., Ivanov S., Bold T.D., Zaitsev K., Williams J.W. Trem2 promotes foamy macrophage lipid uptake and survival in atherosclerosis. Nat Cardiovasc Res. 2023;2:1015–1031. doi: 10.1038/s44161-023-00354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Piollet M., Porsch F., Rizzo G., Kapser F., Schulz D.J.J., Kiss M.G., Schlepckow K., Morenas-Rodriguez E., Sen M.O., Gropper J., Roesch M., Göderle L., Hladik A., Knapp S., Colonna M., Martini R., Haass C., Zernecke A., Binder C.J., Cochain C. TREM2 limits necrotic core formation during atherogenesis by controlling macrophage survival and efferocytosis. bioRxiv. 2023 doi: 10.1101/2023.05.15.539977. [Preprint] [DOI] [Google Scholar]
- 113.Burke A.P., Farb A., Malcom G., Virmani R. Effect of menopause on plaque morphologic characteristics in coronary atherosclerosis. Am Heart J. 2001;141 Suppl 2:S58–S62. doi: 10.1067/mhj.2001.109946. [DOI] [PubMed] [Google Scholar]
- 114.Diez Benavente E., Karnewar S., Buono M., Mili E., Hartman R.J.G., Kapteijn D., Slenders L., Daniels M., Aherrahrou R., Reinberger T., Mol B.M., de Borst G.J., de Kleijn D.P.V., Prange K.H.M., Depuydt M.A.C., de Winther MPJ, Kuiper J., Björkegren J.L.M., Erdmann J., Civelek M., Mokry M., Owens G.K., Pasterkamp G., den Ruijter H.M. Female gene networks are expressed in myofibroblast-like smooth muscle cells in vulnerable atherosclerotic plaques. Arterioscler Thromb Vasc Biol. 2023;43:1836–1850. doi: 10.1161/ATVBAHA.123.319325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.de Jager S.C.A., Meeuwsen J.A.L., van Pijpen F.M., Zoet G.A., Barendrecht A.D., Franx A., Pasterkamp G., van Rijn B.B., Goumans M.-J., den Ruijter H.M. Preeclampsia and coronary plaque erosion: manifestations of endothelial dysfunction resulting in cardiovascular events in women. Eur J Pharmacol. 2017;816:129–137. doi: 10.1016/j.ejphar.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 116.Ståhl P.L., Salmén F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., Giacomello S., Asp M., Westholm J.O., Huss M., Mollbrink A., Linnarsson S., Codeluppi S., Borg Å., Pontén F., Costea P.I., Sahlén P., Mulder J., Bergmann O., Lundeberg J., Frisén J. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 117.Sun J., Singh P., Shami A., Kluza E., Pan M., Djordjevic D., Michaelsen N.B., Kennbäck C., van der Wel N.N., Orho-Melander M., Nilsson J., Formentini I., Conde-Knape K., Lutgens E., Edsfeldt A., Gonçalves I. Spatial transcriptional mapping reveals site-specific pathways underlying human atherosclerotic plaque rupture. J Am Coll Cardiol. 2023;81:2213–2227. doi: 10.1016/j.jacc.2023.04.008. [DOI] [PubMed] [Google Scholar]
- 118.Langley S.R., Willeit K., Didangelos A., Matic L.P., Skroblin P., Barallobre-Barreiro J., Lengquist M., Rungger G., Kapustin A., Kedenko L., Molenaar C., Lu R., Barwari T., Suna G., Yin X., Iglseder B., Paulweber B., Willeit P., Shalhoub J., Pasterkamp G., Davies A.H., Monaco C., Hedin U., Shanahan C.M., Willeit J., Kiechl S., Mayr M. Extracellular matrix proteomics identifies molecular signature of symptomatic carotid plaques. J Clin Invest. 2017;127:1546–1560. doi: 10.1172/JCI86924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goltsev Y., Samusik N., Kennedy-Darling J., Bhate S., Hale M., Vazquez G., Black S., Nolan G.P. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968–981.e15. doi: 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schneider M.K., Wang J., Kare A., Adkar S.S., Salmi D., Bell C.F., Alsaigh T., Wagh D., Coller J., Mayer A., Snyder S.J., Borowsky A.D., Long S.R., Lansberg M.G., Steinberg G.K., Heit J.J., Leeper N.J., Ferrara K.W. Combined near infrared photoacoustic imaging and ultrasound detects vulnerable atherosclerotic plaque. Biomaterials. 2023;302 doi: 10.1016/j.biomaterials.2023.122314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Taylor M.J., Lukowski J.K., Anderton C.R. Spatially resolved mass spectrometry at the single cell: recent innovations in proteomics and metabolomics. J Am Soc Mass Spectrom. 2021;32:872–894. doi: 10.1021/jasms.0c00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li W., Luo J., Peng F., Liu R., Bai X., Wang T., Zhang X., Zhu J., Li X.-Y., Wang Z., Liu W., Wang J., Zhang L., Chen X., Xue T., Ding C., Wang C., Jiao L. Spatial metabolomics identifies lipid profiles of human carotid atherosclerosis. Atherosclerosis. 2023;364:20–28. doi: 10.1016/j.atherosclerosis.2022.11.019. [DOI] [PubMed] [Google Scholar]
- 123.Greco F., Quercioli L., Pucci A., Rocchiccioli S., Ferrari M., Recchia F.A., McDonnell L.A. Mass spectrometry imaging as a tool to investigate region specific lipid alterations in symptomatic human carotid atherosclerotic plaques. Metabolites. 2021;11:250. doi: 10.3390/metabo11040250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moerman A.M., Visscher M., Slijkhuis N., Van Gaalen K., Heijs B., Klein T., Burgers P.C., De Rijke Y.B., Van Beusekom H.M.M., Luider T.M., Verhagen H.J.M., Van der Steen A.F.W., Gijsen F.J.H., Van der Heiden K., Van Soest G. Lipid signature of advanced human carotid atherosclerosis assessed by mass spectrometry imaging. J Lipid Res. 2021;62 doi: 10.1194/jlr.RA120000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Seeley E.H., Liu Z., Yuan S., Stroope C., Cockerham E., Rashdan N.A., Delgadillo L., Finney A.C., Kumar D., Das S., Razani B., Liu W., Traylor J., Orr A.W., Rom O., Pattillo C.B., Yurdagul A., Jr. Spatially resolved metabolites in stable and unstable human atherosclerotic plaques identified by mass spectrometry imaging. Arterioscler Thromb Vasc Biol. 2023;43:1626–1635. doi: 10.1161/ATVBAHA.122.318684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Deng Y., Bartosovic M., Ma S., Zhang D., Kukanja P., Xiao Y., Su G., Liu Y., Qin X., Rosoklija G.B., Dwork A.J., Mann J.J., Xu M.L., Halene S., Craft J.E., Leong K.W., Boldrini M., Castelo-Branco G., Fan R. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature. 2022;609:375–383. doi: 10.1038/s41586-022-05094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thrupp N., Sala Frigerio C., Wolfs L., Skene N.G., Fattorelli N., Poovathingal S., Fourne Y., Matthews P.M., Theys T., Mancuso R., de Strooper B., Fiers M. Single-nucleus RNA-Seq is not suitable for detection of microglial activation genes in humans. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Elishaev M., Hodonsky C.J., Ghosh S.K.B., Finn A.V., von Scheidt M., Wang Y. Opportunities and challenges in understanding atherosclerosis by human biospecimen studies. Front Cardiovasc Med. 2022;9 doi: 10.3389/fcvm.2022.948492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ma W.F., Turner A.W., Gancayco C., Wong D., Song Y., Mosquera J.V., Auguste G., Hodonsky C.J., Prabhakar A., Ekiz H.A., van der Laan S.W., Miller C.L. PlaqView 2.0: a comprehensive web portal for cardiovascular single-cell genomics. Front Cardiovasc Med. 2022;9 doi: 10.3389/fcvm.2022.969421. [DOI] [PMC free article] [PubMed] [Google Scholar]