Abstract
The perception of light signals by the phytochrome family of photoreceptors has a crucial influence on almost all aspects of growth and development throughout a plant’s life cycle. The holistic regulatory networks orchestrated by phytochromes, including conformational switching, subcellular localization, direct protein-protein interactions, transcriptional and posttranscriptional regulations, and translational and posttranslational controls to promote photomorphogenesis, are highly coordinated and regulated at multiple levels. During the past decade, advances using innovative approaches have substantially broadened our understanding of the sophisticated mechanisms underlying the phytochrome-mediated light signaling pathways. This review discusses and summarizes these discoveries of the role of the modular structure of phytochromes, phytochrome-interacting proteins, and their functions; the reciprocal modulation of both positive and negative regulators in phytochrome signaling; the regulatory roles of phytochromes in transcriptional activities, alternative splicing, and translational regulation; and the kinases and E3 ligases that modulate PHYTOCHROME INTERACTING FACTORS to optimize photomorphogenesis.
Keywords: phytochrome, PHYTOCHROME INTERACTING FACTOR, PIF, light signaling, E3 ligase, kinase, light-regulated alternative splicing
1. INTRODUCTION
Light affects almost every major developmental stage of plants, from germination through flowering, and plays a particularly prominent role during seedling establishment. To respond to light, plants require sophisticated sensing of light’s intensity, duration, and wavelength. The perception of light is mediated by a group of photoreceptors that convert information contained in external light to biological signals. Plants possess phototropins and cryptochromes (CRYs) to perceive blue (B) and UV-A light, UV resistant locus 8 to perceive UV-B light, and phytochromes to perceive red/far-red (R/FR) light signals (24, 97). Phytochromes are encoded by five different genes (PHYA–PHYE) in Arabidopsis (75, 110) and are responsible for regulating various light-dependent responses, including seed germination, seedling photomorphogenesis, shade avoidance, and flowering time (97). Among the five phytochromes in Arabidopsis, phyA and phyB are the most well understood. The photoreceptor phyA is classified as a type I photolabile phytochrome (33) and plays a major role in seedling development during the transition from dark to light and under shade conditions (114). By contrast, phyB-phyE are classified as type II phytochromes and are relatively stable under prolonged light exposure. Among the type II phytochromes, phyB, with overlapping functions from phyC-phyE, plays a prominent role in light-grown plants (54, 131). Phytochromes exist in two spectral forms: the inactive Pr state, which photoconverts to the active Pfr state upon R light absorption. Pfr is inactivated upon FR light absorption or through a process of temperature-dependent thermal relaxation called thermal reversion. This allows phytochromes to act as a switch that is turned on by R light and turned off by FR light.
Upon light perception, active phytochromes together with other photoreceptors orchestrate multiple signaling pathways to optimize plant light responses. Seedlings germinated under dark conditions undergo skotomorphogenesis and display closed cotyledons, apical hooks, and elongated hypocotyls. Once exposed to light, seedlings proceed with photomorphogenesis, or de-etiolation, characterized by termination of rapid elongation of hypocotyls, expanded green cotyledons, and initiation of true leaves. It is a pivotal developmental switch that allows the seedlings to undergo anatomical changes for optimal photosynthetic activity and vegetative growth. In environments with high plant density, where the R/FR ratio drops substantially, reduced phytochrome activity also triggers the shade-avoidance response, by which plant anatomy changes drastically (17, 18, 139). The different expression levels, protein structure, protein stability, subcellular localization, or a combination thereof of phytochromes contribute to the light-regulated responses and developmental transitions (1, 43, 69, 109, 113). Light-induced changes in all these properties enable phytochromes to interact with their downstream signaling partners, including transcription factors (TFs) and enzymes such as kinases, phosphatases, and E3 ligases that regulate phytochromes, resulting in large-scale transcriptional reprograming (23, 73, 104). Phytochromes also regulate chromatin remodeling, precursor messenger RNA (pre-mRNA) splicing, and translation through mechanisms that are not yet completely understood.
Another important physiological process tightly controlled by phytochromes is the circadian clock. Phytochromes introduce the R/FR and other light-quality signals to entrain the circadian clock, which greatly affects diurnal metabolic changes, growth, and flowering time (44, 128). Moreover, phytochromes regulate temperature responses, as high ambient temperature promotes morphological changes similar to those of the shade-avoidance responses (e.g., stem and petiole elongation) (19). phyB also functions as a thermosensor and governs temperature responses such as thermomorphogenesis in response to the surrounding ambient temperature (19). However, owing to space constraints, we summarize in this article the present knowledge of phytochrome signaling in response to light, emphasizing work performed mostly with the model plant Arabidopsis.
2. REGULATORY MACHINERY OF PHYTOCHROMES
2.1. Modular Structures of Phytochromes
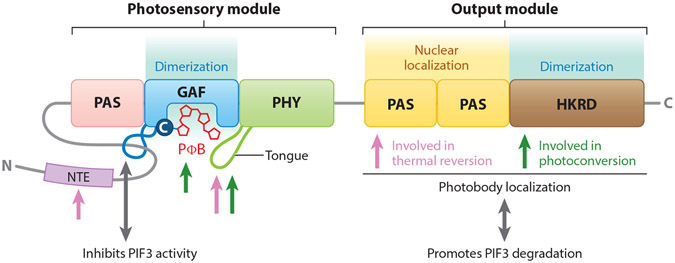
Phytochromes are dimeric chromoproteins in which the two apoproteins are covalently bound to phytochromobilin, a linear tetrapyrrole chromophore, forming a holoprotein. Upon exposure to R light, the cytosolic Pr form of phytochromes undergoes a reversible allosteric conformational change to the active Pfr form, which then translocates into the nucleus (69). Over the past few decades, studies involving domain mapping and mutational analyses have deciphered the structure and functional domains of phytochromes. Together, studies of the crystal structure of the phyB N-terminal photosensory module (PSM), solid-state nuclear magnetic resonance analyses of the Avena sativa phyA PSM in the Pr and Pfr forms, and the studies of structures of bacterial and cyanobacterial phytochromes have revealed presumptive structural information about higher-plant phytochromes (12, 13, 88, 129). The monomeric apoproteins of phytochromes harbor the PSM, which binds to the bilin, and a C-terminal output module (OPM), which mediates dimerization and signal transmission to the downstream effectors (4, 13, 110). The PSM sequentially consists of a variable N-terminal extension that differs among plant phytochrome isoforms; a Period/Arnt/Single-Minded (PAS) domain; a cGMP phosphodiesterase/adenylyl cyclase/FhlA (GAF) domain, which binds to bilin; and a phytochrome-specific (PHY) domain. The OPM includes two tandem PAS domains termed PRD (PAS-repeat domain) and a histidine kinase-related domain (HKRD) (13) (Figure 1). Using the intrinsic lyase activity in the GAF domain, the phytochrome apoprotein covalently attaches a chromophore to a conserved cysteine (Cys357 in AtphyB) in the GAF domain (13, 88). Various nuclear magnetic resonance analyses and spectroscopic studies suggested that light perception triggers a Z-to-E isomerization at the C15-C16 double bond of phytochromobilin, which results in a series of conformational changes in the protein (13, 88). Structural studies of the PAS, GAF, and PHY domains of bacteriophytochromes (42, 134, 157) and the Arabidopsis PHYB proposed that the hairpin loop protrusion or tongue in the PHY domain, which is close to the bilin-binding pocket of GAF, undergoes a β sheet-to-α helix transition during the Pr-to-Pfr photoactivation. Moreover, this structural conversion also affects the light-sensing knot lasso, which is a figure-eight knot between the PAS and GAF domains that pulls them together and then tugs on the PHY domain (13, 42, 137, 157). This ultimately leads to an effective toggle in the position/activity of the sister OPMs (3, 7, 14, 135), thus resulting in differential interactions with other proteins necessary for nuclear localization and interactions with several nuclear proteins, including TFs of the PHYTOCHROME INTERACTING FACTOR (PIF) family and ubiquitin E3 ligase complexes (64, 71, 94, 99, 104, 106, 108).
Figure 1.
Structural domains of phytochromes and their roles in perception of environmental signals and downstream signaling. Light sensing is mediated by the N-terminal photosensory module of phytochrome through the bilin chromophore in the GAF domain and subsequent conformational changes of the tongue. The knot lasso of the N-terminal module interacts with PIFs upon photoactivation and induces light signaling by repressing the transcriptional activity of PIFs. Both the GAF domain and HKRD are responsible for dimerization between each monomer. The C-terminal output module directly interacts with PIF and mediates its degradation. The PRD mediates the nuclear accumulation of phytochrome, and the entire output module is required for photobody localization. Pink arrows indicate domains involved in thermal reversion, and green arrows indicate domains involved in photoconversion. Adapted with permission from Reference 13. Abbreviations: NTE, N-terminal extension; PΦB, phytochromobilin.
Because Pr/Pfr interconversion is critical to phytochrome signal transduction, the multiple features both proximal and distal to the bilin are crucial for the plant phytochromes to perceive and transduce both light and temperature signals. In the PSM, the N-terminal extension controls phytochrome thermal reversion, and the phosphorylation of the N-terminal extension negatively modulates this reaction (31, 92). Although the function of the N-terminal extension is yet to be elucidated, the sequence variations seem to contribute to the different types of phytochromes. Several residues from the GAF and PHY domains also control Pfr stability and thermal relaxation (11, 13, 72). Within the GAF domain pocket, several amino acids are essential for the formation of Pfr. Substitutions of key bilin-protein contacts either block the Pr-to-Pfr transition or result in an altered thermal reversion rate by up to a millionfold, without significant effect on absorption or photoconversion. The knot lasso interacts directly with PIFs, and this interaction is considered a trigger for PIF3 degradation and sequestration (2, 64, 126). The PHY domain also contains key determinants distinguishing phyA from phyB, which may relate to different rates of thermal relaxation (85, 94, 110).
Many bacterial and cyanobacterial phytochromes have a histidine kinase domain in their OPM and are bona fide histidine kinase sensors, whereas plant phytochromes have a HKRD but show serine/threonine kinase activity (110, 127). The OPM of phyB forms a dimer (86, 137) and the HKRD in both phyA and phyB was proposed as a dimerization domain (30, 32, 40, 137). Besides dimerization, the OPM plays important roles in nuclear import and photobody localization (26, 27, 86). Photobodies are poorly understood subnuclear structures hypothesized to be the storage sites that stabilize the active form of phytochromes (16). The PRD mediates phyB’s nuclear localization, and the entire OPM is required for photobody formation (27, 86). The D1040V mutation in phyB HKRD is sufficient to abrogate the dimerization of HKRD and thus attenuates the early signaling functions of phyB in nuclear accumulation and photobody localization. Moreover, it is the OPM, not the PSM, that plays a direct and essential signaling-output role in interaction with PIF3 to mediate PIF3 degradation (108). In addition to regulating PIF3, the activity of the OPM domain was shown in 2015 to be posttranslationally regulated by SUMOylation, limiting the ability of active phyB to interact with downstream signaling targets, thereby limiting light responses (112). While the OPM domain modulates Pfr phytochrome levels and the HKRD promotes dimerization, nuclear import, and photobody localization, the PRD domain promotes thermal reversion (11). For a comprehensive review of phytochrome structure and thermal reversion, please refer to References 13 and 68.
2.2. Regulation of Phytochrome Signaling by Interacting Proteins
Because phytochromes do not possess any biochemical activities other than serine/threonine kinase activity, their downstream signaling cascades are believed to be regulated by interacting partner proteins. Here, we summarize most of the phytochrome signaling components that physically or genetically interact with phytochromes (Table 1). Most of these proteins are either positive or negative regulators, although a few of them play both positive and negative roles in phytochrome signaling pathways.
Table 1.
Summary of phytochrome signaling components and their biological functions in Arabidopsis
| Component | Positive regulators | Biological function(s) |
|---|---|---|
| Photoreceptors | phyA, phyB, phyC, phyD, phyE, cry1, cry2 | Light signaling, thermomorphogenesis, flowering time, circadian clock, hormone response |
| Coregulators | PCH1, PCHL, HEMERA, RCB, NCP, FHY1, FHY3, FHL | Light signaling |
| Transcription factors | HY5, ELF3, ELF4, HFR1, FHY3, FAR1, FRS, TZP, LAF1, BBX4, BBX20, BBX21, BBX22, HECs, PAR1 | Light signaling, flowering time, circadian clock |
| Splicing factors | RRC1, SFPS | Light signaling |
| Phosphatases | PP5.2, FyPP, PP2A2C, PAPP5 | Light signaling |
| Chromatin remodeler | HDA15 | Light signaling |
| Component | Negative regulators | Biological function(s) |
| Ligases | COP1, HOS1, SPAs, LRB, BOPs, EBF1, EBF2, CTG10 | Light signaling, thermomorphogenesis, hormone response |
| Kinases | SPAs, PPKs, PKS1, CK2, BIN2, MPK6 | Light signaling, hormone response |
| Transcription factors | PIF1, PIF3, PIF4, PIF5, PIF6, PIF7, PIF8, MYB30, BBX24, BBX25, EIN3, EIL1, HLS1 | Light signaling, thermomorphogenesis, flowering time, circadian clock, hormone response |
| RNA association factor | PNT1 | Light signaling |
| Chromatin remodelers | PKL | Light signaling |
| Component | Dual-function regulators | Biological function(s) |
| Ligases | COP1, SPAs | Light signaling |
These components interact genetically or physically with phytochromes. Positive regulators, negative regulators, and regulators with dual functions are shown.
The activity of phytochromes greatly relies on their localization to the nucleus (57, 86). The partitioning of phyA and phyB-phyE to the nucleus is controlled by different mechanisms. Nuclear import of phyA depends on the interactions with nuclear localization signal–containing proteins, FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and FHY1-LIKE (FHL). FHY1 and FHL interact with the Pfr form of phyA in the cytoplasm and rapidly shuttle it to the nucleus to initiate downstream signaling (45, 51). Once in the nucleus, phyA undergoes another cycle of photoactivation to initiate light responses and then is degraded in a light-dependent manner (109). Although the absorption and the ratio of active phyA to total phyA are maximal under R light, phyA is physiologically more active under FR light (109). Using mathematical modeling and experimental validation, Rausenberger et al. (109) showed that the dissociation rate of the phyA-FHY1/FHL nuclear import complex is the principal determinant of the phyA action peak, which means phyA’s activity relies on specific molecular interactions rather than on intrinsic changes to phyA’s spectral properties. The import and export of FHY1 also depend on its phosphorylation state. Under R light, phyA mediates rapid phosphorylation of FHY1 and might thus reduce its interaction and nuclear transport, suggesting a fine-tuning mechanism in response to R light (117). In contrast to the nuclear localization of phyA, the nuclear localization of phyB-phyE is proposed to be mediated through a nuclear localization signal present within the C terminus of the protein. For phyB, the nuclear localization signal within the PRD is masked by interactions with the GAF and PHY domains and unmasked by the light-dependent conformational changes (27). Although phyB nuclear localization is not mediated by FHY1 and FHL, phyB might still translocate to the nucleus through physical interactions with PIFs (102) and possibly other nuclear localization signal-containing phyB-interacting proteins. phyC-phyE are thought to enter the nucleus through similar dynamics (1). Phytochrome activity is also regulated by PHYTOCHROME ASSOCIATED PHOSPHATASE 5 (PAPP5). PAPP5 interacts with both phyA and phyB and dephosphorylates their Pfr form (111). The dephosphorylation increases the stability of the Pfr by preventing phyA and phyB from interacting with the E3 ligases and subsequent degradation.
2.3. The Role of Photobodies
An important feature of phytochrome signaling is the ability of Pfr to aggregate into discrete subnuclear foci known as photobodies (67, 152), where phytochromes and downstream signaling components are colocalized (23). The formation of phyB photobodies is directly regulated by light quality and quantity and is positively correlated with phyB-mediated responses, such as the degradation of phyB targets, inhibition of PIF transcriptional activity, and phenotypes related to photomorphogenesis (2, 5, 16, 136). The assembly and stability of phyB photobodies are controlled by at least five proteins: PHOTOPERIODIC CONTROL OF HYPOCOTYL1 (PCH1), PCH1-LIKE (PCHL), HEMERA (HMR), NUCLEAR CONTROL OF PEP ACTIVITY (NCP), and REGULATOR OF CHLOROPLAST BIOGENESIS (RCB) (25, 41, 55, 56, 155, 159). Mutations in any of these genes affect photobody size and number. PCH1, which preferentially binds to the Pfr form of phyB, promotes phyB photobody accumulation and prevents phyB thermal reversion in vivo. It is required for regulating multiple phyB-controlled physiological processes, including seed germination, hypocotyl gravitropism, chlorophyll biosynthesis, and thermomorphogenesis (28, 55). PCH1 and PCHL physically interact with PIF1 and negatively regulate PIF1 abundance, DNA-binding ability, and transcriptional activity. Moreover, PCH1 and PCHL are posttranslationally regulated by CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and undergo degradation through the 26S proteasome pathway in the dark (28, 41, 55). HMR was identified based on phyB-GFP (green fluorescent protein) mislocalization screening (25). In hmr mutants phyB-GFP fails to localize to large photobodies. In addition, the degradation of PIF1 and PIF3 is significantly impaired in hmr mutants. Photoactivated phytochromes through their photosensory domain physically interact with HMR and promote HMR accumulation, which then promotes the formation of photobodies as well as the degradation of PIFs to establish photomorphogenesis. Furthermore, HMR promotes PIF4-dependent induction of temperature-responsive genes andPIF4 accumulation. Like HMR, NCP and RCB are dual-targeted nuclear/plastidial proteins. They also promote photobody formation and the degradation of PIF1 and PIF3 by direct phytochrome interaction (25, 155, 159). HMR, NCP, and RCB control both plastid-encoded RNA polymerase assembly and chloroplast gene expression primarily from the nucleus by promoting PIF-mediated phytochrome signaling, revealing a distinct nucleus-to-plastid signaling pathway adopted from phytochrome signaling (25, 155, 159).
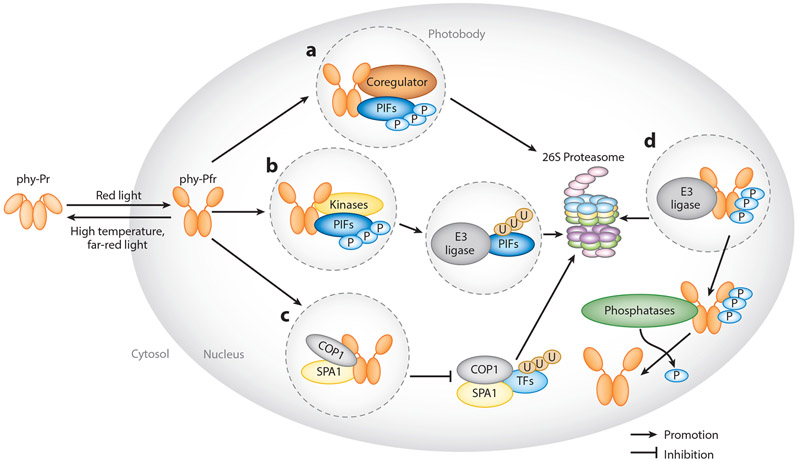
The exact functions of the photobodies in phytochrome signaling are not yet fully understood. So far, most observations have focused on the formation of phyB photobodies, and they are thought to play multiple roles depending on the functions of phytochrome-interacting proteins (Figure 2). First, multiple coregulators (or signaling components), such as PCH1, PCHL, HMR, NCP, and RCB, interact with phyB in the photobodies, where they act as storage sites of phyB-Pfr that preserve or stabilize phyB-Pfr from converting back to the Pr state (16, 41, 55) (Figure 2a). Second, PIFs such as PIF1 and PIF3 interact with phyA and phyB upon light perception and colocalize to photobodies, where they undergo phosphorylation and subsequent ubiquitination and degradation (2, 5, 116) (Figure 2b). Photobodies here are important sites for biochemical reactions required for phytochrome signal transduction. Third, negative regulators of phytochrome signaling, SUPPRESSOR OF phyA-105 1 (SPA1) and CONSTITUTIVE PHOTOMORPHOGENIC1/DEETIOLATED/FUSCA (COP1/DET/FUS) E3 ligase complex, interact with phyA and phyB in response to light within photobodies (81, 115) (Figure 2c). This interaction prevents the formation of the COP1/SPA1 complex and thereby stabilizes the transcriptional regulators that promote light signaling (52). Here, photobodies act as sites to sequester proteins to inhibit their E3 ligase activity. Photobodies are also sites of gene regulation and possibly sites for phytochromes phosphorylation and ubiquitination (Figure 2d), which is discussed in more detail in Section 3.
Figure 2.
Models of phytochrome signaling pathways regulated by their interacting proteins. (a) Phytochrome signaling proteins regulate the size and stability of photobodies. Formation of these photobodies (shown inside dashed circles) are promoted independently by coregulators such as PCH1, PCHL, HMR, NCP, and RCB. (b) Photobodies are also sites for the degradation of PIFs. phyB,PIFs, and kinases, such as PPKs and SPAs, colocalize within the photobodies, resulting in the phosphorylation of PIFs. PIFs are subsequently ubiquitinated and degraded. (c) phyA and phyB colocalize with SPA1 within the photobodies to sequester SPA1 from COP1, suppressing COP1 activity. (d) Phosphorylated phytochromes are also targets of the COP1/SPA1 complex and the E3 ligase LRB. Phytochromes are subsequently ubiquitinated and degraded. Phosphorylated phytochromes can be dephosphorylated by phosphatases, such as FyPP and PAPPs. Abbreviations: FR, far red; FyPP, FLOWER-SPECIFIC PHYTOCHROME-ASSOCIATED PROTEIN PHOSPHATASE; phy, phytochrome; R, red; TF, transcription factor.
Hahm et al. (48) reported in 2020 that high ambient temperature disassembles phyB from individual photobodies with the least thermostability. The thermostability of photobodies relies on phyB’s PSM. Surprisingly, phyB has opposite effects in the hypocotyl and cotyledon tissues in response to rising temperature despite inducing similar photobody dynamics, indicating that tissue-or organ-specific temperature signaling regulation is either downstream of photobody dynamics or independent of phyB (48). This study provides cellular evidence of photobody dynamics but also suggests its distinct tissue-specific regulation.
3. POSTTRANSLATIONAL REGULATION BY PHYTOCHROMES
3.1. Reciprocal Modulation of Positive and Negative Regulators Under Dark and Light Conditions
Phytochromes adopt posttranslational protein modifications as a central tactic to initiate downstream signaling cascades upon light activation. Reversible posttranslational protein modifications are considered rapid and dynamic responses, which is why phytochromes can modulate activities of target proteins with greater plasticity. Therefore, many protein-modifying enzymes, such as kinases, phosphatases, and E3 ligases, play a pivotal role in phytochrome-mediated responses. In this section, we discuss how phytochromes modulate the activities of these enzymes to promote photomorphogenesis.
3.1.1. Phytochrome-mediated inhibition of COP1 activity.
Within the highly sophisticated strategies employed by plant phytochromes sits a well-studied ubiquitin E3 ligase, COP1. Although COP1 functions as a central repressor of phytochrome signaling by destabilizing multiple positively acting TFs in the dark, phytochromes also counteract COP1 activity to release the repression by the COP1 E3 ligase activity (49, 150). COP1 is retained in the nucleus and forms functional complexes with the SPA1–SPA4 protein family in the dark (162). The COP1/SPA E3 ligase complex actively ubiquitinates TFs such as ELONGATED HYPOCOTYL 5 (HY5), HY5 homolog (HYH), Long Hypocotyl in Far-Red 1 (HFR1), HECATEs (HECs), and B-BOX zinc-finger protein family (BBX), among many others, and mediates their degradation through the 26S proteasome pathway (49, 63, 77, 130, 150). The light-activated phytochromes, however, rapidly enter the nucleus and inactivate COP1 activity by dual mechanisms. As mentioned in Section 2.3 (Figure 2), at the early stage, phytochromes directly interact with the COP1/SPA complex and reorganize the protein complex to inactivate E3 ligase activity. The exact stoichiometry of the re-organization has not been elucidated, but it is suggested that COP1 is no longer in direct contact with the SPA proteins, resulting in reduction of the activity Under prolonged light exposure, phytochromes induce exclusion of COP1 from the nucleus, thus stabilizing multiple COP1-targeting TFs that promote photomorphogenesis (95, 132).
3.1.2. Nontranscriptional roles of PIFs as cofactors of E3 ligase.
In addition to the COP1/SPA E3 ligase complex, the basic helix-loop-helix (bHLH) PIFs compose another key class of negatively acting TFs in the phytochrome signaling cascade. PIFs interact directly with the Pfr form of phytochromes and function as central repressors of phytochrome signaling. PIFs can activate or repress hundreds of their downstream target genes, fulfilling their function as transcriptional regulators. However, they also have nontranscriptional roles as a cofactor of COP1 E3 ligase. The COP1/SPA complex and PIFs function synergistically to repress photomorphogenesis (150). Genetic analysis showed that a cop1 pif1 double mutant or a spa1 spa2 spa3 pif1 quadruple mutant displayed stronger constitutive photomorphogenic phenotypes compared with their respective parents (149). Moreover, by directly interacting with PIFs, COP1 exhibits stronger substrate recognition and ubiquitination activity (63, 148, 149). Conversely, COP1 is also required for PIF stability in the dark in a HFR1-dependent manner, suggesting an additional unknown connection between PIFs and COP1 (38, 78, 103, 105, 121). Therefore, two major classes of negatively acting factors in phytochrome signaling, PIF TFs and the E3 ligase, orchestrate to inhibit phytochrome-mediated gene expression, while phytochromes negatively regulate their activities to promote photomorphogenesis.
3.2. Kinases in Phytochrome Signaling Pathways
One of the earliest posttranslational modifications triggered by phytochromes is phosphorylation. The activated phytochromes are indispensable for the rapid phosphorylation and degradation of PIFs through the 26S proteasome pathway (Figure 2b). One of the leading candidate kinases is the phytochrome itself, as it possesses a HKRD at its C terminus (127). Earlier studies have shown serine/threonine kinase activity for phyA purified from A. sativa (50, 158). More recently, Arabidopsis phyA and phyB purified from Pichia pastoris and Saccharomyces cerevisiae also displayed serine/threonine kinase activity (50, 96, 127). A more in-depth mutational analysis also identified residues necessary for kinase activity and biological function of A. sativa phyA (127). However, these residues are located at the N terminus (PAS, GAP, and PHY domains), which has no homology to any kinase domain as opposed to the HKRD present at the C terminus of phytochromes, raising concerns that these mutations might affect the structural integrity of A. sativa phyA as opposed to a specific kinase active site. Thus, phytochrome kinase activity remains debatable.
In the absence of conclusive evidence for phytochrome kinase activity, different laboratories have embarked on identifying other kinases using various genetics- and proteomics-based approaches. These searches have identified multiple kinases that function in the early phytochrome-mediated phosphorylation of PIFs. Photoregulatory Protein Kinases (PPK1–PPK4), which were previously known as MUT9-Like Kinases (MLKs), phosphorylate PIF3 in vitro (89). In ppk1 ppk2 ppk3 or ppk1 ppk2 ppk4 triple mutants, phytochrome-induced PIF3 phosphorylation was still observed, albeit at a significantly lower level. However, ppk1 ppk2 ppk3 ppk4 quadruple mutants pre-sumably showed lethality, suggesting a more general function of PPKs in early developmental processes. ppk mutants displayed hypersensitive phenotypes under R light as opposed to hyposensitive phenotypes expected for PIF kinases. This is due to a prominent role of PPKs in controlling phyB-PIF3 co-degradation, resulting in higher levels of phyB under R light in ppk mutants, causing hypersensitive phenotypes. SPA1, which was previously known to be a COP1 E3 ligase cofactor, possesses serine/threonine kinase activity toward PIF1 and PIF4 (70, 96). A null mutation in four SPA family genes (SPA1–SPA4) resulted in a lack of light-induced phosphorylation of PIF1 in vivo, suggesting the importance of SPAs in early light-induced phosphorylation of PIF1. However, SPA1 along with phyB failed to demonstrate light-induced phosphorylation of PIF1 in vitro, suggesting additional kinase(s) or factors in this response may be required as previously hypothesized (9).
In addition to PPKs and SPAs, other protein kinases, including BIN2, CK2, and mitogen-activated protein kinase (MAPK) MPK6, have been implicated in PIF phosphorylation and subsequent UPS-mediated degradation (6, 10, 78, 146). BIN2, a key component of the brassinosteroid (BR) signaling pathway, can phosphorylate and mediate the degradation of PIF4. However, this is probably specific not to phytochrome-mediated PIF regulation but rather to the light-BR signaling crosstalk, which is important for regulating plant growth (6). Whether BIN2 activity is under the regulation of phytochromes is unclear, as no direct phytochrome-BIN2 interaction was reported. CK2 phosphorylates at least seven serine/threonine residues present in PIF1 in vitro (10). However, PIF1 was still phosphorylated in response to light, suggesting that CK2 is probably not a light-regulated kinase. MPK6 also directly phosphoryates PIF3 and controls its abundance to regulate cotyledon opening in response to R light (146). Unlike BIN2 and CK2, MPK6 activity is stimulated by R light through MKK10, a MAP kinase kinase (MAPKK). Thus, the MAPK pathway regulates early steps in photomorphogenesis by controlling the level of PIF3.
Overall, the present model involving multiple kinase families may suggest the complexity and importance of the early phytochrome-initiated phosphorylation events. It is also possible that multiple kinases are sequentially phosphorylating specific target residues on PIFs. A specific phosphorylation event may serve as a prime event for subsequent phosphorylation by other kinases, which ultimately might lead to the robust phosphorylations necessary to degrade PIFs. Further studies are needed to clarify the significance of multiple kinases in phytochrome signaling pathways.
3.3. E3 Ligases in Phytochrome-Mediated Light Signaling
E3 ubiquitin ligases play central roles in many plant signaling cascades, including phytochrome signaling pathways. These enzymes covalently attach ubiquitin moieties to the lysine residues on the target protein and the polyubiquitinated proteins undergo degradation mediated by the 26S proteasome. In chronological order, phosphorylation of PIFs leads to rapid polyubiquitination and subsequent degradation (Figure 2b-d). This is partly because of the higher affinity of the phosphorylated PIFs toward the E3 ligases. Although it is well known that multiple serine/threonine phosphorylations in PIFs lead to rapid polyubiquitination, a direct causal relationship between phosphorylation on a specific serine/threonine residue and polyubiquitination on a specific lysine residue is yet to be fully demonstrated.
Similar to the kinases, multiple E3 ligases from all three Arabidopsis CULLIN (CUL) RING UBIQUITIN LIGASE (CRL) families are involved in light-dependent polyubiquitination of PIFs. Light-Response Bric-a-Brac/Tramtrack/Broad 1–3 (LRB1-LRB3), which form CUL3LRB complexes, were first discovered in Arabidopsis seedlings by PIF3 affinity purification and mass spectrometry (90). LRBs were originally reported to affect phyB level; however, it was later suggested that LRBs trigger ubiquitination of both the receptor phytochrome and the PIF3 through a mutually assured destruction mechanism (90). A separate study showed that EIN3-BINDING F BOX PROTEIN 1 (EBF1) and EBF2, which form the CUL1EBF1 and CUL1EBF2 complexes, respectively, were involved in the light-induced ubiquitination and degradation of PIF3 (36). These studies suggested that the EBFs more specifically target PIF3 for ubiquitination under a wide range of light intensities, and the LRBs are responsible for ubiquitination of both phyB and PIF3 under conditions of high light intensity (Figure 5a). Several hormone signaling mediators, such as BIN2 and EBFs, are also involved in phytochrome-induced PIF posttranslational modifications, suggesting an intricate connection between light and hormone signaling cascades.
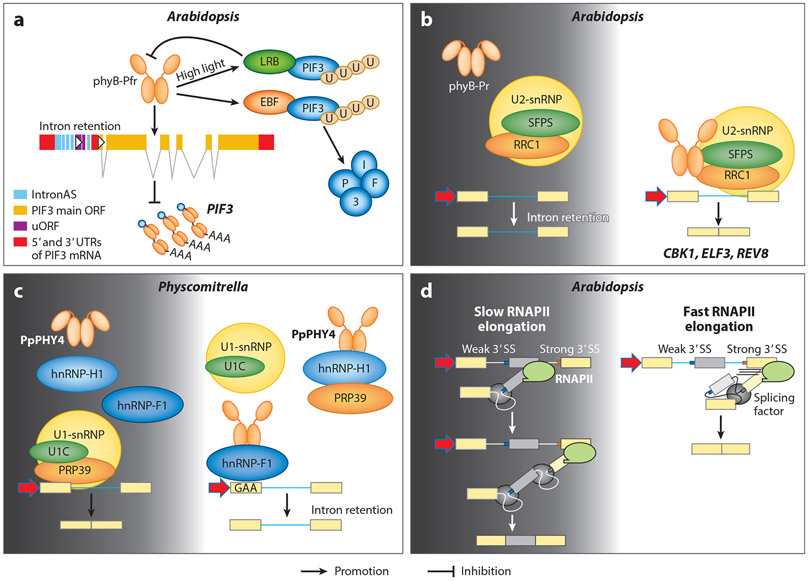
Figure 5.
Present models of light-regulated alternative splicing of pre-mRNAs. Dark conditions are shown by a dark gray background, and light conditions are shown by a white background. (a) phyB can induce alternative splicing in light-grown plants that results in the retention of intronAS within the UTR of PIF3 mRNA. The uORFs inside the intronAS sequence inhibit translation of the PIF3 main ORF. Panel a adapted with permission from Reference 35. (b) SFPS and RRC1 directly interact with phyB and regulate pre-mRNA splicing. (c) In Physcomitrella patens, phytochromes directly regulate alternative splicing by interacting with splicing regulators in the spliceosome. Panel c adapted with permission from Reference 123. (d) Light regulates alternative splicing through the control of transcriptional elongation. Plants exposed to light show faster gene transcription than plants in the dark. This serves as a control for alternative mRNA splicing decisions. Panel d adapted with permission from Reference 46. Abbreviations: hnRNP, heterogenous nuclear ribonucleoprotein; intronAS, intron alternative splicing; phy, phytochrome; pre-mRNA, precursor messenger RNA; RNAPII, RNA polymerase II; SS, splice site; snRNP, small nuclear ribonucleoprotein; uORF, upstream open reading frame; UTR, untranslated region.
As discussed above, another key E3 ligase in early phytochrome signaling is the CUL4COP1/SPA complex. The COP1/SPA complex functions as a central repressor of photomorphogenesis in the dark; however, it can also trigger rapid degradation of PIF1 and PIF5 in response to light (103, 163). PIF1 is a master negative regulator of phytochrome-induced seed germination. Thus, in the presence of light or active phytochromes, the COP1/SPA complex can act as a positive regulator of phytochrome signaling through the ubiquitination and degradation of PIF1 and PIF5. In addition, the COP1/SPA complex and PIFs function synergistically to repress photomorphogenesis in the dark. However, during the dark-to-light transition, the COP1/SPA complex induces degradation of its cofactor PIFs. The exact molecular basis and kinetics for the dual function of the COP1/SPA complex are not known. However, phytochromes may act as a molecular switch to turn the cofactor PIF1 into a substrate of COP1 in the presence of light. This might lead to a gradual derepression of photomorphogenesis under light conditions.
Three additional E3 ligases directly interact with PIFs. An F-box protein called COLD TEMPERATURE-GERMINATING 10 (CTG10), which forms the CUL1CTG10 complex, regulates seed germination by controlling the level of PIF1 (83). BLADE-ON-PETIOLE (BOP), originally identified as a CUL3-based E3 ligase (CUL3BOP) that regulates leaf and flower development, ubiquitinates PIF4 (160). In addition, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1) also directly interacts with PIF4. HOS1 only inhibits PIF4 transcriptional activity but cannot trigger polyubiquitination and degradation (65). Further studies are needed to decipher whether BOPs and HOS1 are acting specifically in phytochrome signaling pathways or are involved in crosstalk between light and other signaling pathways.
4. FINE-TUNING GENE EXPRESSION BY PHYTOCHROMES
4.1. Phytochrome-Mediated Transcriptional Regulation
Light signals stimulate large-scale transcriptional reprogramming during developmental transitions (such as photomorphogenesis and flowering) through the action of photoreceptors, TFs, and signaling components. Many reviews have summarized and discussed light-regulated transcription (59, 71, 73, 143). Here, we focus on more recent progress in phytochrome-mediated transcriptional regulation. As shown in Table 1, several families of TFs, including bHLH TFs (PIFs, HFR1, HEC, and PAR1), basic leucine zipper TFs (HY5 and HYH), transposon-derived novel TFs [FAR-RED ELONGATED HYPOCOTYL3 (FHY3), FAR-RED IMPAIRED RESPONSE1 (FAR1), and FAR1-Related Sequence (FRS)], myb domain–containing TFs (MYB30 and LAF1), B-BOX-containing TFs (BBX4, BBX20, BBX21, BBX22, BBX24, and BBX25), tandem zinc finger PLUS3 (TZP), and plant-specihc ETHYLENE-INSENSITIVE 3 (EIN3) and EIN3-LIKE 1 (EIL1), play crucial roles in light-regulated plant development. Within the same family, many of these TFs are in a negative feedback regulatory loop. For example, PIFs promote the expression of HFR1, HECs, and PAR1, whereas these atypical HLHs sequester PIFs from DNA binding, thereby inhibiting their transcriptional activities (53, 104, 122, 164). BBX proteins also interact with each and regulate each other’s activities (130). TFs of different families also interact and regulate each other’s activities. For example, BBX20, BBX21, and BBX22 interact with HY5 and act as crucial coregulators for HY5 transcriptional activity (15). HMR and ELF3 interact with PIFs and act as transcriptional coregulators without directly binding to DNA (91, 107), while ELF4 acts in a complex with ELF3 and LUX (93). Moreover, three families of TFs (PIFs, HY5, and EIN3) reprogram transcription during the dark-to-light transition at the seedling stage (120). Light-regulated and tissue-specific expression of microproteins (miP1a and miP1b) regulates PIF3 and EIN3 activities to regulate photomorphogenesis (142). In addition, Small Auxin Up RNAs (SAURs) and LAZY genes exhibit specific temporal-spatial expression patterns, which are regulated by multiple TFs, including TCP4, PIFs, HY5, AUXIN RESPONSE FACTOR 7 (ARF7), and ARF19. The asymmetric expression of SAUR and LAZY genes promotes the gravitropism and phototropism of plant hypocotyls and roots (37, 133, 138, 156). Oligomerization of HOOKLESS 1 (HLS1) also plays a role in differential cell growth. Light-activated phyB interacts with HLS1, disrupting the self-association of HLS1 to initiate hook unfolding (82). Thus, tissue-specific transcriptional reprogramming is crucial for the differential effects of light on different organs.
Many of the TFs colocalize with phytochromes and other signaling components to the nuclei, especially in the photobodies, suggesting active modulation for transcriptomic adjustment, including the modification of chromatin structures and changes in transcriptional activities. Phytochromes regulate downstream TFs and chromatin remodeling through multiple mechanisms, as shown in Figure 3. First, phyB interacts with TFs such as PIFs and HLS1 to sequester or block their DNA-binding activity to inhibit expression of downstream genes (82, 100) (Figure 3a). By contrast, phytochromes also interact with SPAs and disrupt the direct interaction between SPA1 and COP1, as mentioned in Figure 2c. Without the activation of SPA, positively acting TFs such as HY5, HFR1, and LAF1 accumulate and promote photomorphogenesis. As discussed above, PIFs directly interact with COP1 and SPA1 as well as HY5 in the dark (149), promoting the recruitment of HY5 to the COP1/SPA complex and the ubiquitination and degradation of these TFs to repress photomorphogenesis. This reciprocal regulation of PIFs and positively acting TFs is central to the transcriptional control by phytochromes.
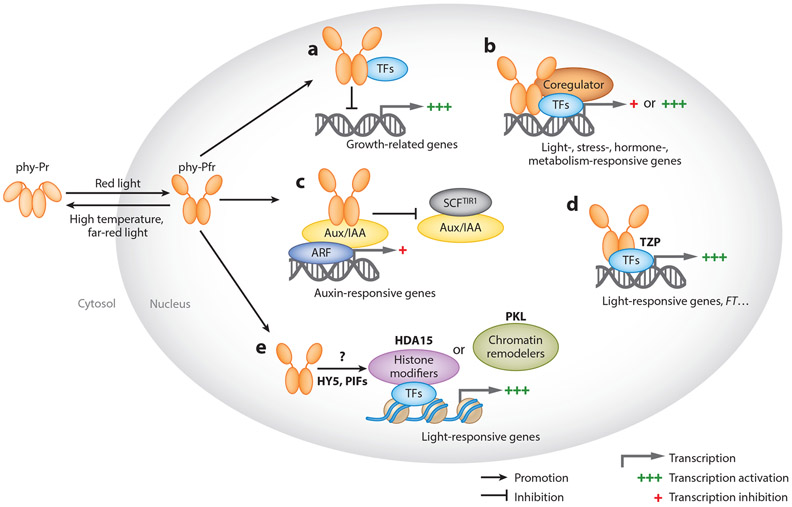
Figure 3.
Present models of phytochrome-mediated transcriptional regulation. (a) Phytochromes interact with transcription factors (e.g., PIFs) to sequester or block their DNA-binding domain to inhibit downstream gene expression. (b) Phytochromes interact with transcription factors such as PIFs and regulate their transcriptional activities with the aid of coregulators. (c) Light triggers interactions between photoreceptors and Aux/IAA, interfering with the auxin-induced degradation of Aux/IAA by the E3 ligase SCFTIR and thereby promoting ARF activity and related auxin signaling. (d) Phytochromes interact with transcriptional activators, such as TZP, and enhance their transcriptional activity. (e) Light-signaling-related transcription factors recruit chromatin-remodeling proteins and histone-modifying factors to regulate light-responsive gene expression. It is not known how photoreceptors regulate this recruitment. The histone-modifying factors establish or read histone marks at the chromatin, whereas the chromatin remodelers alter histone-DNA contacts, leading to a new state, with the binding of light-signaling-related transcription factors occurring at light-responsive elements. Abbreviations: FT, FLOWERING TIME; phy, phytochrome; TF, transcription factor.
Second, phytochromes interact with TFs such as PIFs and regulate their transcriptional activities with the aid of other coregulators (Figure 3b). A central mechanism by which phytochromes rapidly regulate gene expression is to induce the posttranslational modifications and degradation of TFs such as PIFs (74), which has been described in Section 3. Both phyBG111D, which abolishes sequestration activity of PIFs without affecting their degradation activity, and phyB990G767R, which does the opposite, are equally capable of inducing light responses. This finding suggests that phyB requires both sequestration and degradation of PIFs to modulate optimal light responses (99). Direct phytochrome control of TFs for EIN3 and the bHLH protein BRI1-EMS-SUPPRESSOR1 (BES1), a central player in BR signaling, was also reported. phyB interacts with EIN3 and facilitates EIN3-SCFEBF1/EBF2 complex formation and subsequent EIN3 ubiquitination and degradation (119). BES1 activity is also inhibited following interaction with phyB (141). In addition, MYB30 promotes PIF4 and PIF5 protein reaccumulation under prolonged R light irradiation by directly binding to their promoters to induce their expression. MYB30 represses photomorphogenesis by inhibiting PIF-phytochrome interaction and thus PIF degradation (153). Yet another mechanism of phytochromes that regulate TFs is to abolish their DNA-binding abilities or their transcriptional activities by recruiting coregulators such as HMR or PCH1. A 2020 study has shown that the phyB-interacting PCH1 and PCHL could reduce the DNA-binding ability and transcriptional activity of PIF1 (28). HMR, NCP, and RCB also affect transcriptional activity of PIFs in addition to their positive role in PIF degradation (107, 155, 159).
Third, light triggers interactions between phytochromes and AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) proteins, interfering with the auxin-induced degradation of Aux/IAA by E3 ligase SCFTIR1/AFB complexes, thereby repressing ARF activity and related auxin signaling. Several Aux/IAA regulators of auxin-controlled gene expression are modulated through interaction with phyA or phyB (Figure 3c). In these cases, both phyA and phyB could interact with Aux/IAA proteins and protect them from auxin-regulated proteolytic degradation by SCFTIR1/AFB complexes (147, 154). However, phyA and phyB seem to adopt the same mechanism in response to different light conditions to optimize growth. Under normal light conditions, phyB competes with TIR1 to interact with Aux/IAA to inhibit its degradation, thereby repressing ARF activity and auxin signaling to restrict hypocotyl elongation (147). Generally, under shade conditions, dephosphorylated PIF7 promotes auxin biosynthesis and hypocotyl elongation. Under deep shade, however, accumulated phyA also competes with TIR1 to interact with Aux/IAA and prevent its degradation, thus suppressing shade-induced hypocotyl elongation (154). Furthermore, phytochromes interact with a transcriptional activator, such as TZR to enhance or activate their transcriptional activities and thus turn on downstream gene expression. TZP colocalizes to photobodies with phyB under R light to regulate the expression of the flower inducer FLOWERING LOCUS T to promote flowering initiation under long-day conditions (62) (Figure 2d), thus integrating light and photoperiodic signaling. In these cases, phytochromes could independently affect the transcriptional activity of the interacting TFs.
Fourth, light-signaling-related TFs recruit chromatin-remodeling proteins and histone-modifying factors to regulate light-responsive gene expression (Figure 3e). For example, HISTONE DEACETYLASE 15 (HDA15) interacts with PIF3 and PIF1 to suppress the expression of chlorophyll biosynthetic genes and regulates light-initiated seed germination. In addition, PIF1 recruits HDA15 to the chromatin regions (specific light-responsive element sequences) of target genes and represses their expression by decreasing histone acetylation levels, repressing seed germination (47, 80). By contrast, the chromatin remodeler PICKLE (PKL) is recruited to the promoter regions of cell-elongation-related genes by both HY5 and HYH to regulate skotomorphogenesis. PKL represses H3K27 trimethylation levels, thus leading to the expression of these genes and cell elongation (60). Moreover, PKL functions as a key integration node of BR, gibberellin, and light signaling pathways by directly interacting with key signaling components, such as PIF3 and BZR1, that coregulate skotomorphogenesis by repressing the deposition of H3K27 trimethylation on target genes (161). It is not yet known how phytochromes regulate their recruitment; however, phytochromes may play an important role in recruiting these chromatin remodelers or histone modifiers to affect gene expression.
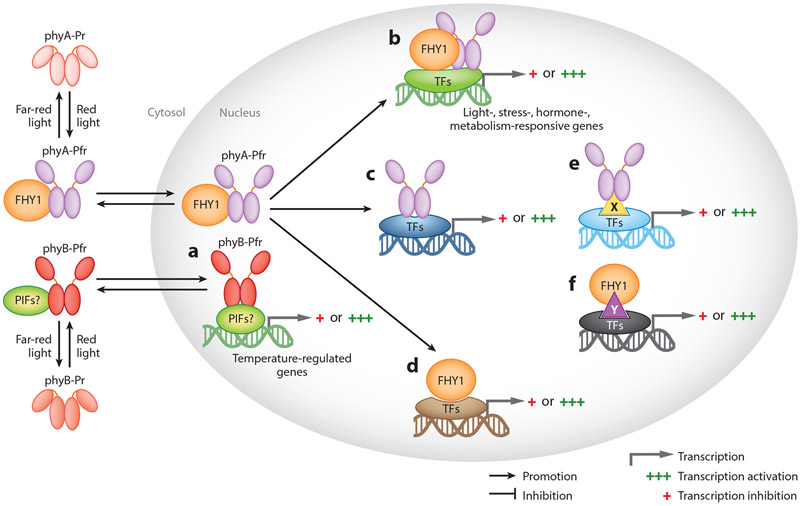
Finally, both phyA and phyB bind to chromatin and directly target specific promoter regions of many genes, as originally hypothesized for phyB-PIF3 (84). As discussed in Section 1, phyB also acts as a temperature sensor and associates with more target gene promoters at low ambient temperature (Figure 4a), where relatively more active phyB is present, than at high ambient temperature, where phyB is converted to an inactive conformer due to a rapid dark reversion (61). Similarly, phyA binds to chromatin in a FHY1-dependent and FHY1-independent manner and modulates gene expression possibly by associating with various TFs (20, 21) (Figure 4b-e). Using chromatin immunoprecipitation sequencing and RNA sequencing (RNA-seq) analyses, Chen et al. (20) identified many genes that are regulated either by both phyA and FHY1 or by phyA or FHY1 alone. As shown in Figure 4, phyA and FHY1 together can bind to different TFs through unique and specific conformations and thus regulate the transcriptional activity of the TFs to their target genes (Figure 4b). In some cases, phyA or FHY1 alone can bind to TFs and regulate their activity (Figure 4c,d). In other cases, phyA or FHY1 can bind to specific TFs with the help of unknown coregulators (Figure 4e,f). Although these models explain how plants rapidly fine-tune their growth upon changes in the light environment by escorting photoreceptors to the promoters of hormone- and/or stress-responsive genes for individualized regulation, FHY1 and FHL are not required for phyA nuclear signaling (87). Thus, further studies are needed to conclude whether phytochromes can bind to chromatin and control gene expression more directly.
Figure 4.
Phytochromes directly associate with chromatin through TFs, coregulators, or both and control gene expression. (a) phyB is enriched to temperature-regulated genes at lower temperature possibly through interaction with PIFs, other TFs, or both. (b) phyA and FHY1 together associate with TFs to regulate their specific binding to different cis elements. (c) phyA and (d) FHY1 interact with TFs to regulate their specific binding to different cis elements independently. (c) phyA and (f) FHY1 interact with TFs and regulate their specific binding with the aid of unknown factors. Different DNA colors indicate different cis elements. X and Y indicate unknown factors that are involved in specific associations. Abbreviations: phy, phytochrome; TF, transcription factor.
4.2. Light-Regulated Posttranscriptional Control (Pre-mRNA Splicing)
A light signal has a profound effect on gene expression not only at the transcriptional level but also at the posttranscriptional RNA processing steps, particularly alternative splicing in plants (29). Moreover, several RNA-seq analyses coupled with genetic and biochemical evidence clearly established that phytochromes play a major role in modulating alternative splicing in plants (35, 76, 123, 125,144, 145). Alternative splicing involves the use of variable splice site selection, generating two or more mRNA isoforms from the same pre-mRNA, and is categorized as exon skipping, intron retention, alternative 5′ donor splice site, or alternative 3′ acceptor splice site. Alternative splicing is executed by the spliceosome machinery, which is a dynamic multi-megadalton ribonuclear protein complex consisting of approximately 200 proteins and 5 small nuclear ribonucleoproteins (U1-snRNP U2-snRNP U4-snRNP U5-snRNP and U6-snRNP). In addition, auxiliary splicing regulatory proteins such as serine/arginine-rich proteins and heterogenous nuclear ribonucleoproteins (hnRNPs) play defining roles in pre-mRNA processing (29). Pre-mRNAs containing at least one intron consist of intron-defining splice site consensus sequences including 5′ splice sites and 3′ splice sites, while most pre-mRNAs destined for alternative splicing also consist of additional auxiliary splicing regulatory cis elements, such as exonic and intronic splicing enhancers and silencers (29). Spliceosome complex assembly on pre-mRNA is guided by splice site consensus sequences, and splicing regulators play a critical role in splice site selection by perceiving and interacting with different splicing regulatory cis elements. Therefore, the final outcome of alternative splicing is invariantly determined by the auxiliary splicing regulators through upstream direct protein-protein and downstream protein-RNA interactions. We summarize some of the phytochrome-mediated alternative splicing events below (Figure 5).
By performing elaborate and systematic RNA-seq analyses, Shikata et al. (124) first identified the extensive roles of R-light-activated phytochromes in the genome-wide regulation of alternative splicing. Subsequent reports ascertained either distinct events or specific factors such as splicing factors and regulators involved in phytochrome-mediated regulation of alternative splicing. Overaccumulation of phyB can selectively induce intron retention in the 5′ untranslated region of PIF3 mRNA and the retained intron inhibits PIF3 protein synthesis (35) (Figure 5a). However, the detailed mechanism of the splicing factors or regulators involved in the process have not yet been identified.
One of the first bona fide phytochrome-interacting pre-mRNA splicing factors identified in Arabidopsis is SPLICING FACTOR FOR PHYTOCHROME SIGNALING (SFPS), a potential ortholog to the Drosophila and human splicing factor 45 (SPF45) (145). SFPS colocalizes and physically interacts with phyB in response to R light, and the sfps mutant seedlings displayed hyposensitive phenotypes specifically under light conditions. Moreover, SFPS colocalizes with U2-snRNP-associated factors and modulates pre-mRNA splicing (both constitutive and alternative splicing) in Arabidopsis (Figure 5b). Although all forms of alternative splicing are defective in sfps, the intron retention events are greatly enriched. Furthermore, the gene ontology analyses identified in sfps various categories of gene ontology including light stimulus, circadian clock, transcription activity, and photosynthesis, indicating that SFPS might preferentially regulate the pre-mRNA splicing of genes involved in light and circadian clock signaling through direct interaction with phyB (145). A follow-up study showed that SFPS interacts with the splicing factor REDUCED RED-LIGHT RESPONSES IN CRY1CRY2 BACKGROUND 1 (RRC1), a serine/arginine-rich-like protein (125, 144). RRC1 also interacts with phyB and U2-snRNP-associated factors in a R-light-dependent and R-light-independent manner, respectively. RNA-seq analysis identified that RRC1 also regulates genome-wide pre-mRNA splicing and coregulates hundreds of splicing events with SFPS, suggesting that they might function in part in the same complex. Given that SFPS and RRC1 regulate pre-mRNA splicing under both dark and light conditions, it is possible that the R-light-dependent interaction with phyB might provide specificity to selectively modulate pre-mRNA splicing of genes involved in light signaling, circadian rhythm, and photosynthesis. However, further studies are needed to strengthen this conclusion.
Similar to Arabidopsis phytochromes, phytochromes in the model moss species Physcomitrella patens (PpPHY) also directly participate in the regulation of alternative splicing of pre-mRNA in a R-light-dependent manner. In response to R light, PpPHY preferentially promotes intron retention events in transcripts of genes involved in light signaling pathways (140). Two splicing regulators, PphnRNP-H1 and PphnRNP-F1, were identified in 2019 as interacting partners of PpPHY (123). PphnRNP-H1 directly interacts with PpPRP39-1 (pre-mRNA-processing factor 39-1), a core component of U1-snRNP, in a PpPHY-dependent manner. PpPRP39-1-mediated intron retention events largely overlap with both PpPHY4 and PphnRNP-H1 under R light (123), suggesting a coordinated regulation of R-light-modulated pre-mRNA splicing by all three proteins (Figure 5c). The other splicing factor, PphnRNP-F1, also directly interacts with R-light-activated PpPHY4 within the nucleus, and together they coregulate approximately 70% of R-light-mediated intron retention events (76). A motif search of the 5′ and 3′ flanking regions of donor and acceptor sites of retained introns coregulated by PpPHY4 and PphnRNP-F1 indicated the purine-rich GAA cis-regulatory motif is overrepresented in the adjacent exonic region of the retained intron (76), leading to the conclusion that the exonic cis-regulatory GAA motif is probably selectively involved in the recruitment of PphnRNP-F1 to the RNA forming the RNA-protein complex to suppress pre-mRNA splicing and intron retention.
One common feature observed in these studies is that the differential pre-mRNA splicing patterns are modulated by all the splicing factors/regulators even in the dark, a condition under which these splicing factors/regulators do not interact with the inactive Pr form of phyB or PpPHY4. One possibility for this feature is that these splicing factors/regulators function as general splicing modulators under dark (or different) conditions, whereas in response to R light, the activated phytochromes/PpPHYs recruit them to target a specific set of pre-mRNAs for alternative splicing. However, the molecular or biochemical events leading to the altered target selection by these splicing factors upon interaction with the Pfr form of phyB/PpPHY4 are yet to be described.
These studies also highlight one of the main differences in phytochrome-mediated pre-mRNA splicing between Arabidopsis and P. patens related to snRNPs. Arabidopsis splicing factors (SFPS and RRC1) colocalize with U2-snRNPs, potentially targeting the 3′ splice site, whereas the P. patens splicing factors (PphnRNP-H1 and PphnRNP-F1) coordinate with U1-snRNPs, potentially targeting the 5′ splice site. It is possible that homologous splicing factors are yet to be identified in both Arabidopsis and P. patens. Alternatively, light-regulated pre-mRNA processing might have evolved independently in higher plants. Nevertheless, these studies highlight that phytochromes from across different species regulate the R-light-mediated pre-mRNA splicing by directly interacting with auxiliary splicing factors.
In addition, the transcription elongation dynamics play a critical role in determining the patterns of pre-mRNA alternative splicing. Molecular and biochemical evidence support that slower rate of transcription elongation leads to both higher exon inclusions in some mRNAs and higher exon skipping in others (39). A 2019 report showed that light promotes the elongation of RNA polymerase II in some genes and thereby controls their pre-mRNA splicing patterns (46) (Figure 5d). Moreover, it also demonstrated that the light-dark conditions do not affect overall mRNA accumulation but rather only influence alternative splicing pattern. However, as this study utilized a broad-spectrum white light source instead of a monochromatic light source, pinpointing which specific receptor is involved in the regulation of RNA polymerase II dynamics awaits further study.
5. TRANSLATIONAL REGULATION BY PHYTOCHROMES
5.1. Global Regulation of Translation by Light
Over the past decades, transcriptomic profiling has been widely used to examine how light regulates mRNA levels and gene expression on a genome-wide scale. Before the great effect of light on translation was first demonstrated, few lines of evidence indicated that light might regulate the translation efficiency of specific mRNAs. A mutation in eukaryotic translation initiation factor 3 subunit H1 (eIF3h) or an overexpressing eIF3e caused defects in skotomorphogenic development (66, 98, 151). Paik et al. (98) also showed that both phyA and phyB interact with the cytosolic PENTA1 (PNT1) protein and repress the translation of protochlorophyllide reductase A (PORA) mRNAs. By contrast, light also enhances the translation of photosynthetic genes, such as ferredoxin 1 (34, 101) and photosystem I genes (118). The regulatory effect of light on translation was not clear until 2012, when Liu et al. (79) demonstrated that light greatly enhances global translation efficiency at the early stage of seedling development. Compared with the transcriptomic changes, translational control targets thousands of more genes during photomorphogenesis. Moreover, translational control favors proteins for the organization and function of chloroplasts and ribosomes, suggesting the need for massive translation during photosynthesis. The authors also proposed that shorter and more stable transcripts are preferentially regulated at the translational level, which is an evolutionarily conserved mechanism. Ever since their demonstration, elucidating the underlying molecular mechanism for the massive translation triggered by light signals in de-etiolating Arabidopsis seedlings has become a popular research area. Understanding the translatome triggered by light has become not only a tool for dissecting the molecular machinery of translational regulation but also a new strategy for discovering novel light-responsive genes that might have been previously missed.
5.2. Selective Translation by Phytochrome-Mediated Signaling
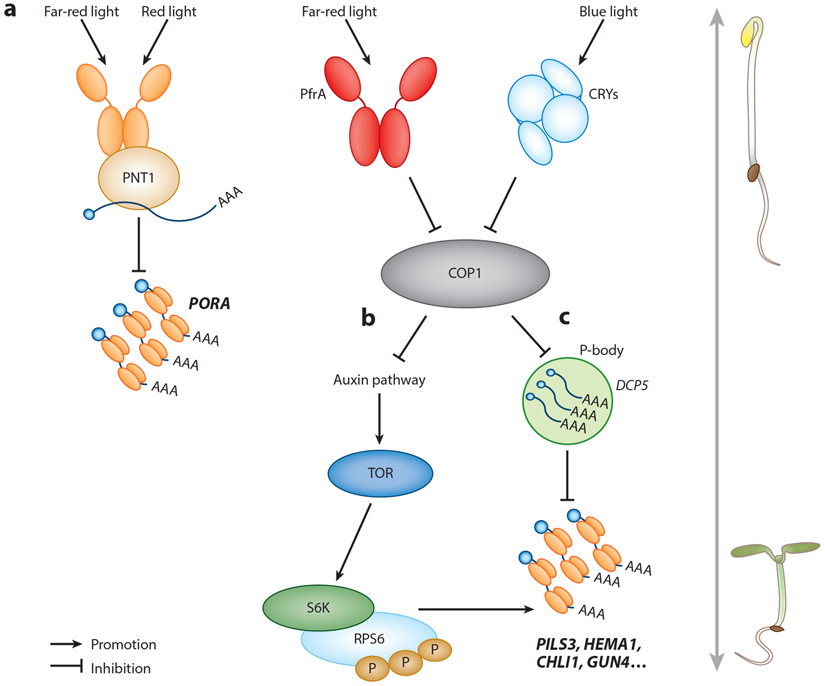
The exact mechanism responsible for the dramatic translational change triggered by light is yet to be demonstrated, and how phytochromes and their signaling components are involved in this phenomenon remains poorly understood. In the past few years, several lines of evidence show that photoreceptors, phytochromes especially, play important roles in this process (Figure 6). As mentioned above, active phyA interacts with cytosolic protein PNT1 and sequesters the mRNA encoding PORA, thus inhibiting its translation (98) (Figure 6a). This was perhaps the only evidence that phytochrome directly regulates the translation of a gene. The rest of the known mechanisms still depend mostly on the important phytochrome signaling negative regulator, COP1. Chen et al. (22) showed that in de-etiolating Arabidopsis seedlings active phyA and cryptochromes could indirectly modulate translation by inactivating COP1. The inhibition of COP1 leads to the target of rapamycin (TOR)-dependent phosphorylation of ribosomal S6 kinase (S6K) (22) (Figure 6b). Mutations in TOR, RIBOSOMAL PROTEIN S6A (RPS6A), or RPS6B exhibited delayed cotyledon opening, representing weaker translational activity in these mutants. Although COP1 seems to be a master switch, turning off translation in the dark, the direct mechanism regulating auxin and TOR signaling still awaits to be clarified. Whether phyA or other signaling components, such as SPAs and PIFs, play distinctive roles in regulating translational repression is still unknown.
Figure 6.
Present models of translational control by phytochromes. (a) Active phytochrome (Pfr form) interacts with the cytosolic protein PNT1 and inhibits the translation of PORA mRNA. (b) TOR and RPS6 transmit light signals to enhance protein translation in de-etiolating Arabidopsis seedlings. (c) P-bodies control the selective translation for optimal development of young Ambidopsis seedlings. Abbreviations: CRY, cryptochrome; P-body, processing body.
Apart from controlling translational efficiency, light also inhibits processing body (p-body) formation, thus allowing the translation of the mRNAs sequestered inside (58). P-bodies are cytoplasmic granules that balance the storage, degradation, and translation of mRNAs in diverse organisms. The mRNAs present in p-bodies are in a translationally repressed state (5 8) (Figure 6c). An Arabidopsis mutant defective in p-body formation [Decapping 5 (dcp5-l)] showed premature translation of specific mRNAs in the dark, including those encoding enzymes for protochlorophyllide synthesis and PIN-LIKES3 (PLS3) for auxin-dependent apical hook opening. Conversely, cop1-6, a constitutively photomorphogenic mutant, failed to form p-bodies, suggesting that COP1 might play a negative role in translational suppression in the dark. Upon light exposure, active phytochromes could inactivate COP1 and release mRNAs for translation. Although the evidence showing light signaling components regulate translation is solid, whether phytochromes are more directly involved in this process awaits further investigation.
6. CONCLUDING REMARKS AND FUTURE DIRECTIONS
Ever since Borthwick et al. (8) first discovered phytochromes in 1952, the phytochrome signaling pathway has become one of the most popular research areas in plant biology. However, despite many decades of study, important questions still remain to be answered. At the molecular level, the structures of bacterial phytochromes and their light-to-dark transition have provided us a template, but the higher-plant phytochrome structures are still incomplete. Crystal structures of full-length plant phytochromes along with their interacting partners, especially PIFs, would provide insights into early light signaling steps. Moreover, these studies might explain why phytochromes can interact with PIF proteins through both their PSM and OPM domains and how these interactions lead to different modes of PIF regulations.
With the technical advancement in liquid chromatography–mass spectrometry, new phytochrome-interacting partners with critical functions, such as PCH1 and PCHL, have been revealed. At the cellular level, the formation of photobodies and the relationship between their size and functions have been linked as sites for PIF degradation, transcriptional control, maintenance of active phytochromes, and even for alternative splicing of pre-mRNAs. However, these conclusions are mostly corelative at this stage, and the dynamic composition and the exact biochemical roles of photobodies are still unknown. Moreover, the N-terminal fragment of phyB was sufficient to activate its photosensory and regulatory activities, even though it fails to form photobodies, indicating that photobodies are dispensable (86). Thus, the importance and functions of photobodies in phytochrome signaling remain unresolved.
Even though phytochrome-dependent phosphorylation and degradation of PIFs play a central role in light signaling, the kinases responsible for such rapid multisite phosphorylation have always been a long-standing question. Several kinases, including the known repressor SPA1, which functions as a kinase for PIF phosphorylation, have been described to date. Whether these kinases act more specifically to a certain PIF or whether they function additively or sequentially during the dark-to-light transition is still unknown. Whether SPA1 regulates only PIF1 or has other substrates and regulates their abundance or activity awaits further study.
Although phytochromes regulate at multiple levels, including at transcriptional, posttranscriptional, translational, and posttranslational stages, direct involvement of phytochromes in epigenetic regulation of gene expression has not yet been demonstrated. Several epigenetic factors interact with light signaling components and regulate photomorphogenesis. However, how phytochromes regulate their activity in response to light is still not clear. In addition, how these regulations are coordinated at multiple levels for optimal photomorphogenesis remains unknown.
SUMMARY POINTS.
Structure-function analyses of phytochromes reveal specific functions for both the photosensory module and the output module, in which both modules are closely linked and contribute to regulation of phytochrome activity and downstream signaling.
The phytochrome-PIF signaling module plays a central role in our understanding of the biochemical mechanisms of early phytochrome signaling.
Identification of new phytochrome-interacting proteins reveals important signaling components such as PCH1 and HMR, which regulate the thermal reversion of phytochromes and the formation of photobodies.
The negative regulator, the COP1/SPA complex, displays both negative and positive roles under dark conditions and during the dark-to-light transition, respectively.
Phytochromes regulate transcriptional and posttranscriptional gene expression by directly interacting with splicing factors in response to light.
In the dark, COP1 plays a negative role in translational repression by regulating auxin, TOR signaling, and processing body formation through yet unknown mechanisms. By contrast, phytochromes repress COP1 activity to stimulate translation under light conditions.
ACKNOWLEDGMENTS
We thank Lin-Chen Huang in the M.-C.C. lab for designing the art used in the figures. This work was supported by grants from the National Science Foundation (MCB-2014408) and the National Institutes of Health (GM-114297) to E.H., and by the Young Scholar Fellowship Einstein Program from the Taiwan Ministry of Science and Technology (MOST 109-2636-B-002-011) to M.-C.C.
Glossary
- Phytochrome
bilin-binding photosensory receptor that detects red/far-red light in plants, bacteria, and fungi
- PHYTOCHROME INTERACTING FACTOR (PIF)
the family of basic helix-loop-helix transcription factors that negatively regulate photomorphogenesis
- PHOTOPERIODIC CONTROL OF HYPOCOTYL1 (PCH1)
binds to the Pfr form of phyB, promotes phyB photobody accumulation, and prevents phyB thermal reversion
- HEMERA (HMR)
binds to phyB, promotes phyB photobody accumulation, and is required for PIF4-dependent induction of temperature responses
- CONSTITUTIVE PHOTOMOR-PHOGENIC1 (COP1)
critical E3 ligase involved in protein ubiquitination and degradation of phytochrome signaling
- SUPPRESSOR OF phya-105 1 (SPA1)
E3 ligase involved in protein ubiquitination; also a kinase for PIF phosphorylation
- SPLICING FACTOR FOR PHYTOCHROME SIGNALING (SFPS)
plicing factor subunit that directly interacts with phyB and regulates pre-mRNA splicing in Arabidopsis
- REDUCED RED-LIGHT RESPONSES IN CRY1CRY2 BACKGROUND 1 (RRC1)
interacts with SFPS and forms a complex and coordinately controls pre-mRNA splicing
- PENTA1 (PNT1)
a cytosolic protein that interacts with phytochromes and represses the translation of protochlorophyllide reductase mRNAs
- Target of rapamycin (TOR)
a serine/threonine protein kinase that regulates sensing of energy status, nitrogen mobilization, glucose utilization, stresses, and hormone coordination
- RIBOSOMAL PROTEIN S6 (RPS6)
a substrate of S6K; together with TOR it functions downstream of COP1 for translational control
- DECAPPING 5 (DCP5)
an Arabidopsis p-body component critical for p-body formation
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Ádám É, Kircher S, Liu P, Mérai Z, González-Schain N, et al. 2013. Comparative functional analysis of full-length and N-terminal fragments of phytochrome C, D and E in red light-induced signaling. New Phytol. 200:86–96 [DOI] [PubMed] [Google Scholar]
- 2.Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. 2006. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23:439–46 [DOI] [PubMed] [Google Scholar]
- 3.Anders K, Daminelli-Widany G, Mroginski MA, von Stetten D, Essen L-O. 2013. Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. J. Biol. Chem 288:35714–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae G, Choi G. 2008. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol 59:281–311 [DOI] [PubMed] [Google Scholar]
- 5.Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, et al. 2004. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16:1433–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernardo-García S,de Lucas M,Martínez C,Espinosa-Ruiz A, Daviere J-M, Prat S. 2014. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28:1681–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Björling A, Berntsson O, Lehtivuori H, Takala H, Hughes AJ, et al. 2016. Structural photoactivation of a full-length bacterial phytochrome. Sci. Adv 2:e1600920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borthwick HA, Hendricks SB, Parker MW, Toole EH, Toole VK. 1952. A reversible photoreaction controlling seed germination. PNAS 38:662–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bu Q, Zhu L, Huq E. 2011. Multiple kinases promote light-induced degradation of PIF1. Plant Sig. Behav 6:1119–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bu Q, Zhu L, Yu L, Dennis M, Lu X, et al. 2011. Phosphorylation by CK2 enhances the rapid light-induced degradation of PIF1. J. Biol. Chem 286:12066–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgie ES, Bussell AN, Lye S-H, Wang T, Hu W, et al. 2017. Photosensing and thermosensing by phytochrome B require both proximal and distal allosteric features within the dimeric photoreceptor. Sci. Rep 7:13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burgie ES, Bussell AN, Walker JM, Dubiel K, Vierstra RD. 2014. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. PNAS 111:10179–84 Proposes a 3D structural detail for the dark-to-light transition of PSM in phytochromes.
- 13.Burgie ES, Vierstra RD. 2014. Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell 26:4568–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgie ES, Zhang J, Vierstra RD. 2016. Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24:448–57 [DOI] [PubMed] [Google Scholar]
- 15.Bursch K, Toledo-Ortiz G, Pireyre M, Lohr M, Braatz C, Johansson H. 2020. Identification of BBX proteins as rate-limiting cofactors of HY5. Nat. Plants 6(8):921–28 [DOI] [PubMed] [Google Scholar]
- 16.Buskirk EKV, Reddy AK, Nagatani A, Chen M. 2014. Photobody localization of phytochrome B is tightly correlated with prolonged and light-dependent inhibition of hypocotyl elongation in the dark. Plant Physiol. 165:595–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buti S, Hayes S, Pierik R. 2020. The bHLH network underlying plant shade-avoidance. Physiol. Plant 169:312–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casal JJ. 2013. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol 64:403–27 [DOI] [PubMed] [Google Scholar]
- 19.Casal JJ, Balasubramanian S. 2019. Thermomorphogenesis. Annu. Rev. Plant Biol 70:321–46 [DOI] [PubMed] [Google Scholar]
- 20.Chen F, Li B, Demone J, Charron J-B, Shi X, Deng XW. 2014. Photoreceptor partner FHY1 has an independent role in gene modulation and plant development under far-red light. PNAS 111:11888–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen F, Li B, Li G, Charron J-B, Dai M, et al. 2014. Arabidopsis phytochrome A directly targets numerous promoters for individualized modulation of genes in a wide range of pathways. Plant Cell Online 26:1949–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen G-H, Liu M-J, Xiong Y, Sheen J, Wu S-H. 2018. TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. PNAS 115:12823–28 Demonstrates that phytochrome regulates translation through the COP1-mediated auxin signaling pathway.
- 23.Chen M, Chory J. 2011. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21:664–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen M, Chory J, Fankhauser C. 2004. Light signal transduction in higher plants. Annu. Rev. Genet 38:87–117 [DOI] [PubMed] [Google Scholar]
- 25.Chen M, Galvão RM, Li M, Burger B, Bugea J, et al. 2010. Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141:1230–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M, Schwab R, Chory J. 2003. Characterization of the requirements for localization of phytochrome B to nuclear bodies. PNAS 100:14493–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M, Tao Y, Lim J, Shaw A, Chory J. 2005. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol 15:637–42 [DOI] [PubMed] [Google Scholar]
- 28.Cheng M-C, Enderle B, Kathare PK, Islam R, Hiltbrunner A, Huq E. 2020. PCH1 and PCHL directly interact with PIF1, promote its degradation and inhibit its transcriptional function during photomorphogenesis. Mol. Plant 13:499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Y-L, Tu S-L. 2018. Alternative splicing and cross-talk with light signaling. Plant Cell Physiol. 59:110–4–10 [DOI] [PubMed] [Google Scholar]
- 30.Cherry JR, Hondred D, Walker JM, Keller JM, Hershey HP, Vierstra RD. 1993. Carboxy-terminal deletion analysis of oat phytochrome A reveals the presence of separate domains required for structure and biological activity. Plant Cell 5:565–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherry JR, Hondred D, Walker JM, Vierstra RD. 1992. Phytochrome requires the 6-kDa N-terminal domain for full biological activity. PNAS 89:5039–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clack T, Shokry A, Moffet M, Liu P, Faul M, Sharrock RA. 2009. Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21:786–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clough RC, Vierstra RD. 1997. Phytochrome degradation. Plant Cell Environ. 20:713–21 [Google Scholar]
- 34.Dickey LF, Petracek ME, Nguyen TT, Hansen ER, Thompson WF. 1998. Light regulation of Fed-1 mRNA requires an element in the 5′ untranslated region and correlates with differential polyribosome association. Plant Cell 10:475–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dong J, Chen H, Deng XW, Irish VF, Wei N. 2020. Phytochrome B induces intron retention and translational inhibition of PHYTOCHROME-INTERACTING FACTOR3. Plant Physiol. 182:159–66 Proposes a reversible regulatory mechanism that phyB induces alternative splicing of PIF3 and fine-tunes its protein level
- 36.Dong J, Ni W, Yu R, Deng XW, Chen H, Wei N. 2017. Light-dependent degradation of PIF3 by SCFEBF1/2 promotes a photomorphogenic response in Arabidopsis. Curr. Biol 27:2420–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong J, Sun N, Yang J, Deng Z, Lan J, et al. 2019. The transcription factors TCP4 and PIF3 antagonistically regulate organ-specific light induction of SAUR genes to modulate cotyledon opening during de-etiolation in Arabidopsis. Plant Cell 31:1155–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong J,Tang D, Gao Z, Yu R, Li K, et al. 2014. Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell Online 26:3630–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dujardin G, Lafaille C, de la Mata M, Marasco LE, Muñoz MJ, et al. 2014. How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell 54:683–90 [DOI] [PubMed] [Google Scholar]
- 40.Edgerton MD, Jones AM. 1992. Localization of protein-protein interactions between subunits of phytochrome. Plant Cell 4:161–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Enderle B, Sheerin DJ, Paik I, Kathare PK, Schwenk P, et al. 2017. PCH1 and PCHL promote photomorphogenesis in plants by controlling phytochrome B dark reversion. Nat. Commun 8:2221. Demonstrates that PCH1/PCHL promotes photomorphogenesis by preventing phyB thermal reversion.
- 42.Essen LO, Mailliet J, Hughes J. 2008. The structure of a complete phytochrome sensory module in the Pr ground state. PNAS 105:14709–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franklin KA, Quail PH. 2009. Phytochrome functions in Arabidopsis development. J. Exp. Bot 61:11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galvāo VC, Fiorucci A-S, Trevisan M, Franco-Zorilla JM, Goyal A, et al. 2019. PIF transcription factors link a neighbor threat cue to accelerated reproduction in Arabidopsis. Nat. Commun 10:4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Genoud T, Schweizer F, Tscheuschler A, Debrieux D, Casal JJ, et al. 2008. FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLOS Genet. 4:e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Godoy Herz MA, Kubaczka MG, Brzyżek G, Servi L, Krzyszton M, et al. 2019. Light regulates plant alternative splicing through the control of transcriptional elongation. Mol. Cell 73:1066–74.e3 [DOI] [PubMed] [Google Scholar]
- 47.Gu D, Chen C-Y, Zhao M, Zhao L, Duan X, et al. 2017. Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res. 45:7137–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hahm J, Kim K, Qiu Y, Chen M. 2020. Increasing ambient temperature progressively disassembles Arabidopsis phytochrome B from individual photobodies with distinct thermostabilities. Nat. Commun 11:1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han X, Huang X, Deng XW 2020. The photomorphogenic central repressor COP1: conservation and functional diversification during evolution. Plant Commun. 1:100044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han Y-J, Kim H-S, Kim Y-M, Shin A-Y, Lee S-S, et al. 2010. Functional characterization of phytochrome autophosphorylation in plant light signaling. Plant Cell Physiol. 51:596–609 [DOI] [PubMed] [Google Scholar]
- 51.Hiltbrunner A, Viczián A, Bury E, Tscheuschler A, Kircher S, et al. 2005. Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol 15:2125–30 [DOI] [PubMed] [Google Scholar]
- 52.Hoecker U 2017. The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol 37:63–69 [DOI] [PubMed] [Google Scholar]
- 53.Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. 2009. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28:3893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hua W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, Lagarias JC. 2013. Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. PNAS 110:1542–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang H, McLoughlin KE, Sorkin ML, Burgie ES, Bindbeutel RK, et al. 2019. PCH1 regulates light, temperature, and circadian signaling as a structural component of phytochrome B-photobodies in Arabidopsis. PNAS 116:8603–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang H, Yoo CY, Bindbeutel R, Goldsworthy J, Tielking A, et al. 2016. PCH1 integrates circadian and light-signaling pathways to control photoperiod-responsive growth in Arabidopsis. eLife 5:e13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huq E, Al-Sady B, Quail PH. 2003. Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 35:660–64 [DOI] [PubMed] [Google Scholar]
- 58. Jang G-J, Yang J-Y, Hsieh H-L, Wu S-H. 2019. Processing bodies control the selective translation for optimal development of Arabidopsis young seedlings. PNAS 116:6451–56 Demonstrates that phytochromes regulate the p-body-controlled selective translation for adaptation.
- 59.Jiao Y, Lau OS, Deng XW. 2007. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet 8:217–30 [DOI] [PubMed] [Google Scholar]
- 60.Jing Y, Zhang D, Wang X, Tang W, Wang W, et al. 2013. Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25:242–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung J-H, Domijan M, Klose C, Biswas S, Ezer D, et al. 2016. Phytochromes function as thermosensors in Arabidopsis. Science 354:886–89 [DOI] [PubMed] [Google Scholar]
- 62.Kaiserli E, Páldi K, O’Donnell L, Batalov O, Pedmale UV, et al. 2015. Integration of light and photoperiodic signaling in transcriptional nuclear foci. Dev. Cell 3 5:311–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kathare PK, Xu X, Nguyen A, Huq E. 2020. A COP1-PIF-HEC regulatory module fine-tunes photomorphogenesis in Arabidopsis. Plant J. 104:113–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kikis EA, Oka Y, Hudson ME, Nagatani A, Quail PH. 2009. Residues clustered in the light-sensing knot of phytochrome B are necessary for eonformer-specific binding to signaling partner PIF3. PLOS Genet. 5:e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim J-H, Lee H-J, Jung J-H, Lee S, Park C-M. 2017. HOS1 facilitates the phytochrome B-mediated inhibition of PIF4 function during hypocotyl growth in Arabidopsis. Mol. Plant 10:274–84 [DOI] [PubMed] [Google Scholar]
- 66.Kim T-H, Kim B-H, Yahalom A, Chamovitz DA, von Arnim AG. 2004. Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 16:3341–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, et al. 2002. Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14:1541–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klose C, Nagy F, Schäfer E. 2020. Thermal reversion of plant phytochromes. Mol. Plant 13:386–97 [DOI] [PubMed] [Google Scholar]
- 69.Klose C, Viezián A, Kireher S, Schäfer E, Nagy F. 2015. Molecular mechanisms for mediating light-dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytol. 206:965–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee S, Paik I, Huq E. 2020. SPAs promote thermomorphogenesis by regulating the phyB-PIF4 module in Arabidopsis. Development 147:dev189233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Legris M, Ince YÇ, Fankhauser C. 2019. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun 10:5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Legris M, Klose C, Burgie ES, Costigliolo C, Neme M, et al. 2016. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354:897–900 [DOI] [PubMed] [Google Scholar]
- 73.Leivar P, Monte E. 2014. PIFs: systems integrators in plant development. Plant Cell 26:56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leivar P, Quail PH. 2011. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li F-W, Melkonian M, Rothfels CJ, Villarreal JC, Stevenson DW, et al. 2015. Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nat. Commun 6:7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin B-Y, Shih C-J, Tu S-L. 2020. Phytochrome coordinates with a hnRNP to regulate alternative splicing via an exonic splicing silencer. Plant Physiol. 182:243–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin F, Jiang Y, Li J, Yan T, Fan L, et al. 2018. BBX28 negatively regulates photomorphogenesis by repressing HY5 activity and itself undergoes COP1-mediated degradation in Arabidopsis. Plant Cell 30:2006–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ling J-J, Li J, Zhu D, Deng XW. 2017. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. PNAS 114:3539–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu M-J, Wu S-H, Chen H-M, Wu S-H. 2012. Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol. Syst. Biol 8:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu X, Chen C-Y, Wang K-C, Luo M, Tai R, et al. 2013. PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25:1258–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu X-D, Zhou C-M, Xu P-B, Luo Q, Lian H-L, Yang H-Q. 2015. Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8:467–78 [DOI] [PubMed] [Google Scholar]
- 82.Lyu M, Shi H, Li Y, Kuang K, Yang Z, et al. 2019. Oligomerization and photo-deoligomerization of HOOKLESS1 controls plant differential cell growth. Dev. Cell 51:78–88.e3 [DOI] [PubMed] [Google Scholar]
- 83.Majee M, Kumar S, Kathare PK, Wu S, Gingerich D, et al. 2018. KELCH F-BOX protein positively influences Arabidopsis seed germination by targeting PHYTOCHROME-INTERACTING FACTOR1. PNAS 115:E4120–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martínez-García JF, Huq E, Quail PH. 2000. Direct targeting of light signals to a promoter element-bound transcription factor. Science 288:859–63 [DOI] [PubMed] [Google Scholar]
- 85.Mathews S 2010. Evolutionary studies illuminate the structural-functional model of plant phytochromes. Plant Cell 22:4–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsushita T, Mochizuki N, Nagatani A. 2003. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424:571–74 [DOI] [PubMed] [Google Scholar]
- 87.Menon C, Klose C, Hiltbrunner A. 2020. Arabidopsis FHY1 and FHY1-LIKE are not required for phytochrome A signal transduction in the nucleus. Plant Commun. 1:100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nagano S 2016. From photon to signal in phytochromes: similarities and differences between prokaryotic and plant phytochromes. J. Plant Res 129:123–35 [DOI] [PubMed] [Google Scholar]
- 89.Ni W, Xu S-L, González-Grandío E, Chalkley RJ, Huhmer AFR, et al. 2017. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun 8:15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ni W, Xu S-L, Tepperman JM, Stanley DJ, Maltby DA, et al. 2014. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344:1160–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nieto C, López-Salmerón V, Davière J-M, Prat S. 2014. ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr. Biol 25:187–93 [DOI] [PubMed] [Google Scholar]
- 92.Nito K, Wong CCL, Yates JR 3rd, Chory J. 2013. Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Rep. 3:1970–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, et al. 2011. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475:398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Oka Y, Ono Y, Toledo-Ortiz G, Kokaji K, Matsui M, et al. 2012. Arabidopsis phytochrome A is modularly structured to integrate the multiple features that are required for a highly sensitized phytochrome. Plant Cell 24:2949–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pacín M, Legris M, Casal JJ. 2014. Rapid decline in nuclear COSTITUTIVE PHOTOMORPHO-GENESIS1 abundance anticipates the stabilization of its target ELONGATED HYPOCOTYL5 in the light. Plant Physiol. 164:1134–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Paik I, Chen F, Ngoc Pham V,Zhu L,Kim J-I,Huq E. 2019. A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat. Commun 10:4216. Proposes a new role for SPA1 as a kinase phosphorylating PIF to promote photomorphogenesis
- 97.Paik I, Huq E. 2019. Plant photoreceptors: multi-functional sensory proteins and their signaling networks. Semin. Cell Dev. Biol 92:114–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Paik I, Yang S, Choi G. 2012. Phytochrome regulates translation of mRNA in the cytosol. PNAS 109:1335–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Park E, Kim Y, Choi G. 2018. Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions. Plant Cell 30:1277–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park E, Park J, Kim J, Nagatani A, Lagarias JC, Choi G. 2012. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72:537–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Petracek ME, Dickey LF, Huber SC, Thompson WF. 1997. Light-regulated changes in abundance and polyribosome association of ferredoxin mRNA are dependent on photosynthesis. Plant Cell 9:2291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pfeiffer A, Nagel M-K, Popp C, Wüst F, Bindics J, et al. 2012. Interaction with plant transcription factors can mediate nuclear import of phytochrome B. PNAS 109:5892–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pham VN, Kathare PK, Huq E. 2018. Dynamic regulation of PIF5 by COP1-SPA complex to optimize photomorphogenesis in Arabidopsis. Plant J. 96:260–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pham VN, Kathare PK, Huq E. 2018. Phytochromes and phytochrome interacting factors. Plant Physiol. 176:1025–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pham VN, Xu X, Huq E. 2018. Molecular bases for the constitutive photomorphogenic phenotypes in Arabidopsis. Development 145: dev169870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Podolec R, Ulm R. 2018. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol 45:18–25 [DOI] [PubMed] [Google Scholar]
- 107.Qiu Y, Li M, Pasoreck EK, Long L, Shi Y, et al. 2015. HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORS in Arabidopsis photomorphogenesis. Plant Cell 27:1409–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qiu Y, Pasoreck EK, Reddy AK, Nagatani A, Ma W, et al. 2017. Mechanism of early light signaling by the carboxy-terminal output module of Arabidopsis phytochrome B. Nat. Commun 8:1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rausenberger J, Tscheuschler A, Nordmeier W, Wüst F, Timmer J, et al. 2011. Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146:813–25 [DOI] [PubMed] [Google Scholar]
- 110.Rockwell NC, Lagarias JC. 2020. Phytochrome evolution in 3D: deletion, duplication, and diversification. New Phytol. 225:2283–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ryu JS, Kim J-I, Kunkel T, Kim BC, Cho DS, et al. 2005. Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120:395–406 [DOI] [PubMed] [Google Scholar]
- 112.Sadanandom A, Ádám É, Orosa B, Viczián A, Klose C, et al. 2015. SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana. PNAS 112:11108–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sánchez-Lamas M, Lorenzo CD, Cerdán PD. 2016. Bottom-up assembly of the phytochrome network. PLOS Genet. 12:e1006413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sheerin DJ, Hiltbrunner A. 2017. Molecular mechanisms and ecological function of far-red light signalling. Plant Cell Environ. 40:2509–29 [DOI] [PubMed] [Google Scholar]
- 115.Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, et al. 2015. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27:189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shen H, Ling Z, Castillon A, Majee M, Downie B, Huq E. 2008. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME INTERACTING FACTOR 1 depends upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20:1586–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shen YP, Zhou ZZ, Feng SH, Li JG, Tan-Wilson A, et al. 2009. Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 21:494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sherameti I, Nakamura M, Yamamoto YY, Pfannschmidt T, Obokata J, Oelmuller R. 2002. Polyribosome loading of spinach mRNAs for photosystem I subunits is controlled by photosynthetic electron transport. Plant J. 32:631–39 [DOI] [PubMed] [Google Scholar]
- 119.Shi H, Liu R, Xue C, Shen X, Wei N, et al. 2016. Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr. Biol 26:139–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shi H, Lyu M, Luo Y, Liu S, Li Y, et al. 2018. Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. PNAS 115:6482–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shi H, Wang X, Mo X, Tang C, Zhong S, Deng XW. 2015. Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. PNAS 112:3817–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shi H, Zhong S, Mo X, Liu N, Nezames CD, Deng XW 2013. HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25:3770–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Shih C-J, Chen H-W, Hsieh H-Y, Lai Y-H, Chiu F-Y, et al. 2019. Heterogeneous nuclear ri-bonucleoprotein H1 coordinates with phytochrome and the U1-snRNP complex to regulate alternative splicing in Physcomitrella patens. Plant Cell 31:2510–24 Demonstrates that P. patens phytochrome regulates alternative splicing by interacting with the spliceosome complex.
- 124.Shikata H, Hanada K, Ushijima T, Nakashima M, Suzuki Y, Matsushita T. 2014. Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. PNAS 111:18781–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shikata H, Shibata M, Ushijima T, Nakashima M, Kong S-G, et al. 2012. The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. Plant J. 70:727–38 [DOI] [PubMed] [Google Scholar]
- 126.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. 2002. A light-switchable gene promoter system. Nat. Biotechnol 20:1041–44 [DOI] [PubMed] [Google Scholar]
- 127.Shin A-Y, Han Y-J, Baek A, Aim T, Kim SY, et al. 2016. Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun 7:11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Somers DE, Devlin PF, Kay SA. 1998. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282:1488–90 [DOI] [PubMed] [Google Scholar]
- 129.Song C, Mroginski MA, Lang C, Kopycki J, Gärtner W, et al. 2018.3D structures of plant phytochrome A as Pr and Pfr from solid-state NMR: implications for molecular function. Front. Plant Sci 9:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Song Z, Bian Y, Liu J, Sun Y, Xu D. 2020. B-box proteins: pivotal players in light-mediated development in plants. J. Integr. Plant Biol 62:1293–309 [DOI] [PubMed] [Google Scholar]
- 131.Strassera B, Sánchez-Lamasa M, Yanovsky MJ, Casal JJ, Cerdán PD. 2010. Arabidopsis thaliana life without phytochromes. PNAS 107:4776–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Subramanian C, Kim B-H, Lyssenko NN, Xu X, Johnson CH, von Arnim AG. 2004. The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer. PNAS 101:6798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sun N, Wang J, Gao Z, Dong J, He H, et al. 2016. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. PNAS 113:6071–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Takala H, Björling A, Berntsson O, Lehtivuori H, Niebling S, et al. 2014. Signal amplification and transduction in phytochrome photosensors. Nature 509:245–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Takala H, Björling A, Linna M, Westenhoff S, Ihalainen JA. 2015. Light-induced changes in the dimerization interface of bacteriophytochromes. J. Biol. Chem 290:16383–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Trupkin SA, Legris M, Buchovsky AS, Tolava Rivero MB, Casal JJ. 2014. Phytochrome B nuclear bodies respond to the low red to far-red ratio and to the reduced irradiance of canopy shade in Arabidopsis. Plant Physiol. 165:1698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. 2005. A light-sensing knot revealed by the structure of the chromophore binding domain of phytochrome. Nature 438:325–31 [DOI] [PubMed] [Google Scholar]
- 138.Wang X, Yu R, Wang J, Lin Z, Han X, et al. 2020. The asymmetric expression of SAUR genes mediated by ARF7/19 promotes the gravitropism and phototropism of plant hypocotyls. Cell Rep. 31:107529. [DOI] [PubMed] [Google Scholar]
- 139.Wit MD, Galvão VC, Fankhauser C. 2016. Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol 67:513–37 [DOI] [PubMed] [Google Scholar]
- 140.Wu H-P, Su Y-S, Chen H-C, Chen Y-R, Wu C-C, et al. 2014. Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol. 15:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu J, Wang W, Xu P, Pan J, Zhang T, et al. 2018. phyB interacts with BES1 to regulate brassinosteroid signaling in Arabidopsis. Plant Cell Physiol. 60:353–66 [DOI] [PubMed] [Google Scholar]
- 142.Wu Q, Kuang K, Lyu M, Zhao Y, Li Y, et al. 2020. Allosteric deactivation of PIFs and EIN3 by microproteins in light control of plant development. PNAS 117:18858–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu S-H. 2014. Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annu. Rev. Plant Biol 65:311–33 [DOI] [PubMed] [Google Scholar]
- 144. Xin R, Kathare PK, Huq E. 2019. Coordinated regulation of pre-mRNA splicing by the SFPS-RRC1 complex to promote photomorphogenesis. Plant Cell 31:2052–69 Together with Reference 145 demonstrates that phytochrome regulates alternative splicing by directly interacting with splicing factors.
- 145.Xin R, Zhu L, Salomé PA, Mancini E, Marshall CM, et al. 2017. SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. PNAS 114:E7018–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xin X, Chen W, Wang B, Zhu F, Li Y, et al. 2017. Arabidopsis MKK10-MPK6 mediates red-light-regulated opening of seedling cotyledons through phosphorylation of PIF3. J. Exp. Bot 69:423–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xu F, He S, Zhang J, Mao Z, Wang W, et al. 2018. Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol. Plant 11:523–41 [DOI] [PubMed] [Google Scholar]
- 148.Xu X, Kathare PK, Pham VN, Bu Q, Nguyen A, Huq E. 2017. Reciprocal proteasome-mediated degradation of PIFs and HFR1 underlies photomorphogenic development in Arabidopsis. Development 144:1831–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Xu X, Paik I, Zhu L, Bu Q, Huang X, et al. 2014. PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell 26:1992–2006 Proposes the nontranscriptional roles of PIFs as cofactors of E3 ligase.
- 150.Xu X, Paik I, Zhu L, Huq E. 2015. Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 20:641–50 [DOI] [PubMed] [Google Scholar]
- 151.Yahalom A, Kim T-H, Roy B, Singer R, Von Arnim AG, Chamovitz DA. 2008. Arabidopsis eIF3e is regulated by the COP9 signalosome and has an impact on development and protein translation. Plant J. 53:300–11 [DOI] [PubMed] [Google Scholar]
- 152.Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. 1999. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol 145:437–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yan Y, Li C, Dong X, Li H, Zhang D, et al. 2020. MYB30 is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell 32:2196–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yang C, Xie F, Jiang Y, Li Z, Huang X, Li L. 2018. Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev. Cell 44:29–41.e4 [DOI] [PubMed] [Google Scholar]
- 155.Yang EJ, Yoo CY, Liu J, Wang H, Cao J, et al. 2019. NCP activates chloroplast transcription by controlling phytochrome-dependent dual nuclear and plastidial switches. Nat. Commun 10:2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang P, Wen Q, Yu R, Han X, Deng XW, Chen H. 2020. Light modulates the gravitropic responses through organ-specific PIFs and HY5 regulation of LAZY4 expression in Arabidopsis. PNAS 117:18840–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yang X, Kuk J, Moffat K. 2009. Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. PNAS 106:15639–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yeh KC, Lagarias JC. 1998. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. PNAS 95:13976–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Yoo CY, Pasoreck EK, Wang H, Cao J, Blaha GM, et al. 2019. Phytochrome activates the plastid-encoded RNA polymerase for chloroplast biogenesis via nucleus-to-plastid signaling. Nat. Commun 10:2629. Proposes a nucleus-to-plastid anterograde signaling pathway by which phytochrome signaling in the nucleus controls plastidial transcription.
- 160.Zhang B, Holmlund M, Lorrain S, Norberg M, Bakó L, et al. 2017. BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. eLife 6:e26759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang D, Jing Y, Jiang Z, Lin R. 2014. The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 26:2472–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhu DM, Maier A, Lee J-H, Laubinger S, Saijo Y, et al. 2008. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20:2307–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhu L, Bu Q, Xu X, Paik I, Huang X, et al. 2015. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun 6:7245. [DOI] [PubMed] [Google Scholar]
- 164.Zhu L, Xin R, Bu Q, Shen H, Dang J, Huq E. 2016. A negative feedback loop between PHYTOCHROME INTERACTING FACTORS and HECATE proteins fine tunes photomorphogenesis in Arabidopsis. Plant Cell 28:855–74 [DOI] [PMC free article] [PubMed] [Google Scholar]