Abstract
Retroviral reverse transcriptase-associated RNase H enzymes are responsible for degradation of viral RNA, including removal of the tRNA primer after plus-strand strong-stop synthesis and cleavage of the polypurine tract primer. These activities are required for the complex viral replication and result in generation of the long terminal repeats. The human immunodeficiency virus type 1 (HIV-1) RNase H domain has been expressed independently of the polymerase domain and possesses Mn2+-dependent activity with a hexahistidine tag. The isolated domain maintains the ability to specifically remove a tRNA primer mimic. In this study, the substrate determinants for recognition of the cognate tRNA3Lys are defined. Model substrates were constructed which mimic the RNA-DNA hybrid obtained from plus-strand strong-stop synthesis. Deletion substrates containing only 12, 9, or 6 positions of the tRNA primer were capable of being cleaved by the isolated RNase H domain. Mismatch and bromodeoxyuridine mutagenesis analysis indicated that positions 2, 3, 4, and 6, when mutated, affected the specificity of RNase H activity. Substitution substrates indicated that positions 4 and 6 within the RNA primer were important for recognition and cleavage by the HIV-1 isolated RNase H domain. Moloney murine leukemia virus-HIV-1 hybrid substrates were constructed which demonstrated that changes to HIV-1 sequences at positions 4 and 6 were sufficient but not optimal for regaining cleavage by the isolated HIV-1 RNase H domain. Optimal site-specific cleavage between the terminal ribonucleotide A and ribonucleotide C requires additional sequences beyond the first six positions but less than nine.
Retroviruses contain an RNA genome which is reverse transcribed into a DNA copy by the viral reverse transcriptase (RT). RT is a multifunctional enzyme containing RNA- and DNA-dependent DNA polymerase activities and RNase H activity. Replication of the virus requires RNase H activity (20, 24, 33, 34) to remove the RNA within RNA-DNA hybrid intermediates formed during the polymerization process. The double-stranded DNA product is subsequently randomly integrated into the host’s genome (2).
Although the viral RNase H is required for the removal of the RNA throughout the replicating genome, two specific cleavages are critical. The first involves the generation of the plus-strand primer (21). The second is the removal of the tRNA primer used in minus-strand DNA synthesis (4, 14). During viral replication, the first 18 nucleotides of the tRNA are reverse transcribed, regenerating the primer binding site (PBS) during synthesis of plus-strand strong-stop DNA. Removal of the tRNA from the RNA-DNA hybrid by RNase H exposes the repeat sequence required for the second strand transfer reaction. Imprecise removal of the tRNA primer could result in long terminal repeat termini, required for the subsequent integration of the viral DNA, being incorrect. The removal of the tRNA primer has been characterized for human immunodeficiency virus type 1 (HIV-1) and Moloney murine leukemia virus (M-MuLV) RTs. For HIV-1, the RT-RNase H cleaves the tRNA3Lys between the terminal ribonucleotide A and ribonucleotide C (18, 32), leaving a terminal ribonucleotide which is stable in vivo (8, 9, 12, 16, 30). Interestingly, M-MuLV RT initially cleaves the tRNAPro at the identical position, between the terminal ribonucleotide A and ribonucleotide C of the tRNA (27, 28). In contrast, this terminal ribonucleotide is subsequently removed and is not found associated in the circle junctions isolated from infected cells (28).
HIV-1 RT consists of a heterodimer of p66 and p51 subunits. The RNase H domain is encoded at the C terminus of p66, and the polymerase domain is at the N terminus. Evidence of the interactions between these domains is established. RNase H activity has been characterized as being either polymerase dependent or polymerase independent (1). In the polymerase-dependent mode, footprinting analysis has determined that the RNase H active site lags approximately 18 to 20 nucleotides behind the actively synthesized polymerase domain (38–40). Mutations within the primer grip region of the HIV-1 polymerase domain have been shown to have decreased RNase H activity and specificity, indicating the interrelations of these two domains (6, 15, 17). Conversely, alterations in RNase H have been shown to destabilize the interactions between RT and the primer template (7, 19).
Isolated retroviral RNase H domains separated from the polymerase domains have been constructed. This allows for the direct analysis of RNase H cleavages independent of the polymerase activity. The isolated RNase H domain of M-MuLV was not capable of specific RNase H cleavages in the absence of the polymerase domain (26, 41). A series of HIV-1 isolated RNase H domains which are active in manganese have been constructed with or without a histidine tag (29, 31). The isolated HIV-1 RNase H domain cleaves model substrates for tRNA3Lys primer removal at the same specific position as the RT-associated RNase H domain (31).
This study focuses on characterizing polymerase-independent RNase H activity catalyzed by an isolated HIV-1 RNase H domain. Current research shows that an isolated HIV-1 RNase H domain is capable of distinguishing a tRNAPro mimic from a tRNA3Lys mimic for primer removal. Model substrates have been constructed possessing an RNA primer consisting of the first 18 nucleotides of tRNA3Lys. This substrate models the viral replication intermediate after plus-strand strong-stop DNA synthesis. Deletion substrates have been constructed to determine the minimal sequences necessary for removal by the HIV-1 RNase H. Mismatch, substitution, and hybrid substrates were constructed to characterize the specific sequences important for recognition. The data indicates that sequences outside the scissile bond, through the first 8 nucleotides of the tRNA, are important for optimal and specific removal of the tRNA.
MATERIALS AND METHODS
Enzymes and nucleotides.
T4 polynucleotide kinase was purchased from New England Biolabs; recombinant RNasin was purchased from Promega. Exonuclease(−) Klenow polymerase was purchased from United States Biochemical. HIV-1 RT was obtained from Jeffrey Culp and Christine Debouch, Department of Protein Biochemistry, SmithKline Beecham Pharmaceuticals. HIV-1 isolated RNase H (NY427) was purified from Escherichia coli containing the plasmid pET-NY427 (31). NY427 encodes an N-terminal hexahistidine tag and contains Mn2+-dependent RNase H activity. [γ-32P]ATP was purchased from ICN.
Oligonucleotides.
The RNA-DNA oligonucleotides 17 mer (5′ GUUCGGGCGCCACTGCT 3′), 14 mer (5′ CGGGCGCCACTGCT 3′), and 11 mer (5′ GCGCCACTGCT 3′) were synthesized by Integrated DNA Technologies (RNA sequences are indicated in boldface). The RNA-DNA oligonucleotides position 3 (5′ GUUCGGGCGUCACTGCT 3′), position 6 (5′ GUUCGUCGCCACTGCT 3′), position 4 and 6 (5′ GUUCGGUCUCCACTGCT 3′), MuLV-HIV hybrid 4&6 (5′ GACGAGGCGCCATTACT 3′), MuLV-HIV hybrid 4,6,&8 (5′ GACGGGGCGCCATTACT 3′), and MuLV-HIV hybrid 4,6,8,&9 (5′ GACCGGGCGCCATTACT 3′) were purchased from the DNA Synthesis Laboratory, Department of Biochemistry, University of Medicine and Dentistry of New Jersey (UMDNJ). The RNA oligonucleotides MPR-1 (5′ ATCCCGGACGAGCCCCCA 3′) and HPR-1 (5′ UCCCUGUUCGGGGCGCCA 3′) were synthesized by Integrated DNA Technologies. The DNA oligonucleotides 5331 (5′ AGCAGTGGCGCCCGAACGCGGGGCTTGTCCCT 3′), 6899 (5′ TCATTTGGCGCCCCGTC 3′), 6956 (5′ TCATTTGGCGCCCGGTC 3′), 7900 (5′ TCATTTGGCGCCTCGTC 3′), 6583 (5′ AGCAGTGGCGTCCGAAC 3′), 6582 (5′ AGCAGTGTCGCCCGTTC 3′), 6523 (5′ AGCAGTGGTGTCCGTTC 3′), 5193 (5′ GTCAGCGGGGGTCTTTCATTTGGGGGCTCGTCCGGGAT 3′), and HTD-1 (5′ GTGTGGAAAATCTCTAGCAGTGGCGCCCCGAACAGGGA 3′) were synthesized by the DNA Synthesis Laboratory, Department of Biochemistry, UMDNJ. The following BrdU oligonucleotides were purchased from the DNA Synthesis Laboratory, Department of Biochemistry, UMDNJ (the position mutated to BrdU is indicated by an X): 6429 (5′ AGCAGXGGCGCCCGAAC 3′), 6435 (5′ AGCAGTGXCGCCCGAAC 3′), 6428 (5′ AGCAGTGGXGCCCGAAC 3′), 6427 (5′ AGCAGTGGCGXCCGAAC 3′), 6426 (5′ AGCAGTGGCGCXCGAAC 3′), and 6425 (5′ AGCAGTGGCGCCXAAC 3′). The complementary DNA strands utilized in the mismatch assays were 6388 (5′ AGCAGTAGCGCCCGAAC 3′), 6341 (5′ AGCAGTGACGCCCGAAC 3′), 6387 (5′ AGCAGTGGAGCCCGAAC 3′), 6342 (5′ AGCAGTGGCACCCGAAC 3′), 6386 (5′ AGCAGTGGCGACCGAAC 3′), and 6385 (5′ AGCAGTGGCGCACGAAC 3′).
tRNA removal substrate preparation.
Substrates were prepared as previously described (28). Briefly, 20 pmol of each substrate was 5′ end labeled with [γ-32P]ATP, gel isolated, and eluted overnight (28). The labeled substrate was annealed to 20 pmol of the indicated annealing strand in a 30-μl reaction mixture containing 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 8 mM MgCl2, and 2 mM dithiothreitol (DTT). The mismatch substrates were prepared in the same manner. The 17 mer RNA-DNA hybrid was 5′ labeled and annealed to an oligonucleotide which would create a mismatch at the indicated position. The bromodeoxyuridine (BrdU)-mutagenized substrates were chemically synthesized to contain a BrdU at the indicated position. These DNA substrates were then annealed to the 17 mer RNA-DNA hybrid substrate as described above.
RNase H cleavage assays.
The annealed, RNA-DNA hybrid substrates were incubated with either HIV-1 RT, M-MuLV RT, or NY427 (an isolated RNase H domain) (31). Reaction mixtures (20 μl) contained approximately 4 pmol of substrate. The HIV-1 RT and M-MuLV RT reaction mixtures contained 50 mM Tris-HCl, pH 7.5, 50 mM KCl, 2 mM DTT, and 8 mM MnCl2. NY427 reaction mixtures contained N-morpholinoethanesulfonic acid (MES), pH 6.4, 0.2 mM DTT, and 8 mM MnCl2. For each experiment, 1 pmol of enzyme was used unless otherwise indicated. The reaction mixtures were all incubated at 37°C. Aliquots (3 μl) were removed at 0, 2, 5, 15, and 30 min. The reactions were stopped by the addition of formamide stop buffer. The reaction products were separated on 20% denaturing polyacrylamide gels.
RESULTS
Sequence-specific tRNA removal by an isolated RNase H domain.
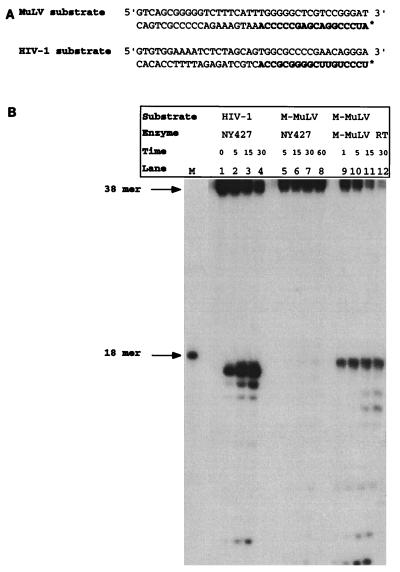
Figure 1A illustrates the model substrates utilized to characterize the specific cleavages of the isolated HIV-1 RNase H domain, NY427. The substrates model an intermediate of reverse transcription after plus-strand strong-stop synthesis. At this point, the first 18 nucleotides of the tRNA has been reverse transcribed, yielding an RNA-DNA hybrid. The tRNA primer is then removed by the RNase H domain of HIV-1 RT. The two model substrates utilized in this assay mimic either the M-MuLV intermediate containing tRNAPro or an HIV-1 intermediate, which utilizes tRNA3Lys. The RNA oligonucleotides are 5′ labeled, annealed to the complementary DNA strand (38 nucleotides long), and extended with Exonuclease(−) Klenow polymerase.
FIG. 1.
Sequence-specific tRNA removal by an isolated RNase H domain. (A) Model substrates utilized to mimic intermediates of M-MuLV and HIV-1 reverse transcription. The RNA portion of each substrate is indicated in boldface. The asterisk denotes the 5′ label. The RNA oligonucleotide was 5′ labeled with [γ-32P]ATP, annealed to a complementary DNA oligonucleotide, and extended with Exonuclease (−) Klenow polymerase. The resultant RNA-DNA hybrid was used in an RNase H cleavage assay. (B) RNase H cleavage assay of NY427, an HIV-1 isolated RNase H domain, with M-MuLV and HIV-1 model substrates. Lanes 1 to 5, NY427 with HIV-1 model substrate; lanes 6 to 10, NY427 incubated with M-MuLV model substrate; lanes 11 to 15, M-MuLV model substrate incubated with M-MuLV RT. Lane M contains the RNA marker, which is an 18-mer. Time points are indicated in minutes above each lane.
The recognitions of these RNA-DNA hybrid substrates by the NY427 RNase H domain (31) of HIV-1 RT were compared (Fig. 1B). Previous analysis of NY427 indicated that this isolated RNase H domain removes the HIV-1 tRNA primer with the same specificity as the p66-p51 full-length RT heterodimer (31), with the initial cleavage occurring between the 3′ C and A of the RNA moiety. With the model substrates shown in Fig. 1A, this specific cleavage would release a 5′-labeled 17-mer product. Figure 1B, lanes 1 to 4, shows a time course of the isolated NY427 with the cognate HIV-1 substrate. The 17 mer oligoribonucleotide is released as the initial and predominant product. This corresponds to cleavage between the 3′ C and A. In contrast, the HIV-1 isolated RNase H domain was incapable of cleaving the MuLV tRNAPro mimic model substrate (Fig. 1B, lanes 5 to 8). Control experiments indicated that the MuLV tRNAPro mimic substrate can be recognized by the MuLV RT-RNase H (Fig. 1B, lanes 9 to 12), verifying that the substrate is a functional RNA-DNA hybrid. The additional cleavage products observed represent the subsequent degradation of the RNA primer by the respective enzymes (Fig. 1B, lanes 3 to 4 and 11 to 12) (25). These results indicate that the isolated HIV-1 RNase H can discriminate between model substrates for the cognate and heterologous tRNAs.
Deletion analysis of model tRNA primer.
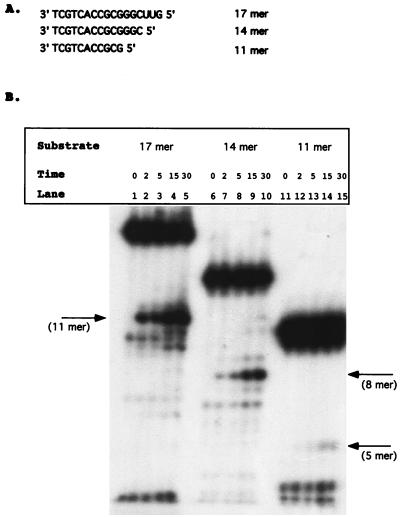
The ability of the isolated RNase H domain, NY427, to distinguish the first 18 nucleotides of the tRNA3Lys from those of tRNAPro implies a sequence of structural recognition of the substrate. The substrate requirements were therefore further defined. Previous data has shown that specific tRNA cleavage requires that the RNA be linked to DNA through a phosphodiester bond (32), with 5 DNA nucleotides being sufficient for specific removal of the tRNA primer mimic. Substrates were constructed which varied in the length of the RNA portions. Substrates contained either 12, 9, or 6 bases of the RNA primer plus 5 nucleotides of DNA and were termed 17 mer, 14 mer, and 11 mer, respectively (Fig. 2A). The RNA-DNA strands in these experiments were chemically synthesized, and the RNA portions were 5′ labeled with [γ-32P]ATP and annealed to the complementary strands. This protocol greatly facilitates the synthesis of the substrates and eliminates the need for extension with DNA polymerases.
FIG. 2.
Deletion analysis of model tRNA primer. (A) Substrates constructed to analyze the effects of deleting the PBS. The substrates are termed either 17 mer, 14 mer, or 11 mer. Deletion substrates were synthesized as RNA-DNA hybrids. The substrates were 5′ labeled with [γ-32P]ATP and annealed to oligonucleotide 5331 (see Materials and Methods). (B) Time course analysis performed with 1 pmol of the HIV-1 isolated RNase H domain, NY427. Lanes 1 to 5, 17 mer substrate; lanes 6 to 10, 14 mer substrate; lanes 11 to 15, 11 mer substrate. Time points are indicated in minutes above each lane. The arrows indicate the initial cleavage product of each deletion construct.
Figure 2B represents a time course of NY427 with the RNA deletion substrates. The 17 mer was capable of being cleaved by the isolated RNase H domain and produced a specific cleavage product between the first and second ribonucleotides (Fig. 2B, lanes 1 to 5). The 14 mer released an 8-mer-size product with kinetics similar to those of the 17 mer, corresponding with cleavage between the terminal ribonucleotides C and A (Fig. 2B, lanes 6 to 10). The kinetics of cleavage of 11 mer was much slower than those of 14 mer or 17 mer (Fig. 2B, lanes 11 to 15). A low level of product corresponding in length to 5-mer nucleotides was released after 15 min. This indicates that the six ribonucleotides are sufficient, but not optimal, for recognition by the isolated RNase H domain. HIV-1 RT was able to cleave these substrates at identical positions (data not shown). The site of cleavage was confirmed by comparison with E. coli RNase H digestion, which cleaves 1 nucleotide downstream from an RNA-DNA junction (3). Both E. coli RNase H and NY427 yielded identical initial products corresponding to the expected sizes according to an RNA ladder (data not shown). The deletion substrates were also incubated for 30 min in Mn2+ reaction buffer; the breakdown products are identical to those found at the zero time point (data not shown).
Cleavage analysis of mismatch substrates.
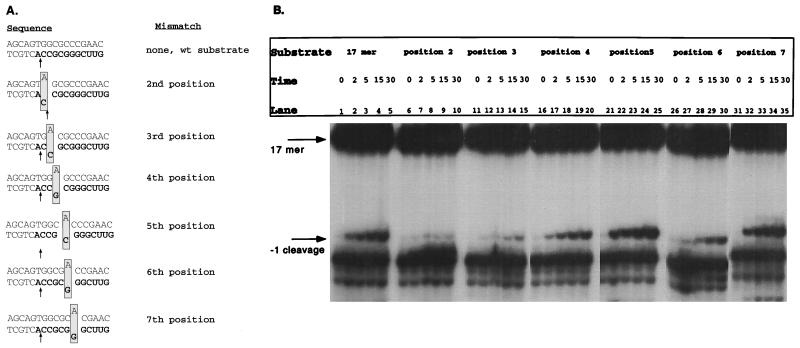
The deletion substrate analysis indicated that, minimally, the first six ribonucleotides were important for recognition and cleavage by the isolated HIV-1 RNase H domain, NY427; however, the first 9 nucleotides were required for optimal RNase H activity. These deletion studies, along with the comparison of HIV-1 and M-MuLV substrates (Fig. 1), indicate there may be particular positions which were key in recognition and cleavage by the HIV-1 isolated RNase H domain. To further investigate this possibility, mismatch substrates were constructed at positions 2 through 7 of the tRNA primer. The substrates were constructed by annealing a 5′ 32P-labeled 17 mer RNA-DNA oligonucleotide substrate with DNA oligonucleotides containing an adenine (A) at position 2, 3, 4, 5, 6, or 7. Mismatches were generated at these positions, which contain either guanidine (G) or cytosine (C) and would not form Watson-Crick base pairs with adenine (A) (Fig. 3A). Figure 3B shows the time course reactions of the mismatch substrates with the HIV-1 isolated RNase H domain, NY427. Incubation of NY427 with the wild-type substrate releases the expected 11 mer product, indicative of cleavage occurring between the terminal ribonucleotide A and ribonucleotide C (Fig. 3B, lanes 1 to 5). Mismatches at positions 5 (Fig. 3B, lanes 21 to 25) and 7 (Fig. 3B, lanes 31 to 35) had no effect on RNase H activity; the release of the product was similar to that with the wild-type substrate. Mismatches at positions 4 (Fig. 3B, lanes 16 to 20) and 6 (Fig. 3B, lanes 26 to 30) resulted in reductions in the overall yield of the initial cleavage product compared to the wild-type RNase H activity. It is of interest that within the first 6 nucleotides of the tRNA, tRNAPro and tRNA3Lys differ at positions 4 and 6 (Fig. 1A). Mismatches at positions 2 (Fig. 3B, lanes 6 to 10) and 3 (Fig. 3B, lanes 11 to 15) greatly altered RNase H activity. Cleavage normally occurs at the 3′ OH of the RNA at position 2. The mismatch at this position shifted the major cleavage site 1 nucleotide upstream, releasing an RNA product 1 nucleotide shorter than that released with the wild-type substrate. This product migrates slightly more slowly than a breakdown product of the substrate, present in the zero-time-point lanes in all of the assays (Fig. 3B, lanes 1, 6, 11, 16, 21, 26, and 31). The substrate containing a mismatch at position 3 resulted in highly diminished RNase H cleavage at the predicted cleavage site (Fig. 3B, lanes 11 to 15). These results indicated that changes at positions 2 and 3 are extremely detrimental to specific removal of the tRNA primer in an in vitro assay. These positions correspond to the CC of the CCA region, which is inherent to all tRNA primers. Along with positions 2 and 3, positions 4 and 6 produced an inhibitory effect on RNase H activity when they were in a mismatch orientation.
FIG. 3.
Cleavage analysis of mismatch substrates. (A) Wild-type (wt) and mismatch substrates utilized in this RNase H cleavage assay. The RNA-DNA hybrid substrates were 5′ labeled within the RNA portion (boldface). The hybrid was then annealed to a complementary DNA strand, which created a mismatch at a specific position. The mismatches are boxed. The annealing DNA oligonucleotides possess an adenine in position 2, 3, 4, 5, 6, or 7. The RNase H predominant cleavage observed for each substrate is indicated with an arrow. (B) RNase H cleavage assays of the mismatch substrates with the HIV-1 isolated RNase H domain, NY427 (1 pmol). Time courses were performed and are indicated in minutes above each lane. Mismatch activities (lanes 6 to 35) were compared to wild-type substrate activities (lanes 1 to 5).
BrdU mutagenesis of the tRNA primer.
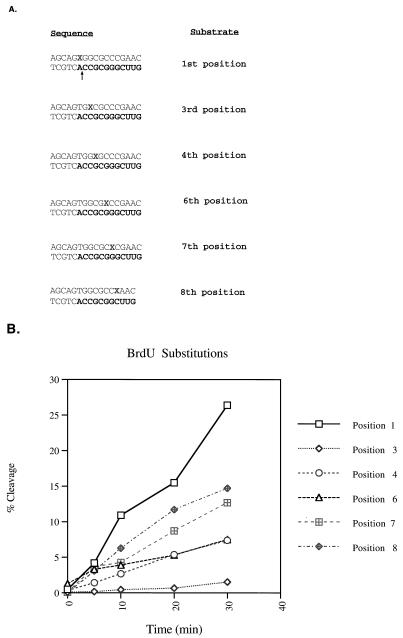
To further analyze the sequence requirements for removal of the model tRNA primer, BrdU mutagenesis was performed on positions 1, 3, 4, 6, 7, and 8. This approach similarly generates mismatches and allows for the comparison of an adenine mismatch to a uridine mismatch. The 17 mer RNA-DNA oligonucleotide substrate was 5′ labeled with [γ-32P]ATP and annealed to the complementary oligonucleotides synthesized with BrdU in either of the positions indicated in Fig. 4A. All of the BrdU substrates produced mismatches, with the exception of the one with BrdU at position 1, which can base pair with the complementary adenine. Analysis of the RNase H with BrdU at position 1 yielded maximal activity (Fig. 4B). All of the BrdU substitutions resulted in release of the 11-mer RNA product, which was quantitated as shown in Fig. 4B. The mismatch containing BrdU at position 3 had the most detrimental effect, yielding less than 6% of the cleavage of the similar substitution at position 1. Individual BrdU substitutions at positions 4 and 6 were hindered in their cleavages. Substitutions at positions 7 and 8 maintained at least 50% of the cleavage of the complementary substitution at position 1. These results confirm the mismatch results presented in Fig. 3, supporting the roles of positions 3, 4, and 6 in the recognition of the cognate tRNA-DNA substrate.
FIG. 4.
BrdU mutagenesis of the tRNA primer. (A) BrdU substrates. The substrates were synthesized with a BrdU in each position indicated by a boldface X. The BrdU is within the annealing strand and is annealed to a 5′-labeled RNA-DNA hybrid, 17 mer (Fig. 2). Mismatches were constructed at all positions except the second. The RNA portion of each substrate is in boldface. (B) Graphs of time course reactions of BrdU substrates assayed with the HIV-1 isolated RNase H domain, NY427. The initial cleavage products were quantified by phosphorimager analysis, and the percentage of the input substrate cleaved was determined. The legend at the right indicates the positions bearing the BrdU substitutions.
Substitution analysis of positions within the tRNA primer binding region.
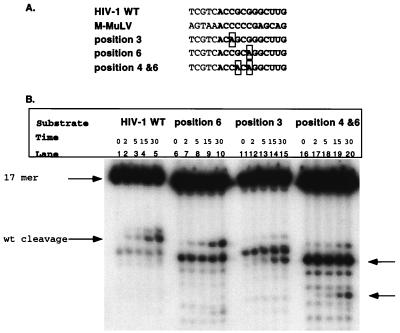
The mismatch data indicated that positions 2, 3, 4, and 6 were important in recognition and cleavage by the isolated RNase H domain. Further analysis aimed at creating complementary base substitutions rather than generation of mismatches. Initial analysis of substitutions of adenine, cytosine, and uracil for guanidine at position 4 indicated no changes in RNase H activity (data not shown). Therefore, substitution substrates were constructed at positions 3 and 6 and double-substitution substrates were prepared at positions 4 and 6. These substrates are illustrated in Fig. 5A. At each of the indicated positions, a uracil was substituted in the RNA portion and an adenine was substituted in the DNA portion of the substrate. This substitution in the RNA regenerates the Watson-Crick base pairing lost in the mismatch studies (Fig. 3) with base substitutions distinct from the wild-type sequences. These substrates were incubated with HIV-1 RNase H, and the initial and predominant cleavage products were compared with the wild-type substrate.
FIG. 5.
Substitution analysis of positions within the tRNA primer. (A) Substitution substrates. The boxed areas indicate the positions which are changed from the wild-type (WT) sequence. The substrates were prepared as described in the legend to Fig. 4. The RNA portion of each substrate is in boldface. (B) Time course reactions of the substitution substrates. The cleavage patterns are compared to that of the wild-type substrate (lanes 1 to 5). The reactions were performed as described in Materials and Methods. Lanes 6 to 10, substitution at position 6; lanes 11 to 15, substitution at position 3; lanes 16 to 20, changes at positions 4 and 6. The time is indicated in minutes above each lane. The arrows on the right indicate altered cleavage sites, with the lower arrow indicating random cleavage events. wt, wild type.
Figure 5B represents time course reactions of each substituted substrate incubated with NY427. A change at position 3 from cytosine to uracil greatly affected RNase H-specific cleavage, with a predominant cleavage product between the ribonucleotides A and G, 3 nucleotides downstream of the RNA-DNA junction (Fig. 5B, lanes 11 to 15). This indicates that position 3 is very significant for specific removal of the tRNA primer between the terminal ribonucleotide A and ribonucleotide C. The alteration at position 6 (Fig. 5B, lanes 6 to 10) had no affect on specific RNase H activity. This is similar to the single base changes at position 4, which also had no affect on RNase H activity. The double-substitution substrate at positions 4 and 6 had the most dramatic effect on specific RNase H activity (Fig. 5B, lanes 16 to 20). No specific cleavages were detected, only random cleavage events. This data indicates that positions 4 and 6 combined are important for recognition and cleavage by the isolated RNase H domain; however, individually they have no effect on RNase H activity.
M-MuLV–HIV-1 hybrid analysis.
Results presented in Fig. 1 indicated that the isolated HIV-1 RNase H, NY427, was incapable of recognizing the M-MuLV model substrate, despite the conservation of the CCA region containing the scissile bond. Further characterization of the HIV-1 substrate had indicated a role of positions 4 and 6 in the specific recognition of the tRNA3Lys mimic. It was therefore of interest to identify substitutions in the MuLV substrate which could support the recognition of this substrate by the HIV-1 RNase H.
Deletion substrate analysis (Fig. 2) indicated that the optimal recognition of the HIV substrate was within the first 12 nucleotides of the RNA. Also, the 14 mer construct possessed kinetics similar to those of the 17 mer construct. Substitution studies as well as mismatch studies had indicated the importance of positions 4 and 6. Hybrid MuLV-HIV substrates were therefore generated in which HIV sequences were introduced into the MuLV RNA at sites between positions 3 and 9 of the RNA.
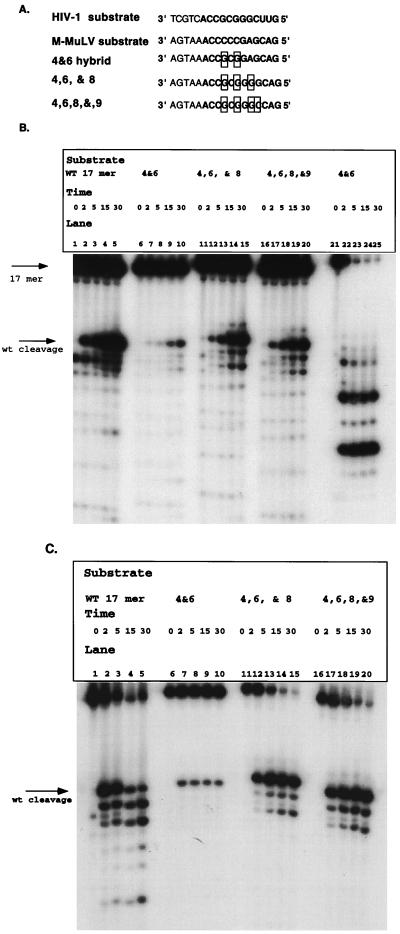
The prior substitution analysis had been performed with substrates that were 17-mers; therefore, hybrid analysis was performed with substrates of this size. Figure 6A shows the hybrid substrates constructed. The 4&6 hybrid was synthesized as a 17-mer to determine whether it would be sufficient to regain cleavage. Two additional hybrids were synthesized, 4,6,&8 and 4,6,8,&9. This allowed us to determine the optimal sequence requirements for specific recognition and removal of the RNA primer. These substrates were prepared in the same manner as the 17 mer substrate in the deletion studies.
FIG. 6.
Extensive M-MuLV–HIV-1 hybrid analysis with 17 mer construct. (A) Hybrid substrates constructed. The boxed regions are those which have been changed from the M-MuLV sequence to the HIV-1 sequence. The substrates were prepared as described in the legends to Fig. 1 to 5. The RNA portion of each substrate is in boldface. (B) Time course analysis of M-MuLV–HIV-1 hybrid substrates 4&6 (lanes 6 to 10), 4,6,&8 (lanes 11 to 15), and 4,6,8&9 (lanes 16 to 20) with the HIV-1 isolated RNase H domain. Time points (in minutes) and constructs are indicated above the lanes. Initial cleavage products, amounts, and specificities were compared to those of the wild-type (WT) 17 mer (lanes 1 to 5). Lanes 21 to 25 represent hybrid 4&6 digested with E. coli RNase H. (C) Analysis of RNase H cleavage of M-MuLV–HIV-1 hybrid substrate with HIV-1 RT. The hybrid substrates were incubated with 1 pmol of HIV-1 RT for 0, 2, 5, 15, and 30 min. The times and the hybrids analyzed are indicated above the lanes. wt, wild type.
Figure 6B shows an analysis of reactions of these hybrid substrates with the isolated RNase H domain. These reaction products were compared with those of wild-type 17 mer substrate. Each of the hybrid substrates was specifically recognized and cleaved by the isolated RNase H domain. The 4,6,&8 (lanes 11 to 15) and 4,6,8,&9 hybrids (Fig. 6B, lanes 16 to 20) possessed cleavage patterns identical to that of the wild-type 17 mer substrate (Fig. 6B, lanes 1 to 5). The 4&6 hybrid (Fig. 6B, lanes 6 to 10) was cleaved kinetically more slowly than the wild-type construct, with kinetics similar to those with 11 mer (Fig. 2). The 4&6 hybrid was confirmed to be a hybrid by digestion with E. coli RNase H (Fig. 6B, lanes 21 to 25).
The recognition of these substrates by the RT-associated RNase H domain was also tested. Previous data had shown that HIV-1 RT cleaves M-MuLV model substrate at the RNA-DNA junction (28), whereas it cleaves its cognate substrate between the terminal ribonucleotide A and ribonucleotide C. Reactions were performed in which these hybrid substrates were incubated with HIV-1 RT to determine if the initial cleavage product produced is 1 base larger for the hybrid substrate than for the wild-type 17 mer substrate. These reactions are shown in Fig. 6C. Hybrid 4&6 (Fig. 6C, lanes 6 to 10), hybrid 4,6,&8 (Fig. 6C, lanes 11 to 15), and hybrid 4,6,8,&9 (Fig. 6C, lanes 16 to 20) produced initial cleavage products at the same position as did the wild-type HIV-1 substrate (Fig. 6C, lanes 1 to 5). The efficiency of cleavage of 4&6 alone is lower than those of the other substrates, and no additional RNase H cleavage products were detected. Interestingly, none of the substrates produced cleavage products indicative of cleavage occurring at the RNA-DNA junction. This indicates that positions 4 and 6 in combination contribute to the recognition and cleavage of the tRNA3Lys mimic.
DISCUSSION
With model substrates for plus-strand strong-stop products, the data illustrates that the HIV-1 isolated RNase H domain, NY427, is capable of selectively recognizing and cleaving substrates containing tRNA3Lys sequences from those containing tRNAPro sequences. Mismatch analysis and BrdU mutagenesis provided a scanning mechanism to determine the positions important for recognition by the isolated RNase H domain. These positions were further investigated by creating substitution and hybrid substrates to directly determine the sites required to retain initial cleavage between the terminal ribonucleotide A and ribonucleotide C. Positions 2, 3, 4, and 6 were determined to be crucial in recognition. Positions 4 and 6 differ between tRNA3Lys and tRNAPro.
The isolated HIV-1 RNase H appears distinct in its ability to specifically recognize its cognate tRNA for primer removal. Expression of the M-MuLV RNase H domain was reported to have little specificity (26, 41). This parallels the characteristics of the enzymes in vivo. For M-MuLV, the site of the initial cleavage is not the final cleavage product; the terminal ribonucleotide A from the tRNA is ultimately removed before completion of the viral replication (28). In contrast, the initial site of tRNA removal catalyzed by the HIV-1 RNase H is extremely stable. The terminal ribonucleotide A remains associated with the viral DNA and can be amplified within the circle junctions isolated from infected cells (8, 9, 12, 16, 30). The site of cleavage by the isolated RNase H domain is identical to that of the full-length protein. This implies an innate recognition within the RNase H domain for the sequence and/or the structure of the tRNA-DNA hybrid. This recognition cannot be ascribed to the histidine tag, since the same enzyme can differentiate between the two tRNA mimic substrates. Further studies are required to identify the domain of the protein involved in this recognition.
These studies cannot address whether it is sequence or structural recognition by the RNase H. The structure of these RNA-DNA junctions may be an important determining factor in the recognition and cleavage mechanism of RNase H enzymes. Structural analyses of related RNA-DNA hybrids indicate that the hybrids are not classic A or B structures but rather a combination of both forms, particularly around the RNA-DNA junctions (5). Studies have also shown that there is diversity in the sugar conformations at the RNA-DNA junction which may be affecting RNase H activity (23). It is quite possible that sequence can influence structure, particularly at a transition junction point. It is of interest that the identified positions which alter the cleavage recognition are at a distance from the scissile bond, up to 5 nucleotides away. This implies either that a secondary region of the RNase H rather than the active site interacts with these nucleotides or that these sequences greatly influence the structure of the junction. For both tRNAPro and tRNA3Lys, the 6 base pairs 5′ of the cleavage site are within GC base pairs. The substrates utilize an RNA primer which mimics the first 18 nucleotides of tRNA3Lys. The effect of any of these changes within the context of the natural tRNA3Lys for tRNA removal is being explored.
The first 3 nucleotides of tRNAPro and tRNA3Lys are encoded by the CCA added posttranscriptionally to all tRNAs. Alterations within this region, at position 2 or 3, either by mismatch or base substitution, altered the cleavage site or greatly reduced the efficiency of cleavage by RNase H. The CCA sequence is present in all tRNA primers, and these alterations do not explain the inability of the isolated HIV-1 RNase H domain to cleave the M-MuLV model substrate. Positions 4 and 6 defined the switch between the cognate and heterologous tRNA substrates. These were the positions which differed between M-MuLV and HIV-1 tRNAs, within the first 7 nucleotides; tRNA3Lys encodes G at both positions 4 and 6, while tRNAPro contains C at both sites. In fact, insertion of AU base pairs at both these positions resulted in completely random RNase H cleavages. Single substitutions at either position 4 or 6 were tolerated. Initial studies of position 4 indicated that changes from a CG base pair to any other combination did not produce an effect. This was not systematically explored with respect to position 6.
The mismatch, BrdU mutagenesis, and substitution analyses were done with NY427 as well as HIV-1 RT. Studies with the HIV-1 RT (data not shown) have indicated that positions 3 and 6 yield alternative cleavage patterns with mismatch, substitution, and BrdU mutagenesis. Cleavage occurred predominantly between positions 3 and 4 within the tRNA primer. Substitutions at both 4 and 6 yielded cleavage at an alternative site. BrdU substitution at position 4 decreased the yield of cleavage at the predicted site. As controls, mismatches at position 5 or position 7 did not alter recognition by the full-length HIV-1 RT. These results indicate that alterations in the RNA sequences encoded in the tRNA alter the recognition of both the isolated RNase H domain and the full-length HIV-1 RT.
Hybrid studies were also performed to determine what was necessary to gain recognition and cleavage of the M-MuLV substrate by the HIV-1 isolated RNase H domain. Hybrid substrates were constructed based on the results with the deletion substrates and the substitution substrates. Other studies analyzing initiation of reverse transcription indicated that the first 6 nucleotides of the tRNA primer were required (22). Therefore, a 4&6 hybrid, as well as 4,6,&8 and 4,6,8,&9 hybrids, was constructed to determine the optimal and sufficient substrates for RNase H activity. The deletion data indicated an 11-mer was sufficient but the 14-mer was optimal. The hybrid data confirmed these results; a 4&6 hybrid was cleavable, but the 4,6,&8 and 4,6,8,&9 hybrids were cleaved more efficiently. This would indicate that for the 17 mer constructs, positions 4, 6, and 8 are important for optimal recognition and cleavage, whereas positions 4 and 6 are sufficient.
Currently, many studies point to the interactions between the polymerase and RNase H domains. Primer grip mutants which have altered RNase H activity have been isolated (6, 15, 17, 19). It would be of interest to determine if these mutations affect the specificity of the RNase H domain on small defined substrates, such as those described in this study for tRNA removal. It is possible that these mutants have diminished RNase H activity due to the inability to position the substrate within the primer-template groove. This may not alter the ability of the small targeted RNase H substrates to bind independently of the polymerase domain. Although the polymerase domain does not affect the RNase H cleavage of the cognate tRNA, the influence on a heterologous substrate is evident. The isolated HIV-1 RNase H is not capable of recognizing the M-MuLV substrate, yet in the presence of the polymerase domain, the HIV-1 RT cleavage occurs at the RNA-DNA junction (27, 28).
Studies have been performed to assess the selection of the tRNA for initiation of reverse transcription (10, 11, 35–37). These studies have highlighted the importance of the PBS as well as a region complementary to the anticodon loop within U5. Modified viruses with altered PBSs and U5 A loops complementary to tRNAPro, tRNATrp, tRNAMet, and tRNAHis have been found which support initiation (10, 11, 35). Slow reversion is still detected for tRNAPro and tRNATrp, indicating that the A loop and PBS are not the sole determinants for selection of specific tRNA for viral replication (11). It is interesting that of these alternative tRNAs, tRNAHis and tRNAMet are the most stable (10, 35), and they contain 3′ termini, predicted by this study to be efficiently removed by the HIV-1 RNase H.
RNase H cleavages can occur in a polymerase-dependent or -independent mode (13, 38, 39). In the polymerase dependent mode, the RNase H cleavage lags 18 to 20 nucleotides behind the 3′ OH position in the polymerization site. In the studies described here, the cleavages occur independently of the presence of the polymerase domain. In addition, several of the truncated substrates maintain distances from the potential 3′ OH, through the RNA-DNA hybrid, shorter than the distance between the polymerase and RNase H active sites. These truncated substrates can be cleaved by the full-length HIV-1 RT (data not shown), indicating that the cleavages may occur in a polymerase-independent mode for the wild-type HIV-1 RT.
ACKNOWLEDGMENTS
This work was supported by NIH grant RO1-GM51151 and the NSF international program NSF-INT 9408501/Fundacion Andes (travel grant).
REFERENCES
- 1.Arts E J, Le Grice S F. Interaction of retroviral reverse transcriptase with template-primer duplex during replication. Prog Nucleic Acid Res Mol Biol. 1998;58:339–393. doi: 10.1016/s0079-6603(08)60041-0. [DOI] [PubMed] [Google Scholar]
- 2.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumor viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 3.Berkower I, Leis J, Hurwitz J. Isolation and characterization of an endonuclease from Escherichia coli specific for ribonucleic acid in ribonucleic acid-deoxyribonucleic acid hybrid structures. J Biol Chem. 1973;248:5914–5921. [PubMed] [Google Scholar]
- 4.Champoux J J, Gilboa E, Baltimore D. Mechanism of RNA primer removal by the RNase H activity of avian myeloblastosis virus reverse transcriptase. J Virol. 1984;49:686–691. doi: 10.1128/jvi.49.3.686-691.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Federoff O Y, Salazar M, Reid B R. Structural variation among retroviral primer-DNA junctions: solution of the HIV-1 (−)-strand Okazaki fragment r(gcca)d(GCAGTGGC) Biochemistry. 1996;35:11070–11080. doi: 10.1021/bi9607822. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh M, Jacques P S, Rodgers D W, Ottman M, Darlix J, LeGrice S F. Alterations in the primer grip of p66 HIV-1 reverse transcriptase and their consequences for primer utilization. Biochemistry. 1996;35:8553–8562. doi: 10.1021/bi952773j. [DOI] [PubMed] [Google Scholar]
- 7.Guo J, Wu W, Yuan Z Y, Post K, Crouch R J, Levin J G. Defects in primer-template binding, processive DNA synthesis, and RNase H activity associated with chimeric reverse transcriptases having the murine leukemia virus polymerase domain joined to the Escherichia coli RNase H. Biochemistry. 1995;34:5018–5029. doi: 10.1021/bi00015a013. [DOI] [PubMed] [Google Scholar]
- 8.Hong T, Drlica K, Pinter A, Murphy E. Circular DNA of human immunodeficiency virus: analysis of circle junction nucleotide sequences. J Virol. 1991;65:551–555. doi: 10.1128/jvi.65.1.551-555.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jurriaans S, Ronde A D, Dekker J, Goudsmit J, Cornellissen M. Analysis of human immunodeficiency virus type 1 LTR-LTR junctions in peripheral blood mononuclear cells of infected individuals. J Gen Virol. 1992;73:1537–1541. doi: 10.1099/0022-1317-73-6-1537. [DOI] [PubMed] [Google Scholar]
- 10.Kang S-M, Zhang Z, Morrow C D. Identification of a sequence within U5 required for human immunodeficiency virus type 1 to stably maintain a primer binding site complementary to tRNAMet. J Virol. 1997;71:207–217. doi: 10.1128/jvi.71.1.207-217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang S M, Wakefield J K, Morrow C D. Mutations in both the U5 region and the primer-binding site influence the selection of the tRNA used for the initiation of HIV-1 reverse transcription. Virology. 1996;222:401–404. doi: 10.1006/viro.1996.0437. [DOI] [PubMed] [Google Scholar]
- 12.Kulkosky J, Katz R A, Skalka A M. Terminal nucleotides of the preintegrative linear form of HIV-1 DNA deduced from the sequence of circular DNA junctions. J Acquired Immune Defic Syndr. 1990;3:852. [PubMed] [Google Scholar]
- 13.Metzger W, Hermann T, Schatz O, LeGrice S F J, Heumann H. Hydroxyl radical footprint analysis of human immunodeficiency virus reverse transcriptase-template primer complexes. Proc Natl Acad Sci USA. 1993;90:5909–5913. doi: 10.1073/pnas.90.13.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omer C A, Faras A J. Mechanism of release of the avian retrovirus tRNA Trp primer molecule from viral DNA by ribonuclease H during reverse transcription. Cell. 1982;30:797–805. doi: 10.1016/0092-8674(82)90284-7. [DOI] [PubMed] [Google Scholar]
- 15.Palaniappan C, Wisniewski M, Jacques P S, LeGrice S F, Fay P J, Bambara R A. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J Biol Chem. 1997;272:11157–11164. doi: 10.1074/jbc.272.17.11157. [DOI] [PubMed] [Google Scholar]
- 16.Pauza C D. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology. 1990;179:886–889. doi: 10.1016/0042-6822(90)90161-j. [DOI] [PubMed] [Google Scholar]
- 17.Powell M D, Ghosh M, Jacques P S, Howard K J, LeGrice S F, Levin J G. Alanine-scanning mutations in the “primer grip” of p66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J Biol Chem. 1997;272:13262–13269. doi: 10.1074/jbc.272.20.13262. [DOI] [PubMed] [Google Scholar]
- 18.Pullen K A, Ishimoto L K, Champoux J J. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1992;66:367–373. doi: 10.1128/jvi.66.1.367-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rausch J W, LeGrice S F J. Substituting a conserved residue of the ribonuclease H domain alters substrate hydrolysis by retroviral reverse transcriptase. J Biol Chem. 1997;272:8602–8610. doi: 10.1074/jbc.272.13.8602. [DOI] [PubMed] [Google Scholar]
- 20.Repaske R, Hartley J W, Kavlick M F, O’Neill R R, Austin J B. Inhibition of RNase H activity and viral replication by single mutations in the 3′ region of Moloney murine leukemia virus reverse transcriptase. J Virol. 1989;63:1460–1464. doi: 10.1128/jvi.63.3.1460-1464.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Resnick R, Omer C A, Faras A J. Involvement of retrovirus reverse transcriptase-associated RNase H in the initiation of strong-stop (+) DNA synthesis and the generation of the long terminal repeat. J Virol. 1984;51:813–821. doi: 10.1128/jvi.51.3.813-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhim H, Park J, Morrow C D. Deletions in the tRNALys primer-binding site of human immunodeficiency virus type 1 identify essential regions for reverse transcription. J Virol. 1991;65:4555–4564. doi: 10.1128/jvi.65.9.4555-4564.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salazar M, Champoux J J, Reid B R. Sugar conformations at hybrid duplex junctions in HIV-1 and Okazaki fragments. Biochemistry. 1993;32:739–744. doi: 10.1021/bi00054a002. [DOI] [PubMed] [Google Scholar]
- 24.Schatz O, Cromme F V, Gruninger-Leitch F, LeGrice S F J. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989;237:311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- 25.Schatz O, Mous J, LeGrice S F J. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3′-5′ exonuclease activity. EMBO J. 1990;9:1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz S J, Champoux J J. RNase H domain of Moloney murine leukemia virus reverse transcriptase retains activity but requires the polymerase domain for specificity. J Virol. 1996;70:8630–8638. doi: 10.1128/jvi.70.12.8630-8638.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz S J, Whiting S H, Champoux J J. Cleavage specificities of Moloney murine leukemia virus RNase H implicated in the second strand transfer during reverse transcription. J Biol Chem. 1995;270:24135–24145. doi: 10.1074/jbc.270.41.24135. [DOI] [PubMed] [Google Scholar]
- 28.Smith C M, Potts III W B, Smith J S, Roth M J. RNase H cleavage of tRNAPro mediated by M-MuLV and HIV-1 reverse transcriptases. Virology. 1997;229:437–446. doi: 10.1006/viro.1997.8454. [DOI] [PubMed] [Google Scholar]
- 29.Smith J S, Gritsman K, Roth M J. Contributions of DNA polymerase subdomains to the RNase H activity of HIV-1 reverse transcriptase. J Virol. 1994;68:5721–5729. doi: 10.1128/jvi.68.9.5721-5729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith J S, Kim S, Roth M J. Analysis of long terminal repeat circle junctions of human immunodeficiency virus type 1. J Virol. 1990;64:6286–6290. doi: 10.1128/jvi.64.12.6286-6290.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith J S, Roth M J. Purification and characterization of an active human immunodeficiency virus type 1 RNase H domain. J Virol. 1993;67:4037–4049. doi: 10.1128/jvi.67.7.4037-4049.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith J S, Roth M J. Specificity of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H in removal of the minus-strand primer, tRNALys3. J Biol Chem. 1992;267:15071–15079. [PubMed] [Google Scholar]
- 33.Tanese N, Goff S P. Domain structure of Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci USA. 1988;85:1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tisdale M, Schulze T, Larder B A, Moelling K. Mutations within RNase H domain of HIV-1 reverse transcriptase abolish virus infectivity. J Gen Virol. 1991;72:59–66. doi: 10.1099/0022-1317-72-1-59. [DOI] [PubMed] [Google Scholar]
- 35.Wakefield J K, Morrow C D. Mutations within the primer binding site of the human immunodeficiency virus type 1 define sequence requirements essential for reverse transcription. Virology. 1996;220:290–298. doi: 10.1006/viro.1996.0317. [DOI] [PubMed] [Google Scholar]
- 36.Wakefield J K, Rhim H, Morrow C D. Minimal sequence requirements of a functional human immunodeficiency virus type 1 primer binding site. J Virol. 1994;68:1605–1614. doi: 10.1128/jvi.68.3.1605-1614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitcomb J M, Ortiz-Conde B A, Hughes S H. Replication of avian leukosis viruses with mutations at the primer binding site: use of alternative tRNAs as primers. J Virol. 1995;69:6228–6238. doi: 10.1128/jvi.69.10.6228-6238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wohrl B M, Ehresman B, Keith G, LeGrice S F. Nuclease footprinting of human immunodeficiency virus/tRNA(Lys-3) complex. J Biol Chem. 1993;268:13617–13624. [PubMed] [Google Scholar]
- 39.Wohrl B M, Georgiadis M M, Telesnitsky A, Hendrickson W A, LeGrice S F. Footprint analysis of replicating murine leukemia virus reverse transcriptase. Science. 1995;267:96–99. doi: 10.1126/science.7528942. [DOI] [PubMed] [Google Scholar]
- 40.Wohrl B M, Tantillo C, Arnold E, LeGrice S F. An expanded model of replicating human immunodeficiency virus reverse transcriptase. Biochemistry. 1995;34:5343–5356. doi: 10.1021/bi00016a005. [DOI] [PubMed] [Google Scholar]
- 41.Zhan X, Crouch R J. The isolated RNase H domain of murine leukemia virus reverse transcriptase. Retention of activity with concomitant loss of specificity. J Biol Chem. 1997;272:22023–22029. doi: 10.1074/jbc.272.35.22023. [DOI] [PubMed] [Google Scholar]