Abstract
An alternatively spliced form of c-myb exists that encodes an additional 120 amino acids in chicken and 121 amino acids in human and mouse. These amino acids are encoded by an additional exon, termed exon 9A. This exon is not present in v-myb, and proteins containing these amino acids have never been tested for oncogenic transformation. A series of myb constructs was therefore created in order to compare the functions of Myb proteins on the basis of their inclusion or exclusion of the amino acids encoded by exon 9A (E9A). We found that the presence of E9A resulted in a robust increase in transactivation for full-length c-Myb (CCC), as well as the singly truncated derivatives dCC and CCd, while doubly truncated Myb proteins v-Myb (dVd) and dCd did not exhibit any differences in transactivation. The increase in transactivation requires the Myb DNA-binding domain. When the leukemic transformation by the Myb proteins was tested, it was found that cells transformed by dVd resembled monoblasts, while cells transformed by CCC and its derivatives, dCd, dCC, and CCd, resembled myelomonoblasts. Despite differences in the morphology of the hematopoietic cells, the cell surface phenotypes and cell cycle profiles of transformed cells did not change for each pair of Myb proteins in the presence or absence of E9A. Thus, there was no direct correlation between the level of transcriptional activation and the strength of leukemic transformation.
Myb proteins were discovered through studies of avian myeloblastosis virus (AMV), a retrovirus that causes acute monoblastic leukemia in chickens. It was demonstrated that AMV contained an oncogene, v-myb, whose counterpart in the host cell was the proto-oncogene c-myb (reviewed in reference 43). Another retrovirus, E26, causes erythroid leukemia in vivo and transforms multipotent hematopoietic precursors in culture; this retrovirus contains v-myb fused in frame to a second oncogene, v-ets. In mice, infection with murine leukemia viruses can lead to myeloid leukemias that result from viral insertions into the c-myb locus.
The proto-oncogene c-myb encodes a 75-kDa nuclear protein with homologs in organisms including human, mouse, chicken, sea urchin, and Drosophila. The protein contains three functional domains: (i) the DNA-binding domain, (ii) the transactivation domain, and (iii) the negative regulatory domain (48). The DNA-binding domain is located at the N terminus and consists of three imperfect repeats, each of which folds into a helix-turn-helix motif (45). Downstream of the DNA-binding domain is the transactivation domain which contains acidic amino acids, as well as other conserved motifs (8, 37, 64). The C-terminal portion of c-Myb contains the negative regulatory domain including a heptad leucine repeat (35). However, sequences around this leucine repeat are required for transcriptional activation and leukemic transformation by v-Myb (23). It has been postulated that inter- and intramolecular interactions involving the negative regulatory domain are required for the modulation of Myb function (11, 14, 20).
The v-Myb oncoprotein from AMV contains N- and C-terminal truncations relative to c-Myb. The N-terminal truncation removes the first 71 amino acids of the protein including the majority of the first repeat of the DNA-binding domain. The C-terminal truncation removes the last 199 amino acids of the protein including much of the negative regulatory domain, although the heptad leucine repeat remains intact (Fig. 1). Ten amino acid substitutions are also present in v-Myb. Although these substitutions determine the lineage of the transformed cell, they are not required for transformation (12, 31, 60). Overexpression of c-Myb in hematopoietic cells, as well as truncated forms of c-Myb, are also sufficient for transforming different cell lineages in vitro, albeit with variable efficiency (13, 21, 22, 24). One possible explanation for these observed differences may be that the presence of the truncations and/or amino acid substitutions in v-Myb and/or c-Myb allows them to regulate distinct sets of target genes. Thus far, the mim-1 gene, which is expressed in immature granulocytes, is the best-characterized target gene for v-Myb (from E26 but not AMV) as well as c-Myb (43, 44). Recently, GBX2, tom-1, and the adenosine receptor 2B gene were identified as regulatory targets for v-Myb (6, 36, 65). Other genes that are targets for regulation by c-Myb include c-kit, CD34 (Sca-1), ADA (adenosine deaminase), and CD4 (18, 29, 41, 57).
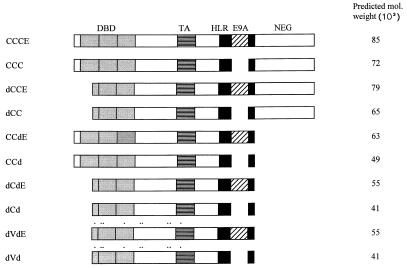
FIG. 1.
Schematic diagrams of Myb proteins used in this study. Comparisons are made between two sets of proteins: one that includes the amino acids encoded by exon 9A, and one that excludes the amino acids encoded by exon 9A. The three repeats of the DNA-binding domain (DBD) are shown in light gray; the horizontally striped box represents the acidic region of the transactivation domain (TA); the heptad leucine repeat region (HLR) is shown as a black box; the E9A sequence is shown as a hatched box. The remaining Myb sequences, including the negative regulatory domain (NEG), are shown as white boxes. CCCE refers to full-length c-Myb that includes E9A. Also shown here are four other pairs of proteins that correspond to the presence or absence of E9A. The proteins dVd (v-Myb) and dVdE contain nine amino acid substitutions (marked by black dots); dCd contains the same N- and C-terminal truncations as dVd but lacks the amino acid substitutions; dCC and CCd contain the N-terminal and C-terminal truncations, respectively, and also do not contain the amino acid substitutions.
Although c-Myb expression has been reported in the skin, colon, respiratory tract, retina, testes, primary fibroblasts, and endothelial cells, the protein is most readily detected in immature hematopoietic precursors at high levels. These levels of c-Myb decrease as cells progress toward terminal differentiation (17, 27, 54, 63). Alterations in levels of Myb proteins result in perturbations in hematopoietic development. Constitutive expression of c-Myb has been shown to block differentiation of erythroid and myeloid cell lines, implying that down-regulation of c-Myb expression is necessary for terminal differentiation of these cell types (4, 5, 9, 53, 59, 62). In vivo studies demonstrate that mice that are homozygous mutants for c-Myb die at day 15 of embryogenesis. The embryos suffer from a lack of erythrocytes and myeloid cells in the fetal liver and blood; no other developmental abnormalities were observed (42). Transgenic mice expressing a dominant negative form of c-Myb in T cells undergo severe disruption of T-cell development and have far fewer mature T lymphocytes than do nontransgenic mice (2). Conversely, overexpression of v-Myb in T cells of transgenic mice leads to lymphomas that are made up predominantly of immature T cells (3). Thus, the importance of c-Myb in regulatory events involved with hematopoietic differentiation and proliferation has been established both in vitro and in vivo.
Although the effects of Myb proteins in hematopoiesis have been shown in a number of different studies, the mechanisms by which the Myb proteins regulate cellular events are still unclear. Intracellular pathways containing upstream factors responsible for regulating c-Myb, as well as downstream c-Myb target genes, have not been well characterized. The role of c-Myb in hematopoietic differentiation became even more complex with the discovery of an alternatively spliced form of c-Myb in mice that encodes a larger protein of ∼89 kDa (15, 46, 56). This alternatively spliced form of c-Myb is also found in human and chicken cells (10, 51). The transcript encoding the larger protein contains an additional exon, termed exon 9A (E9A), which is located downstream of the transactivation domain, disrupts the heptad leucine repeat, and encodes an additional 121 amino acids in humans and mice or 120 amino acids in chickens (Fig. 1). Interestingly, the E9A sequence is not present in AMV or E26. Sequences related to E9A are also found in other Myb-related proteins including A-Myb and B-Myb from vertebrates and Myb proteins from sea urchin and Drosophila. Based on Northern and Western blots of primary yolk sac cells, bone marrow cells, and established cell lines, it was demonstrated that c-Myb containing E9A is coexpressed at a level of 15 to 20% compared to the more dominant form of c-Myb lacking E9A (15, 50–52, 56). Both protein isoforms localize to the nucleus, but the specific function of the protein encoded by the alternatively spliced form of c-myb is unknown.
The mechanism of alternative splicing to create proteins with contrasting functions plays important roles in regulating the development and growth of various organisms. The isoforms of some proteins are expressed at constant levels, but more commonly, expression of these isoforms is regulated at specific points in the development of a particular cell. For example, in Drosophila melanogaster alternatively spliced forms of the doublesex (dsx) gene produce proteins that differ in their carboxyl-terminal regions and determine sexual differentiation in somatic tissues. The doublesex protein in males represses transcription, whereas the doublesex protein in females activates transcription, even though both isoforms recognize the same DNA-binding site in the same target genes. Consequently, expression of sex-specific genes, such as the yolk protein genes, is activated by doublesex (female) and repressed by doublesex (male) (reviewed in reference 39). One could imagine that the alternatively spliced forms of c-Myb also have different functions; the two isoforms may modulate the development of distinct lineages of cells by regulating different sets of target genes. It is also possible that the functions of both isoforms overlap at one point in development, which could result in one isoform influencing the expression level and activity of the other isoform. Therefore, we conducted experiments to determine how the presence of the amino acids corresponding to E9A in various Myb proteins affects transcriptional activation and hematopoietic differentiation.
MATERIALS AND METHODS
Plasmid constructions.
DNA restriction and DNA-modifying enzymes were purchased from New England Biolabs (Beverly, Mass.). To create sp73Iccd, plasmid sp73Iccc (which contains an encephalomyocarditis virus internal ribosomal entry site [IRES] element and the complete chicken c-Myb open reading frame) was digested with MscI/BamHI, and a 1,070-bp fragment corresponding to the C-terminal portion of c-Myb was removed. The plasmid was then treated with Klenow enzyme in order to fill in the BamHI overhang, and the plasmid was then self-ligated to create sp73Iccd; this plasmid was then digested with SalI/XbaI in order to isolate a 375-bp fragment encoding the newly modified 3′ end. This fragment was then ligated into the SalI/XbaI sites of sp73Idcd and sp73Idvd so that all myb constructs containing a C-terminal truncation had identical 3′ ends. To isolate the E9A fragment, the bursas from chicken embryos were used as the source of RNA, which was isolated by the Trizol extraction procedure. Poly(A)+ mRNA was then selected by using an Oligotex mRNA kit (Qiagen Inc., Chatsworth, Calif.), and reverse transcription-PCR was carried out to amplify the 770-bp fragment which contains E9A sequence flanked by c-myb sequence. PCR primers were designed such that the Bsu36I and MscI sites flanking E9A on either side were preserved. This fragment was then cloned into a pCR-Script SK(+) plasmid by using a pCR-Script SK(+) cloning kit (Stratagene, La Jolla, Calif.). The E9A sequence was verified with a Sequenase kit (United States Biochemical Corp., Cleveland, Ohio). A 542-bp fragment, encoding E9A and flanking c-myb sequences, was isolated after digestion with Bsu36I and MscI; this fragment was cloned into the Bsu36I/MscI site of the plasmid sp73Iccc (full-length c-myb) to construct sp73Iccce. An MscI/BamHI deletion was then made to construct sp73Iccde. A SalI-XbaI fragment from sp73Iccce that contains part of c-myb along with E9A was isolated and cloned into sp73Idcc to create sp73Idcce. A SalI/XbaI fragment from sp73Iccde that contains the C-terminally truncated c-myb along with E9A was isolated and cloned into sp73Idcd and sp73Idvd to create sp73Idcde and sp73Idvde. The different IRES-myb coding sequences from these plasmids were then isolated by using ClaI and cloned into the ClaI site of the viral plasmid, N-Cla, to create NIccce, NIdcce, NIccde, NIdcde, and NIdvde, as well as NIccc, NIdcc, NIccd, NIdcd, and NIdvd (Fig. 1). To construct the GAL4-Myb fusion constructs, the SmaI/ClaI fragments from sp73Icmyb, sp73Iccce, sp73Iccd, and sp73Iccde were isolated and cloned into the SmaI/ClaI site of plasmid pSG424-Cla, which contains the GAL4 DNA-binding domain [GAL4(DBD)]. The luciferase gene (luc) reporter plasmids, polyA-EW5 and polyA-GAL4, have been previously described (23).
Cell culture.
QT6 quail fibroblasts were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with glucose (4.5 g/liter), 1× minimal essential medium nonessential amino acids, 1 mM sodium pyruvate, 2 mM glutamine, streptomycin (100 μg/ml), penicillin (100 U/ml), and 5% fetal calf serum. Yolk sac cells isolated from day 12 to 13 chicken embryos were maintained in Iscove’s medium supplemented with 10% fetal calf serum, 5% heat-inactivated chicken serum (56°C for 1 h), 1× minimal essential medium vitamins, and the same concentrations of the other supplements described above. QT6 fibroblasts were grown in a humidified 10% CO2–90% air 37°C incubator, and the yolk sac cells were maintained in a 5% CO2–95% air 37°C incubator.
Transcriptional activation assay.
Transient transfections were performed by a modified calcium phosphate precipitation method (7, 30). The myb-expressing plasmid (3 μg), luc reporter plasmid (1 μg), β-galactosidase (β-Gal)-expressing plasmid (CMVβ-gal; 0.5 μg), and tRNA (5.5 to 6 μg) were transfected into 106 QT6 fibroblasts per 10-cm-diameter plate; CMVβ-gal was added as an internal control. Forty-eight hours after transfection, the cells were washed with 5 ml of phosphate-buffered saline (PBS), scraped, and resuspended in 1 ml of PBS. Each sample was then divided in half and pelleted by centrifugation (5 min, 400 × g). One half was resuspended in 100 μl of 0.25 M Tris buffer (pH 7.5), lysed by freezing and thawing three times, and assayed for luciferase activity and β-Gal activity (1, 49). β-Gal activity was then used to normalize for the variation in transfection efficiency among different plates. The other half of each sample was dissolved in 100 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Normalized volumes of each sample based on β-Gal activity were then used to determine Myb expression by immunoblot (see below). All transfection experiments were performed four times.
Transformation assay.
To convert proviral plasmids into infectious viruses, 10 μg of each proviral construct, carrying the myb and neo genes, was cotransfected with 1 μg of replication-competent helper-virus plasmid (pMAVdX) into QT6 fibroblasts. G418-resistant cells were selected in standard QT6 medium supplemented with G418 (Gibco-BRL) at a final concentration of 200 μg/ml for 2 weeks. The virus-producing QT6 cells were then treated with mitomycin C (10 μg/ml) and used as a feeder layer in cocultivations for 24 h with primary hematopoietic cells isolated from day 12 or 13 chicken embryonic yolk sacs. The next day, the nonadherent hematopoietic cells were transferred to fresh plates. These infected cells were monitored by microscopy and fed with 3 ml of fresh medium every 2 to 3 days. On day 5, 105 cells from each plate were seeded into 4 ml of 0.8% methocel (HCC-4100; Stem Cell Technologies Co., Vancouver, Canada) supplemented with 1× Iscove’s medium, 10% fetal calf serum, and 5% heat-inactivated chicken serum and then incubated at 37°C for 2 to 3 weeks. Each batch of cells was also infected with a virus (N-Cla) containing the neo gene alone as a negative control to examine the effect of the vector itself and of endogenous viruses from chicken embryos in this assay. The viral supernatants from each plate of QT6 fibroblasts used for cocultivation were saved and used to reinfect 106 fresh QT6 fibroblasts. After G418 selection, viral titers were determined, and immunoblot analyses were performed to confirm Myb protein expression. At least two independent transformation assays were performed for each construct.
Immunoblotting.
Extracts were prepared by lysing transfected QT6 fibroblasts of transformed yolk sac cells in SDS loading buffer; the lysates were then boiled for 5 min. Normalized volumes of lysates (based on internal control β-Gal activity for transient transfections or equivalent cell numbers for transformed yolk sac cells) were subjected to SDS-PAGE (10% polyacrylamide gel), and then the proteins were transferred to a nitrocellulose membrane (BA-S83; Schleicher & Schuell). Myb expression was detected by using a mixture of Myb monoclonal antibodies (5E, 2.2, and 2.7) (19, 58). Blots were developed as specified by the manufacturer by using goat anti-mouse immunoglobulin G conjugated to alkaline phosphatase (Promega), 5-bromo-4-chloro-3-indolylphosphate (BCIP), and nitroblue tetrazolium.
Cytocentrifugation and fluorescence-activated cell sorter (FACS) analyses.
Approximately 5 × 104 transformed yolk sac cells were spun onto glass slides with a cytocentrifuge at 400 × g for 5 min (Cytospin 2; Shandon), air dried, fixed with methanol, and stained with a modified Wright-Giemsa stain (Diff-Quick; Baxter). The cells were photographed under a magnification of ×1,000.
For analyses of cell surface markers by FACS, 106 transformed yolk sac cells were resuspended in 1 ml of cold DMEM–10% fetal calf serum–25 mM HEPES (pH 7.4) and centrifuged at 4°C (400 × g, 5 min). The supernatant was removed, and 50 to 100 μl of the primary antibody of interest (HLO72 or 1C3) was then added to the cells. After incubation on ice for 30 min, the cells were washed twice with 1 ml of cold medium as described above and washed once with Hank’s balanced salt solution (HBSS). Each sample was then evenly dispersed into 30 μl of anti-mouse secondary antibodies conjugated to fluorescein or phycoerythrin (Pharmingen, San Diego, Calif.) diluted 125-fold with HBSS, covered with foil, and incubated on ice for 30 min. Stained cells were then washed twice with 1 ml of HBSS and resuspended in 500 μl of cold HBSS-propidium iodide (PI; 1 μg/ml). As a negative control, each type of cell was similarly stained in the absence of primary antibody. The data were collected after gating out PI-stained, nonviable cells.
For analyses of DNA content by FACS, 106 cells were spun down and resuspended in 500 μl of PBS. The cells were then fixed with 70% ethanol (2 to 4 ml), which was slowly added to the cells; the cells were gently vortexed to prevent clumping. The cells were left in this fixative for 30 min at 4°C then centrifuged (400 × g, 5 min), and resuspended in the PI-RNase A solution (PI, 10 μg/ml; RNase A, 250 μg/ml). The cells were incubated at 37°C for 30 min and then analyzed by FACS. All FACS samples were analyzed with Vantage flow cytometer (Becton Dickinson, San Jose, Calif.). The data were analyzed with CellQuest software (Becton-Dickinson).
RESULTS
Transactivation by Myb proteins that differ in the presence/absence of E9A.
This study focused on detecting any difference in function between Myb proteins that contain and those that lack E9A. We constructed two sets of myb-expressing proviral plasmids that differed only in the inclusion/exclusion of exon 9A (Fig. 1). The parent plasmid contains two retroviral long terminal repeats and the neo gene, which encodes a phosphatase that confers neomycin and G418 resistance. Downstream of the neo gene is an IRES element from encephalomyocarditis virus, followed by different forms of the myb gene. When transcribed, the IRES element forms a multistem loop structure that promotes ribosomal binding and translation of the Myb protein (32). Also, IRES-containing retroviruses are more efficient at expressing two genes than those utilizing differential splicing or internal promoters (25).
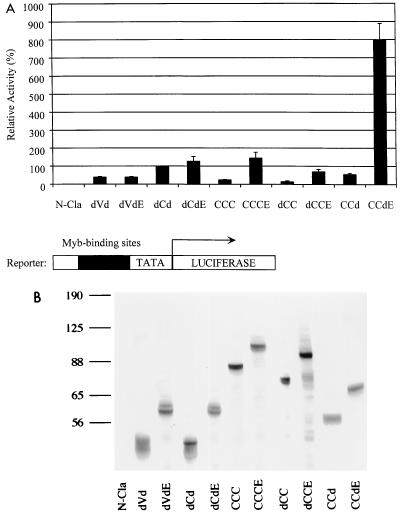
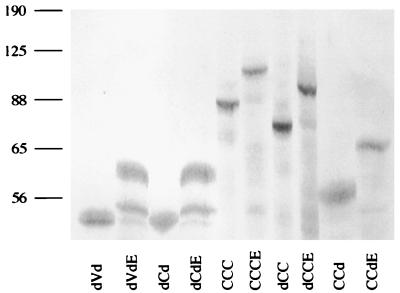
To test for differences in function between the Myb proteins in the presence/absence of E9A, we carried out transcriptional activation assays in which the proviral plasmid containing the myb construct, a luciferase reporter plasmid (EW5), and a β-Gal-expressing plasmid (CMVβ-gal) were cotransfected into QT6 quail fibroblasts. QT6 quail fibroblasts were convenient cells to use for transactivation assays since they lack endogenous c-Myb expression. The reporter plasmid contains five tandem copies of a high-affinity Myb-binding site from the promoter of the mim-1 gene (44), followed by the TATA box from the adenovirus E1B gene and the luc gene. Forty-eight hours after transfection, the cells were analyzed for luciferase activity, which reflects the ability of the Myb protein to up-regulate or (transactivate) the gene expression. Results of the luciferase assays are shown in Fig. 2A. Luciferase activities were normalized for transfection efficiency by using β-Gal expression as an internal control and then compared to the value for dCd. A fivefold increase was seen between CCCE and CCC (c-Myb), as well as dCCE and dCC. A 15-fold increase was seen with CCdE compared to CCd. However, the presence of E9A did not lead to significant differences in transactivation of the doubly truncated proteins, dCdE and dVdE, compared to dCd and dVd, respectively (Fig. 2A). Immunoblot analysis of cell extracts normalized for transfection efficiency confirmed that proteins of the expected sizes were produced for all samples. Similar levels of Myb protein expression were observed for each pair of constructs in the presence and absence of E9A (Fig. 2B). These results demonstrated that the presence of E9A results in an increase in transactivation when the N terminus and/or C terminus of the Myb protein is also present. In contrast, no enhancement in transactivation by E9A is seen when the Myb protein contains truncations at both the N and C termini. This finding suggests that the presence of E9A in a Myb protein with at least one intact terminus (CCCE, CCdE, or dCCE) allows the resulting Myb protein to adopt a different three-dimensional structure that is not observed in doubly truncated Myb proteins (dVdE and dCdE).
FIG. 2.
Transcriptional activation (A) Relative transactivation by different Myb proteins. Each Myb protein was tested for its ability to activate transcription from a reporter plasmid containing five Myb-binding sites (black box), shown at the bottom. TATA, E1B TATA box; LUCIFERASE, luc gene. Reporter and activator plasmids were cotransfected into QT6 fibroblasts, and activities were determined as described in Materials and Methods. Activities are shown relative to that of dCd, which is assigned a value of 100%. The data are represented as the mean of four experiments; the error bars represent the average deviations from the mean. (B) Immunoblot analysis of Myb proteins in transfected cell extracts. Normalized volumes of samples were analyzed by SDS-PAGE, and Myb proteins were detected with a mixture of Myb monoclonal antibodies (2.2, 2.7, and 5E) as described in Materials and Methods. The relative mobilities of molecular weight markers are indicated to the left in kilodaltons.
The Myb DNA-binding domain is required for E9A function.
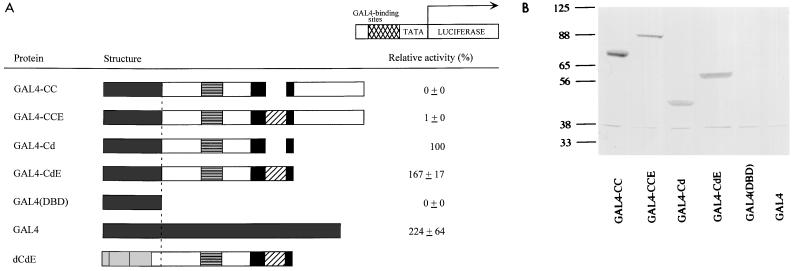
To determine if increased transcriptional activation due to E9A requires the Myb DNA-binding domain, we constructed a series of heterologous fusion proteins in which the Myb DNA-binding domain of each construct was replaced with the GAL4DNA-binding domain. The resulting fusion proteins were named GAL4-CC, GAL4-CCE, GAL4-Cd, and GAL4-CdE. Transactivation assays were then performed with a GAL4 reporter containing GAL4-binding sites. The results of these assays using the wild-type GAL4 protein as a positive control and a construct containing the GAL4 DNA-binding domain alone as a negative control are shown in Fig. 3A. It was previously shown that GAL4-Cd displays activity due to the absence of the negative regulatory domain, whereas GAL4-CC does not activate transcription (14). The presence of E9A did not result in a substantial increase in transactivation by GAL4-Myb fusion proteins relative to those lacking E9A. Immunoblot analysis of cell extracts normalized for transfection efficiency showed similar levels of Myb protein expression for each pair of constructs in the presence and absence of E9A (Fig. 3B). Therefore, these data suggest that the regulatory function of exon 9A is specific for the Myb DNA-binding domain.
FIG. 3.
(A) Relative transactivation by GAL4-Myb fusion proteins. The schematic shows various GAL4-Myb fusion proteins; dCdE is included for comparison. For explanation of Myb sequences, see the legend to Fig. 1. A dashed line indicates the locations of fusions relative to that of dCdE. The reporter plasmid is shown at the top. GAL4-binding sites are indicated by a cross-hatched box. TATA, E1B TATA box; LUCIFERASE, luc gene. The transactivation by different activators is shown relative to GAL4-Cd, which is assigned a value of 100%. The data are represented as the mean of four experiments with average deviations of the mean. (B) Immunoblot of the GAL4-Myb fusion proteins in transfected QT6 fibroblasts. Normalized volumes of cell extracts from each sample were analyzed by SDS-PAGE and detected by a mixture of Myb monoclonal antibodies (2.2 and 2.7). The relative mobilities of molecular weight markers are indicated to the left in kilodaltons.
Transformation by Myb proteins in the presence/absence of E9A.
To study the function of E9A in oncogenic transformation, both sets of constructs were used for leukemia transformation assays in culture. Individual proviral plasmids containing the neo and myb genes were cotransfected into QT6 fibroblasts with a plasmid (pMAVdX) encoding a helper virus, in order to convert the proviral plasmids into infectious viruses. G418 selection for neo expression was completed to isolate stably transfected cells. These virus-producing QT6 fibroblasts were subsequently cocultivated with yolk sac cells isolated from day 12 to 13 chicken embryos. Yolk sac cells are rich in myelomonocytic progenitor cells at this stage in development. When infected with a negative control corresponding to a virus lacking a myb construct (N-Cla), the primary yolk sac cells differentiate within 3 weeks and cease dividing, whereas yolk sac cells infected with a v-myb-expressing virus fail to differentiate and continue to proliferate in liquid culture. Another property of transformed cells is their ability to form colonies in a semisolid methocel matrix. The number of colonies formed is an indication of the number of target cells successfully transformed by the virus, and the size of the colonies is an indication of the growth rate of the transformed cells. Thus, the cell concentration in liquid culture and the methocel assays are both used as measurements of the transformation ability of the different Myb proteins.
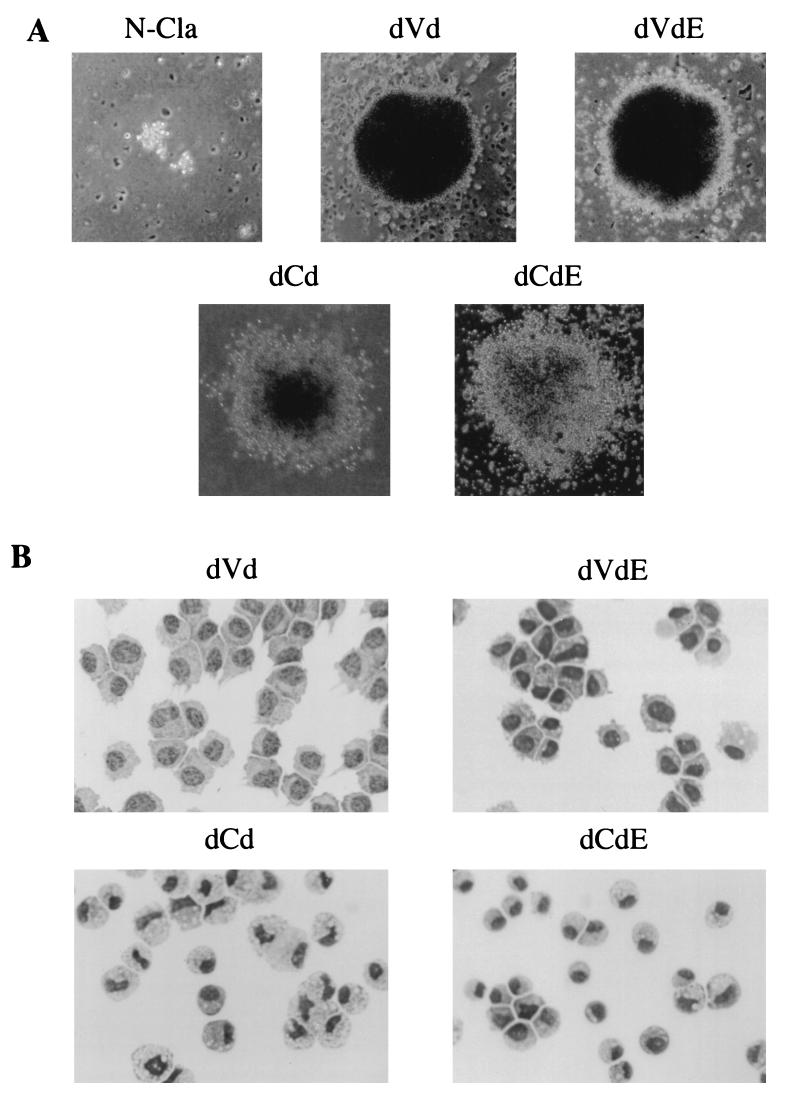
Table 1 contains the results of the outgrowth in liquid culture and the methocel assays. In liquid culture, obvious outgrowth was observed 3 weeks after infection by all myb viruses relative to the negative control, N-Cla. The doubly truncated proteins, dVd and dCd, along with dVdE and dCdE, displayed the strongest transformational efficiencies. The N-terminally truncated protein, dCC, and its counterpart, dCCE, displayed moderate levels of transformation. The C-terminally truncated protein, CCd, and its counterpart, CCdE, as well as CCC and CCCE, were found to be weakly transforming. When the transformed cells were counted on day 21, it was found that relative to N-Cla, the cell concentrations were 300- to 600-fold higher for strongly transforming proteins but only 10- to 20-fold higher for weakly transforming proteins. The results of the methocel assays corroborate the cell numbers found in liquid culture. All transforming constructs gave rise to a larger number of colonies than the negative control (Table 1). Colonies corresponding to dVd and dVdE were extremely compact and possessed distinct borders. Colonies corresponding to dCd/dCdE, as well as all other derivatives of CCC with and without E9A, had compact centers surrounded by a more diffuse border of cells, suggesting increased differentiation along the monocyte/macrophage lineage (Fig. 4A).
TABLE 1.
Summary of transformation assays of Myb proteins in the presence/absence of E9A
| Protein | Transformation ability
|
Expression of cell surface marker in transformed hematopoietic cellsa
|
||
|---|---|---|---|---|
| Liquid culture outgrowth (no. of cells/ml on day 21)b | Methocel assay (no. of colonies)c | HLO72 | 1C3 | |
| N-Cla | <104 | 3 | ND | ND |
| dVd | 6.3 × 106 | 117 | + | − |
| dVdE | 5.1 × 106 | 109 | + | − |
| dCd | 7.7 × 106 | 128 | + | + |
| dCdE | 5.6 × 106 | 119 | + | + |
| dCC | 3.7 × 106 | 71 | + | + |
| dCCE | 3.1 × 106 | 59 | + | + |
| CCC | 2.1 × 105 | 27 | + | + |
| CCCE | 1.8 × 105 | 26 | + | + |
| CCd | 1.6 × 105 | 23 | + | + |
| CCdE | 1.3 × 105 | 20 | + | + |
Cell surface markers HLO72 (a monoblast-specific marker [38]) and 1C3 (a granulocyte-specific marker [40]) were assayed by FACS analyses using the relevant antibodies. ND, not determined; −, not detectable; + detectable (≥85% of the cells stained positively for the cell surface marker).
Transformed cells were counted on day 21. Data from one representative experiment are shown.
105 cells from each culture were seeded into 0.8% methocel on day 5 of the transformation assays, and colonies were counted after 2 weeks. Data from one representative experiment are shown.
FIG. 4.
Growth and morphology of Myb-transformed cells. (A) Morphology of colonies of cells infected by either transforming (dVd, dVdE, dCd, or dCdE) or nontransforming (N-Cla) virus. Methocel assays were performed as described in Materials and Methods. Photographs were taken after 3 weeks with a phase-contrast microscope at a magnification of ×100. (B) Morphology of hematopoietic cells transformed by dVd, dVdE, dCd, and dCdE. The transformed cells were cytocentrifuged and stained with Diff-Quik as described in Materials and Methods. Photographs were taken at a magnification of ×1,000.
The morphologies and cell surface phenotypes of transformed cells from both sets of Myb proteins were further analyzed by using a modified Wright-Giemsa stain and FACS analysis. The presence/absence of E9A did not result in differences in cellular morphology for dVd and dVdE, and both types of cells expressed the monoblast-specific marker HLO72; the cells did not express 1C3, a granulocyte-specific marker (Fig. 4B; Table 1) (38, 40). The morphologies of cells transformed by CCC, as well as its singly and doubly truncated derivatives (dCd, dCC, and CCd), did exhibit differences in comparison to their counterparts containing E9A. Absence of E9A gave rise to cells that were larger in size and contained bilobed nuclei, a characteristic of differentiating cells. In contrast, the presence of E9A gave rise to cells that were smaller and contained nuclei that were rounder, suggesting a lesser degree of differentiation (Fig. 4B). Despite these differences in morphology, all cells were positive for both HLO72 and 1C3, regardless of the presence/absence of E9A. Protein expression was confirmed by immunoblot analysis (Fig. 5). As observed previously with v-Myb, the transformed cells did not appear to express the endogenous c-Myb protein. To look for any differences between the cell cycle profiles of the various transformed cells, we used FACS analysis to measure the DNA content of the cells; we found that the presence/absence of E9A in the Myb proteins did not alter the DNA profiles of the transformed cells (data not shown).
FIG. 5.
Immunoblot analysis of Myb-transformed cells. To test for expression of Myb proteins in hematopoietic cells transformed by the proteins outlined in Fig. 1, 106 cells from each sample were lysed and separated by SDS-PAGE, and proteins were detected with a mixture of Myb monoclonal antibodies (2.2, 2.7, and 5E). The relative mobilities of molecular weight markers are indicated to the left in kilodaltons.
DISCUSSION
In this study, we constructed two sets of viruses to test the functional properties of Myb proteins with or without the E9A and then analyzed the resulting proteins in transactivation and transformation assays. An increase in transactivation was observed in the presence of E9A, provided that at least one terminus of the c-Myb protein was intact. This observation suggests that the amino acids encoded by E9A allow for conformational changes that require either the N or C terminus of the Myb protein. This modified protein structure would thus enable the Myb protein to increase the level of transactivation through the formation of novel intramolecular interactions and/or intermolecular interactions with other proteins. We observed a 15-fold increase between CCd and CCdE but only a 5-fold increase between dCC and dCCE and between CCC and CCCE. This difference is probably due to the absence of the negative regulatory regions contained within the C terminus (14, 33, 48). Evidence exists for proteins that interact with the negative regulatory domain as a means of regulating Myb function (11, 20). In contrast, no significant difference in transactivation was observed with doubly truncated proteins dVd/dVdE and dCd/dCdE. This finding suggests that truncation of both termini resulted in a conformation of Myb proteins that was not influenced by the presence of E9A.
The dependence of E9A function on the Myb DNA-binding domain was investigated by creating GAL4-Myb fusion proteins that contained the GAL4(DBD) fused to Myb sequences. It had already been observed that the absence of the negative regulatory domain in the GAL4-Cd fusion protein results in significant transcriptional activity relative to GAL4-CC (14). We found that the presence of E9A in these GAL4-Myb fusion constructs did not substantially alter their transactivational activity relative to fusion proteins lacking E9A. This finding demonstrates that amino acids encoded by E9A do not constitute a nonspecific transactivation domain. Rather, the GAL4-Myb transactivation studies imply that the function of E9A is specific for the Myb protein and requires the Myb DNA-binding domain for increased activation. These results reinforce the idea that E9A serves to specifically alter the three-dimensional conformation of Myb proteins through the N and C termini.
To study the biological function of E9A, leukemic transformation assays were performed. It was thought that the c-Myb isoform containing E9A might be differentially expressed in a subset of immature hematopoietic cells. Differences in expression of Myb isoforms might then determine the developmental fate of the cell toward distinct lineages. Performing the transformation assays with two sets of Myb proteins that differed in the presence/absence of E9A allowed us to determine that the presence of E9A was compatible with transformation. We observed no differences between each pair of Myb proteins in terms of transformation efficiency and DNA content. The proteins dVd/dVdE and dCd/dCdE conferred the highest efficiencies in transformation, as demonstrated by methocel assays and cell outgrowth in liquid culture. The proteins dCC/dCCE displayed moderate transformation efficiency, while CCC/CCCE and CCd/CCdE displayed only weak levels of transformation. Cells transformed by dVd and dVdE exhibited similar morphologies typical of monoblasts and expressed the monoblast-specific marker HLO72 but not the granulocyte marker 1C3. Comparisons among c-Myb (CCC) and its derivatives (dCC, CCd, and dCd) with their counterparts that contain E9A also showed similarities in transformation phenotypes. All of these transformed cells expressed HLO72 and 1C3. The only differences exhibited by c-Myb-derived proteins with and without E9A pertained to the morphologies of the transformed cells. Myb proteins lacking E9A transformed cells that were larger and contained bilobed nuclei. In contrast, Myb proteins containing E9A transformed cells that were smaller and contained rounded nuclei. These differences in morphology suggest that Myb proteins containing E9A (dCdE, dCCE, CCCE, and CCdE) transform a more immature progenitor than their counterparts lacking E9A (dCd, dCC, CCC, and CCd).
It had previously been observed that a truncation of the N-terminal portion of c-Myb (dCC) conferred moderate transforming ability to the resulting protein; a C-terminal truncation (CCd) or a double truncation (dCd) of c-Myb resulted in weak transformation (13, 28). In addition, it was thought that dCC and CCd transformed promyelocytes, dCd transformed more immature myelomonoblasts, and dVd (v-Myb) transformed monoblasts. The results here show that the phenotypes of transformed cells expressing dCd, CCd, and dCC are in fact fairly similar. All cell types exhibited similar myelomonoblastic morphologies. The cells expressed similar levels of HLO72 and 1C3 on their surfaces. It seems that the presence of the single or double truncations in c-Myb modulates the development of the transformed cells toward similar pathways. Only cells transformed by v-Myb, which contains the amino acid substitutions in addition to the N- and C-terminal truncations, displayed a contrasting cellular phenotype, expressing only the monoblast-specific marker HLO72. It was found that the transformation efficiency of dCd mimicked that of dVd (v-Myb), as shown by the methocel colony formation and cell outgrowth in liquid culture. Differences in transformation ability and cellular phenotype between our results and other published findings may be due to differences in experimental protocol. In some of the previous studies, chicken monocytic growth factor (cMGF) was added to the liquid media and might have influenced the differentiation of the treated cells (28). Because cMGF is most similar to mammalian granulocyte colony-stimulating factor, it seems likely that the absence of exogenous cMGF in our transformation assays allowed the cells to adopt a less committed phenotype than previously observed. Additionally, the inclusion of the IRES element in the proviruses may have resulted in more efficient expression of the myb gene than the spliced retroviruses previously used (25).
Existence of an alternatively spliced form of c-Myb was first observed in mice. Myeloid tumors were generated by retroviral insertional mutagenesis in mice, and cell lines derived from those tumors were analyzed. The cell line ABPL-2 was found to contain two isoforms of c-myb corresponding to the presence/absence of E9A (47). Initially, it was believed that the larger c-myb isoform was unique to transformed cells. Subsequently, it was demonstrated that both isoforms were coexpressed in various tumor cell lines, as well as primary cells harvested from murine tissues (15, 16, 26, 46, 55, 56). The existence of E9A was also found in chickens and humans (10, 51). Although c-myb including E9A was detected in avian bursa, thymus, spleen, and bone marrow, the largest amount of this transcript was observed in yolk sac cells (52). The transformation assays described in this study demonstrate that both Myb isoforms appear to function similarly in oncogenic transformation. Furthermore, no correlation was found between the transactivation and transformation studies. Other experiments have also shown that increased transactivation levels do not necessarily result in increases in transformation (12, 13). Perhaps the elevated transactivation levels seen with a Myb protein that contains E9A is important early in hematopoietic differentiation. It remains possible that one Myb isoform with stronger transactivation ability is needed to establish expression of key target genes in early progenitor cells.
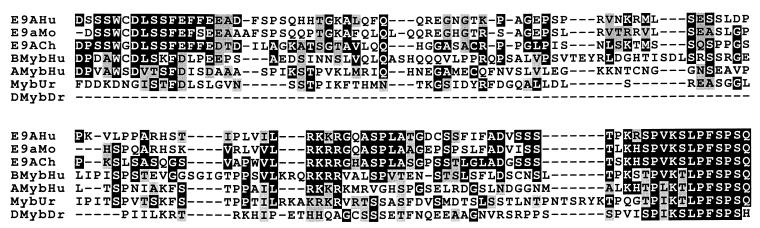
In addition to c-Myb, alternative splicing of a sequence related to exon 9A has also been reported for the murine B-myb gene (34). When the E9A sequence is compared with sequences of other Myb proteins in different organisms, significant regions of homology can be found among the sequences (Fig. 6). The last 12 amino acids in E9A, in particular, are highly conserved among Myb proteins from organisms including human, mouse, chicken, sea urchin, and Drosophila. The conservation of E9A sequences in such a diverse class of organisms attests to the importance of preserving E9A during evolution of the Myb family of proteins. Further studies of Myb will allow delineation of the role this protein has in development of progenitor cells.
FIG. 6.
Alignment of E9A sequences from c-Myb with homologous sequences in Myb proteins from various species. All sequences were obtained from GenBank. The alignments were performed with the computer program ClustalW (61). The most highly conserved residues are highlighted and were determined by using the computer program Boxshade. Dark shading indicates identity; light shading indicates similarity; dashes indicate gaps in the alignment. Dmyb, Drosophila Myb; Hu, human; Mo, mouse; Ch, chicken; Ur, sea urchin; Dr, Drosophila.
ACKNOWLEDGMENTS
We thank members of our laboratory for helpful discussions.
This work was supported by USPHS grant RO1 CA56509. C.H.W. was supported by a Markey Fellowship in Molecular Mechanisms of Disease. L.S. was supported by Public Health Service training grant T32-AI07290.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R C, Moore D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 2.Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- 3.Badiani P A, Kioussis D, Swirsky D M, Lampert I A, Weston K. T-cell lymphomas in v-Myb transgenic mice. Oncogene. 1996;13:2205–2212. [PubMed] [Google Scholar]
- 4.Beug H, Blundell P A, Graf T. Reversibility of differentiation and proliferative capacity in avian myelomonocytic cells transformed by tsE26 leukemia virus. Genes Dev. 1987;1:277–286. doi: 10.1101/gad.1.3.277. [DOI] [PubMed] [Google Scholar]
- 5.Bies J, Mukhopadhyaya R, Pierce J, Wolff L. Only late, nonmitotic stages of granulocyte differentiation in 32Dcl3 cells are blocked by ectopic expression of murine c-myb and its truncated forms. Cell Growth Differ. 1995;6:59–68. [PubMed] [Google Scholar]
- 6.Burk O, Worpenberg S, Haenig B, Klempnauer K H. tom-1, a novel v-Myb target gene expressed in Amv- and E26-transformed myelomonocytic cells. EMBO J. 1997;16:1371–80. doi: 10.1093/emboj/16.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C H, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen R H, Fields S, Lipsick J S. Dissociation of transcriptional activation and oncogenic transformation by v-Myb. Oncogene. 1995;11:1771–1779. [PubMed] [Google Scholar]
- 9.Clarke M F, Kukowska-Latallo J F, Westin E, Smith M, Prochownik E V. Constitutive expression of a c-myb cDNA blocks Friend murine erythroleukemia cell differentiation. Mol Cell Biol. 1988;8:884–892. doi: 10.1128/mcb.8.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasgupta P, Reddy E P. Identification of alternatively spliced transcripts for human c-myb: molecular cloning and sequence analysis of human c-myb exon 9A sequences. Oncogene. 1989;4:1419–1423. [PubMed] [Google Scholar]
- 11.Dash A B, Orrico F C, Ness S A. The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev. 1996;10:1858–1869. doi: 10.1101/gad.10.15.1858. [DOI] [PubMed] [Google Scholar]
- 12.Dini P W, Eltman J T, Lipsick J S. Mutations in the DNA-binding and transcriptional activation domains of v-Myb cooperate in transformation. J Virol. 1995;69:2515–24. doi: 10.1128/jvi.69.4.2515-2524.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dini P W, Lipsick J S. Oncogenic truncation of the first repeat of c-Myb decreases DNA binding in vitro and in vivo. Mol Cell Biol. 1993;13:7334–7348. doi: 10.1128/mcb.13.12.7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubendorff J W, Whittaker L J, Eltman J T, Lipsick J S. Carboxy-terminal elements of c-Myb negatively regulate transcriptional activation in cis and in trans. Genes Dev. 1992;6:2524–2535. doi: 10.1101/gad.6.12b.2524. [DOI] [PubMed] [Google Scholar]
- 15.Dudek H, Reddy E P. Identification of two translational products for c-myb. Oncogene. 1989;4:1061–1066. [PubMed] [Google Scholar]
- 16.Dudek H, Reddy E P. Murine myeloid leukemias with aberrant myb loci show heterogeneous expression of novel myb proteins. Oncogene. 1989;4:1489–1495. [PubMed] [Google Scholar]
- 17.Duprey S P, Boettiger D. Developmental regulation of c-myb in normal myeloid progenitor cells. Proc Natl Acad Sci USA. 1985;82:6937–6941. doi: 10.1073/pnas.82.20.6937. . (Erratum, 83:2281, 1986.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ess K C, Whitaker T L, Cost G J, Witte D P, Hutton J J, Aronow B J. A central role for a single c-Myb binding site in a thymic locus control region. Mol Cell Biol. 1995;15:5707–5715. doi: 10.1128/mcb.15.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evan G I, Lewis G K, Bishop J M. Isolation of monoclonal antibodies specific for products of avian oncogene myb. Mol Cell Biol. 1984;4:2843–2850. doi: 10.1128/mcb.4.12.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Favier D, Gonda T J. Detection of proteins that bind to the leucine zipper motif of c-Myb. Oncogene. 1994;9:305–311. [PubMed] [Google Scholar]
- 21.Ferrao P, Macmillan E M, Ashman L K, Gonda T J. Enforced expression of full length c-Myb leads to density-dependent transformation of murine haemopoietic cells. Oncogene. 1995;11:1631–1638. [PubMed] [Google Scholar]
- 22.Fu S L, Lipsick J S. Constitutive expression of full-length c-Myb transforms avian cells characteristic of both the monocytic and granulocytic lineages. Cell Growth Differ. 1997;8:35–45. [PubMed] [Google Scholar]
- 23.Fu S L, Lipsick J S. FAETL motif required for leukemic transformation by v-Myb. J Virol. 1996;70:5600–5610. doi: 10.1128/jvi.70.8.5600-5610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrido C, Grasser F, Lipsick J S, Stehelin D, Saule S. Protein truncation is not required for c-myb proto-oncogene activity in neuroretina cells. J Virol. 1992;66:6773–6776. doi: 10.1128/jvi.66.11.6773-6776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghattas I R, Sanes J R, Majors J E. The encephalomyocarditis virus internal ribosome entry site allows efficient coexpression of two genes from a recombinant provirus in cultured cells and in embryos. Mol Cell Biol. 1991;11:5848–5859. doi: 10.1128/mcb.11.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonda T J, Cory S, Sobieszczuk P, Holtzman D, Adams J M. Generation of altered transcripts by retroviral insertion within the c-myb gene in two murine monocytic leukemias. J Virol. 1987;61:2754–2763. doi: 10.1128/jvi.61.9.2754-2763.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonda T J, Metcalf D. Expression of myb, myc and fos proto-oncogenes during the differentiation of a murine myeloid leukaemia. Nature. 1984;310:249–251. doi: 10.1038/310249a0. [DOI] [PubMed] [Google Scholar]
- 28.Grasser F A, Graf T, Lipsick J S. Protein truncation is required for the activation of the c-myb proto-oncogene. Mol Cell Biol. 1991;11:3987–3996. doi: 10.1128/mcb.11.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogg A, Schirm S, Nakagoshi H, Bartley P, Ishii S, Bishop J M, Gonda T J. Inactivation of a c-Myb/estrogen receptor fusion protein in transformed primary cells leads to granulocyte/macrophage differentiation and down regulation of c-kit but not c-myc or cdc2. Oncogene. 1997;15:2885–2898. doi: 10.1038/sj.onc.1201472. [DOI] [PubMed] [Google Scholar]
- 30.Ibanez C E, Lipsick J S. Structural and functional domains of the myb oncogene: requirements for nuclear transport, myeloid transformation, and colony formation. J Virol. 1988;62:1981–1988. doi: 10.1128/jvi.62.6.1981-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Introna M, Golay J, Frampton J, Nakano T, Ness S A, Graf T. Mutations in v-myb alter the differentiation of myelomonocytic cells transformed by the oncogene. Cell. 1990;63:1289–1297. doi: 10.1016/0092-8674(90)90424-d. [DOI] [PubMed] [Google Scholar]
- 32.Jang S K, Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosome entry site, and involvement of a cellular 57 KDa RNA-binding protein. Genes Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- 33.Kalkbrenner F, Guehmann S, Moelling K. Transcriptional activation by human c-myb and v-myb genes. Oncogene. 1990;5:657–661. [PubMed] [Google Scholar]
- 34.Kamano H, Burk B, Noben-Trauth K, Klempnauer K H. Differential splicing of the mouse B-myb gene. Oncogene. 1995;11:2575–2582. [PubMed] [Google Scholar]
- 35.Kanei-Ishii C, MacMillan E M, Nomura T, Sarai A, Ramsay R G, Aimoto S, Ishii S, Gonda T J. Transactivation and transformation by Myb are negatively regulated by a leucine-zipper structure. Proc Natl Acad Sci USA. 1992;89:3088–3092. doi: 10.1073/pnas.89.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowenz-Leutz E, Herr P, Niss K, Leutz A. The homeobox gene GBX2, a target of the myb oncogene, mediates autocrine growth and monocyte differentiation. Cell. 1997;91:185–195. doi: 10.1016/s0092-8674(00)80401-8. [DOI] [PubMed] [Google Scholar]
- 37.Lane T, Ibanez C, Garcia A, Graf T, Lipsick J. Transformation by v-myb correlates with trans-activation of gene expression. Mol Cell Biol. 1990;10:2591–2598. doi: 10.1128/mcb.10.6.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J L, Klein P A, Moscovici M G, Moscovici C. Monoclonal antibodies recognizing normal and retrovirus-transformed chicken hematopoietic cells. Virology. 1992;189:583–91. doi: 10.1016/0042-6822(92)90581-9. [DOI] [PubMed] [Google Scholar]
- 39.Lopez A J. Developmental role of transcription factor isoforms generated by alternative splicing. Dev Biol. 1995;172:396–411. doi: 10.1006/dbio.1995.8050. [DOI] [PubMed] [Google Scholar]
- 40.Mandi Y, Veromaa T, Baranji K, Miczak A, Beladi I. Granulocyte-specific monoclonal antibody inhibiting cytotoxicity reactions in the chicken. Immunobiology. 1987;174:292–299. doi: 10.1016/s0171-2985(87)80004-9. [DOI] [PubMed] [Google Scholar]
- 41.Melotti P, Ku D H, Calabretta B. Regulation of the expression of the hematopoietic stem cell antigen CD34: role of c-myb. J Exp Med. 1994;179:1023–1028. doi: 10.1084/jem.179.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mucenski M L, McLain K, Kier A B, Swerdlow S H, Schreiner C M, Miller T A, Pietryga D W, Scott W J, Jr, Potter S S. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 43.Ness S A. The Myb oncoprotein: regulating a regulator. Biochim Biophys Acta. 1996;1288:F123–F139. doi: 10.1016/s0304-419x(96)00027-3. [DOI] [PubMed] [Google Scholar]
- 44.Ness S A, Marknell A, Graf T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- 45.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 46.Ramsay R G, Ishii S, Nishina Y, Soe G, Gonda T J. Characterization of alternate and truncated forms of murine c-myb proteins. Oncogene Res. 1989;4:259–269. [PubMed] [Google Scholar]
- 47.Rosson D, Dugan D, Reddy E P. Aberrant splicing events that are induced by proviral integration: implications for myb oncogene activation. Proc Natl Acad Sci USA. 1987;84:3171–3175. doi: 10.1073/pnas.84.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakura H, Kanei-Ishii C, Nagase T, Nakagoshi H, Gonda T J, Ishii S. Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc Natl Acad Sci USA. 1989;86:5758–5762. doi: 10.1073/pnas.86.15.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 50.Schuur E R, Baluda M A. Expression of MYB proteins in avian hematopoietic tissues. Oncogene. 1991;6:1923–1929. [PubMed] [Google Scholar]
- 51.Schuur E R, Dasgupta P, Reddy E P, Rabinovich J M, Baluda M A. Alternative splicing of the chicken c-myb exon 9A. Oncogene. 1993;8:1839–1847. [PubMed] [Google Scholar]
- 52.Schuur E R, Rabinovich J M, Baluda M A. Distribution of alternatively spliced chicken c-myb exon 9A among hematopoietic tissues. Oncogene. 1994;9:3363–3365. [PubMed] [Google Scholar]
- 53.Selvakumaran M, Liebermann D A, Hoffman-Liebermann B. Deregulated c-myb disrupts interleukin-6- or leukemia inhibitory factor-induced myeloid differentiation prior to c-myc: role in leukemogenesis. Mol Cell Biol. 1992;12:2493–2500. doi: 10.1128/mcb.12.6.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheiness D, Gardinier M. Expression of a proto-oncogene (proto-myb) in hemopoietic tissues of mice. Mol Cell Biol. 1984;4:1206–1212. doi: 10.1128/mcb.4.7.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen-Ong G L. Alternative internal splicing in c-myb RNAs occurs commonly in normal and tumor cells. EMBO J. 1987;6:4035–4039. doi: 10.1002/j.1460-2075.1987.tb02748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen-Ong G L, Luscher B, Eisenman R N. A second c-Myb protein is translated from an alternatively spliced mRNA expressed from normal and 5′-disrupted myb loci. Mol Cell Biol. 1989;9:5456–5463. doi: 10.1128/mcb.9.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siu G, Wurster A L, Lipsick J S, Hedrick S M. Expression of the CD4 gene requires a Myb transcription factor. Mol Cell Biol. 1992;12:1592–1604. doi: 10.1128/mcb.12.4.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sleeman J P. Xenopus A-myb is expressed during early spermatogenesis. Oncogene. 1993;8:1931–1941. [PubMed] [Google Scholar]
- 59.Smarda J, Lipsick J S. c-Myb prevents TPA-induced differentiation and cell death in v-Myb transformed monoblasts. Oncogene. 1994;9:237–245. [PubMed] [Google Scholar]
- 60.Stober-Grasser U, Lipsick J S. Specific amino acid substitutions are not required for transformation by v-myb of avian myeloblastosis virus. J Virol. 1988;62:1093–1096. doi: 10.1128/jvi.62.3.1093-1096.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Todokoro K, Watson R J, Higo H, Amanuma H, Kuramochi S, Yanagisawa H, Ikawa Y. Down-regulation of c-myb gene expression is a prerequisite for erythropoietin-induced erythroid differentiation. Proc Natl Acad Sci USA. 1988;85:8900–8904. doi: 10.1073/pnas.85.23.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Westin E H, Gallo R C, Arya S K, Eva A, Souza L M, Baluda M A, Aaronson S A, Wong-Staal F. Differential expression of the amv gene in human hematopoietic cells. Proc Natl Acad Sci USA. 1982;79:2194–2198. doi: 10.1073/pnas.79.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weston K, Bishop J M. Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell. 1989;58:85–93. doi: 10.1016/0092-8674(89)90405-4. [DOI] [PubMed] [Google Scholar]
- 65.Worpenberg S, Burk O, Klempnauer K H. The chicken adenosine receptor 2B gene is regulated by v-myb. Oncogene. 1997;15:213–221. doi: 10.1038/sj.onc.1201179. [DOI] [PubMed] [Google Scholar]