ABSTRACT
Carbapenem resistance due to metallo-β-lactamases (MBLs) such as the Verona integron-encoded metallo-β-lactamase (VIM) is particularly problematic due to the limited treatment options. We describe a case series of bacterial infections in a tertiary care hospital due to multi-species acquisition of a VIM gene along with our experience using novel β-lactam antibiotics and antibiotic combinations to treat these infections. Four patients were treated with the combination of ceftazidime-avibactam and aztreonam, with no resistance to the combination detected. However, cefiderocol-resistant Klebsiella pneumoniae isolates were detected in two out of the five patients who received cefiderocol within 3 weeks of having started the antibiotic. Strain pairs of sequential susceptible and resistant isolates from both patients were analyzed using whole-genome sequencing. This analysis revealed that the pairs of isolates independently acquired point mutations in both the cirA and fiu genes, which encode siderophore receptors. These point mutations were remade in a laboratory strain of K. pneumoniae and resulted in a significant increase in the MIC of cefiderocol, even in the absence of a beta-lactamase enzyme or a penicillin-binding protein 3 (PBP3) mutation. While newer β-lactam antibiotics remain an exciting addition to the antibiotic armamentarium, their use must be accompanied by diligent monitoring for the rapid development of resistance.
KEYWORDS: carbapenem resistance, VIM, cefiderocol, burn unit, ceftazidime-avibactam and aztreonam
INTRODUCTION
The incidence of carbapenem resistance (CR) among Gram-negative bacteria has increased dramatically over the past three decades (1). This includes an increase in the variety and incidence of carbapenemases, enzymes that hydrolyze carbapenem antibiotics. The genes encoding these enzymes are frequently found on mobile genetic elements, such as plasmids, transposons, and integrons, which can provide additional opportunities for the rapid spread of antibiotic resistance among different bacterial strains or even different species (2, 3). Among carbapenemases, the Ambler class B β-lactamases, also known as metallo-β-lactamases (MBLs), present the greatest treatment challenge due to the limited availability of options for antibiotic treatment. Several families of MBLs have been identified including the Verona integron-encoded metallo-B-lactamases (VIM). These enzymes have a very broad ability to hydrolyze β-lactams and are not inhibited by currently available β-lactamase inhibitors (4). In addition, many β-lactamases genes are found on plasmids that carry resistance elements against other non-β-lactam antibiotics, further limiting antibiotic choices (5). Although there have been several novel β-lactam combination antibiotics developed in the past decade, most of these agents lack activity against MBLs. A notable exception is cefiderocol, which is not only less susceptible to hydrolysis by carbapenemases, including MBLs, but is also novel in its mechanism to promote active uptake by hijacking bacterial iron transport systems (6, 7). The combination of aztreonam and ceftazidime-avibactam has also attracted significant attention due to its success both in vitro and in vivo in treating infections due to organisms producing MBLs (8, 9). This combination provides synergy as MBLs have low affinity for aztreonam, and avibactam can inhibit other β-lactamases, such as extended-spectrum β-lactamases (ESBLs) that would typically inactivate aztreonam, if present.
While these novel antibiotic strategies have shown great promise, experience with their real-world use is still limited. In this study, we describe a case series of complicated skin and soft tissue infections in a tertiary care burn center caused by VIM-producing carbapenem-resistant organisms (VIM-CROs) across multiple species. These infections were treated by a variety of antibiotic approaches including both cefiderocol and the combination of aztreonam with ceftazidime-avibactam. Despite our overall success in resolving the VIM-CRO infections, we experienced rapid development of cefiderocol resistance in two patients following treatment with this antibiotic. Further investigation into the cause of this resistance revealed the independent acquisition of mutations in two siderophore receptors. These mutations contributed to an increase in the MIC of cefiderocol when recreated in a laboratory-adapted Klebsiella pneumoniae strain, independent of any β-lactamase activity.
MATERIALS AND METHODS
Study design
Patients treated in the Vanderbilt University Medical Center (VUMC) Burn Center, a 25-bed level I burn unit, with a documented history of a VIM-CRO isolated from blood or tissue cultures taken directly from an infected site between November 2021 and May 2022 were identified as cases based on a review of culture data. A retrospective chart review was subsequently conducted on all patients meeting this case definition.
Data collection
Data regarding the patients’ hospitalizations were manually extracted from the electronic health record and entered into secure REDcap (REDCap, Nashville TN) forms. Clinical data regarding patient age, demographics, co-morbidities, and entire hospital courses were collected. In addition, information necessary to calculate a Pitt bacteremia score (10) was recorded for the date of the patient’s first culture containing a VIM-CRO. This study was approved by the VUMC institutional review board.
Antimicrobial susceptibility profiling of clinical isolates
Routine antimicrobial susceptibility testing was performed on the Phoenix (BD, Sparks, MD) using the NMIC-306 panels, which evaluated ertapenem and meropenem MICs. Resistance was defined by current Clinical and Laboratory Standards Institute (CLSI) and Food and Drug Administration (FDA) standards as an MIC ≥2 µg/mL for ertapenem and ≥4 µg/mL for meropenem. All carbapenem MICs were confirmed by reference broth microdilution testing according to CLSI protocols (11). Cefiderocol testing was initially performed using a Sensititre panel (ThermoFisher, Lenexa, KS) and confirmed by reference broth microdilution performed according to CLSI standards.
Carbapenemase testing was performed using the Carba-5 lateral flow assay (Hardy, Santa Ana, CA) and, for blood isolates, by testing on the ePlex BCID-GN (Roche, Carlsbad, CA), and later confirmed by the results of whole-genome sequencing. Clinical isolates were tested for synergistic susceptibility to the combination of aztreonam and ceftazidime-avibactam by reference broth microdilution method, whereby ceftazidime was held constant at 4 µg/mL, avibactam at 4 µg/mL, and aztreonam was tested at 0.25–64 µg/mL. The results of this testing were compared to CLSI aztreonam breakpoints (i.e., ≤4 µg/mL was considered susceptible).
Genetic sequencing
Genomic DNA was extracted from the clinical isolates and whole-genome sequencing was performed using Illumina MiSeq (Illumina, CA). Genome assembly and variant analysis were performed using the tools available from the Bacterial and Viral Bioinformatics Resource Center (BV-BRC; https://www.bv-brc.org/) (12). Variants were identified using the BWA-mem-strict aligner and FreeBayes SNP caller. Phylogenetic analysis was done using EpiSeq CS (bioMérieux, Inc., NC). The relatedness of isolates was analyzed using whole-genome multi-locus sequence typing where allelic profile was used to calculate similarities between samples.
Construction of Klebsiella pneumoniae mutants
Siderophore receptor mutants were made in the K. pneumoniae laboratory strain KPPR1S (13) using the pKAS46 plasmid for allelic exchange similar to as previously described (14). Briefly, pKAS46 was linearized by PCR. Primers were designed with homology to regions approximately 1 kb upstream and downstream of the single nucleotide mutations found in the cefiderocol-resistant clinical isolates. Additional primers along with their corresponding reverse complements were designed to have homology overlapping the single nucleotide mutation. These primers were used with KPPR1S genomic DNA as a template to produce two overlapping PCR products approximately 1 kb in length, both of which contained the targeted single nucleotide mutation. These PCR products were joined with the linearized pKAS46 backbone using NEBuilder HiFi DNA Assembly (New England Biolabs, Rowley, MA). The plasmids resulting from the assembly of the DNA fragments were transformed into E. coli strain S17-1 λpir (15) and selected for by plating on LB agar with carbenicillin 75 µg/mL. The plasmids were subsequently transferred into KPPR1S via conjugation. K. pneumoniae isolates containing the plasmid were screened by plating on LB agar with rifampin 30 µg/mL and kanamycin 25 µg/mL. Individual colonies were streaked out on LB agar with streptomycin 2.5 mg/mL and then streptomycin-resistant colonies were restreaked on LB agar with streptomycin 500 µg/mL. Resulting colonies were screened to ensure they were resistant to rifampin and susceptible to kanamycin. The presence of the correct corresponding clinical mutations was confirmed both by Sanger sequencing (Azenta, Burlington, MA) and whole-genome sequencing (Seqcoast, Portsmouth, NH). The plasmids and strains created for this study along with the PCR primers used are further described in Tables S1 and S2.
Cefiderocol susceptibility testing of Klebsiella pneumoniae mutants
The above Klebsiella pneumoniae mutants harboring point mutations identical to those found in the cefiderocol-resistant clinical isolates were tested to determine whether there were changes in their susceptibility to cefiderocol. All isolates were tested using both a disk diffusion method (Hardy Diagnostics, Santa Maria, CA) and a broth microdilution method according to the CLSI M100 (11). Transposon insertion mutants in the KPPR1 background with insertions in cirA and fiu from a previously described K. pneumoniae transposon library were used as reference strains in cefiderocol testing, as well (16).
Growth of Klebsiella pneumoniae in iron-limited conditions
To test the effects of siderophore receptor mutations on the ability to grow in iron-restricted conditions, overnight cultures of the K. pneumoniae clinical isolates and mutant strains generated above were inoculated in round-bottom polypropylene tubes containing LB broth and incubated overnight night for 16 hours at 37°C in a shaking incubator at 180 rpm. Overnight cultures were diluted to a final OD600 of 0.01 in LB containing either 1,200 µM or 600 µM 2,2-dipyridyl (Sigma-Aldrich, Inc, St. Louis, MO) or a vehicle control. Bacteria were grown in a 96-well plate at 37°C with continuous orbital shaking and OD600 was measured at 30-minute intervals for 20 hours using a BioTek plate reader (Agilent, Santa Clara, CA).
RESULTS
Between November 2021 and May 2022, nine patients were found to have complicated skin and soft tissue infections with VIM-CROs isolated in tissue cultures from an infected wound, four of which (44%) also had blood cultures positive for the same VIM-CRO. Full characteristics of these patients are shown in Table 1 with further details of their hospital courses included in File S1. These patients had a median age of 38 and relatively few comorbidities or pre-existing risk factors for a CRO infection (Table S3). No patients were immunocompromised. However, all patients had substantial burn injuries encompassing an average (median) of 40% of total body surface area. Consistent with the severity of their burn injuries, patients had prolonged hospital stays with a median hospitalization length of 75 days. Patients were hospitalized a median of 25 days prior to isolation of a VIM-CRO. Despite all patients receiving parenteral antibiotics for either treatment of an infection or peri-operative surgical prophylaxis prior to initial isolation of a VIM-CRO, none received a carbapenem before isolation of a VIM-CRO.
TABLE 1.
Characteristics of burn patients with wound infections due to VIM-producing carbapenem-resistant organisms (n = 9)
| Characteristic | Value |
|---|---|
| Median age in years (range) | 38 (19–81) |
| Sex, n (%) | |
| Male | 7 (78) |
| Female | 2 (22) |
| Median TBSA of burn (range) | 40% (13%–65%) |
| Median LOS in days (range) | 75 (32–157) |
| Median LOS prior to isolation of CRO in days (range) | 25 (8–45) |
| Median number of surgeriesa during hospitalization (range) | 12 (1-16) |
| Associated bacteremia due to a CRO, n (%) | 4 (44) |
| VIM-CRO Isolated, n (%)b | |
| Klebsiella pneumoniae | 6 (67) |
| Serratia marcescens | 4 (44) |
| Acinetobacter baumannii | 1 (11) |
| Enterobacter cloacae | 1 (11) |
| Pseudomonas aeruginosa | 1 (11) |
| >1 species of VIM-producing CROc | 3 (33) |
| Mean Pitt bacteremia score at the time of initial CRO isolation (range)d | 2.3 (0–6) |
| Among surviving patients (n = 8) | 1.9 (0–4) |
| Treatment, n (%) | |
| Surgery alonee | 2 (22) |
| Surgery + antibiotics | 6 (67) |
| Antibiotics alonef | 1 (11) |
| Mortality, n (%) | |
| Within 30 days of initial CRO isolation | 1 (11) |
| Within 30 days of hospital discharge | 1 (11) |
| Among patients with bacteremia due to a CRO (n = 4) | 1 (25) |
Includes only amputation or debridement surgeries occurring in an operating room.
Percentages do not equal 100 due to multiple different VIM-CRO being isolated from some patients.
This includes one patient with co-culture of VIM-producing K. pneumoniae and S. marcescens, one patient with separate infections due to VIM-producing P. aeruginosa and K. pneumoniae during their hospitalization, and one patient with three separate episodes of infection due to VIM-producing K. pneumoniae, S. marcescens, and A. baumannii.
Highest Pitt bacteremia score calculated for the 24-hour period during which the initial VIM-CRO was isolated from a patient.
Refers to patients who did not receive any antibiotics expected to be active against the VIM-CRO isolate.
Refers to patients who did not have any further surgery on or after the date of their final culture containing a VIM-CRO. LOS: length of stay, TBSA: total body surface area.
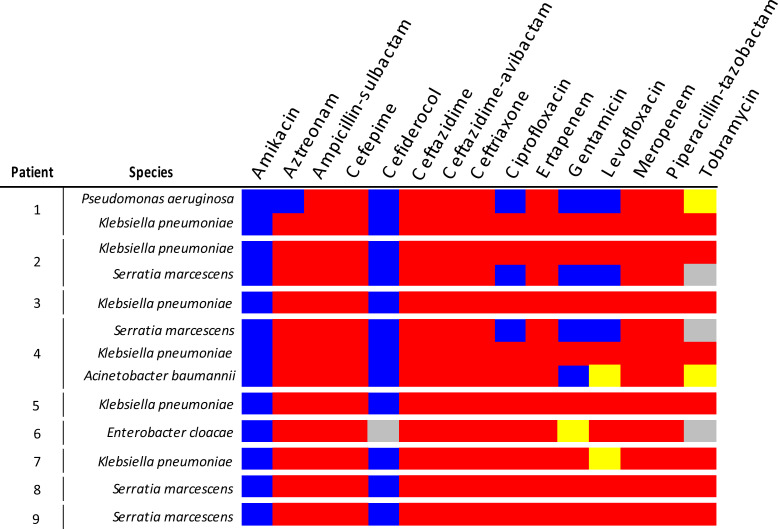
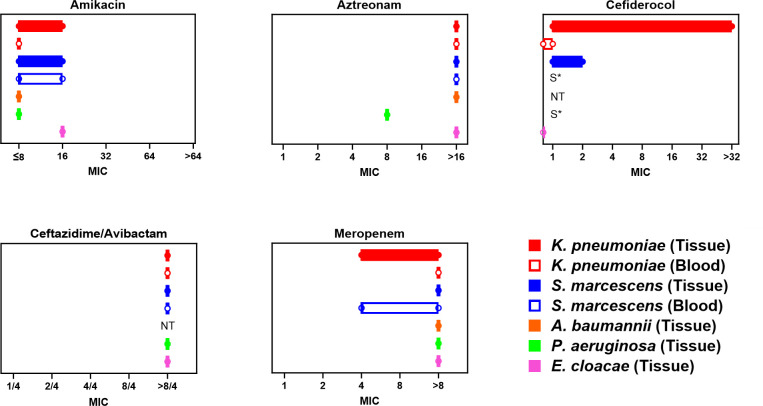
A total of five different bacterial species were found to express the VIM enzyme during this outbreak, with three patients having infections due to multiple species of VIM-CROs. Klebsiella pneumoniae was the most common VIM-CRO isolated (6/9) followed by Serratia marcescens (4/9). VIM-expressing Pseudomonas aeruginosa, Enterobacter cloacae, and Acinetobacter baumannii were all isolated from one patient each. The susceptibility pattern of the initial VIM-expressing isolate from each patient is shown in Fig. 1. The range of MIC values for all the VIM-CROs with available antimicrobial resistance testing data is shown in Fig. 2.
Fig 1.
Susceptibility pattern of initial VIM-CROs isolates from each patient. The susceptibility to the antibiotics listed is shown for the first VIM-CRO isolated from each patient. If a patient had multiple species of VIM-producing CROs isolated, then the first VIM-CRO isolates for each species is shown. Blue indicates susceptibility, yellow indicates intermediate susceptibility, and red indicates resistance to a given antibiotic. The gray boxes represent antibiotic susceptibility data that were unavailable. Amikacin susceptibility was determined prior to the 2023 CLSI update to amikacin breakpoints.
Fig 2.
MIC values among VIM-CROs for select antibiotics. The range (min-max) of MICs (µg/mL) for all VIM-CRO isolates with antimicrobial susceptibility testing results available from all nine patients included in this study. MICs are shown for amikacin, aztreonam, cefiderocol, ceftazidime-avibactam, and meropenem. For A. baumannii, P. aeruginosa, and E. cloacae, MIC values are reflective of a single isolate each. For K. pneumoniae and S. marcescens, isolates are separated by culture source, either blood or tissue/wound. Results include multiple isolates from the same patient in some cases. S*- susceptible but determined by disk diffusion method. NT: not tested.
Treatment of the VIM-CRO-infected patients involved a combination of antimicrobial therapy and surgical debridement. Two patients improved with debridement and treatment with antibiotics without activity against the isolated VIM-CROs (ciprofloxacin and piperacillin-tazobactam, respectively). Among the other seven patients, antibiotic therapy targeting VIM-CROs included cefiderocol (n = 5 patients) and the combination of aztreonam and ceftazidime-avibactam (n = 4) with two patients receiving each treatment option at separate points in their hospitalization (Table 2). Among the five patients who were treated with cefiderocol, four patients also received concurrent amikacin. Two of those patients (40% of those receiving cefiderocol) had cefiderocol-resistant K. pneumoniae isolated from tissue cultures 11 and 17 days after the initiation of cefiderocol (2 and 4 days after discontinuing cefiderocol, respectively). Antibiotic therapy was switched to salvage therapy with the combination of aztreonam, ceftazidime-avibactam, and amikacin in one case and a combination of amikacin and minocycline in the other. Doses of antibiotics used along with renal adjustments are included in Table S4. The dose of cefiderocol did not appear to be related to the development of resistance as the one patient who received every 8 hours dosing of cefiderocol instead of every 6 hours did not develop cefiderocol resistance.
TABLE 2.
Patient characteristics and outcomes of infections treated with cefiderocol or ceftazidime-avibactam + aztreonam
| Patients treated with cefiderocol | |||||||
|---|---|---|---|---|---|---|---|
| Patient | Age, Sex | Co-morbidities | VIM infection site | Pathogens identified | Duration of cefiderocol treatment (d) | Co-administration of amikacin | Treatment outcome (determined by) |
| 1 | 38, female | None | Bacteremia, neck wound | Klebsiella pneumoniae (VIM), Pseudomonas aeruginosa (wound only) | 17 | Yes (15 d) | Success (clearance of bacteremia, resolution of fevers) |
| 2a | 30, male | None | Bacteremia, R arm | Klebsiella pneumoniae (VIM), Serratia marcescens (VIM) | 9 | Yes (4 d) | Failure (persistent fevers, persistent bacteremia, recovery of cefiderocol resistant-Klebsiella pneumoniae from blood) |
| 3 | 67, male | HTN, diabetes | L lower extremity | Klebsiella pneumoniae (VIM), Ochrobactrum anthropi | 12 | Yes (6 d) | Failure (persistent fevers, recovery of cefiderocol resistant-Klebsiella pneumoniae from the same site) |
| 4b | 30, male | None | R lower extremity | Klebsiella pneumoniae (VIM), Pseudomonas aeruginosa | 19 | No | Success (resolution of fevers, clinical assessment of wound, subsequent tissue culture) |
| 5 | 23, male | Diabetes | Bacteremia, neck wound | Klebsiella pneumoniae (VIM) | 10 | Yes (9 d) | Success (clearance of bacteremia) |
Patient 2 is included in both lists as he was treated with cefiderocol and amikacin followed by aztreonam, ceftazidime-avibactam, and amikacin for salvage therapy. Although his infection improved with salvage therapy, he subsequently developed candidemia and an Aspergillus wound infection. He passed away after a transition of goals of care.
Patient 4 is included in both lists as he had multiple separate infections due to VIM-CROs during his hospitalization and received both cefiderocol and the combination of ceftazidime-avibactam and aztreonam at different times.
Patient 8 had two infections due to VIM-CROs, isolated 21 days apart, during his hospitalization for which he received two separate courses of ceftazidime-avibactam and aztreonam.
One patient died within 30 days of the initial isolation of a CRO. This patient developed cefiderocol resistance but had documented clearance of the VIM-CRO infection (based on follow-up cultures) after the initiation of aztreonam, ceftazidime-avibactam, and amikacin salvage therapy. This patient succumbed to a subsequent invasive fungal infection. The Pitt bacteremia score on the first day that a VIM-CRO was isolated (10) was 6 for the patient who died, while the other eight patients had a mean score of 1.9 (range 0–4).
At the time of this outbreak, in vitro synergy testing for susceptibility to the combination of ceftazidime-avibactam and aztreonam was not clinically available. BMD susceptibility testing of VIM-producing isolates from the patients treated with ceftazidime-avibactam and aztreonam combination therapy along with the isolates with cefiderocol resistance was retrospectively performed for this study. All strains tested were susceptible to the combination of aztreonam and ceftazidime-avibactam with an MIC of ≤0.25/4/4 (aztreonam/ceftazidime/avibactam, data not shown), despite these isolates having high levels of resistance to both ceftazidime-avibactam and aztreonam individually (Fig. 2).
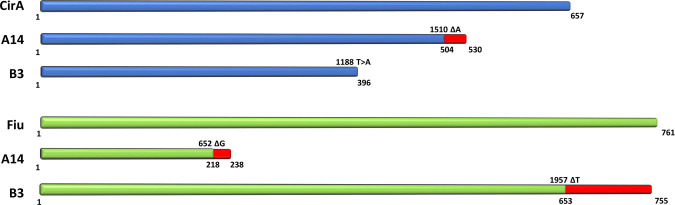
To understand the mechanisms underlying the cefiderocol resistance seen in this outbreak, whole-genome sequencing was performed on isolates from the patients who developed cefiderocol-resistant K. pneumoniae. Pairs of isolates immediately prior to the detection of cefiderocol resistance and once cefiderocol resistance was detected were subjected to whole-genome sequencing and compared to identify variations. Isolates identified as A12 and B2 were collected from patient 2 and patient 3 (File S1), respectively, prior to the detection of cefiderocol resistance. These were paired with isolates A14 and B3, both of which were collected from the sample patients after completing cefiderocol treatment. A third isolate from patient 3 (identified as B4), collected 13 days after the first cefiderocol-resistant isolate was identified, was also sequenced. There were no other changes in the susceptibility, neither increases nor decreases in MICs, to any other antibiotics tested besides cefiderocol in these strain pairs. Comparison of strain pairs A12/A14 and B2/B3 showed numerous non-synonymous variations within predicted open reading frames, the complete lists of which are shown in Tables S5 and S6, respectively. High-impact SNPs were identified in two siderophore receptors in both pairs of isolates. The genes encoding catecholate siderophore receptors CirA and Fiu (17) were both found to have single nucleotide deletions in the cefiderocol-resistant isolates. However, the specific mutations were not the same between the two cefiderocol-resistant isolates, suggesting they arose spontaneously and independently of each other. Furthermore, phylogenetic analysis of these five isolates suggests A12 and B2 share a common ancestor while B3 and B4 are more closely related to B2 (Fig. S1). The identified mutations along with their predicted effect on their respective protein products are shown in Fig. 3. Given the presence of mutations affecting siderophore receptors, the ability of these strains to grow in iron-restricted conditions was tested. When grown in LB, there was no difference in the growth of the strain pairs (Fig. S2A and B). No difference in growth was observed between A12 and A14 when iron was restricted by the addition of 600 µM 2,2-dipyridyl, and A14 actually showed slightly improved growth in the presence of 1,200 µM 2,2-dipyridyl when compared to A12 (Fig. S2A). In the same iron-limiting conditions, there was a mild growth defect in B3 versus B2; however, the growth of B4 was comparable and even slightly enhanced versus wild-type levels (Fig. S2B). On analysis of the whole-genome sequence of B4, it was revealed that B4 contained the same cirA and fiu mutations that were found in B3. However, B4 had an additional three non-synonymous mutations in predicted open reading frames, as shown in Table S7.
Fig 3.
Siderophore receptor mutations identified in clinical isolates with cefiderocol resistance. The schematic shows the representative full-length protein products of cirA and fiu along with the truncated versions that result from the mutations found in each clinical isolate. Red lines indicate the missense region of the protein. Numbers above the line indicate the single nucleotide deletion present while numbers below each line indicate the amino acid where the missense mutation or termination of the protein occurs.
Given that the WGS detected mutations in siderophore receptors were likely to affect their function, we tested whether these mutations affected the susceptibility of K. pneumoniae to cefiderocol. Since there were many other mutational differences between the paired cefiderocol susceptible and resistant isolates, including mutations in a porin and a class A β-lactamase (SHV-7), we sought to confirm the effect of the siderophore receptor mutations in a clean genetic background. To do this, the identical point mutations that were found in the clinical isolates were introduced into a laboratory strain of K. pneumoniae, KPPR1S. The mutations found in A14, cirAK504fs-1 and fiuE218fs-1 mutations, were recreated both alone and in combination to make a double siderophore receptor mutant as were the mutations found in B3, cirAY396* and fiuS653fs-1. These strains were subsequently tested for their susceptibility to cefiderocol both by broth microdilution and disk diffusion assays (Table 3). While all of the strains remained susceptible to cefiderocol (defined as an MIC ≤4 µg/mL or zone of inhibition (ZOI) ≥ 16 mm), there was an increase in the MIC and a decrease in the ZOI for all of the mutants compared to their parent strain. Both double siderophore receptor mutants had 16-fold increases in cefiderocol MICs over the wild type. Similar to what was seen with the clinical isolates, these mutations did not affect the growth of these strains in LB even in iron-limiting conditions (Fig. S2C and D). Together, these results suggest the potential for rapid selection of cefiderocol resistance during antibiotic treatment without a trade-off in bacterial fitness.
TABLE 3.
Cefiderocol susceptibility testing of siderophore receptor mutants
| Strain | Cefiderocol resistance testing | ||
|---|---|---|---|
| BMD MIC (µg/mL) | Fold Increase in BMD MIC over WT | Disk susceptibility ZOI | |
| KPPR1S | 0.0625 | - | 29 mm |
| KPPR1S cirAK504fs-1 | 0.25 | 4 | 23 mm |
| KPPR1S cirAY396* | 0.125 | 2 | 24 mm |
| KPPR1S fiuE218fs-1 | 0.5 | 8 | 28 mm |
| KPPR1S fiuS653fs-1 | 0.25 | 4 | 28 mm |
| KPPR1S cirAK504fs-1, fiuE218fs-1 | 1 | 16 | 19 mm |
| KPPR1S cirAY396*, fiuS653fs-1 | 1 | 16 | 21 mm |
| Clinical Isolate A12 | 1 | - | 22 mm |
| Clinical Isolate A14 | >32 | N/A | 6 mm |
| Clinical Isolate B2 | 0.5 | - | 21 mm |
| Clinical Isolate B3 | >32 | N/A | 6 mm |
DISCUSSION
As noted in this case series, MBL-producing CROs can be particularly challenging to treat in a burn unit where there is frequent antibiotic exposure and potential for multi-species spread, even in the absence of specific carbapenem selection pressure. Outbreaks of MBLs have been described in burn units where patients are disproportionally vulnerable to environmental sources and the spread of resistance mechanisms by horizontal gene transfer due to their extended duration of open wounds (18). Given that VIM is an unusual carbapenemase at our institution, we suspect this outbreak is also plasmid associated, although this is currently being confirmed by long-read sequencing.
Despite a high percentage of bacteremia (44%) among the patients included in this study, there was only one death within 30 days, representing a mortality of 25% among patients with VIM-CRO bacteremia and 11% mortality overall among patients with VIM-CRO infections. This is on the lower end of the estimated mortality among patients with infections due to CRO (10%–40%), especially given that mortality is usually higher in cases of bacteremia due to carbapenemase-producing organisms (19, 20). The observed survival rate may be related to the patient population in this study or the ability to obtain a high degree of source control through repetitive surgical debridement.
While differentiating between colonization and true infection can be challenging in patients with burns, we felt confident in classifying these nine patients as having true infections for several reasons. Four out of the nine patients had the same VIM-CRO isolated from their bloodstream at some point during their hospital stay, which clearly represents an infection. Among the other five patients, clinical records all document concern for an infection at the time of collection of the cultures in question. In addition, four out of the five patients were febrile at the time of culture collection and their fevers resolved shortly after further debridement or initiation of antibiotics (File S1). The one possible exception is patient 7 who also had a KPC-producing Enterobacter cloacae isolated from a urine culture the day after they had a VIM-producing Klebsiella pneumoniae isolated from a wound culture. A Pseudomonas aeruginosa isolate that was susceptible to carbapenems and fluoroquinolones was also recovered from this wound culture. This patient was empirically started on ciprofloxacin and defervesced despite the Klebsiella pneumoniae isolate being resistant to ciprofloxacin. Therefore, it is possible that the non-CRO pathogens present were responsible for this patient’s wound infection and fever and the VIM-CRO merely reflected colonization, in this case.
One limitation of this study is the use of now outdated amikacin breakpoint points. Since this outbreak occurred prior to the 2023 revision to the CLSI breakpoints for amikacin, MIC values for amikacin were not determined below 8 µg/mL. This means that isolates from patients who received amikacin as part of their antibiotic treatment may be reclassified as having intermediate susceptibility under the revised amikacin breakpoints (11). However, none of the patients treated with amikacin had VIM-CRO isolates with amikacin MICs that fall in the resistant range based on the revised breakpoints.
At the time of this outbreak, our facility did not have a standard method in place to predict whether the combination of aztreonam and ceftazidime-avibactam would be effective against a CRO. The use of this combination was empiric in all cases, as the isolates were resistant to both ceftazidime-avibactam and aztreonam alone. However, good clinical responses were observed in the patients treated with the combination of aztreonam and ceftazidime-avibactam (Table 2; File S1). Although AST or ALT elevations occurred in more than half of patients receiving this combination as part of a Phase I study (21), we did not observe any such elevations in the patients treated with aztreonam and ceftazidime-avibactam. Subsequent in vitro synergy testing confirmed the susceptibility of the respective isolates to combination therapy. In addition to reference broth microdilution testing, CLSI has endorsed the use of a ceftazidime-avibactam and aztreonam broth disk elution test which represents another potential option for clinical laboratories to evaluate susceptibility to the combination of ceftazidime-avibactam and aztreonam.
Although there have been multiple antibiotics brought to market in the past decade to counteract the rising incidence of CROs, resistant isolates have been detected almost as soon as novel agents are put into clinical use. This includes cefiderocol, which despite its novel mechanism of action has already been shown to be vulnerable to resistance developing in the clinical setting (22). There have been multiple reports of cirA mutations contributing to cefiderocol resistance in K. pneumoniae (23, 24). However, this is the first example, to our knowledge, of a fiu mutation arising in any clinical Enterobacterales isolate. Furthermore, this appears to be the first report of multiple siderophore receptor mutations arising in a single clinical isolate. Early studies of cefiderocol demonstrated that targeted deletion of both fiu and cirA resulted in a synergistic increase in the MIC in an E. coli laboratory strain (6). Given that two mutations were required to significantly increase the MIC, this was considered to be a strength of cefiderocol. Likewise, neither fiu nor cirA mutations were found in an analysis of clinical isolates with reduced susceptibility to cefiderocol in the initial trials leading to its approval (25).
Despite a limited duration of cefiderocol use, we saw a rapid emergence of cefiderocol-resistant K. pneumoniae in 40% of the cases (2/5) treated with cefiderocol in this case study. Both cases of cefiderocol resistance were found within 4 days of discontinuing cefiderocol. In one of these cases, cefiderocol was discontinued and antibiotics were empirically switched due to presumed treatment failure while in the other case, the patient remained febrile even after completion of the full course of cefiderocol. Even more surprising was the fact that both cases of cefiderocol resistance involved the acquisition of different sets of mutations in both fiu and cirA, suggesting they arose completely independently. The rapid selection of resistance may be explained by the pre-existence of heteroresistance among the bacterial populations, a previously described, yet under-appreciated occurrence among clinical isolates (26). In this context, it is possible that a subpopulation of bacteria with pre-existing siderophore receptor mutations existed within the infected wound environment, but at levels that initially evaded clinical detection by standard susceptibility testing. As opposed to Choby et. al., who found the mechanism of cefiderocol resistance in their clinical isolates to be the acquisition of multiple copies of a carbapenemase gene on a plasmid through gene replication, our study identified single nucleotide mutations within siderophore receptors as the mechanism responsible for resistance to cefiderocol. While we cannot rule out the possibility that increased expression or duplication of the carbapenemase gene is contributing to the cefiderocol resistance seen in our clinical isolates, re-creating the mutations found in our clinical isolates in a laboratory K. pneumoniae strain caused a 16-fold increase in the cefiderocol MICs, even in the absence of any β-lactamase (Table 3). Presumably, the addition of a β-lactamase, such as the VIM gene found in the clinical isolates, would have led to further increases in cefiderocol MICs in these strains, and possibly resulted in resistance, although this was not tested. Interestingly, in one of the two strain pairs, a third mutation in a siderophore receptor was found (Table S5), with a single amino acid change in the product of the fhuA gene (fhuAP399S). This mutation was not investigated further in this study but it may also contribute to the cefiderocol resistance seen in this strain. This strain also acquired a premature stop codon in the MutL DNA mismatch repair protein which may explain why there were so many mutational differences between the two strains in this pair.
Interestingly, only one of these isolates had a growth deficiency in iron-limiting conditions (Fig. S2B). However, a subsequent cefiderocol-resistant isolate from that patient recovered 13 days after the detection of the first resistant isolate did not have a growth defect. This isolate, B4, still had the same cirA and fiu mutations found in B3, suggesting that the isolates were related and not due to the acquisition of a new K. pneumoniae isolate. B4 had additional mutations in several genes including a carbohydrate porin gene (Table S7), although it was not readily apparent what was responsible for the reversal of the low iron growth restriction. Furthermore, the creation of the cirA and fiu mutations found in B3 and B4 in KPPR1S did not result in restricted growth in low iron conditions (Fig. S2D). This suggests that these mutations could be easily selected for as they are not detrimental to growth in iron-restricted conditions even though they lead to increased resistance to cefiderocol (Table 3).
As demonstrated in this study, the treatment of infections due to MBL-producing CROs is challenging. Fortunately, the development of new antibiotics with activity against MBL-producing CROs is ongoing, with cefepime-taniborbactam and the combination of aztreonam-avibactam two recent examples of β-lactam/β-lactamase inhibitors to finish phase III trials. Treatment regimens require personalization based on consideration of not only the antibiotic resistance pattern but also the characteristics of the patient. While new antimicrobials provide additional options in some of the most difficult-to-treat infections, they inherently come with a lack of experience in their use. While we found good success with the combination of aztreonam and ceftazidime-avibactam, our clinical failure rate with cefiderocol was disappointing. Our findings suggest that patients treated with cefiderocol should be monitored closely for the development of resistance, particularly in the setting of early clinical signs of treatment failure.
ACKNOWLEDGMENTS
Plasmid pKAS46 and strains KPPR1S and S17-1 λpir were graciously provided by Dr. Ammar Zafar. JAF was supported by funding from the National Institute of Allergy and Infectious Diseases at the National Institutes of Health under grant numbers T32AI007474 and F32AI169905 and from the VUMC Faculty Research Scholars program. LAM received funding from the American Heart Association under grant number 23CDA1056712. This work was funded by R01AI101171 to EPS.
Contributor Information
Jeffrey A. Freiberg, Email: Jeffrey.freiberg@vumc.org.
Ryan K. Shields, University of Pittsburgh, Pittsburgh, Pennsylvania, USA
DATA AVAILABILITY
Raw sequencing reads for whole-genome sequencing of clinical Klebsiella pneumoniae isolates were deposited to the NCBI SRA archive under BioProject PRJNA1058428.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aac.01507-23.
Supplemental Figures S1 and S2 and Tables S1 to S7.
Supplemental file with an overview of the clinical course of the patients found to be harboring VIM+ Gram-negative isolates.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202 [DOI] [PubMed] [Google Scholar]
- 2. van Duin D, Doi Y. 2017. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8:460–469. doi: 10.1080/21505594.2016.1222343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis 11:381–393. doi: 10.1016/S1473-3099(11)70056-1 [DOI] [PubMed] [Google Scholar]
- 5. Orlek A, Anjum MF, Mather AE, Stoesser N, Walker AS. 2023. Factors associated with plasmid antibiotic resistance gene carriage revealed using large-scale multivariable analysis. Sci Rep 13:2500. doi: 10.1038/s41598-023-29530-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ito A, Sato T, Ota M, Takemura M, Nishikawa T, Toba S, Kohira N, Miyagawa S, Ishibashi N, Matsumoto S, Nakamura R, Tsuji M, Yamano Y. 2018. In vitro antibacterial properties of cefiderocol, a novel siderophore cephalosporin, against Gram-negative bacteria. Antimicrob Agents Chemother 62:e01454-17. doi: 10.1128/AAC.01454-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ito A, Nishikawa T, Matsumoto S, Yoshizawa H, Sato T, Nakamura R, Tsuji M, Yamano Y. 2016. Siderophore cephalosporin cefiderocol utilizes ferric iron transporter systems for antibacterial activity against Pseudomonas aeruginosa . Antimicrob Agents Chemother 60:7396–7401. doi: 10.1128/AAC.01405-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Falcone M, Daikos GL, Tiseo G, Bassoulis D, Giordano C, Galfo V, Leonildi A, Tagliaferri E, Barnini S, Sani S, Farcomeni A, Ghiadoni L, Menichetti F. 2021. Efficacy of ceftazidime-avibactam plus aztreonam in patients with bloodstream infections caused by metallo-β-lactamase-producing Enterobacterales. Clin Infect Dis 72:1871–1878. doi: 10.1093/cid/ciaa586 [DOI] [PubMed] [Google Scholar]
- 9. Marshall S, Hujer AM, Rojas LJ, Papp-Wallace KM, Humphries RM, Spellberg B, Hujer KM, Marshall EK, Rudin SD, Perez F, Wilson BM, Wasserman RB, Chikowski L, Paterson DL, Vila AJ, van Duin D, Kreiswirth BN, Chambers HF, Fowler VG, Jacobs MR, Pulse ME, Weiss WJ, Bonomo RA. 2017. Can ceftazidime-avibactam and aztreonam overcome β-lactam resistance conferred by metallo-β-lactamases in Enterobacteriaceae? Antimicrob Agents Chemother 61:e02243-16. doi: 10.1128/AAC.02243-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Henderson H, Luterbach CL, Cober E, Richter SS, Salata RA, Kalayjian RC, Watkins RR, Doi Y, Kaye KS, Evans S, Fowler VG, Bonomo RA, Harris A, Napravnik S, Van Duin D. 2020. The Pitt bacteremia score predicts mortality in nonbacteremic infections. Clin Infect Dis 70:1826–1833. doi: 10.1093/cid/ciz528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. CLSI . 2023. Performance standards for antimicrobial susceptibility testing. In CLSI supplement M100, 33rd ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 12. Olson RD, Assaf R, Brettin T, Conrad N, Cucinell C, Davis JJ, Dempsey DM, Dickerman A, Dietrich EM, Kenyon RW, et al. 2023. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res 51:D678–D689. doi: 10.1093/nar/gkac1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palacios M, Broberg CA, Walker KA, Miller VL. 2017. A serendipitous mutation reveals the severe virulence defect of a Klebsiella pneumoniae fepB mutant. mSphere 2:e00341-17. doi: 10.1128/mSphere.00341-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skorupski K, Taylor RK. 1996. Positive selection vectors for allelic exchange. Gene 169:47–52. doi: 10.1016/0378-1119(95)00793-8 [DOI] [PubMed] [Google Scholar]
- 15. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR . J Bacteriol 170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mike LA, Stark AJ, Forsyth VS, Vornhagen J, Smith SN, Bachman MA, Mobley HLT. 2021. A systematic analysis of hypermucoviscosity and capsule reveals distinct and overlapping genes that impact Klebsiella pneumoniae fitness. PLoS Pathog 17:e1009376. doi: 10.1371/journal.ppat.1009376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miethke M, Marahiel MA. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71:413–451. doi: 10.1128/MMBR.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meradji S, Barguigua A, Bentakouk M cherif, Nayme K, Zerouali K, Mazouz D, Chettibi H, Timinouni M. 2016. Epidemiology and virulence of VIM-4 metallo-beta-lactamase-producing Pseudomonas aeruginosa isolated from burn patients in eastern Algeria. Burns 42:906–918. doi: 10.1016/j.burns.2016.02.023 [DOI] [PubMed] [Google Scholar]
- 19. Lodise TP, Bassetti M, Ferrer R, Naas T, Niki Y, Paterson DL, Zeitlinger M, Echols R. 2022. All-cause mortality rates in adults with carbapenem-resistant Gram-negative bacterial infections: a comprehensive review of pathogen-focused, prospective, randomized, interventional clinical studies. Expert Rev Anti Infect Ther 20:707–719. doi: 10.1080/14787210.2022.2020099 [DOI] [PubMed] [Google Scholar]
- 20. Tamma PD, Goodman KE, Harris AD, Tekle T, Roberts A, Taiwo A, Simner PJ. 2017. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis 64:257–264. doi: 10.1093/cid/ciw741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lodise TP, O’Donnell JN, Raja S, Guptill JT, Zaharoff S, Schwager N, Fowler VG Jr, Beresnev T, Wall A, Wiegand K, Serti Chrisos E, Balevic S, Chambers HF, Antibacterial Resistance Leadership Group . 2022. Safety of ceftazidime-avibactam in combination with aztreonam (COMBINE) in a phase I, open-label study in healthy adult volunteers. Antimicrob Agents Chemother 66:e0093522. doi: 10.1128/aac.00935-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karakonstantis S, Rousaki M, Kritsotakis EI. 2022. Cefiderocol: systematic review of mechanisms of resistance, heteroresistance and in vivo emergence of resistance. Antibiotics 11:723. doi: 10.3390/antibiotics11060723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lan P, Lu Y, Chen Z, Wu X, Hua X, Jiang Y, Zhou J, Yu Y. 2022. Emergence of high-level cefiderocol resistance in carbapenem-resistant Klebsiella pneumoniae from bloodstream infections in patients with hematologic malignancies in China. Microbiol Spectr 10:e0008422. doi: 10.1128/spectrum.00084-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moon SH, Udaondo Z, Jun S-R, Huang E. 2022. Cefiderocol heteroresistance in Klebsiella pneumoniae is linked to mutations in the siderophore receptor cirA and β-lactamase activities. Int J Antimicrob Agents 60:106635. doi: 10.1016/j.ijantimicag.2022.106635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nordmann P, Shields RK, Doi Y, Takemura M, Echols R, Matsunaga Y, Yamano Y. 2022. Mechanisms of reduced susceptibility to cefiderocol among isolates from the CREDIBLE-CR and APEKS-NP clinical trials. Microb Drug Resist 28:398–407. doi: 10.1089/mdr.2021.0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Choby JE, Ozturk T, Satola SW, Jacob JT, Weiss DS. 2021. Widespread cefiderocol heteroresistance in carbapenem-resistant Gram-negative pathogens. Lancet Infect Dis 21:597–598. doi: 10.1016/S1473-3099(21)00194-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures S1 and S2 and Tables S1 to S7.
Supplemental file with an overview of the clinical course of the patients found to be harboring VIM+ Gram-negative isolates.
Data Availability Statement
Raw sequencing reads for whole-genome sequencing of clinical Klebsiella pneumoniae isolates were deposited to the NCBI SRA archive under BioProject PRJNA1058428.