Abstract
p53 protein was able to block human and bovine papillomavirus DNA amplificational replication while not interfering with Epstein-Barr virus oriP once-per-cell cycle replication. Oligomerization, intact DNA-binding, replication protein A-binding, and proline-rich domains of the p53 protein were essential for efficient inhibition, while the N-terminal transcriptional activation and C-terminal regulatory domains were dispensable for the suppressor activity of the p53 protein. The inhibition of replication was caused neither by the downregulation of expression of the E1 and E2 proteins nor by cell cycle block or apoptosis. Our data suggest that the intrinsic activity of p53 to suppress amplificational replication of the papillomavirus origin may have an important role in the virus life cycle and in virus-cell interactions.
Human papillomaviruses (HPVs) are small DNA viruses clearly associated with the induction of cancer. The papillomavirus life cycle can be divided into three stages (7, 20). First, following initial entry, the papillomavirus genome is amplified in the nucleus and viral copy number is increased up to 1,000 per haploid cell genome. During the second, maintenance stage, the viral DNA replicates in synchrony with the cellular DNA, at a constant copy number per cell. The third, vegetative replication stage of the viral genome occurs in the terminally differentiated cells. Papillomaviruses have developed an efficient system for modulating the activity of cellular tumor suppressor genes. HPV type 16 (HPV-16) and HPV-18 E6 proteins are capable of interacting with p53 and directing its degradation (50), while the E7 protein forms a complex with retinoblastoma protein (pRB) (15). These events lead to the loss of cell control over crucial events—DNA replication, repair and apoptosis—therefore creating favorable conditions for rapid viral DNA amplification and establishment of infection. In addition, expression of the E6 and E7 proteins may be an indication that some stages of papillomavirus replication during the three-step life cycle are susceptible to the action of p53 or pRB.
The tumor suppressor protein p53 is believed to be one of the key players in the control of the genomic stability of the cells (25, 27, 32). It is structured as a typical eukaryotic transcription activator which contains DNA-binding and transactivation domains and is able to activate or repress the transcription of certain genes (for a review, see reference 25). Exposure of normal cells to different stress conditions induces both an intracellular increase in the steady-state level of p53 and direct activation of the protein (23). As a result, the transition of cells in the cell cycle may be prevented, and apoptotic death of the cells with damaged DNA may be induced (reviewed in reference 32).
Several studies found that the mutation or loss of one or both alleles of p53 was sufficient to allow gene amplification to occur in the cells (36, 67), thus indicating that the p53 protein is involved in the control of events leading to the amplification of genomic sequences. The p53 protein seems to be directly involved in the control of DNA replication and repair (for reviews, see references in reference 25). It has been demonstrated that the p53 protein is capable of interacting with several proteins and enzymes involved in DNA repair or replication, such as single-stranded DNA (ssDNA)-binding replication protein A (RPA) (14, 33), cellular DNA helicases (47), and homologous recombination factor RAD51/RecA (53). The p53 protein lacking its C-terminal regulatory part blocks nuclear DNA replication in the transcription-free Xenopus egg extracts (13). Immunostaining studies show colocalization of the p53 protein with proliferating cell nuclear antigen (PCNA), DNA polymerase α, DNA ligase, and RPA at the sites of DNA replication in herpes simplex virus-infected cells (62). Replication of simian virus 40 (SV40) DNA can be prevented by binding to and inactivating the large T antigen by the p53 protein (52, 60). Replication of the polyomavirus origin is inhibited by p53 in vitro when up to 16 copies of the p53-specific binding sites have been inserted into the plasmid (39), while replication of the polyomavirus origin in vivo is activated by the same protein in a sequence-dependent manner (22).
We studied the effect of the p53 protein on the replication of papillomavirus origins in vivo in different cell lines and found that the p53 protein is a potent repressor of bovine and human papillomavirus amplificational replication. The repression of replication was dependent on the p53 protein concentration in the cells. We show that the intact central DNA-binding domain and the oligomerization domain of the p53 protein, as well as a part of the N-terminal domain containing the RPA-binding and proline-rich sequences, are essential for this activity. In the same time, the p53 protein and its mutants were unable to interfere with the once-per-cell cycle replication of Epstein-Barr virus (EBV) oriP. Repression of papillomavirus DNA amplification is neither an indirect consequence of the p53-dependent cell cycle block or apoptosis nor mediated by the transactivation or transrepression activities of the p53 protein. Possible implications of the observed phenomena on virus-cell interactions will be discussed.
MATERIALS AND METHODS
Plasmids.
Bovine papillomavirus type 1 (BPV-1) E1 expression vector pCGEag, E2 expression vector pCGE2, minimal replication origin plasmid pUCAlu, HPV-11 E1 expression vector pMT2-E1, HPV-11 E2 expression vector pMT2-E2, and HPV-11 upstream regulatory region (URR)-containing plasmid p7072-99 have been described previously (11, 57). pNeoBgl40 contains the BPV-1-URR from nucleotides 6946 to 63 and has been described previously (44). The BPV-1 origin constructs pUC12B and pUC18A have been described previously (55). The HPV-18 E1 and E2 expression vector pCGE1B and origin plasmid pLCR have been reported earlier (45). Plasmid p994 harboring the EBV latent oriP is a kind gift of B. Sugden (24). Bcl-2 expression plasmid pcDBCL2 has been described by Mah et al. (37).
Human p53 cDNAs were cloned into expression vector pCG (54). pCGwtp53 and pCGtrp248 encode wild-type (wt) and Arg248Trp mutant p53 proteins, respectively. The mutant Arg248Trp p53 cDNA was kindly provided by Bert Vogelstein. All deletion mutants were created by PCR and expressed from the pCG vector. pCGΔN39 encodes wt p53 protein with deletion of the first 39 amino acids. pCGΔC362 and pCGΔ305 encode truncated proteins with stop codons at positions 362 and 305, respectively. pCGΔ324-355 encodes p53 with deletion of amino acids at residues 324 to 355. pCGΔN39ΔC362 and pCGΔN39ΔC362trp248 encode wild-type or Arg248Trp mutant p53 starting from amino acid 40 and containing a stop codon at position 362. ΔN61ΔC362 and ΔN92ΔC362 lack the first 61 and 92 N-terminal amino acids, respectively, and contain a stop codon at position 362. ΔProΔC362 and ΔN39ΔProΔC362 lack amino acids 63 to 91 and contain the stop codon at position 362; ΔN39ΔProΔC362 lacks also the N-terminal 39 amino acids. The correctness of the endpoints and all mutated sites of the p53 coding regions were verified by sequencing.
Cells and transfections.
The cell line CHO and its derivatives CHO4.15 (expressing BPV-1 E1 and E2 proteins), CHOBgl40 (in addition containing latent BPV-1 origin plasmid), and CHO212 (expressing BPV-1 E1) (44) were maintained in Ham’s F12 medium supplemented with 10% fetal calf serum. Human osteosarcoma 143 (66), Cos7, and SAOS-2 cells were maintained in Iscove’s modified Dulbecco’s medium with 10% fetal bovine serum. Electroporation experiments were carried out as described earlier, using an Invitrogen ElectroPorator at capacitance setting 960 μF. Voltage settings were 230 V for CHO, CHO4.15, and CHOBgl40 cells, 170 V for human osteosarcoma 143 cells, 180 V for Cos7 cells, and 210 V for SAOS-2 cells. Transfection efficiencies were determined by in situ staining of the cells transfected in parallel with the β-galactosidase-expressing plasmid pON260 (56). Transient replication assays were performed as described previously (56).
Immunoblotting and DNA binding assays.
The expression level of p53 mutant proteins was estimated by Western blot analysis of CHO4.15 cells transfected with 500 ng of p53 expression plasmid and processed 24 or 48 h after transfection according to standard methods (48). Equal amounts of total protein were analyzed in each experiment. Antibodies pAb240, pAb421, and pAb1801 were used for detection of p53 proteins. The E2 protein level in CHO4.15 cells in the presence of expressed p53 constructs was analyzed in the same way, using a mixture of purified monoclonal E2-specific antibodies 1E2, 3F12, 1H10, 1E4, and 3C1 (2). Goat anti-mouse antibody conjugated to alkaline phosphatase was used as a secondary antibody.
The effect of p53 expression on E2-specific DNA-binding activity in CHO4.15 cells was measured as described earlier (2). Analysis was performed 48 h after transfection with p53 expression plasmids. p53-specific DNA binding was tested by an analogous protocol, using the artificial p53-binding double-stranded oligonucleotide 5′AGACATGCCTAGACATGCCT3′ (21). Monoclonal antibodies pAb421 and 3F12 were added for supershifting the p53-specific and E2-specific complexes, respectively. Monoclonal antibody HO7.1 was used for p53 deletion mutants lacking the pAb421 epitope.
Northern blotting of E1 mRNA.
CHO4.15 cells were transfected with 500 ng of p53 expression constructs, and 48 h later the total RNA was extracted by using an RNeasy Total RNA kit supplied by Qiagen. Northern blot analysis of the extracted RNA was performed according to standard methods (48). E1-specific radioactive probe was generated by random priming using the 1.8-kb XbaI-Eco91I BPV-1 E1-encoding fragment from pCGEag (57) as a template. E1-specific signals were quantitated on a PhosphorImager SI (Molecular Dynamics), and the results were normalized to S7- and β-actin-specific signals. Human ribosomal protein S7 (3) and β-actin cDNA plasmids used as a templates to generate radioactive probes were kind gifts of Tarmo Annilo and Mati Reeben, respectively.
Analysis of cell cycle distribution and sub-G1 DNA content of p53-transfected CHO4.15 cells.
Both floating and adherent cells were collected 48 h posttransfection, washed once with phosphate-buffered saline (PBS), and fixed in 5 ml of ice-cold 70% ethanol for flow cytometric analysis. The propidium iodide fluorescent staining of nuclei was analyzed in an ATC3000 flow cytometer (Odam-Brucker, Wissembourg, France) equipped with a Spectraphysics argon laser. Cells were pelleted prior to the analysis, washed once in PBS, suspended in 500 μl of PBS with 1 mM MgCl2 and 30 μg of RNase A per ml, and incubated at 37°C for 1 h to digest cellular RNA. Propidium iodide was added to a final concentration of 10 μg/ml, and samples were incubated on ice for at least 15 min to stain the nuclear DNA. The signals from 50,000 cells were collected from each sample and analyzed by the method of Dean and Jett (13a), using the standard software provided by the manufacturer of the flow cytometer. Cells for the parallel replication assay were processed as described above. The terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed as described in reference 17.
RESULTS
p53 protein inhibits amplificational replication of papillomavirus origins.
We have developed an efficient model system to study the replication of papillomavirus origins in tissue culture (11, 44, 45, 56). To determine whether the p53 protein has any effect on the replication, we performed transient replication assays in CHO-K1 cells, where BPV-1 and HPV origin-containing plasmids replicate in the presence of homologous and heterologous E1 and E2 proteins (11, 57). CHO cells have been used extensively for DNA amplification studies and have been shown to carry the defective p53 gene with substitution Thr211Lys (28). Inspection of these cells with a mixture of p53-specific antibodies did not reveal any detectable endogenous expression of the p53 protein in our hands (data not shown).
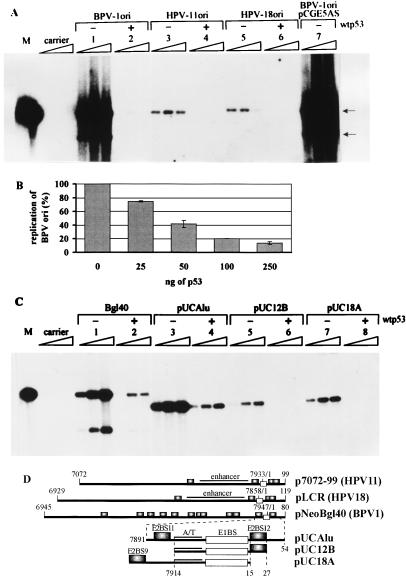
Cotransfection of the BPV-1 E1 and E2 expression plasmids with the BPV-1 origin plasmid into CHO cells resulted in robust replication (Fig. 1A, lane 1). Coexpression of human wt p53 protein suppressed BPV-1 origin replication almost completely in this system (Fig. 1A, lane 2). The extent of suppression was proportional to the amount of introduced p53 expression plasmid (Fig. 1B) and was detected at 25 ng of the cotransfected plasmid DNA. The effects of p53 protein expression on the replication of the HPV-11 (Fig. 1A, lanes 3 and 4) and HPV-18 (lanes 5 and 6) origin plasmids in the presence of the homologous E1 and E2 replication proteins were identical. The replication signal of the HPV origins in CHO cells decreased for the third time point (96 h posttransfection), possibly as a result of the less intense replication and the loss of E1 and E2 expression plasmids from the cells upon cell division. Cotransfection with the vector carrying no p53 sequences did not affect replication of the papillomavirus origin (Fig. 1A, lane 7), which indicates that the block of replication is not caused by promoter competition between the p53, E1, and E2 expression cartridges. Experiments carried out with mouse wt p53 protein gave identical results (data not shown).
FIG. 1.
Southern blot analyses. p53 suppresses the amplificational replication of different papillomavirus replication origins. Episomal DNA was extracted from cells at 48, 72, and 96 h after transfection and digested with restriction endonucleases PstI and DpnI. Filters were probed with radiolabeled HPV-11 URR containing plasmid p7072-99. M, 200 pg of the linear HPV-11 origin plasmid marker; carrier, mock-transfected cells. Arrows indicate the bands generated after digestion of the episomal BPV-1 origin plasmid with PstI. (A) Effect of wt p53 expression on the transient replication of BPV-1, HPV-11, and HPV-18 full-length origin plasmids in CHO cells. In this assay, 100 ng of BPV-1 origin pNeoBgl40 (lanes 1 and 2), HPV-11 origin p7072-99 (lanes 3 and 4), or HPV-18 origin pLCR (lanes 5 and 6), with (+) or without (−) 100 ng of wt p53 expression construct pCGwtp53, was transfected. pCGE5AS, control with plasmid producing no p53 (lane 7). Amounts of E1 and E2 expression vectors used were 250 ng for BPV-1 (pCGEag and pCGE2), 500 ng for HPV-11 (pMT2-E1 and pMT2-E2), and 650 ng for HPV-18 (pCGE1B). (B) The inhibition of replication of the BPV-1 replication origin is proportional to the amount of introduced p53. The replication signals of two independent experiments (72 h posttransfection) were quantified with a PhosphorImager, and signals from cells transfected with origin plasmid only were used as a control to normalize the results. (C) Effect of wt p53 expression on the transient replication of plasmids containing different BPV-1 origin constructs in CHO4.15 cells. In this assay, 100 ng of wt p53 expression plasmid and 100 ng of each BPV-1 replication origin construct were transfected into the cells. Lanes: 1 and 2, replication of full-length BPV-1 origin plasmid pNeoBgl40; 3 and 4, origin plasmid pUCAlu; 5 and 6, origin plasmid pUC12B; 7 and 8, origin plasmid pUC18A. (D) Schematic representation of the papillomavirus replication origin inserts used. The specific transcription enhancer region in HPV replication origins (enhancer), the BPV-1 origin A/T-rich region (A/T), and E1 protein (E1BS; unfilled boxes)- and E2 protein (E2BS9, E2BS11, and E2BS12; shadowed boxes)-binding sites are indicated. Numbers indicate positions on the HPV-11, HPV-18, or BPV-1 nucleotide sequence.
In the next step, we studied the effect of p53 on the replication of different BPV-1 origin deletion mutants in the cell line CHO4.15. This cell line exhibits constitutive expression of BPV-1 E1 and E2 replication proteins from the integrated expression vectors (44). Figure 1C represents replication of the BPV-1 full-length origin plasmid pNeoBgl40 and of origin deletion variants pUCAlu, pUC12B, and pUC18A in the absence and presence of overexpressed p53 protein. Our data show that the replication of plasmids pNeoBgl40, pUCAlu, pUC12B, and pUC18A (depicted schematically in Fig. 1D) is efficiently blocked by the overexpressed p53 protein (Fig. 1C, lanes 2, 4, 6, and 8) and suggest that there are no defined p53-specific cis elements in the BPV-1 origin of replication that could mediate the effect, unless it is the minimal replication origin itself: A/T-rich region and E1- and E2-binding sites.
Structural determinants of the p53 protein responsible for inhibition of amplificational replication of the BPV-1 origin.
To map the domains of the p53 protein responsible for the inhibition of papillomavirus replication, a set of p53 mutants was constructed (schematically depicted in Fig. 2A). The stability, expression level, and activity of the mutant proteins were tested in CHO4.15, Cos7, and SAOS-2 cell lines by Western blot and specific DNA band shift analysis. The mutant proteins with the deleted N-terminal activation domain were expressed at an approximately fivefold-higher level than proteins with the intact N terminus, wt p53, ΔC305, ΔC362, and ΔProΔC362. The N-terminal activation domain contains the binding site of the Mdm2 protein, which has been shown to facilitate degradation of the p53 protein in vivo and therefore reduce the half-life and steady-state level of the p53 protein in cells (19, 26). All of the mutants except those with point mutation Trp248 and deletions ΔC305 and Δ324-355, gave a specific complex with the double-stranded oligonucleotide corresponding to the artificial p53-binding site (21). The intensity of the band shift correlated with the expression level of the p53 proteins in the extract (data not shown).
FIG. 2.
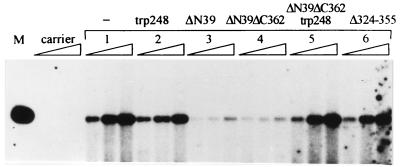
Mapping of the p53 domains necessary for suppression of papillomavirus amplificational replication. (A) Schematic representation of p53 mutants. Numbers indicate positions on the amino acid sequence. (B) Southern blot analysis of the transient replication of BPV-1 origin plasmid pNeoBgl40 in the presence of different p53 mutants in the CHO4.15 cell line. Episomal DNA was extracted from cells at 48, 72, and 96 h after transfection and digested with restriction endonucleases PstI and DpnI. Filters were probed with radiolabeled HPV-11 URR containing plasmid p7072-99; 100 ng of pNeoBgl40 together with 250 ng of p53 expression plasmid was transfected into the cells. Lanes 1 to 8 correspond to the transfections with p53 mutants in the same order as depicted in panel A. Carrier, mock-transfected cells; BPV1 ori, control with no added p53. (C) Relative inhibition of replication of the BPV-1 replication origin by different p53 mutants. The replication signals of three independent experiments (72 h posttransfection) were quantified with a PhosphorImager and signals from the cells transfected with origin plasmid only were used as a control to normalize the results. (D) Southern blot analysis of transient replication of the BPV-1 origin plasmid pUCAlu in the presence of additional N-terminal p53 deletion mutants in the CHO4.15 cell line. Episomal DNA was extracted from cells at 72 and 96 h after transfection and digested with restriction endonucleases PstI and DpnI. Filters were probed with radiolabeled pUCAlu plasmid. Lanes 1, 7, 9, 10, and 11 correspond to transfections with p53 mutants as depicted in panel A.
The BPV-1 origin plasmid and the different mutant p53 protein expression plasmids were cotransfected into CHO4.15 cells; episomal DNA was harvested and analyzed by Southern blotting (Fig. 2B). wt p53, the C-terminal regulatory domain-defective mutant ΔC362, the N-terminal deletion mutant ΔN39 lacking the transcription activation domain, and the double-deletion mutant ΔN39ΔC362 all retained the ability to suppress replication (Fig. 2B; compare lanes 1, 2, 4, and 7 with lane BPV1 ori). The replication signals from three independent experiments were measured with a PhosphorImager, and the data are presented in Fig. 2C. The mutants with a deleted oligomerization domain (Δ324-355) or the whole C-terminal part of the protein up to amino acid 305 (ΔC305) (Fig. 2B, lanes 3 and 5; Fig. 2C) had little or no effect on replication. The point mutation Arg248Trp in the DNA-binding domain of the p53 protein abolished the suppressor activity of the full-size p53 protein (Fig. 2B, lane 6) and even seemed to convert the double-deletion mutant ΔN39ΔC362 to an activator of replication (Fig. 2B, lane 8; Fig. 2C). These data indicate that intact DNA-binding and oligomerization domains are both necessary for the p53 protein activity to suppress papillomavirus DNA amplificational replication, while the N-terminal transcription activation and C-terminal regulatory domains are dispensable for this activity.
The active p53 deletion mutant ΔN39ΔC362 contains, in addition to the DNA-binding core region (residues 100 to 300), flexible linker region (residues 301 to 320), and oligomerization domain (residues 320 to 360) (25), also the RPA-binding domain (residues 40 to 60) (1, 14, 30) and a proline-rich putative binding site for proteins with the SH3 domain (residues 61 to 91) (59). We constructed four additional p53 deletion variants and tested their stability and DNA-binding activity. The constructed mutants were stable in CHO4.15 cells and bound DNA sequence specifically, as measured by DNA gel shift assay (data not shown). These mutants were used for the suppression of replication of the minimal origin plasmid pUCAlu in CHO4.15 cells (Fig. 2D). None of the newly constructed deletion mutants was able to block replication of the pUCAlu origin plasmid comparably to wt p53 or ΔN39ΔC62. These data indicate that four domains of the p53 protein—oligomerization (residues 320 to 360), DNA-binding (residues 100 to 300), proline-rich (residues 61 to 92), and RPA-binding (residues 40 to 61) domains—are necessary for the replication suppressor activity of the protein.
p53 protein suppresses only amplificational DNA replication.
The action of p53 and its mutants on different replication modes was studied in human osteosarcoma cell line 143. The 143 cell line expresses constitutively EBNA-1, the only viral protein necessary for the replication of EBV latent oriP. These cells are also permissive for the E1- and E2-dependent replication of the papillomavirus origin. In contrast to papillomaviruses, which quickly amplify their genome after viral entry into the cell, EBV oriP probably makes use of the cellular control mechanisms that guarantee once-per-cell cycle replication (65). We cotransfected the plasmids encoding p53 and HPV-11 E1 and E2 proteins together with the HPV-11 origin plasmid and EBV oriP plasmid into the 143 cells and studied their replication by Southern blot analysis. The replication assay conditions were adjusted so that relative replication signals of oriP and HPV-11 origin had comparable intensities on the same Southern blot. Once-per-cell cycle replication of the oriP-containing plasmid was not suppressed by wt p53, while amplificational replication of the papillomavirus origin was abolished in the same cells (Fig. 3; compare lanes 1 and 2). The mutant p53 proteins affected papillomavirus replication similarly in the 143 cells and CHO cells. Mutants ΔN39 and ΔC362, which suppressed replication of the BPV-1 full-length origin in CHO4.15 cells, also blocked replication of the HPV-11 origin in the 143 cell line and at the same time had little effect on the replication of oriP (lanes 3 and 5). Mutants ΔC305 and Δ324-355 influenced the replication of neither HPV-11 origin nor oriP (lanes 4 and 6).
FIG. 3.
Southern blot analysis of coreplication of oriP and HPV-11 origin plasmids in human osteosarcoma 143 cells. p53 blocks replication of the papillomavirus origin but not EBV oriP. Episomal DNA was extracted at 48, 72, and 96 h posttransfection, digested with BamHI and DpnI, and probed with radiolabeled origin plasmid p7072-99. One microgram of oriP plasmid p994 and 250 ng of HPV-11 origin plasmid p7072-99 together with HPV-11 E1 and E2 expression plasmids pMT-E1 and pMT-E2 (1 μg of each) were transfected into the cells; 250 ng of wt or mutant p53 expression plasmid was added as indicated (lanes 2 to 6). Other lanes: oriP and HPV-11 ori, 200 pg of the marker plasmids linearized with BamHI; carrier, negative control with carrier DNA only; 1, positive control with no added p53.
p53 inhibits amplificational replication of the BPV-1 origin in SAOS-2 cells.
Replication of the papillomavirus origin was tested also in human osteosarcoma SAOS-2 cells that lack endogenous p53 and pRB expression. The expression of exogenous wt p53 and several transactivation-competent mutants in SAOS-2 cells is sufficient to lead the cells to apoptosis (10, 68). To avoid these side effects, we used p53 mutants deficient in transcription activation activity. Cotransfection of the BPV-1 E1 and E2 expression plasmids together with the replication origin and p53 expression plasmids into SAOS-2 cells and subsequent analysis of the episomal DNA showed that mutants ΔN39 and ΔN39ΔC362 inhibited replication of the papillomavirus origin in SAOS-2 cells (Fig. 4, lanes 3 and 4), while mutants Trp248, ΔN39ΔC362 Trp248, and Δ324-355 (lanes 2, 5, and 6, respectively) had no effect on replication. These data are similar to the results of experiments with the cell lines CHO4.15 (using BPV-1 origin) and 143 (using HPV-11 origin) and suggest that the suppression of papillomavirus replication is a direct intrinsic property of the exogenously expressed p53 protein and is neither influenced by the endogenous p53 nor achieved through the pRB-regulated pathways.
FIG. 4.
Southern blot analysis of the BPV-1 origin plasmid pUCAlu in SAOS-2 cells (radiolabeled origin plasmid pUCAlu used as a probe). p53 mutant proteins inhibit replication of the BPV-1 minimal origin in SAOS-2 cells lacking endogenous p53 and pRB proteins. BPV-1 minimal origin plasmid pUCAlu (500 ng) together with BPV-1 E1 and E2 expression plasmids pCGEag and pCGE2 (1 μg of each) was transfected into the cells; 500 ng of p53 mutant proteins was cotransfected as indicated (lanes 2 to 6). Other lanes: M, 200 pg of the pUCAlu marker linearized with PstI; carrier, control transfection with carrier DNA only; 1, positive control with no p53 construct added.
p53 does not cause downregulation of expression of the E1 and E2 proteins.
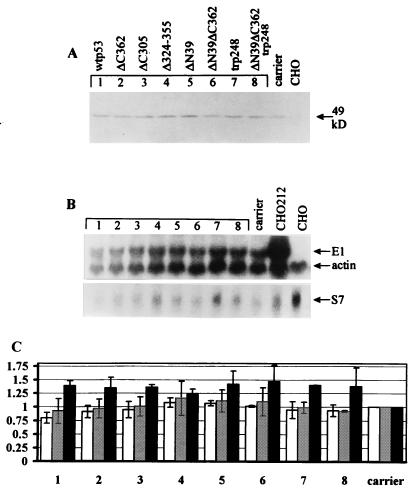
The E1 and E2 proteins are absolutely necessary for papillomavirus replication. The p53 protein has been shown to possess transcription repressor activity in certain cases. Therefore, the inhibition of papillomavirus replication could, in principle, be achieved by downregulation of the expression level or activity of these proteins. We studied the expression level and activity of the BPV-1 replication proteins in CHO4.15 cells in the presence of the overexpressed wt and mutant p53 proteins. E2 protein expression is directed by the HSP70 promoter, and E1 protein expression is directed by the SRα promoter in CHO4.15 cells (44). These cells are very efficiently transfected by electroporation (about 70%, based on parallel β-galactosidase expression vector pON260 transfections), and this fact served as a rationale for the measurements described below. The transfected CHO4.15 cells were studied for the expression level of the E2 protein by Western blot analysis of the cell lysates. Transfection efficiencies were determined in parallel in all experiments. Western blot analysis did not reveal any reproducible effects of the expression of wt or mutant p53 proteins on the steady-state level of the E2 protein in CHO4.15 cells (Fig. 5A). A possibility remained that p53 could modulate the activities of the E2 protein, for example, the ability to bind DNA.
FIG. 5.
Expression of p53 does not affect the level of E1 and E2 expression. A p53 expression construct (500 ng) was electroporated into CHO4.15 cells. In panels B and C, lanes and columns 1 to 8 represent transfections with different p53 mutants in the same order as in panel A. carrier, control with carrier DNA only; CHO, mock-transfected CHO cells (lacking both E1 and E2 expression). All analyses were performed 48 h posttransfection. (A) Western blot analysis of the E2 protein levels in p53-transfected CHO4.15 cells, using a mixture of five different E2-specific monoclonal antibodies (see Materials and Methods). (B) Northern blot analysis of the endogenous E1 mRNA levels in total RNA preparations from transfected CHO4.15 cells. CHO212, total RNA from E1-expressing cell line CHO212. The same filter was probed first with radiolabeled E1- and β-actin-specific probes and then reprobed with ribosomal protein S7-specific probe. Approximate lengths for mRNAs are 700 bp for S7, 2.0 kb for β-actin and 2.3 kb for E1. (C) Quantitation of the E1 Northern blots and E2 gel shift assay with a PhosphorImager. The E1 mRNA-specific signals in the total RNA preparations were normalized to the β-actin (open columns) and ribosomal protein S7 (shaded columns) mRNA signals in the RNA samples. Black columns represent the E2 gel shift data. The E2-specific signal in the lysates of the mock-transfected cells and the normalized E1 mRNA-specific signal from carrier-transfected control cells were set at 1.0 in each experimental series. Each column represents the average of two independent experiments.
We performed a DNA mobility shift assay of CHO4.15 cell lysates transfected with p53 expression constructs. The lysates were tested for E2-specific DNA binding with the oligonucleotide corresponding to E2-binding site 9 of the BPV-1 genome (34). To increase the specificity of the assay, we supershifted the E2-DNA complex with an excess of the E2-specific monoclonal antibody 3F12. E2-specific radioactive signals were measured with a PhosphorImager, and the results were normalized to the total amount of protein in the lysate, as determined by the Bradford assay (8). As in the case of measuring the steady-state level of the E2 protein, we were unable to detect any significant changes in the levels of active E2 protein in response to the expression of wt or mutant p53 proteins in CHO4.15 cells (Fig. 5C).
The low expression level of the E1 protein in CHO4.15 cells made it impossible to detect E1 by quantitative Western blot analysis or immunoprecipitation. Instead, we performed Northern blot analysis and analyzed the steady-state level of the E1 mRNA in response to p53 expression (Fig. 5B). The transcription level of the E1 protein coding sequence was determined relative to β-actin and ribosomal protein S7 mRNA levels on the same blots (Fig. 5B and C), and E1 mRNA-specific hybridization signals were measured with a PhosphorImager. Quantitation of the E1 mRNA level normalized to β-actin and S7 mRNA levels showed no downregulation of the E1 mRNA level in response to wt and mutant p53 expression in CHO4.15 cells (Fig. 5C).
These data suggest that the effect of p53 on papillomavirus amplificational replication is not caused by downregulation of expression of the E1 or E2 proteins, although these experiments do not exclude the possibility that p53 interferes with E1 or E2 (or both) activities at some stage of initiation or elongation of replication.
The inhibition of papillomavirus replication is not the consequence of p53-induced cell cycle block or apoptosis.
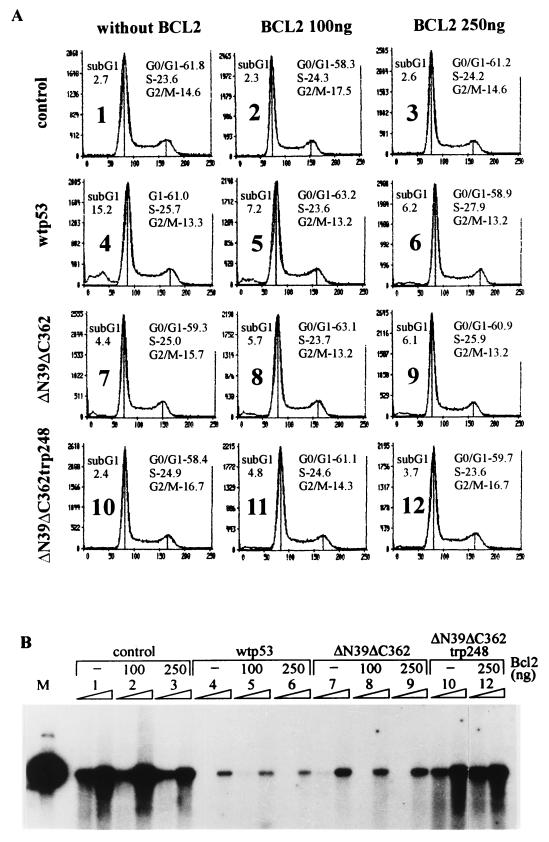
p53 is a mediator of cell cycle block and apoptotic cell death. To examine the possibility that the suppression of papillomavirus amplification is an indirect consequence of any (or both) of these effects, we analyzed the p53-transfected CHO4.15 cells by flow cytometry. Overexpression of wt p53 protein in CHO4.15 cells induced detectable apoptosis in the culture, as shown by the appearance of the sub-G1 DNA-containing fraction in the cell cycle profile 48 h posttransfection (Fig. 6A, panel 4). To examine the possible connection between p53-induced apoptosis and the suppression of replication, we made use of the ability of the Bcl2 protein to prevent the p53-induced apoptosis of cells (51). Increasing amounts of the Bcl2 expression plasmid were transfected into the cells. The expression of Bcl2 considerably reduced the amount of cells in the sub-G1 DNA-containing fraction of the cells transfected with wt p53 (compare panels 4, 5, and 6). The cells transfected with p53 deletion mutant ΔN39ΔC362 as well as with the same deletion mutant with the Arg248Trp point mutation had some small sub-G1 fraction, probably induced by electroporation, which was not influenced by the expression of Bcl2 in the cells (panels 7 to 12). The percentage of the apoptotic cells in these experiments was measured also by the TUNEL assay (Table 1), which gave essentially the same result and showed that Bcl2 expression in CHO4.15 cells reduced considerably the number of the p53-induced apoptotic cells in the culture. The distribution of CHO4.15 cells in G1/G0, S, and G2/M stages of the cell cycle was not influenced by the expression of Bcl2 or p53. We also analyzed if the Bcl2 rescues the replication suppression induced by p53 or its mutants. Expression of Bcl2 in CHO4.15 cells did not influence the replication of the BPV-1 origin itself, nor did it abrogate the inhibitory effects of wt p53 and deletion mutant ΔN39ΔC362 on replication of the origin (Fig. 6B, lanes 1 to 9). These data support the conclusion that the effect of the p53 protein on papillomavirus amplificational replication is not an indirect consequence of cell cycle block or apoptotic cell death.
FIG. 6.
Suppression of BPV-1 amplificational replication by p53 proteins is not the consequence of the p53-induced apoptosis or cell cycle block. (A) Flow cytometric analysis of the cell cycle distribution and the sub-G1 DNA-containing apoptotic fraction of the p53-transfected CHO4.15 cells. In this assay, 250 ng of the p53 expression constructs without Bcl2 or together with 100 or 250 ng of the Bcl2 expression plasmid pcDBCL2 was transfected into the CHO4.15 cells; 100 ng of BPV-1 full-length origin plasmid pNeoBgl40 was used in each transfection. control, cells with no p53 expression constructs added. Cells were fixed 48 h after transfection. The percentage of apoptotic sub-G1 DNA-containing signals and the calculated percentages of cells in G0/G1, S, and G2/M phases (from total of 50,000 cells) are indicated on the each graph. y axis, cell number; x axis, DNA content. The sub-G1 DNA fraction was not considered in the cell cycle calculations. Standard software provided by the manufacturer (Odam-Brucker) was used for the cell cycle calculations. (B) Southern blot analysis of the episomal DNA in the cells cotransfected with p53, Bcl2, and the BPV-1 origin plasmid pNeoBgl40. Episomal DNA was extracted at 72 and 96 h after transfection, digested with HindIII and DpnI, and probed with radiolabeled origin plasmid pUCAlu. Lanes: M, 200 pg of the marker plasmid linearized with HindIII; 1 to 12, transfections 1 to 12 in panel A.
TABLE 1.
Apoptotic fraction in total population of CHO4.15 cells transfected with p53 and Bcl2 constructs (as measured by TUNEL assay)
| Cells | Apoptotic fraction (%)
|
|
|---|---|---|
| No Bcl2 cotransfected | 250 ng of Bcl2 cotransfected | |
| Control (no p53 transfected) | NDa | 2.9 |
| Transfected with p53 construct: | ||
| wt p53 | 19.2 | 6.0 |
| ΔN39ΔC362 | 2.3 | 6.7 |
| ΔN39ΔC362Trp248 | 4.8 | ND |
ND, not determined.
DISCUSSION
p53 as a suppressor of papillomavirus amplificational replication: possible implications for virus-cell interactions.
Amplificational replication of papillomavirus DNA is initiated after entry of the viral genome into the cell nucleus, which leads to a rapid increase in copy number of the virus genome during S phase (20). Papillomaviruses rely on cellular replication factors and enzymes (40) and coordinate the initiation of replication by two viral origin recognition and initiation proteins, E1 and E2 (11, 45, 56, 57, 64). The same viral proteins are used at the following latent replication stage. The mechanism of switching from amplificational to controlled-maintenance replication is unknown. Our data show that amplificational replication of bovine and different human papillomaviruses in the short-term replication assay can be suppressed by the p53 protein in all cell lines studied. It seems not to require any response elements in the origin of replication. It also does not require any activities carried by the C-terminal regulatory and N-terminal transactivation domains of the p53 protein, including the ability to activate transcription. The DNA-binding domain of p53 has been shown to be the target for most of the missense mutations which inactivate the tumor suppressor function of this protein in cells (12). Incidentally, the very same mutations inactivated p53 in the replication system studied.
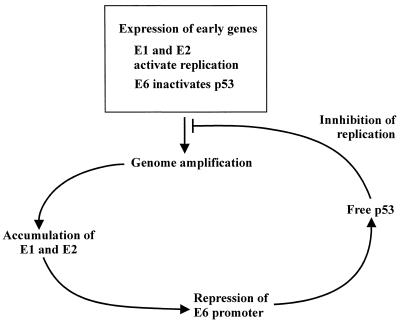
p53 has been shown to block the replication of SV40 by interacting with large T antigen. The binding of SV40 large T antigen by the p53 protein downregulates the helicase function of the T antigen (52); in addition, p53 competes with DNA polymerase α for the binding of SV40 large T antigen at the initiation of SV40 DNA synthesis (16). Mouse polyomavirus replication was shown not to be inhibited by the p53 protein (22, 39) unless additional (up to 16) p53-specific RGC sites were included in the plasmid (39). This shows that sensitivity of the viruses within the papovavirus family to the action of tumor suppressor protein p53 is variable and obviously reflects the differences in the viral life cycles and different strategies for the utilization of cellular control mechanisms by these viruses. Papillomaviruses must infect basal epithelial cells in order to establish productive infection of basal and suprabasal epithelial cells. Amplificational replication of the viral genome in these cells is essential for the establishment of infection. The oncoproteins encoded by the E5, E6, and E7 open reading frames of papillomaviruses are essential for providing the cellular environment for the replication of viral DNA. However, amplificational replication has to be controlled in order to avoid overreplication and unscheduled death of basal or suprabasal cells, because the synthesis of late genes and the production of infectious particles takes place only in the terminally differentiated epithelial cells. It is tempting to speculate that the ability of p53 to block the papillomavirus amplificational replication characterized in the model system studied is actually used by the virus to control the productive infection of basal cells. The E6 proteins of the high-risk (50) and low-risk (35) HPVs have been shown to interact with p53; however, only E6 from the high-risk HPVs directs p53 to degradation (50). It can be speculated that the binding of p53 by the E6 proteins of either high-risk or low-risk human and animal viruses reduces the replication suppressor activity of p53. Other important players in this regulatory mechanism are the replication proteins E1 and E2, which determine the efficiency of initiation of replication. The expression level of these proteins would certainly depend on the copy number of the viral genome, therefore providing the positive feedback for amplification. The papillomavirus replication proteins E1 and/or E2 have been shown to repress the promoter which is closest to the replication origin and directs E6 expression (31, 38, 49, 58). Therefore, higher levels of the E1 and E2 proteins would reduce the level of E6, which in turn results in the higher level of the active p53 protein capable of suppressing replication. These interrelationships among p53, E6, E1, and/or E2 proteins could provide a regulatory loop which can be used by some papillomaviruses to keep viral genome amplification in optimal limits (Fig. 7). The proposed regulatory loop could further serve as one of the mechanisms for the copy number control of the replication of papillomavirus genome during the latent infection of the basal cells. However, the mechanism may be different with different papillomavirus types, as, for example, attempts to find any interaction between BPV-1 E6 and p53 have appeared to be unsuccessful. It is still possible that in this case some other step in the cellular control pathways, up- or downstream of p53 itself, may be neutralized by viral regulatory proteins.
FIG. 7.
A putative p53-controlled regulatory loop in the amplificational replication step of the papillomavirus life cycle.
The putative mechanism of action of the p53 protein.
The p53 protein, in principle, could suppress papillomavirus DNA replication in vivo by a number of different mechanisms, such as by arresting the cell cycle, inducing apoptotic death of cells, downregulating the expression or activity of the E1 and E2 proteins, or interfering with viral and cellular replication proteins at the stages of initiation or elongation of DNA replication.
The induction of apoptosis or cell cycle block is an unlikely mechanism for the apparent suppression of replication by p53 or its mutants in the cells studied, as shown by the measurement of apoptosis and distribution of cells in the cell cycle. In addition, the efficient rescue of CHO4.15 cells from the wt p53-induced apoptosis by Bcl2 expression did not affect the suppression of BPV-1 origin replication in the same cells. Mutant ΔN39ΔC362 efficiently blocked replication of the papillomavirus origin in all of the studied cells but was unable to induce any detectable apoptosis. Additional convincing data come from the coreplication assay of the EBV and HPV-11 origins, which show that in the same cells two origins have differing sensitivity to the expression of p53 or its mutants. Replication of EBV oriP (65) and the papillomavirus origin (18) takes places during the S phase of the cell cycle, and intensive apoptotic death of the cells or cell cycle block should have also considerably reduced the replication of EBV oriP. Therefore, these experiments exclude several indirect and obvious explanations for the observed suppression of papillomavirus amplificational replication. It also seems unlikely in the light of these data that the replication block could have been achieved through the inactivation of general replication factors such as RPA, PCNA, and others by the expression of p53 or its mutants, because those factors are presumably used for the replication of EBV oriP and chromosomal DNA as well.
Another simple explanation is that the p53-induced suppression of papillomavirus replication could have been achieved through negatively modulating the activity of essential viral replication proteins (similarly to the case of SV40 virus) or through downregulating the expression of these proteins. However, we could not detect any significant p53-induced drop of the expression level and DNA-binding activity of the E2 protein and transcription level of E1 in CHO4.15 cells. Also, there are no data in the literature showing the interaction of the p53 protein with the E1 or E2 proteins of any papillomaviruses or demonstrating the modulation of activities of these proteins by p53. Therefore, the p53 protein has to act at later stages of replication initiation process, i.e., loading of the replication complex on the origin, unwinding of DNA, or elongation of the replication fork.
Our findings are substantiated by the fact that the C-terminally truncated form of the p53 protein (analogous to our mutant ΔC362) is able to block nuclear DNA replication in vitro in the transcription-free DNA replication extract from Xenopus laevis activated eggs (13). As for the suppression of amplificational replication of papillomavirus origin in the somatic cells, the DNA-binding activity of p53 was needed for the block of nuclear DNA replication in the transcription-free Xenopus extracts. It is possible that these two replication systems have similar p53-sensitive steps. Mapping of the p53 protein domains necessary for the repression of papillomavirus amplificational replication demonstrated that the intact DNA-binding core and oligomerization domains are clearly necessary. Several activities have been mapped to the core domain, including the sequence-specific DNA-binding (6, 43, 61), ssDNA-binding (4), and 3′-to-5′ exonuclease (41) activities of the p53 protein. All these activities, as well as the ability to suppress papillomavirus amplificational replication, are inactivated by point mutations which either abolish the direct contact of the protein with DNA or induce inactive conformation of the protein (12, 41). It is unlikely that the sequence-specific double-stranded DNA-binding function of p53 could be responsible for the suppression of amplificational replication, while sequence-nonspecific ssDNA-binding activity could be used by the p53 protein in this process.
Full-length p53 protein DNA-binding activity is regulated, sterically or allosterically, by the C-terminal domain of the protein (for a review, see reference 25). In addition, the C-terminal domain binds to DNA bulges resulting from DNA deletion/insertion mismatches (29) and also to the ends of short ssDNA molecules (5), promoting the reannealing of complementary strands (9, 42). Deletion of the last 30 residues, which has previously been shown to remove the above-mentioned activities of the p53 protein, did not affect its ability to suppress papillomavirus amplification in our assays. However, it is possible that both the ssDNA-binding and reannealing functions of the C terminus additionally contribute to the amplification suppressor activity of p53 in the case of the full-length protein.
Core and oligomerization domains, though necessary, are not sufficient for the replication suppressor activity. An additional N-terminal sequence that has been shown to contain two intriguing determinants, RPA-binding (residues 40 to 60) and proline-rich (residues 61 to 90) domains, is also needed. Deletion of any or both of these domains crippled the p53 protein in the replication suppression assay. It is highly likely that p53 coordinates its replication suppressor activity with other proteins bound on the ssDNA, such as through the interaction with RPA (14). RPA facilitates DNA unwinding and DNA synthesis in the initiation and elongation stages of DNA replication (63). The interaction of p53 with RPA could be important in two respects. First, ssDNA-bound RPA could be the target for p53 action, and its interaction with p53 could sequester RPA from the ssDNA; second, interaction between RPA and p53 on the stabilized ssDNA facilitates recognition of the amplifying DNA by p53. Interaction of p53 and RPA in solution does not require an intact DNA-binding domain (1, 14, 30), while it is needed for the suppression of replication. This suggests the possibility that p53-RPA interaction takes place on the ssDNA. Deletion mapping of p53 activity showed that also the proline-rich putative signalling domain in the N-terminal part of the protein is required for the suppression of papillomavirus replication. This domain contains several copies of the PXXP motif (P represents proline; X represents any amino acid), which constitute a binding site for the proteins with the SH3 domain (59). It has been suggested that this domain plays a critical role in the transmission of transactivation-independent antiproliferative signals and presumably links p53 directly to the appropriate signal transduction pathways (46, 59).
However, despite the findings provided here pointing to an attractive putative mechanism, additional experimental data are needed to determine in detail the mechanism of action of p53 in the suppression of papillomavirus amplificational replication.
ACKNOWLEDGMENTS
D.L. and I.I. contributed equally to this work.
We thank Bill Sugden for providing human osteosarcoma cell line 143 and EBV oriP plasmid; Bert Vogelstein for providing the Trp248 mutant of p53 cDNA; Jo Milner and Thierry Soussi for p53 antibodies; Anne Kalling, Jevgeni Popov, and Ilvi Rimm for excellent technical assistance; and Alar Karis, Juhan Sedman, Tõnis Örd, Richard Villems, and Tanel Tenson for critical reading of the manuscript.
This study was supported by grants 2496, 2497, 2315, and 2316 from the Estonian Science Foundation, grant HHMI 75195-541301 from the Howard Hughes Medical Institute, and grants and CIPA930257, ERBCIPD 94002, and CIPA-CT94-0154 from the EU.
REFERENCES
- 1.Abramova N A, Russell J, Botchan M, Li R. Interaction between replication protein A and p53 is disrupted after UV damage in a DNA repair-dependent manner. Proc Natl Acad Sci USA. 1997;94:7186–7191. doi: 10.1073/pnas.94.14.7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abroi A, Kurg R, Ustav M. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J Virol. 1996;70:6169–6179. doi: 10.1128/jvi.70.9.6169-6179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annilo T, Laan M, Stahl J, Metspalu A. The human ribosomal protein S7-encoding gene: isolation, structure and localization in 2p25. Gene. 1995;165:297–302. [PubMed] [Google Scholar]
- 4.Bakalkin G, Selivanova G, Yakovleva T, Kiseleva E, Kashuba E, Magnusson K P, Szekely L, Klein G, Terenius L, Wiman K G. p53 binds single-stranded DNA ends through the C-terminal domain and internal DNA segments via the middle domain. Nucleic Acids Res. 1995;23:362–369. doi: 10.1093/nar/23.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakalkin G, Yakovleva T, Selivanova G, Magnusson K P, Szekely L, Kiseleva E, Klein G, Terenius L, Wiman K G. p53 binds single-stranded DNA ends and catalyzes DNA renaturation and strand transfer. Proc Natl Acad Sci USA. 1994;91:413–417. doi: 10.1073/pnas.91.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bargonetti J, Manfredi J J, Chen X, Marshak D R, Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993;12:2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- 7.Botchan M R, Berg L, Reynolds J, Lusky M. The bovine papillomavirus replicon. In: Evered D, Clark S, editors. Papillomaviruses. New York, N.Y: John Wiley & Sons; 1986. pp. 53–67. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Brain R, Jenkins J R. Human p53 directs DNA strand reassociation and is photolabelled by 8-azido ATP. Oncogene. 1994;9:1775–1780. [PubMed] [Google Scholar]
- 10.Chen X, Ko L J, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 11.Chiang C M, Ustav M, Stenlund A, Ho T F, Broker T R, Chaw L T. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc Natl Acad Sci USA. 1992;89:5799–5803. doi: 10.1073/pnas.89.13.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 13.Cox L S, Hupp T, Midgley C A, Lane D P. A direct effect of activated human p53 on nuclear DNA replication. EMBO J. 1995;14:2099–2105. doi: 10.1002/j.1460-2075.1995.tb07201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Dean P N, Jett J H. Mathematical analysis of DNA distributions derived from flow microfluorometry. J Cell Biol. 1974;60:523–527. doi: 10.1083/jcb.60.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta A, Ruppert S M, Aster J C, Winchester E. Inhibition of DNA replication factor RPA by p53. Nature. 1993;365:79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- 15.Dyson N, Howley P M, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 16.Gannon J V, Lane D P. p53 and DNA polymerase alpha compete for binding to SV40 T antigen. Nature. 1987;329:456–458. doi: 10.1038/329456a0. [DOI] [PubMed] [Google Scholar]
- 17.Gavrieli Y, Sherman Y, Ben-Sasson S A. Identification of programmed cell-death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert D M, Cohen S N. Bovine papillomavirus plasmids replicate randomly in mouse fibroblasts throughout S-phase of the cell cycle. Cell. 1987;50:59–68. doi: 10.1016/0092-8674(87)90662-3. [DOI] [PubMed] [Google Scholar]
- 19.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 20.Howley P M. Papillomavirinae: the viruses and their replication. In: Fields B N, et al., editors. Virology. 2nd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2045–2076. [Google Scholar]
- 21.Hupp T, Meek D, Midgley C, Lane D. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 22.Kanda T, Segawa K, Ohuchi N, Mori S, Ito Y. Stimulation of polyomavirus DNA replication by wild-type p53 through the DNA-binding site. Mol Cell Biol. 1994;14:2651–2663. doi: 10.1128/mcb.14.4.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 24.Kirchmaier A L, Sugden B. Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J Virol. 1995;69:1280–1283. doi: 10.1128/jvi.69.2.1280-1283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–72. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 26.Kubbutat M, Jones S, Vousden K. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 27.Lane D P. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 28.Lee H, Larner J M, Hamlin J L. Cloning and characterization of Chinese hamster p53 cDNA. Gene. 1997;184:177–183. doi: 10.1016/s0378-1119(96)00592-6. [DOI] [PubMed] [Google Scholar]
- 29.Lee S, Elenbaas B, Levine A, Griffith J. p53 and its 14 kDa C-terminal domain recognize primary DNA damage in the form of insertion/deletion mismatches. Cell. 1995;81:1013–1020. doi: 10.1016/s0092-8674(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 30.Leiter L M, Chen J, Marathe T, Tanaka M, Dutta A. Loss of transactivation and transrepression function, and not RPA binding, alters growth suppression by p53. Oncogene. 1996;12:2661–2668. [PubMed] [Google Scholar]
- 31.Le Moal M A, Yaniv M, Thierry F. The bovine papillomavirus type 1 (BPV1) replication protein E1 modulates transcriptional activation by interacting with BPV1 E2. J Virol. 1994;68:1085–1093. doi: 10.1128/jvi.68.2.1085-1093.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine A J. P53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 33.Li R, Botchan M R. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 34.Li R, Knight J D, Bream G, Stenlund A, Botchan M R. Specific recognition nucleotides and their DNA context determine the affinity of E2 protein for 17 binding sites in the BPV-1 genome. Genes Dev. 1989;3:510–526. doi: 10.1101/gad.3.4.510. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Coffino P. High-risk human papillomavirus E6 protein has two distinct binding sites within p53, of which only one determines degradation. J Virol. 1996;70:4509–4516. doi: 10.1128/jvi.70.7.4509-4516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Altered cell-cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 37.Mah S P, Zhong L T, Liu Y, Roghani A, Edwards R H, Bredesen D E. The protooncogene bcl-2 inhibits apoptosis in PC12 cells. J Neurochem. 1993;60:1183–1186. doi: 10.1111/j.1471-4159.1993.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 38.McBride A A, Romanczuk H, Howley P M. The papillomavirus E2 regulatory proteins. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- 39.Miller S D, Farmer G, Prives C. p53 inhibits DNA replication in vitro in a DNA-binding-dependent manner. Mol Cell Biol. 1995;15:6554–6560. doi: 10.1128/mcb.15.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller F, Seo Y-S, Hurwitz J. Replication of bovine papillomavirus type 1 origin-containing DNA in crude extracts and with purified proteins. J Biol Chem. 1994;269:17086–17094. [PubMed] [Google Scholar]
- 41.Mummenbrauer T, Janus F, Müller B, Wiesmüller L, Deppert W, Grosse F. P53 protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85:1089–1099. doi: 10.1016/s0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- 42.Oberosler P, Hloch P, Ramsperger U, Stahl H. p53-catalyzed annealing of complementary single-stranded nucleic acids. EMBO J. 1993;12:2389–2396. doi: 10.1002/j.1460-2075.1993.tb05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pavletich N P, Chambers K A, Pabo C D. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993;12:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 44.Piirsoo M, Ustav E, Mandel T, Stenlund A, Ustav M. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 1996;15:1–11. [PMC free article] [PubMed] [Google Scholar]
- 45.Remm M, Brain R, Jenkins J R. The E2 binding sites determine the efficiency of replication for the origin of human papillomavirus type 18. Nucleic Acids Res. 1992;20:6015–6021. doi: 10.1093/nar/20.22.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruaro E M, Collavin L, Del Sal G, Haffner R, Oren M, Levine A J, Schneider C. A proline-rich motif in p53 is required for transactivation-independent growth arrest as induced by Gas1. Proc Natl Acad Sci USA. 1997;94:4675–4680. doi: 10.1073/pnas.94.9.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakurai T, Suzuki M, Sawazaki T, Ishii S, Yoshida S. Anti-oncogene product p53 binds DNA helicase. Exp Cell Res. 1994;215:57–62. doi: 10.1006/excr.1994.1314. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 49.Sandler A B, Vande Pol S B, Spalholz B A. Repression of bovine papillomavirus type 1 transcription by the E1 replication protein. J Virol. 1993;67:5079–5087. doi: 10.1128/jvi.67.9.5079-5087.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 51.Strasser A, Harris A W, Jacks T, Cory S. DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell. 1994;79:329–339. doi: 10.1016/0092-8674(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 52.Stürzbecher H-W, Brain R, Maimets T, Addison C, Rudge K, Jenkins J R. Mouse p53 blocks SV40 DNA replication in vitro and downregulates T antigen DNA helicase activity. Oncogene. 1988;3:405–413. [PubMed] [Google Scholar]
- 53.Stürzbecher H-W, Donzelmann B, Henning W, Knippschild U, Buchhop S. P53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 55.Ustav E, Ustav M, Szymanski P, Stenlund A. The bovine papillomavirus origin of replication requires a binding site for the E2 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:898–902. doi: 10.1073/pnas.90.3.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ustav M, Ustav E, Szymanski P, Stenlund A. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 1991;10:4321–4329. doi: 10.1002/j.1460-2075.1991.tb05010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vande Pol S B, Howley P M. Negative regulation of the bovine papillomavirus E5, E6, and E7 oncogenes by the viral E1 and E2 genes. J Virol. 1994;69:395–402. doi: 10.1128/jvi.69.1.395-402.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker K K, Levine A J. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang E H, Friedman P N, Prives C. The murine p53 protein blocks replication of SV40 DNA in vitro by inhibiting the initiation functions of SV40 large T antigen. Cell. 1989;57:379–392. doi: 10.1016/0092-8674(89)90913-6. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y, Reed M, Wang P, Stenger J E, Mayr G, Anderson M E, Schwedes J F, Tegtmeyer P. p53 domains: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 62.Wilcock D, Lane D P. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature. 1991;349:429–431. doi: 10.1038/349429a0. [DOI] [PubMed] [Google Scholar]
- 63.Wold M S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Bichem. 1997;66:61–91. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 64.Yang L, Li R, Mohr I J, Clark R, Botchan M R. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991;353:628–632. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 65.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 67.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 68.Yonish-Rouach E, Deguin V, Zaitchouk T, Breugnot C, Mishal Z, Jenkins J R, May E. Transcriptional activation plays a role in the induction of apoptosis by transiently transfected wild-type p53. Oncogene. 1996;12:2197–2205. [PubMed] [Google Scholar]