Abstract
Cobalamin (B12), an essential nutrient and growth cofactor for many living organisms on Earth, can be fully synthesized only by selected prokaryotes in nature. Therefore, microbial communities related to B12 biosynthesis could serve as an example subsystem to disentangle the underlying ecological mechanisms balancing the function and taxonomic make‐up of complex functional assemblages. By anchoring microbial traits potentially involved in B12 biosynthesis, we depict the biogeographic patterns of B12 biosynthesis genes and the taxa harboring them in the global ocean, despite the limitations of detecting de novo B12 synthesizers via metagenomes alone. Both the taxonomic and functional composition of B12 biosynthesis genes were strongly shaped by depth, differentiating the epipelagic zones from the mesopelagic layers. Functional genes related to B12 biosynthesis were relatively stably distributed across different oceans, but the taxa harboring them varied considerably, showing clear functional redundancy among microbial systems. Microbial taxa carrying B12 biosynthesis genes in the surface water were influenced by environmental factors such as temperature, oxygen, and nitrate. However, the composition of functional genes was only weakly associated with these environmental factors. Null model analyses demonstrated that determinism governed the variations in B12 biosynthesis genes, whereas a higher degree of stochasticity was associated with taxonomic variations. Significant associations were observed between the chlorophyll a concentration and B12 biosynthesis, confirming its importance in primary production in the global ocean. The results of this study reveal an essential ecological mechanism governing the assembly of microbes in nature: the environment selects for function rather than taxonomy; functional redundancy underlies stochastic community assembly.
Keywords: B12 biosynthesis, community assembly, functional genes, functional redundancy, ocean primary production
Impact statement
A central question in ecology is how a galaxy of microbial taxa is assembled and distributed across space and through time, executing essential ecosystem functions. By anchoring microbial functional traits potentially involved in B12 biosynthesis and their carrying microbial taxa in the global ocean, this study addresses essential ecological questions from functional and taxonomic angles. Integrating multiple lines of evidence, we show that the ecosystem selects functional traits rather than taxonomic groups, and functional redundancy underlies stochastic taxonomic community assembly. Also, microbial communities potentially involved in B12 biosynthesis are significantly associated with chlorophyll a concentration, demonstrating their importance in global ocean primary production. This study provides valuable mechanistic insights into the complex microbial community assembly in ecosystems.
INTRODUCTION
As the home to a galaxy of life forms 1 , the global ocean accounts for roughly 97% of the water on Earth, provides 50% of the oxygen and plays an irreplaceable role in impacting the global climate 2 , 3 . Microbial communities, the unseen majority 4 , are of fundamental importance in maintaining the functionality and stability of the global ocean's ecosystems. They not only drive the global biogeochemical cycling of various nutrients and elements and maintain multiple functions in the ecosystem 5 , 6 , but also provide essential nutrients to other organisms, including both prokaryotes and eukaryotes 7 . One such example is B12, an essential nutrient and growth cofactor that is utilized extensively by prokaryotes and eukaryotes for numerous metabolic functions 8 , 9 , 10 , 11 . In natural ecosystems, B12 biosynthesis is energetically extremely expensive, which imposes a high metabolic burden upon B12 producers 12 . Only a small cohort of prokaryotes holds the genetic potential to accomplish such a complex process, while the others have to rely on exogenous supply, forming the “corrinoid economy” 13 . Therefore, B12 auxotrophs may establish close mutualistic interactions with B12 producers, offsetting the cost of B12 biosynthesis to ensure sustainable sources 14 . Such interactive relationships have significant impacts on the composition and structure of marine microbial communities. Two distinct pools of B12 analogs were found in the ocean: the B12 pool produced by a few prokaryotes such as Thaumarchaeota and alpha‐/gamma‐proteobacterial lineages (e.g., Rhodobacterales, Rhizobiales, and most members of the Rickettsiales) 7 , 11 , 14 , 15 , and the pseudocobalamin pool produced by Cyanobacteria as representatives 11 , 14 . In recent years, the importance of B12 has been widely recognized. It influences the growth rate of phytoplankton in the ocean 16 , affects the size and diversity of microbial communities in terrestrial ecosystems 17 , and affects the health status of gut microbes in the human intestinal system 18 , 19 . In addition, the availability of B12 has critical impacts on both cellular‐level metabolic processes (e.g., methionine synthesis) 20 and system‐level biogeochemical cycling (e.g., photosynthesis, aerobic nitrogen cycle) 7 , 21 , 22 . As one of the highly limited nutrients and growth factors controlled by a minority of microbes, B12 can be considered as a “hard currency” in the global ocean ecosystem.
Several studies have focused on the importance of marine B12 biosynthesis in recent years. For example, most of the eukaryotic phytoplanktons in the surface ocean are B12 auxotrophs 9 , and their growth rate can be limited by B12 availability, which further affects their primary productivity. In addition, stoichiometric studies of diatoms in the Subarctic Pacific showed that the carbon:phosphorus (C:P) ratios of B12‐limited cells are significantly lower in comparison with B12‐replete cells 23 . This phenomenon is becoming more pronounced with the significantly increased partial pressure of CO2 caused by anthropogenic activities and global climate change. For example, the C:P ratio gap between B12‐replete and B12‐limited cells was found to gradually widen as the carbon dioxide partial pressure (pCO2) increased, reaching about 40% at 670 ppm pCO2 23 . Recent studies have also demonstrated that the growth rate and primary productivity of phytoplankton are affected by B12 availability 22 , 23 , 24 . Although B12 is of critical importance, the diversity, distribution, and underlying ecological mechanisms shaping the patterns of microbial communities involved in B12 biosynthesis in the global ocean remain largely unexplored. Studies focused on this topic will not only provide a clearer understanding of this subset of prokaryotes in the global ocean but also shed light on the consequential global ocean ecosystem function. Importantly, the Tara Oceans Expedition 25 , 26 , 27 provides a valuable resource that includes comprehensive data sets at the global scale, covering a total of eight ocean regions and three ocean depth ranges, making it possible to investigate the global patterns of various microbial (sub)communities, including the microbial taxa related with B12 biosynthesis.
In this study, by utilizing the Tara Oceans shotgun metagenome sequencing data sets, we surveyed the diversity patterns and ecological importance of microbial traits (functional genes and the corresponding taxonomic groups) potentially involved in B12 biosynthesis in the global ocean ecosystem. Community‐level investigations were mainly performed because of the limitations of identifying de novo B12 synthesizers from metagenomes alone. The following essential ecological questions were addressed: (i) How are B12 biosynthesis traits distributed globally? (ii) What ecological mechanism drives and maintains the diversity patterns of B12 biosynthesis traits? (iii) How do microbial B12 biosynthesis traits contribute to the functions of the global ocean ecosystem, for example, the ocean's primary production? Because of their critical importance to the global ocean, microbial functional genes involved in B12 biosynthesis were expected to show a relatively stable abundance and distribution across the global ocean. However, because of functional redundancy among microbial systems 28 , the microbial taxonomic groups carrying them may vary across different oceanic regions and depths. Determinism, therefore, should be mainly responsible for the diversity patterns of functional traits. However, compared with functional traits, microbial taxonomic groups would be more strongly influenced by stochastic processes, due to functional redundancy among microbial systems. Our results support the above hypotheses and show that B12 biosynthesis traits are significantly associated with the chlorophyll a concentration, confirming their important role in primary production in the global ocean.
RESULTS
Overall diversity of potential B12 biosynthesis traits in the global ocean
Only a small fraction of prokaryotes can fully synthesize B12 7 , 15 because of the multiple enzymatic steps involved (Figure S1). By applying VB12Path 29 to the Tara Oceans shotgun metagenome data set, an average of 0.2% reads per sample were identified to encode gene families potentially involved in B12 biosynthesis pathways. Consistent with the result of the Tara Oceans study that microbial communities significantly differ between the mesopelagic layer (MES) and the epipelagic zones 26 , the same pattern was observed for microbial taxa carrying B12 biosynthesis genes. Compared with microbial communities in the epipelagic zone, those potentially involved in B12 biosynthesis in the MES showed significantly higher taxonomic and functional diversity as well as dramatically different composition (Figures 1A, S2–S4 and Table S1). Surprisingly, the evenness of B12 biosynthesis functional traits and their carrying taxa were negatively correlated, leading to a negative correlation between community diversity (Shannon–Wiener index) (Figure S5). The negative correlation is likely because only a small fraction of microbial taxa carry a (nearly) full set of gene families involved in B12 biosynthesis; therefore, the even distribution of microbial taxa resulted in an uneven distribution of functional traits.
Figure 1.
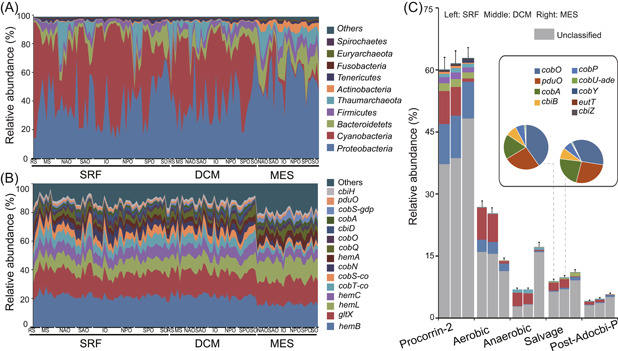
Composition of microbial taxonomic groups and functional traits related to B12 biosynthesis in the global ocean. (A) Composition of microbial taxa carrying B12 biosynthesis genes across different samples. (B) Composition of microbial functional traits potentially involved in B12 biosynthesis across different samples. (C) Relative abundance of microbial phyla carrying genes in different B12 biosynthesis pathways and different ocean layers. Pie charts show the relative abundance of functional traits related to the salvage pathway in the epipelagic zone. The same scaling color code is used in (A) and the stacked bar chart in (C). The figure shows major microbial taxa and functional traits. DCM, deep chlorophyll maximum layer; MES, mesopelagic zone; SRF, surface water layer.
At the pathway level, microbial functional traits potentially involved in precorrin‐2 synthesis (63.84%) and aerobic B12 biosynthesis (24.48%) pathways exhibited the highest relative abundance in the Tara Oceans samples, while anaerobic (9.26%) and post‐Adocbi‐P (4.87%) pathways were less abundant (Figure 1C). At the functional gene level, gene families related to the aerobic B12 biosynthesis pathway were generally more abundant in the epipelagic zones, while the ones related to the anaerobic pathway were more abundant in the MES (Figure 1C and Table S4). Most importantly, consistent with our expectations, the relative abundance of functional genes related to B12 biosynthesis was relatively stable in the global ocean (Figure 1B), while the taxonomic composition was highly variable. This pattern was observed for microbial communities sampled from different depth intervals and oceanic regions (Figure 1A). These results pinpointed an essential microbial ecological discipline that taxonomically highly varied microbial communities still executed similar ecosystem functions.
Microbial taxa carrying B12 biosynthesis genes in the global ocean
Among the identified microbial taxa containing B12 biosynthesis genes, Proteobacteria were abundantly detected in all samples, whereas Cyanobacteria were dominant in the epipelagic zones and dramatically depleted in the MES. Compared with their abundance in the epipelagic zones, Thaumarchaeota was significantly enriched in the MES, and harbored genes related to the anaerobic pathway of B12 biosynthesis (specifically, nine cbi genes were detected) (Figure 1A,C and Table S3). Different modules of the B12 biosynthesis pathway were featured by different microbial taxonomic groups (Figure 1C). This was especially evident for taxa in the MES. Microbial taxa belonging to Thaumarchaeaota and Bacteroidetes were, respectively, dominantly observed with genes belonging to anaerobic and salvage pathways. This result agreed with those of previous studies suggesting that B12 in the surface ocean may be primarily the result of de novo synthesis by heterotrophic bacteria or via modification of pseudocobalamin produced by Cyanobacteria, whereas Thaumarchaeota may be the major B12 producers at depth 14 . Despite the high abundance of Bacteroidetes in the MES, studies have shown that only 0.6% of Bacteroidetes harbor complete B12 synthesis pathways 15 . Gene families (e.g., cobO, pduO, and cobA) belonging to the salvage pathway were dominantly carried by Cyanobacteria, more specifically Prochlorococcus (Figure 1C and Table S2). A quick BLAST searching these gene families against Prochlorococcus genomes in the NCBI database suggested that these gene families are widespread among Prochlorococcus (data not shown). While Cyanobacteria are generally pseudocobalamin synthesizers 30 , the fact that Prochlorococcus carries gene families involved in the salvage pathway indicated the potential of this genus to remodel B12 precursors/analogs under certain conditions. Notably, a recent genomic study also detected gene families involved in the salvage pathway in Synechococcus genomes, possibly due to horizontal gene transfer events or loss of function (of de novo B12 biosynthesis) during evolution 31 . In addition, a high portion of microbial taxa carrying B12 biosynthesis genes belonged to unclassified taxonomic groups, especially in the MES, suggesting that much remains to be further explored for the B12 biosynthesis genes and the taxa that harbor them in the deep ocean.
Microbial taxa potentially involved in B12 biosynthesis in the global ocean were further investigated (Table S2) by selecting the putative key B12 synthesis gene families identified in previous investigations 7 , 30 . Although B12 biosynthesis genes were detected in many microbial taxa, those carrying complete de novo B12 biosynthesis pathways were rarely found, possibly due to inadequate sequencing depth to detect these genes and/or because of the rarity of microbial taxa containing complete B12 biosynthesis pathways. Overall, microbial species including Prochlorococcus marinus, Candidatus Nitrosopelagicus brevis, Candidatus Nitrosomarinus catalina, and Synechococcus sp. CC9902 were the taxa carrying a large number of key B12 biosynthesis gene families. Although B12 biosynthesis genes have been detected in some microbial families (e.g., Synechococcaceae, Prochlorococcaceae, and Pelagibacteraceae), these taxa are considered to be auxotrophic because they lack the gene families necessary for 5,6‐dimethylbenzimidazole (DMB) synthesis, such as bluB 32 , 33 , and for DMB activation, such as cobT 30 . For example, members of the genus Synechococcus contain many genes belonging to B12 biosynthesis pathways but lack key genes for DMB synthesis (Table S2) and have been shown to be B12 auxotrophic 30 . Therefore, detection of B12 biosynthesis genes in microbial taxa does not necessarily imply the capacity of de novo biosynthesis of this cofactor. Further experimental evidence is required to validate such a capacity. These results also highlighted the challenges in identifying potential B12 synthesizers using metagenomic approaches, on the basis that the majority of microbial taxa were unknown and metagenomic recovery of rare microbial taxa was almost impossible.
Latitudinal diversity patterns and distance–decay relationships (DDR)
We also investigated whether microbial communities potentially involved in B12 biosynthesis followed typical biogeographic patterns such as a latitudinal diversity gradient (LDG) and/or DDR, which are well‐recognized ecological patterns for both microbial and macrobial communities 34 , 35 . Discordant patterns between the composition of microbial taxonomic groups and the composition of functional genes were observed in this study (Figure 1A,B). Although B12 biosynthesis serves as an essential ecosystem function and shall be stably maintained in the global ocean, the microbial taxa carrying these functional traits are influenced by various environmental conditions. We expected clear LDG and DDR patterns for microbial taxa carrying B12 biosynthesis genes, but weaker or even nonexistent patterns for the functional genes. Consistent with our expectation, a weak LDG pattern was detected for the functional genes at the surface water layer (SRF) (P = 0.02), but not at the deep chlorophyll maximum layer (DCM) and the MES. No significant DDR pattern was detected for B12 biosynthesis genes at all three pelagic zones. For microbial taxa carrying B12 biosynthesis genes, a strong LDG pattern was observed at the SRF (P = 0.007), whereas DDR was observed at all three pelagic zones (P ≤ 0.001) (Figure 2A,B). Such distinct biogeographic patterns of functional genes and taxonomic groups again pointed to an essential microbial ecology principle, that is, microbial functional genes executing essential ecosystem functions are prevalently distributed, whereas their carrying microbial taxa may vary dramatically.
Figure 2.
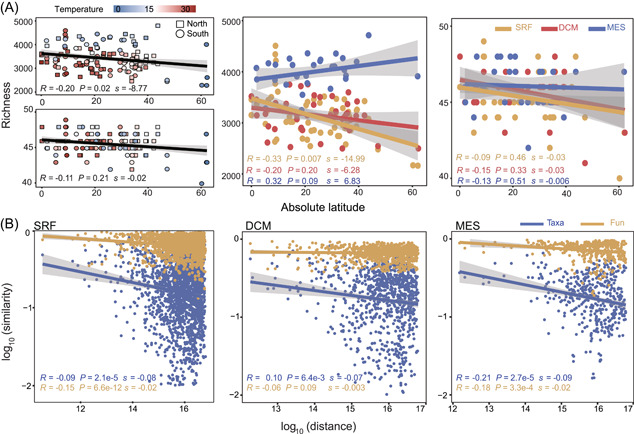
Biogeographic patterns of potential B12 biosynthesis traits in the global ocean. (A) Latitudinal diversity gradient (LDG) patterns for B12 biosynthesis traits in the global ocean. (B) Distance–decay relationship (DDR) for B12 biosynthesis traits in the global ocean. Patterns of taxonomic groups and functional traits were investigated. Fun, functional composition; Taxa, taxonomic composition.
Environmental factors associated with variations in potential B12 biosynthesis traits
Next, we investigated the associations between B12 biosynthesis traits and environmental factors (Figure S6). Since both the functional and taxonomic compositions of B12 biosynthesis genes dramatically differ by depth, the associations with geo‐environmental factors were analyzed for a given range of water depths, thereby eliminating the effects of depth and depth‐correlated environmental factors. As a result, weakened effects of environmental factors on the taxonomic compositions were observed from the SRF to the MES. In the SRF, the concentrations of dissolved oxygen and nitrate availability were significantly associated with the taxonomic composition. Such effects, however, were gradually diminished in the DCM and MES layers. Interestingly, no significant associations were detected between environmental factors and the functional composition of B12 biosynthesis genes in all three oceanic layers, suggesting that environmental conditions mainly affected the taxonomic composition.
The associations between environmental factors and community diversity were also investigated. Significant associations between environmental factors and community diversity could be observed (Figure S7A). However, such effects were weakened or even diminished when looking at individual pelagic zones (Figure S7B–D), suggesting that depth differences from the SRF to the MES and their correlations with environmental factors were mainly responsible for such “pseudo‐associations.” Surprisingly, the effects of temperature on the B12 biosynthesis functional trait diversity differed dramatically among oceanic layers. Temperature was positively associated with functional gene diversity in the epipelagic layers (Figure S7B,C), but negatively in the MES (Figure S7D), leading to a nonsignificant association across the whole upper ocean (Figure S7A). Such opposite patterns were also observed for other environmental factors such as oxygen, nitrite and nitrate concentration (NO2NO3), and nitrate, although some of them were not statistically significant (P ≥ 0.05).
Ecological mechanisms governing the assembly of B12 biosynthesis traits
Considering the critical roles that B12 plays in the ecosystem, we expected that the assembly of microbial functional traits would be highly deterministic. To examine this hypothesis, we quantified the relative importance of deterministic and stochastic processes in governing the assembly of functional traits potentially involved in B12 biosynthesis. In this study, the null model analysis was employed to characterize the ratio of stochasticity to determinism by comparing the observed and null model community β‐diversity (Figure 3A). Consistent with our hypothetical expectations, the stochastic ratio suggested that both the assembly of microbial functional genes and their carrying taxa were highly deterministic. Compared with the functional traits, the taxonomic groups had higher stochastic ratios, especially in the MES, suggesting that the assembly of taxonomic groups was more stochastic than functional traits. Such patterns of stochastic ratios between functional traits and taxonomic groups were consistent in different oceanic layers.
Figure 3.
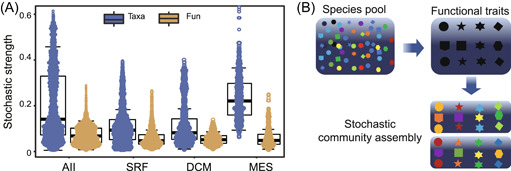
Mechanisms governing assembly of B12 biosynthesis traits in the ocean ecosystem. (A) Stochasticity of community assembly as revealed by null model analysis. (B) Ecological model explaining community assembly of microbial functional groups in the ocean ecosystem. According to the model, the environment selects microbial functional traits rather than taxonomic groups, and functional redundancy underlies stochastic community assembly. In the ecological model, different colors represent different microbial taxa, whereas different shapes represent different functional traits.
We hypothesized that deterministic factors should govern the assembly of microbial functional traits and that the assembly of microbial taxa shall be relatively more stochastic than functional traits. All the results described above, for example, the stable distribution of functional traits versus highly varied taxonomic groups (Figure 1A,B), stronger biogeographic patterns for taxonomic groups than for functional traits (Figures S6 and S7), and the relative importance of deterministic and stochastic processes (Figure 3A), provided evidence to support our hypotheses for community assembly of B12 biosynthesis traits. Integrating all lines of evidence, we proposed a functional trait‐based ecological model to explain complex microbial community assembly in natural ecosystems (Figure 3B). Variations in geo‐environmental factors such as depth, temperature, and oxygen form multiple ecological niches in the oceanic ecosystem (e.g., the epipelagic zones and the MES). Microorganisms capable of living in these ecological niches comprise the species pools. To maintain fundamental ecosystem functions, microorganisms carrying essential functional traits are selected. Therefore, it is the function, rather than taxonomy that the environment truly selects 36 . However, owing to functional redundancy among microbial systems 28 , different taxonomic groups carry the same functional traits. Meanwhile, stochastic processes such as drift and dispersal are associated with microbial taxa. Stochastic community assembly occurs simultaneously with the selection of functional traits. As a result, varied taxonomic compositions come with comparable combinations of functional traits, as observed in multiple ecosystems 37 , 38 , 39 . For microbial traits potentially involved in B12 biosynthesis, both taxonomic groups and functional traits were governed by deterministic processes, and functional redundancy of microbial taxonomic groups led to higher stochasticity in community assembly.
Ecological importance of potential B12 biosynthesis traits in the global ocean
Finally, we investigated the ecological roles of potential B12 biosynthesis traits in the oceanic ecosystem, such as their effects on B12‐dependent microorganisms and their contribution to the ocean's primary production 7 , 9 , 14 , 24 . To investigate whether B12 biosynthesis traits are potentially associated with B12‐dependent microbial communities and global ocean primary productivity, we investigated the associations between the community diversity of B12 biosynthesis traits and the relative abundance of the metH gene family (encoding B12‐dependent methionine synthase) and the chlorophyll a concentration. First, a significant association was observed between the relative abundance of the metH gene family and B12 biosynthesis trait diversity (Figure S8), confirming the importance of B12 biosynthesizing‐members to B12‐dependent members in the oceanic ecosystem. Second, the concentration of chlorophyll a in the epipelagic zone was also significantly associated with B12 biosynthesis trait diversity (P ≤ 0.005) (Figure 4A). Notably, the concentration of chlorophyll a was positively correlated with the taxonomic diversity but negatively correlated with functional gene diversity of B12 biosynthesis traits. Such an opposite pattern was attributed to the negative correlation between the evenness of B12 biosynthesis genes and the taxa harboring them (Figure S8). To exclude the potential influence of the whole microbial community and further confirm the significant correlation between the chlorophyll a concentration and B12 biosynthesis traits, we also evaluated the association between the chlorophyll a concentration and the diversity of the prokaryotic community (taxonomic and Kyoto Encyclopedia of Genes and Genomes [KEGG] orthologous groups). The strength of the association between the chlorophyll a concentration and prokaryotic community diversity was either nonsignificant or much weaker than that of the association with B12 biosynthesis traits (Figure 4B). Finally, a random forest machine learning approach was employed to further verify the importance of B12 biosynthesis traits by predicting the chlorophyll a concentration from B12 community profiles. The results demonstrated that both the taxonomic and functional profiles of B12 biosynthesis traits can well predict the concentration of chlorophyll a in the ocean (Figure 4C,D). This also held true when using SRF microbial data as the training data set to predict the chlorophyll a concentration in the DCM layer, or vice versa (Figure S9).
Figure 4.
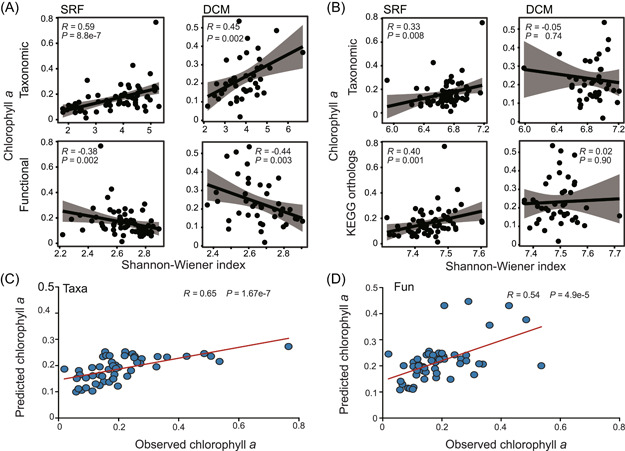
Association between microbial community diversity and chlorophyll a concentration in the global ocean. (A) Association (Spearman's ρ) between B12 biosynthesis trait diversity (taxonomic and functional trait) and chlorophyll a concentration. (B) Association (Spearman's ρ) between overall prokaryotic community diversity (taxonomic and KEGG orthologous groups) and chlorophyll a concentrations. (C) Chlorophyll a concentrations predicted from microbial taxa carrying B12 biosynthesis genes. (D) Chlorophyll a concentrations predicted from B12 biosynthesis functional trait profiles. KEGG, Kyoto Encyclopedia of Genes and Genomes.
DISCUSSION
Focusing on “who is doing what, where, and how?” this study investigated the ecological mechanisms driving the patterns of diversity of microbial traits potentially involved in B12 biosynthesis and their ecological importance in the global ocean. Because of the limitations of the rarity of the targeted microbial taxa and current technologies, it was difficult to confidently infer specific de novo B12 synthesizers. Therefore, community‐level investigations were performed in this study. Similar to what has been observed for the global ocean microbiome 26 , both the taxonomic and functional gene composition related to B12 biosynthesis differed by depth instead of oceanic regions. Multiple factors such as depth, light, temperature, and other associated environmental factors are responsible for such patterns. This suggests that there are completely different niche preferences of B12 biosynthesis traits in different oceanic layers. We also noticed that the evenness of B12 biosynthesis genes and their carrying taxa were negatively correlated, suggesting that an even distribution of microbial taxa may not lead to an even distribution of functional traits. This negative correlation is due to the fact that only a small fraction of microbial taxa contain (near) complete B12 biosynthesis pathways in their genomes, and an even distribution of microbial taxa does not reflect even functional traits.
Microbial taxa carrying B12 biosynthesis genes in the ocean ecosystem were also investigated at a refined taxonomic resolution. However, limited information was gained in this analysis. First, the taxonomy of the majority of B12 biosynthesis genes remained unclassified, even when searched against taxonomic databases built from the most recent NCBI database. This was especially the case for microbial taxa in the MES. Such a shortage of taxonomic information is mainly because of the limitations of current genomic databases 40 , the fact that the majority of microbial taxa in nature remain uncultured 41 , and the potential limitations of read‐based analyses. This result also suggests that there is still much to learn about this tiny group of microorganisms on Earth, especially in the deep ocean. Second, consistent with our current knowledge 14 , only a few microbial genera in the ocean were found to have the potential to synthesize B12 de novo, judging by the gene families linked to the microbial taxa. However, comparative genomic analyses of sequenced microbial genomes from NCBI RefSeq suggest that 37% of prokaryotic microbial species have the potential to biosynthesize cobamides de novo, although complete pathways are not always detected 15 . Among these, 57% of Actinobacteria are predicted to biosynthesize cobamides, whereas only 0.6% of Bacteroidetes have the complete pathway 15 . Such inconsistencies between metagenomic and genomic studies are due to the rarity and unknown properties of de novo B12 synthesizers in the ocean and because current sequencing technologies and depth may not capture them well. Third, identifying de novo B12 synthesizers is challenging and requires further attention. Rhodobacteraceae, Rhizobiales, and a subset of Cyanobacteria were found to be the most important candidates as B12 prototrophs in neritic ecosystems in metatranscriptomic and metaproteomic analyses 42 . However, one needs to be aware that the lower ligand must be DMB to produce B12 and not pseudocobalamin. Perhaps judgment based on key genes related to the synthesis and activation of DMB, for example, bluB 32 , 33 and cobT 30 , is also an option. Cyanobacteria strains release pseudo‐B12 into the media at a high rate, so it has been speculated that Cyanobacteria may be the main providers of (pseudo‐)B12 in algal metabolism 43 . Similarly, genes potentially involved in B12 biosynthesis have been frequently detected in cyanobacterial genera such as Synechococcus and Prochlorococcus, which may only produce pseudocobalamin because adenine is the lower ligand instead of DMB, consistent with previous studies 7 , 11 , 14 . In certain cases, microbial taxa (e.g., Dehalococcoides mccartyi strain 195, Chlamydomonas reinhardtii) may remodel nonfunctional cobamides (e.g., pseudocobalamin) to B12 under suitable environmental conditions such as at the presence of DMB or its intermediate α‐ribazole 11 , 30 , 44 . Interestingly, bluB and cobT were detected from P. marinus at a high taxonomic level (Table S2), and previous studies also mentioned that the P. marinus SS120 genome may encode the full set of enzymes in the heme B12 biosynthetic pathway 45 . In the marine ecosystem, Rhodobacterales are the major alphaproteobacterial B12 producers, but we did not detect bluB from them (e.g., Epibacterium mobile). Therefore, even if these key B12 biosynthesis gene families are detected, further experimental validation is needed to confirm their function in the ecosystem.
This study also revealed important implications in terms of the ecological roles that B12 biosynthesis traits play in the oceanic ecosystem. Previous studies have suggested that eukaryotic phytoplankton in the surface ocean are B12 auxotrophs 9 , 30 , and their growth rate may be limited by B12 availability, further affecting ocean primary productivity 16 , 24 , 46 , 47 . The requirements of these eukaryotic algae for B12 are primarily mediated by methionine synthase 9 , 48 , a key enzyme in cellular one‐carbon metabolism responsible for catalyzing the conversion of homocysteine and 5‐methyl‐tetrahydrofolate to tetrahydrofolate and methionine 49 , 50 . Although B12‐independent methionine synthase (MetE) and B12‐dependent methionine synthase (MetH) are capable of completing this reaction 9 , 48 , MetE is approximately 100‐fold less catalytically efficient than MetH 51 , and this inefficiency further results in an approximately 30‐ to 40‐fold increase in nitrogen and zinc requirements for MetE compared with MetH 52 . Consistent with previous studies, we detected significant correlations between B12 biosynthesis traits and metH encoding B12‐dependent methionine synthase, and between B12 biosynthesis traits and the chlorophyll a concentration. This suggests that B12 biosynthesis traits exert strong effects on the chlorophyll a concentration, demonstrating the importance of this microbial group to the global ocean's primary production.
Our results reveal the diversity patterns of B12 biosynthesis traits in the oceanic ecosystem. The microbial subcommunities also served as an example to reveal an intriguing functional trait‐based ecological mechanism explaining complex microbial community assembly in nature. Both deterministic and stochastic processes govern microbial community assembly, and a major question is which one is more important 53 , 54 , 55 . Considering that B12 biosynthesis is an essential ecosystem function and shall be stably maintained in the global ocean 7 , 15 , 56 , we speculated that strong determinism should govern the assembly of potential B12 biosynthesis traits. However, microbial communities are usually functionally redundant 28 , that is, multiple different microbial taxa may execute the same function. Similar to previous studies on the ocean's microbiome 26 , 57 , 58 , high functional redundancy was also observed in this study. A previous study suggested that the ecosystem tends to select microbial functional traits rather than taxonomic groups 36 . In addition, stochastic processes such as drift and dispersal are associated with microbial taxa 59 . As multiple microbial taxa carry the same functional traits, a certain degree of randomness is associated with microbial taxa in the ecosystem. Consistent with our expectations, higher stochasticity was observed in the assembly of microbial taxa than in functional traits. To summarize, the environment selects microbial functional traits rather than taxonomic groups 36 , and functional redundancy 28 underlies stochastic microbial community assembly, thereby maintaining essential ecosystem function and stability 60 . In addition, we urge that mechanistic studies on microbial community ecology should not only focus on microbial taxonomic groups but also on the functional genes that they carry. Whenever possible, microbial functional genes and taxonomy should be equally considered in microbial systems.
In conclusion, using the B12 biosynthesis subsystem as an example, this study investigated the diversity, biogeographic patterns, and ecological drivers of this specific microbial functional group in the global ocean. Comparative analyses of the patterns of B12 biosynthesis genes and the microbial taxa that harbor them revealed an important microbial ecological mechanism, elucidating the relationship between natural ecosystems and complex microbial communities from the functional angle. Also, B12 biosynthesis traits were significantly associated with the chlorophyll a concentration, demonstrating the importance of this function in primary production in the global ocean. The results of this study provide valuable mechanistic insights into complex microbial community assemblies in natural ecosystems.
MATERIALS AND METHODS
Tara Oceans shotgun metagenomes and geo‐environmental factors
A total of 359 shotgun metagenomes targeting 138 samples covering three oceanic layers, including the SRF (5–10 m), DCM (17–180 m), and MES (250–1000 m), were downloaded from the European Bioinformatics Institute (EBI) repository under project ID ERP001736 26 . Forward and reverse reads were merged into longer sequences by the program PEAR (version 0.9.6, ‐q 30) 61 . An average of 208,881,758 merged reads per sample were obtained. Geo‐environmental factors, the overall taxonomical profiles, and KEGG orthologous group profiles associated with the shotgun metagenome data were downloaded from http://ocean-microbiome.embl.de/companion.html. Metadata for chlorophyll a concentrations in these Tara Oceans samples were obtained from the ZENODO website under the record number 7739198 (https://zenodo.org/record/7739198) according to a previous study 62 .
Metagenomic profiling of marine functional genes potentially involved in B12 biosynthesis
To keep the fidelity of taxonomic and functional profiles and get more usable information from the metagenomic data set 63 , read‐based analysis was performed. Considering the accuracy of gene definition and computational efficiency, VB12Path 29 , a specific functional gene database for metagenomic profiling of gene families involved in B12 biosynthesis pathways, was employed. Although this database is relatively small, both targeted gene families and their homologs from large public databases (e.g., KEGG, eggNOG, and COG) are integrated, minimizing false positive assignments. Briefly, merged metagenomic reads were searched against VB12Path. A total of 54 gene families involved in five modules of B12 biosynthesis pathway as previously described 29 , including precorrin‐2 synthesis processes, aerobic pathway, anaerobic pathway, salvage and remodeling pathway, and post‐Adocbi‐P pathway, are targeted in the database. The program DIAMOND (version 0.9.25, option: ‐k 1 ‐e 0.0001) 64 was used to search nucleotide sequences against VB12Path using the blastx mode. Sequences matching VB12Path were retrieved to generate functional profiles targeting gene families involved in marine B12 biosynthesis using the PERL script provided in VB12Path. To minimize bias associated with sequence number variations across different samples, rarefaction was applied to each metagenome by a random subsampling effort of 100,000,000 sequences. Four samples were excluded from further analysis due to insufficient sequences.
To obtain taxonomic profiles for microbial taxa carrying B12 biosynthesis genes, merged metagenomic sequences belonging to targeted gene families in VB12Path were extracted by the seqtk program (https://github.com/lh3/seqtk). Extracted sequences were then subjected to taxonomic assignment by Kraken2 65 . A standard Kraken2 database was built locally based on the most recent NCBI database at the time this study was carried out. Taxonomic profiles were generated at multiple taxonomic levels based on the Kraken2 report files. After obtaining the functional and taxonomic profiles, the Kruskal–Wallis test was conducted to estimate statistical differences in relative abundances of potential B12 biosynthesis taxonomic groups and functional traits between the epipelagic (SRF/DCM) zone and MES. The false discovery rate approach was employed to adjust the P value to control for false positives using the “stats” package in R. All gene families of the B12 biosynthetic pathway, and the microbial taxa containing B12 biosynthetic gene families are collectively referred to as B12 biosynthesis traits in the context.
Diversity indices
Various diversity indices were calculated by the “vegan” package 66 in R (software version 4.0.3). Specifically, the richness, Shannon–Wiener index, and Pielou's evenness index were calculated for within‐sample diversity, that is, alpha diversity. The Bray–Curtis dissimilarity was calculated to represent between sample diversity, that is, community dissimilarity or beta diversity. The complement of community dissimilarity (1−dissimilarity) was calculated to quantify community similarity. Both within‐sample and between‐sample diversity indices were calculated for functional and taxonomic profiles. Compositional variance among samples in different layers and oceans, as well as epipelagic zone and MES, was calculated using Bray–Curtis dissimilarities and explored by principal coordinates analysis (PCoA), of which the first two axes were extracted for visualization. Three different nonparametric analyses, including permutational multivariate analysis of variance, analysis of similarity, and multiresponse permutation procedure, were performed to evaluate the statistical significance of compositional variations among SRF, DCM, and MES layers.
LDG and DDR
Two major biogeographic patterns, including the LDG and DDR, were analyzed to investigate the diversity trend of B12 biosynthesis traits. For LDG, the relationship between community richness (species and functional traits) and absolute latitude was analyzed. For DDR, the relationship between community similarity and geographic distance was analyzed. The geographic distance between different samples was calculated by the Vincenty Ellipsoid formula based on the latitude and longitude coordinates using the “geosphere” package in R 67 . Community similarity values (Bray–Curtis indices) were obtained by subtracting community dissimilarity from 1. For DDR analyses, both the geographic distance and community similarity values were logarithmically transformed. For both LDG and DDR, linear regression analysis was carried out to visualize the diversity trendline. Values including correlation coefficients, slope, and significance P values were calculated. Analyses were performed for samples in three different layers.
Correlating environmental factors with the diversity and composition of microbial communities
To identify the potential environmental factors shaping the variations of B12 community diversity and composition, the partial Mantel test was performed by correcting geographic distance. Bray–Curtis dissimilarity was selected to characterize the community distance for both taxonomic and functional trait profiles. The Euclidean distance method was used to characterize the distance between environmental factors. A permutation time of 9999 was set for the partial Mantel test. A total of 11 environmental variables were recruited, including latitude, longitude, depth, temperature, oxygen, mean nitrates concentration, NO2, nitrite and nitrate concentration (NO2NO3), phosphate (PO4), salinity, and silica (Si). To analyze the associations between environmental factors and community diversity, redundancy analysis was used to evaluate the collinearity between environmental variables and the taxonomic and functional trait composition. After excluding variables with high collinearity, a total of six geo‐environmental variables were retained, including depth, temperature, oxygen, nitrates, NO2NO3, and PO4. Then, linear regression analyses were conducted to investigate the relationships between each remaining individual environmental variable and community diversity (Shannon–Wiener index). Spearman's rank coefficient of correlation was calculated. All of the above statistical analyses were performed using the “vegan” package 66 in R.
Correlating metH gene abundance and chlorophyll a concentrations with B12 biosynthesis trait diversity
To disentangle the potential effects of B12 biosynthesis traits on B12‐dependent microbial communities and the ocean's primary productivity, the metH gene relative abundance and chlorophyll a concentration were correlated with the community diversity of B12 biosynthesis traits. Of these, metH gene was selected for its encoding of B12‐dependent methionine synthase, a pivotal enzyme of cellular one‐carbon metabolism and DNA synthesis 48 . Positive associations were expected between metH communities and B12 biosynthesis functional genes. Chlorophyll a was selected as a proxy for phytoplankton biomass to further approximate primary productivity. Linear regression analysis was used to explore the relationship between metH relative abundance, the chlorophyll a concentration, and B12 biosynthesis trait diversity. To eliminate the potential impact on the whole prokaryotic community and confirm the importance of B12 biosynthesis traits, linear regression analysis was also carried out between the whole prokaryotic microbial community and chlorophyll a concentration. Both the taxonomic profiles and functional profiles (KEGG orthologous groups) were analyzed. The analyses were carried out for samples in different layers. Spearman's rank coefficient of correlation was calculated. Correlation coefficients with significance P < 0.005 were termed as significant correlation.
In addition to linear regression analyses, the machine learning approach random forest was also employed to verify the importance of B12 biosynthesis traits on chlorophyll a concentration by predicting chlorophyll a concentrations using the functional and taxonomic profiles of B12 biosynthesis traits. In this study, half of the microbial data from epipelagic zones were randomly selected for developing a random forest training model, which was used to predict chlorophyll a concentration using the remaining microbial data in epipelagic zones. In addition, individual layers were validated, using samples from one layer (SRF/DCM) as the training set to predict the chlorophyll a concentration in the other layer. The relationship between predicted and observed chlorophyll a concentration was analyzed to evaluate the importance of B12 communities. The random forest analysis was performed using the “randomForest” package 68 in R.
Community assembly mechanisms
The null model analysis was employed to investigate the potential ecological mechanisms governing the compositional variations of B12 biosynthesis traits. Since the taxonomic and functional trait profiles for B12 biosynthesis genes were obtained by extracting targeted sequences from the shotgun metagenomic data set, phylogenetic markers for these profiles were not applicable. Therefore, the approach proposed by Zhou et al. was employed in this study 38 , 69 . In the analysis, stochastic strength was calculated via null models to characterize the relative importance of deterministic and stochastic processes in driving the assembly of B12 biosynthesis traits. The within‐sample (local) and across‐sample (regional) richness were constrained to produce null models, to rule out the potential influence of local and regional species richness on beta diversity 70 . A dissimilarity matrix was calculated based on the Bray–Curtis index. The complementary similarity matrix was obtained by (1−dissimilarity). This procedure was repeated 1000 times to generate a total of 1000 null models, based on which an average similarity matrix was obtained. Community assembly stochasticity was estimated by comparing the observed and randomized community similarity, according to a modified method as described previously 53 , 71 . The stochastic ratio was calculated considering two scenarios: (i) communities are governed by deterministic factors that produce more similar communities. In such a case, the observed community similarity () between the i‐th and j‐th communities would be larger than the null expectations (). (ii) Communities are governed by deterministic factors making communities more dissimilar. As such, would be smaller than . As a result, the observed dissimilarity () would be larger than the null model dissimilarity (). Hence, the following functions can be used to evaluate the stochastic ratio:
The null model analysis was carried out for both taxonomic and functional profiles. R packages including vegan 66 , bioenv 72 , and NST 38 were used in the analysis.
AUTHOR CONTRIBUTIONS
Jiayin Zhou: Formal analysis (lead); investigation (equal); visualization (lead); writing—original draft (lead). Wei Qin: Conceptualization (supporting); formal analysis (supporting); writing—review and editing (supporting). Xinda Lu: Conceptualization (supporting); writing—review and editing (supporting). Yunfeng Yang: Writing—review and editing (supporting). David Stahl: Conceptualization (supporting); writing—review and editing (supporting). Nianzhi Jiao: Writing—review and editing (supporting). Jizhong Zhou: Conceptualization (supporting); writing—review and editing (supporting). Jihua Liu: Writing—review and editing (supporting). Qichao Tu: Conceptualization (lead); formal analysis (equal); funding acquisition (lead); writing—review and editing (lead).
ETHICS STATEMENT
This study does not contain any studies with human participants or animals performed by any of the authors.
CONFLICT OF INTERESTS
The authors declare no conflict of interests.
Supporting information
Supplemental information.
Supplemental information.
Supplemental information.
Supplemental information.
ACKNOWLEDGMENTS
This study was supported by National Key Research and Development Program of China (2020YFA0607600 and 2019YFA0606700), the National Natural Science Foundation of China (Nos. 31971446, 92051110, and 32371598), the Natural Science Foundations of Shandong Province (2020ZLYS04 and ZR2020YQ21), the Taishan Young Scholarship of Shandong Province, and the Distinguished Young Scholarship of Shandong University.
Zhou J, Qin W, Lu X, Yang Y, Stahl D, Jiao N, et al. The diversity and ecological significance of microbial traits potentially involved in B12 biosynthesis in the global ocean. mLife. 2023;2:416–427. 10.1002/mlf2.12095
Edited by Yong‐Guan Zhu, Institute of Urban Environment, Chinese Academy of Sciences, China
Contributor Information
Jihua Liu, Email: liujihua1982@foxmail.com, Email: tuqichao@sdu.edu.cn.
Qichao Tu, Email: liujihua1982@foxmail.com, Email: tuqichao@sdu.edu.cn.
DATA AVAILABILITY
Sequences belonging to the B12 biosynthesis traits extracted from the Tara Oceans shotgun metagenome datasets are deposited at the ZENODO website under the record number 7520550.
REFERENCES
- 1. Locey KJ, Lennon JT. Scaling laws predict global microbial diversity. Proc Natl Acad Sci USA. 2016;113:5970–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Doney SC, Ruckelshaus M, Emmett Duffy J, Barry JP, Chan F, English CA, et al. Climate change impacts on marine ecosystems. Ann Rev Mar Sci. 2012;4:11–37. [DOI] [PubMed] [Google Scholar]
- 3. Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmerman AE, Keeling PJ. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science. 2015;347:1257594. [DOI] [PubMed] [Google Scholar]
- 4. Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. [DOI] [PubMed] [Google Scholar]
- 6. Fuhrman JA. Microbial community structure and its functional implications. Nature. 2009;459:193–199. [DOI] [PubMed] [Google Scholar]
- 7. Sañudo‐Wilhelmy SA, Gómez‐Consarnau L, Suffridge C, Webb EA. The role of B vitamins in marine biogeochemistry. Ann Rev Mar Sci. 2014;6:339–367. [DOI] [PubMed] [Google Scholar]
- 8. Akduman N, Lightfoot JW, Röseler W, Witte H, Lo W‐S, Rödelsperger C, et al. Bacterial vitamin B12 production enhances nematode predatory behavior. ISME J. 2020;14:1494–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Croft MT, Lawrence AD, Raux‐Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. [DOI] [PubMed] [Google Scholar]
- 10. Stubbe J. Binding site revealed of nature's most beautiful cofactor. Science. 1994;266:1663–1664. [DOI] [PubMed] [Google Scholar]
- 11. Wienhausen G, Dlugosch L, Jarling R, Wilkes H, Giebel HA, Simon M. Availability of vitamin B12 and its lower ligand intermediate α‐ribazole impact prokaryotic and protist communities in oceanic systems. ISME J. 2022;16:2002–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raux E, Schubert HL, Roper JM, Wilson KS, Warren MJ. Vitamin B12: insights into biosynthesis's mount improbable. Bioorg Chem. 1999;27:100–118. [Google Scholar]
- 13. Sonnenburg ED, Sonnenburg JL. Gut microbes take their vitamins. Cell Host Microbe. 2014;15:5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heal KR, Qin W, Ribalet F, Bertagnolli AD, Coyote‐Maestas W, Hmelo LR, et al. Two distinct pools of B12 analogs reveal community interdependencies in the ocean. Proc Natl Acad Sci USA. 2017;114:364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shelton AN, Seth EC, Mok KC, Han AW, Jackson SN, Haft DR, et al. Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J. 2019;13:789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bertrand EM, McCrow JP, Moustafa A, Zheng H, McQuaid JB, Delmont TO, et al. Phytoplankton‐bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc Natl Acad Sci USA. 2015;112:9938–9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu X, Heal KR, Ingalls AE, Doxey AC, Neufeld JD. Metagenomic and chemical characterization of soil cobalamin production. ISME J. 2020;14:53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Degnan PH, Taga ME, Goodman AL. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 2014;20:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. Human gut microbes use multiple transporters to distinguish vitamin B₁₂ analogs and compete in the gut. Cell Host Microbe. 2014;15:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Romine MF, Rodionov DA, Maezato Y, Anderson LN, Nandhikonda P, Rodionova IA, et al. Elucidation of roles for vitamin B12 in regulation of folate, ubiquinone, and methionine metabolism. Proc Natl Acad Sci USA. 2017;114:E1205–E1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite‐oxidizing bacteria. Proc Natl Acad Sci USA. 2010;107:13479–13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bertrand EM, Allen AE. Influence of vitamin B auxotrophy on nitrogen metabolism in eukaryotic phytoplankton. Front Microbiol. 2012;3:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. King AL, Sañudo‐Wilhelmy SA, Leblanc K, Hutchins DA, Fu F. CO2 and vitamin B12 interactions determine bioactive trace metal requirements of a subarctic Pacific diatom. ISME J. 2011;5:1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koch F, Marcoval MA, Panzeca C, Bruland KW, Sañudo‐Wilhelmy SA, Gobler CJ. The effect of vitamin B12 on phytoplankton growth and community structure in the Gulf of Alaska. Limnol Oceanogr. 2011;56:1023–1034. [Google Scholar]
- 25. Sunagawa S, Acinas SG, Bork P, Bowler C, Acinas SG, Babin M, et al. Tara Oceans: towards global ocean ecosystems biology. Nat Rev Microbiol. 2020;18:428–445. [DOI] [PubMed] [Google Scholar]
- 26. Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, et al. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. [DOI] [PubMed] [Google Scholar]
- 27. Bork P, Bowler C, de Vargas C, Gorsky G, Karsenti E, Wincker P. Tara Oceans studies plankton at planetary scale. Science. 2015;348:873. [DOI] [PubMed] [Google Scholar]
- 28. Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O'Connor MI, et al. Function and functional redundancy in microbial systems. Nat Ecol Evol. 2018;2:936–943. [DOI] [PubMed] [Google Scholar]
- 29. Zhou J, Yu X, Liu J, Qin W, He Z, Stahl D, et al. VB12Path for accurate metagenomic profiling of microbially driven cobalamin synthesis pathways. mSystems. 2021;6:e00497‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Helliwell KE, Lawrence AD, Holzer A, Kudahl UJ, Sasso S, Kräutler B, et al. Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12 . Curr Biol. 2016;26:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Włodarczyk A, Selão TT, Norling B, Nixon PJ. Newly discovered Synechococcus sp. PCC 11901 is a robust cyanobacterial strain for high biomass production. Commun Biol. 2020;3:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taga ME, Larsen NA, Howard‐Jones AR, Walsh CT, Walker GC. BluB cannibalizes flavin to form the lower ligand of vitamin B12 . Nature. 2007;446:449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Campbell GRO, Taga ME, Mistry K, Lloret J, Anderson PJ, Roth JR, et al. Sinorhizobium meliloti bluB is necessary for production of 5,6‐dimethylbenzimidazole, the lower ligand of B12 . Proc Natl Acad Sci USA. 2006;103:4634–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hillebrand H. On the generality of the latitudinal diversity gradient. Am Nat. 2004;163:192–211. [DOI] [PubMed] [Google Scholar]
- 35. Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. [DOI] [PubMed] [Google Scholar]
- 36. Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA. 2011;108:14288–14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Louca S, Jacques SM, Pires AP, Leal JS, Srivastava DS, Parfrey LW, et al. High taxonomic variability despite stable functional structure across microbial communities. Nat Ecol Evol. 2016;1:0015. [DOI] [PubMed] [Google Scholar]
- 38. Ning D, Deng Y, Tiedje JM, Zhou J. A general framework for quantitatively assessing ecological stochasticity. Proc Natl Acad Sci USA. 2019;116:16892–16898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, et al. A phylogeny‐driven genomic encyclopaedia of bacteria and archaea. Nature. 2009;462:1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Steen AD, Crits‐Christoph A, Carini P, DeAngelis KM, Fierer N, Lloyd KG, et al. High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 2019;13:3126–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gómez‐Consarnau L, Sachdeva R, Gifford SM, Cutter LS, Fuhrman JA, Sañudo‐Wilhelmy SA, et al. Mosaic patterns of B‐vitamin synthesis and utilization in a natural marine microbial community. Environ Microbiol. 2018;20:2809–2823. [DOI] [PubMed] [Google Scholar]
- 43. Bonnet S, Webb EA, Panzeca C, Karl DM, Capone DG, Wilhelmy SAS. Vitamin B12 excretion by cultures of the marine Cyanobacteria crocosphaera and Synechococcus . Limnol Oceanogr. 2010;55:1959–1964. [Google Scholar]
- 44. Yi S, Seth EC, Men YJ, Stabler SP, Allen RH, Alvarez‐Cohen L, et al. Versatility in corrinoid salvaging and remodeling pathways supports corrinoid‐dependent metabolism in Dehalococcoides mccartyi . Appl Environ Microbiol. 2012;78:7745–7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V, et al. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc Nat Acad Sci USA. 2003;100:10020–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bertrand EM, Saito MA, Rose JM, Riesselman CR, Lohan MC, Noble AE, et al. Vitamin B12 and iron colimitation of phytoplankton growth in the Ross Sea. Limnol Oceanogr. 2007;52:1079–1093. [Google Scholar]
- 47. Browning TJ, Achterberg EP, Rapp I, Engel A, Bertrand EM, Tagliabue A, et al. Nutrient co‐limitation at the boundary of an oceanic gyre. Nature. 2017;551:242–246. [DOI] [PubMed] [Google Scholar]
- 48. Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol. 2011;28:2921–2933. [DOI] [PubMed] [Google Scholar]
- 49. Warren MJ, Raux E, Schubert HL, EscalanteSemerena JC. The biosynthesis of adenosylcobalamin (vitamin B12). Cheminform. 2002;19:390–412. [DOI] [PubMed] [Google Scholar]
- 50. Roth J, Lawrence J, Bobik T. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. [DOI] [PubMed] [Google Scholar]
- 51. Gonzalez JC, Banerjee RV, Huang S, Sumner JS, Matthews RG. Comparison of cobalamin‐independent and cobalamin‐dependent methionine synthases from Escherichia coli: two solutions to the same chemical problem. Biochemistry. 1992;31:6045–6056. [DOI] [PubMed] [Google Scholar]
- 52. Bertrand EM, Moran DM, McIlvin MR, Hoffman JM, Allen AE, Saito MA. Methionine synthase interreplacement in diatom cultures and communities: implications for the persistence of B12 use by eukaryotic phytoplankton. Limnol Oceanogr. 2013;58:1431–1450. [Google Scholar]
- 53. Zhou J, Ning D. Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev. 2017;81:e00002–e00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Logares R, Deutschmann IM, Junger PC, Giner CR, Krabberød AK, Schmidt TSB, et al. Disentangling the mechanisms shaping the surface ocean microbiota. Microbiome. 2020;8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Milke F, Wagner‐Doebler I, Wienhausen G, Simon M. Selection, drift and community interactions shape microbial biogeographic patterns in the Pacific Ocean. ISME J. 2022;16:2653–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sultana S, Bruns S, Wilkes H, Simon M, Wienhausen G. Vitamin B12 is not shared by all marine prototrophic bacteria with their environment. ISME J. 2023;17:836–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353:1272–1277. [DOI] [PubMed] [Google Scholar]
- 58. Tully BJ, Wheat CG, Glazer BT, Huber JA. A dynamic microbial community with high functional redundancy inhabits the cold, oxic subseafloor aquifer. ISME J. 2018;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stegen JC, Lin X, Fredrickson JK, Chen X, Kennedy DW, Murray CJ, et al. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013;7:2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Biggs CR, Yeager LA, Bolser DG, Bonsell C, Dichiera AM, Hou Z, et al. Does functional redundancy affect ecological stability and resilience? A review and meta‐analysis. Ecosphere. 2020;11:e03184. [Google Scholar]
- 61. Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina paired‐end reAd mergeR. Bioinformatics. 2014;30:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Salazar G, Paoli L, Alberti A, Huerta‐Cepas J, Ruscheweyh HJ, Cuenca M, et al. Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell. 2019;179:1068–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhou J, Song W, Tu Q. To assemble or not to assemble: metagenomic profiling of microbially mediated biogeochemical pathways in complex communities. Brief Bioinform. 2023;24:1–10. [DOI] [PubMed] [Google Scholar]
- 64. Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. [DOI] [PubMed] [Google Scholar]
- 65. Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–930. [Google Scholar]
- 67. Karney CFF. Algorithms for geodesics. J Geodesy. 2013;87:43–55. [Google Scholar]
- 68. Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 69. Zhou J, Deng Y, Zhang P, Xue K, Liang Y, Van Nostrand JD, et al. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc Natl Acad Sci USA. 2014;111:E836–E845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chase JM, Kraft NJB, Smith KG, Vellend M, Inouye BD. Using null models to disentangle variation in community dissimilarity from variation in α‐diversity. Ecosphere. 2011;2:art24. [Google Scholar]
- 71. Guo X, Feng J, Shi Z, Zhou X, Yuan M, Tao X, et al. Climate warming leads to divergent succession of grassland microbial communities. Nat Clim Change. 2018;8:813–818. [Google Scholar]
- 72. Clarke K, Ainsworth M. A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser. 1993;92:205–219. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental information.
Supplemental information.
Supplemental information.
Supplemental information.
Data Availability Statement
Sequences belonging to the B12 biosynthesis traits extracted from the Tara Oceans shotgun metagenome datasets are deposited at the ZENODO website under the record number 7520550.


