Abstract
The antiviral activity of a CD8+ cytotoxic T-lymphocyte (CTL) clone (TCC108) directed against a newly identified HLA-B14-restricted epitope, human immunodeficiency virus type 1 (HIV-1) Rev(67-75) SAEPVPLQL, was analyzed with respect to its kinetics of target cell lysis and inhibition of HIV-1 production. Addition of TCC108 cells or CD8+ reverse transcriptase-specific CTLs to HLA-matched CD4+ T cells at different times after infection with HIV-1 IIIB showed that infected cells became susceptible to CTL-mediated lysis before peak virus production but after the onset of progeny virus release. When either of these CTLs were added to part of the infected cells immediately after infection, p55 expression and virus production were significantly suppressed. These data support a model in which CTLs, apart from exerting cytolytic activity which may prevent continued virus release, can interfere with viral protein expression during the eclipse phase via noncytolytic mechanisms. TCC108-mediated inhibition of virus replication in peripheral blood mononuclear cells caused rapid selection of a virus with a mutation (69E→K) in the Rev(67-75) CTL epitope which abolished recognition by TCC108 cells. Taken together, these data suggest that both cytolytic and noncytolytic antiviral mechanisms of CTLs can be specifically targeted to HIV-1-infected cells.
Identification of immune responses that may limit progression toward AIDS and may eliminate infected cells that persist despite effective antiviral therapy (6, 20, 30) is a major goal for current research aimed at the development of vaccines and immunotherapies against AIDS (1). It is generally assumed that an effective vaccine against human immunodeficiency virus type 1 (HIV-1) should elicit an antiviral immune response which includes virus-specific major histocompatibility complex class I-restricted CD8+ cytotoxic T lymphocytes (CTL), because their presence is associated with the control of primate lentivirus replication and they have been detected in individuals exposed to but apparently uninfected with HIV (reviewed in reference 8). Furthermore, CTL have been shown to exert pressure on virus replication in vivo (2, 9) and in vitro (3, 27, 32). CTL clones directed against the late viral proteins Gag, reverse transcriptase (RT), and Env, have been shown to lyse HIV-1-infected cells before peak virus production (31) and to suppress HIV-1 replication in immortalized CD4+ T-cell lines such as H9 and T1 (32). Env-specific CTL have been shown to eliminate HIV-1-infected CD4+ peripheral blood mononuclear cells (PBMC) and H9 cells (32), indicating that inhibition of HIV-1 replication involved cytolytic mechanisms. In addition to exerting cytolytic activity, HIV-specific CTL have been shown to suppress virus replication by the excretion of soluble factors (3, 32). Although CTL against late viral proteins do exhibit antiviral activity, elimination of nonproductively infected cells with still incomplete protein expression (18) and suppression of low-level virus replication (20) may require CTL directed against the regulatory viral proteins Tat and Rev. These proteins are translated early in the replication cycle of HIV-1 and are necessary for transcription (Tat) or expression of the intermediate and late proteins (Rev) (14, 15). Consistent with such a protective role of CTL against Rev and Tat, we have shown previously that CTL with these specificities were preferentially found in individuals who experienced a long-term asymptomatic course of disease progression (28). In contrast, CTL responses against the late proteins Gag and RT did not correlate with the rate of disease progression (28).
Here we present a detailed analysis of the Rev-specific CTL response in one individual who has been infected for more than 12 years without developing symptoms. Rev-specific CTL clones were generated, and a minimal epitope as well as the HLA class I restriction of its recognition were identified. It is shown that both Rev- and RT-specific CTL can suppress HIV-1 production before they exert cytolytic activity and that Rev-specific CTL-mediated inhibition of virus replication in PBMC leads to the rapid selection of virus mutated in the CTL epitope.
MATERIALS AND METHODS
Cells.
All cells were maintained in RPMI 1640 (BioWhittaker, Verviers, Belgium) supplemented with l-glutamine (2 mM), penicillin (100 U/ml), streptomycin (10 μg/ml), and 10% pooled human serum (R10H) for PBMC and T cells or 10% fetal bovine serum (BioWhittaker) (R10F) for B-LCL cells. CTL clones and the CD4+ TCL2H7 cells were stimulated at 2-week intervals with phytohemagglutinin-L (PHA-L) (1 μg/ml; Boehringer Mannheim, Germany) and gamma-irradiated (3,000 rads) allogenic feeder cells and maintained in R10H containing recombinant interleukin-2 (rIL2) (50 U/ml; Eurocetus, Amsterdam, The Netherlands). International Histocompatibility Workshop B-LCL cells were obtained from the European Collection of Cell Cultures (Salisbury, United Kingdom).
Generation of CTL clones.
HIV-1 Rev-specific T-cell lines obtained from individual L709, HLA-A1,28;−B14,57, were seeded at 0.3, 1, and 3 cells per well in 60-well Terasaki plates (Greiner, Alphen a/d Rijn, The Netherlands) in a final volume of 20 μl per well containing PHA-L (1 μg/ml), rIL2 (50 U/ml), and irradiated allogenic feeder cells. The plates were incubated at 37°C in a humidified chamber with 5% CO2. After 10 to 14 days, growing cell cultures from plates showing growth in 10 to 15% of wells were restimulated. Cell samples from these cultures were analyzed for Rev-specific CTL activity on autologous B-LCL cells infected with recombinant vaccinia virus (rVV) containing the HIV-1LAI rev gene (TG4113; rVV-rev) in chromium release assays as described below. The Rev-specific TCC108 clone expressed T-cell receptor VB14S1 uniformly (data not shown), confirming its clonality. Since the five Rev-specific CTL clones were restricted by the same HLA class I allele, recognized the same 20-mer peptide, and were established from the same Rev-specific cell line, these clones most likely originated from the same cell. For practical reasons, further analyses were performed with one of these CTL clones, TCC108.
Chromium release assays.
Target cells were labelled for 1 h with 100 μCi of Na2 51CrO4 (Amersham, Buckinghamshire, United Kingdom), washed three times, and adjusted to a concentration of 2 × 105 cells per ml in R5F. A volume of 50 μl (104 cells) was plated in 96-well V-bottom plates. Effector cells were added at ratios of between 3:1 and 10:1 in a final volume of 150 μl. The spontaneous and maximum chromium releases were determined by the incubation of the target cells with R5F only or with 5% Triton X-100, respectively. Triplicate incubations were performed in all assays. After incubation at 37°C for 4 h, supernatants were harvested with a harvesting device (Skatron, Oslo, Norway), and radioactivity was counted in a gamma counter (LKBWallac, Turku, Finland). The percent specific lysis was calculated as (experimental release − spontaneous release)/(maximum release − spontaneous release) × 100. To ensure sufficient HIV-1-infected cells at each time point, TCL2H7 cells were infected at a multiplicity of infection (MOI) of approximately 1.0.
Flow cytometry.
The expression of CD4, CD8, and HLA-A2 was analyzed by incubation of viable cells with CD4-fluorescein isothiocyanate (CD4-FITC) or CD8-phycoerythrin (Becton Dickinson, Leiden, The Netherlands) or HLA-A2 specific monoclonal antibody BB7.2 (kindly provided by W. Biddison). FITC-conjugated goat anti-mouse immunoglobulin (Becton Dickinson) was used as a second antibody for detection of expression of HLA-A2. For detection of HIV-1 p55 expression, cells were incubated subsequentially with paraformaldehyde–lyso-lecithin, cold absolute methanol, Nonidet P-40 (Sigma), and FITC- or phycoerythrin-labelled anti-HIV-1 p55 monoclonal antibody (clone KC57) according to the instructions of the manufacturer (Coulter, Mijdrecht, The Netherlands).
Peptides.
The 20-mer peptides with 10 residues of overlap, together spanning the entire HIV-1 Rev sequence were kindly provided by H. Holmes (Medical Research Council, South Mimms, Potters Bar, United Kingdom). For the preparation of target cells, 0.5 × 106 to 1 × 106 B-LCL cells were incubated with these peptides at 20 μM in 100 μl of R0. After 1 h, 900 μl of R5F was added and cells were incubated overnight. The peptides used for fine mapping of the CTL epitope recognized by TCC108 cells were manufactured at the European Veterinary Laboratory (Woerden, The Netherlands). Chromium-labelled B-LCL cells were incubated with these shorter peptides for 1 h at concentrations ranging from 1 × 10−9 to 3 × 10−4 M, washed twice, and used as target cells.
Virus stocks.
rVV-rev and rVV containing the polylinker without insert (186-poly; rVV-control) were kindly provided by M. P. Kieny (Transgène, Strasbourg, France). Stocks were prepared on RK13 cells and stored at concentrations of 1 × 108 to 3 × 108 PFU/ml at −70°C. For the preparation of rVV-infected target cells, B-LCL cells were incubated with 10 PFU per cell at 107 cells per ml for 1 h. Subsequently, the cells were diluted to 106/ml with R10F and incubated overnight.
An HIV-1 IIIB stock was prepared on freshly infected PHA-activated CD4+ TCC cells and stored at −70°C. The RT activity of this stock was 105 RT cpm/ml. The infectious viral titer was determined by infection of PHA-activated PBMC or TCL2H7 cells with serial fivefold dilutions of this stock in quadruplicate. With both cell types an estimated titer of 105 infectious particles per ml was found.
RT assay.
RT activity was assayed in a microassay as previously described by Gregersen et al. (10) and adapted by Siebelink et al. (25). Culture supernatants were precipitated with 32% polyethylene glycol 6000–1.5 M NaCl. The pellets were resuspended in 15 μl of lysis buffer (50 mM Tris [pH 8.3], 20 mM dithiothreitol, 0.25% Triton X-100) and mixed with 35 μl of H2O and 50 μl of RT cocktail [100 mM Tris (pH 7.9), 150 mM KCl, 10 mM MgCl2, 4 mM dithiothreitol, 0.6 U of poly(rA)-oligo(dT), 60 μCi of [3H]TTP per ml]. After incubation at 37°C for 1 h, the DNA was precipitated with 20 μl of 120 mM Na4P2O7 · 10H2O in 60% trichloroacetic acid for 15 min at 4°C. The DNA was harvested on glass fiber filters with a Skatron cell harvester and washed with 12 mM Na4P2O7 ·10H2O in 5% trichloroacetic acid. The filters were dried at 80°C, and [3H]TTP incorporation was measured in a beta scintillation counter (LKBWallac).
Kinetics of target cell recognition.
Three days after their most recent stimulation with PHA-L, 1.2 × 106 CD4+ TCL2H7 cells were incubated with IIIB at 1.2 × 106 RT cpm (MOI of approximately 1) for 3 h at 37°C. The cells were washed three times and cultured at 3 × 105 cells per ml in R10H supplemented with rIL2 (50 U/ml). At various time points, a volume of 0.6 ml, including cells, was harvested. After centrifugation, the supernatants were stored at −70°C for RT assays, part of the cells were fixed for flow cytometric analysis of p55, CD4, and CD8 expression, and part of the cells were labelled with chromium for analysis in chromium release assays.
Coculture of CTL and acutely infected PBMC.
PBMC (2 × 107) isolated from a buffy coat were incubated with HIV-1 IIIB at 105 RT cpm for 3 h at 37°C. The cells were washed three times and cultured at 5 × 106 cells per 10 ml of R10H supplemented with PHA-L (1 μg/ml) and rIL2 (50 U/ml) in 25-cm2 flasks. Effector cells were added at a ratio of 0.1:1. At various time points, 2 ml of the culture was harvested and centrifuged (250 × g). The supernatants were stored at −70°C for analyses of RT activity and viral RNA sequences, and the cells were fixed with paraformaldehyde–lyso-lecithin for flow cytometric analysis of HIV-1 p55, CD4, CD8, and HLA-A2 expression. At days 6 and 11 postinfection, part of the cells were not fixed but were separated into a CD8+ fraction and a CD8− fraction with an anti-CD8 monoclonal antibody covalently conjugated to magnetic beads (Becton Dickinson) according to the manufacturer’s instructions. After treatment of the CD8+ fraction with DetachaBead (Becton Dickinson) and overnight incubation at 37°C, these cells were analyzed for cytolytic activity against rVV-rev- and rVV-control-infected autologous B-LCL cells. CD8+ PBMC could be discriminated from TCC108 and TCC112 cells by flow cytometric analysis of the expression of HLA-A2, which was present on the PBMC only.
Sequencing.
Viral RNA was isolated from supernatants harvested from cultures of HIV-1-infected PBMC with and without the Rev-specific TCC108 cells. The second exon of rev was amplified by RT-PCR. After reverse transcription with random primers, cDNA was amplified with primers GTACTTTCTATAGTGAATAGAGTTAGGC and CCTATCTGTCCCCTCAGCTACT. PCR conditions were as follows: 30 s at 95°C, 30 s at 50°C, and 30 s at 72°C for 30 cycles and then 7 min of extension at 72°C. Amplified fragments were sequenced directly on both strands by using the PCR primers with the Taq Dye Deoxy Terminator sequencing kit on a 373A sequencing system from Applied Biosystems (Foster City, Calif.). Sequences were analyzed with Geneworks (Intelligenetics, Mountain View, Calif.).
RESULTS
HLA restriction and fine specificity of Rev-specific CTL clones.
To analyze the Rev-specific CTL response of a long-term asymptomatic individual in more detail, we cloned a Rev-specific T-cell line generated in a previous study from PBMC of individual L709 (28). Five CD4− CD8+ Rev-specific CTL clones, TCC102, TCC104, TCC106, TCC108, and TCC110, and one CD4− CD8+ non-HIV-specific CTL clone, TCC112, were obtained (Tables 1 and 2).
TABLE 1.
Specificities of CD8+ CTL clones from individual L709
| CTL clone | % Specific lysisa
|
|
|---|---|---|
| rVV-control | rVV-rev | |
| TCC102 | 1 ± 3 | 61 ± 5 |
| TCC104 | 1 ± 2 | 59 ± 3 |
| TCC106 | 0 ± 0 | 86 ± 9 |
| TCC108 | −1 ± 0 | 78 ± 5 |
| TCC110 | 0 ± 1 | 79 ± 5 |
| TCC112 | 0 ± 1 | 3 ± 1 |
Average (± standard error; triplicates) percentages of CTL-mediated chromium release by autologous rVV-infected B-LCL cells.
TABLE 2.
Lysis of HIV-1 IIIB-infected cells by CD8+ T cells used in this studya
| CD8+ T-cell line | Specificity
|
Effector/target ratio | % Specific lysis (mean ± SD)
|
||
|---|---|---|---|---|---|
| Protein | Epitope | Mock infection | HIV-1 IIIB infection | ||
| TCC108 | Rev | SAEPVPLQL | 10 | 5 ± 4 | 48 ± 6 |
| 3 | 0 ± 3 | 37 ± 1 | |||
| TCC112 | NDb | ND | 10 | −1 ± 1 | 4 ± 2 |
| 3 | −4 ± 1 | −8 ± 1 | |||
| TCL1C11 | RT | IVLPEKDSW | 10 | −1 ± 1 | 63 ± 8 |
| 3 | −1 ± 1 | 62 ± 9 | |||
Target cells were CD4+ TCL2H7 cells 10 days after mock or HIV-1 IIIB infection (MOI of approximately 0.05).
ND, not determined.
The HLA restriction of the Rev-specific CTL clones was determined with a panel of partially HLA class I-matched B-LCL cells infected with rVV-rev or rVV-control in standard chromium release assays (Fig. 1A). All Rev-specific CTL clones lysed the four rVV-rev-infected heterologous target cells that shared HLA-B14 (Fig. 1A shows the results obtained with TCC108 cells). Heterologous target cell lines that were matched for HLA-A1 (four cell lines), HLA-A28 (three cell lines), or HLA-B57 (three cell lines) were not lysed by the Rev-specific CTL clones (Fig. 1A). These data indicate that recognition of Rev by the CTL clones was restricted by HLA-B14.
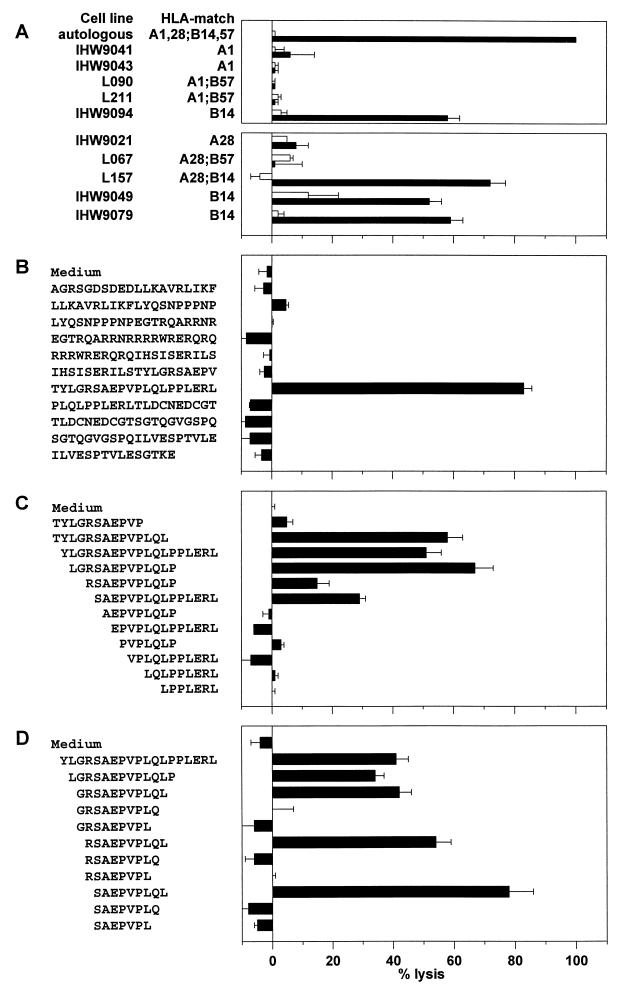
FIG. 1.
HLA restriction and fine specificity of HIV-1 Rev-specific CTL clone TCC108. (A) Autologous and partially HLA class I-matched B-LCL cells were infected with rVV-rev (filled bars) or rVV-control (open bars) and analyzed for recognition by TCC108 cells in a standard chromium release assay (upper panel). Additional B-LCL cells were analyzed in a separate assay to confirm HLA-B14 restriction (lower panel). (B to D) Peptide-pulsed autologous B-LCL cells were analyzed for recognition by TCC108 cells in standard chromium release assays. Chromium-labelled target cells were incubated overnight with one of the 11 20-mer peptides together spanning the entire Rev sequence (B) or for 1 h with the N- and C-terminally truncated peptides before the addition of effector cells (C and D, respectively). Effector-to-target ratios were between 3:1 and 10:1 in all assays. The average percent specific lysis (with standard error) for triplicates is shown. Results similar to those presented in panels A and B were obtained with the Rev-specific CTL clones TCC102, TCC104, TCC106, and TCC110 (data not shown). The non-Rev-specific clone TCC112 did not lyse any of the rVV-infected or peptide-pulsed target cells (data not shown).
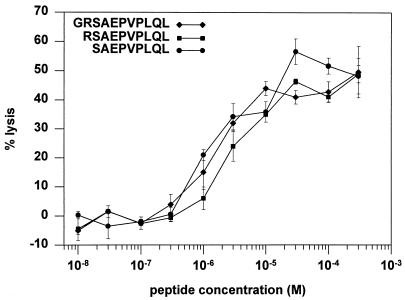
The location of the CTL epitope within the Rev protein was estimated by using 11 20-mer peptides with 10 amino acid residues of overlap, together spanning the entire Rev sequence. All five clones specifically lysed autologous B-LCL cells pulsed with the peptide Rev(62-81) TYLGRSAEPVPLQLPPLERL but not those pulsed with the other peptides (Fig. 1B). Truncated peptides lacking the N-terminal residues 62T, 63Y, 64L, 65G, and 66R were recognized by TCC108 cells, but those without 67S were not (Fig. 1C), indicating that 67S defines the N terminus of the minimal epitope. TCC108 cells recognized peptides truncated C terminally at 75L but not peptides truncated at 74Q or 73L (Fig. 1D). These data show that Rev(67-75) SAEPVPLQL is the minimal epitope recognized by TCC108 cells. The amino acid arginine (R) has been described to serve as an anchor at position 2 in HLA-B14 binding peptides (5), and it flanks the minimal epitope at position 66 of Rev. Titration of the peptides SAEPVPLQL, RSAEPVPLQL, and GRSAEPVPLQL on autologous B-LCL cells revealed that all three peptides required a concentration of at least 1 μM to be recognized by TCC108 cells and showed a similar increase in specific lysis with increasing concentrations (Fig. 2). Thus, the additional residues 66R and 65G did not contribute to the optimal recognition of the CTL epitope.
FIG. 2.
Titration of peptides recognized by the Rev-specific clone TCC108. Chromium-labelled autologous B-LCL cells were incubated with the peptide Rev(65-75) GRSAEPVPLQL, Rev(66-75) RSAEPVPLQL, or Rev(67-75) SAEPVPLQL for 1 h at the concentrations indicated. Subsequently, the target cells were washed and cocultivated with TCC108 cells for 4 h. The effector-to-target cell ratios were 10:1. The average percent specific lysis (with standard error) for triplicates is shown. No lysis of B-LCL cells without peptide was observed (data not shown).
Kinetics of target cell lysis and suppression of HIV-1 production by Rev- and RT-specific CTL.
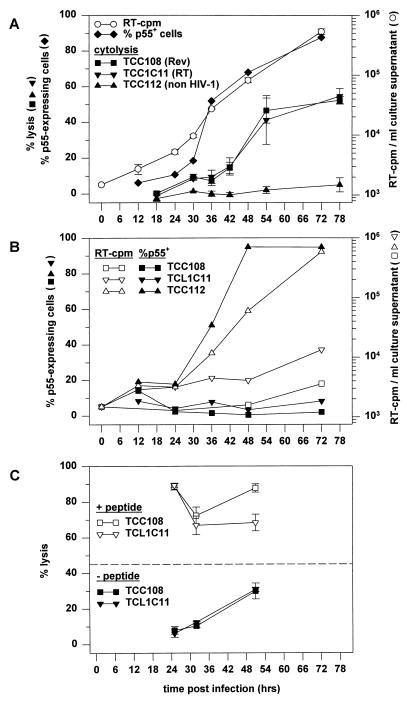
To evaluate the temporal relationship between protein expression in infected cells and the antiviral activity of CTL, we compared the kinetics of cytolysis of infected cells by CTL against the early protein Rev (TCC108 cells) and by CTL against the late protein RT [TCL1C11 cells; specific for RT(244-252) IVLPEKDSW in the context of HLA-B57 (29)]. Both TCC108 and TCL1C11 cells were shown to lyse an HIV-1 IIIB-infected CD4+ T-cell line, TCL2H7, obtained from individual L709 (Table 2). At 12 h after infection of TCL2H7 cells with HIV-1 IIIB (MOI of approximately 1.0), no significant population of p55-expressing cells was detected (Fig. 3A). The percentage of p55-expressing cells increased slightly between 12 and 24 h and increased rapidly thereafter: from 18% at 30 h to 50% at 36 h, 68% at 48 h, and 89% at 72 h. Significant virus production was found at 30 h after infection, in agreement with previous reports on the replication cycle of HIV-1 (14, 21), and production was increased at 36, 48, and 72 h (Fig. 3A). These data indicate that all cells expressing detectable levels of p55 at 48 h (here 68% of the cells) had been infected by the initial inoculum. Since Rev-encoding mRNA and Rev protein have been detected at between 16 and 18 h after infection in IIIB-infected CD4+ T cells (14, 22), we expected to find significant specific lysis by TCC108 cells added between 26 and 30 h or between 32 and 36 h after infection. Although the percentage of specific lysis was higher at these time points than at 14 to 18 h, it did not exceed 10% (Fig. 3A). At 38 to 42 h it was still only 15%, and at 50 to 54 h it was 42%, reaching a maximum of 55% after 74 to 78 h (Fig. 3A). The RT-specific CTL lysed the infected cells at similar levels and with similar kinetics as the Rev-specific CTL (Fig. 3A). As expected, the infected cells were not lysed by the non-HIV-specific TCC112 cells at any time (Fig. 3A). In a parallel experiment, part of the infected TCL2H7 cells were cocultured with the CTL immediately after infection. In cultures containing TCC108 or TCL1C11 cells, p55 expression and virus production were significantly suppressed at all times (Fig. 3B). The presence of the TCC112 cells did not affect p55 expression or virus production (Fig. 3B).
FIG. 3.
Kinetics of HIV-1 production and lysis of infected cells by Rev- and RT-specific CTL. (A) CD4+ TCL2H7 cells were infected as described in Materials and Methods, and culture supernatants were harvested at the indicated times for analysis of virus production. The TCL2H7 cells were analyzed for p55 expression and for susceptibility to CTL-mediated lysis by Rev-specific clone TCC108, RT-specific clone TCL1C11, and non-HIV-specific clone TCC112. The effector-to-target ratios were 10:1. Lysis of uninfected CD4+ TCL2H7 cells was below 5% in all assays (data not shown). The chromium release data are plotted as the average (with standard error) for triplicates at the time point at which the chromium release assay was terminated, i.e., 6 hours after the addition of chromium. This time was required for the chromium labelling (1 h), washing of the target cells and preparing the cocultures of the effector and target cells (1 h), and incubation (4 h). (B) p55 expression (closed symbols) and virus production (open symbols) by TCL2H7 cells in the presence of TCC108 cells, TCL1C11 cells, or TCC112 cells. Effector and target cells were discriminated by flow cytometric analyses of CD8 and CD4 expression, respectively. The population of p55-expressing TCL2H7 cells is expressed as a percentage of the CD8− cells and not of the CD4+ cells, since CD4 was down-regulated in a major fraction of the infected cells. (C) Infected TCL2H7 cells were analyzed in chromium release assays after incubation without peptide or with the relevant peptides at 10 μM: SAEPVPLQL for TCC108 cells and IVLPEKDSW for TCL1C11 cells. The average specific lysis (with standard error) for triplicates is shown. The dashed line shows the percentage of p55-expressing cells at 48 h after infection. Lysis of uninfected TCL2H7 cells without peptides was always below 5% (data not shown), and peptide-pulsed uninfected TCL2H7 cells were lysed as efficiently as peptide-pulsed infected cells (data not shown).
That TCC108 and TCL1C11 cells were cytolytic at all time points was verified in a separate assay: early after infection, target cells loaded with the relevant synthetic peptides were indeed lysed efficiently (Fig. 3C). Later, at 47 to 51 h, specific lysis of infected cells not loaded with the peptides also was observed (Fig. 3C), confirming the results shown in Fig. 3A.
Together these data indicate that infected cells cultured in the absence of HIV-specific CTL are not susceptible to cytolysis before they produce virus. Furthermore, they suggest that mechanisms other than cytolysis caused the observed CTL-mediated inhibition of virus production in the cocultures of effector and target cells.
In vitro inhibition of HIV-1 replication by TCC108 cells.
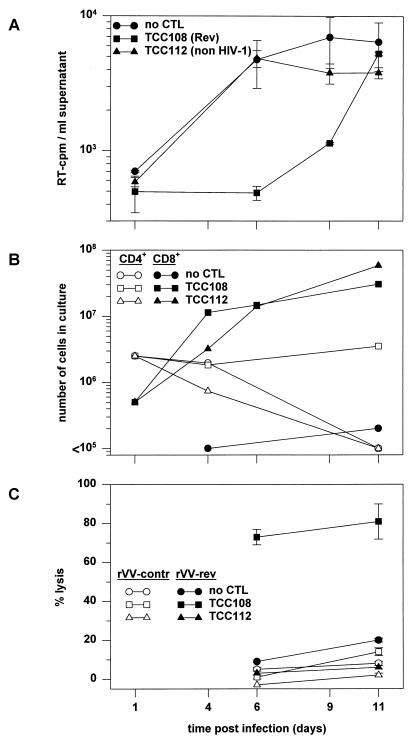
Subsequently, we investigated the longevity of the TCC108-mediated antiviral activity with freshly isolated PBMC. HLA-B14-matched PBMC were infected with HIV-1 IIIB (MOI of approximately 0.05) and cocultured with TCC108 cells or the non-HIV-1-specific TCC112 cells for 11 days. The TCC-to-CD4 cell ratio was 0.2 at the start of the experiment. Virus production was detected in cultures without TCC108 cells on days 6, 9, and 11 (Fig. 4A). In the coculture with TCC108 cells, only a low level of RT activity was detected on day 9. On day 11 this level was similar to that in the control cultures. Flow cytometric analyses showed that the number of TCC108 and TCC112 cells increased 100-fold during the culture period (Fig. 4B). Furthermore, CD8+ cells, recovered from the culture containing TCC108 cells on days 6 and 11 by magnetic bead selection, showed significant Rev-specific CTL activity (Fig. 4C). These data indicate that the lack of control of virus replication could not have been due to the disappearance of the clone from the culture or to impairment of CTL function.
FIG. 4.
Inhibition of HIV-1 replication by Rev-specific CTL. HLA-B14-expressing PBMC from an HIV-seronegative individual were infected with HIV-1 IIIB as indicated in Materials and Methods. PBMC (5 × 106) were cocultivated without CTL, with 5 × 105 Rev-specific TCC108 cells, or with 5 × 105 non-HIV-specific TCC112 cells. Both types of TCC cells had been stimulated 7 days before addition to the PBMC. (A) Virus production was analyzed by quantification of the RT activity in culture supernatants. The average RT activity (with standard error) for triplicates is shown. (B) The fates of the CD4+ PBMC and the CD8+ TCC108 and TCC112 cells were determined by counting of the cells and flow cytometric analysis of membrane-expressed CD4, CD8, and HLA-A2. HLA-A2 was included to discriminate between CD8+ PBMC (expressing HLA-A2) and the added TCC108 and TCC112 cells (both expressing HLA-A1 and -A28). (C) CD8+ cells were recovered from the cultures on days 6 and 11 by magnetic bead selection and analyzed for Rev-specific CTL activity on autologous B-LCL cells infected with rVV-rev or rVV-control. The average percent specific lysis (with standard error) for triplicates is shown.
To test whether the virus had escaped CTL recognition by mutation in the epitope, we sequenced the second exon of Rev directly on the amplicons from the total virus pool produced in the presence or absence of TCC108 cells. Sequencing was performed on samples from day 11 only, since amplifications carried out with earlier samples did not yield PCR products. Only the virus from the coculture with TCC108 cells was found to have a mutation, 69E→K, located in the third residue of the minimal epitope recognized by TCC108 cells (Fig. 5). The sequence signal was uniform, indicating that >90% of the virus population contained the mutation. The mutant peptide SAKPVPLQL was not recognized by TCC108 cells at concentrations ranging from 10 nM to 300 μM (data not shown), indicating that the new virus was indeed an escape variant.
FIG. 5.
Sequence analysis of the second-exon Rev from virus cultured in the absence or presence of Rev-specific TCC108 cells. Viral RNA was isolated from culture supernatants from the experiment shown in Fig. 4 on day 11 postinfection. The second-exon Rev sequences of virus cultured in the absence or presence of TCC108 cells are shown below the sequences of three known IIIB clones (19) for reference purposes. The CTL epitope region is in boldface.
To assess whether escape from a Rev-specific CTL response had occurred in vivo, we attempted to amplify the plasma virus of individual L709 at different times after seroconversion. Primers and PCR conditions were the same as for the amplification of the in vitro-cultured virus. However, no PCR products were obtained, most likely due to the low viral load in this individual.
DISCUSSION
The fine specificity, kinetics of target cell lysis, and capacity to inhibit HIV-1 replication of Rev-specific CTL from an individual infected with HIV-1 for more than 12 years without developing symptoms were analyzed. CTL clones generated from this individual’s PBMC recognized the 9-mer peptide Rev(67-75) SAEPVPLQL as their minimal epitope in the context of HLA-B14. The presence of residue L75 at position 9 is consistent with the reported motif for HLA-B14 binding peptides (5). Other predicted anchor residues were not present. Two longer peptides containing a potential anchor, 66R, were recognized at similar levels of efficiency as the minimal epitope, indicating that this residue did not enhance presentation to the CTL. Analysis of the interactions between HIV-1-specific CTL and HIV-1-infected target cells, i.e., immortalized polyclonal CD4+ T cells (TCL2H7 cells) infected with HIV-1 IIIB, showed target cell lysis by both Rev- and RT-specific CTL. This indicates that their respective epitopes, as defined by rVV and synthetic peptide analyses, were indeed generated in these HIV-1-infected cells. The kinetics of Rev- and RT-specific CTL-mediated cytolysis, first observed well before peak virus production, indicate that CTL may prevent a significant quantity of virus from being produced. This is in agreement with the kinetics of Gag-, RT-, and Env-specific CTL-mediated lysis of HIV-1-infected immortalized H9 and T1 cells, as reported by Yang et al., who sampled at 24, 48, 72, and 96 h after infection (31). As a result of frequent sampling between 24 and 48 h after infection, we were able to extend their findings by showing that infected cells, in the absence of CTL, produce virus before they become susceptible to CTL-mediated lysis. A peptide-pulsing experiment revealed that limited target cell lysis during the first 42 h after infection could not be explained by insufficient effector cell numbers or impaired effector cell function. Also, the possibility that limiting antigen levels had affected the efficiency of cytolysis (26) is probably not relevant for our system, considering the extent of viral protein expression and virus production observed at 30 to 36 h after infection. The similarity in the kinetics of cytolysis targeted at the early protein Rev and at the late protein RT suggests that HIV-1 infection had interfered, transiently, with a general aspect of the antigen processing and presentation pathway. A 20 to 50% reduction of HLA class I surface expression after HIV-1 infection has been reported (13, 23, 31). This down-regulation was shown to decrease cytolysis by HLA-specific CTL (13, 23) but had no appreciable effect on the capacity of infected cells to present synthetic peptides (31). The latter finding is consistent with our observation that TCL2H7 cells were susceptible to CTL-mediated lysis early after infection when pulsed with the relevant peptides. The presentation of endogenous epitopes, however, may be affected when the intracellular expression of new HLA class I molecules is impaired. Indeed, recently it has been shown that HIV-1 Nef is involved in protecting infected cells from specific CTL-mediated lysis by affecting the HLA class I surface expression (4). Also, other mechanisms, such as Tat-mediated interference with the proteasome function (24), may have impeded the generation of HLA class I presentable peptides early after infection.
If lysis of infected cells were the only inhibitory mechanism of the CTL, one would expect a steady increase of the levels of p55 expression and virus production by infected cells, even in the presence of CTL, until the time at which they became susceptible to CTL-mediated lysis. However, when the Rev- or RT-specific CTL were added to the infected cells immediately after infection, viral protein expression and virus production were suppressed without delay during the entire coculture period. These results suggest that early antiviral activity involved noncytolytic mechanisms. Indeed, CD8+ T cells have been shown to inhibit HIV-1 replication by the production of soluble factors (3, 16, 32), to inhibit hepatitis B virus gene expression by a noncytolytic mechanism (11), and to exert antiviral effects against murine rotavirus and VV by perforin- and Fas-independent mechanisms (7, 12). Because the Rev- and RT-specific CTL were added to the target cells after infection, factors that prevent binding or entry of HIV-1 could not have been involved in the observed suppression of HIV-1 production. It is possible that the suppression was mediated by CD8+-T-cell-derived factors that interfere with viral transcription, like IL-16 (17, 33) or CD8+ T-cell antiviral factor (16). If noncytolytic antiviral mechanisms of TCC108 cells also contributed to the observed suppression of viral replication in HLA-B14-matched PBMC, they must have been specifically targeted toward the infected cells expressing the appropriate HLA epitope complex, since the replication of virus that had escaped CTL recognition by a mutation in the HLA-B14-restricted epitope was not significantly affected. Noncytolytic interference with transcription and translation may, like cytolysis, require that CTL are targeted to infected cells via major histocompatibility complex-epitope complexes, for this would be a safeguard against harming bystander cells. To determine the relative contributions of CTL against different proteins to the control of viral replication, further characterization of (i) CTL against late proteins with respect to their capacity to inhibit viral replication, (ii) the affinities of the different epitopes for their HLA restriction elements, and (iii) the affinity of each clone for its HLA peptide complex is required.
In summary, the present data show that CTL against early and late viral proteins can lyse acutely HIV-1-infected cells efficiently, but only after the production of progeny virus has started. Yet, virus replication in freshly isolated PBMC was significantly suppressed by CTL against the Rev protein, which resulted in the rapid selection of CTL escape virus in vitro. It is important to realize that the antiviral effect of CTL responses in vivo is determined by multiple factors, including the breadth of the CTL response. Such factors will be difficult to address in vitro with a limited set of CTL clones. However, studies on the kinetics of viral protein-mediated interference with target cell killing and on the relative contributions of CTL against early and late viral proteins to the inhibition of viral replication will shed new light on complications which the immune system encounters in clearing virus and accordingly may contribute to the development of vaccines and immunotherapies against AIDS.
ACKNOWLEDGMENTS
We acknowledge the participants of the Amsterdam Cohort Studies on AIDS; O. Pontesilli for providing cell cultures; K. Sintnicolaas, E. F. Prsespolewsky, and the donors of the Rotterdam Blood Bank for providing HLA-typed PBMC; R. S. de Swart, G. M. G. M. Verjans, and F. C. G. M. UytdeHaag for stimulating discussions; C. S. A. Guillon, H. W. Vos, B. van’t Land, and M. E. M. Dings for technical support; and C. W. H. M. Kruyssen, G. J. G. M. Osterop, J. M. Rimmelzwaan, and G. L. van der Water for continued management support.
This work was supported by grant no. 1314 from the Dutch AIDS Foundation.
REFERENCES
- 1.Bloom B R. A perspective on AIDS vaccines. Science. 1996;272:1888–1890. doi: 10.1126/science.272.5270.1888. [DOI] [PubMed] [Google Scholar]
- 2.Borrow P, Lewicki H, Wei X, Horwitz M S, Peffer N, Meyers H, Nelson J A, Gairin J E, Hahn B H, Oldstone M B A, Shaw G M. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 3.Buseyne F, Fevrier M, Garcia S, Gougeon M L, Riviere Y. Dual function of a human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte clone: inhibition of HIV replication by noncytolytic mechanisms and lysis of HIV-infected CD4+ cells. Virology. 1996;225:248–253. doi: 10.1006/viro.1996.0597. [DOI] [PubMed] [Google Scholar]
- 4.Collins K L, Chen B K, Kalams S A, Walker B D, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 5.DiBrino M, Parker K C, Margulies D H, Shiloach J, Turner R V, Biddison W E, Coligan J E. The HLA-B14 peptide binding site can accommodate peptides with different combinations of anchor peptides. J Biol Chem. 1994;269:32426–32434. [PubMed] [Google Scholar]
- 6.Finzi D, Hermankova M, Pierson T, Carruth L, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 7.Franco M A, Tin G, Rott L S, VanCott J L, McGhee J R, Greenberg H B. Evidence for CD8+ T-cell immunity to murine rotavirus in the absence of perforine, Fas and gamma interferon. J Virol. 1997;71:479–486. doi: 10.1128/jvi.71.1.479-486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotch F M, Koup R A, Safrit J T. New observations on cellular immunoresponses to HIV and T-cell epitopes. AIDS. 1997;11:S99–S107. [PubMed] [Google Scholar]
- 9.Goulder P J R, Phillips R E, Colbert R A, McAdam S, Ogg G, Nowak M, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael A J, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 10.Gregersen J P, Wege H, Preiss L, Jentsch K D. Detection of human immunodeficiency virus and other retroviruses in cell culture supernatants by a reverse transcriptase microassay. J Virol Methods. 1988;19:161–168. doi: 10.1016/0166-0934(88)90159-0. [DOI] [PubMed] [Google Scholar]
- 11.Guidotti L G, Ando K, Hobbs M V, Ishikawa T, Runkel L, Schreiber R D, Chisari F V. Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc Natl Acad Sci USA. 1994;91:3764–3768. doi: 10.1073/pnas.91.9.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kagi D, Ledermann B, Burki K, Zinkernagel R M, Hengartner H. Molecular mechanisms of lymphocyte-mediated cytotoxicity and their role in immunological protection and pathogenesis in vivo. Annu Rev Immunol. 1996;14:207–232. doi: 10.1146/annurev.immunol.14.1.207. [DOI] [PubMed] [Google Scholar]
- 13.Kerkau T, Schmitt Landgraf R, Schimpl A, Wecker E. Downregulation of HLA class I antigens in HIV-1-infected cells. Aids Res Hum Retroviruses. 1989;5:613–620. doi: 10.1089/aid.1989.5.613. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Byrn R, Groopman J, Baltimore D. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J Virol. 1989;63:3708–3713. doi: 10.1128/jvi.63.9.3708-3713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingsman S M, Kingsman A J. The regulation of human immunodeficiency virus type-1 gene expression. Eur J Biochem. 1996;240:491–507. doi: 10.1111/j.1432-1033.1996.0491h.x. [DOI] [PubMed] [Google Scholar]
- 16.Levy J A, Mackewicz C E, Barker E. Controlling HIV pathogenesis: the role of the noncytotoxic anti-HIV response of CD8+ T cells. Immunol Today. 1996;17:217–224. doi: 10.1016/0167-5699(96)10011-6. [DOI] [PubMed] [Google Scholar]
- 17.Maciaszek J W, Parada N A, Cruikshank W W, Center D M, Kornfeld H, Viglianty G A. IL-16 represses HIV-1 promoter activity. J Immunol. 1997;158:5–8. [PubMed] [Google Scholar]
- 18.McCune J M. Viral latency in HIV disease. Cell. 1995;82:183–188. doi: 10.1016/0092-8674(95)90305-4. [DOI] [PubMed] [Google Scholar]
- 19.Meyers G, Foley B, Mellors J W, Korber B, Jeang K, Wain-Hobson S. Human retroviruses and AIDS 1996: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. [Google Scholar]
- 20.Pantaleo G. How immune-based interventions can change HIV therapy. Nat Med. 1997;3:483–486. doi: 10.1038/nm0597-483. [DOI] [PubMed] [Google Scholar]
- 21.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 22.Ranki A, Lagerstedt A, Ovod V, Aavik E, Krohn K J. Expression kinetics and subcellular localization of HIV-1 regulatory proteins Nef, Tat and Rev in acutely and chronically infected lymphoid cell lines. Arch Virol. 1994;139:365–378. doi: 10.1007/BF01310798. [DOI] [PubMed] [Google Scholar]
- 23.Scheppler J A, Nicholson J K A, Swan D C, Ahmed-Ansari A, McDougal J S. Down-modulation of MHC-I in CD4+ T cell line CEM-E5, after HIV-1 infection. J Immunol. 1989;143:2858–2866. [PubMed] [Google Scholar]
- 24.Seeger M, Ferrell K, Franck R, Dubiel W. HIV-1 Tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation. J Biol Chem. 1997;272:8145–8148. doi: 10.1074/jbc.272.13.8145. [DOI] [PubMed] [Google Scholar]
- 25.Siebelink K H, Chu I H, Rimmelzwaan G F, Weijer K, Osterhaus A D, Bosch M L. Isolation and partial characterization of infectious molecular clones of feline immunodeficiency virus obtained directly from bone marrow of DNA of a naturally infected cat. J Virol. 1992;66:1091–1097. doi: 10.1128/jvi.66.2.1091-1097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsomides T J, Aldovini A, Johnson R P, Walker B D, Young R A, Eisen H N. Naturally processed viral peptides recognized by cytotoxic T lymphocytes on cells chronically infected by human immunodeficiency virus type 1. J Exp Med. 1994;180:1283–1293. doi: 10.1084/jem.180.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsubota H, Lord C I, Watkins D I, Morimoto C, Letvin N L. A cytotoxic T lymphocyte inhibits acquired immunodeficiency syndrome virus replication in peripheral blood lymphocytes. J Exp Med. 1989;169:1421–1434. doi: 10.1084/jem.169.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Baalen C A, Pontesilli O, Huisman R C, Geretti A M, Klein M R, De Wolf F, Miedema F, Gruters R A, Osterhaus A D M E. Human immunodeficiency virus type 1 Rev- and Tat-specific cytotoxic T lymphocyte frequencies inversely correlate with rapid progression to AIDS. J Gen Virol. 1997;78:1913–1918. doi: 10.1099/0022-1317-78-8-1913. [DOI] [PubMed] [Google Scholar]
- 29.Van der Burg S H, Klein M R, Pontesilli O, Holwerda A M, Drijfhout J W, Kast W M, Miedema F, Melief C J M. HIV-1 reverse transcriptase-specific CTL against conserved epitopes do not protect against progression to AIDS. J Immunol. 1997;159:3648–3654. [PubMed] [Google Scholar]
- 30.Wong J K, Hezareh M, Günthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 31.Yang O O, Kalams S A, Rosenzweig M, Trocha A, Jones N, Koziel M, Walker B D, Johnson R P. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. J Virol. 1996;70:5799–5806. doi: 10.1128/jvi.70.9.5799-5806.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang O O, Kalams S A, Trocha A, Cao H, Luster A, Johnson R P, Walker B D. Suppression of human immunodeficiency virus type 1 replication by CD8+ cells: evidence for HLA class I-restricted triggering of cytolytic and noncytolytic mechanisms. J Virol. 1997;71:3120–3128. doi: 10.1128/jvi.71.4.3120-3128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou P, Goldstein S, Devadas K, Tewari D, Notkins A L. Human CD4+ cells transfected with IL-16 cDNA are resistant to HIV-1 infection: inhibition of mRNA expression. Nat Med. 1997;3:659–664. doi: 10.1038/nm0697-659. [DOI] [PubMed] [Google Scholar]