Nearly 90% of the disease risk-associated variants identified by genome-wide association studies are in non-coding regions of the genome. The annotations obtained by analyzing functional genomics assays can provide additional information to pinpoint causal variants, which are often not the lead variants identified from association studies. However, the lack of available annotation tools limits the use of such data. To address the challenge, we previously built the ‘RegulomeDB database’ to prioritize and annotate variants in non-coding regions1, which has been a highly utilized resource for the research community (Supplementary Fig. 1).
Here we present an update of the RegulomeDB web server, RegulomeDB v.2 (http://regulomedb.org). RegulomeDB annotates a variant by intersecting its position with genomic intervals identified from functional genomic assays and computational approaches. It also incorporates variant hits into a heuristic ranking score, representing its potential to be functional in regulatory elements. We improve and boost annotation power by incorporating thousands of newly processed data from functional genomic assays in GRCh38 assembly and include probabilistic scores from the SURF algorithm that was the top performing non-coding variant predictor in the Fifth Critical Assessment of Genome Interpretation (CAGI-5)2.
The update of RegulomeDB now includes more than 650 million and 1.5 billion genomic intervals in hg19 and GRCh38, respectively — a fivefold increase compared with the previous version (Supplementary Fig. 2). We included approximately 5,000 chromatin immunoprecipitation followed by sequencing experiments targeting transcription factors (TF ChIP–seq), and chromatin accessibility experiments from the ENCODE project3, the Roadmap Epigenomics program4, and the Genomics of Gene Regulation project. We also produced a comprehensive set of footprint predictions using over 800 chromatin accessibility experiments and 591 transcription factor motifs in GRCh38 using the TRACE pipeline5. In addition, we refined the included transcription factor motifs by using the non-redundant vertebrates set from the JASPAR database6. We also integrated approximately 71 million variant–gene pairs in expression quantitative trait loci (eQTL) studies from the GTEx project7, and 450,000 chromatin-accessibility QTLs (caQTLs) from 9 recent publications (Supplementary Information). Finally, we included chromatin state annotations known as from chromHMM in EpiMap for 833 biosamples8.
RegulomeDB accepts any query variants genome-wide in either GRCh38 or hg19 genome assembly by rsID or genome coordinates. The query variants can then be prioritized by functional prediction scores shown in a sortable table. For any variant of interest, an information page on five types of supported genomic evidence, as well as a genome browser view is displayed. Each of the six sections can be clicked to show more detail for functionality exploration (Supplementary Figs. 3-5).
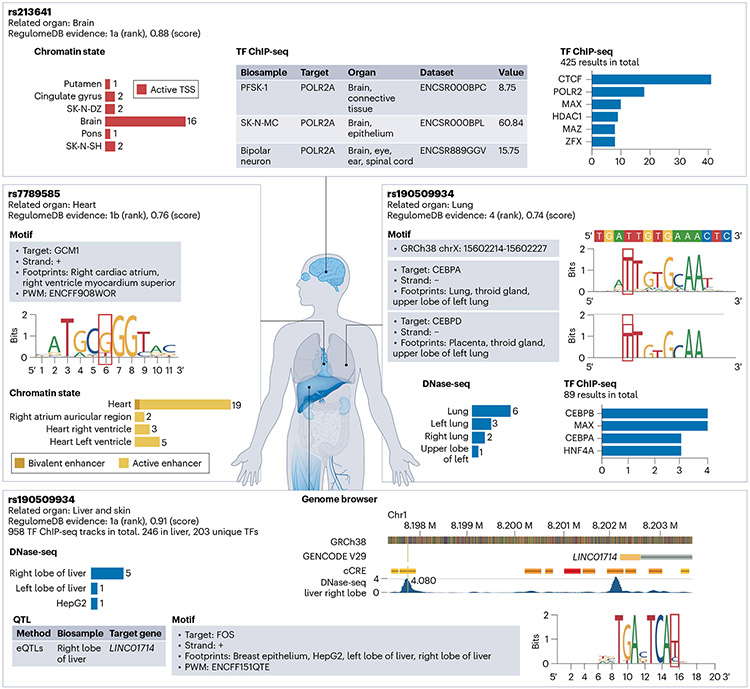
RegulomeDB enables researchers to quickly separate functional variants from a large pool of variants and assign tissue or organ specificity for each variant. Here we showcase this using four verified variants from recent literature9-13, and demonstrate the applicability of RegulomeDB to annotate those variants based on various sources of data (Fig. 1).
Fig. 1 ∣. Prioritization of functional variants with RegulomeDB version 2.
Four example variants with verified functions in related organs from recent literature. Various sources of evidence in RegulomeDB are indicated by gray boxes. RegulomeDB heuristic ranking score and probability score summarized all evidence.
Transcription factor motifs and ChIP–seq data together provide evidence about how a variant is likely to affect phenotype in a cell-specific context. For example, rs213641 is known to affect behavioral responses to fear and anxiety stimuli9. The POLR2A binding and the active transcriptional start site (TSS) state in the brain indicate that rs213641 is likely to function in the brain by disrupting the TSS of STMN1. We also examined rs7789585, in which RegulomeDB transcription factor motif evidence suggests that mutation to the reference allele G would disrupt the binding of GCM1, which may interrupt the active enhancer state at the locus in the heart. Hocker et al.10 recently confirmed this hypothesis using reporter assays, and discovered that rs7789585 disrupts a KCNH2 enhancer and affects cardiomyocyte electrophysiologic function.
DNase-seq assays and underlying footprint predictions identify open chromatin regions with mapped transcription factor binding sites in hundreds of biosamples and can also be used to assign putative function to variants. rs190509934 has been associated with the risk of COVID-19 infection by affecting ACE2 expression11. RegulomeDB shows hits to several DNase-seq peaks in lung-related biosamples. Furthermore, RegulomeDB extends this tissue effect with the hypothesis that ACE2 expression may be regulated by CEBP by its overlap with DNase footprints in the lung found in the upstream promoter region of ACE212. In addition, eQTL studies provide correlation evidence between the variants and their target genes. For example, rs72635708 is predicted as a regulatory variant by RegulomeDB with a high probability of 0.91 due to its locus overlapping with DNase and ChIP–seq peaks, footprints, and it is an eQTL that associates with LINC01714 gene expression in the right lobe liver. Because rs72635708 lies in the FOS motif, it is likely to be a functional variant in the liver by modulating the binding of the AP-1 complex13.
In summary, RegulomeDB provides a user-friendly tool to annotate and prioritize variants in non-coding regions of the human genome, which can aid variant function interpretation and guide follow-up experiments. We welcome user feedback through regulomedb@mailman.stanford.edu.
Supplementary Material
Acknowledgements
We thank the RegulomeDB users and the scientific community for producing and sharing functional genomic experiments. We also thank all members in the Cherry and Boyle laboratories for constructive feedbacks. This research was supported by US National Institutes of Health (NIH) grants U24 HG009293 (A.P.B. and J.M.C).
Footnotes
Competing interests
The authors have no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41588-023-01365-3.
Reporting Summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RegulomeDB v.2 can be accessed through the web server at https://regulomedb.org. All datasets collected in RegulomeDB are accessible through the ENCODE portal https://www.encodeproject.org/search/?internal_tags=RegulomeDB_2_2.
Code availability
The code RegulomeDB uses is available on GitHub repository at https://github.com/ENCODE-DCC/regulome-encoded/releases/tag/v2.2 and https://github.com/ENCODE-DCC/genomic-data-service/releases/tag/v2.2.
References
- 1.Boyle AP et al. Genome Res. 22, 1790–1797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong S & Boyle AP Hum. Mutat 40, 1292–1298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ENCODE Project Consortium. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roadmap Epigenomics Consortium. Nature 518, 317–330 (2015).25693563 [Google Scholar]
- 5.Ouyang N & Boyle AP Genome Res. 30, 1040–1046 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fornes O. et al. Nucleic Acids Res. 48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.GTEx Consortium. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boix CA, James BT, Park YP, Meuleman W & Kellis M Nature 590, 300–307 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brocke B. et al. Am. J. Med. Genet. B Neuropsychiatr. Genet 153B, 243–251 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Hocker JD et al. Sci. Adv 7, eabf1444 (2021).33990324 [Google Scholar]
- 11.Horowitz JE et al. Nat. Genet 54, 382–392 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beacon TH, Delcuve GP & Davie JR Genome 64, 386–399 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Kubota N & Suyama M BMC Med. Genomics 13, 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RegulomeDB v.2 can be accessed through the web server at https://regulomedb.org. All datasets collected in RegulomeDB are accessible through the ENCODE portal https://www.encodeproject.org/search/?internal_tags=RegulomeDB_2_2.
The code RegulomeDB uses is available on GitHub repository at https://github.com/ENCODE-DCC/regulome-encoded/releases/tag/v2.2 and https://github.com/ENCODE-DCC/genomic-data-service/releases/tag/v2.2.